Abstract
Cutaneous papillomas and squamous cell carcinoma (SCC) are common in captive North American snow leopards (Panthera uncia). Our objective was to determine if these lesions are potentially associated with papillomavirus(es). PCR was performed on 3 cutaneous papillomas using degenerate primers for papillomaviruses. A putatively novel papillomavirus was identified that shared 76% sequence identify to Felis catus papillomavirus 2. Specific PCR for this virus was performed on 5 cutaneous SCC samples and 7 normal skin samples, which were all positive. In situ hybridization for this novel virus was performed, which revealed strong hybridization signals within hyperplastic cells in cutaneous papillomas (n=3) and within neoplastic cells in cutaneous squamous cell carcinoma samples (n=5). No hybridization signals were identified within normal skin. Ultimately, identification of a causal viral agent in the development of papillomas and SCC in snow leopards will help guide therapeutic intervention and lay the foundation for development of prophylactic vaccines.
Keywords: Panthera uncia, snow leopards, papillomavirus, squamous cell carcinoma, viral papillomas
Snow leopards (SLs) (Panthera uncia) are iconic animals in many North American (NA) zoos and are currently listed as vulnerable by the IUCN.7 Information gained about diseases that impact SLs will aid in the long-term management of the captive population and may have global implications for the conservation of this species.
In a 20-year mortality review of captive NA SLs, malignant neoplasia affected approximately 46% of the population.16 Squamous cell carcinoma (SCC) was the most common, affecting 10% of the population.16 The oral cavity, specifically the sublingual mucosa, and the skin were the most reported sites.16 In domestic cats, papillomaviruses are associated with a subset of cutaneous SCC due to neoplastic transformation from viral plaques.9,11–14 Sublingual and cutaneous plaques occur frequently in captive NA SLs affecting up to 12% of the population (Timothy Georoff, personal communication). Panthera uncia papillomavirus 1 (PuPV1) has been sequenced from oral plaques; however, a viral association with cutaneous lesions and their transformation to neoplasia has yet to be established.4,5,8
Our objective was to determine if papillomaviruses are associated with cutaneous papillomas (CPs) and SCC in captive NA SLs.
