Abstract
Background
Artemisinin-based combination therapy (ACT) is currently recommended for treatment of uncomplicated malaria. However, the emergence and spread of partial artemisinin resistance threatens their effectiveness for malaria treatment in sub-Saharan Africa where the burden of malaria is highest. Early detection and reporting of validated molecular markers (pfk13 mutations) in Plasmodium falciparum is useful for tracking the emergence and spread of partial artemisinin resistance to inform containment efforts.
Methods
Genomic surveillance was conducted at 50 surveillance sites across four regions of Uganda in Karamoja, Lango, Acholi and West Nile from June 2021 to August 2023. Symptomatic malaria suspected patients were recruited and screened for presence of parasites. In addition, dried blood spots (DBS) were collected for parasite genomic analysis with PCR and sequencing. Out of 563 available dried blood spots (DBS), a random subset of 240 P. falciparum mono-infections, confirmed by a multiplex PCR were selected and used for detecting the pfk13 mutations by Sanger sequencing using Big Dye Terminator method. Regional variations in the proportions of pfk13 mutations were assessed using the chi square or Fisher’s exact tests while Kruskal–Wallis test was used to compare absolute parasite DNA levels between wild type and mutant parasites.
Results
Overall, 238/240 samples (99.2%) contained sufficient DNA and were successfully sequenced. Three mutations were identified within the sequenced samples; pfk13 C469Y in 32/238 (13.5%) samples, pfk13 A675V in 14/238 (5.9%) and pfk13 S522C in (1/238 (0.42%) samples across the four surveyed regions. The prevalence of pfk13 C469Y mutation was significantly higher in Karamoja region (23.3%) compared to other regions, P = 0.007. The majority of parasite isolates circulating in West Nile are of wild type (98.3), P = 0.002. Relative parasite DNA quantity did not differ in samples carrying the wild type, C469Y and A675V alleles (Kruskal–Wallis test, P = 0.6373).
Conclusion
Detection of validated molecular markers of artemisinin partial resistance in multiple geographical locations in this setting provides additional evidence of emerging threat of artemisinin partial resistance in Uganda. In view of these findings, periodic genomic surveillance is recommended to detect and monitor levels of pfk13 mutations in other regions in parallel with TES to assess potential implication on delayed parasite clearance and associated treatment failure in this setting. Future studies should consider identification of potential drivers of artemisinin partial resistance in the different malaria transmission settings in Uganda.
Keywords: Genomic surveillance, Plasmodium falciparum, pfk13 mutations, Emergence, Artemisinin resistance
Background
Malaria remains a public health problem in Uganda. The country contributed 5.4% of the 249 million malaria cases reported globally making it the 3rd largest contributor to the global malaria burden in 2022 [1]. The entire country’s population remains at risk of infection. Transmission is highly heterogeneous between regions, sub-national levels and among specific sub-populations [2]. Uganda’s national malaria programme has achieved substantial success in implementation of the core interventions such as mass campaigns for long-lasting insecticidal nets (LLIN), indoor residual spraying (IRS), integrated community case management (iCCM), intermittent preventive treatment in pregnancy (IPTp) as well as case management. The programme has also invested substantially in the private sector in form of co-payments to subsidize anti-malarial drugs making them accessible to the population [2].
However, despite the investments and milestones achieved in intervention scale-up and coverage, the country’s progress against malaria has levelled-off and many districts and regions appear to be losing ground with potential for reversal [3]. Overall, the recent malaria programme review 2019–2020 has shown limited effect of interventions on reducing malaria burden in all districts. Recently, malaria control in Uganda is further threatened by the reports of emerging biological and genetic changes in malaria parasites and vectors. While artemisinin-based combination therapy (ACT) remains efficacious against malaria, presence of partial artemisinin resistance has been reported for the first time on the African continent with reports of delayed parasite clearance. [4, 5]. Continuous monitoring of ACT efficacy is needed to inform treatment policies in malaria-endemic countries, and to ensure early detection of, and response to, drug resistance [6–8].
Uganda’s Malaria Programme adopted the World Health Organization (WHO) recommendation on the use of ACT as the first- and second-line treatment for uncomplicated Plasmodium falciparum malaria [9, 10]. The country’s current treatment policy for uncomplicated malaria recommends three artemisinin-based combinations that combine an artemisinin derivative (artesunate, artemether and dihydroartemisinin) with a partner drug (amodiaquine, lumefantrine and piperaquine), respectively. The role of the artemisinin compound is to reduce the number of parasites during the first 3 days of treatment (reduction of parasite biomass), while the role of the partner drug is to eliminate the remaining parasites (cure). Artemisinin partial resistance first emerged in Cambodia as delayed parasite clearance after treatment with artemisinin derivatives and increased survival of ring stage parasites following exposure to artemisinin derivatives [9]. In vitro and in vivo studies have shown that mutations in the PfKelch13 propeller domain (pfk13 gene) are associated with this delayed parasite clearance [8, 9]. The molecular markers based on mutations of the pfk13 gene are classified as either validated, associated or candidate markers. In order to classify a marker as a candidate or associated pfk13 markers of artemisinin partial resistance, there must be a statistically significant association (p < 0.05) between that pfk13 mutation and parasite clearance half-life > 5 h, or day 3 parasitaemia on a sample of at least 20 clinical cases, or survival of > 1% using the RSA0–3 h in at least five individual isolates with a given mutation, or a statistically significant difference (p < 0.05) in the RSA0–3 h assay between culture-adapted recombinant isogenic parasite lines, produced using transfection and gene editing techniques, which express a variant allele of pfk13 as compared with the wild-type allele. The pfk13 mutations are classified as a validated marker of artemisinin partial resistance if it satisfies both of the above requirements [11].
Particularly more concerning are recent reports of emergence of artemisinin partial resistance in Africa particularly in Rwanda, Uganda, Tanzania and the Horn Africa [5, 8, 12–15].
