Abstract
Neural stem cells have intact innate immune responses that protect them from virus infection and cell death. Yet, viruses can antagonize such responses to establish neuropathogenesis. Using a forebrain organoid model system at two developmental time points, we identified that neural stem cells, in particular radial glia, are basally primed to respond to virus infection by upregulating several antiviral interferon-stimulated genes. Infection of these organoids with a neuropathogenic Enterovirus-D68 strain, demonstrated the ability of this virus to impede immune activation by blocking interferon responses. Together, our data highlight immune gene signatures present in different types of neural stem cells and differential viral capacity to block neural-specific immune induction.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12974-024-03275-5.
Keywords: EV-D68, Innate immunity, Central nervous system, Forebrain organoids, Neurons, Neural stem cell
Introduction
Enterovirus-D68 (EV-D68) is a re-emerging, single-stranded, positive-sense RNA virus that was first isolated from four children with respiratory disease in the 1960s [1]. Intriguingly, since 2014, EV-D68 has been associated with a polio-like paralysis called acute flaccid myelitis (AFM) as biennial AFM cases have coincided with EV-D68 outbreaks in the United States [2]. Epidemiological surveillance and virus genome sequencing have confirmed the divergence of EV-D68 strains from Fermon, a prototypic strain from the 1960s, suggesting virus evolution and differential strain tropism [3]. Studies to determine the capacity of contemporary and prototypic EV-D68 strains to infect cells of the central nervous system (CNS) are conflicting [4, 5]. Given the emergence of respiratory viruses associated with neurological disease, understanding the neural tropism and viral factors associated with pathogenesis is important.
Brown et al. showed that strain VR1197, representative of a prehistoric EV-D68 strain similar to the prototypic strain Fermon, replicated at low levels in differentiated neuroblastoma cells and primary human fetal brain-derived neurons [4]. However, several other studies have suggested that EV-D68 neurotropism is not a recently acquired phenotype as contemporary circulating EV-D68 strains and two 1962 strains (Fermon and Rhyne) replicate in various murine and human neuronal systems [5, 6]. In addition to the inconclusive host cellular tropism of EV-D68, the viral receptor remains under investigation. Thus, understanding how the different EV-D68 strains interact within the cellular environment to dictate virus strain differences is important towards our goal of understanding the role immune responses play in protecting from neurological disease.
Throughout CNS development, the human brain is largely protected from virus infection through antiviral innate immune signaling and protective barriers [7]. Given the relative post-mitotic nature of neurons and other cells of the CNS, immune signaling serves a vital role in protecting these limited cells from excessive inflammation and cellular death. Neural stem cells have been proposed to serve as gatekeepers within the cortex, guarding the developing brain. However, neurotropic viruses may bypass such defenses to enter the CNS and cause neurological disease [8, 9]. CNS-specific immunity and viral immune evasion mechanisms are only beginning to be uncovered.
We hypothesized that type I antiviral signaling within cells of the CNS could contribute to EV-D68 strain differences. To model the heterogeneous complexity of the CNS, we utilized a recently established forebrain organoid model at two developmental time points and identified immune regulation within different neural stem cell subtypes by different EV-D68 strains. We found that neural stem cells basally express higher levels of several interferon stimulated genes (ISG); neuropathogenic EV-D68 can block upregulated IFITM1 defenses; and that different neural lineages contain, diverse immune gene signatures, which may protect against virus infection.
Results
SH-SY5Y cells are permissive to neuropathogenic and non-neuropathogenic EV-D68 strains
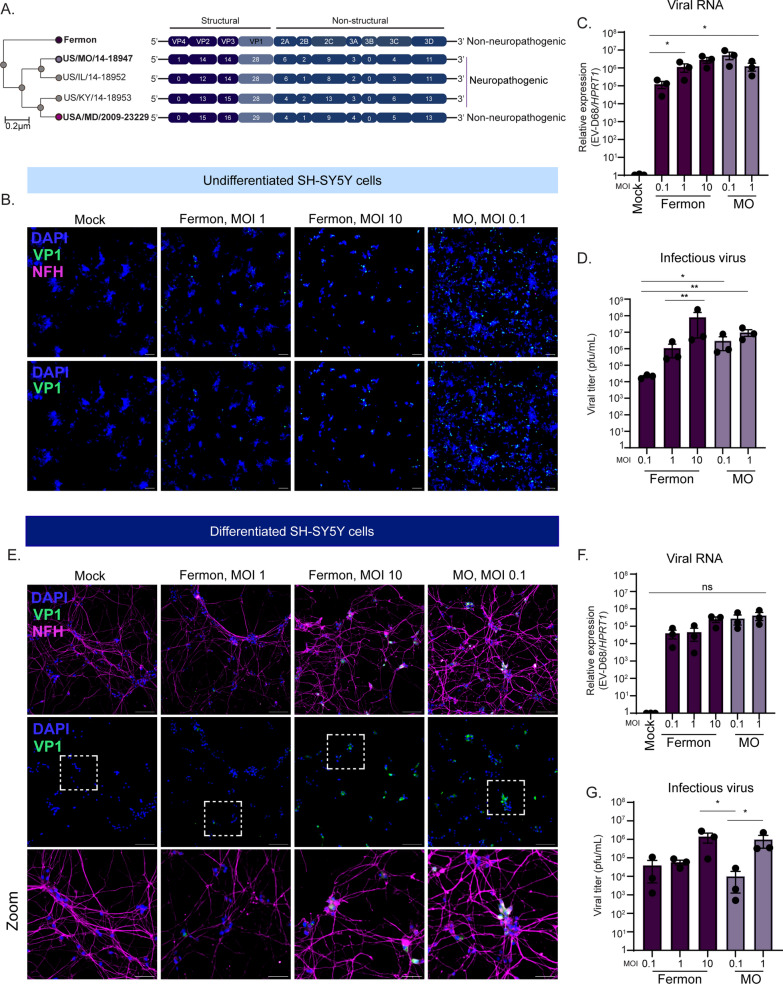
Several studies have sequenced circulating EV-D68 strains in many countries, identifying virus mutations that have diverged from the historic Fermon strain (illustrated in Fig. 1A). However, whether EV-D68 mutations have conferred CNS permissiveness remains unclear. Given the inconsistent replication kinetics reported in the literature, we sought to determine whether Fermon and a contemporary neurotropic EV-D68 strain, MO/14-18947 (defined throughout as MO), could infect either undifferentiated or differentiated SH-SY5Y cells. SH-SY5Y cells have historically been used to investigate EV-D68 replication dynamics [4]. To differentiate SH-SY5Y cells, we utilized a previously established 18-day protocol dependent upon addition of retinoic acid and gradual depletion of fetal bovine serum [10]. Temperature is known to influence EV-D68 replication kinetics with 33 °C, that of the upper respiratory tract, being more suitable; however, there is still infectious virus production at 37 °C [11, 12]. Our investigation sought to examine the mechanisms underlying EV-D68 replication and immune regulation within the CNS, where the human temperature is 37 °C. Thus, we performed our infections at 37 °C. To determine virus permissiveness, we first infected undifferentiated SH-SY5Y cells at varying multiplicities of infection (MOI) and measured viral VP1 protein expression via immunofluorescence (Fig. 1B), viral RNA copies (Fig. 1C), and viral titer (Fig. 1D). We found that both Fermon and MO could infect and replicate in undifferentiated SH-SY5Y cells, even at the lowest MOI. Interestingly, we did not observe a stark MOI-dependent response in MO-infected undifferentiated SH-SY5Y cells, likely due to an increased in cellular death at the higher MOI. We next differentiated SH-SY5Y cells and infected them with Fermon and MO at varying MOIs. Similar to the undifferentiated cells, we found that both Fermon and MO expressed viral VP1 protein (Fig. 1E), replicated to detectable RNA copies (Fig. 1F), and produced infectious virus titer (Fig. 1G). Together, these data support the notion that SH-SY5Y cells are not ideal models to recapitulate the EV-D68 strain differences in the CNS as non-neuropathogenic Fermon was capable of infecting and replicating within them.
Fig. 1.
