Abstract
Background
The effectiveness of adipose-derived stem cells (ADSCs) in therapy diminishes with age. It has been reported that transcription factors (TFs) play a crucial role in the aging and functionality of stem cells. Nevertheless, there is limited understanding regarding the involvement of TFs in the aging mechanism of ADSCs.
Methods
RNA sequencing (RNA-seq) was utilized to discern the differentially expressed genes in ADSCs obtained from donors of varying ages. TFs exhibiting significant variations across age groups were identified and subsequently validated. ADSCs were manipulated to exhibit either enhanced expression or reduced levels of HES1 and STAT1 via lentivirus transfection and small interfering RNA (siRNA) techniques. The impact of these genetic alterations on ADSCs’ proliferation, migration, and cellular senescence was assessed using EdU, transwell, and senescence-activated β-galactosidase (SA-β-gal) staining assays. The DNA sequences bound by HES1 were investigated through the CUT & Tag assay. Lastly, the therapeutic efficacy of aged ADSCs with HES1 overexpression was evaluated in skin injury model of male Sprague-Dawley rats.
Results
678 genes showed differential expression between ADSCs obtained from young and old donors (Y-ADSCs and O-ADSCs), with 47 of these genes being TFs. Notably, the expression of the TF hairy and enhancer of split 1 (HES1) was notably reduced in ADSCs from old donors. Introducing HES1 overexpression in aged ADSCs resulted in improved cellular function and the suppression of cellular senescence, while reducing HES1 levels in young ADSCs had the opposite effect. Mechanistically, HES1 was found to interact with the promoter region of another TF, signal transducer and activator of transcription 1 (STAT1), to inhibit its transcription. Knocking down STAT1 could fully reverse the negative effects caused by decreased HES1 in ADSCs, leading to a reduction in the secretion of pro-inflammatory cytokines such as TNF-α, IL-6, and IL-8. Ultimately, restoring HES1 expression in aged ADSCs demonstrated enhanced therapeutic potential in promoting skin wound healing.
Conclusion
HES1 acts as an inhibitor of cellular senescence in the aging progression of ADSCs through the modulation of STAT1 expression, suggesting a promising avenue for rejuvenating senescent ADSCs and improving wound healing.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13287-024-04002-w.
Keywords: Adipose stem cell, Aging, HES1, STAT1, Transcription factors
Introduction
Adipose-derived stem cells (ADSCs) play a crucial role in cell-based therapies for a variety of human ailments, including skin wound healing, spinal cord injuries, stroke, myocardial damage, and peripheral nerve injury [1–4]. These cells are easily obtainable and exhibit rapid proliferation, facilitating extensive research and widespread clinical application [3]. However, research has shown that the aging of donors and the in vitro expansion process can negatively impact these cells, leading to reduced proliferative capacity and an increased proportion of senescent cells, thereby impeding their clinical utility [5, 6]. The mechanisms underlying stem cell dysfunction with aging are intricate and involve disruptions in proteostasis, DNA damage, epigenetic modifications, and organelle dysfunction [7]. Previous studies have indicated that the modulation of key miRNAs, lincRNAs, and genes can partially mitigate the detrimental effects of aging on adipose-derived stem cells [8–10]. Nevertheless, the manipulation of individual molecules may not fully restore the therapeutic potential of aged stem cells to that of their younger counterparts. Therefore, further investigations are still warranted to identify suitable candidate factors for rejuvenating stem cells.
Transcription factors (TFs) in humans are responsible for identifying specific genomic binding sites to regulate the transcription of genetic material [11]. These TFs are widely recognized for their involvement in stem cell rejuvenation and age-related ailments [12]. Certain members of the sry-related HMG box (SOX) gene family exhibit modified expression patterns as individuals age, contributing to the preservation of stem cell characteristics and the continual self-renewal of adult stem cells [13]. For example, the interaction of SOX5 has been shown to elevate the expression of high mobility group box 2 (HMGB2) by amplifying enhancer activity, thereby promoting the revitalization of human stem cells [14]. Another TF, early growth response 1 (EGR1), undergoes modulated expression during the aging process and could potentially serve as a target for rejuvenating hematopoietic stem cells [15]. Furthermore, forkhead box P1 (FOXP1), a constituent of the FOX family of TFs, has been identified as a factor that enhances the self-renewal potential of cardiac myocytes and bone marrow-derived mesenchymal stem cells (BMMSCs) while shielding them from senescence [16, 17]. Moreover, the aging of mesenchymal stem cells (MSCs) has been associated with reduced levels of the TF, activating transcription factor 6 (ATF6), which is instrumental in governing the gene expression network linked to the homeostatic regulation of membrane organelles [18]. Nevertheless, there remains a dearth of knowledge regarding the precise role of TFs in the aging progression of ADSCs.
In the current investigation, a comprehensive analysis using high-throughput RNA sequencing (RNA-seq) was carried out on ADSCs obtained from donors of varying ages. The study revealed a notable decrease in the expression of the transcription factor HES1 in ADSCs from older individuals. HES1 primarily functions as a transcriptional repressor involved in regulating the self-renewal and quiescence of stem cells. Reinstating HES1 expression levels was observed to enhance the proliferation and migration of aged ADSCs, diminish cell senescence, and improve their capacity for wound healing. Mechanistically, HES1 was found to interact with the promoter region of another transcription factor, STAT1, thereby inhibiting its transcriptional activity. This inhibition led to reduced expression of the senescence-associated marker p21, as well as decreased secretion of pro-inflammatory cytokines such as TNF-α, IL-6, and IL-8. Consequently, these results propose a novel mechanism underlying the dysfunction of aged ADSCs and suggest potential therapeutic targets for rejuvenating the regenerative capabilities of aged stem cells.
Materials and methods
ADSCs isolation and culture
This research was approved by the Institutional Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology, China, and followed the principles set forth in the Declaration of Helsinki regarding the handling of patient samples. Consent was obtained from the parents of young donors and old donors before they took part in the study. The detailed information of these donors has been described in our previous study [5]. Samples of adipose tissue were taken from patients who had skin-flap transplantation surgery and did not have any additional systemic illnesses. The ADSCs were cultured in Human Adipose-derived Mesenchymal Stem Cells Medium (OriCell, China), following a set procedure. Cells from passages 2 to 7 were used for the experiments.
Multilineage differentiation assay
ADSCs subjected to gene transfection were employed to evaluate both chondrogenic and adipogenic differentiation, in accordance with the standardized protocol provided by the manufacturer (Oricell, China). The capacity for adipogenesis was evaluated following a two-week adipogenic culture period through oil red staining, whereas the capacity for chondrogenesis was assessed after a three-week chondrogenic culture period using alcian blue staining.
RNA sequencing
Total RNA was isolated from ADSCs at passage 3 or 4 using the Tissue & Cell Total RNA Kit from HY Cezmbio in China. The quality and quantity of RNA were assessed through the Bioanalyzer 2100 from Agilent in CA, USA, and the NanoDrop ND-1000 from NanoDrop in Wilmington, DE, USA. mRNA containing poly(A) tails was enriched using dynabeads oligo (dT) from Thermo Fisher in the USA, followed by fragmentation with the NEBNextR Magnesium RNA Fragmentation Module from NEB in the USA. Subsequently, a standard cDNA library was prepared and sequenced on the Illumina NovaseqTM 6000 platform. Raw data underwent filtration to obtain clean data, which was then aligned to the reference genome using Hisat2 [19]. The expression levels of mRNAs were quantified using the Fragments Per Kilobase Million (FPKM) value calculated by StringTie [20]. Genes showing significant dysregulation were identified based on the criteria of fold changes ≥ 2 or ≤ -2 and a p value < 0.05.