Available evidence shows that the selection of pfk13 mutations in Uganda was at a comparable rate to that observed in South-East Asia (SEA) in samples collected between 2016 and 22, suggesting that pfk13 mutations may continue to increase quickly in Uganda [16]. Several studies previously conducted in Uganda have reported the presence of pfk13 469Y, C469F, P574L and A675V as common mutations in Northern and other regions of Uganda [14, 17] while others reported non-synonymous and synonymous pfk13 single nucleotide polymorphisms (SNPs) [18, 19]. Other studies either reported lower prevalence or detected no pfk13 mutations in Ugandan parasite isolates [20–26]. Among the pfk13 mutations reported in previous studies in Uganda, the C469Y, A675V and P574L are all classified by the WHO as validated markers of partial artemisinin resistance. Recent therapeutic study in Uganda (unpublished 2023) has shown decreased artemether/lumefantrine efficacy of 84.6% (95% CI 76.5–90.3) and 82.9% (95% CI 74.5–88.9) at Busia and Arua sites, respectively.
Elsewhere on the African continent similar markers of resistance have been reported in Rwanda, Sudan, Tanzania, Ethiopia and Eritrea suggesting widespread potential for artemisinin resistance in parasite population on the continent [15, 27–30]. Previous reports suggest independent and spontaneous emergence of parasites with pfk13 mutations in multiple location based on evidence of unrelatedness between mutant strains collected in different geographical locations [13, 31]. Delayed parasite clearance has been detected in returning travellers in the UK and other non-malaria endemic territories posing threat to global health security [32–34]. In an effort to manage and contain the threat, the WHO has recently developed a strategy for artemisinin resistance management for Africa [8]. Due to the changing dynamics of malaria transmission and parasite related biological changes, the molecular epidemiology and the extent of spread of artemisinin resistant parasites as well as its effect on malaria control in northern regions of Uganda is unclear. Widespread artemisinin resistance, as observed in Southeast Asia, has the potential to disrupt and reverse significant gains that have been achieved against the malaria burden in Africa [8, 35, 36]. Timely molecular surveillance and detection of pfk13 mutations is a useful tool for tracking the emergence and containment of the spread of artemisinin resistance [10, 11]. The purpose of this study was to assess the presence and distribution of pfk13 polymorphisms in P. falciparum parasites at fifty surveillance sites in Northern Uganda representing a wide range of transmission setting.
Methods
Study design and setting
A total of 563 dried blood spots (DBS) were available from a primary study for pfhrp2 and pfhrp3 deletion surveillance [37]. In the primary study, all the 563 available DBS samples had been confirmed with multiplex qPCR for presence of P. falciparum parasite DNA out of which 73.9% (416/563) gave valid PCR results. The current study utilized a random sample of 240 dried blood spots (selected from the 416) that were sequenced to assess the presence of pfk13 mutations across the surveillance regions. Selection of 240 DBS for sequencing ensured even distribution across the four survey regions (60 per region).
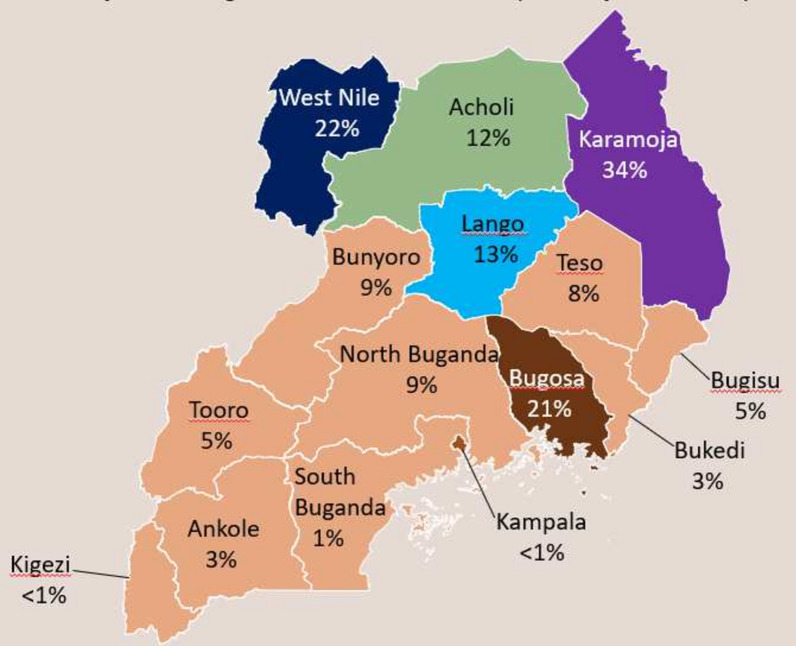
Details of the study population, settings, DBS collection and sampling were published under the primary study where the samples originated [37]. Briefly, capillary blood was collected from malaria symptomatic patients seeking care across 50 surveillance sites in the regions of Acholi, Karamoja, W. Nile and Lango in northern Uganda. Capillary blood collected from eligible patients screened and enrolled was spotted onto filter paper (Whatman No. 903). The four regions are well designed demographic health survey (DHS) clusters or enumeration areas that are periodically used for the national malaria indicator surveys in the country [38] and serve a total population of 10 million people that are at risk of malaria infection based on the recent national housing and population census [39]. Malaria transmission across all the four regions is intense and stable at holoendemic levels, with a test positivity rate (TPR) ranging from 50 to 70% at health facility and parasite prevalence of 13–34% as determined by blood smear microscopy (Fig. 1) [38].
Fig. 1.
