ABSTRACT
Immune checkpoint inhibitors in combination with tyrosine kinase inhibitors (TKIs) are improving the response rates of advanced renal cancer patients. However, many treated patients do not respond, making novel immune checkpoint-based immunotherapies potentially clinically beneficial only for specific groups of patients. We detected high expression of the immune checkpoint protein B7-H3 in clear cell renal cell carcinomas (ccRCCs) and evaluated B7-H3 immunohistochemistry staining in tissue microarray samples from two distinct renal cancer cohorts. B7-H3 was highly expressed in approximately 50% of primary tumors and in 30% of metastatic lesions. B7-H3 expression in primary tumors correlated with tumor necrosis, sarcomatoid transformation, disease-free survival, and synchronous metastasis, while B7-H3 expression in metastasis correlated with metastases to the lymph nodes. Gene expression analysis revealed the association of B7-H3 expression with gene expression scores involved in T cell exhaustion and myeloid immune evasion, as well as with PI3K/AKT and JAK/STAT pathways. Furthermore, knocking down B7-H3 expression in renal cancer cells by siRNA and CRISPR/Cas resulted in lower 2D and 3D cell proliferation and viability as well as increased sensitivity to TKI axitinib. Together, our findings suggest a pro-oncogenic and immune evasive role for B7-H3 in ccRCC and highlight B7-H3 as an actionable novel immune checkpoint protein in combination with TKI in advanced renal cancer.
KEYWORDS: Clear cell renal cell carcinoma (ccRCC), immune checkpoint protein B7-H3 (CD276), renal cancer
Introduction
Human renal cancers, including clear cell renal cell carcinoma (ccRCC), chromophobe renal cell carcinomas (ChRCCs), and papillary renal cell carcinomas (PRCCs), are of relatively high incidence in the population.1 Immunotherapy using immune checkpoint inhibitory monoclonal antibodies is an approach that has increased the survival of renal cancer patients over the past decade. The therapeutic landscape in renal cancer is a field in progress, with several clinical trials testing immune checkpoint inhibitors (CPIs) targeting PD-1/PD-L1 in combination with targeted therapies, and there is still an urgent need for novel and efficient anti-cancer targets. In this context, deeper biological and clinical knowledge of alternative immune checkpoint proteins might be exploited in the therapeutic landscape of renal cancer.2
Renal cancer is considered an immunogenic cancer form and it shows relative high response rates upon treatment with CPIs. However, this response has not been linked to either the accumulation of neo-antigens no tumor mutational burden.3,4 Current approved CPIs in the treatment of advanced RCC include CPIs blocking the protein–protein binding between PD-1 and its ligands PD-L1 (B7-H1) and PD-L2 (B7-DC) (anti-PD-1 (nivolumab and pembrolizumab) and anti-PD-L1 (avelumab)) and between CTLA-4 and its ligands B7-1, B7-2 and B7-H2 (anti-CTLA-4 ipilimumab).5 The CPIs are approved in combination (nivolumab and ipilimumab) or combined with tyrosine kinase inhibitors (TKI) (axitinib and lenvantinib) or with mTOR inhibitors (everolimus and temsirolimus).6 The response rate of CPI-alone treated patients is around 25–40%,7,8 while the combination with TKIs increases the response rate to 50–60%.9–12 Despite these improvements, still around 40% of patients experience little benefit, high toxicity, or develop resistance to therapy, emphasizing the need for finding novel and effective treatments alternative to the current PD-1/PD-L1 and CTLA-4 based therapies.13 Synergistic or compensatory functions from other related co-immunoregulatory B7-family proteins could account for the non-durable benefits obtained from current CPIs.
B7-family proteins consist of 10 structurally related proteins with immune-regulatory and pro-oncogenic functions,14 including several good candidates for therapeutic targeting and as biomarkers in ccRCC.15–18 B7-H3 is considered a pan-cancer antigen and it is highly overexpressed in many cancers, including renal cancer.14,19 B7-H3 has been proposed as a renal cancer marker17 and as an enhancer of cell invasion and metastasis in renal cancer.20 Overexpression of B7-H3 in monocytes is associated with renal cell carcinoma progression.21 In ccRCC, lower microRNA-187 expression is associated with lower patient survival, and this mRNA inhibits cell growth and migration in association with regulating B7-H3.22 Expression of B7-H3 in tumor and tumor vasculature is associated with higher grade and poorer survival outcomes in patients with ccRCC and metastatic ccRCC.23–27 B7-H3 targeting of renal cancers has been validated in preclinical studies using B7-H3 monoclonal and drug-conjugated antibodies,28,29 and it is currently in clinical trials as a novel immune checkpoint inhibitor for renal cancer treatment.
Here, we have performed molecular studies on the role of B7-H3 in ccRCC using renal cells and ccRCC tumor samples. Our results unveil high expression of B7-H3 mRNA and protein in ccRCC, in correlation with poor prognosis, synchronous and lymph node metastasis, T cell exhaustion and myeloid immune evasion pathways. We propose a pro-oncogenic and metastatic role for B7-H3 in ccRCC modulated through JAK/STAT and PI3K/AKT signaling pathways.
Material and methods
mRNA expression analysis
In silico mRNA expression analysis of B7-family genes: normal kidney tissue expression was from publicly available data sets at NCBI Gene Resource30 (https://www.ncbi.nlm.nih.gov/gene/), and ccRCC expression was from the publicly available TCGA dataset retrieved from the Protein Atlas31 (https://www.proteinatlas.org/). Validation of B7-H3 (CD276) co-expression analysis was retrieved from the ccRCC dataset from TCGA (mRNA expression (log2); RNA Seq V2 RSEM (417 samples)32 using cBioportal (https://www.cbioportal.org/); and GEPIA (http://gepia.cancer-pku.cn).33
Immunohistochemical staining and scoring
The anti-B7-H3 antibody used for immunohistochemistry was AF1027 (1:1000, R&D). Antigen retrieval was performed by PT link (Agilent Technologies, Santa Clara, CA). Immunostaining of B7-H3 was performed with EnVision FLEX and Dako Autostainer Link 48 (Agilent) as previously described.34 The B7-H3 antibody was incubated for 30 min, followed by secondary antibody incubation for 15 min using secondary polyclonal rabbit anti-goat Ig/HRP (1:200; in EnVision FLEX Antibody diluent Dako), EnVision FLEX/HPR for 20 min, and EnVision FLEX DAB/Sub Chromo for 10 min and finally counterstaining with EnVision FLEX hematoxylin. Slides were dehydrated through incubations with sequentially increasing alcohol concentration, followed by xylene incubation and cover-slipping. TMAs were evaluated by an experienced uropathologist (JIL). Scoring of B7-H3 immunostaining was made following the immunoreactivity scoring system (IRS). Immunostaining for the tumor cells: low/no: <10% positive cells and 0–1 intensity; and high: >10% positive cells and 2–4 moderate to strong intensity.
