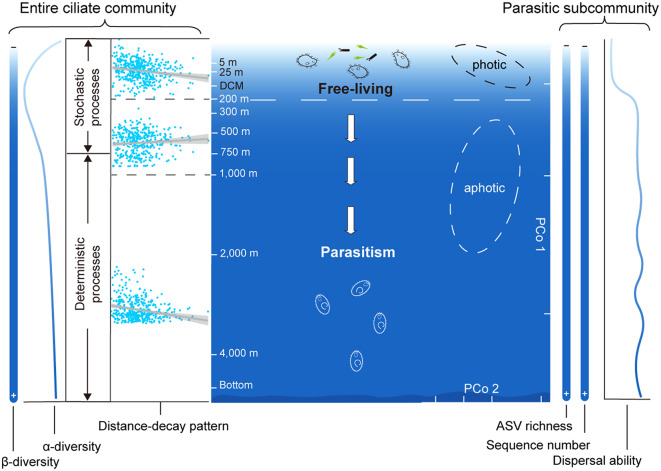
Abstract
Background
An increase in upper-ocean thermal stratification is being observed worldwide due to global warming. However, how ocean stratification affects the vertical profile of plankton communities remains unclear. Understanding this is crucial for assessing the broader implications of ocean stratification. Pelagic ciliates cover multiple functional groups, and thus can serve as a model for studying the vertical distribution and functional strategies of plankton in stratified oceans. We hypothesize that pelagic ciliate communities exhibit vertical stratification caused by shifts in functional strategies, from free-living groups in the photic zone to parasitic groups in deeper waters.
Results
306 samples from the surface to the abyssopelagic zone were collected from 31 stations in the western Pacific and analyzed with environmental DNA (the V4 region of 18 S rDNA) metabarcoding of pelagic ciliates. We found a distinct vertical stratification of the entire ciliate communities, with a boundary at a depth of 200 m. Significant distance-decay patterns were found in the photic layers of 5 m to the deep chlorophyll maximum and in the 2,000 m, 3000 m and bottom layers, while no significant pattern occurred in the mesopelagic layers of 200 m – 1,000 m. Below 200 m, parasitic Oligohymenophorea and Colpodea became more prevalent. A linear model showed that parasitic taxa were the main groups causing community variation along the water column. With increasing depth below 200 m, the ASV and sequence proportions of parasitic taxa increased. Statistical analyses indicated that water temperature shaped the photic communities, while parasitic taxa had a significant influence on the aphotic communities below 200 m.
Conclusions
This study provides new insights into oceanic vertical distribution, connectivity and stratification from a biological perspective. The observed shift of functional strategies from free-living to parasitic groups at a 200 m transition layer improves our understanding of ocean ecosystems in the context of global warming.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40793-024-00630-0.
Keywords: Pacific Ocean, Plankton, High-throughput sequencing, Deep-sea, Community connectivity, Functional strategy
Background
Ocean stratification has a significant impact on the marine biogeochemical cycle [1]. Global warming has increased the surface ocean temperature, resulting in increasing stratification in the upper ocean worldwide [2]. Furthermore, climate model predictions in phases 3 and 5 of the Coupled Model Intercomparison Project (CMIP) indicate that ocean stratification will continue to intensify throughout this century [3–6]. In the tropical Pacific, the thermocline usually occurs around 200 m [7], coinciding with the boundary between the photic and aphotic zones. While most studies focus on ocean stratification from a physical perspective, a comprehensive understanding from a biological standpoint is required to develop more effective strategies for managing and mitigating the effects of global warming.
Upper-ocean stratification can directly influence the availability of light for photosynthesis and the supply of nutrients to deeper waters, both of which are crucial for primary productivity [8]. Primary productivity, in turn, directly or indirectly affects marine organisms at higher trophic levels through the food web. Additionally, changes in key environmental variables, such as light, temperature, oxygen, and food supply along the water column can significantly impact plankton diversity and community structure [9–13]. In both the western Pacific and northeastern Atlantic, strong fluctuations in the vertical distribution of plankton communities have been observed in relation to ocean stratification [14, 15]. However, the causes of such biological stratification and the specific taxa responsible for these vertical shifts remain largely unknown.
Pelagic ciliates serve as important trophic intermediaries between primary producers and consumers at higher trophic levels, employing a wide range of functional strategies, including autotrophy, heterotrophy, mixotrophy, and symbiosis including parasitism [16–19]. Heterotrophic ciliates play a key role in oceanic ecosystems by grazing on bacteria and algae. Their feeding behavior can transfer energy to higher trophic levels and significantly alter the composition, activity, and physiology of their prey [16, 20, 21]. Given their ecological importance and diverse trophic strategies, pelagic ciliates can be used as a model to study the vertical distribution and transition of functional strategies among plankton in the deep sea.
However, our understanding on ciliate communities and their functional strategies in aphotic zones remains limited, and the vertical diversity patterns of ciliates are still blurry due to conflicting findings from previous studies. For instance, Countway et al. observed more ciliate OTUs (operational taxonomic units) in the photic layers compared to aphotic layers [22]. In contrast, Grattepanche et al. [23] and Hu et al. [24] reported an increase in diversity and activity of ciliate communities in aphotic zones. Additionally, Sun et al. found no significant change in alpha diversity between the mesopelagic layers and surface water [25].
Current research on ciliate distribution patterns has rarely extended to the deep sea [26–28], and studies on the driving factors of pelagic ciliate communities are even scarcer. Depth had been identified as the most important factor influencing variations in the ciliate community structure [25, 29]. Schoenle et al. highlighted a high and unique diversity of deep-sea ciliates, underscoring the significance of deep-sea microbial communities [30]. While water temperature and chlorophyll a have been reported as the primary factors affecting the surface-water protistan communities [31], how the deep-sea communities are regulated remains unknown. Previous studies have found certain parasitic ciliates to be unexpectedly active under high-pressure conditions [32]; and the limited export of organic carbon may contribute to the parasitic life strategies of deep-sea ciliates [33, 34]. In fact, the diversity of parasitic ciliates has also been reported to increase with depth [29, 35, 36], suggesting a potentially larger role for parasitic ciliates in the deep-sea communities.
In this study, we hypothesize that the pelagic ciliate communities exhibit vertical stratification driven by shifts in functional strategies that are prone to parasitism in deep water. To test this hypothesis, we collected a total of 306 samples from 31 stations in the western Pacific (Fig. 1), covering the full water column from the surface layer to the abyssopelagic zone, reaching depths of nearly 6,000 m. This large-scale sampling provides a possibility to verify our hypothesis. Using the environmental DNA metabarcoding approach, we analyzed the diversity, community composition and potential connections of the ciliate communities between different water layers, and their driving factors as well. The shifts in the ciliate functional strategies and the effect of parasitic taxa on the vertical communities were further revealed. This study aims to provide insights into oceanic stratification from a biological perspective and to enhance our understanding of the deep-sea microbial community under global warming.
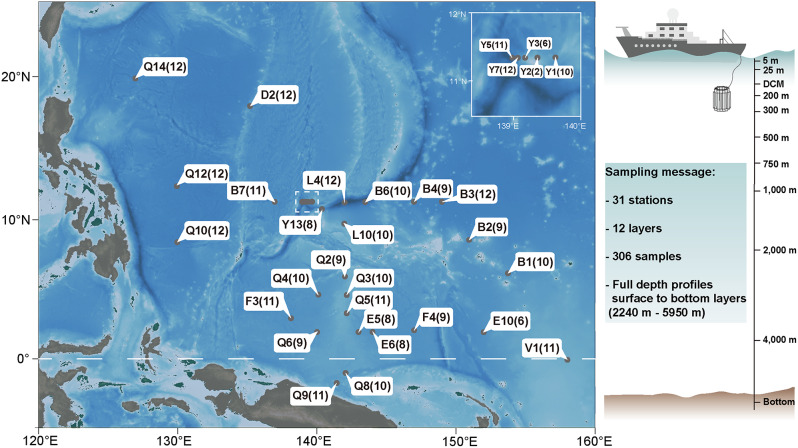
Fig. 1.