Prototypic Fermon strain replicates in SH-SY5Y cells. A Alignment and phylogenetic tree of several EV-D68 non-neuropathogenic and neuropathogenic strains, with the prototypic Fermon strain serving as reference strain. This schematic was adapted from [11]. Phylogenetic tree from viral genome sequences was done using the NCBI phylogenetic tree software, and the protein sequences were aligned using Clustal Omega. B Representative immunofluorescence images of undifferentiated SH-SY5Y cells mock-infected or infected with Fermon or MO viruses at the indicated multiplicity of infection (MOI) at 24 h post infection (hpi). Cells were stained for DAPI (nuclei, blue), VP1 (green), and neurofilament-H (NFH, magenta). Scale bar—100 µm. C RT-qPCR analysis of RNA harvested from undifferentiated SH-SY5Y cells mock-infected or infected with Fermon or MO at the indicated MOIs for 24 h. The data are normalized to mock-infected cells and are presented as relative expression of EV-D68 to HPRT1 with Mock set to 1. Data are presented as means ± SEM (n = three biological replicates). D Plaque-forming assay of supernatants from undifferentiated SH-SY5Y cells infected with either Fermon or MO at the indicated MOIs for 24 h. Data are presented as means ± SEM (n = three biological replicates). N.D. = not detected E Representative immunofluorescence images of differentiated SH-SY5Y cells mock-infected or infected with Fermon or MO viruses at the indicated multiplicity of infection (MOI) at 24 hpi. Cells were stained for DAPI (nuclei, blue), VP1 (green), and phosphorylated neurofilament-H (NFH, magenta). Scale bar—100 µm. F RT-qPCR analysis of RNA harvested from differentiated SH-SY5Y cells mock-infected or infected with Fermon or MOI at the indicated MOIs for 24 h. The data are normalized to mock-infected cells and are presented as relative expression of EV-D68 to HPRT1 with Mock set to 1. Data are presented as means ± SEM (n = three biological replicates). N.D. = not detected G Plaque-forming assay of supernatants from differentiated SH-SY5Y cells infected with either Fermon or MO at the indicated MOIs for 24 h. Data are presented as means ± SEM (n = three biological replicates). Statistical analysis performed with one-way analysis of variance (ANOVA)
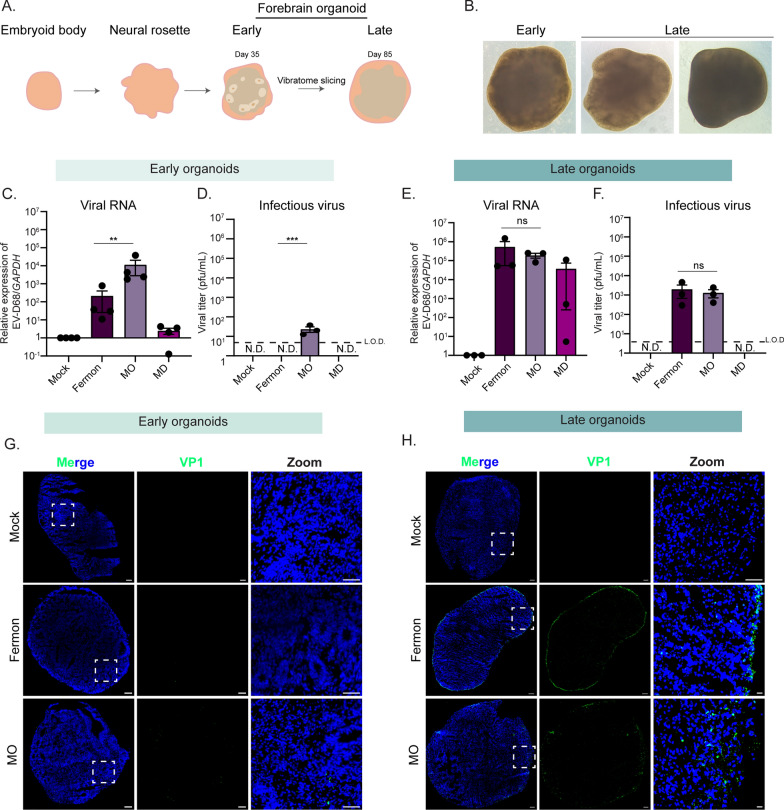
Developmentally less mature forebrain organoids are selectively permissive to neuropathogenic EV-D68
To attempt to better model EV-D68 strain differences observed in the human population in vitro, we turned to the recently established forebrain organoid model system. Forebrain organoids display the complex cellular environment observed in the developing human cortex, with sustained maintenance in culture permitting development of more mature neuronal subtypes [13] (Fig. 2A). To model a heterogeneous cell population and emergence of maturing neuronal cells, we cultured forebrain organoids until either day in vitro (DIV) 35 (early) or 85 (late) from the human induced pluripotent stem cells [14–16]. Early organoids are characterized by the appearance of cellular niches of stem and progenitor cells, or neural rosettes, that line the periphery of the organoid (Fig. 2B). On the other hand, late organoids contain few neural rosette structures (Fig. 2B). Both early and late organoids were mock, Fermon-, or MO-infected for 24 h. At 24 h post-infection (hpi), infection inoculum was removed, and media was changed daily until 7 days post infection (dpi), at which time both organoids and culture supernatant were harvested for analysis. We observed viral RNA copies (Fig. 2C) in both Fermon- and MO-infected organoids, but infectious viral titer (Fig. 2D) was limited to MO-infected organoids only. At the late time point, however, we found that both Fermon and MO are capable of replicating (Fig. 2E) and producing infectious titer (Fig. 2F). We next examined viral VP1 protein via immunofluorescence of early and late organoids and saw that only MO-infected early organoids contained VP1-positive cells (Fig. 2G) whereas both Fermon and MO-infected late organoids expressed viral antigen (Fig. 2H). To address the concern of reproducibility amongst various human iPSCs, we validated findings with forebrain organoids generated from another iPSC line, WTC11[17], and found similar phenotypes (Supplemental Fig. 1). These data establish forebrain organoids as an in vitro system that phenocopies the viral tropism observed in the human population.
Fig. 2.
Differential Enterovirus-D68 replication in early and late forebrain organoids. A Schematic of organoid structures. B Representative bright field images taken at 40X from organoids at day in vitro (DIV) 35 (early) and DIV85 (late) (C–H). Early and late organoids were infected mock-infected or infected with Fermon, MO, and MD viruses for 7 days. At 7 dpi, lysates and organoids were harvested for downstream analysis. C RT-qPCR analysis of DIV35 organoids. The data are normalized to mock-infected cells and are presented as relative expression of EV-D68 to GAPDH with Mock set to 1. Data are presented as means ± SEM (n = four biological replicates). D Plaque-forming assay of supernatants from DIV35 organoids. Data are presented as means ± SEM (n = three biological replicates). ND = not detected; LOD = limit of detection E RT-qPCR analysis of RNA harvested of DIV85 organoids. The data are normalized to mock-infected cells and are presented as relative expression of EV-D68 to GAPDH with Mock set to 1. Data are presented as means ± SEM (n = three biological replicates). F Plaque-forming assay of supernatants harvested from DIV85 organoids. Data are presented as means ± SEM (n = three biological replicates). G Representative immunofluorescence images of DIV35 forebrain organoids mock-infected or infected with either MO or Fermon viruses harvested at 7 dpi. Dashed boxes indicate the location of the zoom images. DAPI (blue) stains nuclei and VP1 (green, virus protein) indicates infected cells. H Immunofluorescence images of DIV85 forebrain organoids mock-infected or infected with either MO or Fermon viruses harvested at 7 dpi. Dashed boxes indicate the location of the zoom images. DAPI (blue) stains nuclei and VP1 (green, virus protein) indicates infected cells. Scale bar—100 µm. Statistical analysis performed with one-way ANOVA
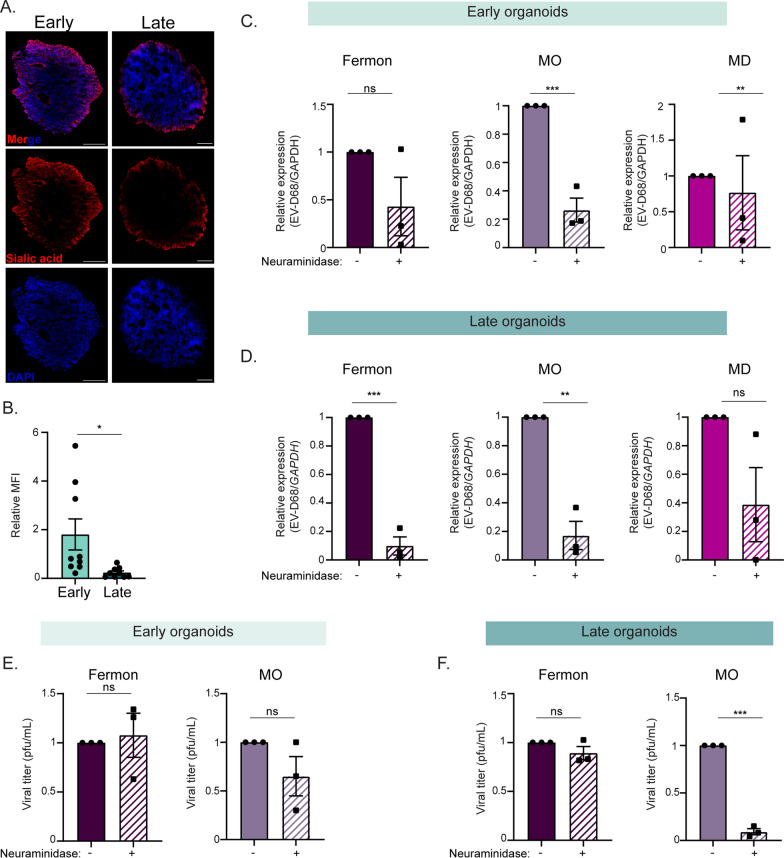
Neuropathogenic and non-neuropathogenic EV-D68 strains rely on sialic acid for entry into forebrain organoids
Although an EV-D68 cellular receptor is still undefined, sialic acid or a sialylated glycoprotein has been proposed to be an entry receptor [18]. To determine whether differential levels of sialic acid expression could explain the divergent virus susceptibility between early and late organoids, we performed click chemistry. We found that both early and late forebrain organoids sialic acid (Fig. 3A). Interestingly, quantification of the mean fluorescence intensity of the click chemistry images (total sialic acid / total DAPI) revealed that late organoids express less total sialic acid compared to the early organoids (Fig. 3B). To demonstrate if EV-D68 entry into the forebrain organoids is dependent upon sialic acid, we treated organoids with neuraminidase, a sialic acid cleaving enzyme, for one hour prior to infection and then mock-infected or infected the organoids with either Fermon, MO, or MD-09/23229 (MD) viruses. MD is an EV-D68 strain that was isolated from a patient with respiratory illness in 2009, prior to the first reported AFM outbreaks in 2014, and represents a non-neuropathogenic strain in circulation independent of reported AFM cases [11, 19]. At 24 hpi, we harvested lysates and supernatants to measure virus entry. We found that in the early organoids, treatment with neuraminidase reduced viral copies (Fig. 3C) of MO and MD strains. When we treated and infected late organoids, viral copies of both Fermon and MO were reduced (Fig. 3D). Further, only infectious viral titer of the MO strain was impacted by treatment with neuraminidase in either early or late organoids (Fig. 3E, F). We also tested for the presence of an additional proposed EV-D68 receptor, intracellular adhesion molecule-5 (ICAM-5) [20]. We detected minimal ICAM-5 protein via immunofluorescence at either time point, and there was no statistical detectable difference in ICAM-5 transcripts between the time points (Supplemental Fig. 2). Although forebrain organoids at both time points express sialic acid, the presence of sialic acid alone does not explain the strain differences observed.