Clinical samples
Two separate cohorts with renal cancers and with metastatic ccRCCs were analyzed, consisting of 129 renal cancer patients and 52 metastatic ccRCC patients, respectively, treated with surgery at the Cruces University Hospital (Spain) between 1997 and 2001. The first cohort included 96 ccRCC, 17 ChRCC, and 16 PRCC. The second metastatic ccRCC cohort included primary and matching metastasis tissue samples. 29% of the patients had synchronous metastasis at the time of surgery, while the rest developed metastasis in a time range between 6 and 204 months. The median diameter of the tumors in the first cohort was 6.8 mm and in the second cohort was 8.5 mm. An experienced pathologist (JIL) selected tumor areas with well-preserved tissue representative of the whole tumor from formalin-fixed and paraffin-embedded tissue blocks from these patients, and TMA blocks were made from these areas. 4 µm sections were made from the TMA blocks, one of which was stained with hematoxylin and eosin to verify the presence of tumor content. Both renal cancer cohorts have been previously described.35,36 Ethical approvals have been obtained for clinical material (CEIC 2015/060, CEIC PI2022116, and CEIC PI2022085).
Gene expression analysis
Total RNA was isolated from twelve 10 μM-thick ccRCC whole tissue sections from the ccRCCs that were selected from 6 whole tumor tissue sections with high B7-H3 immunostaining and from 6 whole tumor tissue sections with low B7-H3 immunostaining. The adjacent 4 -μM whole tissue section was hematoxylin- and eosin-stained, and tumor areas were marked by an uropathologist (JIL). Tumor tissue was scraped off and deparaffinized with deparaffinization solution (Qiagen), and RNA was isolated with an AllPrep DNA/RNA FFPE kit (Qiagen). RNA concentration and quality were determined by NanoDrop (Thermo Scientific) and BioAnalyzer (Agilent), respectively. Gene expression was analyzed on the Immune Exhaustion Panel and PanCancer Pathways Panel (NanoString Technologies, covering 785 genes involved in immune exhaustion (previously described by Nunes-Xavier et al.37) and 770 genes involved in canonical cancer pathways). Analysis and quality check were performed at the Genomics Core Facility at the Oslo University Hospital (Norway).
Cell lines, transfection and luciferase assay
The 786-O human renal cancer cell line and nonmalignant RC-124 renal cells were grown in RPMI, and A498 and Caki-1 human renal cancer cell lines were grown in EMEM and McCoy-5A medium, respectively. HEK293 cell line (human embryonic kidney) was cultured in DMEM (Dulbecco’s Modified Eagle’s Medium). All media were supplemented with 10% FBS, 1% L-glutamine and 1% penicillin–streptomycin. PMA (phorbol 12-myristate 13-acetate, Sigma-Aldrich). Plasmids were pcDNA6 B7-H3 cDNA38 and pSG5 HA-AKT1.39 The GenJet™ DNA In Vitro Transfection Reagent (SignaGen Laboratories) protocol was used to transfect HEK293 cells, as described previously,40 with pcDNA6 B7-H3, and pSG5 HA-AKT1 cDNA, and STAT3 Cignal Reporter Assay (CCS-9028 L, Qiagen). STAT3 Reporter Assay was measured using the Dual-Luciferase® Reporter Assay System (Promega), and the luminescence signal was measured immediately after the addition of the substrate solution using an Infinite® M plex (TECAN) plate reader.
RNA interference and CRISPR/Cas
B7-H3 knockdown in Caki-1 and 786-O cells was performed using the transfection reagents Lipofectamine RNAiMAX (Invitrogen) and PepMute (SignaGen Laboratories) according to the manufacturer protocol. siRNAs for the human B7-H3 gene were from FlexiTube GeneSolution (GeneGlobe ID: SI03113341 and SI02645944; Qiagen), and nonspecific (siNS) RNAs were from AllStars Neg. siRNA (Qiagen). Cells were transfected with 20 nM siRNAs. For CRISPR/Cas knockout experiments, 786-O cells were transfected with the p01 U6-gRNA:CMV-Cas9-2a-tGFP plasmid using Lipofectamine 2000 (Thermo Fisher Scientific). 48 h post transfection, cells were GFP-sorted, using a FACS Aria II cell sorter (BD Biosciences) at the Flow Cytometry Core Facility at the Oslo University Hospital (Norway). CRISPR/Cas knockout was verified by DNA sequencing at the Genetics-Genomics Core Facility at the Biobizkaia Health Research Institute (Spain).
Western blot
Total cell lysates were obtained lysing cells in the M-PER mammalian protein extraction reagent (Thermo Fisher Scientific) supplemented with phosphatase and protease inhibitor cocktails (PhosSTOP and cOmplete; Roche). Immunoblot analysis of B7-H3 (anti-B7-H3 AF1027, 1:500, R&D systems), pAKT (anti-pAKT #9271, 1:1000, Cell Signaling), α-tubulin (anti-α-tubulin DM1A, 1:5000, Merck Millipore), and GAPDH (anti-GAPDH 6C5, sc-32233, 1:500, Santa Cruz Biotechnology) was performed in 10% SDS-PAGE gels as previously described.41
2D and 3D cell proliferation and viability assay
Caki-1 and 786-O cells were plated in 96 well plates (Corning), transfected with siRNAs, and treated the following day with the TKI axitinib (10 µM, Selleck Chemicals). For 3D-spheroid viability, cells were plated in low-attachment 96 well plates (Corning). Cells were treated for 72 h before cell proliferation was measured using a CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega) for 2D cell growth and a CellTiter-Glo® 3D Cell Viability Assay (Promega) for 3D-spheroid viability according to manufacturer’s protocols. Absorbance was measured for 2D assays at 490 nm using a Mark™ Microplate Absorbance Reader (Bio-Rad), and luminescent signals in 3D-spheroid assays were measured using the Infinite® M plex (TECAN) plate reader. 2D and 3D data are represented as the mean from at least two biological replicates, each one with at least three technical replicates. Statistically significant results (p < 0.05) are marked with an asterisk.