Overview of the sampling stations in the Pacific. Stations in the white dotted box are presented in the upper right corner of the map; number in parentheses indicates the number of layers taken at that station
Methods
Sampling station and collection
Water samples were collected from 31 stations across a horizontal distance of 3,454 km in the tropical western Pacific Ocean from November 2018 through January 2019. Depending on the maximum water depth (ranging from 2,240 m to 5,950 m) at each station, water samples were collected from the surface (5 m), 25 m, deep chlorophyll maximum (DCM), 200 m, 300 m, 500 m, 750 m, 1,000 m, 2,000 m, 3,000 m, 4,000 m, and the bottom layer, reaching depths of nearly 6,000 m (Fig. 1). At each station, 20 L of seawater was collected from each layer using Niskin bottles coupled to a Sea-bird SBE911 CTD system, by which the salinity and temperature were measured. After collection, water samples were pre-filtered through a 200 μm sieve and then filtered through a polycarbonate filter with a pore size of 0.22 μm. The filters were quickly frozen in liquid nitrogen and then stored at − 80 °C until further processing. In situ probes were used to directly monitor of salinity and water temperature.
DNA extraction, PCR, and illumina sequencing
Environmental DNA was extracted from each sample using the All Prep DNA/RNA Mini Kit (Qiagen, Germany), following the manufacturer’s recommended protocol. The hypervariable V4 region of the ciliate 18S rRNA gene was amplified using a nested PCR approach with three DNA subsamples from each sample [37]. First, the ciliate 18S rRNA gene was amplified with the CilF and CilR I-III primers [38]. This was followed by a second PCR using eukaryote-specific primers targeting the V4 region (EukF: 5’ CCAGCASCYGCGGTAATTCC3’, EukR: 5’ ACTTTCGTTCTTGATYRA3’) [39], with purified PCR products from the first reaction as the template. To minimize PCR bias, three replicate PCRs were conducted for each sample and the PCR products were pooled before proceeding to the subsequent steps. Sequencing libraries were conducted using the NEB Next® Ultra™ II FS DNA PCR-free Library Prep Kit (New England Biolabs, USA) and sequenced on an Illumina NovaSeq 6000 platform with 250 bp paired-end reads by Novogene Bio-informatics Technology Co., Ltd.
Bioinformatics and statistical analysis
Raw sequencing data were processed using the Easy Amplicon pipeline [40]. Primers and low-quality reads were first removed with VSEARCH (v.2.15.2), followed by a denoising step with USEARCH (v.10.0.240) to obtain representative high-quality sequences for amplicon sequence variants (ASVs). Taxonomic annotation was performed using the PR2 (v.4.14.0) reference database. The final ASV table was randomly subsampled to guarantee an equal number of sequences in each sample. Supplementary figures and tables, metadata (with environmental information), the unrarefied ASV table, and the corresponding taxonomic assignments are available in Additional files 1, 2, 3 and 4, respectively.
To ensure comparability of taxonomic richness and statistics across samples, the number of sequences per sample was rarefied to the smallest sample size (n = 1026). Statistical analyses were performed using R (v.4.1.3), with data visualization provide by the package ggplot2. The R codes used for statistical analysis is included in Additional file 5.
Taxonomic diversity (TDAlpha) and phylogenetic diversity (PDAlpha) were calculated using the R package vegan. Taxonomic diversity refers to the ASV richness, which was measured by counting the number of ASVs. In contrast, phylogenetic diversity incorporates the evolutionary distances between species, and it measures the evolutionary history shared by a set of species. It is defined as the minimum total length of all the phylogenetic branches required to connect a given set of taxa on a phylogenetic tree [41].
Principal Coordinate Analysis (PCoA) was conducted using the function `beta_pcoa()` in the R package amplicon (v.1.11.1) [40]. The Bray-Curtis dissimilarity (BC) index, computed with the `vegdist()` function (vegan package v.2.6.4) [42], was used to calculate pairwise dissimilarities of ciliate communities across all stations. In this context, dissimilarities (referring to beta diversity) range from 0 to 1, with a value of 1 indicating no shared ASVs between paired stations, and a value of 0 indicating identical communities. ‘Similarity’ in this context is defined as 1 – BC, where a value of 1 indicates identical communities, and vice versa. Additionally, a linear model was constructed to reveal the relationship between the parasitic subcommunity and the entire ciliate community. The R-squared value was used to assess the goodness of fit of the model; a lower R-squared value indicates greater dissimilarity between two communities, and vice versa.
Temperature, salinity, depth, longitude, and latitude were used to construct the environmental difference matrix based on Euclidean distance using the vegan package. Oceanic distance was used to evaluate the influence of dispersal, with the oceanic distance matrix calculated using the `distm()` function from in the geosphere package (v.1.5.18). To assess the potential correlations between variations in the ciliate community composition and environmental factors and oceanic distances as well, a Mantel test was performed using the `mantel()` function of the vegan package (permutations = 999) [43]. Each matrix included pairwise distances or dissimilarities for all stations. The correlation coefficient was calculated using the `corr.test()` function from psych package.
In the vertical connectivity analysis, each ASV was categorized with the water depth where it was initially detected from the surface to the abyssopelagic zone [44]. For the analysis of the ASV’s contribution at each depth, the ASVs from all stations were considered together.
We evaluated the potential role of stochastic processes in shaping the ciliate communities by using the neutral community model (NCM) [45]. The observed occurrence frequencies and the mean relative abundance of ASVs in the metacommunity were fit into the neutral community model, with the immigration rate (m) used as a metric for dispersal limitation [45]. Normalized stochasticity ratios (NST) were also employed to quantify the relative importance of deterministic and stochastic processes in community assembly [46], using a cutoff of 0.5 to distinguish between more deterministic (NST < 0.5) and more stochastic (NST > 0.5) processes. The calculation of dispersal ability, which was defined as passive transported by water movements, followed Yeh et al. [47].
Data availability
The 18 S rRNA gene sequencing raw data have been submitted in the Genome Sequence Archive in the BIG DATA Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences (https://ngdc.cncb.ac.cn/gsa) under the accession number CRA012724 at https://bigd.big.ac.cn/gsa/browse/CRA012724. The raw sequencing data were also submitted to the National Center for Biotechnology Information (NCBI) database under the accession number PRJNA1172926.
Results
Overview of sequencing data
A total of 7,257,615 ciliate sequences were retrieved from 306 samples collected at 31 stations in the Pacific (Fig. 1). The number of sequences per sample ranged from 1,026 (F49) to 104,384 (B111), with an average of 23,718 sequences per sample. The rarefaction analysis indicated near-saturated sampling profiles for all samples (Additional file 1: Fig. S1). A total of 3,300 ASVs were obtained and were classified into eight ciliate classes, namely Spirotrichea, Oligohymenophorea, Nassophorea, Colpodea, Phyllopharyngea, Prostomatea, Litostomatea, and Heterotrichea. Overall, the mean similarity of all detected ciliate ASVs to formally described sequences was 99.5%. Despite the high similarity, many ASVs belonged to environmental clades, which are composed exclusively of taxa detected by molecular surveys. Among these, NASSO_1, PHYLL_4, and OLIGO_5 were the most notable environmental clades, representing 97.6%, 96.4%, and 29.1% of the Nassophorea, Phyllopharyngea and Oligohymenophorea sequences, respectively.