Fig. 3.
Enterovirus-D68 replication in forebrain organoids is partially dependent upon sialic acid expression. A Representative immunofluorescence images of DIV35 or DIV85 forebrain organoids for sialic acid protein expression using the Click-iT reaction. Scale bar—200 µm (B). Quantification of the relative mean fluorescence intensity (MFI, alkyne-555/DAPI) of the immunofluorescence images from (A). Three sections from three independent biological replicates for either early or late organoids were quantified, and data are presented as the means ± SEM (n = three biological replicates, 9 total replicates). Statistical analysis performed with unpaired student’s t-test. C RT-qPCR analysis of lysates harvested from DIV35 forebrain organoids treated with neuraminidase for 1 h and then infected with Fermon, MO, or MD viruses for 24 h. D RT-qPCR analysis of lysates harvested from DIV85 forebrain organoids treated with neuraminidase for 1 h and then infected with Fermon, MO, or MD viruses for 24 h. The data are normalized to untreated organoids and are presented as relative expression of EV-D68 to GAPDH with untreated set to 1. Data are presented as means ± SEM (n = three biological replicates). E, F Plaque-forming assay of supernatants harvested at 1 dpi from forebrain organoids mock-infected or infected with either Fermon, MO, or MD at DIV35 (E) or DIV85 (F). Data are presented as means ± SEM (n = three biological replicates). Statistical analysis was performed with unpaired student’s t-test
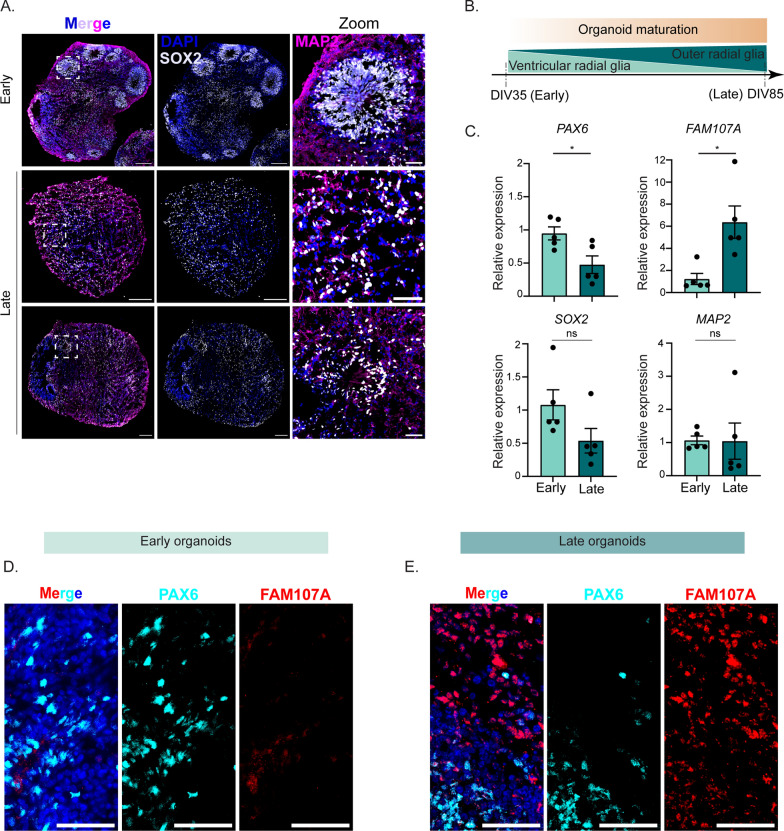
Early and late forebrain organoids contain distinct neural stem cell populations
Mammalian cortex development begins in utero as distinct laminations, consisting of six layers with marked neural cell types emerging throughout each layer [15]. Forebrain organoids recapitulate this anatomical lamination and cellular heterogeneity observed in the human brain, and thus serve as an ideal model to interrogate host responses in different neural contexts [13].
Early in forebrain organoid development, neural stem cells reside along the periphery of the organoid in a cellular niche called a neural rosette, analogous to the neural tube in the developing human cortex. As forebrain organoids mature, neural rosettes decrease, as developmentally mature neurons increase (Fig. 4A). To determine whether early and late forebrain organoids display differences in morphological features, namely the presence of these neural rosettes and mature neurons, we immunostained for SOX2 and MAP2, respectively. We found that early forebrain organoids display clusters of neural rosettes positive for SOX-2, a broad marker of neural stem cells (Fig. 4A). However, in late forebrain organoids, localization of SOX-2-positive cells shifts with a marked change in distinct pockets of accumulation. (Fig. 4A). Both early and late forebrain organoids contain MAP2-positive cells, suggesting that differences between early and late development timepoints are largely centered around localization and accumulation of neural stem cells (Fig. 4A). Indeed, in our forebrain organoids, we did not detect any GFAP-positive astrocytes in our early organoids, in contrast to some visible GFAP-positive staining in late organoids (Supplemental Fig. 3), supporting what has previously been reported [17, 21, 22] Since we observed increased viral copies and infectious virus in the late organoids, but minimal GFAP-positive cells, immune signaling by astrocytes cannot account for the viral strain differences.
Fig. 4.
Early and late forebrain organoids display different morphology, protein expression, and protein localization. A Representative immunofluorescence images of early and late forebrain organoids. DAPI (blue) stains nuclei, MAP2 (magenta) stains maturing neurons, and SOX2 (grey) indicates neural stem cells. Scale bar of the whole organoid—200 µm and scale bar of zoom—50 µm B Schematic of cell type expression as organoids mature from DIV35 to DIV85 (C). RT-qPCR analysis of RNA harvested from uninfected forebrain organoids at DIV35 (early) and DIV85 (late). The data are normalized to the early samples and are presented as relative expression of the respective genes to GAPDH. Data are presented as means ± SEM (n = five biological replicates). Statistical analysis was performed with unpaired student’s t-test. D, E Representative immunofluorescence images of early (D) and late (E) uninfected forebrain organoids. DAPI (blue) stains nuclei, PAX6 (cyan) marks ventricular radia glia cells, and FAM107A (red) indicates outer radial glia cells. Scale bar—50 µm
The developing human cortex consists of several types of neural stem cells, named after the distinct cortical regions from which they arise. Important within the cortex are radial glia neural stem cells [23, 24]. Radial glia cells emanating from the ventricular zone are called ventricular radial glia cells (vRG) and those appearing in the outer subventricular zone are called outer radial glia (oRG) [25]. Interestingly, oRGs are human-specific, as rodent species do not contain them [26, 27]. oRGs emerge following vRGs, as the outer subventricular zone emerges later in development from the ventricular zone [28, 29]. As such, it has been shown that as forebrain organoids further develop in vitro, they begin to express more oRGs compared to vRGs and vice versa early in development ([30] and Fig. 4B). To confirm whether our early forebrain organoids contain increased vRGs compared to oRGs, we isolated RNA from uninfected early and late forebrain organoids and performed RT-qPCR for mRNA transcripts of the established vRG marker, PAX6 [25, 31, 32]. We found that early organoids contained higher PAX6 transcripts compared to late organoids (Fig. 4C). Conversely, we found that late organoids express higher amounts of FAM107A, a known marker for oRGs [25, 33, 34] (Fig. 4C). Consistent with our immunofluorescence images, we did not observe a significant difference in RNA expression of MAP2 or SOX2, although SOX2 trended towards increased RNA expression in the early organoids (Fig. 4C). Orthogonally, we performed immunofluorescence for PAX6 and FAM107A protein. We observed that early organoids express PAX6 protein in some neural rosettes (Fig. 4D), with minimal FAM107A protein present. Late forebrain organoids, however, expressed FAM107A protein radially emerging outward from some PAX6-positive neural rosettes (Fig. 4D). Taken together, these data implicate the presence of vRGs in early forebrain organoids, with oRGs appearing more in late forebrain organoids.