Malaria transmission and parasite prevalence in the survey regions. Figure shows the population-based parasite prevalence per survey region including the four regions of Karamoja, Lango, Acholi and West Nile from where filter papers samples were collected
(The figure is extracted from the most recent Uganda national malaria indicator survey report [38])
Parasite DNA extraction and quality control
Genomic DNA extracted previously [37] for the detection of pfhrp2/3 deletions was stored at − 20 °C to preserve its integrity and was available for use in this study to amplify and sequence for pfk13 mutations. The presence of parasite DNA in the samples was confirmed using a multiplex qPCR assay adapted from Grignard et al. [40], which detects human tubulin, parasite lactate dehydrogenase (pLDH), and P. falciparum histidine rich proteins 2 (pfhrp2) and 3 (pfhrp3) genes. Samples positive for both human tubulin and pLDH were deemed eligible for selection for pfk13 analysis, regardless of their pfhrp2 and pfhrp3 status. Quantity of parasite DNA relative to human blood volume in each sample was calculated based on ΔCq values of pLDH-human tubulin as an indicator for parasite density.
PCR amplification of pfk13 propeller region
Amplification of the pfk13 propeller region (849 bp (1329-to-2178 nucleotides) was achieved through nested PCR as described previously by Ariey et al. [41] using primers K13-1 5′-cggagtgaccaaatctggga-3′ and K13-4 5′-gggaatctggtggtaacagc-3′ in the primary PCR as well as K13-2 5′-gccaagctgccattcatttg-3′ and K13-3 5′-gccttgttgaaagaagcaga-3′ in the nested PCR. Both the primary and nested PCR reactions consisted of 1 μl of DNA template, 1 μM of each primer, 0.2 mM dNTPs (Invitrogen, USA) and 2 U Taq DNA polymerase (Invitrogen, USA) in a volume of 25 μl. The cycling programme was as follows: 5 min at 94 °C, followed by 40 cycles of 30 s at 94 °C, 90 s at 60 °C, 90 s at 72 °C and final extension of 10 min at 72 °C on a Mastercycler X50s (Eppendorf, Germany).
Sequencing of pfk13 propeller region
PCR products were confirmed using gel electrophoresis on a 2% agarose gel stained with GelRed® Nucleic Acid Gel Stain (Biotium, USA). Positive samples were purified with the DNA Clean & Concentrator-5 PCR purification kit (Zymo, USA) and sequenced from both directions. A mixture containing 3 μl purified PCR product, 2 μl unidirectional primer (K13-2 or K13-3) and 5 μl nuclease-free water was submitted to QIMR Berghofer Medical Research Institute (Brisbane, Queensland, Australia) for sequencing with Big Dye Terminator v3.1 (Applied Biosystems, USA). Sequences were aligned using ClustalW in MEGA 11 with the corresponding region from parasite clone Pf3D7 used as a reference. Sequencing trace of samples that had both wild type and mutant sequence peaks at the same location were classified as mixed wild type and mutant alleles. Proportions of mutant in a sample was determined by peak height of mutant allele relative to wild type allele as 10–29%, 30–59%, 60–89% and 90–100%. Mutant allele was determined as a dominant or non-dominant allele when its peak height relative to wild type was > 60% or < 60%, respectively.
Data analysis
Electronic excel database of the previous study that linked study identification numbers of samples to their respective demographics was accessed for data extraction. Data quality checks were done to assess its quality and consistency. Data analysis was done with STATA Version 14 (College Station, TX, USA: StataCorp LP). Descriptive analysis was done to describe the baseline characteristics of samples and determine proportion, distribution and spread of pfk13 mutations in the four surveillance regions. Statistical analysis was done using Chi-square or Fisher’s exact test to assess regional variation in proportions of pfk13 mutations while Kruskal–Wallis test was used to compare absolute parasite DNA levels between wild type and mutant parasites with a p value < 0.05 considered statistically significant.
Ethical approval
The study was approved by the Makerere University School of Public Health Research and Ethics committee (#REC REF SPH-2021-135) and the Uganda National Council of science and technology (Ref No. HS1911ES). In the study surveys from where samples were collected, participants provided written informed consent for future use of biological samples for molecular analysis.
Results
Baseline characteristics of dried blood spots from where the sequenced samples originated
Overall, 390 (69.4%) and 173 (30.6%) were from symptomatic individuals of ≥ 5 and < 5 years old respectively. Samples were evenly distributed across all the four regions of Karamoja 24.0% (135), West Nile 23.7% (133), Acholi 24.8% (140) and Lango 27.4% (155) Table 1. Across the four surveillance regions, a total population of 10,000,000 people is at risk of malaria infection.
Table 1.
Characteristics of samples (N = 563) from where the K13 sequenced samples originated
| Variables | Frequency (n) | Proportion (%) |
|---|---|---|
| Age (years) | ||
| < 5 | 173 | 30.6 |
| ≥ 5 | 390 | 69.4 |
| Sex | ||
| Male | 220 | 39.0 |
| Female | 343 | 60.7 |
| Samples collected per survey region | ||
| Acholi | 140 | 24.9 |
| Lango | 155 | 27.4 |
| W. Nile | 133 | 23.7 |
| Karamoja | 135 | 24.0 |
Parasite DNA amplification and pfk13 mutations detected in the samples
Out of a random sample of 240 PCR positive DBS, 238/240 (99.2%) were successfully sequenced and returned good sequences. Three (mutations) polymorphisms in the pfk13 propeller region were identified in these 238 samples: C469Y, S522C and A675V. Mutation S522C was detected in only one sample from Lango (1/238, 0.42%). The most prevalent mutation, C469Y, was detected in 32/238 samples (13.6%), and majority (31/32) were detected in Acholi, Karamoja and Lango regions. Furthermore, 21/32 (65.6%) of the samples with C469Y mutation had mixed sequences of both the wild type and mutant alleles. 11/15 (73.3%) of samples where the mutant is dominant (> 60%) originated in Karamoja, whereas non-dominant (< 60%) mutations were more common in Lango (8/9, 88.9%) and in Acholi (5/8, 62.5%). pfk13 A675V was detected in 14/238 (5.9%) valid sequences which were spread evenly across Acholi (4/14, 28.6%), Karamoja (5/14, 35.7%) and Lango (5/14, 35.7%). The wild type allele only was detected in 191/238 samples (80.3%), majority of which were in West Nile. An additional 21 samples (21/238, 8.8%) contained mixed pfk13 C469Y mutations i.e. both the wild type and mutant alleles were detected (Table 2).
