Abstract
The human ATP-binding cassette (ABC) transporter ABCA1 plays a critical role in lipid homeostasis as it extracts sterols and phospholipids from the plasma membrane for excretion to the extracellular apolipoprotein A-I and subsequent formation of high-density lipoprotein (HDL) particles. Deleterious mutations of ABCA1 lead to sterol accumulation and are associated with atherosclerosis, poor cardiovascular outcomes, cancer, and Alzheimer’s disease. The mechanism by which ABCA1 drives lipid movement is poorly understood, and a unified platform to produce active ABCA1 protein for both functional and structural studies has been missing. In this work, we established a stable expression system for both a human cell-based sterol export assay and protein purification for in vitro biochemical and structural studies. ABCA1 produced in this system was active in sterol export and displayed enhanced ATPase activity after reconstitution into a lipid bilayer. Our single-particle cryo-EM study of ABCA1 in nanodiscs showed protein induced membrane curvature, revealed multiple distinct conformations, and generated a structure of nanodisc-embedded ABCA1 at 4.0-Å resolution representing a previously unknown conformation. Comparison of different ABCA1 structures and molecular dynamics simulations demonstrate both concerted domain movements and conformational variations within each domain. Taken together, our platform for producing and characterizing ABCA1 in a lipid membrane enabled us to gain important mechanistic and structural insights and paves the way for investigating modulators that target the functions of ABCA1.
Keywords: ABC transporter, Cholesterol, Cryo-electron microscopy, Membrane Transport, Membrane transporter reconstitution
The eukaryotic Plasma Membrane (PM) houses thousands of Membrane Proteins (MPs), which perform countless necessary functions for cellular survival and proliferation 1. Importantly, these MPs reside within the context and environment of the PM and require the surrounding lipid bilayer to maintain their native, functionally active conformations. ATP-Binding Cassette (ABC) transporters are canonical MPs that mediate the passage of various substances across the lipid bilayer 2,3. They do so by coupling the movement of substrates through the transmembrane domain (TMD; i.e., lipids, drugs, sterols) to the binding and hydrolysis of adenosine triphosphate (ATP) 4–6. ATP binds at the interface of two cytosolic nucleotide binding domains (NBDs); closure of NBDs propagates structural alterations to the TMDs which change conformation to mediate the passage of the substrate across the bilayer. The allosteric linkage between the TMDs and NBDs requires interfacial helices and likely depends on native TMD-lipid interactions 7.
One particularly challenging class of substrates for passage across the PM are the components which comprise the lipid bilayer itself. The eukaryotic plasma membrane is composed of a variety of structural lipids, including those with phosphatidyl-choline (PC), phosphatidyl-ethanolamine (PE), and phosphatidyl-serine (PS), among others 8. Maintenance of this asymmetric PM is required for cell survival, and phospholipid trafficking deficiencies are a hallmark of several cancers 9. In addition to phospholipids, the PM of human cells contains approximately 30% cholesterol – cholesterol trafficking is particularly critical as this sterol affects the local bilayer rigidity and phospholipid/MP packing 8. For cholesterol shuttling between cells and tissues, it must first be extracted from the PM: sterol removal from lipid membranes requires overcoming the immense energetic barrier of pulling this highly insoluble moiety from the hydrophobic core of the bilayer 10. However, this process occurs readily within the human body, as the flux of cholesterol throughout tissues is quite high (i.e., 10 mg/day/kg) – aberrant cholesterol efflux and metabolism are associated with poor atherosclerotic health, along with cancer 11–13. The mechanistic pathway for the removal of cholesterol from the PM for subsequent secretion from the body is termed the reverse cholesterol transport (RCT) pathway in which the ABC transporter ABCA1 plays a central role 14.
After the initial discovery of the abca1 gene, ABCA1 protein expression was quickly found to be inversely correlated with cellular cholesterol levels 6,15,16. It has been well established through cell-biology based characterization that ABCA1 mediates the efflux of cholesterol and phospholipids with PC headgroups from the membrane to the extracellular acceptor protein, apolipoprotein A-I (apoA-I) 16–27. Together with apoA-I, sterols and phospholipids form nascent high-density lipoprotein (nHDL). ABCA1 and apoA-I are vital in the RCT as they mediate sterol efflux from macrophages, which is essential for clearance of cardiovascular plaques; therefore ABCA1 function is directly linked to atherosclerosis 28,29. Genetic mutations to the abca1 gene are naturally occurring and are associated with Tangier’s Disease, which results in impaired cholesterol clearance 30,31. ABCA1 function has further been implicated in neurodegenerative disorders, such as Alzheimer’s Disease 32. ABCA1 has long been suggested as a potential drug target 30,31,33. Thus, elucidating how the ABCA1 TMD drives lipid export and how this process can be pharmacologically regulated is of immense biomedical importance.
The overall domain architecture of ABCA1 was elucidated by a previously reported cryo-EM structure of ABCA1 in detergent 34. While native MPs reside within the complex PM, detergents are a common lipid mimetic utilized for solubilization and biochemical characterization of MPs 35. Detergents are known to affect the biochemical activity of MPs, which seems particularly true for detergent-solubilized ABC transporters as a variety of deleterious effects on ATP-hydrolysis activities have been reported in the literature 34–36,37,41,42. In line with this notion, the ABCA1 protein in the detergent digitonin used for previous structural studies exhibited minimal ATP hydrolysis activity 34. In addition to the TMDs and NBDs which are present in all ABC transporters, ABCA1 contains a strikingly large extra-cellular domain (ECD) which interacts with apoA-I 19,21,22,34. The ECD contains approximately 850 amino acid residues and its pinnacle reaches 100 Å above the membrane plane. Naturally occurring mutations in the ABCA1 ECD have shown impaired apoA-I-dependent cholesterol efflux 24,37. However, the exact function of the ECD and how this activity is coupled to the TMDs and NBDs to coordinate lipid movement from the membrane bilayer to apoA-I are unknown.
One notable limitation in previous ABCA1 investigations is that various studies have used protein from distinct sources and in different membrane environments, thus preventing reliable correlation of the multiple activities and dynamic conformations of ABCA1. To overcome this technical hurdle, we have established an inducible ABCA1 expression system and a unified workflow of characterizing ABCA1 in nanodiscs containing a defined lipid system. Taking advantage of this platform, we utilized a combination of cell-based, biochemical, structural, and computational approaches to gain mechanistic insights into ABCA1 functions.
ABCA1 stably expressed in Human Embryonic Kidney (HEK) cells is active in sterol transport
Previous studies of human ABCA1 have been performed in various membrane mimetics and environments 35. While the in vivo function of ABCA1 has been extensively investigated in a variety of physiologically relevant cell types, including J774 macrophages and human skin fibroblasts 21,38,39, one previously reported structural investigation of ABCA1 utilized protein purified from Spodoptera frugiperda (Sf9) insect cells 34. Insect cells are unable to synthesize cholesterol 40, a key transport substrate for ABCA1 34. ABCA1 extracted from Sf9 cells was subsequently reconstituted in the digitonin detergent which has deleterious effects on the ATP hydrolysis function of ABCA1 34. Therefore, a missing link exists between the in vivo and biochemical characterization of functional ABCA1 and the structural information on biologically relevant protein conformations.
Such limitations highlight the necessity to establish a consistent protein production strategy for both functional assays and structure determination. To this end, we utilized the Sleeping Beauty transposon system to engineer a stable line of HEK cells with inducible expression of ABCA1 41 (Supplementary Fig. 1A). Notably, this expression platform is dual function: controlled expression of ABCA1 can be used for cell-based sterol transport activity measurements, and purification of overexpressed ABCA1 permits for in vitro activity quantification and cryo-EM studies in lipid nanodiscs with distinct lipid compositions.