Radial glia cells express high transcript levels of antiviral genes
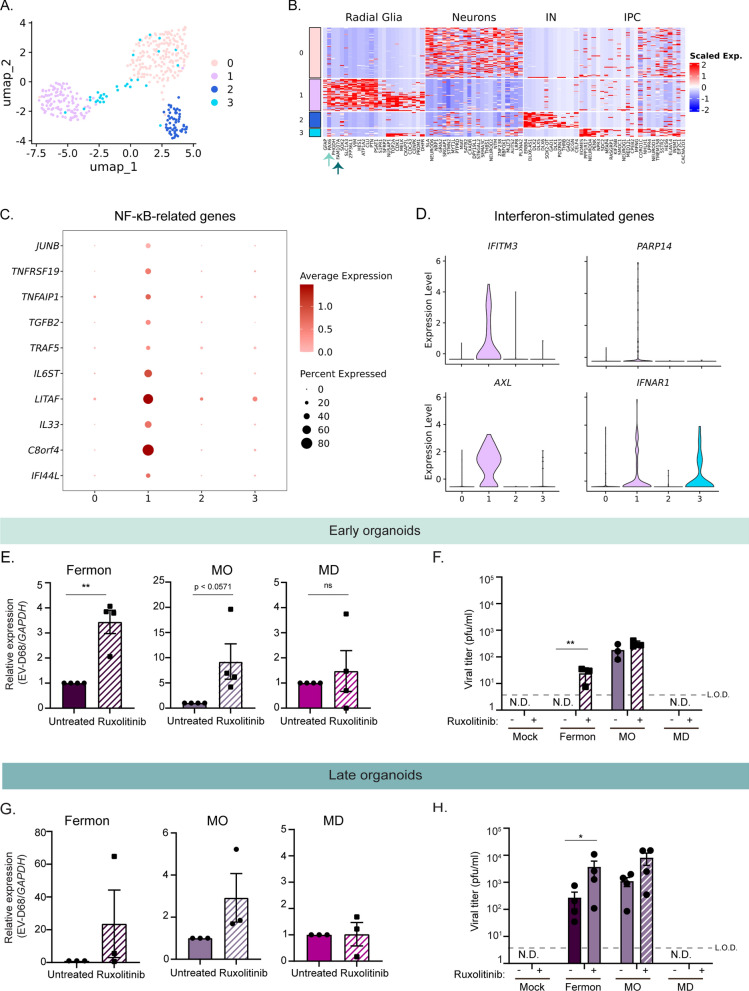
The immune capacity of vRGs and oRGs has not been explored. Given that only the MO EV-D68 strain produced infectious titer in early forebrain organoids while infectious virus production from Fermon and MO was observed in late organoids, we hypothesized that early forebrain organoids are protected from EV-D68 infection by elevated levels of antiviral genes. Further, this primed antiviral gene state may be driven by radial glia cells present in the early forebrain organoids. We sought to evaluate the potential for antiviral priming in radial glia using a previously published single cell RNA sequencing dataset. Pollen and colleagues isolated cells from the ventricular and subventricular zones of human fetal tissue [25]. We re-clustered these data and identified/found four distinct clusters (Fig. 5A). Using gene lists marking distinct neural subtypes [25], we defined the clusters as neurons (Cluster 0), radial glia (Cluster 1), interneurons (Cluster 2), and intermediate progenitor cells (Cluster 3) (Fig. 5B). Corroborating our analysis, the radial glia markers PAX6 and FAM107A were among the top expressed in the radial glia cluster (Fig. 5B, arrows). We next performed differential expression analysis to identify genes unique to the radial glial cluster compared to the other cell clusters. Interestingly, we found several immune genes that were upregulated in the radial glial cluster. Specifically, NF-κB-related genes were highly abundant, including IL6ST, TNFRSF19, LITAF (Fig. 5C). Further, we also identified interferon-stimulated genes predominantly present in the radial glia cluster, including IFITM3 and AXL (Fig. 5D). These data suggest that radial glia cells are primed to be in an antiviral state by basally expressing key antiviral immune genes.
Fig. 5.
Distinct antiviral signaling pathways are basally expressed in early forebrain organoids. A Uniform manifold approximation and projection (UMAP) of previously published single cell transcriptomics data representative of 393 cells from ventricular and sub-ventricular zone human fetal brain samples [25]. B Heatmap displaying distinct neural cell populations. Arrows highlight radial glia specific genes. C, D Dot plot displaying NF-kB genes (C) and violin plots displaying interferon stimulated genes (ISGs) (D) identified using differential gene expression analysis of cluster 1 (radial glia) compared to other clusters (cutoff = log2 fold-change = 1.9) (E). RT-qPCR analysis of lysates harvested at 7 dpi from DIV35 forebrain organoids untreated or treated with Ruxolitinib and simultaneously infected with Fermon, MO, or MD viruses. The data are normalized to untreated organoids and are presented as relative expression of EV-D68 to GAPDH with untreated set to 1. Data are presented as means ± SEM (n = four biological replicates). Statistical analysis performed with unpaired student’s t-test. F Plaque-forming assay of supernatants harvested at 7 dpi from forebrain organoids mock-infected or infected with either Fermon, MO, or MD at DIV35. Data are presented as means ± SEM (n = three biological replicates). Statistics performed with two-way ANOVA. N.D. = not detected (G). RT-qPCR analysis of lysates harvested at 7 dpi from DIV85 forebrain organoids untreated or treated with Ruxolitinib and simultaneously infected with Fermon, MO, or MD viruses. The data are normalized to untreated organoids and are presented as relative expression of EV-D68 to GAPDH with untreated set to 1. Data are presented as means ± SEM (n = three biological replicates). Statistical analysis performed with unpaired student’s t-test. (H). Plaque-forming assay of supernatants harvested at 7 dpi from forebrain organoids mock-infected or infected with either Fermon, MO, or MD at DIV85. Data are presented as means ± SEM (n = four biological replicates). Statistical analysis performed with two-way ANOVA. N.D. = not detected
Immune activation limits EV-D68 infectious virus production in early and late forebrain organoids
Based on our identification of genes from multiple antiviral immune pathways present in radial glia cells and the previously established importance of the immune transcription factor STAT3 in neural stem cell maintenance and outer radial glia identity [25], we hypothesized that immune activation within forebrain organoids may limit EV-D68 infection. To this end, we utilized the JAK1/JAK2 inhibitor Ruxolitinib to broadly suppress immune activation. We simultaneously mock-infected or infected early forebrain organoids with either Fermon, MO, or MD viruses and treated with Ruxolitinib, at a concentration that has previously been used in other brain-derived organoids [35]. We found that JAK1/JAK2 inhibition resulted in increased virus RNA copies of Fermon and MO, but not that of MD (Fig. 5E). We also found that infectious virus production increased with Ruxolitinib treatment, which is most evident during Fermon infection, in which infectious virus production is now detected and above the limit of detection (Fig. 5F). We performed the same infections and Ruxolitinib treatment in late forebrain organoids. Surprisingly, we found that viral copies in late organoids were not significantly increased during either Fermon, MO, or MD viruses (Fig. 5G). Instead, we observed an increase in infectious virus production in the late organoids (Fig. 5H). These data emphasize the importance of evading neural stem cell immune activation, which may in turn, facilitate pathogenesis.
Ventricular radial glia stem cells express antiviral proteins
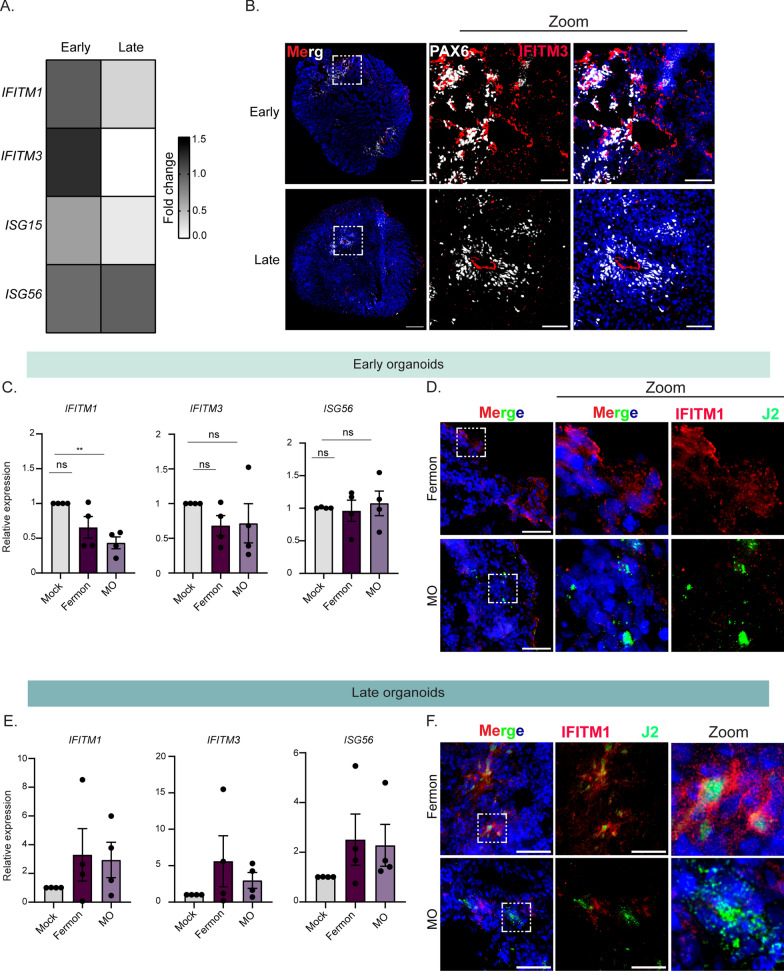
Neural stem cells play an important role as the initiators of other neural cell lineages. As humans age, neural stem cell pools become depleted as a result of the emergence of mature neurons or glial cells. As such, protection of this limited cellular supply from viral infection, cellular death, and inflammation is paramount. One mechanism by which stem cells defend against pathogen insult is by upregulating various interferon-stimulated genes. Wu and colleagues found that neural stem cells basally express higher levels of IFITM family member proteins. We thus hypothesized that there may be additional ISGs that are basally upregulated in our early forebrain organoids. We extracted RNA from mock-infected early or late forebrain organoids and performed RT-qPCR for known antiviral ISGs [36]. We found that in addition to IFITM1, several other ISGs are upregulated in early organoids compared to late organoids (Fig. 6A). Interestingly, RNA expression of ISG56 did not change in late organoids, suggesting that it may not mediate the differential virus infectivity between the early and late organoids (Fig. 6A). We next hypothesized that our early forebrain organoids would express IFITM3 protein in PAX6- positive regions compared to our late forebrain organoids. We found IFITM3 protein expression in early organoids by both PAX6 expressing and not-expressing cells, while only PAX6-positive cells exhibited IFITM3 expression in late organoids (Fig. 6B). In early organoids, we observed IFITM3 protein expression in PAX6-positive cells and also in PAX6-negative neural rosettes. In late organoids, IFITM3 protein expression was restricted to PAX6-positive cells (Fig. 6B). These data suggest that vRGs, which express PAX6 protein and are more abundant in early forebrain organoids, serve to defend against virus infection by upregulating IFITM3 expression. Further, a decrease in PAX6-expressing cells, which are immune active radial glia cells, results in an increase in virus infectivity.