Table 2.
Prevalence and distribution of the PfK13 mutations (S522C, A675V, C469Y) and C469Y mutant allele or mixed with wild-type allele across the survey regions
| Region | Valid sequences | Wild Type Only | S522C | A675V | C469Ya | C469Y | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10–29%b | 30–59%b | 60–89%b | 90–100%c | ||||||||||||||
| n | % | n | % | n | % | N | % | n | % | N | % | n | % | n | % | ||
| Acholi | 59 | 47 | 79.7 | 0 | 0 | 4 | 6.8 | 8 | 13.6 | 5 | 8.5 | 0 | 0.0 | 1 | 1.7 | 2 | 3.4 |
| West Nile | 59 | 58 | 98.3 | 0 | 0 | 0 | 0.0 | 1 | 1.7 | 0 | 0.0 | 1 | 1.7 | 0 | 0.0 | 0 | 0.0 |
| Karamoja | 60 | 41 | 68.3 | 0 | 0 | 5 | 8.3 | 14 | 23.3 | 2 | 3.3 | 1 | 1.7 | 3 | 5.0 | 8 | 13.3 |
| Lango | 60 | 45 | 75.0 | 1 | 1.7 | 5 | 8.3 | 9 | 15.0 | 3 | 5.0 | 5 | 8.3 | 0 | 0.0 | 1 | 1.7 |
| Total | 238 | 191 | 80.3 | 1 | 0.4 | 14 | 5.9 | 32 | 13.4 | 10 | 4.2 | 7 | 2.9 | 4 | 1.7 | 11 | 4.6 |
aIncludes mixed parasite populations with C469Y mutant allele and wild-type alleles
b% mutant in sample
cPrevalence of single clone mutation
Regional variation of pfk13 mutations
The pfk13 mutation C469Y was higher in proportion in Karamoja region (23.3%) compared to other surveillance regions (P = 0.007). The largest proportion of parasite populations circulating in West Nile was of wild type (58/59, 98.3%), significantly higher than other regions (P = 0.002). In contrast, the proportions of the S522C and A675V were not significantly different across the study regions (Table 3).
Table 3.
Regional variation of Pfk13 mutations
| Mutation | Status | Acholi (n = 59) (%) | Karamoja (n = 60) (%) | Lango (n = 60) (%) | West Nile (n = 59) (%) | P value |
|---|---|---|---|---|---|---|
| Wild Type | Present | 47 (79.6) | 41 (68.3) | 45 (75.0) | 58 (98.3) | 0.002$ |
| Absent | 12 (20.3) | 19 (31.7) | 15 (25.0) | 1 (0.0) | ||
| C469Y | Present | 8 (13.6) | 14 (23.3) | 9 (15.0) | 1 (1.7) | 0.007$ |
| Absent | 51 (86.4) | 46 (76.7) | 51 (85.0) | 58 (98.0) | ||
| S522C | Present | 0 (0.0) | 0 (0.0) | 1 (1.7) | 0 (0.0) | 1.000* |
| Absent | 59 (100.0) | 60 (100.0) | 59 (98.3) | 59 (100.0) | ||
| A675V | Present | 4 (6.8) | 4 (6.7) | 5 (8.3) | 0 (0.0) | 0.095* |
| Absent | 55 (93.2) | 56 (93.3) | 55 (91.7) | 59 (100.0) |
$Chi square P value, *Fishers exact test P value
Relative quantity of parasite DNA
Relative parasite DNA quantity (as a surrogate for parasite density in peripheral blood) in samples carrying the wild type, C469Y and A675V alleles was compared between wild type and mutants based on ΔCq (pLDH-Human tubulin). There was no significant difference (Kruskal–Wallis test, p = 0.6373) in DNA concentrations across the three pfk13 alleles (WT, C469Y, A675V) suggesting comparable peripheral parasite densities between parasites carrying a wild type and a mutant pfk13 (Fig. 2).
Fig. 2.
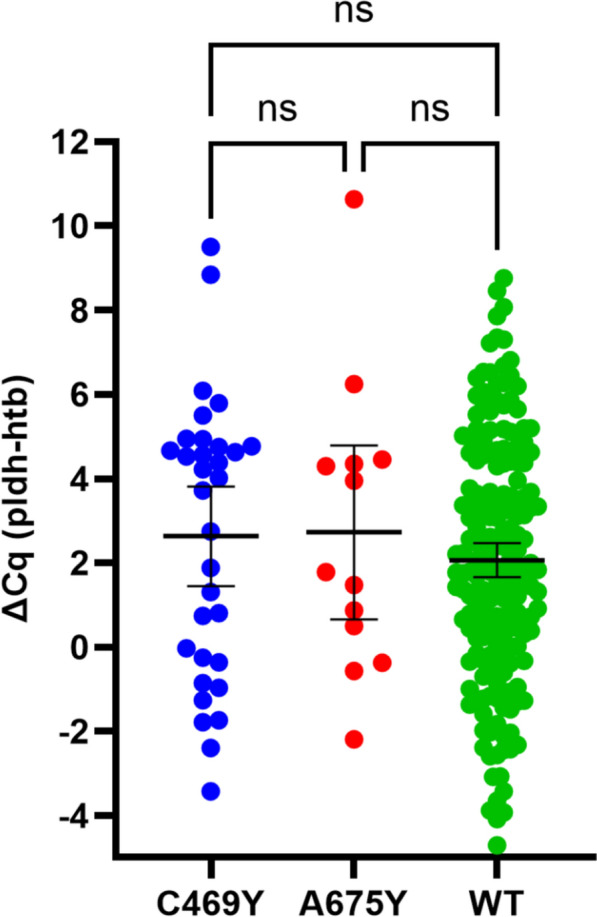
Comparison of relative parasite DNA quantity between wild type (WT) samples and those with C469Y or A675V mutations. Ns not significant, wt wild type, pldh parasite lactate dehydrogenase, cq quantification cycle (cycle number at which the curve first rises above the threshold level), htb human tubulin
Discussion
Genomic surveillance data for malaria parasites and vectors can provide key information for implementation and control efforts in malaria endemic regions including timely detection and response to biological threats such as the development of resistance to drugs and insecticides. Molecular markers of partial artemisinin resistance (pfk13 mutations) have been used as early pointers of partial artemisinin resistance to inform early response and containment efforts [8]. Historically, Uganda introduced and adopted the use of ACT as treatment of choice for uncomplicated malaria in early 2000’s. Reports of development of partial artemisinin resistance are coming on board close to two decades after the initial introduction of ACT in Uganda. The detection of validated markers of artemisinin resistance pfk13 C469Y and pfk13 A675V in Northern Uganda adds to the existing body of evidence reporting potential development of partial artemisinin resistance in Africa [8, 13]. Similarly, the C469Y and 675V had been previously reported in Central region of Uganda [14]. On the contrary, different types of pfk13 mutations 469F and 561H were detected at high prevalence in southwestern Uganda near the Uganda-Rwanda border [14, 26]. The 561H had been detected at high prevalence in Rwanda where partial artemisinin resistance has been confirmed [15, 42]. The P441L was also detected at several sites in far western Uganda [14].