We first assessed the sterol efflux activity of ABCA1 as follows – cells were initially incubated with the fluorescent cholesterol analog, dehydroergosterol (DHE), followed by the addition of lipid-poor apoA-I to the culture medium 42. DHE efflux from the cells to apoA-I was subsequently measured. Our results show that DHE export from the PM was dependent on both apoA-I and ABCA1 (Supplementary Fig. 2A–C). As a comparison with the wild-type (WT) ABCA1, we included the well-studied C1477R mutant, which has diminished efflux capacity due to a deleterious ECD mutation 20,24. In our assays (Fig. 1A), the C1477R mutant showed minimal sterol efflux activity, which is consistent with previously reported cell- and liposome-based cholesterol efflux assay 43,44. Thus, our inducible expression system enables interrogation of the sterol efflux activity of WT and mutants of ABCA1 under various conditions.
Figure 1. ABCA1 stably expressed in HEK cells is functionally active.
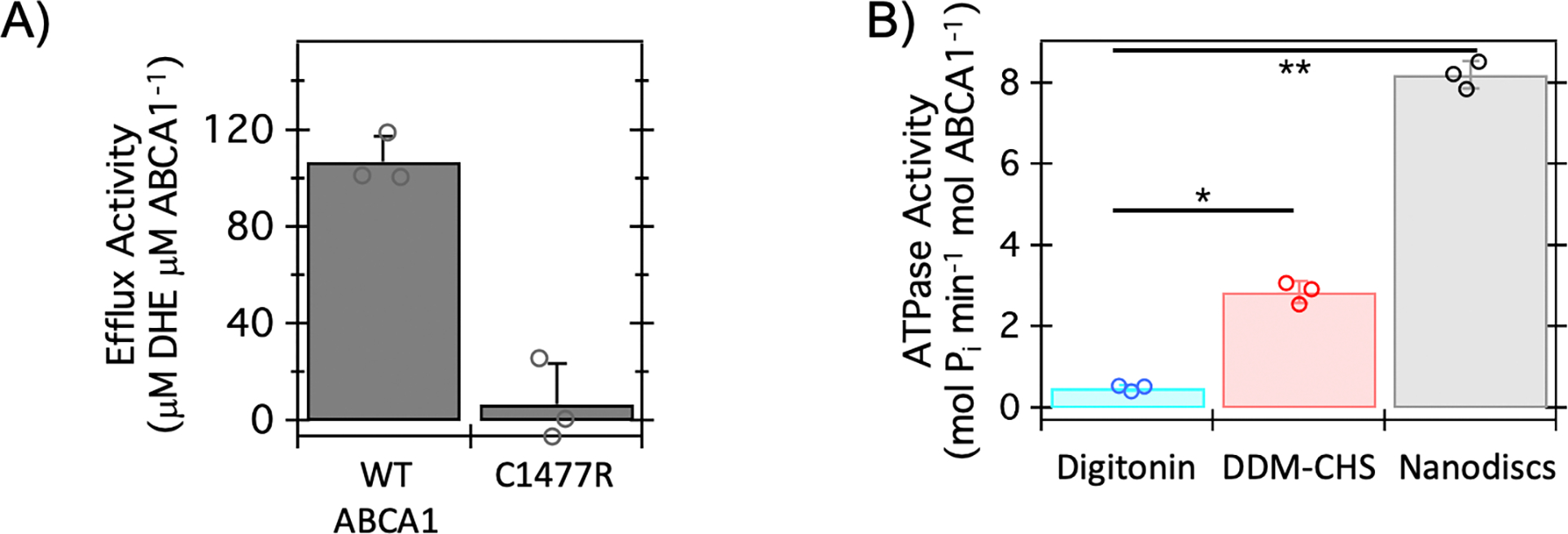
A) Cell-based efflux activity of ABCA1 for the cholesterol analog, dehydroergosterol (DHE) is shown for wild-type (WT) ABCA1 and the well-characterized ECD mutant: C1477R. Modulating the ECD conformation (i.e., C1477R) reduces the measured sterol efflux of ABCA1. Data represent the average of three biological replicate experiments and error bars indicate the standard deviation; individual data points are shown. The probability associated with a Student’s paired t-test with a two-tailed distribution for comparison of these data is < 0.025. Briefly, DHE efflux was monitored in HEK Expi293F cells stably expressing WT-NGFP-ABCA1 or C1477R with the addition of exogeneous His-apoA-I. B) ATPase activity of ABCA1 was measured in two detergents: digitonin and Dodecyl-β-Maltoside with cholesteryl hemisuccinate (DDM-CHS), along with nanodiscs prepared with 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (POPG) lipid and the membrane scaffold protein (MSP) E3D1. Data represent the average of three biological replicate experiments and error bars indicate the standard deviation; individual data points are indicated. The probability associated with a Student’s paired t-test with a two-tailed distribution for data demarcated by a single asterisk (*) is < 0.0025. The probability associated with a Student’s paired t-test with a two-tailed distribution for data demarcated by a double asterisk (**) is < 0.0005. Briefly, WT-ABCA1 was purified from HEK Expi293F cells and isolated through affinity chromatography in the respective detergents or reconstituted into nanodiscs containing POPG lipids and the Membrane Scaffold Protein (MSP) E3D1 after cleavage of the N-terminal GFP. ATP hydrolysis was quantified by colorimetric assay of phosphate release.
ABCA1 exhibits enhanced ATPase activity in nanodiscs with POPC or POPG containing lipids
All ABC transporters demonstrate two fundamental activities: substrate translocation and ATP hydrolysis. For ABCA1, the former has been extensively characterized using various cell-based assays and in vivo studies 5, whereas the latter has not been well-characterized due to the technical challenges of obtaining homogeneous ABCA1 in lipid membranes for accurate ATPase activity quantitation. Previous studies have used liposome reconstituted ABCA1 to characterize its ATPase activity 44.While a proteoliposome system allows for studying transporter activities in membranes with various lipid compositions, this measurement is complicated by an unpredictable amount of inside-in ABC transporters with their NBDs inside the liposomes and inaccessible to the exogenously added ATP. Further, membrane proteins in liposomes are not readily amenable for structural determination. In comparison, ABC transporter proteins reconstituted into nanodiscs 45 have their NBDs accessible for ATP binding and hydrolysis and are also feasible targets for high-resolution cryo-EM structure determination 46,47.
For in vitro biochemical and structural studies, ABCA1 stably expressed in HEK cells was purified in Dodecyl-β-Maltoside (DDM) with cholesteryl hemisuccinate (CHS) and subsequently reconstituted into nanodiscs (Supplementary Fig. 1B–D). The resulting nanodisc-embedded ABCA1 demonstrated ATP concentration-dependent activity of ATP hydrolysis (Supplementary Fig. 3A); these experiments are performed in the absence of apoA-I. Importantly, the lipid composition of nanodiscs may be systematically varied to assess the effect of membrane environment on ABCA1 activity. To determine the effect of lipid composition for ABCA1 function, several lipids and lipid mixtures were tested in the nanodisc reconstitution. ABCA1 embedded in the nanodiscs containing pure 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) or 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) demonstrated the highest ATPase activity (Supplementary Fig. 3B). These observations agree with previously reported ATPase activities of ABCA1 in liposomes 44. Notably, the highest activity of ABCA1 in nanodiscs is over 15- and 5-fold greater than the activity of ABCA1 in digitonin and DDM-CHS, respectively (Fig. 1B).