Fig. 6.
Differential interferon-stimulated gene expression during Enterovirus-D68 infection of early and late forebrain organoids. A Heat map of fold change values from RT-qPCR of lysates from early and late forebrain organoids for several interferon-stimulated genes. B Representative immunofluorescence micrographs of early and late forebrain organoids. Organoids were stained for DAPI (nuclei, blue), PAX6 (grey, ventricular radial glia cells), and IFITM3 (red). Dashed boxes indicate the location of the zoom images. Scale bar (tiled image)—200 µm; Scale bar (zoom image)—25 µm. C RT-qPCR analysis of lysates harvested at 7 dpi from DIV35 forebrain organoids mock-infected or infected with Fermon, MO, or MD viruses. The data are normalized to mock-infected organoids and are presented as relative expression of the respective interferon-stimulated gene to GAPDH with mock-infected set to 1. Data are presented as means ± SEM (n = four biological replicates). D Representative immunofluorescence images of early organoids infected with Fermon or MO viruses at 7 dpi. Organoids were stained for DAPI (blue, nuclei), J2 (green, virus), and IFITM1 (red). Scale bar—50 µm. E RT-qPCR analysis of lysates harvested at 7 dpi from DIV85 forebrain organoids mock-infected or infected with Fermon, MO, or MD viruses. Data are normalized to mock-infected organoids and are presented as relative expression of the respective interferon-stimulated gene to GAPDH with mock-infected set to 1. Data are presented as means ± SEM (n = three biological replicates). F Representative immunofluorescence images of late organoids infected with Fermon or MO viruses at 7 dpi. Organoids were stained for DAPI (blue, nuclei), J2 (green, virus), and IFITM1 (red). Scale bar—50 µm
To determine whether virus infection within early forebrain organoids is dependent upon antagonizing this basally elevated interferon-stimulated gene signature, we measured RNA expression of several ISGs during infection with either MO or Fermon viruses. We found that MO reduced the induction and expression of IFITM1, compared to mock or Fermon-infected conditions (Fig. 6C). To identify IFITM1 antagonism on the protein level in single, infected cells, we performed immunofluorescence on MO or Fermon-infected early forebrain organoids staining for IFITM1 protein and an antibody against double-stranded RNA (J2). We found that in MO-infected cells, IFITM1 protein was not present, whereas in Fermon-infected cells, IFITM1 was activated, as indicated by increased IFITM1 protein expression (Fig. 6D). In late organoids, both Fermon and MO viruses induced expression of several ISGs, including IFITM1, IFITM3, and ISG56, with MO-infected organoids trending towards a reduction in IFITM3 expression (Fig. 6E). Since this measured bulk RNA transcripts, we wanted to investigate what was occurring in individually infected cells in late organoids. Interestingly, we found that IFITM1 protein expressed in Fermon-infected cells and neighboring uninfected cells, suggesting a broad immune activation during infection (Fig. 6F). IFITM1 protein expression was reduced in MO-infected cells, indicating immune antagonism during infection with MO virus in late organoids (Fig. 6F). Taken together, these data imply that at steady-state, neural stem cells express an antiviral program to protect the CNS. For EV-D68 to establish a productive CNS infection, it must first overcome this upregulated ISG barrier.
Discussion
In this study, we sought to identify cellular features mediating differential infectivity during EV-D68 infection within the CNS. We found that the canonical in vitro system used to study EV-D68 replication, SH-SY5Y cells, was permissive to both non-neuropathogenic and neuropathogenic EV-D68 strains, underscoring an alternative approach towards understanding EV-D68 infection within the CNS. Further, using forebrain organoids at two developmental time points, we identified that organoids at a later developmental stage were permissive to both neuropathogenic and non-neuropathogenic EV-D68 strains; however, only neuropathogenic EV-D68 strains were able to productively infect forebrain organoids at an early developmental time point. We identified that specific neural stem cells differentially expressed key innate immune signaling pathways that may dictate neuron-specific EV-D68 infection. Notably, the type of neural stem cell is important, as ventricular radial glia cells expressed antiviral immune proteins compared to outer radial glia cells, and EV-D68 regulation of this antiviral program may be critical in establishing neurological disease.
Until recently, the innate immune signaling capacity of neurons and neural stem cells has been woefully underappreciated [37]. Neurons have largely been considered immune signaling incompetent, with a role ascribed to simply responding to cues from microglia or astrocytes [38, 39]. Recent literature has suggested that neural stem cells can mount protective type I IFN signaling responses [37, 40, 41]. Interestingly, several immune genes have been implicated in maintenance of neural stem cells identity as well as brain development, including LIF and STAT3. However, a role in immune function and antiviral signaling capacity within neural stem cells has largely been overlooked. Our work expounds upon the existing literature by providing insight into immune pathways that are differentially expressed in neural stem cells compared to maturing neurons. Our study further illuminates the under-appreciated immune signaling capacity of neurons and how it interplays with susceptibility to virus infection.
The full cellular tropism of EV-D68 is largely unknown. Mouse model studies have demonstrated a preference for EV-D68 infection within spinal cord motor neurons. In vitro studies have also identified EV-D68 infection with cortical neurons and astrocytes, suggesting that EV-D68 could infect more than just spinal cord motor neurons [4, 6] To our knowledge, there is one documented fatal case of EV-D68-associated AFM in the United States, a 5 year old boy who was later found to have detectable EV-D68 viral RNA in the CSF [42]. Later, in a New England Journal of Medicine report, Vogt and colleagues obtained an autopsy sample from that patient and identified EV-D68 viral RNA and protein within the neurons of the anterior horn of the spinal cord [43]. Additionally, a recent study using human iPSC-derived spinal cord organoids found that only contemporary neuropathogenic EV-D68 strains, not Fermon, can infect these organoids at 14 days post-differentiation [44]. An outstanding question is whether Fermon is capable of infecting spinal cord organoids during later stages of development.
Viral determinants and host immune responses, likely together, contribute to the differences in neuropathogenesis of EV-D68. We show that viral factors promote infection within the CNS, as different non-AFM-associated EV-D68 strains (Fermon vs. MD) have vastly different CNS infection capacity. One potential reason why EV-D68 strain MD was not infectious in the forebrain organoids may be the temperature of the CNS (37 °C vs 33 °C). Though this assumption requires further investigation, it underscores the importance of evolutionary pressure in driving adaptation to the host and cellular environment. Given how relatively conserved the different EV-D68 strains are, it is interesting how much variation is observed in terms of replication kinetics and emergence of neuropathogenicity. Perhaps there are viral amino acid residues that mediate this variation or residues that persist despite the evolutionary pressure. Indeed, our amino acid alignment of the various EV-D68 strains (Fig. 1A) reveals that the viral 3B protein sequence is fully conserved amongst many contemporary and historic EV-D68 strains. Additionally, mutations in the viral VP1 protein may also contribute to how EV-D68 establishes neuropathogenesis [45]. Future studies should examine the contributions of mutations in the viral life cycle, entry into the CNS, and immune evasion that allow viral persistence.
We hypothesized that a basally elevated immune gene signature in the early forebrain organoids largely protects against EV-D68 infection compared to the late forebrain organoids. We acknowledge that elevated ISGs may be one of several likely factors contributing to increased viral permissiveness in the late forebrain organoids. Perhaps there are additional host proteins that may function in an antiviral capacity to protein the CNS from viruses. Recently, Dai and colleagues identified the protein TMEFF1 as a neuron-specific restriction factor against herpes simplex virus [46]. Thus, it is likely that additional host cellular proteins may function as restriction factors against EV-D68 infection and may be differentially expressed between early and late forebrain organoids. Future studies should be aimed at identifying these cellular antiviral proteins during EV-D68 infection within the CNS.
Though the host has several mechanisms by which it defends itself against virus infection, viruses have evolved ways to counteract such defenses. Our data suggest that limiting basal levels of immune genes may be one way viruses establish infection within nervous system tissue. Indeed, several viruses have been shown to preferentially infect neural stem cells, with Zika virus being one of the more well-characterized neurotropic viruses in recent years. Additionally, another study found that Japanese Encephalitis virus could replicate in less mature forebrain organoids but not in more mature forebrain organoids, even though there was a decrease in IFN induction in the later organoids [47]. This could suggest that the virus may encode viral proteins capable of immune antagonism. The same may hold true for EV-D68 as we know that EV-D68 can block several immune factors (IRF7, TRIF, and STAT1) in non-CNS cells [48–50].