Of concern is the presence of validated molecular markers of partial artemisinin resistance in this traditionally high malaria transmission setting in northern Uganda. Validated makers are known to improve parasite survival in vitro during the ring-stage survival assay (RSA), as well as delayed parasite clearance in vitro or in clinical cases assessed by day 3 parasitaemia [43]. The pfk13 mutations, such as C580Y and R539T, have been associated with partial artemisinin resistance in the greater Mekong region where the first reports of delayed parasite clearance and treatment failure following ACT occurred [44, 45]. Uganda’s malaria treatment policy recommends the use of artemether in combination with lumefantrine as partner drug for first line treatment of uncomplicated malaria, Artesunate combined with amodiaquine as alternative first line treatment for uncomplicated malaria and dihydroartemisinin combined with piperaquine as partner drug as second line treatment for uncomplicated malaria. The drugs were selected based on evidence on safety and efficacy generated from in-country therapeutic efficacy studies [10]. Although artemisinin resistance data in Uganda is currently limited by coverage, delayed parasite clearance has been reported in therapeutic efficacy studies (TES) at Arua site in West Nile and Busia site in Eastern Uganda [46], which points to potential development of resistance in the country.
The observed heterogeneity in the frequency of pfk13 mutations between regions may suggest differences in the intrinsic factors linked to the parasite, host and drugs used and the environmental factors all of which are documented drivers of artemisinin resistance. Treatment practices (both patient and provider) related drivers include those affecting frequency dose and duration in which parasite population is exposed to the given drug [8]. However, additional studies need to be conducted to establish the actual drivers of partial artemisinin resistance in the Ugandan context. While the malaria treatment policy on first- and second-line drugs for the treatment of uncomplicated and severe malaria applies uniformly in Uganda, previous surveys by ACT Watch have reported the presence and use of unauthorized artemisinin-based combinations circulating in the private sector including monotherapies [47, 48]. Additional ACT surveys conducted under the Affordable Medicines Facility for malaria (AMFm) co-payments have reported similar findings of presence of non-quality assured ACT on the market in Uganda and Tanzania [48, 49]. Unauthorized and non-quality assured ACT are associated with poor quality based on the active pharmaceutical ingredient (API) of artemisinin measurements that are outside the acceptable WHO API range and the associated risks of sub-optimal doses [49]. Based on these survey findings and those from previous studies, it can be seen that the Northern region particularly Acholi and Lango accounts for the higher burden of validated ACT resistance markers [5, 14, 21]. Additional plausible explanation for this trend could be related to the political instability that lasted close to two decades in those regions characterized by population displacements [50]. Political instability is associated with breakdown of health systems affecting health service delivery such as implementation of case management trainings, treatment policies as well as supervision of drug use and regulation both in public and private sector [51]. Uncontrolled drug use and unregulated antimalaria drug supply chains can potentially continuously expose the parasite to sub-optimal doses and hence mounting intense selection pressure leading to resistance. Similar conditions existed in the Mekong region particularly at the Thai-Cambodia border that has been described as the epicenter and origin of all anti-malarial drug resistance [52, 53]. The area is characterized by uncontrolled cross border movements of forest and rubber plantation workers reported to have contributed to chloroquine and later artemisinin resistance in the region [54].
Interestingly, the prevalence of pfk13 C469Y was higher in Karamoja region (23.3%) where malaria transmission is persistently highest over the last 5 years based on the last malaria indicator surveys [38]. The association between presence of molecular markers and the persistent malaria transmission in the Karamoja region needs to be investigated further including consideration for positioning a TES site. Historically, the Karamoja region in inhabited by mobile nomadic and pastoralist populations with uncontrolled activities across the porous border between Karamoja region and North western Kenya which could provide potential favorable conditions for drug resistance development [55]. Similarly, the presence of mutations C469Y and A675V in relative higher frequency in Lango and Acholi may suggest close monitoring to detect any delayed parasite clearance including pharmacokinetic studies to monitor efficacy of the partner drugs. A single sample carrying S522C was identified in Lango indicating its low frequency in the study areas. This mutation had been reported to be associated with delayed parasite clearance, but not considered by the WHO as a validated or candidate mutation due to insufficient evidence [56] and was detected in low frequency in Kenya [57].