GO and KEGG analyses
The identified genes exhibiting differential expression were subjected to additional analysis using the Gene Ontology (GO) database (http://geneontology.org) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (http://www.genome.jp/kegg). The GO categories, encompassing cellular component, molecular function, and biological process information of the candidate genes, were utilized for further characterization. Furthermore, the biological pathways associated with these genes were investigated through the KEGG database. Significantly enriched gene sets were identified using the hypergeometric distribution test, with a p-value of less than 0.05 considered as statistically significant.
CUT & tag assay
The CUT & Tag assay was carried out utilizing a Hyperactive Universal CUT & Tag Assay Kit (TD903, Vazyme, China) in accordance with the provided guidelines. In summary, 60,000 ADSCs were collected, rinsed, and subsequently exposed to Concanavalin A-coated magnetic beads. Subsequently, the cells were treated with either an anti-flag antibody (Cat. 14793 S, Cell Signaling Technology, USA) or an isotype control antibody (Cat. 2729 S, Cell Signaling Technology, USA) for a duration of 2 h, followed by an additional hour with a secondary antibody at ambient temperature. The hyperactive pG-Tn5 transposonase was then introduced to facilitate tagmentation. Subsequently, DNA fragments were isolated and amplified using indexed P5 and P7 primers. The DNA library was sequenced on the Illumina NovaseqTM 6000 platform employing the PE150 model. The initial data underwent filtration to acquire clean data, which was subsequently aligned to the reference genome utilizing bowtie2 (version 2.2.6) with default settings [21]. The distribution of reads upstream and downstream of TSS was depicted using DeepTools (version 2.4.1) [22]. Peaks annotation and peak distribution analysis were conducted using bedtools (version 2.30.0) [23]. Differential peaks were discerned utilizing csaw (version 1.24.3) [24]. To further validate the outcomes of DNA-seq, the DNA fragments were amplified using primers specific to the binding sites of HES1 with STAT1 genome, visualized through agarose gel electrophoresis, and sequenced via Sanger’s sequencing.
Lentiviral transfection
A lentivirus vector containing green fluorescence protein was utilized to create a stable overexpression system for HES1, along with a negative control. These vectors were subsequently transfected into O-ADSCs for research purposes. In short, O-ADSCs at passage 3 were seeded in a six-well culture plate at a density of 1 × 105 cells per well. The next day, the ADSCs medium was supplemented with the virus stock solution and cotransfection reagent, with an appropriate multiplicity of infection. After 12 h of transfection, the medium containing the virus was swapped with ADSCs medium. When the cells reached 70% confluence, puromycin-containing ADSCs medium was employed to isolate the cells that were successfully transfected.
siRNA transfection
The si-HES1, si-STAT1 and negative control si-NC were synthesized by RiboBio, China, and the sequences of these oligonucleotides are listed in Table S1. In short, O-ADSCs at passage 3 were seeded in a six-well culture plate at a density of 1 × 105 cells per well. The next day, the cells were transfected with the oligonucleotides using the riboFECT CP Transfection Kit (RiboBio, China), in accordance with the provided guidelines. The effectiveness of transfection was verified through qRT-PCR and western blot analysis. Following a 72-hour incubation period, the ADSCs with different treatments were gathered for the subsequent experiment.
Real-time PCR analysis
In this study, total RNA was isolated using the Tissue & cell RNA Kit (HYCEZMBIO, China), and the RNA samples were converted into complementary DNA (cDNA) through reverse transcription using the HiScript III RT SuperMix for qPCR (Vazyme, China). Then the qRT-PCR was conducted was carried out in accordance with the manufacturer’s guidelines, using the ChamQ SYBR qPCR Master Mix (Vazyme, China). The qRT-PCR primer sequences are available in Table S2. Gene expression levels were calculated using the 2−ΔΔCt method and were standardized against GAPDH.
Western blot
Total protein was extracted by using RIPA (Epizyme, China) with protease inhibitor, and the protein concentration was measured using the BCA protein assay (HYCEZMBIO, China). 20 µg of total protein was separated using a 10% SDS-PAGE gel, then transferred to PVDF membranes. The membranes were blocked with blocking buffer and left to incubate overnight with primary antibodies specific to HES1((Abcam, ab108937, USA), STAT1 (Abcam, ab109320, USA), β-tubulin (Proteintech, 10094-1-AP, China), GAPDH (Proteintech, 60004-1-Ig, China). Afterward, the membranes were exposed to a secondary antibody tagged with HRP for one hour. Then, they were treated with the immobilon ECL substrate kit for one minute and photographed using an imaging system. Original blots were exhibited in Figure S3.
Cell proliferation and migration assay
In the cell proliferation test, ADSCs with different treatment were grown in 96-well plates with 7000 cells per well, and each group had 5 replicated wells. After incubating overnight, the cells were exposed to the BeyoClick™ EdU-594 kit (Beyotime, China) for 3 h as per the manufacturer’s guidelines. Afterwards, the EdU-labeled ADSCs (colored red) and all cells (colored blue) were photographed with a fluorescent microscope. The rate of cell proliferation was assessed by dividing the count of red-stained cells by the count of blue-stained cells, then multiplying by 100%.
In the cell migration experiment, ADSCs with different treatment were placed in the upper chamber of 24-well transwell insert with 8.0 μm pores (Corning, USA), with each well was seeded 3 × 104 cells in DMEM medium without fetal bovine serum, and the lower chamber contained 650 µl of ADSCs medium. There were 5 replicated wells in each experimental group. After 24 h, the cells that moved to the lower chamber were stained with crystal violet and photographed under a microscope. The quantity of migrated cells was then measured for each group.
β-galactosidase staining assay
ADSCs with different treatment were grown in 48-well plates with 1.5 × 104 cells per well. There were 5 replicated wells in each experimental group. After being left to incubate overnight, the cells were stained with Senescence β-Galactosidase Staining Kit (Beyotime, China). The appearance of blue-stained cells indicated the presence of SA-β-gal, which was visible through a light microscope. The percentage of cells testing positive for SA-β-gal was then calculated for each group in the experiment.
Enzyme-linked immunosorbent assay (ELISA)
The levels of interleukin-6 (IL-6), interleukin-8 (IL-8), and tumor necrosis factor-alpha (TNF-α) in cell culture supernates were determined using commercially available enzyme-linked immunosorbent assay (ELISA) kits (Bioswamp, China) according to the manufacturers’ protocols. The ELISA plates were measured using the plate reader at a wavelength of 450 nm; With the concentration of the standard product as the horizontal coordinate and OD value as the vertical coordinate, the standard curve was obtained use four parameter logistics curve fitting, and then plug the OD value of a sample into the fitting equation to calculate the concentrations of the sample.