Statistical analysis
Statistical analyses were performed using the statistical software SPSS Statistics (IBM, version 23). Spearman’s rho (R) test was used to correlate B7-H3 expression with clinicopathologic parameters. The estimated survival curves were compared using the log-rank test. Univariate and multivariate Cox correlation analyses were used to test the independent effects of variables of interest on survival. A stepwise variable selection for a Cox proportional hazards predictive model was used with an entry criterion of p = 0.05 and a stay value of p = 0.20. All statistical analyses were performed using patients with non-missing values. Viability analysis and gene expression analysis in box plot were calculated using the Graphpad Prism t test calculator, and significance was calculated using a two-tailed student t-test. In the viability analysis, significant viability differences are indicated with an asterisk (*), and error bars represent standard deviation (S.D.). All experiments were performed at least twice, and the results shown are from one representative experiment. A two-sided p value of less than 0.05 was considered significant. Gene expression analyses was made with nSolver Analysis Software 4.0 (nanoString technologies), R (3.3.2), and XQuartz (2.8.1) using the code set name NS_HS_EXHAUSTION_V1.0. Pathway scores, cell type score, and differential expression were performed with nCounter Advanced Analysis (version 2.0.134). Volcano plot analysis was performed with the p-value adjusted to Benjamini-Yekutieli.
Results
Β7-Η3 is highly expressed in renal cancer and correlates with poor outcome in ccRCC
In silico analysis illustrates that B7-H3 is amongst the most abundant B7-family immune checkpoint gene expressed, together with B7-H5 and B7-H7, in ccRCC relative to normal kidney expression (Figure 1a). TCGA data showed statistically higher expression of B7-H3 in ccRCC/KIRC and PRCC/KIRP but not in ChRCC/KICH (Figure 1b). IHC analysis of B7-H3 expression in human renal tumor samples revealed the presence of B7-H3 on tumor and tumor-associated stroma, with a cytosolic/membranous staining. Representative examples of B7-H3 low and high expression in ccRCC are shown in Figure 2a. In a first renal cohort consisting of 96 ccRCC, 17 ChRCC, and 16 PRCC specimens, we explored the correlations between B7-H3 expression, renal cancer subtype, and pathological parameters. In our ccRCC cohort, B7-H3 immunostaining correlated positively with sex (p = 0.038), stage (p = 0.041), diameter (p = 0.012) and overall survival (OS) (p = 0.009). In ChRCCs, B7-H3 expression was only observed in 1 case, which correlated with the stage (p = 0.025) and OS (p = 0.002). In PRCC, B7-H3 expression did not correlate with any clinico-pathological variables (Table 1). Kaplan–Meier plots show the significant association between B7-H3 expression and decreased OS in ccRCC (p = 0.044) (Figure 2b), but not in PRCC (p = 0.863) (Supplementary Figure S1) . Cox univariate and multivariate regression analyses showed that stage (p = 0.019) and grade (p = 0.031) were independent prognostic parameters for the survival of this cohort, but not B7-H3 expression (Table 2).
Figure 1.
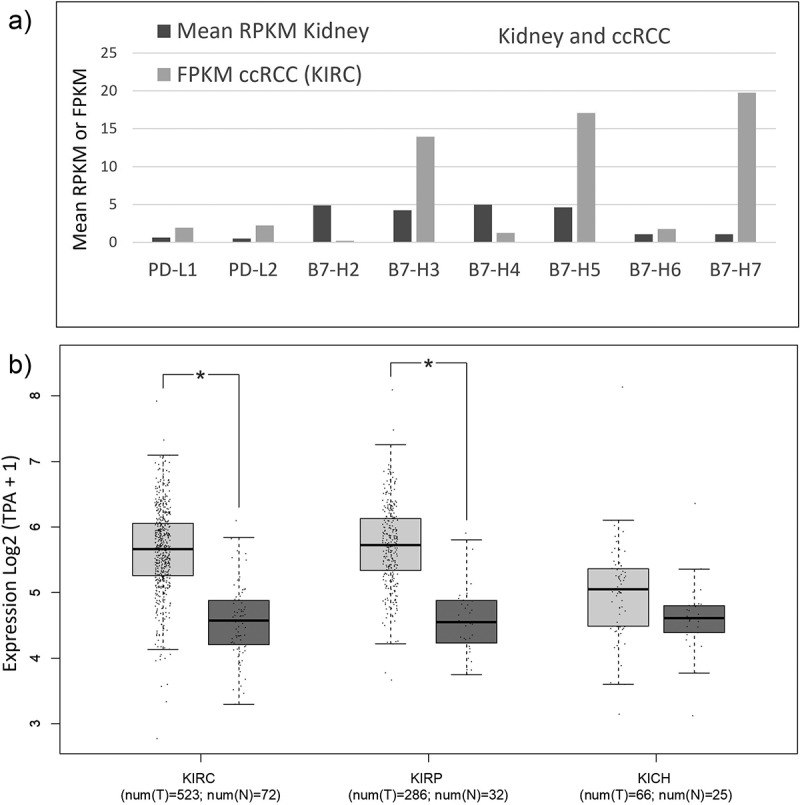
Expression of B7 family genes in normal kidney and kidney cancer.
a) B7 mRNA expression in kidney cells and ccRCCs. Mean mRNA expression of B7 family genes in normal kidney tissue and ccRCC (from KIRC study), as retrieved from https://www.ncbi.nlm.nih.gov/gene/ and https://www.proteinatlas.org/. RNA sequencing data are reported as median expression. RPKM: reads per kilo base per million mapped reads; FPKM: number fragments per kilobase of exon per million reads. Official gene names; PD-L1: CD274, PD-L2: PDCD1LG2, B7-H2: ICOSLG, B7-H3: CD276, B7-H4: VTCN1, B7-H5: VSIR, B7-H6: NCR3LG1, and B7-H7: HHLA2. b) mRNA expression of B7-H3 in normal kidney and kidney cancer. Box plots for B7-H3 in TCGA data sets: ccRCC/KIRC (n = 523) and normal kidney tissue (n = 72), kidney renal clear cell carcinoma; PRCC/KIRP (n = 286) and normal kidney tissue (n = 32), kidney renal papillary cell carcinoma; and chRCC/KICH, kidney chromophobe (n = 66) and normal kidney tissue (n = 25). Plots show mRNA expression in tumor tissue (shown in light gray) in comparison to normal kidney tissue (in dark gray). Data are represented in a logarithmic scale (Log2) and obtained from tumor samples and normal tissue samples. Statistically significant results (p < 0.01) are marked with an asterisk.
Figure 2.