Finally, the impact of pfk13 mutations on parasite fitness remains unclear. While C580Y mutation was shown to confer modest fitness costs when accompanied by multicopy pm2/3, it remains fitness-neutral in the presence of single pm2/3 copy [58]. An assessment was done as part of the current study to check if the parasite isolates carrying the pfk13 mutations had different DNA levels (a proxy of parasite densities) in peripheral blood of patients compared to parasites with wild type pfk13 alleles. A significant difference in parasite densities may provide an indication if pfk13 mutant parasites have likely gained or lost fitness compared to the wild type parasites. Similarities in relative DNA concentrations across the wild type, C469Y and A675V pfk13 alleles between the parasites (Kruskal–Wallis test, p = 0.6373) indicated comparable peripheral parasite densities between parasites carrying a wild type and a mutant pfk13 suggesting comparable parasite fitness during blood stage. Future surveys could consider parasite density and fitness studies between wild type and mutant parasites (with K13 mutations) in longitudinal prospective designs.
Implication for the national malaria programme
The presence of parasites with validated markers of partial artemisinin resistance particularly in this traditionally high malaria transmission setting calls for increased vigilance that may include development of a country specific artemisinin resistance management strategy. Expansion of parasite genomic surveillance to enhance detection of molecular markers of artemisinin resistance in other regions of Uganda as well as monitoring the levels in regions where they have been confirmed is recommended [8]. In areas with confirmed presence of P. falciparum parasites with validated markers of partial artemisinin resistance, consideration for location and positioning of TES sites may be necessary to assess the extent to which presence of markers at those levels translates into delayed parasite clearance or treatment failure in this setting. In view of these findings and similar results reported previously [5, 14, 21, 26, 46, 59, 60], the programmatic implication may include comprehensive evidence review to identify the possible drivers of partial artemisinin resistance in Uganda to inform containment efforts. Based on the current guideline [8], the country specific strategy would aim to intensify genomic surveillance and TES to enhance early detection, delay the emergence of resistance through appropriate treatment practices and drug regulations to avoid exposure of the parasites to sub-optimal doses of ACT and finally, to limit the spread of resistant parasite strains to other areas. For now, only one TES has reported declining efficacy of artemether/lumefantrine at Busia and Arua study sites with PCR-corrected efficacy of AL below the 90% WHO threshold at both sites [46]. The declining efficacy seen from TES coupled with presence of markers calls for increased vigilance to detect and contain emergence of partial artemisinin resistance that could be a potential disaster for malaria control efforts.
Conclusion
Detection of validated molecular markers of artemisinin partial resistance in multiple geographical locations in this high malaria transmission setting provides additional evidence of emerging threat of partial artemisinin resistance in Uganda. In view of these findings, periodic genomic surveillance is recommended to detect and monitor levels of pfk13 mutations in other regions in parallel with TES to assess potential implication on delayed parasite clearance and associated treatment failure in this setting. Future studies should consider identification of potential drivers of artemisinin partial resistance in the different malaria transmission settings in Uganda.
Acknowledgements
We thank the field study teams for leading the field activities and sample collections at the facilities. We are grateful to the study participants who participated in the survey.
Abbreviations
- ACT
Artemisinin-based combination therapy
- Cq
Quantitative cycle
- DBS
Dried blood spots
- DHS
Demographic health program
- HRP2
Histidine rich protein 2
- htb
Human tubulin
- PCR
Polymerase chain reaction
- pfhrp2
Plasmodium falciparum histidine rich protein 2 gene
- pfhrp3
Plasmodium falciparum histidine rich protein 3 gene
- pldh
Parasite lactate dehydrogenase
- qPCR
Quantitative polymerase chain reaction
- RDTs
Rapid diagnostic tests
- RSA
Ring-stage survival assay
- TES
Therapeutic efficacy studies
- WHO
World Health Organization
- WT
Wild type
Author contributions
BBA designed the study. BBA, DS, JT, QC did the genomic and sequencing, BBA, RM, JT, QC supported the data analysis. BBA drafted the manuscript. BBA, SR, SR, MZ, CK, DB, JO, MA, BK, JN, MK, PK, PP, QC reviewed the manuscript. All authors read and approved the final.
Funding
This work was supported, in whole or in part, by the Bill & Melinda Gates Foundation Grant Number: INV-031515. The grant is awarded to BBA as the Principal investigator. The molecular analysis of parasites was funded by the US DoD Armed Forces Health Surveillance Division, Global Emerging Infections Surveillance Branch (AFHSD/GEIS), PROMIS ID P0111_22_AF. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Ethical approval for the study was obtained from the Makerere University School of Public of Health Research and Ethics Committee, the Uganda National Council of Science and Technology.
Consent for publication
All authors read and approved the manuscript for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO. World malaria report. Geneva: World Health Organization; 2022. [Google Scholar]
- 2.NMCD. National malaria control strategic plan, Uganda. 2021.
- 3.DHIS2. Health management information system, Uganda. 2023.
- 4.Agaba BB, Anderson K, Gresty K, Prosser C, Smith D, Nankabirwa JI, et al. Molecular surveillance reveals the presence of pfhrp2 and pfhrp3 gene deletions in Plasmodium falciparum parasite populations in Uganda, 2017–2019. Malar J. 2020;19:300–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balikagala B, Fukuda N, Ikeda M, Katuro OT, Tachibana SI, Yamauchi M, et al. Evidence of artemisinin-resistant malaria in Africa. N Engl J Med. 2021;385:1163–71. [DOI] [PubMed] [Google Scholar]
- 6.WHO. Template protocol for therapeutic efficacy studies (TES). Geneva: World Health Organization. https://www.who.int/teams/global-malaria-programme/case-management/drug-efficacy-and-resistance/tools-for-monitoring-antimalarial-drug-efficacy.
- 7.WHO. Report on antimalarial drug efficacy, resistance and response. Geneva: World Health Organization. https://iris.who.int/bitstream/handle/10665/336692/9789240012813-eng.pdf?sequence=12020.