Tissues from thirteen adult SLs were collected from archived biopsy and necropsy samples which included three biopsy samples of CPs from two animals, five biopsy and necropsy samples of SCC, and seven control skin samples. Demographics for each sample are summarized in Table 1. In Case 2, both CP and SCC were diagnosed on separate biopsies from different sites one year apart with histologic evidence of transition from CP to SCC. In the animals in which the cause of death (COD) was known, SCC was the cause of euthanasia in one case with metastasis to the lung, regional lymph node, and liver. Other known COD were generally unrelated to cutaneous lesions (Table 1). Control skin samples included follicular cysts, hypersensitivity dermatitis, dermal fibrosis, and normal skin.
Table 1:
Fifteen Samples from thirteen captive North American snow leopards
| Case No. | Age At Submit | Sex | Diagnosis | COD | DG Primers | SP Primers | SL-RPL13 | ISH |
|---|---|---|---|---|---|---|---|---|
| 1a | 12y | M | Cutaneous Papilloma | Unknown | + | + | + | + |
| 1b | 11y | Cutaneous Papilloma | + | + | + | + | ||
| 2a | Unk | F | Cutaneous papilloma | Renal Failure | + | + | + | + |
| 2b | 16 | Cutaneous papilloma with transition to SCC | NP | + | + | + | ||
| 3 | 15 | M | Cutaneous SCC | Unknown | NP | + | + | + |
| 4 | Unk | Unk | Cutaneous SCC | Cutaneous SCC with mets to Lung, Liver, and LN | NP | + | + | + |
| 5 | Unk | F | Cutaneous SCC | Gastric carcinoid | NP | + | + | + |
| 6 | Unk | Unk | Cutaneous SCC | Unknown | NP | - | - | + |
| 7 | 12y | F | Eosinophilic and hyperplastic dermatitis | Neurologic disease | + | + | + | - |
| 8 | Unk | M | Dermal fibrosis | Unknown | NP | + | + | - |
| 9 | Unk | Unk | Follicular cyst | Unknown | NP | + | + | - |
| 10 | 13y | M | Nodular dermal fibrosis | Chronic renal disease, lymphoma (spleen, liver) | NP | + | + | - |
| 11 | Unk | Unk | Normal skin | Unknown | NP | + | + | - |
| 12 | Unk | Unk | Follicular cyst | Unknown | NP | + | + | - |
| 13 | 17y | M | Dermal fibrosis | Spondylosis with bladder atony; chronic renal disease | NP | + | + | - |
No.: Number; Submit: Submission; Unk: Unknown, y: years old; M: Male, F: Female; SCC: Squamous cell carcinoma; COD: Cause of Death; Mets: metastasis; LN: Lymph node; DG: Degenerate; SP: Specific; NP: not performed; ISH: in situ hybridization