In vivo wound-healing model
In this study, all animal tests were approved by the Animal Care Committee of Tongji Medical College and followed the ARRIVE guidelines. The experiment was conducted by separate individuals for the design and operation tasks in order to minimize the influence of subjective factors, ensuring strict adherence to the experimental methodology. Twenty-eight healthy male Sprague Dawley rats, 10 weeks old and weighing around 180–200 g, were sourced from the Experimental Animal Center at Tongji Medical College, Huazhong University of Science and Technology. The rats were equipped with numbered ear tags and subsequently assigned to four groups through a digital randomization process: the PBS group (PBS), the oe-NC O-ADSCs group (oe-NC), the HES1 O-ADSCs group (oe-HES1), and the Y-ADSCs group (Y-ADSCs). Based on past experience, we allocated 7 rats per group. Before the experiments, the rats were given anesthesia through intraperitoneal injections of ketamine (0.025 mg/kg) and xylazine (0.25 mg/kg). Then, a circular full-thickness skin wound with 18 mm diameter was created on the back of each rat. Around 1.0 × 106 ADSCs were mixed in 100 µl of PBS and injected at four points around the wound, while the control group was injected with 100 µl of PBS only. To prevent wound infection, all rats were administered intramuscular injections of veterinary antibiotics following the modeling procedure. To minimize potential confounding factors, such as wound bites, each rat was housed individually in a cage and given uniform laboratory food, following a 12-hour day and night cycle. The endpoint of the wound healing experiment was set at 18 days, as previous experience indicates that re-epithelialization can be achieved by this time for most wounds, thereby facilitating analysis of the internal healing process. Images of the injuries were captured on days 0, 3, 7, 10, 14 and 18 after the wounds were inflicted, and the size of the wounds was determined in each image using Image J software. In order to obtain more accurate statistics, the maximum and minimum values of each group’s data were excluded, leaving 5 data points for statistical analysis. The percentage of wound closure was determined by dividing the reduced area by the original wound area and multiplying by 100%. On the 18th day, the rats were euthanized while under anesthesia through cervical dislocation after all samples were collected.
Histological analysis
On the 18th day, the wounds were gathered and then processed through tissue fixation, embedding, and sectioning. The sections were later stained with hematoxylin and eosin (H&E) to examine the wound healing. Furthermore, Masson staining was performed to assess the collagen accumulation.
Immunofluorescence analysis
To assess the density of blood vessels in wound beds, the tissue samples were treated with α-SMA (Abcam, ab7817, USA) and CD31 (Abcam, ab182981) antibody overnight at 4 °C, then exposed to a fluorescein-conjugated secondary antibody (Invitrogen, 710,369, USA) for 1 h, at room temperature. The cell nuclei were stained with DAPI (Sigma-Aldrich, D9542-5MG, USA). Subsequently, the sections were scanned under a fluorescence microscope, and the quantity of blood vessels in the dermal layer of each sample was measured using Image J software.
Immunohistochemistry analysis
To assess cell growth in wound areas, the tissue samples were treated with Proliferating Cell Nuclear Antigen (PCNA) antibody (Abcam, ab7817, USA) overnight at 4 °C. Next, a second antibody linked to horseradish peroxidase (Beyotime, A0181, China) was applied to the samples for 1 h at room temperature. The samples were then scanned and the percentage of PCNA-positive cells in each sample was determined using Image J software.
Statistics analysis
The data analysis was conducted using GraphPad Prism software, version 8.0.2. Unpaired Student’s t-test was used to compare data between two groups, while one- and two-way analysis of variance (ANOVA) test was used to compare three or more groups. Results are shown as mean ± standard deviation (SD), and statistical significance was considered at p < 0.05.
Result
Analyses of differentially expressed genes in O-ADSCs compared with Y-ADSCs
To explore the fundamental processes contributing to the decline in stem cell function associated with aging, a comparative RNA-seq was conducted on a larger cohort comprising five Y-ADSCs and six O-ADSCs (GSE269853). This contrasts with the limited sample size of three Y-ADSCs and three O-ADSCs utilized in our prior investigation (GSE174502). The findings indicated that 316 genes exhibited decreased expression levels, while 362 genes displayed increased expression levels in O-ADSCs in comparison to Y-ADSCs, as depicted in the heatmap and volcano plot (Fig. 1A and B, Data S1). Subsequent analyses using GO and KEGG were performed to further elucidate the roles of the identified differentially expressed genes. The GO analysis revealed that these genes are predominantly localized in the plasma membrane and extracellular region (Fig. 1C), serving as constituents of the extracellular matrix and participating in protein binding (Fig. 1D). Additionally, they are implicated in processes related to cell adhesion and signal transduction pathways (Fig. 1E). KEGG analysis indicated enrichment in pathways such as neuroactive ligand-receptor interaction, calcium signaling, pathways in cancer, ErbB signaling, and cell adhesion molecules (Fig. 1F).
Fig. 1.
Analyses of differentially expressed genes in O-ADSCs compared with Y-ADSCs. (A and B) A heatmap and volcano plot illustrating the distinct patterns of gene expression variation. (C-E) The cellular component, molecular function, and biological process analyses of differentially expressed genes. (F) The KEGG pathway analysis of the differentially expressed genes
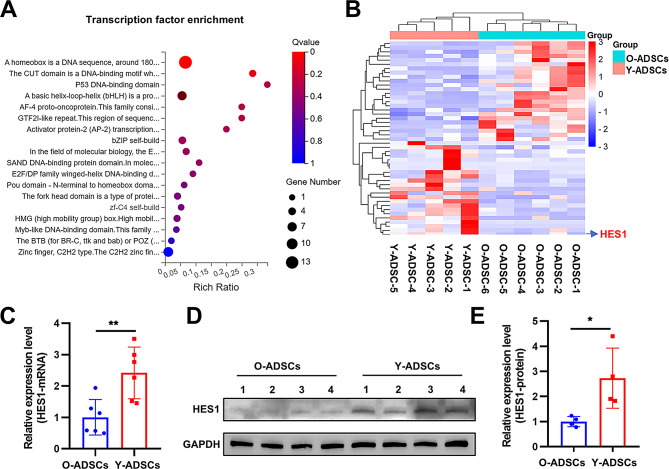
HES1 is a candidate transcription factor down-regulated in O-ADSCs
TFs are known to have significant roles in the differentiation of stem cells and the aging process. In this study, particular attention was given to the TFs that were differentially expressed, as they could potentially serve as targets for rejuvenating the function of aged stem cells. The analysis revealed that 47 TFs (24 upregulated and 23 downregulated) exhibited differential expression in O-ADSCs compared to Y-ADSCs, as illustrated in the heatmap (Fig. 2A, Data S2). These TFs primarily belong to the homeobox, bHLH, and zf-C2H2 families (Fig. 2A). Among these TFs, HES1 was singled out due to its high expression level and low p-value (Fig. 2B). Furthermore, HES1 was also identified as a downregulated gene in O-ADSCs within our prior cohort study (GSE174502, Data S3). Subsequent PCR and western blot analyses confirmed that the expression of HES1 was reduced in O-ADSCs relative to Y-ADSCs (Fig. 2C-E).
Fig. 2.