Diversity and vertical stratification
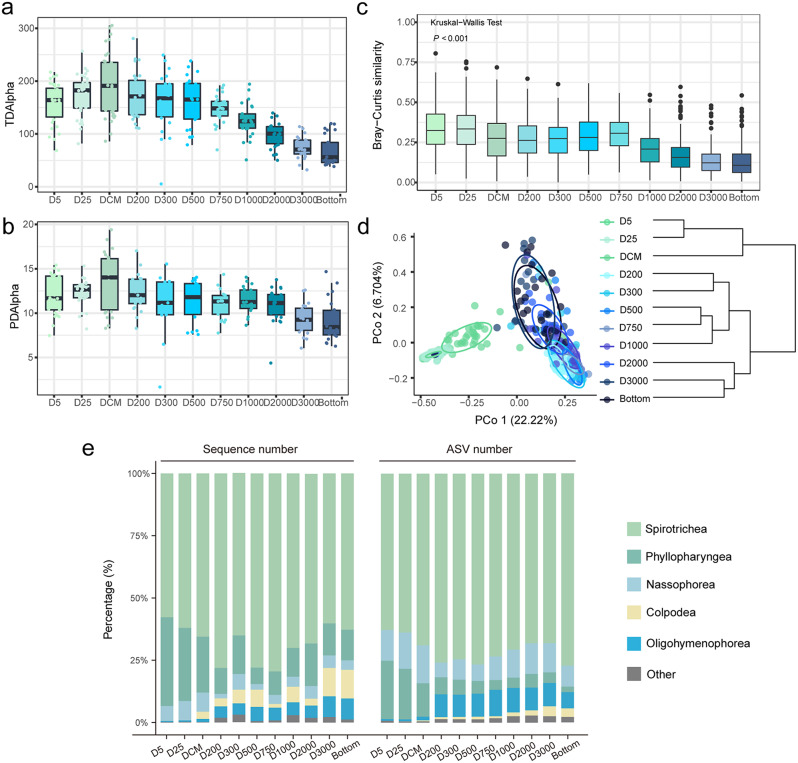
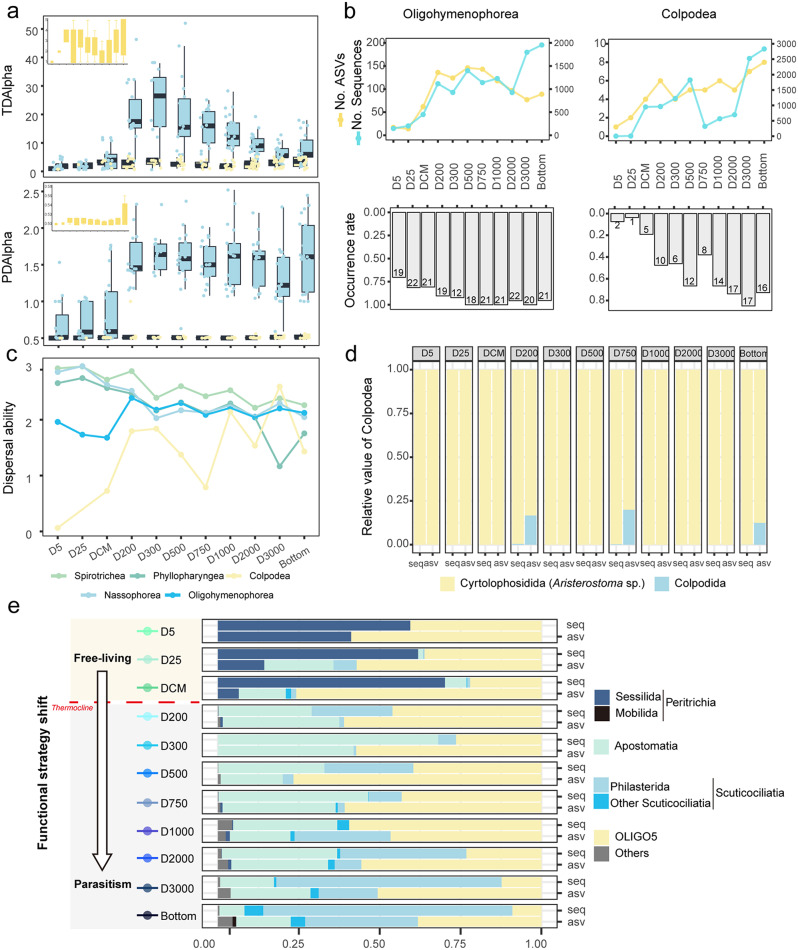
Both the taxonomic (TDAlpha) and phylogenetic diversity (PDAlpha) presented a unimodal pattern, with the highest diversity in the deep chlorophyll maximum (DCM) layer (Fig. 2a, b). The similarity of ciliate communities decreased with increasing depth (Fig. 2c). The horizontal community similarity among pairwise stations was shown in Additional file 1: Fig. S2.
Fig. 2.
Ciliate diversity and community structure in different water layers. (a) Taxonomic (TDAlpha) and (b) phylogenetic alpha diversity (PDAlpha). (c) Similarity boxplot and (d) Principal Co-ordinates Analysis (PCoA) plot of ciliate communities based on Bray-Curtis distances; the tree is based on UPGMA clustering. (e) Taxonomic composition of ciliate communities at the class level
Principal Coordinates Analysis (PCoA) revealed two distinct clusters of ciliate communities, viz., the photic and aphotic clusters, with the 200 m as a transition water layer, as also indicated by the transitions in temperature and salinity (Additional file 1: Fig. S3d). PCoA1 and PCoA2 accounted for 22.2% and 6.7% of the total variability, respectively (Fig. 2d). This result was consistent with those of the clustering analyses based on UPGMA (unweighted pair-group method with arithmetic means) and K-means (Additional file 1: Fig. S3a–c).
Spirotrichea and Phyllopharyngea were the most dominant groups throughout the water columns, followed by Nassophorea and Oligohymenophorea (Fig. 2e). In contrast, Prostomatea, Litostomatea, and Heterotrichea were the least abundant groups, accounting for less than 0.1% of all ASVs. Oligohymenophorea represented 4.1% of the total sequences and 14.1% of the total ASVs. The proportion of oligohymenophorean ASVs in the photic layers was relatively low, averaging 1% (ranging from 0 to 17%). Below 200 m, their proportion increased significantly, averaging 6.5% (ranging from 0 to 44.4%). Additionally, the ASV richness of Oligohymenophorea below the 200 m layer was considerably higher than that in the photic layers. Colpodea accounted for 4.2% of the total sequences and 0.5% of the total ASVs (14 ASVs). The highest sequence and ASV proportions of Colpodea were recorded in the 3,000 m layer (on average 12.3% and 1.2%, respectively) and in the bottom layer (on average 12.6% and 1.2%, respectively).
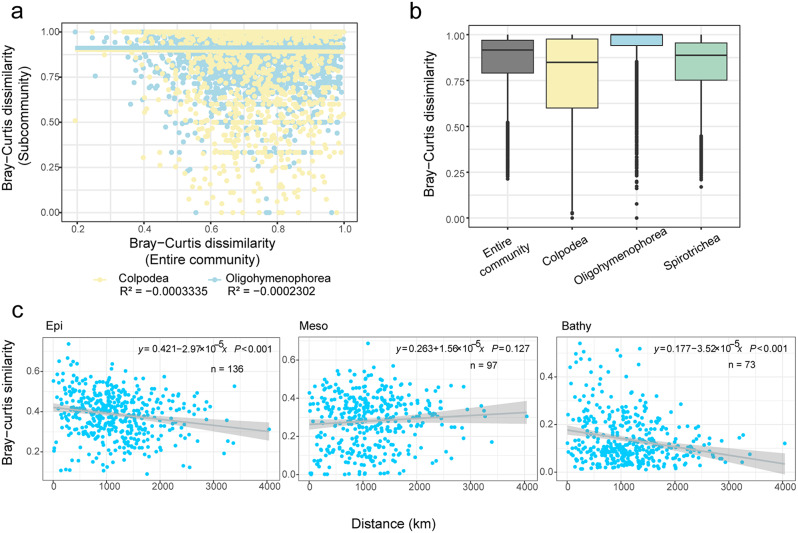
The ASVs of Spirotrichea, Phyllopharyngea, and Nassophorea are mostly heterotrophic. With increasing water depth below 200 m, the proportions of the parasitic ASVs and sequences of Oligohymenophorea and Colpodea generally increased. The Oligohymenophorea and Colpodea communities showed significant dissimilarity between individual paired stations, with minimal overlap with the entire ciliate community, as indicated by lower R-squared values (Fig. 3a, Additional file 1: Fig. S5). According to the Bray-Curtis dissimilarity of all layers (Fig. 3b), Oligohymenophorea showed the highest community dissimilarity throughout the water column, whereas Spirotrichea displayed less community variance. The taxonomic composition of the Oligohymenophorea shifted notably from dominance by the subclass Peritrichia (order Sessilida, family Zoothamniidae) in the photic waters to the subclasses Scuticociliatia (order Philasterida) and Apostomatia (order Apostomatida and Astomatophorida) below 200 m (Fig. 4e). In contrast, the heterotrophic and most abundant group Spirotrichea, demonstrated a much higher modelling fitness (i.e., R2 value) to the overall community (Additional file 1: Fig. S4a).