Hematoxylin and eosin-stained slides were evaluated to confirm the diagnoses. CPs were characterized by exophytic epidermal proliferations with hyperkeratosis. The epidermis exhibited varying degrees of spongiosis, hypergranulosis, and koilocytosis (viral cytopathic change) with variable superficial dermal inflammation (Figure 1). In case 1, in both examined biopsy samples (a/b) submitted a year apart, epithelial cells exhibited normal differentiation with orderly maturation. In Case 2a, epithelial cells exhibited mild disorderly maturation and dysplasia. In the same animal, Case 2b, submitted a year prior, there was histologic evidence of a CP with transition to SCC (Figure 2). In this case, and in three other cases of SCC, the neoplasm was characterized by anastomosing islands and cords of atypical stratified squamous epithelium supported by varying amounts of fibrovascular stroma (Figure 3). Neoplastic cells showed disorganized maturation and prominent intercellular bridging with centralized concentric lamellae of keratin (keratin pearls). Neoplastic cells were polygonal with a moderate amount of eosinophilic cytoplasm, and large round nuclei with vesiculated chromatin and prominent magenta nucleoli (Figure 3). In one animal (Case 4), there were multicentric CPs that progressed to SCC prior to death, however, these skin lesions were not available for review except for a biopsy of the right popliteal lymph node with evidence of metastatic SCC (Figure 4).
Figure 1–4.
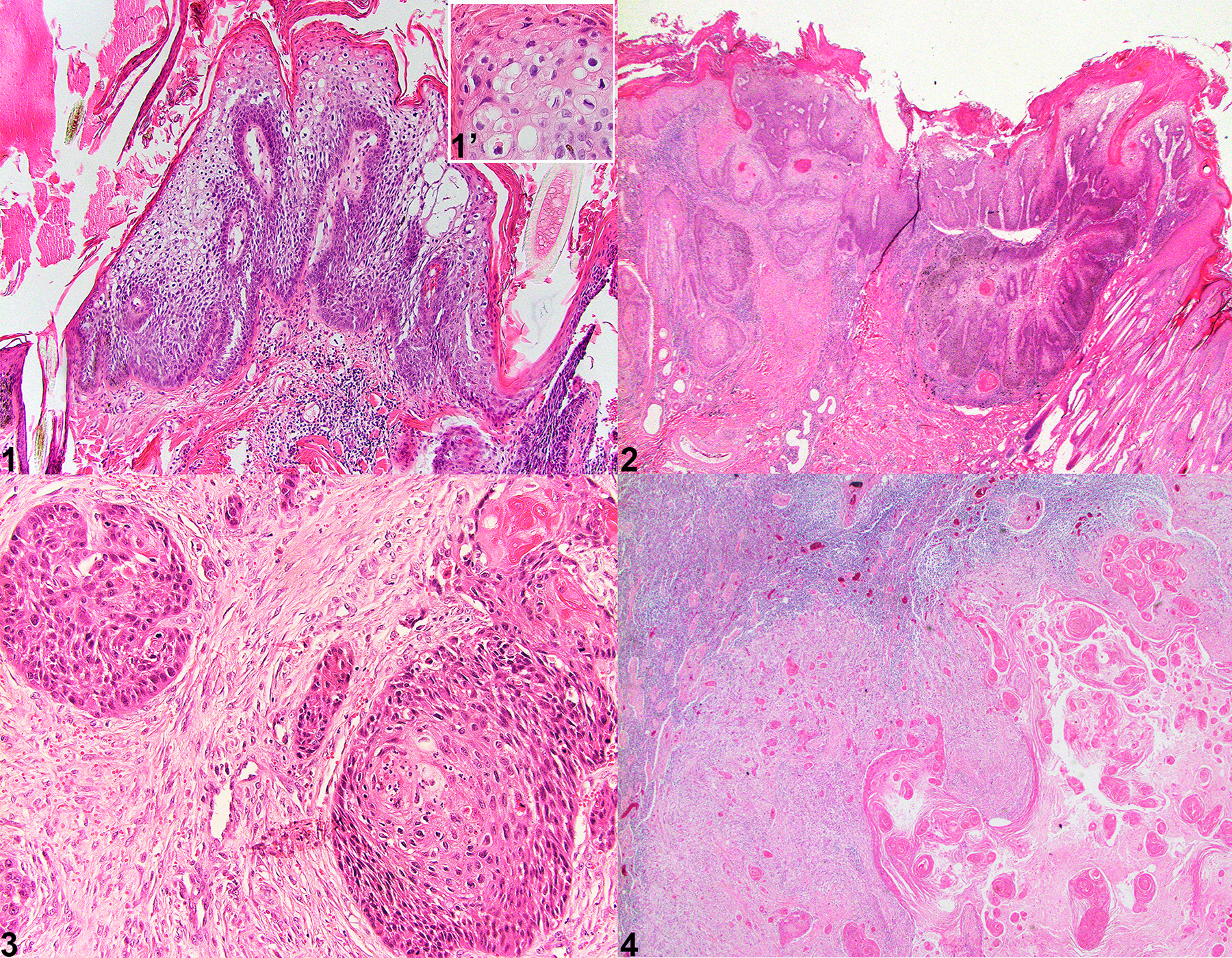
Tissues from captive North American snow leopards. Hematoxylin and Eosin. Figure 1. Case 2a. Haired skin. Cutaneous papilloma with viral cytopathic effect. The epithelium is thrown into multiple papillary projections supported by a fibrovascular stromal core. The epidermis exhibits viral cytopathic effect (Inset, Figure 1) with koilocytosis, clumping of keratin tonofilaments, and spongiosis. Figure 2. Case 2b. Haired skin. Cutaneous papilloma with transition to squamous cell carcinoma. Viral papilloma exhibits epithelial cell dysplasia with transition to invasive squamous cell carcinoma as dysplastic epithelial cells invade through the basement membrane forming islands of neoplastic cells within the superficial dermis. Figure 3. Case 4. Haired skin. Squamous cell carcinoma. Islands and lobules of dysplastic epithelial cells invade the dermis, exhibit disorderly maturation, pleomorphism, and increased mitotic count. Figure 4. Case 3. Lymph node. Cutaneous squamous cell carcinoma with metastasis to the regional lymph node. The lymph node is partially effaced by neoplastic squamous epithelial cells that form concentric lamellated layers of keratin (keratin pearls).