- 8.WHO. Strategy to respond to antimalarial drug resistance in Africa. Geneva: World Health Organization; 2022. https://iris.who.int/bitstream/handle/10665/364531/9789240060265-eng.pdf?sequence=1.
- 9.WHO. Guidelines for malaria. Geneva: World Health Organization; 2023. [Google Scholar]
- 10.Division NMC. Malaria control policy, Uganda. 2021.
- 11.WHO. List of validated, associated and candidate markers of artemisinin resistance, 18th November 2022. Geneva: World Health Organization; 2022. [Google Scholar]
- 12.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Assefa A, Fola AA, Tasew G. Emergence of Plasmodium falciparum strains with artemisinin partial resistance in East Africa and the Horn of Africa: is there a need to panic? Malar J. 2024;23:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conrad MD, Asua V, Garg S, Giesbrecht D, Niaré K, Smith S, Namuganga JF, Katairo T, Legac J, Crudale RM, et al. Evolution of partial resistance to artemisinins in malaria parasites in Uganda. N Engl J Med. 2023;389:722–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uwimana A, Legrand E, Stokes BH, Ndikumana JM, Warsame M, Umulisa N, et al. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat Med. 2020;26:1602–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meier-Scherling CPG, Watson OJ, Asua V, Ghinai I, Katairo T, Garg S, et al. Selection of artemisinin partial resistance Kelch13 mutations in Uganda in 2016–22 was at a rate comparable to that seen previously in South-East Asia. medRxiv. 2024.
- 17.Conrad MD, Nsobya SL, Rosenthal PJ. The diversity of the Plasmodium falciparum K13 propeller domain did not increase after implementation of artemisinin-based combination therapy in Uganda. Antimicrob Agents Chemother. 2019;63:e01234-e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ocan M, Ashaba FK, Mwesigwa S, Edgar K, Kamya MR, Nsobya SL. Prevalence of arps10, fd, pfmdr-2, pfcrt and pfkelch13 gene mutations in Plasmodium falciparum parasite population in Uganda. PLoS ONE. 2022;17: e0268095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ocan M, Bwanga F, Okeng A, Katabazi F, Kigozi E, Kyobe S, et al. Prevalence of K13-propeller gene polymorphisms among Plasmodium falciparum parasites isolated from adult symptomatic patients in northern Uganda. BMC Infect Dis. 2016;16:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maniga JN, Akinola SA, Odoki M, Odda J, Adebayo IA. Limited polymorphism in Plasmodium falciparum artemisinin resistance kelch13-propeller gene among clinical isolates from Bushenyi District, Uganda. Infect Drug Resist. 2021;14:5153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asua V, Vinden J, Conrad MD, Legac J, Kigozi SP, Kamya MR, et al. Changing molecular markers of antimalarial drug sensitivity across Uganda. Antimicrob Agents Chemother. 2019;63:e01818-e1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balikagala B, Mita T, Ikeda M, Sakurai M, Yatsushiro S, Takahashi N, et al. Absence of in vivo selection for K13 mutations after artemether–lumefantrine treatment in Uganda. Malar J. 2017;16:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tumwebaze P, Tukwasibwe S, Taylor A, Conrad M, Ruhamyankaka E, Asua V, et al. Changing antimalarial drug resistance patterns identified by surveillance at three sites in Uganda. J Infect Dis. 2016;215:631–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawkes M, Conroy AL, Opoka RO, Namasopo S, Zhong K, Liles WC, et al. Slow clearance of Plasmodium falciparum in severe pediatric Malaria, Uganda, 2011–2013. Emerg Infect Dis. 2015;21:1237–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper RA, Conrad MD, Watson QD, Huezo SJ, Ninsiima H, Tumwebaze P, et al. Lack of artemisinin resistance in Plasmodium falciparum in Uganda based on parasitological and molecular assays. Antimicrob Agents Chemother. 2015;59:5061–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conrad MD, Bigira V, Kapisi J, Muhindo M, Kamya MR, Havlir DV, et al. Polymorphisms in K13 and falcipain-2 associated with artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan children. PLoS ONE. 2014;9: e105690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jalei AA, Na-Bangchang K, Muhamad P, Chaijaroenkul W. Monitoring antimalarial drug-resistance markers in Somalia. Parasites Hosts Dis. 2023;61:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mihreteab S, Platon L, Berhane A, Stokes BH, Warsame M, Campagne P, et al. Increasing prevalence of artemisinin-resistant HRP2-negative malaria in Eritrea. N Engl J Med. 2023;389:1191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohamed AO, Hussien M, Mohamed A, Suliman A, Elkando NS, Abdelbagi H, et al. Assessment of Plasmodium falciparum drug resistance molecular markers from the Blue Nile State, Southeast Sudan. Malar J. 2020;19:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moser KA, Madebe RA, Aydemir O, Chiduo MG, Mandara CI, Rumisha SF, et al. Describing the current status of Plasmodium falciparum population structure and drug resistance within mainland Tanzania using molecular inversion probes. Mol Ecol. 2021;30:100–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siddiqui FA, Liang X, Cui L. Plasmodium falciparum resistance to ACTs: emergence, mechanisms, and outlook. Int J Parasitol Drugs Drug Resist. 2021;16:102–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shahinas D, Lau R, Khairnar K, Hancock D, Pillai DR. Artesunate misuse and Plasmodium falciparum malaria in traveler returning from Africa. Emerg Infect Dis. 2010;16:1608–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casanova D, Baptista V, Costa M, Freitas B, Pereira MDNI, Calçada C, et al. Artemisinin resistance-associated gene mutations in Plasmodium falciparum: a case study of severe malaria from Mozambique. Travel Med Infect Dis. 2024;57: 102684. [DOI] [PubMed] [Google Scholar]
- 34.UK Health Agency. Treatment resistant malaria reported in the UK in traveller returning from Uganda. 2023.