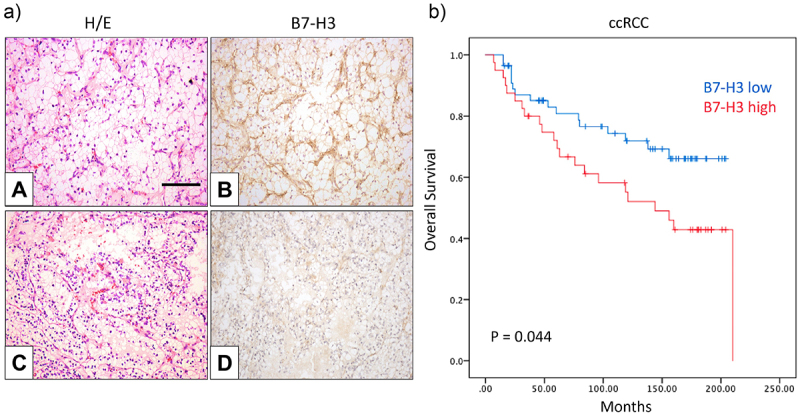
B7-H3 protein expression and Kaplan–Meier survival curves of ccRCC patients.
a) Expression of B7-H3 in ccRCC specimens. Representative immunohistochemical staining of B7-H3 expression in positive (B) and low/no (D) specimens. Hematoxylin and eosin staining (A and C). Magnification in all pictures X250; scale bar represents 100 μm. b) Overall survival of ccRCC patients related to B7-H3 expression. Kaplan–Meier curves for B7-H3 expression and overall survival in ccRCC (p = 0.044).
Table 1.
Correlation between clinical and pathological variables and B7-H3 protein expression in renal cancers.
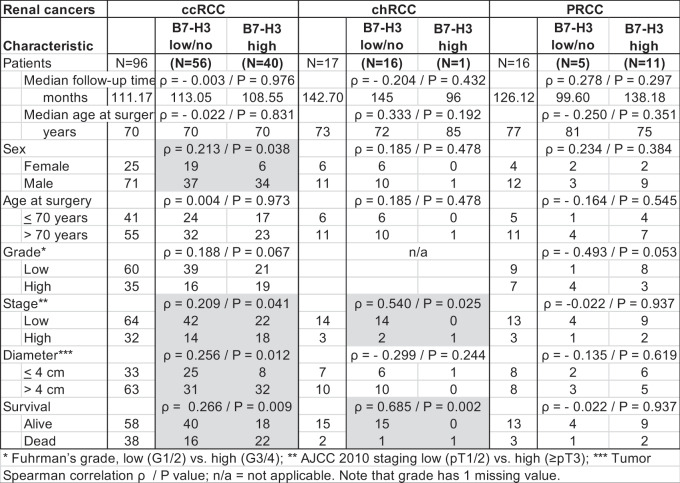 |
Table 2.
Univariate and Multivariate analyses to predict overall survival of ccRCC patients.
| Univariate Analysis ccRCC | |||||
|---|---|---|---|---|---|
| Variable |
Description |
Point Estimate |
95% Wald Confidence Limits |
P-value Log-rank |
|
| Grade | Low vs high | 5.929 | 2.934 | 11.980 | <0.0001 |
| Stage | Low vs high | 6.632 | 3.309 | 13.293 | <0.0001 |
| Diameter | <4 cm >4 cm |
4.528 | 1.760 | 11.654 | 0.002 |
| B7-H3 |
Low/no vs high |
1.927 |
1.005 |
3.695 |
0.048 |
| Multivariate Analysis | |||||
| Variable |
Description |
Point Estimate |
95% Wald Confidence Limits |
P-value Cox |
|
| Stage | Low vs high | 2.930 | 1.191 | 7.204 | 0.019 |
| Grade | Low vs high | 2.607 | 1.091 | 6.226 | 0.031 |
| Diameter | <4 cm >4 cm |
1.639 | 0.551 | 4.881 | 0.375 |
| B7-H3 | Low/no vs high | 1.482 | 0.765 | 2.872 | 0.261 |
Univariate and multivariate Cox proportional hazards test/point estimate = hazard ratio.
Β7-Η3 expression associates with synchronous metastases and metastases to lymph nodes in ccRCC
In a second cohort of 52 metastatic ccRCC cases, we analyzed B7-H3 expression in both primary and matching metastatic tumor samples (Table 3 and Figure 3a). Approximately half of primary tumors with metastasis expressed B7-H3 at the primary site, whereas 30% of the metastatic tumors were positive for B7-H3 expression (Table 3). B7-H3 expression in primary tumors correlated positively with higher histological grade (p = 0.026), tumor necrosis (p = 0.009), sarcomatoid transformation (p = 0.021), and synchronous metastasis (p = 0.039) (Table 3), while B7-H3 expression at the metastatic lesion correlated with metastases to the lymph nodes (p = 0.009) (Table 3). Kaplan–Meier plots were performed to study the 10-year overall survival of metastatic ccRCC patients. The curves showed a significant correlation between B7-H3 and overall survival in ten years (p = 0.020) (Figure 3b). Cox regression univariate and multivariate analysis, including statistically significant variables and variables previously shown to be clinically relevant, revealed that the temporal development of metastasis was the only independent prognostic parameter for the ten-year survival of patients in this cohort (p = 0.002) (Table 4). The sarcomatoid transformation variable was excluded from the analysis due to the lack of effect in the predictive model, and B7-H3 expression (p = 0.140) and tumor necrosis (p = 0.127) were shown as determinant factors for overall survival prognosis (Table 4).
Table 3.
Correlation between clinical and pathological variables and B7-H3 protein expression in primary tumor and metastatic lesion in ccRCC.