To determine if there is an association with papillomavirus, genomic DNA (gDNA) from each sample of CPs and SCCs was extracted from two 25μm scrolls of formalin fixed, paraffin embedded tissue blocks using a commercially available kit (DNeasy Blood and Tissue Kit, Qiagen) as previously described.2 In order to develop primers specific to a SL reference gene, PCR was run on SL samples using primers specific for canine RPL13A, K9RPL13A-For 5’ - TGG GCC GGA AGG TTG TAG TCG T-3’ and K9RPL13A-Rev 5’ - TTG CGG AGG AAG GCC AGG TAA TTC A - 3’. Using the amplified sequence, specific primers were designed using Primer3 (v. 0.4.0) design software (https://bioinfo.ut.ee/primer3-0.4.0/ and are as follows: SL-RPL13-For, 5’—CAAGTGTAAGTTCGGACGTG—3’ and SL-RPL13-Ref, 5’ CCCCAGACACACAAACATC—3’, which amplifies a 156bp product. RPL13A was amplified from all samples except one cutaneous SCC sample (Case 6). This sample was excluded from further PCR assays. An initial PCR was performed on gDNA extracted from three CP samples and one control tissue using two degenerate primer pairs, FAP64/6085 and MY09/MY11, which amplify a highly conserved region of the papillomavirus L1 gene, following previously published protocols.1,6 The positive control consisted of gDNA from a canine CP that contained papillomavirus previously shown to react with these primers. A sample without template DNA was used as the negative control. All PCR products were electrophoresed through 1% agarose, stained with SYBR Safe DNA gel stain (Thermofisher Scientific) and visualized using a BioRad ChemiDoc Imagine System. Using both sets of degenerate primers, papillomavirus DNA was identified in 1/3 of CPs (Case 1a). Positive and negative controls reacted appropriately. PCR amplicons were purified using a commercially available kit (QIAquick PCR purification kit, Qiagen) following manufacturer’s recommended protocol and submitted to a commercial DNA sequencing laboratory (Eton Biosciences, Research Triangle Park, NC). The MY09/11 and FAP6085/64 primer sets amplify different regions of the L1 gene. To obtain a slightly larger sequence of the L1 gene, specific primers were designed that spanned the intervening sequence between the two different primer sets. These primers are as follows: PuPV-3316-For 5’-TCCAATTGAACTCGTTGCTA-3’ and PuPV-3316-Rev 5’-GCGTCATAGGACTCAGGTTT-3’. PCR was then performed on the three CPs samples with the following reaction conditions: 95°C for 15 minutes followed by 45 cycles of 95°C for 1 minute, 57°C for 1 minute, and 72°C for 1 minute, followed by 72°C for 10 minutes. Amplicons from all three samples were purified and sequenced as above and yielded a 1115 bp nucleotide sequence. This sequence