Fig. 3.
Ciliate community dissimilarity and distance-decay pattern. (a) Relationship between the entire ciliate community and the Oligohymenophorea as well as Colpodea subcommunities, considering all water layers. (b) Bray-Curtis dissimilarity boxplot of the entire ciliate community and subcommunities considering all layers. (c) Relationships between the horizontal community similarity and oceanic distance among pairwise stations in Epi (Epipelagic zone, 5 m – DCM), Meso (Mesopelgaic zone, 200 m – 1,000 m) and Bathy (Bathypelagic zone, 2,000 m – bottom). The points show the community similarity values for each distance unit considering all pairwise stations. The relationship between community similarity and oceanic distance is fitted by a linear model (solid line). The smooth shadows represent the 95% confidence level intervals for the predictions of a linear model. A similarity value of 0 indicates that the two stations have no ASV in common, while a value of 1 indicates the two stations have identical communities. n indicates the number of stations. The significance level (P value) is displayed for each of the linear regressions
Fig. 4.
Variations of parasitic groups along the water column. (a) Taxonomic alpha diversity (TDAlpha) and phylogenetic alpha diversity (PDAlpha) of each layer; blue boxes represent Oligohymenophorea, yellow ones represent Colpodea. (b) The number of ASVs and sequences of Oligohymenophorea and Colpodea. The frequence of occurrence indicates the ratio between the stations at which the taxa were detected and the total number of stations. (c) Dispersal ability of the main ciliate groups. (d) Taxonomic composition of Colpodea at the order level. (e) Taxonomic composition of Oligohymenophorea at the order level
Distribution pattern and vertical connectivity
Significant horizontal distance-decay patterns were found in the photic layers (5 m – DCM) and in the aphotic layers of 2,000 m, 3,000 m and bottom, while no significant patterns occurred in the mesopelagic layers of 200 m – 1,000 m. The bathypelagic layers exhibited the highest slope coefficients, suggesting strong dispersal limitation and increased heterogeneity of the entire ciliate communities. (Fig. 3c). The horizontal dispersal ability of Oligohymenophorea and Colpodea were lower than that of other ciliate groups in each water layer, showing a tendency of increased dispersal ability with increasing water depth below 200 m (Fig. 4c).
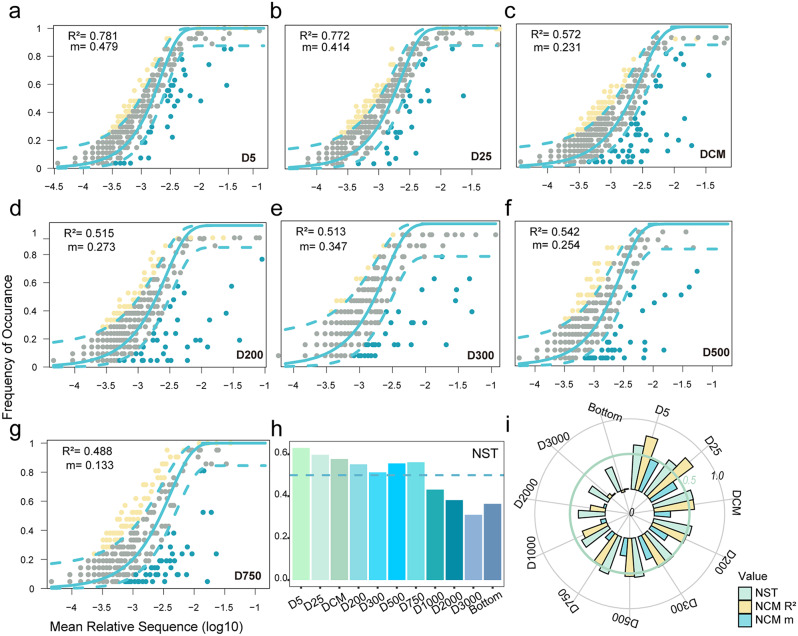
The ciliate communities were fit into a neutral community model (NCM), and the normalized stochasticity ratio (NST) was performed to quantify the role of deterministic and stochastic processes in shaping the ciliate communities (Fig. 5). Overall, the communities in layers above 750 m were predominantly shaped by the stochastic processes, while those in deeper layers were shaped by the deterministic processes (Fig. 5h, i). Specifically, above the 750 m layer, the ciliate communities exhibited a better goodness fit to the model, with an average of 59.8% of community variance explained, compared to those in deeper layers (15.2%). The communities in the 5 m and 25 m layers had relatively high fitness, with values of 78.1% and 77.2%, respectively, indicating significant influences of the stochastic processes in the surface communities. The NST results suggested stochastic process was the main assembly process in communities in layers above 750 m, with an average NST value of 57%. Below 750 m, deterministic process was the main assembly process in communities, with an average NST value of 37.1%. Additionally, the migration rate of ciliate communities decreased with increasing water depth, indicating that dispersal processes have little effect on the entire ciliate communities in the deeper waters.
Fig. 5.
Assembly rules of the ciliate communities. (a–g) Estimation of the neutral processes based on the Sloan neutral model. The parameters R2 and m represent the goodness of fit and migration rate, respectively. The species that occur more and less frequently than predicted are shown in yellow and blue, respectively. Dashed lines represent 95% confidence intervals and the species falling within the confidence intervals are considered neutrally distributed. (h) Normalized stochasticity ratio (NST) of each layer, higher NST value indicates the community assembly is more stochastic. (i) Assembly processes of the ciliate communities in each layer, circular axes are under scale of 0 to 1, with the green grid line represents a value of 0.5
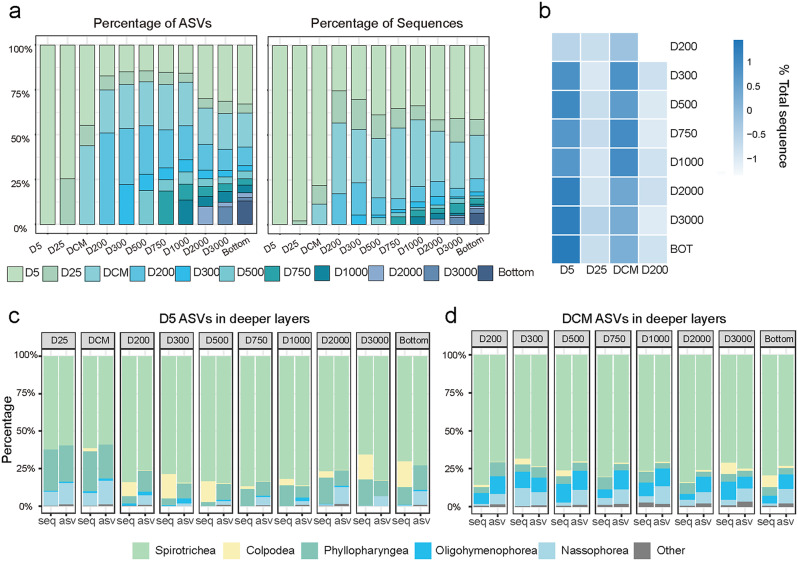
The vertical connectivity analysis showed that the ciliate communities were mainly composed of the photic ASVs which originated from the 5 m, 25 m, and DCM layer. These ASVs accounted for 81.7–94.7% of the sequence proportions in the aphotic communities (Fig. 6a). Among these, Spirotrichea, Phyllopharyngea and Nassophorea in the 5 m ASVs and DCM ASVs contributed the sequence proportions of 65.5%–74.7% to the aphotic communities (Fig. 6, Additional file 1: Fig. S8, Table S2). In addition, the relative sequence contributions of the 25 m, DCM, and 200 m ASVs to deeper communities also increased with increasing depth (Additional file 1: Table S1&S2).
Fig. 6.