|
Metastatic ccRCC |
Primary Tumor |
|
Metastatic Lesion |
|
|||
| Characteristic | B7-H3 low/no | B7-H3 high | B7-H3 low/no | B7-H3 high | |||
| Patients |
|
N=52 |
(N=29) |
(N=23) |
N=52 |
(N=37) |
(N=15) |
| Median follow-up time | ρ = -0.320 / p = 0.021 | ρ = -0.030 / p = 0.834 | |||||
| months | 47.5 | 78 | 31.5 | 47.5 | 59.5 | 36 | |
| Median age at surgery | ρ = -0.150 / p = 0.289 | ρ = 0.140 / p = 0.322 | |||||
| years | 59 | 61 | 55 | 59 | 55.5 | 61 | |
| Sex | ρ = -0.112 / p = 0.430 | ρ = 0.074 / p = 0.604 | |||||
| Female | 13 | 6 | 7 | 13 | 10 | 3 | |
| Male | 39 | 23 | 16 | 39 | 27 | 12 | |
| Age at surgery | ρ = -0.086 / p = 0.547 | ρ = -0.102 / p = 0.476 | |||||
| < 70 years | 45 | 24 | 21 | 44 | 30 | 14 | |
| > 70 years | 6 | 4 | 2 | 6 | 5 | 1 | |
| Grade* | ρ = 0.312 / p = 0.026 | ρ = 0.215 / p = 0.130 | |||||
| Low | 22 | 16 | 6 | 22 | 18 | 4 | |
| High | 29 | 12 | 17 | 29 | 18 | 11 | |
| Stage** | ρ = 0.151 / p = 0.287 | ρ = 0.067 / p = 0.637 | |||||
| Low | 27 | 17 | 10 | 27 | 20 | 7 | |
| High | 25 | 12 | 13 | 25 | 17 | 8 | |
| Diameter*** | ρ = 0.124 / p = 0.380 | ρ = -0.122 / p = 0.389 | |||||
| < 4 cm | 7 | 5 | 2 | 7 | 4 | 3 | |
| > 4 cm | 45 | 24 | 21 | 45 | 33 | 12 | |
| Tumor necrosis | ρ = 0.358 / p = 0.009 | ρ = 0.164 / p = 0.246 | |||||
| No | 24 | 18 | 6 | 24 | 19 | 5 | |
| Yes | 28 | 11 | 17 | 28 | 18 | 10 | |
| Sarcomatoid transformation | ρ = 0.322 / p = 0.021 | ρ = - 0.016 / p = 0.911 | |||||
| No | 47 | 28 | 19 | 47 | 34 | 13 | |
| Yes | 4 | 0 | 4 | 4 | 3 | 1 | |
| Disease free survival | ρ = 0.272 / p = 0.051 | ρ = - 0.122 / p = 0.389 | |||||
| Yes | 11 | 9 | 2 | 11 | 9 | 2 | |
| No | 41 | 20 | 21 | 41 | 28 | 13 | |
| Survival | ρ = 0.78 / p = 0.581 | ρ = 0.017 / p = 0.904 | |||||
| Alive | 18 | 11 | 7 | 18 | 13 | 5 | |
| Dead | 34 | 18 | 16 | 34 | 24 | 10 | |
| Metastasis | ρ = 0.288 / p = 0.039 | ρ = 0.031 / p = 0.829 | |||||
| Metachronous | 37 | 24 | 13 | 37 | 26 | 11 | |
| Synchronous | 15 | 5 | 10 | 15 | 11 | 4 | |
| Metastatic site | |||||||
| Lymph node | ρ = 0.064 / p = 0.654 | ρ = 0.356 / p = 0.009 | |||||
| No | 40 | 23 | 17 | 40 | 32 | 8 | |
| Yes | 12 | 6 | 6 | 12 | 5 | 7 | |
| Endothelial tissue | ρ = - 0.194 / p = 0.169 | ρ = -0.212 / p = 0.131 | |||||
| No | 26 | 12 | 14 | 26 | 16 | 10 | |
| Yes | 26 | 17 | 9 | 26 | 21 | 5 | |
| Soft tissue and sarcoma | ρ = 0.158 / p = 0.264 | ρ = -0.099 / p = 0.483 | |||||
| No | 38 | 23 | 15 | 38 | 26 | 12 | |
| Yes | 14 | 6 | 8 | 14 | 11 | 3 | |
* Fuhrman’s grade, low (G1/2) vs. high (G3/4); ** AJCC 2010 staging low (pT1/2) vs. high (≥pT3); *** tumor diameter, small (≤4 cm) vs. large (>4 cm).
Spearman correlation ρ/p = p value. Note that age, grade, and sarcomatoid transformation have 1 missing value.
Figure 3.
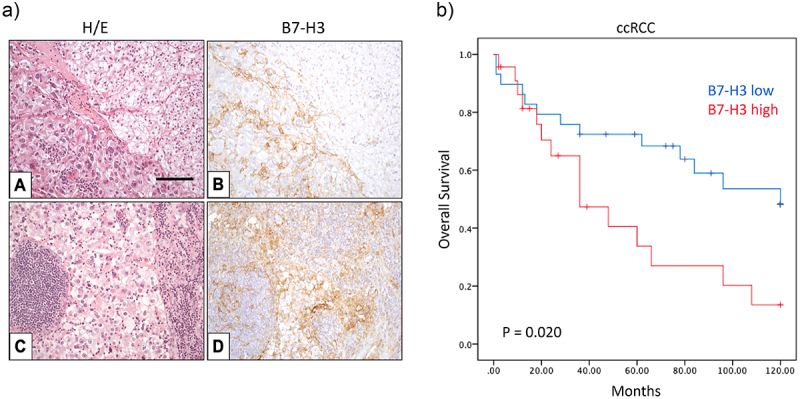
B7-H3 protein expression and Kaplan–Meier survival curves of metastatic ccRCC patients.
a) Expression of B7-H3 in ccRCC specimens. Representative immunohistochemical staining of B7-H3 expression in positive tumor site (B) and matching lymph node metastasis (D). Hematoxylin and eosin staining (A and C). Magnification in all pictures ×250; scale bar represents 100 μm. b) 10-year overall survival in metastatic ccRCC patients related to B7-H3 expression. Kaplan–Meier curves for B7-H3 expression and 10-year overall survival in metastatic ccRCC (p = 0.020).
Table 4.
Univariate and Multivariate analyses to predict overall survival of metastatic ccRCC patients.
| Univariate Analysis metastatic ccRCC | |||||
|---|---|---|---|---|---|
| Variable | Description | Point Estimate | 95% Wald Confidence Limits | P-value Log-rank | |
| Grade | Low vs high | 1.241 | 0.578 | 2.666 | 0.580 |
| Stage | Low vs high | 1.080 | 0.507 | 2.301 | 0.842 |
| Diameter | <4 cm > 4 cm |
1.359 | 0.410 | 4.503 | 0.616 |
| Tumor necrosis | No vs yes | 2.424 | 1.109 | 5.295 | 0.026 |
| Sarcomatoid transformation | No vs yes | 2.547 | 0.573 | 11.315 | 0.219 |
| Metastasis synchronicity | Metachronous vs Synchronous | 0.228 | 0.099 | 0.522 | <0.0005 |
| Metastatic site Lymph node | No vs yes | 1.291 | 0.520 | 3.207 | 0.582 |
| Metastatic site Endothelial tissue | No vs yes | 0.666 | 0.313 | 1.417 | 0.291 |
| Metastatic site Soft tissue, sarcoma | No vs yes | 1.337 | 0.600 | 2.978 | 0.478 |
| B7-H3 primary |
No vs yes |
2.373 |
1.113 |
5.060 |
0.025 |
|
Multivariate Analysis | |||||
|
Variable |
Description |
Point Estimate |
95% Wald Confidence Limits |
P-value Cox |
|
| Tumor necrosis | No vs yes | 1.876 | 0.791 | 4.451 | 0.154 |
| Sarcomatoid transformation | No vs yes | 0.459 | 0.092 | 2.300 | 0.344 |
| Metastasis synchronicity | Metachronous vs Synchronous | 0.217 | 0.084 | 0.563 | 0.002 |
| B7-H3 primary |
No vs yes |
2.205 |
0.949 |
5.124 |
0.066 |
|
Multivariate Analysis (excluding sarcomatoid transformation) | |||||
|
Variable |
Description |
Point Estimate |
95% Wald Confidence Limits |
P-value Cox |
|
| Tumor necrosis | No vs yes | 1.926 | 0.829 | 4.474 | 0.127 |
| Metastasis synchronicity |
Metachronous vs Synchronous | 0.270 | 0.111 | 0.658 | 0.004 |
| B7-H3 primary | No vs yes | 1.854 | 0.817 | 4.205 | 0.140 |
Univariate and multivariate stepwise Cox proportional hazards test/point estimate = hazard ratio.