has been deposited in GenBank, accession number OR355483. This sequence was aligned to known papillomavirus sequences using the National Center for Biotechnology Information Basic Local Alignment Search Tool (BLAST) and was most closely related to Felis catus papillomavirus 2 (FcPV2), which shared 76% sequence homology (Figure 5). This supports identification of a novel snow leopard papillomavirus sequence. A phylogenetic tree was constructed using Geneious Prime software based upon alignment of the L1 gene from 48 different papillomaviruses representing 17 different Genera, and the partial L1 sequence obtained from this study (Figure 5).
Figure 5–8.
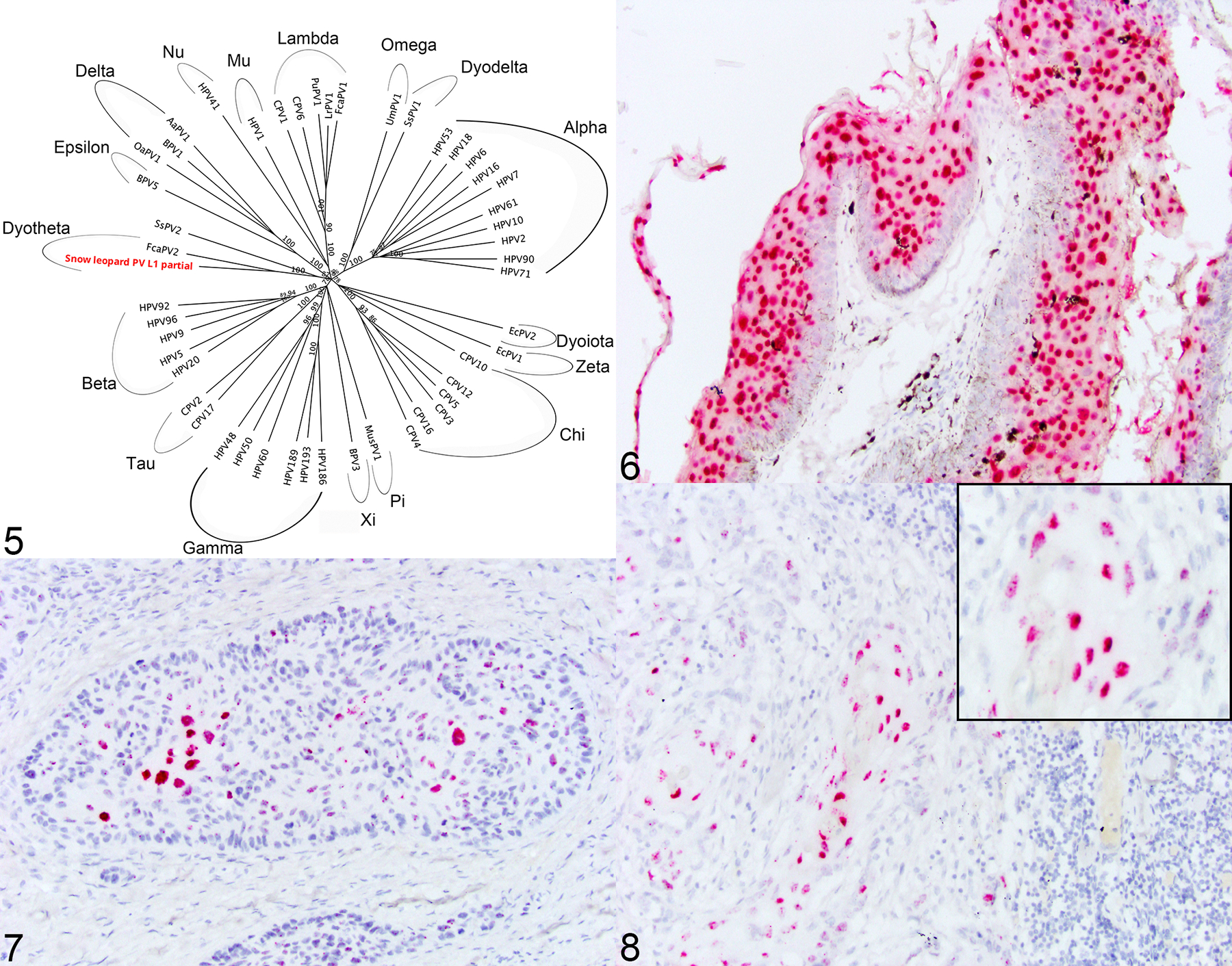
Figure 5. A phylogenetic tree was constructed based upon an alignment of the L1 gene from 48 different papillomaviruses representing 17 different Genera, and the partial L1 sequence obtained from this study (Snow leopard PV L1 partial). The putatively novel papillomavirus shares 76% sequence identity to Felis catus papillomavirus 2. Figure 6–8. Tissue from captive North American snow leopards. In situ hybridization specific for the putatively novel snow leopard papillomavirus. Figure 6. Case 1a. Haired skin. Cutaneous papilloma. Epithelial cells exhibit strong nuclear hybridization signal to probes against the putatively novel snow leopard papillomavirus. Figure 7. Case 4. Haired skin. Squamous cell carcinoma. Neoplastic epithelial cells exhibit strong nuclear hybridization signal to probes against the putatively novel snow leopard papillomavirus. Figure 8. Case 3. Lymph node. Metastatic squamous cell carcinoma. Neoplastic epithelial cells within a metastatic regional lymph node exhibit strong nuclear hybridization signal to probes against the putatively novel snow leopard papillomavirus.
Using the identified putatively novel papillomavirus DNA sequence, additional specific primer pairs were designed using Primer3 (v. 0.4.0) design software (https://bioinfo.ut.ee/primer3-0.4.0/). The primer set is as follows: PuPV-1179-For 5’-CTTTCCCCAATGCTCGCCTA-3’ and PuPV-1179-Rev 5’-TGACAAGGAACGGTTGGTGT-3’, which yields a 234bp amplicon. The PCR reaction conditions are as above for the PuPV-3316 primer set. This specific papillomavirus PCR was performed on all CPs (n=3), SCC (n=5), and control (n=7) tissue samples which identified putatively novel papillomavirus DNA in 3/3 (100%) of CPs, 4/4 (100%) of SCCs including a metastatic site to the popliteal lymph node, and 7/7 (100%) control skin samples. This data suggests that this putatively novel papillomavirus is ubiquitous in this population, being present in all samples and control tissues. The no-template DNA control was negative.