Cryo-EM structure of ABCA1 in lipid nanodiscs
The enhanced ATPase activity of ABCA1 in the above nanodiscs suggests that the conformational transition cycle of ABCA1 is distinct from that in detergent. To understand the impact of membrane environment on ABCA1 at the structural level, we determined the cryo-EM structure of nanodisc-embedded ABCA1 at an overall resolution of 4.0 Å (Fig. 2A–C, Supplementary Fig. 4, Supplementary Table 1). In this cryo-EM map, the TMDs inside the nanodiscs exhibit the best resolution, while the ECD apex (residues 140–250) displays the lowest resolution (Fig. 2D), suggesting heterogeneous conformations or high mobility around the tip of the ECD.
Figure 2. Cryo-EM structure and conformational dynamics of ABCA1 in a lipid bilayer.
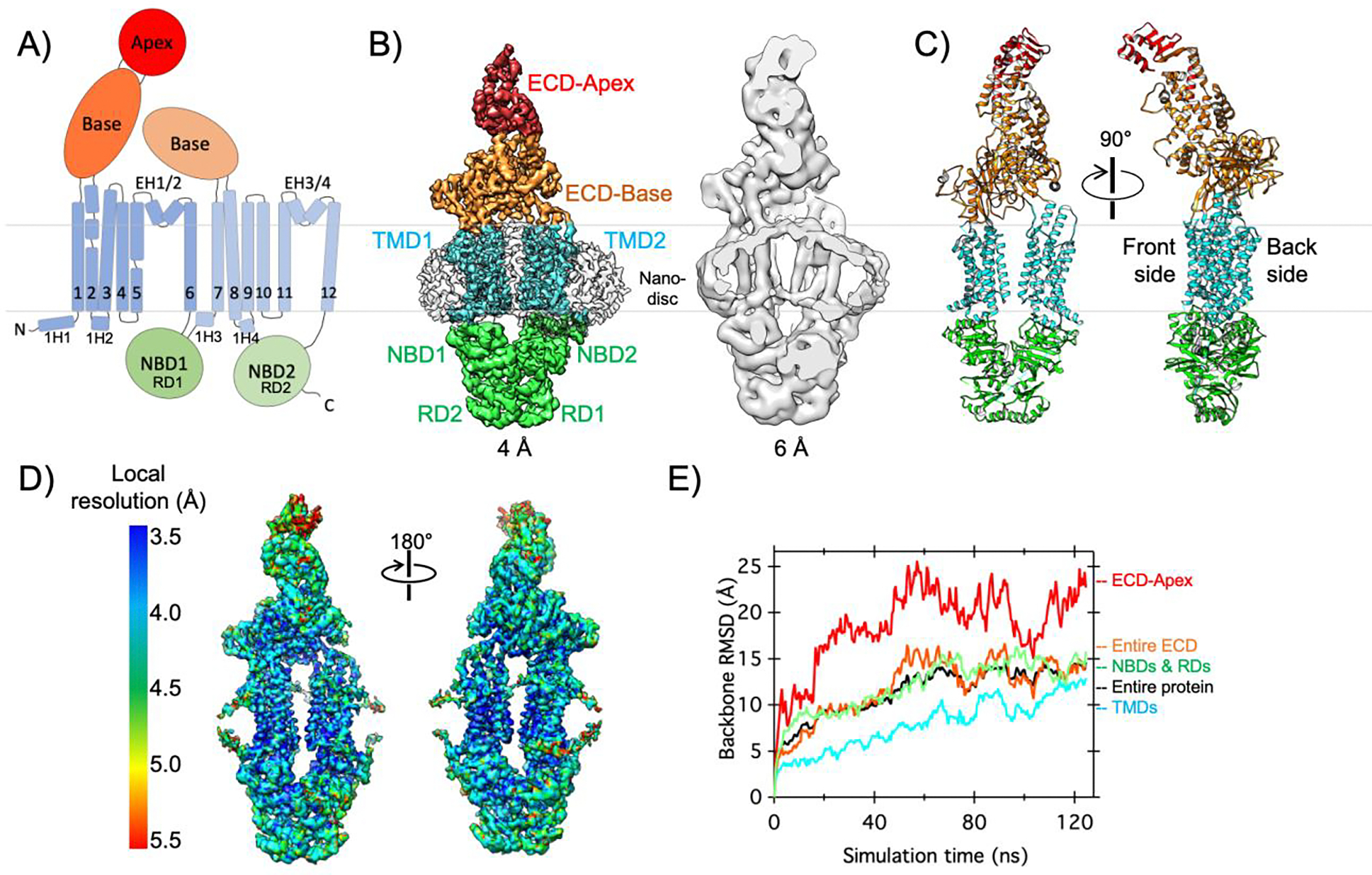
A) The domain architecture and secondary structure of ABCA1 are indicated with the NBDs, TMDs, ECD base, and ECD apex shown in green, cyan, orange, and red, respectively. The boundaries of the inner membrane are indicated by gray lines. B) Side view of cryo-EM map of nanodisc-embedded ABCA1 low-pass filtered at 4.0-Å resolution, with different regions denoted and colored as in A and nanodisc shown as an outline (left). Cross-sectional side view of the same map low-pass filtered at 6.0-Å resolution, to highlight the nanodisc and membrane curvature (right). C) Two side views of the structural model of ABCA1, colored as in A. D) Local resolution of the cryo-EM map of nanodisc-embedded ABCA1. E) All-atom MD simulations of ABCA1 in a lipid bilayer reveal conformational changes. Backbone α-carbon root mean square displacement (RMSD) values are shown for different regions of ABCA1 and colored as in A. Briefly, two datasets collected on nanodisc-embedded ABCA1 were collected for combined processing and 3D reconstruction. All-atom MD simulations were completed on membrane embedded ABCA1 as described in the Supplemental Information.
Combining cryo-EM with nanodisc technology provides a unique opportunity of studying membrane remodeling by MPs. Our cryo-EM map reveals that, instead of being planar, the membrane surrounding ABCA1 is curved particularly on the front side (Fig. 2B and Supplementary Fig. 5). Membrane curvature increases lipid packing defects, which can be recognized and bound by amphipathic peptides 48. ApoA-I is composed of regularly repeating amphipathic helices, and the membrane curvature induced by ABCA1 may play a mechanistic role in ABCA1-apoA-I interaction. Consistent with this notion, apoA-I was suggested to insert into bilayer defects adjacent to ABCA1 to extract lipids and sterols 27,49.
Structural variation of ABCA1 in different membrane environments
Our cryo-EM structure of ABCA1 in nanodiscs allows us to investigate the influence of different membrane environments on the conformation of ABCA1. The structure of nanodisc-embedded ABCA1 exhibits a similar overall architecture as the previously determined digitonin-reconstituted ABCA1 (PDB ID: 5XJY), albeit with notable differences particularly in the TMDs and ECD 34. These changes also lead to different volumes of internal cavities within the protein (Supplementary Fig. 6). Slight differences are also observed in the ECD, while minimal conformational changes occur in the NBDs which are in an open conformation.
The 12 TM helices of ABCA1 constitute two distinct TMDs forming a V-shaped opening towards the extracellular leaflet (Fig. 3A). While the TM helices in TMD1 from both structures superimpose well, those in TMD2 display larger lateral shifts (Fig. 3B and Supplementary Fig. 6C). Relative to the TMD2 in detergent, the TMD2 in nanodiscs moves towards the center and the back side of ABCA1, decreasing the distance between TMs 11 and 8 (in TMD2) and TM5 (in TMD1) and creating a larger space at the front side of the TMDs. While no clear lipid densities were observed at the TMD interface in the cryo-EM map of nanodisc-embedded ABCA1, the observed TM helix rearrangements in the membrane environment likely have functional repercussions for the translocation of lipid substrates.
Figure 3. Conformation of the TMDs of nanodisc-embedded ABCA1.