Vertical connectivity of the ciliate communities. (a) Contribution of ASVs in each layer, the category of each ASV was categorized with the water depth where it was initially detected from the surface to the abyssopelagic zone. (b) Sequence proportion of shared ASVs in the aphotic layers. (c, d) Taxonomic compositon based on the sequence number (seq) and ASV number (asv) of the 5 m (D5) ASVs (c) and DCM ASVs (d) in the deeper water layers
Drivers of vertical community variations
The result of Mantel test showed that the environmental factors, particularly those in the photic zone, had a significant effect on the entire ciliate communities (Mantel r = 0.33, P = 0.001) (Additional file 1: Table S4). Temperature (Mantel r = 0.62, P = 0.001) and depth (Mantel r = 0.33, P = 0.001) were the most important factors. In contrast, salinity had a weak effect on the ciliate communities (Mantel r = 0.04, P = 0.015) (Additional file 1: Table S5). No significant effect was observed for those below the 200 m layer, where parasitic ciliates (members of Oligohymenophorea and Colpodea) generally increased with water depth (Fig. 2d, e).
Further analyses were conducted to reveal the relationship between the entire ciliate communities and the parasitic subcommunities. The results showed that parasitic ciliates had a significant effect (Mantel r = 0.49, P = 0.001) on the entire ciliate communities, particularly in the aphotic zone (Additional file 1: Table S6). The result of single Mantel test showed that the ASV richness (Mantel r = 0.26, P = 0.001) and sequences (Mantel r = 0.22, P = 0.001) of the parasitic Oligohymenophorea and Colpodea had also significant effects on the entire ciliate communities (Additional file 1: Table S5).
Discussion
Our study reveals a distinct vertical stratification and community variation of pelagic ciliates from the surface to the abyssopelagic zone, with a boundary at a depth of 200 m, aligning with the thermocline in the western Pacific Ocean [7]. The vertical biological stratification was associated with the shifts of relative dominance of ciliates from the free-living groups in the photic zone to parasitic groups in deeper waters. Earlier studies have documented an increase in parasitic ciliates with increasing water depth [29, 36]. Our study further showed that the deep-water ciliate communities below the 200 m layer were mainly regulated by parasitic taxa, indicating that the parasitic loop might play a crucial role in deep-sea ecosystems.
Biological stratification in the deep ocean
Ocean stratification refers to the variation in water density between the surface and deep water, a phenomenon influenced mainly by the vertical distribution of the water temperature and salinity [48]. Stratification suppresses the vertical mixing process, thereby influencing the vertical transport of heat, carbon, dissolved oxygen, and nutrients [2, 49]. Due to significant increase in surface ocean temperature caused by global warming, stratification is expected to be notably affected [50, 51]. These changes in vertical stratification and mixing are likely to influence the phytoplanktonic communities [52–55], ultimately affecting the marine biogeochemical cycle through changes in the food web structure and energy transfer.
Our investigations showed a distinct vertical stratification in the ciliate communities at a boundary layer of 200 m depth. This phenomenon coincides with the thermocline around 200 m depth, where environmental factors change drastically in the western Pacific Ocean [7]. The changes of environmental factors may lead to different capacities and trophic modes of microorganisms, driven by environmental selection and adaptation to variations in light, oxygen, nutrients, salinity, water temperature and pressure [56, 57]. In our study, the parasitic groups showed a significant positive correlation with depth, while free-living ciliates exhibited either no correlation or negative correlation with depth. These contrasting correlation patterns indicate that stratification may have a potential role in the vertical distribution and functional shifts of pelagic ciliates.
Shift in functional strategies from free-living to parasitic groups
Pelagic ciliates exhibit diverse morphologies and have various trophic modes, including autotrophy, heterotrophy, mixotrophy, and symbiosis including parasitism [18]. As an indispensable component of food web, ciliates serve as a bridge for primary productivity and energy transfer to higher trophic levels [16, 17]. In the deep sea, parasitic loop regulates host mortality and drive carbon as well as nutrient cycling [19]. By killing hosts, parasites release organic matter, fueling the microbial loop and influencing carbon sequestration. This parasitic activity thus plays a significant role in the carbon cycle of deep-sea ecosystems, where organic matter is more limited and efficiently recycled [19].
In this study, we found a shift in functional strategies from free-living dominance in the photic zone to parasitic dominance in the aphotic zone. Our results showed that with increasing water depth, the ASV richness and relative abundance of the entire ciliate communities decreased, while those of parasitic ciliates distinctly increased below the 200 m water depth. The shift in functional strategies was mainly due to the occurrence of Colpodea and Oligohymenophorea below 200 m. The colpodean ciliates obtained in this study were predominantly Aristerostoma sp., formerly known as a gill parasitic pathogen in farmed Atlantic salmon [58]. Previous pressure experiments suggested that Colpodea might be active in deep-sea environment [32]. Our study not only confirmed the presence of Aristerostoma sp. in the western Pacific Ocean, but also observed distinct increases in the ASV and sequence proportions of the species with increasing depth below 200 m (Fig. 4).
Compared to Colpodea, Oligohymenophorea contributed most to the functional shift. Within the class Oligohymenophorea, members of the subclass Apostomatia are obligate parasites, for instance, members of the family Pseudocolliniidae can establish symbiotic or parasitic relationships with invertebrates, such as krill and copepods [18]. Opalinopsidae (Apostomatia, Astomatophorida) is a family of endoparasitic ciliates previously found in the digestive tract and renal appendages [59]. The subclass Scuticociliatia, which encompasses several parasitic or facultatively parasitic taxa, contributed the second most to the functional shift. Within Scuticociliatia, Pseudocohnilembidae can cause a disease called Scuticociliatosis, primarily affecting aquatic organisms, particularly fish [60].
OLIGO5 are environmental clades of Oligohymenophorea, and have a weak phylogenetic relationship with divergent members of Scuticociliatia [35, 61]. Though it is tempting to speculate that they are parasitic, the OLIGO5 remains poorly understood [62]. In our study, OLIGO5 exhibited high ASV and sequence proportions. However, the environmental sequences (e.g., NASSO1, PHYLL4, and OLIGO5) are rarely assignable to families or lower taxonomic ranks and exhibit low similarity to the recognized morphospecies [29, 35]. This highlights our limited understanding regarding ciliates in open ocean waters, particularly those in the aphotic zone.
Establishing a comprehensive database is a pivotal task for improving taxonomic resolution. Collaborative efforts among taxonomists and molecular ecologists across disciplines and regions could enhance the classification of species with limited molecular information and uncharacterized sequences. Expanding diversity databases by encouraging researchers to submit novel and unclassified sequences to public repositories would help broaden the reference sequences. Metagenomic and single-cell genome assembly techniques could be employed to construct reference genomes, providing deeper insights into the taxonomic placement of these sequences. Additionally, bioinformatics tools, including machine learning algorithms, could aid in predicting taxonomic affiliations based on sequence features and evolutionary relationships.
Factors driven the community variations
Our study showed that parasitic ciliates contributed the most to the vertical variations of ciliate communities below 200 m. The insignificant pattern observed in the mesopelagic zone may be due to the oxygen minimum zone (OMZ, oxygen < 160 µmol/kg, 600–850 m), where parasitic groups may not adapt to the low oxygen environment [63]. Dissolved oxygen is known to be an essential factor shaping marine microbial trophic functions and community structures [33, 63, 64]. In hypoxic conditions, parasites maintain but exhibit reduced mitochondrial respiration, indicating that a low oxygen condition adversely affected their survival or metabolic activity [65]. The low dissolved oxygen in the OMZ layer might prevent strong dominance by any particular species and support a wider range of niches [11], which could explain the insignificant effect of the parasitic taxa on the entire ciliate communities. However, due to limited environmental data, we could not precisely determine how oxygen affects the ciliates, particularly parasitic ones. Future studies could culture ciliates under varying oxygen concentrations to explore the effect of oxygen on the transition of nutritional types in ciliates. Field studies could involve collecting samples along oxygen concentration gradients and combining genome and transcriptome sequencing to reveal the response mechanisms of ciliates and other planktons to changes in oxygen concentrations.
Our results indicate that the dispersal ability of Oligohymenophorea and Colpodea is crucial for the distance-decay pattern in the entire ciliate communities. Their dispersal ability decreased significantly below the 200 m layer, resulting no significant distance-decay pattern in the entire ciliate communities in the mesopelagic zone. These findings indicate that the prevalence of parasitic ciliates in the aphotic zone could represent either active or inactive parasites, which may have been collected as dispersal stages or fragments from deteriorated animal tissues. The association of parasitic ciliates with hosts or sinking particles facilitates their occurrence and activity in the aphotic zone. Host movements across different depth layers can transport parasites, thereby influencing their distribution patterns [66]. After hosts die, parasites may enter a dormant stage and disperse to other areas through water currents or other physical factors [67]. This dispersal mechanism is important in deep-sea environments. Additionally, our results highlight the potential role of parasitic ciliates in recycling carbon stored in animal biomass, suggesting a stronger involvement of parasitic ciliates in the aphotic zone compared to the photic zone.