Gene expression profiling of B7-H3 low and high in ccRCC
Next, we performed pilot experiments of molecular gene profiling analysis by the immune exhaustion pathway panel and pan cancer pathway panel (NanoString) from whole tissue sections from 12 ccRCC patient samples from the first cohort: 5 tumors with low B7-H3 expression and 7 with high B7-H3 expression. Gene expression of B7-H3 demonstrated good correlation between B7-H3 mRNA expression from whole tissue sections and the matching B7-H3 protein expression from TMA sections (Figure 4a). Differentially expressed genes from the immune exhaustion panel in B7-H3 high tumors as compared to the baseline B7-H3 low are shown in the volcano plot (Figure 4b), and significantly differentially expressed genes are listed in Supplementary Table S1. Genes showing significantly higher expression in B7-H3 high tumor samples were insulin receptor substrate 1 (IRS1, p = 0.0026), matrix metalloproteinase-14 (MMP14, p = 0.00882), colony stimulating factor 1 receptor (CSF1R, p = 0.0294), and janus activated kinase 3 (JAK3, p = 0.0315). Among the top lower expressed genes were glycerol kinase (GK, p = 0.013), aldehyde dehydrogenase 1 family member A1 (ALDH1A1, p = 0.0177), bone morphogenetic protein 2 (BMP2, p = 0.013) and hepatitis A virus cellular receptor 2 (HAVCR2, also known as T-cell immunoglobulin and mucin-domain containing-3 TIM3, p = 0.0382) (Figure 4c). All the above mentioned differentially expressed genes were validated and verified in the TCGA dataset containing 417 ccRCC samples (Figure 4d).32
Figure 4.
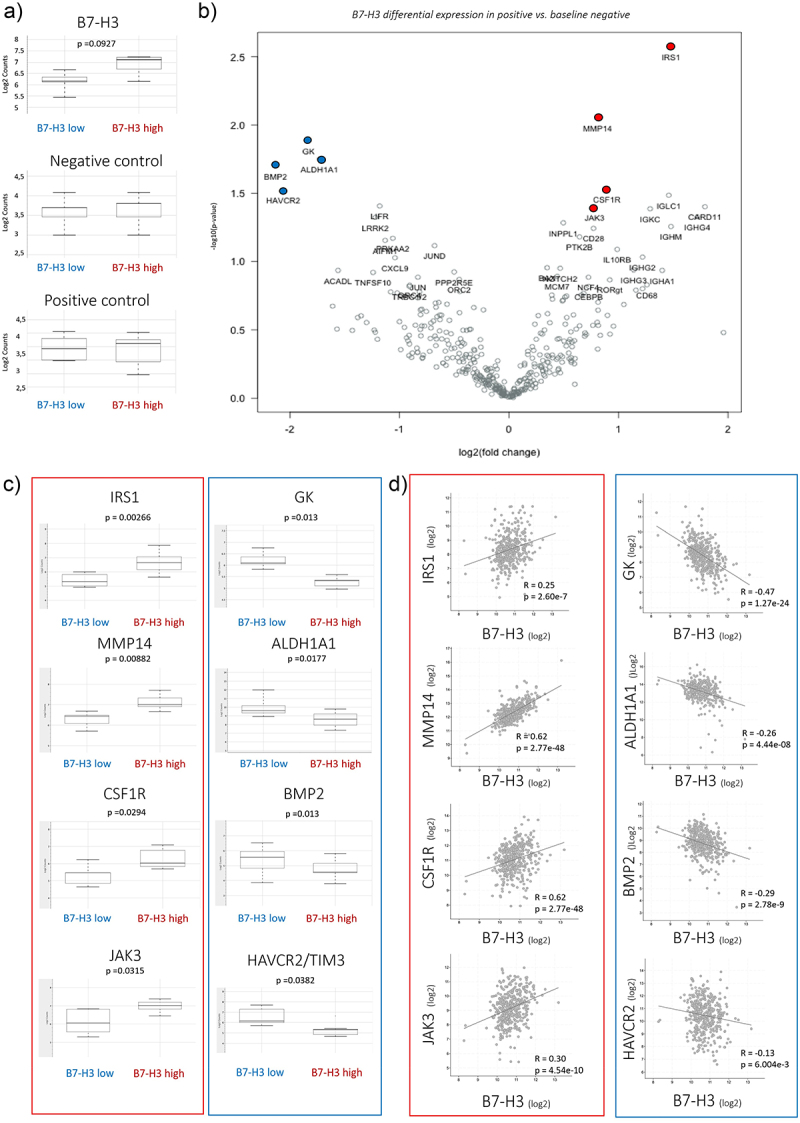
Gene expression of B7-H3 in a panel of ccRCC samples.
a) Gene expression of B7-H3 in a panel of ccRCC samples showed higher B7-H3 mRNA expression in IHC-scored B7-H3 high tumors vs. B7-H3 low tumors. No differences were observed in negative and positive controls. b) Volcano plot of B7-H3 differential expression in positive vs. baseline negative. Volcano plot displaying each gene’s −log10 (p-value) and log2 fold change with the selected covariate. The 40 most statistically significant genes are labeled in the plot. Genes also showed in Figure 4c are colored: top 4 higher expression in B7-H3 high tumor samples (IRS1, MMP14, CSF1R and JAK3) in red and top 4 lower in B7-H3 high tumor samples (GK, ALDH1A1, BMP2, and HAVCR2/TIM3) in blue. c) Gene expression of up- and down-regulated genes. Up-regulated genes in B7-H3 high tumors are shown in red box: IRS1, MMP14, CSF1R and JAK3; and downregulated genes in blue box: GK, ALDH1A1, BMP2 and HAVCR2/TIM3. Values are in log2 scale. d) B7-H3 co-expression analysis was identified from the ccRCC dataset from TCGA (Nature 2013; mRNA expression (log2); RNA Seq V2 RSEM; 417 samples) (32) using cBioportal (https://www.cbioportal.org/). R: Spearman, p values are included. All data are shown on log2.