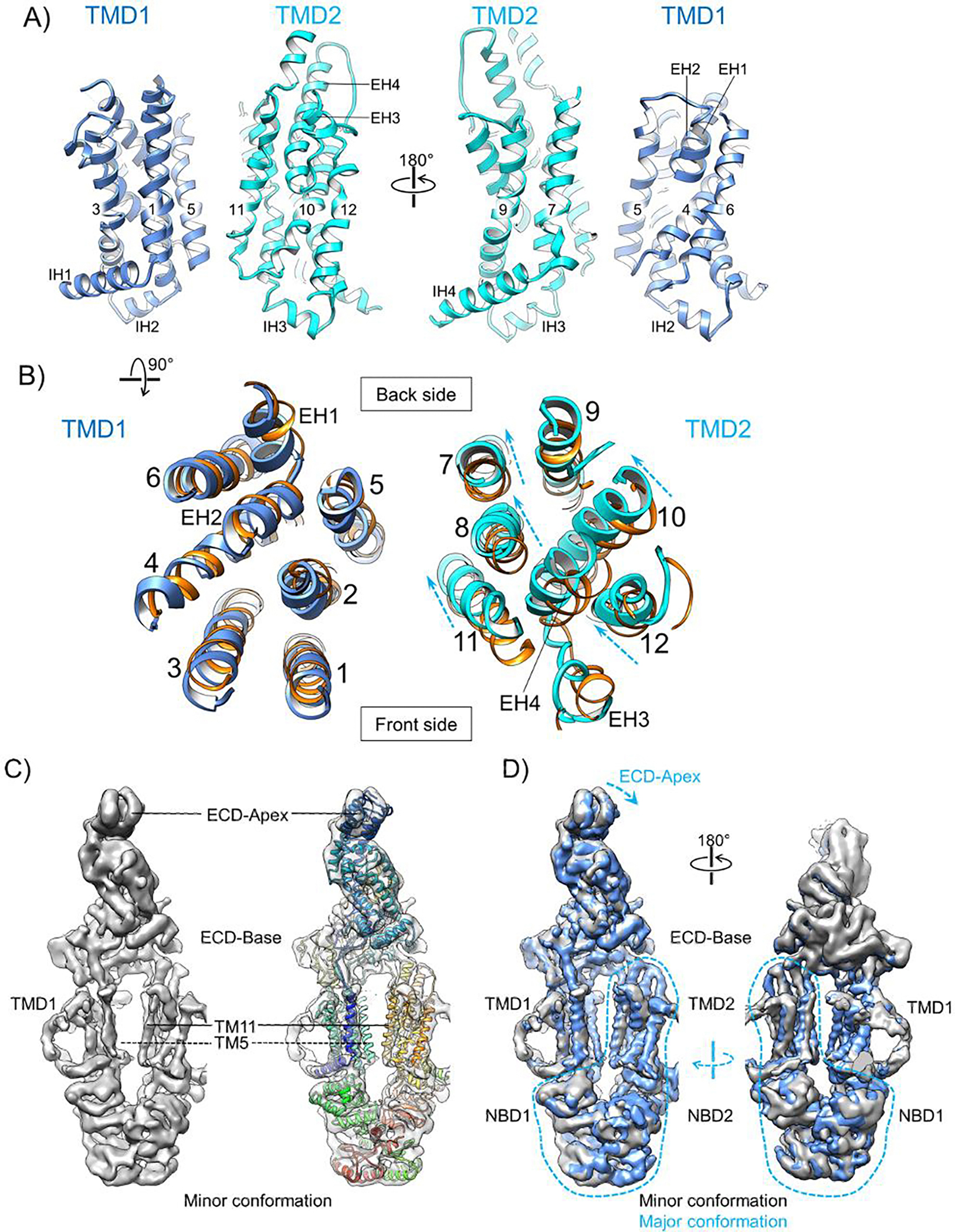
A) Two side views of the TMDs (TMD1 in blue and TMD2 in cyan) with the transmembrane helices indicated by numbers and the other helices denoted. Left panel is the same view as in Fig. 1B and C (left). B) Top-down view of the TMDs. The lateral shifts of individual helices in TMD2 relative to their positions in the previously resolved ABCA1 digitonin conformation (PDB ID: 5XJY, orange) are indicated by dashed arrows. These two ABCA1 structures are aligned using TMD1. C) Side views of the cryo-EM map of nanodisc-embedded ABCA1 in the minor conformation and rigid-body fitting of the structural model of nanodisc-embedded ABCA1 in the major conformation (rainbow). The ECD-Apex and TM11 show clear discrepancies and TM5 is missing from the cryo-EM map, as indicated. D) Front and back side views of the superimposition between the cryo-EM maps of nanodisc-embedded ABCA1 in the major (blue) and minor (gray) conformations. Both maps are low-pass filtered to 4.4-Å resolution, sharpened with −100 Å2 b-factor, and displayed at 7σ contour level. The transitions from the minor to the major conformation in the ECD-Apex and TMD2-NBDs-RDs regions are indicated by blue dashed arrows.
Compared to TMD2, the ECD displays an even greater variation between the two ABCA1 structures. Relative to ABCA1 in detergent, the ECD of nanodisc-embedded ABCA1 rotates downward and towards the side of TMD2 (Fig. 4A). Notably, helices α8 (residues 278–288) and α9 (residues 299–309) shift towards helices α18 (residues 485–504) and helix α-10 (residues 352–364) in our cryo-EM structure, thus slightly closing the opening of the pocket in the ECD base of ABCA1. Taken together, the characteristic movements of TMD2 and the ECD of ABCA1 upon reconstitution into lipid bilayer environment suggest the structural plasticity of these domains and conformational coupling between the TMDs and ECD.
Figure 4. Conformational dynamics of the ECD and its large hydrophobic pocket.