Overall, our work contributes to our understanding of differential immune regulation within neural stem cells, which can explain strain differences of neuropathogenic viruses.
Methods
Cells and viruses
RD cells were grown in Dulbecco’s modification of eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; HyClone), 1% penicillin–streptomycin (Gibco), and 1% Gluta-Max (Gibco). Undifferentiated SH-SY5Y cells were maintained in DMEM/F-12/Glutamax medium (Gibco) supplemented with 10% fetal bovine serum (FBS; Hyclone). Enterovirus-D68 strains (Fermon, US/MO/14-18947, and MD/2009-23229) were purchased from BEI resources (catalog numbers NR-51430, NR-49129 and NR-51994, respectively). EV-D68 strains were propagated at the following temperatures: Fermon at 37 °C, US/MO/14-18947 at 33 °C, and MD/2009-23229 at 33 °C. Human-induced pluripotent stem cell lines C1-2 and WTC11 (Coriell and [17]) were maintained in mTSeR media prior to forebrain organoid generation protocol. All cells were tested and found to be Mycoplasma-free at the Cell Center Services Facility (University of Pennsylvania).
SH-SY5Y cell differentiation
Differentiation of SH-SY5Y cells was performed as previously published for 18 days [10]. Briefly, undifferentiated SH-SY5Y cells were plated in six-well plates, followed by media changes with gradual depletion of FBS and addition of retinoic acid and a two-step cell splitting process. Initially, cells were maintained in differentiation media containing 2.5% heat-inactivated FBS and 10 µM retinoic acid. On day 7, cells were split 1:1 onto six-well plates, and the following day, differentiation media containing 1% heat-inactivated FBS and 10 µM retinoic acid was added. On day 10, cells were split 1:1 onto poly-D-lysine-coated six-well plates. The following day, differentiation media containing 1X B27, 20 mM KCl, 50 ng/mL brain-derived neurotrophic factor, 2 mM dibutyryl cyclic AMP, and 10 µM retinoic acid was added. Media was changed every three days until day 18, at which point the cells were ready for experimental analysis.
Generation of forebrain organoids
Forebrain organoids were generated following a previously established protocol [17]. Induced pluripotent stem cells were cultured with mTeSR Plus complete medium (STEMCELL Technologies) until ~ 90% confluent. On day 0 of cerebral organoid generation, iPSCs were dissociated into a single cell suspension using ReLeSR (STEMCELL Technologies) and resuspended in mTeSR supplemented with Rho-associated protein kinase inhibitor Y27632 (ROCKi) at 10 µM. Non-cell culture treated 96-well U bottom plates were seeded with 200 µL of cell suspension at a concentration of 2.5 × 105 cells/mL (50,000 cells/well). Plates were incubated at 37 °C and 5% CO2 for 2 days to form embryoid bodies (EB). On day 3 of organoid generation, EBs were resuspended and transferred into 6-well plates containing H1 medium (Basal Medium with F-12 nutrient (DMEM/F12), 20% Knockout Serum Replacement (KOSR), 1% glutamine supplement alternative (GlutaMAX), 1% MEM Non-essential amino acid solution (NEAA), 0.182% 2-beta-mercaptoethanol (BME), 1% penicillin/strep (Pen/Strep), 1 µM LDN193189, 5 µM SB431542, and 0.2% heparin solution) supplemented with 10 µM ROCKi. EBs were incubated at 37 °C and 5% CO2 on a shaker at 120 revolutions per minute (rpm). On days 3–6, media was half-changed with H1 medium. On day 7, healthy EBs with round, smooth and bright edges were embedded in a Matrigel (Corning) and F2 medium solution (DMEM/F12, 1% N2 supplement, 1% GlutaMAX, 1% NEAA, 0.182% BME, 1% Pen/Strep, 1 µM SB431542 and 1 µM CHIR99021) and plated on ultra-low attachment 6-well plates. The EB/matrigel solution was incubated for 60 min at 37 °C to solidify. F2 medium was added to the well and half-media changes were completed every other day until day 13 of organoid generation. On day 14, organoids (formerly EBs) were broken out of matrigel and resuspended with H3 medium (47% DMEM/F12, 47% Neurobasal media, 1% N2 supplement, 2% B27 supplement, 1% GlutaMAX, 1% NEAA, 0.182% BME, 1% Pen/Strep and 2.5 µg/mL insulin) in 6-well plates. Plates were incubated at 37 °C and 5% CO2 on a shaker at 120 rpm. Media changes were performed daily with H3 media until organoid harvest. For organoids that were maintained past DIV 70, media changes were performed with F4 medium (Neurobasal media, 2% B27 supplement, 1% GlutaMAX, 1% NEAA, 0.182% BME, 1% Pen/Strep, 0.05 mM cAMP, 0.02 mM ascorbic acid, 20 ng/mL BDNF and GDNF) and were sliced using a Leica VT 1200S vibratome between days 45–60 as previously described [15]. Virus infections were performed in forebrain organoids (2 organoids per condition per biological replicate) at 5 × 103 plaque forming units at 37 °C.
Plaque assay
Viral titer was determined using plaque assays. RD cells were plated on 6-well plates at a concentration that would allow for ~ 80% confluency at the time of assay infection. Supernatants from infected cells or organoids, collected at the conclusion of respective experiments, were serially diluted and plated onto the RD cells. After virus adsorption for 1 h at 37 °C, cells were overlaid with 1% SeaPlaque, 1% SeaKem in MEM and 5% FBS. Plates were incubated at 37 °C for 4 days, fixed with 10% neutral buffered formalin, agarose discs were removed, and plaques were stained with 0.5% crystal violet.
Immunofluorescence
Undifferentiated or differentiated SH-SY5Y cells were plated onto 6-well plates or 12-well plates and infected at varying multiplicities of infection the next day. Twenty-four hours post-infection, cells were fixed in 4% paraformaldehyde, permeabilized with 0.1% TritonX-100, and stained. For imaging of the forebrain organoids, organoids were fixed in 4% paraformaldehyde for 30 min with head-to-tail rotation, washed with 1X PBS, and incubated overnight in a 30% sucrose solution. Following sucrose incubation, the organoids were washed with 1X PBS, and mounted into cryomolds using TissueTek (Sakura Finetek). Organoids were sectioned into 10 µm sections using a Leica CM1950 or a Leica CM3050S cryostat, permeabilized with 0.4% Triton X-100 and PBS, and stained. Primary antibodies used for staining are as follows: Sox2 (1:300, Cell Signaling Technologies), GFAP (1:100, Dako), phosphorylated neurofilament-H (1:100, Biolegend), IFITM1 (1:100, Genetex), IFITM3 (1:100, Proteintech), ICAM-5 (1:00, Abcam), MAP2 (1:3000, Abcam), EV-D68 VP1 (1:100, Genetex), Alexa-fluor 488-conjugated PAX6 (1:200, BD Biosciences), and FAM107A (1:300, Sigma). Images were acquired using a Nikon Ti2E scope. Image processing was conducted using ImageJ software.
Neuraminidase treatment
At either DIV35 or DIV85, organoids were incubated with neuraminidase (Sigma) at 6 units/mL for 1 h. After 1 h, organoids were washed with 1X PBS, and infected with EV-D68 strains or mock-infected. At 24 hpi, organoids were washed with 1X PBS and harvested for downstream analysis.
Click-iT analysis
At DIV35 or 85, organoids were incubated with 25 µM Click-iT® ManNAz (tetraacetylated N-azidoacetyl-d-mannosamine, ThermoFisher Scientific) for 24 h. Following incubation, organoids were washed with 1× PBS, fixed with 4% paraformaldehyde for 30 min—1 h, and incubated overnight in a 30% sucrose solution. Fixed organoids were placed in TissueTek cryomold and sectioned onto glass slides using a cryostat. The Click-iT analysis was performed on the glass slides according to the manufacturer’s protocol. Organoid sections were incubated in a solution of 1× Click-iT cell reaction buffer, CuSO4, Click-iT cell buffer additive, and 1 µM of 555-alkyne secondary antibody (ThermoFisher Scientific) for 30 min. Sections were washed with 1% BSA in PBS, incubated with 1X DAPI in PBS for 10 min, washed again with 1% BSA in PBS, and mounted with cover slips using Prolong Diamond Antifade Mountant (ThermoFisher Scientific). Image analysis on ImageJ was done by setting an organoid sample incubated without a label or copper (negative control) as the background reference and applying the LUTs settings to the organoids with the label. Quantification of total sialic acid was performed using the ROI manager analysis tool on ImageJ by outlining the organoid on the DAPI channel, demarcating that outline as the region of interest (ROI), applying that ROI to the cy5 channel (sialic acid, alkyne-555), and measuring the mean fluorescence intensity (MFI). The MFI for both the DAPI and cy5 channels were obtained, and the relative MFI was calculated as a ratio of MFI-cy5/MFI-DAPI using three sections from three independent biological organoid replicates at either the early or late time point.
RT-qPCR
RNA was extracted from cells or organoids using a Zymo Research Direct-zol RNA MiniPrep kit, and cDNA synthesis was performed on extracted RNA using iScript (Bio-Rad). The resulting cDNA was diluted 1:3 in double-distilled water. RT-qPCR analysis was performed using either the Power SYBR green PCR master mix (ThermoFisher Scientific) or Taqman Fast Advanced master mix (ThermoFisher Scientific) on the QuantStudio 3 RT-PCR system. Primer sequences used are listed in Table 1.
Table 1.