The transcription factor HES1 exhibited reduced levels in O-ADSCs. (A) The bubble chart displayed the transcription factor families that exhibited differential expression between O-ADSCs and Y-ADSCs. (B) A heatmap illustrating the expression profiles of various transcription factors exhibiting differential expression patterns. (C) PCR and Western blot (D and E) analyses of the expression level of HES1 between O-ADSCs and Y-ADSCs; n = 6 for PCR, and n = 4 for Western blot. **p < 0.01
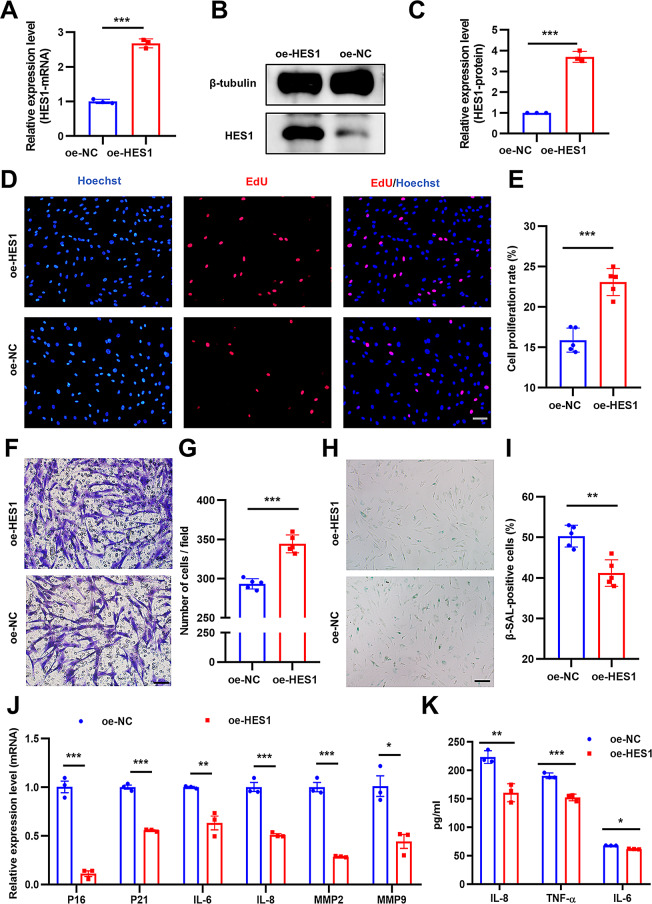
HES1 overexpression promotes O-ADSCs proliferation and migration, yet alleviated cell senescence and cellular inflammation
The overexpression efficiency of HES1 was validated by PCR and western blot assay (Fig. 3A-C), with an approximately three-fold increase following lentiviral vector transfection. To evaluate the retention of mesenchymal stem cell characteristics following genetic modifications, we performed a multilineage differentiation assay encompassing adipogenesis and chondrogenesis. The findings indicated that the overexpression of the HES1 gene did not influence the multidirectional differentiation capabilities of the stem cells, as demonstrated by the results of oil red and alcian blue staining (Figure S1A, 1B). The proliferation rate in the oe-HES1 group was almost 1.4-fold greater than that in the oe-NC group (Fig. 3D and E). Additionally, cell migration was almost 1.2-fold higher compared to the oe-NC group (Fig. 3F and G). Moreover, the proportion of SA-β-gal-positive cells in the oe-HES1 group was 20% less compared to the oe-NC group (Fig. 3H and I). We also assessed the senescence-related markers (p16, p21) and inflammation-related factors (IL-6, IL-8, MMP2, MMP9) level after HES1 overexpression, the results show that HES1 overexpression alleviated the level of p16, p21, IL-6, IL-8, MMP2 and MMP9 (Fig. 3J). In a similar manner, the concentrations of IL-6, IL-8, and TNF-α were evaluated using ELISA on cell culture supernatants subsequent to the overexpression of HES1. The results indicate that the overexpression of HES1 is associated with a decrease in the levels of IL-6, IL-8, and TNF-α (Fig. 3K). Overall, the overexpression of HES1 in O-ADSCs could ameliorate its unsatisfactory phenotype and function.
Fig. 3.
The heightened expression of HES1 led to a revitalization of the functions of O-ADSCs. (A) PCR and Western blot (B and C) analyses were conducted to assess the transfection efficiency of the HES1 overexpression lentiviral vector. (D and E) The proliferation rate of O-ADSCs treated with the HES1 overexpression lentiviral vector (oe-HES1 O-ADSCs) or scramble vector (oe-NC O-ADSCs) was determined using the EdU assay; scale bars: 50 μm. (F and G) Migration of cells in the oe-HES1 O-ADSCs and oe-NC O-ADSCs groups was examined, as well as the presence of SA-β-gal-positive cells (H and I) in these groups; scale bars: 50–100 μm. (J) Expressional changes of genes following HES1 overexpression in O-ADSCs were evaluated through PCR analysis. (K) The levels of IL-6, IL-8, and TNF-α in the supernatant of O-ADSCs subsequent to the overexpression of HES1 were assessed using ELISA. N = 5. *P < 0.05, **P < 0.01, ***P < 0.001
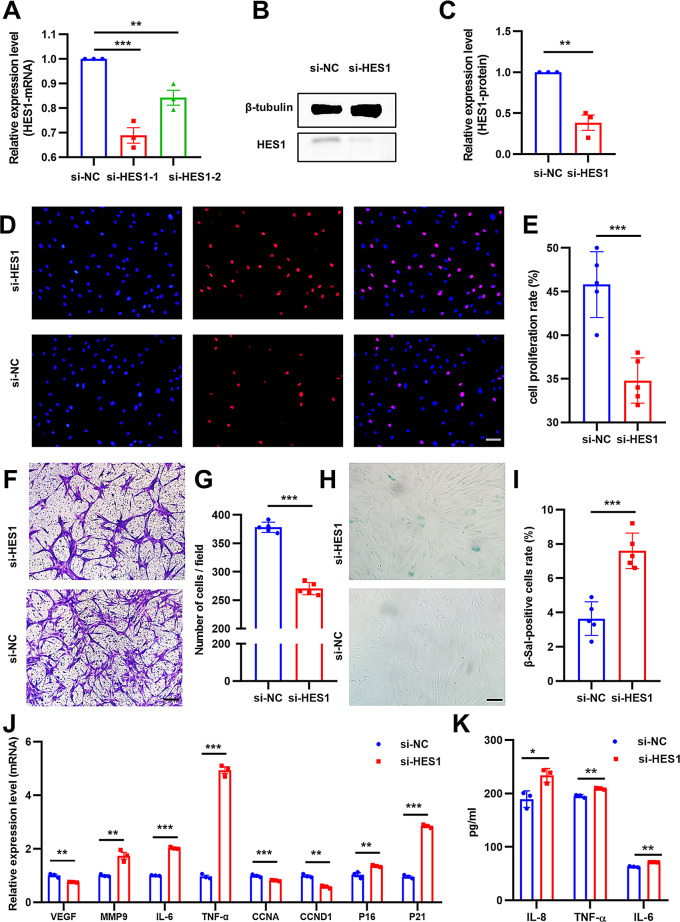
HES1 silencing impairs Y-ADSCs proliferation and migration, yet exacerbated cell senescence and cellular inflammation
The silencing efficiency of HES1 was validated by PCR and western blot assay (Fig. 4A-C), with its level reduced by half using si-HES1-1 oligonucleotides sequence, and si-HES1-1 was selected for the subsequent experiments. Y-ADSCs proliferation and migration were significantly impaired by HES1 inhibition, decreasing by 11% and 31%, respectively (Fig. 4D-G). Moreover, the proportion of SA-β-gal-positive cells in the si-HES1 group was almost 2-fold greater compared to the si-NC group (Fig. 4H and I). PCR analysis showed that HES1 silencing in Y-ADSCs could significantly improve the expression of MMP9, IL-6, TNF-α, p16 and p21, while decrease the expression of growth factors VEGF, and cell cycle dependent elements CCNA1 and CCND2 (Fig. 4J). ELISA showed consistent results that HES1 knockdown could reduce the expression of IL-6, IL-8 and TNF-α (Fig. 4K). Overall, the silencing of HES1 in Y-ADSCs could deteriorate its phenotype and function.