Conclusions
Based on water sample collection from a wide range of latitudes and longitudes in the western Pacific Ocean, we analyzed the horizontal and vertical diversity and distribution of ciliates by using the environmental DNA metabarcoding approach. Significant distance-decay patterns of ciliate communities were found in different water layers of the photic, bathypelagic and abyssopelagic zones, but not in the mesopelagic layers of 200 m – 1,000 m. We found a distinct vertical stratification of ciliate communities in a boundary layer depth of 200 m, coinciding with the thermocline in the western Pacific Ocean. Below 200 m, parasitic Oligohymenophorea and Colpodea became more prevalent, resulting in a distinct shift of relative dominance of ciliates from free-living to parasitic groups. A linear model showed that parasitic taxa were the main groups causing the ciliate community variation along the water column. While temperature influenced the photic communities, parasitic taxa emerged as the primary drivers of the deep-water ciliate communities below 200 m. The observed shift provides new insights into oceanic stratification from a biological perspective, indicating that the parasitic loop might play a crucial role in deep-sea ecosystems.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Additional file 1 Supplementary data and results. Fig. S1 Rarefaction curves of amplicon sequence variants (ASVs). Fig. S2 Frequency of horizontal community similarity values between pairs of stations of each layer. Fig. S3 Clustering results of different groups. (a) Unweighted Pair-Group Method with Arithmetic means (UPGMA) clustering. (b) K-means clusters based on (c) the average silhouette width. (d) Temperature and salinity variations along water column. Fig. S4 Spirotrichean community variations along the water column. (a) relationship of spirotrichean community structure with the entire ciliate community structure of each layer. (b) taxonomic composition of Spirotrichea at the order level. Fig. S5 The relationship of Oligohymenophorea and Colpodea community structure with entire ciliate community structure. Fig. S6 Spearman correlation heatmap of environmental variables and ciliate groups. Fig. S7 Heatmap clustering of the 60 most abundant ASVs. Fig. S8 Vertical contribution of ASVs in each layer. (a) the sequence number < 1 and (b) relative percentage of total sequences < 0.01% of ASVs were deleted from the ASV table. Table S1 Relative proportion of shared ASVs in the 5 m, 25 m, DCM, and 200 m layer, respectively. Table S2 Relative proportion of shared ASVs in the 200 m, 300 m, 500 m, 750 m, 1,000 m, 2,000 m, 3,000 m, and bottom layer, respectively. Table S3 Partial Mantel correlations and MRM results between ciliate community dissimilarity, oceanic distance, and environmental distance. (a) partial Mantel correlations. (b) Multiple Regression on distance Matrices (MRM). Table S4 Mantel correlations between ciliate community dissimilarity and environmental factors. Table S5 Single Mantel correlations between the total ciliate community dissimilarity and each independent variable. Table S6 Mantel correlations between the entire ciliate community and parasitic community dissimilarity.
Supplementary Material 2: Additional file 2 Metadata used in this study.
Supplementary Material 3: Additional file 3 Unrarefied amplicon sequence variant table used for this study.
Supplementary Material 4: Additional file 4 Taxonomic assignments for all amplicon sequence variants in Additional file 3.
Supplementary Material 5: Additional file 5 Codes of statistical analysis performed in R.
Acknowledgements
We thank the crews of the R/V KeXue for their support in sample collection.
Abbreviations
- WPO
Western Pacific Ocean
- ASV
Amplicon sequence variant
- OTU
Operational taxonomic unit
- DCM
Deep chlorophyll maximum
- OMZ
Oxygen minimum zone
- PCoA
Principal coordinate analysis
- NCM
Neutral community model
- NST
Normalized stochasticity ratios
- TDAlpha
Taxonomic alpha diversity
- PDAlpha
Phylogenetic alpha diversity
- UPGMA
Unweighted pair group method with arithmetic mean
Author contributions
YW carried out the samples processing, statistical analyses, and original draft writing, participated in references researching; FZ participated in study designing, statistical analyses, references researching, original draft writing, manuscript revising and provided funding support; SF participated in original draft writing and manuscript revising; AH participated in original draft writing and manuscript revising; RZ carried out the samples processing; KX participated in study designing, original draft writing, manuscript revising and provided funding support. All authors read and approved the final manuscript.
Funding
The research was financially supported by the National Natural Science Foundation of China (No. 42376134, 41930533 and 41976099), the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDB42000000), the Science and Technology Innovation Project of Laoshan Laboratory (LSKJ202203102), and the Youth Innovation Promotion Association CAS (No. 2022206).
Data availability
The 18 S rRNA gene sequencing raw data have been submitted in the Genome Sequence Archive in the BIG DATA Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences (https://ngdc.cncb.ac.cn/gsa) under the accession number CRA012724 at https://bigd.big.ac.cn/gsa/browse/CRA012724. The raw sequencing data were also submitted to the National Center for Biotechnology Information (NCBI) database under the accession number PRJNA1172926.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Feng Zhao, Email: fzhao@qdio.ac.cn.
Kuidong Xu, Email: kxu@qdio.ac.cn.
References
- 1.Cronin M, Bond N, Farrar J, Ichikawa H, Jayne S, Kawai Y, et al. Formation and erosion of the seasonal thermocline in the Kuroshio Extension Recirculation Gyre. Deep Sea Res Part II Top Stud Oceanogr. 2013;85:62–74. [Google Scholar]
- 2.Intergovernmental Panel on Climate Change (IPCC), editor. Observations: Ocean Pages. Climate Change 2013 – the physical science basis: Working Group I contribution to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press; 2014. pp. 255–316. [Google Scholar]
- 3.Cabre A, Marinov I, Leung S. Consistent global responses of marine ecosystems to future climate change across the IPCC AR5 earth system models. Clim Dyn. 2014;45:1–28. [Google Scholar]
- 4.Capotondi A, Alexander M, Bond N, Curchitser E, Scott J. Enhanced upper ocean stratification with climate change in the CMIP3 models. J Geophys Res Oceans. 2012;117:4031. [Google Scholar]
- 5.Fu W, Randerson J, Moore J. Climate change impacts on net primary production (NPP) and export production (EP) regulated by increasing stratification and phytoplankton community structure in the CMIP5 models. Biogeosciences. 2016;13:5151–5070. [Google Scholar]
- 6.Moore J, Fu W, Primeau F, Britten G, Lindsay K, Long M, et al. Sustained climate warming drives declining marine biological productivity. Science. 2018;359:1139–43. [DOI] [PubMed] [Google Scholar]
- 7.Hollstein M, Mohtadi M, Rosenthal Y, Prange M, Oppo DW, Méndez GM, et al. Variations in Western Pacific warm pool surface and thermocline conditions over the past 110,000 years: forcing mechanisms and implications for the glacial walker circulation. Q Sci Rev. 2018;201:429–45. [Google Scholar]
- 8.Yamaguchi R, Suga T. Trend and Variability in Global Upper-Ocean Stratification since the 1960s. J Geophys Res Oceans. 2019;124:8933–48. [Google Scholar]
- 9.Blanco-Bercial L, Parsons R, Bolaños L, Johnson R, Giovannoni S, Curry R. The protist community traces seasonality and mesoscale hydrographic features in the oligotrophic Sargasso Sea. Front Mar Sci. 2022;9. 10.3389/fmars.2022.897140.