Primer sequences used in this study
| Gene name | Forward primer | Reverse primer | Probe |
|---|---|---|---|
| EV-D68 VP1 | TAACCCGTGTGTAGCTTGG | ATTAGCCGCATTCAGGGGC | TTGGCGGCCTACTCATGGTGAAA |
| IFITM1 | ACTCCGTGAAGTCTAGGGACA | TGTCACAGAGCCGAATACCAG | |
| IFITM3 | CTGGGCTTCATAGCATTGGCCT | AGATGTTCAGGCACTTGGCGGT | |
| ISG56 | TACAGCAACCATGAGTACAA | TCAGGTGTTTCACATAGGC | |
| ISG15 | CTCTGAGCATCCTGGTGAGGAA | AAGGTCAGCCAGAACAGGTCGT | |
| GAPDH | GTCTCCTCTGACTTCAACAGCG | ACCACCCTGTTGCTGTAGCCAA | |
| HPRT1 | CATTATGCTGAGGATTTGGAAAGG | CATTATGCTGAGGATTTGGAAAGG | |
| HOPX | TCAACAAGGTCGACAAGCAC | TCTGTGACGGATCTGCACTC | |
| PAX6 | TTGCCCGAGAAA GACTAGCA | TCTCCATTTGGC CCTTCGATTA | |
| ICAM-5 | TTGGCGCGGCAGCTGGT | GCATCAGCTCTACGCGATCT | |
| SOX2 | GCCGAGTGGAAACTTTTGTCG | GCCGAGTGGAAACTTTTGTCG | |
| FAM107A | GCAGCGTGTCCTAGAGCAC | CCGCAGGTTTTCCCTGACT | |
| MAP2 | CTCAGCACCGCTAACAGAGG | CATTGGCGCTTCGGACAAG |
Single cell RNA sequencing analysis
Single cell transcriptomics data representative of cells isolated from the ventricular and sub-ventricular zone human fetal brain samples were obtained from a previously published dataset [25]. Specifically, we analyzed normalized gene expression values expressed as counts per million reads for all genes across 393 cells using R Studio Server. Using the Seurat package, we applied a principal component cut-off of 4 and a clustering resolution of 0.5 to identify 4 clusters. Using the previously defined cell identity markers, we generated a heatmap displaying each neural subtype. We next used Seurat’s FindAllMarkers to conduct differential expression analysis on radial glia and displayed NF-κB-related and interferon stimulated genes using Seurat.
Statistical analysis
All data were analyzed using the appropriate statistical test using GraphPad Prism software. One-way analysis of variance (ANOVA), two-way ANOVA, or unpaired student’s t-test were implemented for statistical analysis of the data using GraphPad Prism Software as indicated. Graphed values are presented as mean ± SEM (n = 3 or greater); *p ≤ 0.05, **p ≤ 0.01, and ***p ≤ 0.001.
Supplementary Information
Supplemental Figure 1. Forebrain organoids derived from a second human induced pluripotent stem cell line also support pathogenic Enterovirus-D68 infection. (A). RT-qPCR analysis of lysates harvested at 7 dpi from DIV35 forebrain organoids mock-infected or infected with Fermon, MO, or MD viruses. Data are normalized to mock-infected organoids and are presented as relative expression of EV-D68 to GAPDH with mock-infected set to 1. Data are presented as means ± SEM (n = three biological replicates). (B). Plaque-forming assay of supernatants harvested at 7 dpi from forebrain organoids mock-infected or infected with either Fermon, MO, or MD at DIV35 (C). RT-qPCR analysis of lysates harvested at 7 dpi from DIV85 forebrain organoids Fermon, MO, or MD viruses. The data are normalized to mock-infected organoids and are presented as relative expression of EV-D68 to GAPDH with mock-infected set to 1. Data are presented as means ± SEM (n = three biological replicates). N.D. = not detected (D). Plaque-forming assay of supernatants harvested at 7 dpi from forebrain organoids mock-infected or infected with either Fermon, MO, or MD at DIV85. Statistical analysis performed with one-way ANOVA. N.D. = not detected.
Supplemental Figure 2. ICAM-5 is minimally expressed in forebrain organoids. Representative immunofluorescence micrographs of early (A). or late (B). forebrain organoids. Organoids were stained for DAPI (nuclei, blue), ICAM-5 (red), and MAP2 (magenta). Scale bar - 100 µm. (C). RT-qPCR analysis of RNA harvested from early or late forebrain organoids. Data are normalized to the early organoid sample and are presented as relative expression of ICAM-5 to HPRT1 with early set to 1. Data are presented as means ± SEM (n =five biological replicates). Statistical analysis performed with unpaired student’s t-test.
Supplemental Figure 3. Late forebrain organoids contain few astrocytes. Representative immunofluorescence images of early or late forebrain organoids. Organoids were stained for DAPI (nuclei, blue) and GFAP (red, astrocytes). Dashed boxes indicate the location of the zoom images. Scale bar (tiled image) - 200 µm; Scale bar (zoom image)—25 µm.
Acknowledgements
We would like to thank members of the Jurado laboratory and Ming laboratory for helpful discussion and manuscript review. We specifically would like to thank Ronni Kurzion, Angela Bongiovanni, and Emma LaNoce for technical support and guidance. We also acknowledge Dr. Scott Hensley and the Hensley laboratory for equipment usage.
Abbreviations
- ANOVA
Analysis of variance
- DAPI
4ʹ,6-Diamidino-2-phenylindole
- DIV
Days in vitro
- DMEM
Dulbecco’s modified eagle’s medium
- Dpi
Days post infection
- EV-D68
Enterovirus-D68
- FAM107A
Family with sequence similarity 107 member A
- GAPDH
Glyceraldehyde-3 phosphate dehydrogenase
- GFAP
Glial fibrillary acidic protein
- Hpi
Hours post infection
- HPRT1
Hypoxanthine phosphoribosyltransferase 1
- ICAM-5
Intracellular adhesion molecule-5
- IFITM
Interferon induced transmembrane protein
- IFN
Interferon
- IL
Interleukin
- iPSC
Induced pluripotent stem cell
- ISG
Interferon-stimulated gene
- LITAF
Lipopolysaccharide induced tumor necrosis factor factor
- MAP2
Microtubule associated protein 2
- NF-κB
Nuclear factor kappa-light-chain enhancer of activated B cells
- PAX6
Paired box 6
- RD
Rhabdomyosarcoma
- rt-qPCR
Quantitative reverse transcription polymerase chain reaction
Author contributions
CV: Data curation, investigation, methodology, formal analysis, study conceptualization, experimental work, manuscript writing; SGN: Experimental work, formal analysis, manuscript review; CDB: Experimental work, manuscript review; SS: Experimental work; G-LM: Methodology, study design, data interpretation, manuscript review; KAJ: Formal analysis, funding acquisition, supervision, study conceptualization, manuscript writing. Lead contact: Further information and requests for reagents should be directed to and will be fulfilled by the lead contact, Dr. Kellie Jurado.
Funding
This work was supported by the University of Pennsylvania Office of the President (KAJ), the University of Pennsylvania Office of the Provost (CV and SGN), and the Burroughs Wellcome Fund (CV).