B7-H3 expression associates with different immune populations and signaling pathways
Immune exhaustion pathway analysis unveiled differences in several immune cells and immune states (Figure 5a). Pathway and cell type score unveiled a higher immune exhausted score in the B7-H3 high tumors. In pathway score analysis, we observed that alterations in the T cell exhaustion (Figure 5b), myeloid immune evasion (Figure 5c), tumor necrosis factor (TNF) signaling (Figure 5d), interleukin-2 (IL-2) signaling (Figure 5e), and NK-κB signaling score (Figure 5f) are higher in B7-H3 high vs B7-H3 low samples. Furthermore, cell type analysis showed a higher macrophage score in B7-H3 high tumors (Figure 5g), indicating a positive correlation between B7-H3 expression and intratumor macrophage (TAM) infiltration.
Figure 5.
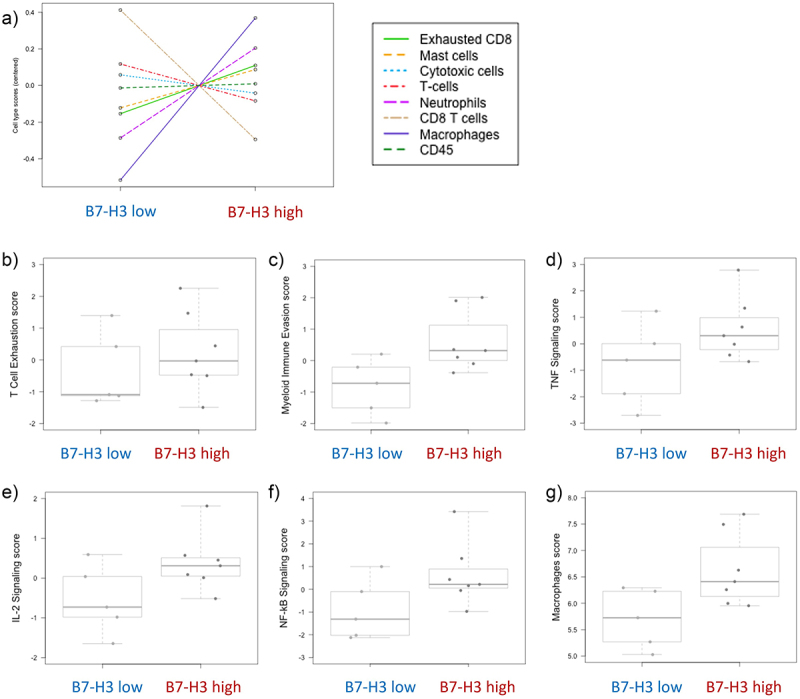
Molecular immune exhaustion panel analysis of ccRCC patients according to B7-H3 expression.
a) Relative cell type measurements vs. B7-H3. Plots show the relative cell type abundance measurements against B7-H3 expression. Pathway analysis of the different cell types: b) T cell exhaustion, c) myeloid immune evasion, d) TNF signaling, e) IL-2 signaling, f) NF-κΒ signaling, and g) macrophages scores. Data are represented in a logarithmic scale (Log2) and obtained from 12 ccRCC tumor samples. Plots pathway scores are represented against B7-H3 expression.
B7-H3 expression impacts PI3K/AKT and JAK/STAT signaling pathways
PanCancer pathway analysis showed differentially expressed signaling pathways in B7-H3 high and B7-H3 low samples (Figure 6a). In particular, higher pathway scores were observed in pathways JAK/STAT and PI3K signaling scores (Figure 6b,c). Next, we validated the B7-H3-related modulations in these pathways in renal cells. Due to low transfection efficiency of renal cancer cells, we validated B7-H3 implication in STAT3 and AKT activation by co-transfecting HEK293-immortalized human embryonic kidney cells. Co-transfection of B7-H3 with the STAT3 reporter assay, or with HA-AKT1, confirmed an increase in STAT3 activation and phospho-AKT (pAKT) levels when B7-H3 was co-transfected with Cignal STAT3 Reporter kit assay (Figure 6d) and with HA-AKT1 (Figure 6e), respectively, as measured by Luciferase and Western blot.
Figure 6.
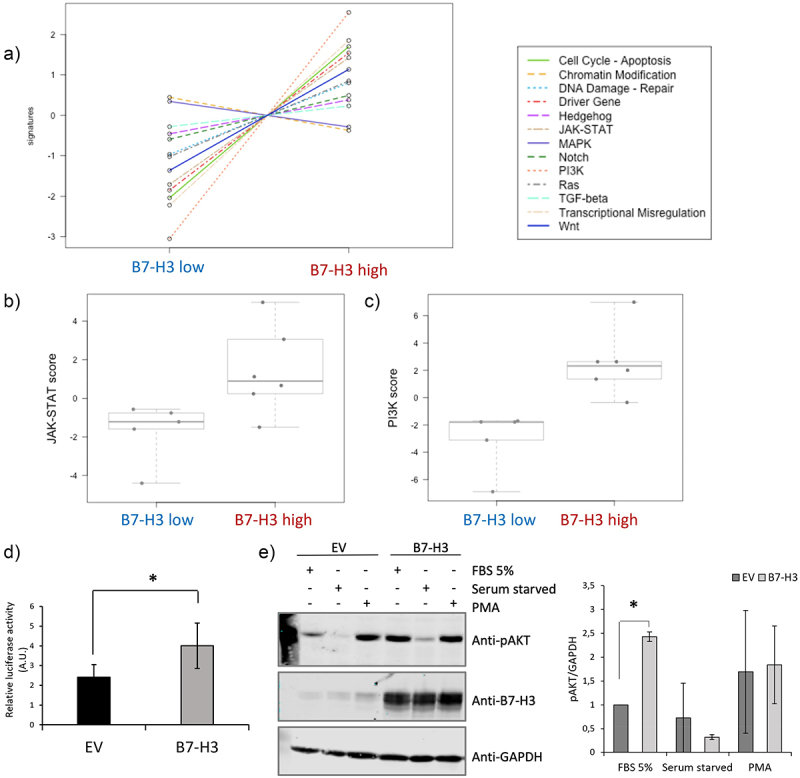
Molecular pan cancer pathway panel analysis of ccRCC tumor samples according to B7-H3 expression.