- 35.Tun KM, Imwong M, Lwin KM, Win AA, Hlaing TM, Hlaing T, et al. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect Dis. 2015;15:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan H, Kong X, Zhang T, Xiao H, Feng X, Tu H, et al. Prevalence of Plasmodium falciparum kelch 13 (PfK13) and ubiquitin-specific protease 1 (pfubp1) gene polymorphisms in returning travelers from Africa reported in Eastern China. Antimicrob Agents Chemother. 2020;64:e00981-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agaba BB, Smith D, Travis J, Pasay C, Nabatanzi M, Arinaitwe E, et al. Limited threat of Plasmodium falciparum pfhrp2 and pfhrp3 gene deletion to the utility of HRP2-based malaria RDTs in northern Uganda. Malar J. 2024;23:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.NMCD. Uganda malaria indicator survey 2018–19. Kampala, Uganda, and Rockville, Maryland, USA: NMCD, UBOS, and ICF, 2020. 2019. https://www.dhsprogram.com/pubs/pdf/MIS34/MIS34. Accessed 23 Aug 2024.
- 39.UBOS. Uganda population and housing census. 2024.
- 40.Grignard L, Nolder D, Sepúlveda N, Berhane A, Mihreteab S, Kaaya R, et al. A novel multiplex qPCR assay for detection of Plasmodium falciparum with histidine-rich protein 2 and 3 (pfhrp2 and pfhrp3) deletions in polyclonal infections. EBioMedicine. 2020;55: 102757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tacoli C, Gai PP, Bayingana C, Sifft K, Geus D, Ndoli J, et al. Artemisinin resistance-associated K13 polymorphisms of Plasmodium falciparum in Southern Rwanda, 2010–2015. Am J Trop Med Hyg. 2016;95:1090–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.WHO. Responding to antimalarial drug resistance. Geneva: World Health Organization. http://www.who.int/malaria/areas/drug_resistance/overview/en/. Accessed 22 Mar 2019.
- 44.Noedl H, Socheat D, Satimai W. Artemisinin-resistant malaria in Asia. N N Engl J Med. 2009;361:540–1. [DOI] [PubMed] [Google Scholar]
- 45.Imwong M, Suwannasin K, Kunasol C, Sutawong K, Mayxay M, Rekol H, et al. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: a molecular epidemiology observational study. Lancet Infect Dis. 2017;17:491–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ebong C, Sserwanga A, Namuganga JF, Kapisi J, Mpimbaza A, Gonahasa S, et al. Efficacy and safety of artemether–lumefantrine and dihydroartemisinin–piperaquine for the treatment of uncomplicated Plasmodium falciparum malaria and prevalence of molecular markers associated with artemisinin and partner drug resistance in Uganda. Malar J. 2021;20:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans DR, Higgins CR, Laing SK, Awor P, Ozawa S. Poor-quality antimalarials further health inequities in Uganda. Health Policy Plan. 2019;34:iii36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Act Consortium Drug Quality Project Team. ACTwatch outlet survey, Uganda. 2015.
- 49.Act Consortium Drug Quality Project Team and the Impact Study Team. Quality of artemisinin-containing antimalarials in Tanzania’s private sector—results from a nationally representative outlet survey. Am J Trop Med Hyg. 2015;92(Suppl 6):75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chi PC, Bulage P, Urdal H, Sundby J. Perceptions of the effects of armed conflict on maternal and reproductive health services and outcomes in Burundi and northern Uganda: a qualitative study. BMC Int Health Hum Rights. 2015;15:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murray CJ, King G, Lopez AD, Tomijima N, Krug EG. Armed conflict as a public health problem. BMJ. 2002;324:346–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Satimai W, Sudathip P, Vijaykadga S, Khamsiriwatchara A, Sawang S, Potithavoranan T, et al. Artemisinin resistance containment project in Thailand. II: responses to mefloquine–artesunate combination therapy among falciparum malaria patients in provinces bordering Cambodia. Malar J. 2012;11:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaijaroenkul W, Na Bangchang K, Mungthin M, Ward SA. In vitro antimalarial drug susceptibility in Thai border areas from 1998–2003. Malar J. 2005;4:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wongsrichanalai C, Sirichaisinthop J, Karwacki JJ, Congpuong K, Miller RS, Pang L, et al. Drug resistant malaria on the Thai-Myanmar and Thai-Cambodian borders. Southeast Asian J Trop Med Public Health. 2001;32:41–9. [PubMed] [Google Scholar]
- 55.Health Journalist Network. Karamoja health services frustrated by insecurity and poor roads, Uganda, 2003. https://hejnu.ug/insecurity-and-poor-roads-frustrate-health-services-in-karamoja/.
- 56.WHO. Artemisinin resistance and artemisinin-based combination therapy efficacy: status report. Geneva: World Health Organization; 2018. https://apps.who.int/iris/handle/10665/274362.
- 57.Schmedes SEPD, Dhal S, Kelley J, Svigel SS, Dimbu P. Plasmodium falciparum kelch 13 mutations, 9 countries in Africa, 2014–2018. Emerg Infect Dis. 2021;27:1902–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mok S, Yeo T, Hong D, Shears MJ, Ross LS, Ward KE, et al. Mapping the genomic landscape of multidrug resistance in Plasmodium falciparum and its impact on parasite fitness. bioRxiv. 2023. [DOI] [PMC free article] [PubMed]
- 59.Fidock DA, Rosenthal PJ. Artemisinin resistance in Africa: how urgent is the threat? Med (N Y). 2021;2:1287–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moyo E, Mhango M, Moyo P, Dzinamarira T, Chitungo I, Murewanhema G. Emerging infectious disease outbreaks in Sub-Saharan Africa: learning from the past and present to be better prepared for future outbreaks. Front Public Health. 2023;11:1049986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.