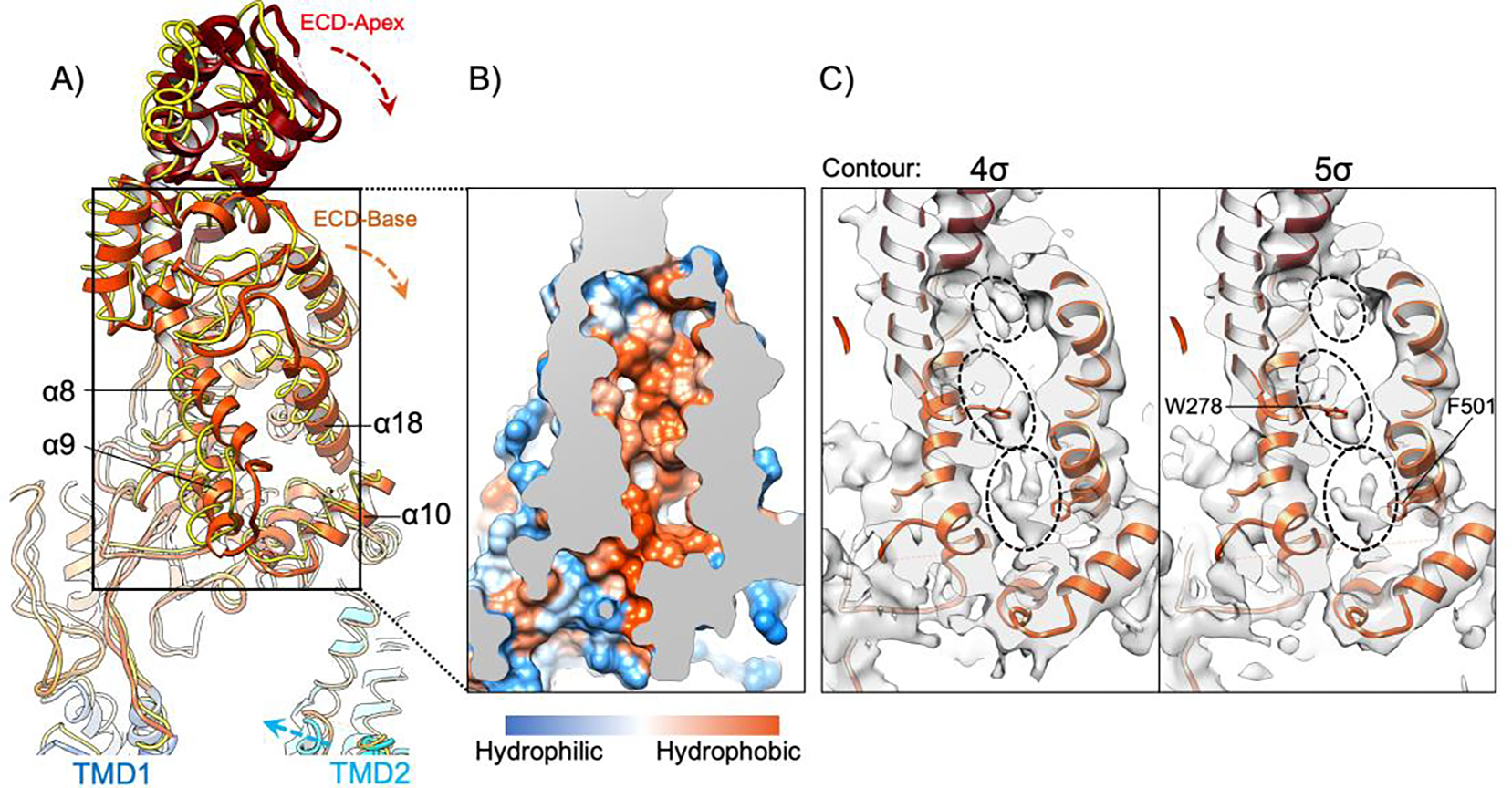
A) Structural model of the ECD in the nanodisc-embedded ABCA1, viewed and colored as Fig. 1C (left) and superimposed with the ECD of ABCA1 in digitonin (PDB ID: 5XJY) (yellow wire). These two structures are aligned using their NBDs and RDs. The movements of TMD2, ECD base, and ECD apex of nanodisc-embedded ABCA1 relative to the ABCA1 in digitonin are indicated by dashed arrows. The important helices to form the hydrophobic pocket in the ECD base are denoted. B) Cross section of the boxed region in A, shown in hydrophobic surface representation. C) Cross section of the boxed region in A, showing superimposition of the structural model of nanodisc-embedded ABCA1 with its cryo-EM map displayed at two different contour levels. Three non-proteinaceous densities are indicated in dashed ovals. Trp278 and Phe501 are in close contact with the non-proteinaceous densities and are denoted.
Cryo-EM analysis reveals multiple conformations of ABCA1 in lipid bilayers
In addition to the above-mentioned cryo-EM structure of nanodisc-embedded ABCA1, our 3D classification of particle images generated another cryo-EM map that represents a distinct conformation (Supplementary Fig. 4C). This is referred to as the minor conformation, as it contains slightly fewer particles and exhibits lower resolution. The cryo-EM map of ABCA1 in the minor conformation shows poorly resolved TMD1 with the TM5 helix completely missing, whereas the other structural elements are more readily traceable (Fig. 3C). Superimposition of the cryo-EM maps of the two conformations demonstrates the greatest conformational changes in the regions of the ECD-Apex, TMD2, NBDs and Regulatory Domains (RDs). A morph between these two cryo-EM maps shows that the TMD2 and NBDs/RDs rotate about an axis roughly perpendicular to the membrane plane and the ECD-Apex tilts towards the TMD2; in comparison, the ECD-Base and TMD1 exhibit smaller changes (Supplementary Movie 1). After rigid-body fitting into the cryo-EM map of ABCA1 in the minor conformation, our structural model of ABCA1 in the major conformation demonstrates clear differences in the regions of ECD-Apex and TMD2 (Fig. 3C and Supplementary Fig. 4G). In contrast, rigid-body fitting of the structure of ABCA1 in digitonin (PDB: 5XJY) over the minor conformation led to almost perfect superimposition (Supplementary Fig. 4G). Thus, ABCA1 in a lipid bilayer environment populates multiple distinct conformations, one of which was stabilized by the digitonin detergent. Such structural plasticity of ABCA1 within the cell membrane is likely essential for its functions and may underpin its higher ATPase activity in lipid bilayers (Fig. 1B).
The ECD is highly dynamic and contains a large lipid-binding pocket
To further interrogate the conformational dynamics of ABCA1 in a membrane environment, we completed all-atom molecular dynamics (MD) simulations of ABCA1 embedded in a lipid bilayer (Fig. 2E and Supplementary Fig. 7A–C). Consistent with the observed lower local resolution of the ECD apex in our cryo-EM map (Fig. 2D), this region demonstrates remarkable mobility with backbone α-C root mean square displacement (RMSD) values greater than 20 Å at times, which is much greater than the mobility in all other domains of ABCA1 (Fig. 2E). These analyses indicate a high level of conformational flexibility of the ECD, particularly near its tip, which is likely relevant to the lipid transport function of ABCA1. Consistent with this notion, many disease mutations in ABCA1 are in the ECD (Supplementary Fig. 8) and the ECD has been suggested to undergo large conformational changes upon ATP binding at the NBDs 50.
The ECD base contains a large hydrophobic pocket with an inner volume of 5316 Å3 (Fig. 4B). Within this hydrophobic pocket, we observe three non-proteinaceous densities that are likely contributed by cholesterol and phospholipids (Fig. 4C). In close contact with these potential lipid densities are several hydrophobic residues with clear side chain densities, such as Trp278 (α8) and Phe501 (α18). Conceivably, the aromatic side chains of these residues form ring stacking interactions with the bound cholesterol molecules. These observations provide additional evidence that the lipids extracted from the PM may be shielded from the aqueous extracellular environment within the ECD for subsequent trafficking to apoA-I 43. While the large hydrophobic pocket in the ECD base was previously described as a “tunnel” or static pathway for lipid passage, it may function more as a dynamic lipid container 34. The structural variations of the ECD as observed in our MD simulations likely reflect the flexible conformation of the ECD hydrophobic pocket and its changing lipid affinity to bind and release substrates. This aligns with the recent report that the ABCA1 ECD acts as a reservoir for lipids extracted from the outer leaflet of the PM by ABCA1 51. Consistent with this notion, the observed structural rearrangements in the ECD result in significant volume changes for the ECD cavity (Supplementary Fig. 7G). Furthermore, the stored lipids may exit the pocket through the opening lined by the helices α8, α18 and α10 (Fig. 4A), if a hydrophobic acceptor such as apoA-I is accessible from outside the pocket.
Comparison with ABCA4 suggests a missing element for ABCA1 lipid transport
ABCA1 and ABCA4 are highly homologous, sharing >50% amino acid identity, even though ABCA4 performs a distinctive lipid import function by moving retinoid from the lumenal leaflet to the cytoplasmic leaflet of disk membranes in the eye 52. Three previously determined ABCA4 structures in the ATP-free state display similar protein conformations and contain substrate lipid densities, revealing the structural basis of substrate recognition and transport 53–55. To obtain more insight into the interaction of ABCA1 with its substrate lipids, our ABCA1 structure was superimposed with a structure of ABCA4 53 (Supplementary Fig. 9A). While the TMDs of ABCA1 and ABCA4 both form a general V-shaped opening towards the ECD, the TMDs in ABCA4 shift drastically towards the front side and the center, closing the space between the TMDs to sandwich several lipids at the levels of both membrane leaflets (Supplementary Fig. 9C, D). Together, this suggests that the wide-open conformation of the TMDs as observed in our ABCA1 structure represents a substrate-free state before capturing a lipid or sterol substrate.