To establish a more specific association of this putatively novel papillomavirus with the development of CPs and SCC, colorimetric in situ hybridization (ISH) was performed on all FFPE tissue samples using the RNAscope ISH method (Advanced Cellular Diagnostics [ACD], Inc). A 7ZZ probed named V-PuPV-1179-L1 was designed and synthesized by ACD. A probe to the bacterial gene dihydrodipicolinate reductase (DapB) served as the negative control and a predesigned probe to the reference gene Ubiquitin C (UBC) served to ensure presence of adequate RNA within each sample (ACD). ISH was performed on 5-μm-thick FFPE sections using the RNAscope 2.5 RED assay kit following the manufacturer’s recommended protocols. As papillomaviruses are double stranded DNA viruses, this probe detects both viral DNA and viral messenger RNA transcripts of the target sequence. Strong hybridization signals for this novel papillomavirus were observed within hyperplastic epithelial cells of 3/3 of CPs (Figure 6) and neoplastic epithelial cells in 5/5 SCC samples including metastatic disease (popliteal lymph node) (Figures 7 and 8). No hybridization signals were observed in adjacent epithelial and stromal cells. No hybridization signals were identified in any of the control tissues (n=7) or using the negative control probe. Hybridization signals for UBC were identified within all samples. Given the specific localization of viral nucleic acid within both CPs and SCC and histologic evidence within one sample of a transition between the two lesion types, we propose a causation between infection with this putatively novel papillomavirus and the development of hyperplastic and neoplastic cutaneous lesions in captive NA SLs.
Over the past several decades, a viral association with CPs and SCC has been speculated within the captive NA SL population and has been name Panthera uncia papillomavirus 2 in some reports; however, the full genomic sequence would be necessary for this virus to be officially named.4–5
In a recent report evaluating free-ranging SL fecal samples for DNA viruses, a sequence was identified most similar to FcPV2, sharing 74% sequence identity to the E1 gene.3 We speculate that this virus may be the same as the putatively novel papillomavirus we identified in captive NA SLs, suggesting that this papillomavirus may occur in free-ranging populations. This information combined with the presence of DNA from this putatively novel papillomavirus within control skin samples also suggests that this virus is common in the captive population and can be present without causing disease, reinforcing the importance of using ISH to demonstrate presence of viral nucleic acid within hyperplastic or neoplastic lesions, and rule out a latent infection or surface contaminant.
In other species, including humans and domestic cats, papillomaviruses are known to express the oncogenes E6 and E7.9,11,13 The E6 oncogene activates telomerase, impairing cell cycle regulation while also inactivating the transformation related protein 53 (p53). 9,11,13 The E7 oncogene promotes cell cycling by inactivating retinoblastoma (Rb) tumor suppression protein. 9,11,13 Future studies to evaluate potential oncogene expression in these cutaneous hyperplastic and neoplastic lesions in SLs are warranted.
Ultimately, identifying a viral association with cutaneous lesions in captive NA SLs will provide guidance for the medical management of this populations and aid in the future development of prophylactic vaccines.
Acknowledgements:
The authors are grateful to the snow leopard species survival plan advisory committee Jay Tetzloff, D. McAloose, Timothy Georoff, Nancy Carpenter, and Kelly Helmick for their support with this project. The authors would like to thank the Santa Barbora Zoo, Woodland Park Zoo, Northeastern Lincoln University, Brookfield Zoo, Los Angeles Zoo, Oregan Zoo, Zoo New England, San Franciso Zoo, and Rosamond Griffon Zoo and well as the NCSU CVM histology laboratory, for their contribution to the project.
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article:
Research reported in this publication was supported by the Office of Research Infrastructure Programs of the NIH under award number K01 OD123219-03.
Footnotes
Conflict of interest: The authors declare no conflicts of interest.
Contributor Information
Mandy Womble, North Carolina State University College of Veterinary Medicine, Raleigh, NC.
Shaina Weingart, North Carolina State University College of Veterinary Medicine, Raleigh, NC.
Susan May, North Carolina State University College of Veterinary Medicine, Raleigh, NC.
Michael Garner, Northwest ZooPath, Monroe, WA.