Fig. 4.
Silencing of HES1 resulted in a reduction in the functionality of Y-ADSCs. (A) PCR and Western blot (B and C) analyses were conducted to assess the effectiveness of si-HES1 knockdown. (D and E) The proliferation rate of Y-ADSCs treated with si-HES1 or si-NC was determined using the EdU assay; scale bars: 50 μm. (F and G) Migration of cells in si-HES1 Y-ADSCs and si-NC Y-ADSCs groups was examined; scale bars: 50 μm. (H and I) The presence of SA-β-gal-positive cells in the aforementioned groups was assessed; scale bars: 100 μm. (J) Expressional changes of genes following HES1 knockdown in Y-ADSCs were evaluated through PCR analysis. (K) The levels of IL-6, IL-8, and TNF-α in the supernatant of Y-ADSCs subsequent to the knockdown of HES1 were assessed using ELISA. N = 5. **P < 0.01, ***P < 0.001
Analyses of differentially expressed genes in O-ADSCs after HES1 overexpression
To investigate the downstream effectors of HES1, an RNA-seq analysis was conducted to examine the differentially expressed genes in O-ADSCs following the overexpression of HES1 (GSE270037). The findings indicated that 448 genes exhibited increased levels, while 865 genes showed decreased levels in the oe-HES1 O-ADSCs compared to the oe-NC O-ADSCs (Fig. 5A and B, Data S4). Subsequently, GO and KEGG analyses were performed to further elucidate the functions of the identified differentially expressed genes. The GO analysis revealed that these genes are primarily localized in the nucleus and cytoplasm (Fig. 5C), and are involved in activities such as protein binding and DNA binding (Fig. 5D). Furthermore, they are associated with processes related to cell adhesion and the regulation of transcription by RNA polymerase II (Fig. 5E). The KEGG analysis indicated enrichment in pathways such as the NOD-like receptor signaling pathway, TNF signaling pathway, IL-17 signaling pathway, and PI3K-Akt signaling pathway (Fig. 5F).
Fig. 5.
Analyses of differentially expressed genes in O-ADSCs after HES1 overexpression. (A and B) A heatmap and volcano plot illustrating the distinct patterns of gene expression variation. (C-E) The cellular component, molecular function, and biological process analyses of differentially expressed genes. (F) The KEGG pathway analysis of the differentially expressed genes
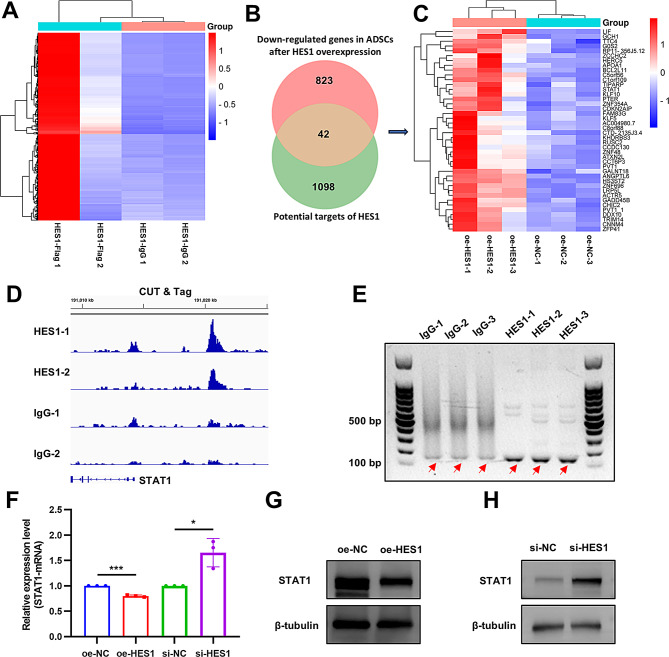
STAT1 is a downstream effector of HES1
To investigate the DNA binding sites associated with HES1, a CUT & Tag assay was conducted utilizing an anti-flag antibody to isolate the DNA sequences that interact with HES1. Subsequently, these DNA sequences were subjected to analysis through DNA-seq (GSE270038). The results indicated that HES1 has the capability to bind to the promotor-transcription start sites of 1140 genes (Fig. 6A, Data S5). Given that HES1 primarily functions as a transcriptional repressor, a comparison was made between these genes and those exhibiting decreased expression in O-ADSCs following HES1 overexpression. Through this comparison, forty-two genes were identified as overlapping (Fig. 6B and C, Data S6), with STAT1 being singled out due to its notably high expression level. Further validation through PCR and Sanger’s sequencing confirmed the binding of HES1 to the promotor-transcription start region of STAT1 (Fig. 6D and E). PCR and Western blot analyses additionally supported the observation that overexpression of HES1 led to a reduction in STAT1 levels in O-ADSCs, while knockdown of HES1 resulted in an increase in STAT1 levels in Y-ADSCs (Fig. 6F-H). In summary, it was determined that STAT1 functions as a downstream factor of HES1 in ADSCs.
Fig. 6.
STAT1 is a downstream effector of HES1. (A) A heatmap displaying the genes whose promoter region has the potential to be specifically bound by HES1. (B) A diagram illustrating the intersection between the potential targets of HES1 and the genes that are down-regulated in ADSCs following HES1 overexpression, as determined through CUT & Tag DNA-seq and RNA-seq analyses. (C) A visual representation in the form of a heatmap depicting the expression patterns of the genes that overlap. (D) The genomic locus of STAT1, which is susceptible to binding by HES1, was visualized utilizing the IGV software. (E) A gel blot assay was conducted to exhibit the interaction between HES1 and STAT1 through PCR analysis. (F) PCR and Western blot (G and H) techniques were employed to assess the relative expression levels of STAT1 subsequent to the overexpression or knockdown of HES1. N = 3. *P < 0.05, ***P < 0.001
STAT1 silencing rescues ADSCs function following HES1 inhibition
To determine whether STAT1 inhibition could improve ADSCs function, we transfected si-STAT1 or si-NC into ADSCs, and verified the efficiency by western blot (Fig. 7K). Next, to determine whether STAT1 silencing could rescue the functional deterioration induced by HES1 inhibition in ADSCs, we transfected si-STAT1 or si-NC into ADSCs that were simultaneously transfected with si-HES1. The results showed that STAT1 silencing partly reversed the functional deterioration of ADSCs induced by HES1 inhibition, with ~ 9% greater proliferation and ~ 25% greater migration in the si-HES1 + si-STAT1 group compared to the si-HES1 group (Fig. 7A-D). STAT1 silencing also reversed the increase in SA-β-gal-positive cells induced by si-HES1 in ADSCs, with a 6% decrease compared to the si-HES1 group (Fig. 7E and F). Additionally, STAT1 silencing could decrease the level of p21, IL-6, IL-8, and TNF-α in si-HES1 + si-STAT1 group compared to the si-HES1 group (Fig. 7G-J), which is consistent with the ELISA results (Figure S2A-C). Besides, the mRNA and protein level of STAT1 were assessed by PCR and western blot, both showing that si-HES1 could increase STAT1 expression, which can be rescued by si-STAT1 (Fig. 7L and M). Overall, STAT1 silencing could rescue the functional deterioration induced by HES1 inhibition in ADSCs.