In the ABCA4 structure, the retinoid lipid substrate is present at the level of the outer membrane leaflet, interacting with not only TMDs but also the “S loop” between the ECD helices α9 and α10 (Supplementary Fig. 9B). This scenario of using a hydrophobic loop to extract specific lipid substrates is reminiscent of bacterial LolCDE, an ABC transporter that uses the elongated “shoulder loop” to recognize and lift triacylated proteins above the membrane surface 56. Among the four residues of ABCA4 in the center of the S loop that mediate lipid interactions, three are conserved in ABCA1: Trp339, Tyr340 and Tyr345 (Supplementary Fig. 9E). Thus, the S loop of ABCA1 likely also plays an important role in lipid transport. Intriguingly, the S loop is completely absent from the cryo-EM maps of ABCA1 in detergent and in nanodiscs. Considering the wide open TMDs and lack of ABCA1 lipid densities at the level of the outer membrane leaflet, the lipid substrates originally bound to ABCA1 were likely removed during protein purification with detergents, leading to destabilization of the S loop and wide opening of the TMDs. Interestingly, electrostatic interactions involving the ABCA1 S-loop were recently shown to play a role in phospholipid extraction to the ECD51.
Comparison with ABCA7 suggests commonalities between ABCA transporter mechanisms
The recently resolved structures of nanodisc-embedded ABCA757 allow for an additional comparison of the novel structure of nanodisc-embedded ABCA1 (Supplementary Fig. 10). Overall, the TM cavity of ABCA1 is relatively closed compared to the ABCA7 conformation which likely restricts phospholipid binding at the cytosolic leaflet of ABCA1. When aligned on TMD2, ABCA1 exhibits concerted movement of the TMD1, NBDs, and the ECD relative to ABCA7, highlighting the connectivity between each structural unit of ABCA1. One notable commonality between ABCA1 and ABCA7 may be the deformations of the surrounding membrane, particularly in the outer leaflet of the PM. The mirrored importance of the local lipid environment for ABCA7 suggests this may be a common theme for function of ABCA family transporters.
Based on the results from our work and previous functional and structural studies, we propose a “lipid container” model for ABCA1-driven lipid transport and nHDL formation (Supplementary Fig. 11). The key of this proposed model is the large ECD hydrophobic pocket functioning as a dynamic lipid container which can both accept lipids extracted from the PM and release lipids in the extracellular space. In this model, the functional cycle of ABCA1 is divided into three states: pre-substrate loading, substrate loading, and substrate expulsion. In the pre-substrate loading state, ABCA1 has relaxed and wide-open TMDs, and the large hydrophobic pocket in the ECD contains lipids loaded from previous functional cycles. In the substrate loading state, cholesterol and phospholipids interact with the TMDs and S loop, closing the TMDs and causing the bound lipids to be elevated to the interface between the ECD and the TMDs (i.e., the level of the outer membrane surface), and local membrane deformations occur to promote apoA-I association with ABCA1. In the substrate expulsion state, ATP binding to the NBDs induces closure of the NBDs and TMDs to squeeze the bound lipids into the ECD hydrophobic pocket, which can push the excess lipids out of the pocket from the side to interact with ApoA-I and form nHDL. ATP hydrolysis leads to the opening of the NBDs and TMDs, and ABCA1 returns to the pre-substrate loading state. According to this model, all currently available ABCA1 structures without ATP binding are in the pre-substrate loading state. The structural basis of lipid recognition and transport by ABCA1 awaits future studies. While our model focuses on the aspect of lipid extraction, it is noteworthy that ABCA1 has been implicated in many other functions, such as phospholipid flipping 44, lateral reorganization of cholesterol rafts 58, phosphatidylinositol 4,5-bisphosphate flipping 59, and apoA-I independent sterol efflux 20,37,60. Future work is also required to relate each of these effects to the conformational states of ABCA1.
Our findings highlight the importance of the membrane environment for ABCA1 activity and structure. Besides the first determined digitonin-reconstituted ABCA1 structure34, additional cryo-EM structures of nucleotide-free ABCA1 have recently been resolved in digitonin and in a lipid nanodisc, along with a catalytically inactive mutant bound to ATP in digitonin 61. In contrast to our results, after nanodisc reconstitution, their protein exhibited similar or lower ATPase activities than detergent samples, and the conformation of ABCA1 in nanodiscs containing mostly PS shows little difference from those in digitonin from both cryo-EM studies34,61. It is unclear whether their similar ABCA1 conformations in nanodiscs and in digitonin are a result of the particular lipid composition of nanodiscs, different protein preparation parameters, or addition of the detergent Fos-choline-8 to their nanodisc sample prior to freezing cryo-EM grids61. The biological relevance of local lipid environment is significant as it may modulate MP structure and function 62. For ABC transporters specifically, the chemical properties of the phospholipid headgroups may affect the allosteric connections between the TMD and NBD and therefore impact ATP hydrolysis and efflux activities.
Additionally, we observe distinctive nanodisc curvature in the cryo-EM analyses and modulations in the bilayer adjacent to ABCA1 in MD simulations (Supplementary Figs. 5 and 7). Amphipathic helices, like those characteristic of apoA-I, have been shown to selectively insert into defects within membrane bilayers 63,64. Following an association with the lipid bilayer interface, apoA-I may further disrupt the lipid bilayer, promoting conformational changes to both the ABCA1 TMDs and ECD – the mobile ECD may undergo further conformational changes to accommodate apoA-I binding and sterol efflux 19. This ECD conformational change is likely associated with allosteric rearrangement of the NBDs as this type of interaction has been previously reported 22.Taken together, these data suggest that ABCA1 may collaborate with the surrounding lipid bilayer in a functionally relevant manner, similar to the mechanism recently proposed for the homologous ABCA757. These findings are consistent with previously proposed models in which ABCA1 forms large complexes, and perhaps large bilayer defects, to recruit apoA-I for lipid and sterol exchange 65.
Our establishment of an inducible expression system for multifaceted investigations of ABC transporters in cell-based assays and parallel biochemical and structural studies provides invaluable tools for future characterizations of ABCA1 and its variants, as well as for understanding the functional and conformational impacts of small molecules and antibodies on ABCA1 in various membrane environments.
Supplementary Material
Acknowledgements
We are grateful to YC Shin for modification of the Sleeping Beauty plasmid vector. We thank the HMS Cryo-EM center for the assistance in collection of EM data included here. We acknowledge the support of the ICCB-Longwood Screening Facility at Harvard Medical School and the Harvard Medical School Center for Macromolecular Interactions. Computational resources were provided by the Harvard Medical School Orchestra O2 cluster and Stampede XSEDE allocations (Allocation Numbers: BIO200018 & BIO210005). We thank all past and present Liao lab members for technical support and helpful discussion. The authors acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources that have contributed to the research results reported within this paper.
Funding and Additional Information
AMPM was funded by an American Heart Association Post-Doctoral Fellowship (AHA Grant Number 18POST34030341); ATC was funded by an NIH F32 Fellowship (Grant Number 1F32GM136092-01); CLMP was funded by an American Cancer Society Postdoctoral Fellowship (Grant Number PF-20-107-01-TBE).
Footnotes
Data Availability
The atomic model for the major ABCA1 conformation has been deposited in the PDB with the accession code 7TDT. EM maps for the major and minor ABCA1 conformations have been deposited in the EMDB with the accession codes: 25838 and 27079 respectively. The authors declare that all other data supporting the findings of this study are available within the paper and its supplementary information files; source data are provided with this paper.