Jennifer Luff, North Carolina State University College of Veterinary Medicine, Raleigh, NC.
References
- 1.Li J, Pan Y, Xu Z, et al. Improved detection of human papillomavirus harbored in healthy skin with FAP6085/64 primers. J Virol Methods. 2013. Nov;193(2):633–8. [DOI] [PubMed] [Google Scholar]
- 2.Luff JA, Affolter VK, Yeargan B, Moore PF. Detection of six novel papillomavirus sequences within canine pigmented plaques. J Vet Diagn Invest. 2012;24(3):576–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johannson O, Ullman K, Lkhagvajav P, Wiseman M, Malmsten J, and Leijon M. 2020. Detection and genetic characterization of viruses present in free-ranging snow leopards using next-generation sequencing. Front. Vet Sci. 2020:7:645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joslin J, Garner M, Collins D, et al. Viral papilloma and squamous cell carcinomas in snow leopards (Uncia uncia): In: Proc Am Assoc Zoo Vet and Internat Assoc Aq Anim Med. 2000:155–58. [Google Scholar]
- 5.Joslin J, Jenson AB, Ghim SJ, et al. In: Proc Am Assoc Zoo Vet and Assoc Rept and Amp Vet and the Assoc Exo Mam Vet, 2012;39–40. [Google Scholar]
- 6.Manos M, et al. The use of polymerase chain reaction amplification for the detection of genital human papillomaviruses In: Furth M, Greaves M, eds. Molecular Diagnostics of Human Cancer. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 1989:209–214. [Google Scholar]
- 7.McCarthy T, Mallon D, Jackson R, Zahler P & McCarthy K 2017. Panthera uncia. The IUCN Red List of Threatened Species 2017: e.T22732A50664030. 10.2305/IUCN.UK.2017-2.RLTS.T22732A50664030.en. Accessed on 15 September 2023. [DOI] [Google Scholar]
- 8.Mitsouras K, Faulhaber EA, Hui G, Joslin JO, Eng C, Barr MC, Irizarry KJ. Development of a PCR assay to detect papillomavirus infection in the snow leopard. BMC Vet Res. 2011;7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munday JS. Papillomaviruses in felids. Vet J. 2013;199:340–347. [DOI] [PubMed] [Google Scholar]
- 10.Munday JS, Gibson I, French AF. Papillomaviral DNA and increased p16CDKN2A protein are frequently present within feline cutaneous squamous cell carcinomas in ultraviolet-protected skin. Vet Dermatol. 2011;22(4):360–6. [DOI] [PubMed] [Google Scholar]
- 11.Munday JS, Kiupel M. Papillomavirus-associated cutaneous neoplasia in mammals. Vet Pathol. 2020;47(2):254–264. [DOI] [PubMed] [Google Scholar]
- 12.Munday JS, Kiupel M, French AF, Howe L. Amplification of papillomaviral DNA sequences from a high proportion of feline cutaneous in situ and invasive squamous cell carcinomas using a nested polymerase chain reaction. Vet Dermatol. 2008;19(5):259–63. [DOI] [PubMed] [Google Scholar]
- 13.Munday JS, Thomson NA, Luff JA. Papillomaviruses in dogs and cats. Vet J. 2017;225:23–31. [DOI] [PubMed] [Google Scholar]
- 14.Sundberg JP, Van Ranst M, Montali R, Homer BL, Miller WH, Rowland PH, Scott DW, England JJ, Dunstan RW, Mikaelian I, Jenson AB. Feline papillomas and papillomaviruses. Vet Pathol. 2000;37:1–10. [DOI] [PubMed] [Google Scholar]
- 15.Terio KA, McAloose D, Mitchell (nee Lane) E. Felidae. In: McAloose D, Terio K, St. Leger J (eds.). Pathology of Zoo and Wildlife. London, United Kingdom: Elsevier; 2018. p. 263–285. [Google Scholar]
- 16.Womble M, Georoff TA, Helmick K, et al. Mortality review for the North American snow leopard (Panthera uncia) zoo population from January 1999 to December 2019. J Zoo Wildl Med. 2021:52(1):145–156. [DOI] [PubMed] [Google Scholar]