Fig. 7.
The functional capabilities of ADSCs after HES1 knockdown can be restored by silencing STAT1. (A and B) The proliferation rate of ADSCs following treatment with si-NC, si-STAT1, si-HES1, or si-HES1 + si-STAT1 was assessed using the EdU assay; scale bars: 50 μm. (C and D) The migratory cells in these groups were also evaluated; scale bars: 50 μm. (E and F) The presence of SA-β-gal-positive cells was examined; scale bars: 100 μm. (G-J) PCR analysis was conducted to determine the changes in genes expression in ADSCs subjected to the different treatments. (K) The knockdown efficiency of si-STAT1 was assessed through Western blot analysis. (L and M) The relative expression of STAT1 in ADSCs treated with the aforementioned conditions was analyzed using PCR and Western blot techniques. N = 3 or 5. *P < 0.05, **P < 0.01, ***P < 0.001
The increased expression of HES1 augments the capacity of O-ADSCs in promoting skin wound healing
An animal model for wound healing was utilized to investigate the impact of HES1 in O-ADSCs. On the 18th day after modeling, all rats were alive when collecting wound samples. Three days after modeling, there was no significant variance in the size of the wound area between oe-NC and oe-HES1 O-ADSCs group. On days 7, 10, and 14 after the injury, the PBS group showed the largest wound area, followed by the oe-NC O-ADSCs group, then the oe-HES1 O-ADSCs group, and the Y-ADSCs group having the smallest wound area. However, there was no significant variance in the size of the wound area between the four groups at day 18 (Fig. 8A and B). Moreover, H&E staining analysis showed that the PBS group exhibited longest wound scar length, followed by oe-NC O-ADSCs group, then the oe-HES1 O-ADSCs group, and the Y-ADSCs group achieved shortest scar length (Fig. 8C and D). In addition, Masson staining and PCNA immunohistochemical staining showed that the Y-ADSCs group achieved the best collagen deposition and cell proliferation, followed by the oe-HES1 O-ADSCs group, then the oe-NC O-ADSCs group, and the PBS group exhibited the worst collagen deposition and cell proliferation (Fig. 8E-H). Lastly, the α-SMA and CD31 immunofluorescence staining showed that the PBS group had lowest level of new blood vessel formation, followed by the NC group with a higher level, then the oe-HES group with even more vessel. The Y-ADSCS group exhibited the highest level of CD31 expression, nevertheless, there is no significant variance in α-SMA level between oe-HES1 O-ADSCs group and Y-ADSCs group (Fig. 8I-K). Overall, Overexpression of HES1 in O-ADSCs greatly enhances its healing potential in wounds by stimulating cell growth, new blood vessel formation, and collagen production, while reducing scar formation.
Fig. 8.
The wound healing potential of O-ADSCs was augmented through the overexpression of HES1. (A) The morphological changes in wounds treated with PBS, oe-NC O-ADSCs, oe-HES1 O-ADSCs, and O-ADSCs at various time intervals. (B) Statistical analysis was conducted to compare wound characteristics among the four groups at different time points. (C) Histological images from H&E staining were analyzed to assess wound length, while Masson staining (E) images were used to evaluate collagen deposition in wounds; scale bars: 2–100 μm. (G) PCNA immunohistochemical staining images were utilized to determine cell proliferation in wounds; scale bars: 100 μm. (I) Additionally, CD31 (green) and α-SMA (red) immunofluorescent staining images were examined to assess blood vessel regeneration in wounds; scale bars: 50 μm. (D, F, H, J, K) Statistical analyses were performed for each of these parameters. One-way ANOVA with Tukey’s multiple comparisons test was used in (D, F, H, J, K). Two-way ANOVA with Tukey’s multiple comparisons test was used in (B). N = 5. *P < 0.05, **P < 0.01, ***P < 0.001
Discussion
The therapeutic effectiveness of stem cells diminishes as donors age, posing significant challenges to their clinical utilization. Various research studies have investigated the impact of TFs on cellular senescence and the aging of stem cells [13, 25, 26]. However, limited research has been conducted on the influence of TFs on the aging process of adipose stem cells. This study revealed a notable decrease in the expression of HES1 in aged ADSCs, and HES1 acts as a transcriptional suppressor that hinders the transcription of another TF, STAT1, a known promoter of senescence. Consequently, the dysregulation of the HES1/STAT1 axis during aging plays a crucial role in the diminished functionality of aged ADSCs, highlighting potential targets for rejuvenating aged stem cell function.
The disruption of crucial TFs has been identified as a contributing factor to the aging process of stem cells. In our current study, we have detected several dysregulated TFs in aged ADSCs through analysis of RNA sequencing data. Notably, HES1 has emerged as a prominent TF that significantly decline in aged ADSCs. HES1 is recognized as a transcriptional repressor, although it can also exhibit transcriptional activation properties under specific conditions, contingent upon the formation of transcriptional complexes with other proteins [27]. Its expression is widespread across various tissues and cell types, particularly in quiescent and stem cells, where it plays a critical role in inhibiting differentiation [28]. Upon exposure to quiescent signals such as contact inhibition, serum-deprivation, loss of adhesion, and mitogen withdrawal, the expression levels of HES1 increase rapidly [29]. Several studies have reported heightened expression of HES1 in various tumor tissues, where it impedes the differentiation of tumor stem cells, allowing them to sustain self-renewal continuously. Conversely, the suppression of HES1 has been demonstrated to reduce tumor stemness markers and the capacity of tumors to form colonies [30]. Furthermore, HES1 has been found to inhibit senescence associated with oncogene activation and cell cycle arrest. Previous investigations have also elucidated the role of HES1 in the regulation of tissue aging. For example, HES1 is a critical transcription factor that undergoes significant down-regulation in skin tissues obtained from aged donors. Knockdown of HES1 led to a decrease in the proportion of ki67 positive cells and an increase in the proportion of SA-β-gal positive cells, while the ectopic overexpression of HES1 had the opposite effect [31]. These findings are consistent with our own observations, which revealed that restoring the expression of HES1 enhanced the proliferative and migratory capacities of aged ADSCs and reversed cellular senescence. In conclusion, HES1 acts as a suppressor of senescence in various cell types and tissues.
The downstream effectors of HES1 demonstrate variability among different types of stem cells. For instance, in breast cancer stem cells, HES1 may target Slug to enhance stemness characteristics [32]. Moreover, HES1 serves a protective function in hematopoietic stem cells by preventing cell exhaustion induced by inflammatory stress through the suppression of genes linked to PPARγ signaling and fatty acid metabolism pathways [33]. Increased HES1 expression can induce dormancy in neural stem cells by repressing Ascl1 [34]. In activated muscle stem cells, Hes1 is expressed in a rhythmic manner and controls the oscillatory expression of MyoD to sustain these cells [35]. By conducting combined analyses of RNA-seq data from aged ADSCs post Hes1 overexpression and CUT & Tag data from HES1-DNA complex pull-down experiments, the potential target STAT1 was identified. Subsequent investigations demonstrated that HES1 binds to the promoter region of STAT1, inhibiting its transcription, and that suppression of HES1 results in a notable increase in STAT1 expression. A prior study indicated that aging-induced inflammation of the subventricular zone (SVZ) niche is characterized by activation of Stat1 signaling on SVZ neural stem cells (NSCs), along with heightened expression of aging markers p21 and P16. Mechanistically, Stat1 can impede the transcription of SOX9, a pro-stemness gene, thereby diminishing the self-renewal capacity of NSCs [36]. Additionally, there is evidence suggesting that BMP9 suppresses the transcription of STAT 1 by activating Smad1, leading to a reduction in P21 expression and pro-inflammatory factors production in osteoblasts, thereby ameliorating age-related osteoporosis [37]. In our investigation, we observed that the detrimental impact of reduced HES1 in aged ADSCs could be fully reversed by the knockdown of STAT1. Consequently, our study reveals a novel mechanism involving the HES1/STAT1 axis in the aging process of stem cells.