Availability of data and materials
All raw data and code will be uploaded to a public repository upon publication, and is available to reviewers by contacting lead author, kellie.jurado@pennmedicine.upenn.edu.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schieble JH, Fox VL, Lennette EH. A probable new human picornavirus associated with respiratory diseases. Am J Epidemiol. 1967;85(2):297–310. [DOI] [PubMed] [Google Scholar]
- 2.Russo MV, McGavern DB. Immune surveillance of the CNS following Infection and Injury. Trends Immunol. 2015;36(10):637–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodcroft EB, Dyrdak R, Andrés C, Egli A, Reist J, García Martínez de Artola D, et al. Evolution, geographic spreading, and demographic distribution of Enterovirus D68. PLoS Pathog. 2022;18(5):e1010515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown DM, Hixon AM, Oldfield LM, Zhang Y, Novotny M, Wang W, et al. Contemporary circulating Enterovirus D68 strains have acquired the capacity for viral entry and replication in human neuronal cells. MBio. 2018;9(5):e01954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenfeld AB, Warren AL, Racaniello VR. Neurotropism of Enterovirus D68 isolates is independent of sialic acid and is not a recently acquired phenotype. MBio. 2019;10(5):e02370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poelaert KCK, van Kleef R, Liu M, van Vliet A, Lyoo H, Gerber LS, et al. Enterovirus D-68 infection of primary rat cortical neurons: entry, replication, and functional consequences. MBio. 2023;14(2):e0024523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Telikani Z, Monson EA, Hofer MJ, Helbig KJ. Antiviral response within different cell types of the CNS. Front Immunol. 2022;13:1044721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nath A, Johnson TP. Mechanisms of viral persistence in the brain and therapeutic approaches. FEBS J. 2022;289(8):2145–61. [DOI] [PubMed] [Google Scholar]
- 9.Vazquez C, Jurado KA. Neurotropic RNA virus modulation of immune responses within the central nervous system. Int J Mol Sci. 2022;23(7):4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shipley MM, Mangold CA, Szpara ML. Differentiation of the SH-SY5Y human neuroblastoma cell line. J Vis Exp. 2016;108:53193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freeman MC, Wells AI, Ciomperlik-Patton J, Myerburg MM, Yang L, Konopka-Anstadt J, et al. Respiratory and intestinal epithelial cells exhibit differential susceptibility and innate immune responses to contemporary EV-D68 isolates. Elife. 2021;10:e066687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith BD, Pekosz A. Contemporary Enterovirus D68 strains show enhanced replication and translation at 37 °C. bioRxiv. 2020;55:1. [Google Scholar]
- 13.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165(5):1238–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan W, Christian KM, Song H, Ming GL. Applications of brain organoids for infectious diseases. J Mol Biol. 2022;434(3): 167243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian X, Su Y, Adam CD, Deutschmann AU, Pather SR, Goldberg EM, et al. Sliced human cortical organoids for modeling distinct cortical layer formation. Cell Stem Cell. 2020;26(5):766-81.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoon KJ, Nguyen HN, Ursini G, Zhang F, Kim NS, Wen Z, et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell. 2014;15(1):79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob F, Pather SR, Huang WK, Zhang F, Wong SZH, Zhou H, et al. Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-CoV-2 neurotropism predominates in choroid plexus epithelium. Cell Stem Cell. 2020;27(6):937-50.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Sheng J, Baggen J, Meng G, Xiao C, Thibaut HJ, et al. Sialic acid-dependent cell entry of human enterovirus D68. Nat Commun. 2015;6:8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trimarco JD, Nelson SL, Chaparian RR, Wells AI, Murray NB, Azadi P, et al. Cellular glycan modification by B3GAT1 broadly restricts influenza virus infection. Nat Commun. 2022;13(1):6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei W, Guo H, Chang J, Yu Y, Liu G, Zhang N, et al. ICAM-5/telencephalin is a functional entry receptor for Enterovirus D68. Cell Host Microbe. 2016;20(5):631–41. [DOI] [PubMed] [Google Scholar]
- 21.Wang M, Zhang L, Novak SW, Yu J, Gallina IS, Xu LL, et al. Morphological diversification and functional maturation of human astrocytes in glia-enriched cortical organoid transplanted in mouse brain. Nat Biotechnol. 2024. [DOI] [PMC free article] [PubMed]
- 22.Petersilie L, Heiduschka S, Nelson JSE, Neu LA, Le S, Anand R, et al. Cortical brain organoid slices (cBOS) for the study of human neural cells in minimal networks. iScience. 2024;27(4):109415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merkle FT, Tramontin AD, García-Verdugo JM, Alvarez-Buylla A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc Natl Acad Sci USA. 2004;101(50):17528–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eze UC, Bhaduri A, Haeussler M, Nowakowski TJ, Kriegstein AR. Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat Neurosci. 2021;24(4):584–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163(1):55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukaszewicz A, Savatier P, Cortay V, Giroud P, Huissoud C, Berland M, et al. G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron. 2005;47(3):353–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaMonica BE, Lui JH, Hansen DV, Kriegstein AR. Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nat Commun. 2013;4:1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fietz SA, Kelava I, Vogt J, Wilsch-Bräuninger M, Stenzel D, Fish JL, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13(6):690–9. [DOI] [PubMed] [Google Scholar]
- 29.Lewitus E, Kelava I, Huttner WB. Conical expansion of the outer subventricular zone and the role of neocortical folding in evolution and development. Front Hum Neurosci. 2013;7:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh RM, Luongo R, Giacomelli E, Ciceri G, Rittenhouse C, Verrillo A, et al. Generation of human cerebral organoids with a structured outer subventricular zone. Cell Rep. 2024;43(4): 114031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25(1):247–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thakurela S, Tiwari N, Schick S, Garding A, Ivanek R, Berninger B, et al. Mapping gene regulatory circuitry of Pax6 during neurogenesis. Cell Discov. 2016;2:15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehman NL, Spassky N, Sak M, Webb A, Zumbar CT, Usubalieva A, et al. Astroblastomas exhibit radial glia stem cell lineages and differential expression of imprinted and X-inactivation escape genes. Nat Commun. 2022;13(1):2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomsen ER, Mich JK, Yao Z, Hodge RD, Doyle AM, Jang S, et al. Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat Methods. 2016;13(1):87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winkler CW, Woods TA, Groveman BR, Carmody AB, Speranza EE, Martens CA, et al. Neuronal maturation reduces the type I IFN response to orthobunyavirus infection and leads to increased apoptosis of human neurons. J Neuroinflammation. 2019;16(1):229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Xu L, Zhang Z, Su X, Wang Z, Wang T. Enterovirus D68 infection upregulates SOCS3 expression to inhibit JAK-STAT3 signaling and antagonize the innate interferon response of the host. Virol Sin. 2023;38(5):755–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Negatu SG, Vazquez C, Bannerman C, Amses KR, Ming G-l, Jurado KA. Neuronal interactions in forebrain organoids lead to protective antiviral responses. bioRxiv. 2024;16:229. [Google Scholar]
- 38.Colonna M, Butovsky O. Microglia function in the central nervous system during health and neurodegeneration. Annu Rev Immunol. 2017;35:441–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasband MN. Glial contributions to neural function and disease. Mol Cell Proteomics. 2016;15(2):355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin JY, Kuo RL, Huang HI. Activation of type I interferon antiviral response in human neural stem cells. Stem Cell Res Ther. 2019;10(1):387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X, Dao Thi VL, Huang Y, Billerbeck E, Saha D, Hoffmann HH, et al. Intrinsic immunity shapes viral resistance of stem cells. Cell. 2018;172(3):423-38.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kreuter JD, Barnes A, McCarthy JE, Schwartzman JD, Oberste MS, Rhodes CH, et al. A fatal central nervous system enterovirus 68 infection. Arch Pathol Lab Med. 2011;135(6):793–6. [DOI] [PubMed] [Google Scholar]
- 43.Vogt MR, Wright PF, Hickey WF, De Buysscher T, Boyd KL, Crowe JE Jr. Enterovirus D68 in the anterior horn cells of a child with acute flaccid myelitis. N Engl J Med. 2022;386(21):2059–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aguglia G, Coyne CB, Dermody TS, Williams JV, Freeman MC. Contemporary enterovirus-D68 isolates infect human spinal cord organoids. MBio. 2023;14(4):e0105823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leser JS, Frost JL, Wilson CJ, Rudy MJ, Clarke P, Tyler KL. VP1 is the primary determinant of neuropathogenesis in a mouse model of enterovirus D68 acute flaccid myelitis. J Virol. 2024;98:e0039724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai Y, Idorn M, Serrero MC, Pan X, Thomsen EA, Narita R, et al. TMEFF1 is a neuron-specific restriction factor for herpes simplex virus. Nature. 2024;632(8024):383–9. [DOI] [PubMed] [Google Scholar]
- 47.Zhang B, He Y, Xu Y, Mo F, Mi T, Shen QS, et al. Differential antiviral immunity to Japanese encephalitis virus in developing cortical organoids. Cell Death Dis. 2018;9(7):719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiang Z, Li L, Lei X, Zhou H, Zhou Z, He B, et al. Enterovirus 68 3C protease cleaves TRIF to attenuate antiviral responses mediated by Toll-like receptor 3. J Virol. 2014;88(12):6650–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang J, Huang M, Li J, Zhang K, Zhu C, Liu S, et al. Enterovirus D68 VP3 targets the interferon regulatory factor 7 to inhibit type I interferon response. Microbiol Spectr. 2023;11(3): e0413822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Guo H, Yang J, Liu X, Li H, Yang W, et al. Enterovirus D68 3C protease antagonizes type I interferon signaling by cleaving signal transducer and activator of transcription 1. J Virol. 2024;98(2): e0199423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Forebrain organoids derived from a second human induced pluripotent stem cell line also support pathogenic Enterovirus-D68 infection. (A). RT-qPCR analysis of lysates harvested at 7 dpi from DIV35 forebrain organoids mock-infected or infected with Fermon, MO, or MD viruses. Data are normalized to mock-infected organoids and are presented as relative expression of EV-D68 to GAPDH with mock-infected set to 1. Data are presented as means ± SEM (n = three biological replicates). (B). Plaque-forming assay of supernatants harvested at 7 dpi from forebrain organoids mock-infected or infected with either Fermon, MO, or MD at DIV35 (C). RT-qPCR analysis of lysates harvested at 7 dpi from DIV85 forebrain organoids Fermon, MO, or MD viruses. The data are normalized to mock-infected organoids and are presented as relative expression of EV-D68 to GAPDH with mock-infected set to 1. Data are presented as means ± SEM (n = three biological replicates). N.D. = not detected (D). Plaque-forming assay of supernatants harvested at 7 dpi from forebrain organoids mock-infected or infected with either Fermon, MO, or MD at DIV85. Statistical analysis performed with one-way ANOVA. N.D. = not detected.
Supplemental Figure 2. ICAM-5 is minimally expressed in forebrain organoids. Representative immunofluorescence micrographs of early (A). or late (B). forebrain organoids. Organoids were stained for DAPI (nuclei, blue), ICAM-5 (red), and MAP2 (magenta). Scale bar - 100 µm. (C). RT-qPCR analysis of RNA harvested from early or late forebrain organoids. Data are normalized to the early organoid sample and are presented as relative expression of ICAM-5 to HPRT1 with early set to 1. Data are presented as means ± SEM (n =five biological replicates). Statistical analysis performed with unpaired student’s t-test.
Supplemental Figure 3. Late forebrain organoids contain few astrocytes. Representative immunofluorescence images of early or late forebrain organoids. Organoids were stained for DAPI (nuclei, blue) and GFAP (red, astrocytes). Dashed boxes indicate the location of the zoom images. Scale bar (tiled image) - 200 µm; Scale bar (zoom image)—25 µm.
Data Availability Statement
All raw data and code will be uploaded to a public repository upon publication, and is available to reviewers by contacting lead author, kellie.jurado@pennmedicine.upenn.edu.