References
- 1.Engel A & Gaub HE Structure and mechanics of membrane proteins. Annu Rev Biochem 77, 127–148 (2008). 10.1146/annurev.biochem.77.062706.154450 [DOI] [PubMed] [Google Scholar]
- 2.Dean M, Rzhetsky A & Allikmets R The human ATP-binding cassette (ABC) transporter superfamily. Genome Res 11, 1156–1166 (2001). 10.1101/gr.184901 [DOI] [PubMed] [Google Scholar]
- 3.Ford RC & Beis K Learning the ABCs one at a time: structure and mechanism of ABC transporters. Biochem Soc Trans 47, 23–36 (2019). 10.1042/BST20180147 [DOI] [PubMed] [Google Scholar]
- 4.Srikant S & Gaudet R Mechanics and pharmacology of substrate selection and transport by eukaryotic ABC exporters. Nat Struct Mol Biol 26, 792–801 (2019). 10.1038/s41594-019-0280-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plummer AM, Culbertson AT & Liao M The ABCs of Sterol Transport. Annu Rev Physiol 83, 153–181 (2021). 10.1146/annurev-physiol-031620-094944 [DOI] [PubMed] [Google Scholar]
- 6.Langmann T et al. Molecular cloning of the human ATP-binding cassette transporter 1 (hABC1): Evidence for sterol-dependent regulation in macrophages. Biochemical and Biophysical Research Communications 257, 29–33 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Wen PC & Tajkhorshid E Conformational coupling of the nucleotide-binding and the transmembrane domains in ABC transporters. Biophys J 101, 680–690 (2011). 10.1016/j.bpj.2011.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Meer G, Voelker DR & Feigenson GW Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9, 112–124 (2008). 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fadeel B & Xue D The ins and outs of phospholipid asymmetry in the plasma membrane: roles in health and disease. Crit Rev Biochem Mol Biol 44, 264–277 (2009). 10.1080/10409230903193307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saad H & Higuchi W Water Solubility of Cholesterol. Journal of Pharmaceutical Sciences 54, 1205–1206 (1965). [DOI] [PubMed] [Google Scholar]
- 11.Dietscshy JM & Turley SD Cholesterol metabolism in the brain. Current Opinion in Lipidology 12, 105–112 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Riscal R, Skuli N & Simon MC Even Cancer Cells Watch Their Cholesterol! Mol Cell 76, 220–231 (2019). 10.1016/j.molcel.2019.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikonen E Mechanisms for cellular cholesterol transport: defects and human disease. Physiol Rev 86, 1237–1261 (2006). 10.1152/physrev.00022.2005 [DOI] [PubMed] [Google Scholar]
- 14.Favari E et al. Cholesterol efflux and reverse cholesterol transport. Handb Exp Pharmacol 224, 181–206 (2015). 10.1007/978-3-319-09665-0_4 [DOI] [PubMed] [Google Scholar]
- 15.Luciani MF, Denizot F, Savary S, Mattei MG & Chimini G Cloning of two novel ABC transporters mapping on human chromosome 9. Genomics 21, 150–159 (1994). [DOI] [PubMed] [Google Scholar]
- 16.Oram JF, Lawn RM, Garvin MR & Wade DP ABCA1 is the cAMP-inducible apolipoprotein receptor that mediates cholesterol secretion from macrophages. J Biol Chem 275, 34508–34511 (2000). 10.1074/jbc.M006738200 [DOI] [PubMed] [Google Scholar]
- 17.Fitzgerald ML et al. ABCA1 and amphipathic apolipoproteins form high-affinity molecular complexes required for cholesterol efflux. J Lipid Res 45, 287–294 (2004). 10.1194/jlr.M300355-JLR200 [DOI] [PubMed] [Google Scholar]
- 18.Smith JD et al. ABCA1 mediates concurrent cholesterol and phospholipid efflux to apolipoprotein A-I. J Lipid Res 45, 635–644 (2004). 10.1194/jlr.M300336-JLR200 [DOI] [PubMed] [Google Scholar]
- 19.Wang S, Gulshan K, Brubaker G, Hazen SL & Smith JD ABCA1 mediates unfolding of apolipoprotein AI N terminus on the cell surface before lipidation and release of nascent high-density lipoprotein. Arterioscler Thromb Vasc Biol 33, 1197–1205 (2013). 10.1161/ATVBAHA.112.301195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaughan AM, Tang C & Oram JF ABCA1 mutants reveal an interdependency between lipid export function, apoA-I binding activity, and Janus kinase 2 activation. J Lipid Res 50, 285–292 (2009). 10.1194/jlr.M800366-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vedhachalam C et al. ABCA1-induced cell surface binding sites for ApoA-I. Arterioscler Thromb Vasc Biol 27, 1603–1609 (2007). 10.1161/ATVBAHA.107.145789 [DOI] [PubMed] [Google Scholar]
- 22.Nagao K et al. ATP hydrolysis-dependent conformational changes in the extracellular domain of ABCA1 are associated with apoA-I binding. J Lipid Res 53, 126–136 (2012). 10.1194/jlr.M019976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chroni A et al. The central helices of ApoA-I can promote ATP-binding cassette transporter A1 (ABCA1)-mediated lipid efflux. Amino acid residues 220–231 of the wild-type ApoA-I are required for lipid efflux in vitro and high density lipoprotein formation in vivo. J Biol Chem 278, 6719–6730 (2003). 10.1074/jbc.M205232200 [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald ML et al. Naturally occurring mutations in the largest extracellular loops of ABCA1 can disrupt its direct interaction with apolipoprotein A-I. J Biol Chem 277, 33178–33187 (2002). 10.1074/jbc.M204996200 [DOI] [PubMed] [Google Scholar]
- 25.Panagotopulos SE et al. The role of apolipoprotein A-I helix 10 in apolipoprotein-mediated cholesterol efflux via the ATP-binding cassette transporter ABCA1. J Biol Chem 277, 39477–39484 (2002). 10.1074/jbc.M207005200 [DOI] [PubMed] [Google Scholar]
- 26.Wang N, Silver DL, Costet P & Tall AR Specific binding of ApoA-I, enhanced cholesterol efflux, and altered plasma membrane morphology in cells expressing ABC1. J Biol Chem 275, 33053–33058 (2000). 10.1074/jbc.M005438200 [DOI] [PubMed] [Google Scholar]
- 27.Chambenoit O et al. Specific docking of apolipoprotein A-I at the cell surface requires a functional ABCA1 transporter. J Biol Chem 276, 9955–9960 (2001). 10.1074/jbc.M010265200 [DOI] [PubMed] [Google Scholar]
- 28.Frambach S et al. Brothers in Arms: ABCA1- and ABCG1-Mediated Cholesterol Efflux as Promising Targets in Cardiovascular Disease Treatment. Pharmacol Rev 72, 152–190 (2020). 10.1124/pr.119.017897 [DOI] [PubMed] [Google Scholar]
- 29.Tall AR, Yvan-Charvet L, Terasaka N, Pagler T & Wang N HDL, ABC transporters, and cholesterol efflux: implications for the treatment of atherosclerosis. Cell Metab 7, 365–375 (2008). 10.1016/j.cmet.2008.03.001 [DOI] [PubMed] [Google Scholar]
- 30.Oram JF & Vaughan AM ATP-Binding cassette cholesterol transporters and cardiovascular disease. Circ Res 99, 1031–1043 (2006). 10.1161/01.RES.0000250171.54048.5c [DOI] [PubMed] [Google Scholar]
- 31.Lawn RM et al. The Tangier disease gene product ABC1 controls the cellular apolipoprotein-mediated lipid removal pathway. J Clin Invest 104, R25–31 (1999). 10.1172/JCI8119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fitz NF et al. Abca1 deficiency affects Alzheimer’s disease-like phenotype in human ApoE4 but not in ApoE3-targeted replacement mice. J Neurosci 32, 13125–13136 (2012). 10.1523/JNEUROSCI.1937-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tietjen I et al. Increased risk of coronary artery disease in Caucasians with extremely low HDL cholesterol due to mutations in ABCA1, APOA1, and LCAT. Biochim Biophys Acta 1821, 416–424 (2012). 10.1016/j.bbalip.2011.08.006 [DOI] [PubMed] [Google Scholar]
- 34.Qian HZX,; Cao P.; Lei J; Yan N; Gong X. Structure of the Human Lipid Exporter ABCA1. Cell 169, 1228–1239 (2017). 10.1016/j.cell.2017.05.020 [DOI] [PubMed] [Google Scholar]
- 35.Seddon AM, Curnow P & Booth PJ Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta 1666, 105–117 (2004). 10.1016/j.bbamem.2004.04.011 [DOI] [PubMed] [Google Scholar]