So far, numerous studies have extensively investigated the therapeutic effects of ADSCs in wound healing [38]. Mechanistically, they mainly act through a paracrine manner, secreting a variety of favorable cytokines to exert anti-inflammatory and pro-healing effects. However, aged and senescent cells exhibit an inflammatory state characterized by an altered secretome, with a higher presence of pro-inflammatory factors [39]. This is commonly referred to as the senescence-associated secretory phenotype (SASP). An in-depth examination of microglial transcriptomic data showed that pro-inflammatory disease associated microglia are controlled by the NFkB, STAT1, and RelA pathways [40]. It has also been reported that the increase of pro-inflammatory immune cells in the intestinal mucosa leads to an increase in aging-related IFN-γ/ STAT1 signaling transduction in intestinal stem cells [41]. In our study, we found that STAT1 knockdown significantly inhibited the expression of pro-inflammatory cytokines IL-6, IL-8 and TNF-α, while HES1 knockdown exerted the opposite effect. Therefore, we hypothesized that HES1 might play an anti-inflammatory role by inhibiting STAT1, promoting wound healing by stimulating cell proliferation and angiogenesis, while reducing inflammatory reactions and scar formation at the wound site. At present, there are still many limitations to stem cell-based treatment approaches, such as the short survival time after stem cell transplantation and unexpected adverse effects caused by changes in single gene expression. Therefore, achieving safer and more effective clinical translation requires further efforts from the scientific research community.
Conclusion
The current investigation illustrated that a decline in the transcription factor HES1 due to aging contributes to the diminished phenotype and functionality of ADSCs obtained from old donors. This phenomenon is attributed to the ability of HES1 to suppress the expression of another transcription factor, STAT1, by binding to its promoter region and inhibiting its transcription. Restoring HES1 levels was found to enhance the proliferation and migration of aged ADSCs, reduce cellular senescence, and improve their therapeutic efficacy in skin wound healing. Conversely, silencing STAT1 also significantly improved the intrinsic capabilities of cells and decreased the secretion of pro-inflammatory cytokines in ADSCs with low HES1 expression. In summary, this research highlights an age-related regulatory axis involving HES1 and STAT1, suggesting potential targets for rejuvenating and enhancing the functionality of dysfunctional adipose stem cells derived from older individuals.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
In this manuscript, we utilized an artificial intelligence for language polishing (https://wordvice.ai/cn/tools/paraphrasing). Apart from that, we did not employ any other artificial intelligence software or websites.
Abbreviations
- ADSCs
Adipose-derived stem cells
- ATF6
Activating transcription factor 6
- ANOVA
Analysis of Variance
- α-SMA
α-smooth muscle actin
- bHLH
Basic helix-loop-helix
- BMMSCs
Bone marrow-derived mesenchymal stem cells
- CCNA1
Cyclin A1
- CCND1
Cyclin D1
- CD31
Platelet endothelial cell adhesion molecule-1
- EGR1 early
Growth Response 1
- ELISA
Enzyme-linked immunosorbent assay
- FOXP1
Forkhead Box P1
- GO
Gene ontology
- HES1
Hairy and enhancer of split 1
- HMGB2
High mobility group box 2
- KEGG
Kyoto encyclopedia of genes and genomes
- lincRNAs
Long non-coding RNAs
- miRNAs
Micro RNAs
- MMP2
Matrix metallopeptidase 2
- MMP9
Matrix metallopeptidase 9
- MSCs
Mesenchymal stem cells
- NSCs
Neural stem cells
- O-ADSCs
ADSCs derived from old donors
- PCNA
Proliferating cell nuclear antigen
- PCR
Polymerase chain reaction
- RNA-seq
RNA sequencing
- SA-β-gal
Senescence-associatedβ-galactosidase
- SASP
Senescence-associated secretory phenotype
- SD
Standard deviation
- siRNA
Small interfering RNA
- SOX
Sry-related HMG box
- STAT1
Signal transducer and activator of transcription 1
- SVZ
Subventricular zone
- TFs
Transcription factors
- TNF-α
Tumor necrosis factor-α
- Y-ADSCs
ADSCs derived from young donors
- ZF-C2H2
Cys2-His2 zinc finger
Author contributions
CL: data collection, assembly, analysis and interpretation, as well as manuscript composition. SR: conception and design. CY: manuscript proofreading; HX, JC, TJ, JG, CW, and YK: ADSCs isolation. JG, SL, and PN: provision of study materials. ZC: financial assistance and the final approval of the manuscript. The final manuscript was read and approved by all authors.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 82172221,82302814).
Data availability
All data generated and/or analyzed during this study are available from the corresponding author upon reasonable request. The raw sequence data have been submitted to the NCBI Gene Expression Omnibus (GEO) datasets with accession number (GSE269853, GSE270037, GSE270038).
Declarations
Ethics approval and consent to participate
The Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology approved the use of human adipose tissue in this study (Title: Research on the Aging and Repair Function of Human Adipose-Derived Stem Cells Approval No. 2022-S220, Date of approval: November 30, 2022). Informed consent was obtained from both the parents of young patients and elderly patients prior to their participation. Additionally, all animal experiments carried out in this research were approved by the Animal Care Committee of Tongji Medical College (Title: HES1 revitalizes the functionality of aged adipose-derived stem cells by inhibiting the transcription of STAT1. Approval No. 3870, Date of approval: November 1, 2023).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chengcheng Li and Sen Ren contributed equally to this work.