- 10.Ollison G, Hu S, Mesrop L, DeLong E, Caron D. Come rain or shine: depth not season shapes the active protistan community at station ALOHA in the North Pacific Subtropical Gyre. Deep Sea Res Part Oceanogr Res Pap. 2021;170:103494. [Google Scholar]
- 11.Schnetzer A, Moorthi S, Countway P, Gast R, Gilg I, Caron D. Depth matters: microbial eukaryote diversity and community structure in the eastern North Pacific revealed through environmental gene libraries. Deep-Sea Res Part -Oceanogr Res Pap - DEEP-SEA RES PT -Ocean RES. 2011;58:16–26. [Google Scholar]
- 12.Sogawa S, Nakamura Y, Nagai S, Nishi N, Hidaka K, Shimizu Y, et al. DNA metabarcoding reveals vertical variation and hidden diversity of Alveolata and Rhizaria communities in the western North Pacific. Deep Sea Res Part Oceanogr Res Pap. 2022;183:103765. [Google Scholar]
- 13.Zhao F, Filker S, Wang C, Xu K. Bathymetric gradient shapes the community composition rather than the species richness of deep-sea benthic ciliates. Sci Total Environ. 2020;755:142623. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y-J, Leung PM, Cook P, Wong W, Hutchinson T, Eate V, et al. Hydrodynamic disturbance controls microbial community assembly and biogeochemical processes in coastal sediments. ISME J. 2021;16:750–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mojica K, van de Poll W, Kehoe M, Huisman J, Timmermans K, Buma A, et al. Phytoplankton community structure in relation to vertical stratification along a north-south gradient in the Northeast Atlantic Ocean. Limnol Oceanogr. 2015;60. 10.1002/lno.10113.
- 16.Azam F, Fenchel T, Field J, Gray JS, Meyer L, Thingstad TF. The ecological role of Water-Column microbes in the Sea. Mar Ecol Prog Ser. 1983;10:257–63. [Google Scholar]
- 17.Caron DA, Countway PD, Jones AC, Kim DY, Schnetzer A. Marine Protistan Diversity. Annu Rev Mar Sci. 2012;4:467–93. [DOI] [PubMed] [Google Scholar]
- 18.Lynn DH. The ciliated protozoa: Characterization, classification, and guide to the literature: Third edition. 2008.
- 19.Worden AZ, Follows MJ, Giovannoni SJ, Wilken S, Zimmerman AE, Keeling PJ. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science. 2015;347:1257594. [DOI] [PubMed] [Google Scholar]
- 20.Sherr E, Sherr B. Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek. 2002; 81(1–4):293–308. [DOI] [PubMed]
- 21.Šimek K, Nedoma PH, Vrba JJP. Community structure, picoplankton grazing and zooplankton control of heterotrophic nanoflagellates in a eutrophic reservoir during the summer phytoplankton maximum. Aquat Microb Ecol. 1997;12:49–63. [Google Scholar]
- 22.Countway P, Vigil P, Schnetzer A, Moorthi S, Caron D. Seasonal analysis of protistan community structure and diversity at the USC Microbial Observatory (San Pedro Channel, North Pacific Ocean). Limnol Oceanogr. 2010;55. 10.4319/lo.2010.55.6.2381.
- 23.Grattepanche J-D, Santoferrara L, Andrade J, Oliverio A, Mcmanus G, Katz L. Distribution and diversity of Oligotrich and choreotrich ciliates assessed by morphology and DGGE in temperate coastal waters. Aquat Microb Ecol. 2014;71:211–21. [Google Scholar]
- 24.Hu D, Wu L, Cai W, Gupta AS, Ganachaud A, Qiu B, et al. Pacific western boundary currents and their roles in climate. Nature. 2015;522:299–308. [DOI] [PubMed] [Google Scholar]
- 25.Sun P, Huang L, Xu D, Warren A, Huang B, Wang Y, et al. Integrated Space-Time dataset reveals high diversity and distinct community structure of Ciliates in Mesopelagic Waters of the Northern South China Sea. Front Microbiol. 2019;10:2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christaki U, Wambeke F, Lefevre D, Lagaria A, Prieur L, Pujo-Pay M, et al. Microbial food webs and metabolic state across oligotrophic waters of the Mediterranean Sea during summer. Biogeosciences. 2011;8:1839–52. [Google Scholar]
- 27.Pitta P, Giannakourou A, Christaki U. Planktonic ciliates in the oligotrophic Mediterranean Sea: longitudinal trends of standing stocks, distributions and analysis of food vacuole contents. Aquat Microb Ecol - AQUAT MICROB ECOL. 2001;24:297–311. [Google Scholar]
- 28.Santoferrara LF, Gómez MI, Alder VA. Bathymetric, latitudinal and vertical distribution of protozooplankton in a cold-temperate shelf (southern Patagonian waters) during winter. J Plankton Res. 2010;33:457–68. [Google Scholar]
- 29.Santoferrara LF, Qureshi A, Sher A, Blanco-Bercial L. The photic‐aphotic divide is a strong ecological and evolutionary force determining the distribution of ciliates (Alveolata, Ciliophora) in the ocean. J Eukaryot Microbiol. 2023;70:e12976. [DOI] [PubMed] [Google Scholar]
- 30.Schoenle A, Duenn M, Hermanns K, Mahé F, de Vargas C, Nitsche F, et al. High and specific diversity of protists in the deep-sea basins dominated by diplonemids, kinetoplastids, ciliates and foraminiferans. Commun Biol. 2021;4:1234567890. 10.1038/s42003-021-02012-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ibarbalz FM, Henry N, Brandão MC, Martini S, Busseni G, Byrne H, et al. Global trends in Marine Plankton diversity across kingdoms of life. Cell. 2019;179:1084–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Živaljić S, Schoenle A, Scherwass A, Hohlfeld M, Nitsche F, Arndt H. Influence of hydrostatic pressure on the behaviour of three ciliate species isolated from the deep-sea floor. Mar Biol. 2020;167:63. [Google Scholar]
- 33.Edgcomb V. Marine protist associations and environmental impacts across trophic levels in the twilight zone and below. Curr Opin Microbiol. 2016;31:169–75. [DOI] [PubMed] [Google Scholar]
- 34.Zoccarato L, Pallavicini A, Cerino F, Fonda-Umani S, Celussi M. Water mass dynamics shape Ross Sea protist communities in mesopelagic and bathypelagic layers. Prog Oceanogr. 2016;149:16–26. [Google Scholar]
- 35.Canals O, Obiol A, Muhovic I, Vaqué D, Massana R. Ciliate diversity and distribution across horizontal and vertical scales in the open ocean. Mol Ecol. 2020;29:2824–39. [DOI] [PubMed] [Google Scholar]
- 36.Zhao F, Filker S, Xu K, Huang P, Zheng S. Patterns and drivers of Vertical distribution of the Ciliate Community from the surface to the Abyssopelagic Zone in the Western Pacific Ocean. Front Microbiol. 2017;8:2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stock A, Edgcomb V, Orsi W, Filker S, Breiner H-W, Yakimov M, et al. Evidence for isolated evolution of deep-sea ciliate communities through geological separation and environmental selection. BMC Microbiol. 2013;13:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lara E, Berney C, Harms H, Chatzinotas A. Cultivation-independent analysis reveals a shift in ciliate 18S rRNA gene diversity in a polycyclic aromatic hydrocarbon-polluted soil. FEMS Microbiol Ecol. 2008;62:365–73. [DOI] [PubMed] [Google Scholar]
- 39.Stoeck T, Bass D, Nebel M, Christen R, Jones MDM, Breiner H-W, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19:21–31. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y-X, Qin Y, Chen T, Lu M, Qian X, Guo X, et al. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell. 2021;12:315–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61(1):1–10. [Google Scholar]
- 42.Legendre P, Legendre L. Numerical Ecology. 3rd edition. Elsevier; 2012.