- 36.Neumann J, Rose-Sperling D & Hellmich UA Diverse relations between ABC transporters and lipids: An overview. Biochim Biophys Acta 1859, 605–618 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Tanaka AR et al. Effects of mutations of ABCA1 in the first extracellular domain on subcellular trafficking and ATP binding/hydrolysis. J Biol Chem 278, 8815–8819 (2003). 10.1074/jbc.M206885200 [DOI] [PubMed] [Google Scholar]
- 38.Duong PT et al. Characterization of nascent HDL particles and microparticles formed by ABCA1-mediated efflux of cellular lipids to apoA-I. J Lipid Res 47, 832–843 (2006). 10.1194/jlr.M500531-JLR200 [DOI] [PubMed] [Google Scholar]
- 39.Sankaranarayanan S et al. A sensitive assay for ABCA1-mediated cholesterol efflux using BODIPY-cholesterol. J Lipid Res 52, 2332–2340 (2011). 10.1194/jlr.D018051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marheinke K, Grunewald S, Christie W & Reilander H Lipid composition of Spodoptera frugiperda (Sf9) and Trichopusia ni (Tn) insect cells used for baculovirus infection. FEBS Lett 441, 49–52 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Kowarz E, Loscher D & Marschalek R Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol J 10, 647–653 (2015). 10.1002/biot.201400821 [DOI] [PubMed] [Google Scholar]
- 42.Mukherjee S, Zha X, Tabas I & Maxfield FR Cholesterol Distribution in Living Cells: Fluorescence Imaging Using Dehydroergosterol as a Fluorescent Cholesterol Analog. Biophysical Journal 75, 1915–1925 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishigami M et al. Temporary sequestration of cholesterol and phosphatidylcholine within extracellular domains of ABCA1 during nascent HDL generation. Sci Rep 8, 6170 (2018). 10.1038/s41598-018-24428-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quazi F & Molday RS Differential phospholipid substrates and directional transport by ATP-binding cassette proteins ABCA1, ABCA7, and ABCA4 and disease-causing mutants. J Biol Chem 288, 34414–34426 (2013). 10.1074/jbc.M113.508812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ritchie TK et al. in Liposomes, Part F Methods in Enzymology 211–231 (2009). [Google Scholar]
- 46.Mi W et al. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 (2017). 10.1038/nature23649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orlando BJ & Liao M ABCG2 transports anticancer drugs via a closed-to-open switch. Nat Commun 11, 2264 (2020). 10.1038/s41467-020-16155-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gimenez-Andres M, Copic A & Antonny B The Many Faces of Amphipathic Helices. Biomolecules 8 (2018). 10.3390/biom8030045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phillips MC Is ABCA1 a lipid transfer protein? J Lipid Res 59, 749–763 (2018). 10.1194/jlr.R082313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sunidhi S, Sacher S, Atul, Garg P, Ray A Elucidating the Structural Features of ABCA1 in its Heterogeneous Membrane Environment. Frontiers in Molecular Biosciences 8, Article 803078 (2022). 10.3389/fmolb.2021.803078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Segrest JP et al. ABCA1 is an extracellular phospholipid translocase. Nat Commun 13, 4812 (2022). 10.1038/s41467-022-32437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Molday RS, Garces FA, Scortecci JF & Molday LL Structure and function of ABCA4 and its role in the visual cycle and Stargardt macular degeneration. Prog Retin Eye Res, 101036 (2021). 10.1016/j.preteyeres.2021.101036 [DOI] [PubMed] [Google Scholar]
- 53.Xie T, Zhang Z, Fang Q, Du B & Gong X Structural basis of substrate recognition and translocation by human ABCA4. Nat Commun 12, 3853 (2021). 10.1038/s41467-021-24194-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu F, Lee J & Chen J Molecular structures of the eukaryotic retinal importer ABCA4. Elife 10 (2021). 10.7554/eLife.63524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scortecci JF et al. Cryo-EM structures of the ABCA4 importer reveal mechanisms underlying substrate binding and Stargardt disease. Nat Commun 12, 5902 (2021). 10.1038/s41467-021-26161-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma S et al. Mechanism of LolCDE as a molecular extruder of bacterial triacylated lipoproteins. Nat Commun 12, 4687 (2021). 10.1038/s41467-021-24965-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le LTM et al. Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms. EMBO J 42, e111065 (2023). 10.15252/embj.2022111065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Landry YD et al. ATP-binding cassette transporter A1 expression disrupts raft membrane microdomains through its ATPase-related functions. J Biol Chem 281, 36091–36101 (2006). 10.1074/jbc.M602247200 [DOI] [PubMed] [Google Scholar]
- 59.Gulshan K et al. PI(4,5)P2 Is Translocated by ABCA1 to the Cell Surface Where It Mediates Apolipoprotein A1 Binding and Nascent HDL Assembly. Circ Res 119, 827–838 (2016). 10.1161/CIRCRESAHA.116.308856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hozoji M, Kimura Y, Kioka N & Ueda K Formation of two intramolecular disulfide bonds is necessary for ApoA-I-dependent cholesterol efflux mediated by ABCA1. J Biol Chem 284, 11293–11300 (2009). 10.1074/jbc.M900580200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Y, Li X Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states. Nature Cardiovascular Research 1, 238–245 (2022). 10.1038/s44161-022-00022-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spector AA, Yorek MA Membrane lipid composition and cellular function. Journal of Lipid Research 26, 1015–1035 (1985). [PubMed] [Google Scholar]
- 63.Gomez-Llobregat J, Elias-Wolff F & Linden M Anisotropic Membrane Curvature Sensing by Amphipathic Peptides. Biophys J 110, 197–204 (2016). 10.1016/j.bpj.2015.11.3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cui H, Lyman E & Voth GA Mechanism of membrane curvature sensing by amphipathic helix containing proteins. Biophys J 100, 1271–1279 (2011). 10.1016/j.bpj.2011.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagata KO, Nakada C, Kasai RS, Kusumi A & Ueda K ABCA1 dimer-monomer interconversion during HDL generation revealed by single-molecule imaging. Proc Natl Acad Sci U S A 110, 5034–5039 (2013). 10.1073/pnas.1220703110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hattori M, Hibbs RE & Gouaux E A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure 20, 1293–1299 (2012). 10.1016/j.str.2012.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guixa-Gonzalez R et al. MEMBPLUGIN: studying membrane complexity in VMD. Bioinformatics 30, 1478–1480 (2014). 10.1093/bioinformatics/btu037 [DOI] [PubMed] [Google Scholar]
- 68.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7 (2018). 10.7554/eLife.42166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Humphrey W, Dalke A & Schulten K VMD - Visual Molecular Dynamics. J. Molec. Graphics 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 70.Sievers F et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539 (2011). 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic model for the major ABCA1 conformation has been deposited in the PDB with the accession code 7TDT. EM maps for the major and minor ABCA1 conformations have been deposited in the EMDB with the accession codes: 25838 and 27079 respectively. The authors declare that all other data supporting the findings of this study are available within the paper and its supplementary information files; source data are provided with this paper.