References
- 1.Chen J, Ren S, Duscher D, Kang Y, Liu Y, Wang C et al. Exosomes from human adipose-derived stem cells promote sciatic nerve regeneration via optimizing Schwann cell function. J Cell Physiol. 2019. [DOI] [PubMed]
- 2.Ren S, Chen J, Guo J, Liu Y, Xiong H, Jing B, et al. Exosomes from adipose stem cells promote Diabetic Wound Healing through the eHSP90/LRP1/AKT Axis. Cells-Basel. 2022;11:3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shukla L, Yuan Y, Shayan R, Greening DW, Karnezis T. Fat therapeutics: the clinical capacity of adipose-derived stem cells and exosomes for Human Disease and tissue regeneration. Front Pharmacol. 2020;11:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kokai LE, Marra K, Rubin JP. Adipose stem cells: biology and clinical applications for tissue repair and regeneration. Transl Res. 2014;163:399–408. [DOI] [PubMed] [Google Scholar]
- 5.Ren S, Xiong H, Chen J, Yang X, Liu Y, Guo J et al. The whole profiling and competing endogenous RNA network analyses of noncoding RNAs in adipose-derived stem cells from diabetic, old, and young patients. Stem Cell Res Ther. 2021;12. [DOI] [PMC free article] [PubMed]
- 6.Zhang H, Cai B, Geng A, Tang H, Zhang W, Li S et al. Base excision repair but not DNA double-strand break repair is impaired in aged human adipose‐derived stem cells. Aging Cell. 2019;19. [DOI] [PMC free article] [PubMed]
- 7.Ermolaeva M, Neri F, Ori A, Rudolph KL. Cellular and epigenetic drivers of stem cell ageing. Nat Rev Mol Cell Bio. 2018;19:594–610. [DOI] [PubMed] [Google Scholar]
- 8.Xiong H, Ren S, Chen J, Yang X, Liu Y, Xu Z et al. Knockdown of long noncoding RNA SAN rejuvenates aged adipose-derived stem cells via miR-143-3p/ADD3 axis. Stem Cell Res Ther. 2023;14. [DOI] [PMC free article] [PubMed]
- 9.Ren S, Li C, Xiong H, Wu Q, Wu X, Xiong Z et al. The rejuvenation and functional restoration of aged adipose stem cells by DUXAP10 Knockdown via the regulation of the miR-214-3p/RASSF5 Axis. Stem Cell Transl Med. 2024. [DOI] [PMC free article] [PubMed]
- 10.Li C, Ren S, Xiong H, Chen J, Jiang T, Guo J et al. MiR-145-5p overexpression rejuvenates aged adipose stem cells and accelerates wound healing. Biol Open. 2024;13. [DOI] [PMC free article] [PubMed]
- 11.Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. Hum Transcription Factors Cell. 2018;172:650–65. [DOI] [PubMed] [Google Scholar]
- 12.Huyghe A, Trajkova A, Lavial F. Cellular plasticity in reprogramming, rejuvenation and tumorigenesis: a pioneer TF perspective. Trends Cell Biol. 2024;34:255–67. [DOI] [PubMed] [Google Scholar]
- 13.Stevanovic M, Lazic A, Schwirtlich M, Stanisavljevic Ninkovic D. The role of SOX Transcription Factors in Ageing and Age-Related diseases. Int J Mol Sci. 2023;24. [DOI] [PMC free article] [PubMed]
- 14.Jing Y, Jiang X, Ji Q, Wu Z, Wang W, Liu Z, et al. Genome-wide CRISPR activation screening in senescent cells reveals SOX5 as a driver and therapeutic target of rejuvenation. Cell Stem Cell. 2023;30:1452–71. [DOI] [PubMed] [Google Scholar]
- 15.Kulkarni R. Early growth response factor 1 in aging hematopoietic stem cells and leukemia. Front Cell Dev Biol. 2022;10:925761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling S, Chen T, Wang S, Zhang W, Zhou R, Xia X, et al. Deacetylation of FOXP1 by HDAC7 potentiates self-renewal of mesenchymal stem cells. Stem Cell Res Ther. 2023;14:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Zheng Y, Wang S, Fan Y, Ye Y, Jing Y, et al. Single-nucleus transcriptomics reveals a gatekeeper role for FOXP1 in primate cardiac aging. Protein Cell. 2023;14:279–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, Hu B, Ding Z, Dang Y, Wu J, Li D, et al. ATF6 safeguards organelle homeostasis and cellular aging in human mesenchymal stem cells. Cell Discov. 2018;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lun AT, Smyth GK. Csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 2016;44:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mariani JN, Mansky B, Madsen PM, Salinas D, Kesmen D, Huynh NPT, et al. Repression of developmental transcription factor networks triggers aging-associated gene expression in human glial progenitor cells. Nat Commun. 2024;15:3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asano N, Takeuchi A, Imatani A, Saito M, Jin X, Hatta W et al. Wnt signaling and aging of the gastrointestinal tract. Int J Mol Sci. 2022;23. [DOI] [PMC free article] [PubMed]
- 27.Rani A, Greenlaw R, Smith RA, Galustian C. HES1 in immunity and cancer. Cytokine Growth F R. 2016;30:113–7. [DOI] [PubMed] [Google Scholar]
- 28.Sang L, Coller HA, Roberts JM. Control of the reversibility of Cellular Quiescence by the Transcriptional Repressor HES1. Science. 2008;321:1095–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sang L, Roberts JM, Coller HA. Hijacking HES1: how tumors co-opt the anti-differentiation strategies of quiescent cells. Trends Mol Med. 2010;16:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z, Dai X, Du B. Hes1: a key role in stemness, metastasis and multidrug resistance. Cancer Biol Ther. 2015;16:353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou Z, Long X, Zhao Q, Zheng Y, Song M, Ma S, et al. A single-cell transcriptomic atlas of human skin aging. Dev Cell. 2021;56:383–97. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Li Y, Du X, Wang X, Guan S, Cao Y, et al. HES1 promotes breast cancer stem cells by elevating slug in triple-negative breast cancer. Int J Biol Sci. 2021;17:247–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu L, Li X, Lin Q, Chowdhury F, Mazumder MH, Du W. FANCD2 and HES1 suppress inflammation-induced PPARɣ to prevent haematopoietic stem cell exhaustion. Brit J Haematol. 2021;192:652–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sueda R, Imayoshi I, Harima Y, Kageyama R. High Hes1 expression and resultant Ascl1 suppression regulate quiescent vs. active neural stem cells in the adult mouse brain. Gene Dev. 2019;33:511–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Lahmann I, Baum K, Shimojo H, Mourikis P, Wolf J, et al. Oscillations of Delta-like1 regulate the balance between differentiation and maintenance of muscle stem cells. Nat Commun. 2021;12:1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imitola J, Hollingsworth EW, Watanabe F, Olah M, Elyaman W, Starossom S, et al. Stat1 is an inducible transcriptional repressor of neural stem cells self-renewal program during neuroinflammation. Front Cell Neurosci. 2023;17:1156802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J, Zhou Y, Zhang L, Chen X, Yang Y, Zhang D, et al. BMP9 reduces age-related bone loss in mice by inhibiting osteoblast senescence through Smad1-Stat1-P21 axis. Cell Death Discovery. 2022;8:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li P, Guo X. A review: therapeutic potential of adipose-derived stem cells in cutaneous wound healing and regeneration. Stem Cell Res Ther. 2018;9:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazini L, Rochette L, Admou B, Amal S, Malka G. Hopes and limits of adipose-derived stem cells (ADSCs) and mesenchymal stem cells (MSCs) in Wound Healing. Int J Mol Sci. 2020;21. [DOI] [PMC free article] [PubMed]
- 40.Rangaraju S, Dammer EB, Raza SA, Rathakrishnan P, Xiao H, Gao T, et al. Identification and therapeutic modulation of a pro-inflammatory subset of disease-associated-microglia in Alzheimer’s disease. Mol Neurodegener. 2018;13:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Omrani O, Krepelova A, Rasa SMM, Sirvinskas D, Lu J, Annunziata F, et al. IFNγ-Stat1 axis drives aging-associated loss of intestinal tissue homeostasis and regeneration. Nat Commun. 2023;14:6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and/or analyzed during this study are available from the corresponding author upon reasonable request. The raw sequence data have been submitted to the NCBI Gene Expression Omnibus (GEO) datasets with accession number (GSE269853, GSE270037, GSE270038).