- 43.Diniz-Filho JA, Soares T, Lima J, Dobrovolski R, Landeiro V, Telles M, et al. Mantel test in population genetics. Genet Mol Biol. 2013;36:475–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mestre M, Ruiz-González C, Logares R, Duarte CM, Gasol JM, Sala MM. Sinking particles promote vertical connectivity in the ocean microbiome. Proc Natl Acad Sci. 2018;115(29):E6799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol. 2006;8:732–40. [DOI] [PubMed] [Google Scholar]
- 46.Ning D, Deng Y, Tiedje J, Zhou J. A general framework for quantitatively assessing ecological stochasticity. Proc Natl Acad Sci. 2019;116:201904623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeh Y-C, Peres-Neto P, Hunag S, Lai Y-C, Tu C-Y, Shiah F, et al. Determinism of bacterial metacommunity dynamics in the southern East China Sea varies depending on hydrography. Ecography. 2014;38:198–212. [Google Scholar]
- 48.Li G, Cheng L, Zhu J, Trenberth KE, Mann ME, Abraham JP, et al. Increasing ocean stratification over the past half-century. Nat Clim Chang. 2020;10:1116–23. [Google Scholar]
- 49.de Lavergne C, Palter J, Galbraith E, Bernardello R, Marinov I. Cessation of deep convection in the open Southern Ocean under anthropogenic climate change. Nat Clim Change. 2014;4:278–82. [Google Scholar]
- 50.Cheng L, Abraham J, Hausfather Z, Trenberth K. How fast are the oceans warming? Science. 2019;363:128–9. [DOI] [PubMed] [Google Scholar]
- 51.Durack P. Ocean Salinity and the Global Water cycle. Oceanogr Wash DC. 2015;28:20–31. [Google Scholar]
- 52.Doney S, Ruckelshaus M, Duffy J, Barry J, Chan F, English C, et al. Climate Change impacts on Marine ecosystems. Annu Rev Mar Sci. 2012;4:11–37. [DOI] [PubMed] [Google Scholar]
- 53.Gregg W, Conkright M, Ginoux P, O’Reilly J, Case N, Casey N. Ocean primary production and climate: global decadal changes. Geophys Res Lett - GEOPHYS RES LETT. 2003;30:1809. [Google Scholar]
- 54.Huisman J, Sharples J, Stroom J, Visser P, Edwin W, Kardinaal A, et al. Changes in turbulent mixing shift competition for light between phytoplankton species. Ecology. 2004;85:2960–70. [Google Scholar]
- 55.Richardson A, Schoeman D. Climate impact on plankton ecosystems in the Northeast Atlantic, Science. Science. 2004;305:1609–12. [DOI] [PubMed] [Google Scholar]
- 56.Quaiser A, Zivanovic Y, Moreira D, López-García P. Comparative metagenomics of bathypelagic plankton and bottom sediment from the sea of Marmara. ISME J. 2010;5:285–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yooseph S, Nealson K, Rusch D, McCrow J, Dupont C, Kim M, et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature. 2010;468:60–6. [DOI] [PubMed] [Google Scholar]
- 58.Dyková I, Tyml T, Kostka M, Peckova H. Strains of Uronema marinum (Scuticociliatia) co-isolated with amoebae of the genus Neoparamoeba. Dis Aquat Organ. 2010;89:71–7. [DOI] [PubMed] [Google Scholar]
- 59.Souidenne D, Furuya H. Protist (Ciliates) and related diseases. In: Gestal C, Pascual S, Guerra Á, Fiorito G, Vieites JM, editors. Handbook of pathogens and diseases in cephalopods. Cham: Springer International Publishing; 2019. pp. 153–8. [Google Scholar]
- 60.Xiong J, Wang G, Cheng J, Tian M, Pan X, Warren A, et al. Genome of the facultative scuticociliatosis pathogen Pseudocohnilembus persalinus provides insight into its virulence through horizontal gene transfer. Sci Rep. 2015;5:15470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boscaro V, Santoferrara LF, Zhang Q, Gentekaki E, Syberg-Olsen MJ, del Campo J, et al. EukRef‐Ciliophora: a manually curated, phylogeny‐based database of small subunit rRNA gene sequences of ciliates. Environ Microbiol. 2018;20:2218–30. [DOI] [PubMed] [Google Scholar]
- 62.Snyder R, Moss J, Santoferrara L, Head M, Jeffrey W. Ciliate microzooplankton from the Northeastern Gulf of Mexico. ICES J Mar Sci. 2021;78. 10.1093/icesjms/fsab002.
- 63.Xu F, Jerlström-Hultqvist J, Kolisko M, Simpson AGB, Roger AJ, Svärd SG, Andersson JO. On the reversibility of parasitism: adaptation to a free-living lifestyle via gene acquisitions in the diplomonad Trepomonas sp. PC1. BMC Biol. 2016;14:62. 10.1186/s12915-016-0284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pernice MC, Giner RG, Logares R, Pererabel-Bel J, Acinas SG, Duarte CM, Gasol JM, Massana R. Large variability of bathypelagic microbial eukaryotic communities across the world’s oceans. ISME J. 2015;10:945–58. 10.1038/ismej.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saraiva FMS, Cosentino-Gomes D, Inacio JDF, Almeida-Amaral EE, Louzada-Neto O, Rossini A, Nogueira NP, Meyer-Fernandes JR, Paes MC. Hypoxia effects on Trypanosoma Cruzi epimastigotes proliferation, differentiation, and energy metabolism. Pathogens. 2022;11(8):897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Foissner W, Chao A, Katz LA. Diversity and geographic distribution of ciliates (Protista: Ciliophora). Biodivers Conserv. 2008;17:345–63. 10.1007/s10531-007-9254-7. [Google Scholar]
- 67.Didi J, Zhishuai Q, Bojue W, David JSM, Xinpeng F, Xiangrui C. Two parasitic ciliates (Protozoa: Ciliophora: Phyllopharyngea) isolated from respiratory-mucus of an unhealthy beluga whale: characterization, phylogeny and an assessment of morphological adaptations. Zool J Linn Soc. 2021;191(4):941–60. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Additional file 1 Supplementary data and results. Fig. S1 Rarefaction curves of amplicon sequence variants (ASVs). Fig. S2 Frequency of horizontal community similarity values between pairs of stations of each layer. Fig. S3 Clustering results of different groups. (a) Unweighted Pair-Group Method with Arithmetic means (UPGMA) clustering. (b) K-means clusters based on (c) the average silhouette width. (d) Temperature and salinity variations along water column. Fig. S4 Spirotrichean community variations along the water column. (a) relationship of spirotrichean community structure with the entire ciliate community structure of each layer. (b) taxonomic composition of Spirotrichea at the order level. Fig. S5 The relationship of Oligohymenophorea and Colpodea community structure with entire ciliate community structure. Fig. S6 Spearman correlation heatmap of environmental variables and ciliate groups. Fig. S7 Heatmap clustering of the 60 most abundant ASVs. Fig. S8 Vertical contribution of ASVs in each layer. (a) the sequence number < 1 and (b) relative percentage of total sequences < 0.01% of ASVs were deleted from the ASV table. Table S1 Relative proportion of shared ASVs in the 5 m, 25 m, DCM, and 200 m layer, respectively. Table S2 Relative proportion of shared ASVs in the 200 m, 300 m, 500 m, 750 m, 1,000 m, 2,000 m, 3,000 m, and bottom layer, respectively. Table S3 Partial Mantel correlations and MRM results between ciliate community dissimilarity, oceanic distance, and environmental distance. (a) partial Mantel correlations. (b) Multiple Regression on distance Matrices (MRM). Table S4 Mantel correlations between ciliate community dissimilarity and environmental factors. Table S5 Single Mantel correlations between the total ciliate community dissimilarity and each independent variable. Table S6 Mantel correlations between the entire ciliate community and parasitic community dissimilarity.
Supplementary Material 2: Additional file 2 Metadata used in this study.
Supplementary Material 3: Additional file 3 Unrarefied amplicon sequence variant table used for this study.
Supplementary Material 4: Additional file 4 Taxonomic assignments for all amplicon sequence variants in Additional file 3.
Supplementary Material 5: Additional file 5 Codes of statistical analysis performed in R.
Data Availability Statement
The 18 S rRNA gene sequencing raw data have been submitted in the Genome Sequence Archive in the BIG DATA Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences (https://ngdc.cncb.ac.cn/gsa) under the accession number CRA012724 at https://bigd.big.ac.cn/gsa/browse/CRA012724. The raw sequencing data were also submitted to the National Center for Biotechnology Information (NCBI) database under the accession number PRJNA1172926.
The 18 S rRNA gene sequencing raw data have been submitted in the Genome Sequence Archive in the BIG DATA Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences (https://ngdc.cncb.ac.cn/gsa) under the accession number CRA012724 at https://bigd.big.ac.cn/gsa/browse/CRA012724. The raw sequencing data were also submitted to the National Center for Biotechnology Information (NCBI) database under the accession number PRJNA1172926.