a) PanCancer Pathway signature in ccRCC tumor samples. b, c) Pan Cancer pathway analysis JAK/STAT and PI3K signaling score. Box plots of the JAK/STAT and PI3K pathway score are shown in high B7-H3 protein expression vs low B7-H3 protein expression tumors. d) B7-H3-dependent STAT3 functional activity in kidney cells. B7-H3 and STAT3 luciferase-based activation assay reporter were co-transfected, and luminescence is shown in arbitrary units (A.U.) from three independent experiments. Statistically significant results (p < 0.05) are marked with *. EV, empty vector. e) B7-H3-dependent AKT functional activity in kidney cells. HEK293 cells were co-transfected with plasmids encoding B7-H3 and HA-AKT1, and the phosphorylation of AKT was monitored by immunoblotting using the anti-pAKT (Ser473) antibody. The expression of B7-H3 and GAPDH (as a loading control) was also monitored using specific antibodies. Cells were either left untreated, serum-starved, or treated with PMA (100 nΜ for 30 min). Quantifications of pAKT/GAPDH are shown in the right panel.
B7-H3 expression affects cell proliferation/viability and TKI sensitivity in ccRCC
We monitored the expression of B7-H3 in human renal cancer cell lines by immunoblot, and we detected high B7-H3 protein expression in Caki-1, A498 and 786-O renal cancer cell lysates, as compared to nonmalignant RC-124 renal cells (Figure 7a). Knocking down of B7-H3 by siRNA in Caki-1 and 786-O renal cancer cells, as confirmed by Western blot (Figure 7b), decreased the cell proliferation in both 2D (Figure 7c) and 3D cell growth cultures (Figure 7d). B7-H3 knockout by CRISPR/Cas in 786-O cells, as confirmed by Western blot (Figure 7e), also decreased 2D and 3D cell proliferation (Figure 7f). Furthermore, silencing or CRISPR/Cas knockout of B7-H3 increased sensitivity to TKI axitinib in Caki-1 and 786-O cells in both 2D and 3D growth assays (Figure 7c,d,f).
Figure 7.
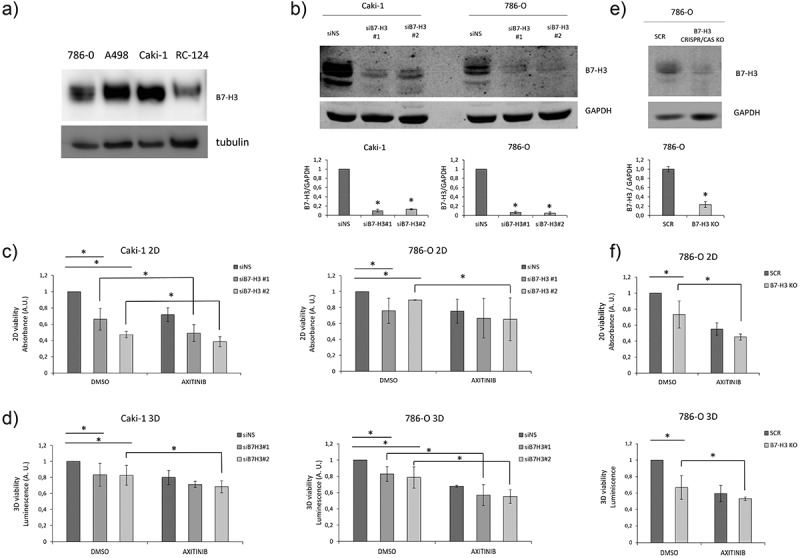
Expression of B7-H3 in renal cancer cells affects 2D and 3D viability.
a) B7-H3 protein expression in renal cancer cell lines. Immunoblot of cell lysates from 786-O, A498 and Caki-1 human renal cancer cells and RC-124 benign human renal cells. b) B7-H3 protein expression in renal cancer cell lines upon B7-H3 knockdown. Immunoblot of cell lysates from Caki-1 and 786-O human renal cancer cells upon B7-H3 knockdown using two different siRNAs. Quantifications of B7-H3/GAPDH are shown in the bottom panel. c, d) Viability of renal cancer cells upon knockdown of B7-H3. c) 2D and d) 3D-spheroid cell viability of Caki-1 and 786-O renal cancer cells upon knockdown of B7-H3 and upon treatment with tyrosine kinase inhibitor axitinib. e) B7-H3 protein expression in 786-O renal cancer cell line upon B7-H3 CRISPR/Cas knockout. Immunoblot of cell lysates from 786-O human renal cancer cells upon scramble (SCR) or B7-H3 CRISPR/Cas knockout (B7-H3 KO). Quantifications of B7-H3/GAPDH are shown in the bottom panel. f) Viability of renal cancer cells upon CRISPR/Cas knockout of B7-H3. 2D (upper panel) and 3D-spheroid (bottom panel) cell viability of 786-O renal cancer cells upon CRISPR/Cas knockout of B7-H3 and upon treatment with axitinib. Data are represented as the mean from at least two biological replicates, each one with at least three technical replicates.
Discussion
Our in vitro and in silico analysis of renal cancer cells, kidney tissue and renal carcinoma, revealed that high expression of B7-H3 was associated with poor outcome, suggesting a potential involvement of B7-H3 in renal cancer progression. We used two separate cohorts of renal cancer, mainly consisting of ccRCC cancers, where B7-H3 expression significantly correlated to various parameters and poor prognosis. In consistence with the prevalence of non-ccRCC cancers to occur,42 in our cohort, 25% cases were non-ccRCC, including 13% ChRCC and 12% PRCC. In PRCC, B7-H3 expression did not correlate with overall survival. This suggests that the pro-oncogenic role of B7-H3 is not the same across all subtypes of renal cancer. In the 17 cases of ChRCC, only one sample was found positive for high B7-H3 expression yet showed significant correlations, making necessary the confirmation of these findings in a larger cohort.
Further, in our analyses, we disclosed correlation between high B7-H3 expression and worse renal cancer patient outcome, specifically with stage and overall survival in ccRCC. Furthermore, B7-H3 correlated with the synchronous metastasis and with lymph node metastasis. Together, these findings support a pro-oncogenic role of B7-H3 in ccRCC and that B7-H3 might have a role in the metastatic potential of ccRCC. Soluble B7-H3 (sB7-H3) in the serum of ccRCC patients has been found to correlate with the clinical stage, as well as with the expression of the soluble interleukin-2 receptor.43 This implies that sB7-H3 from serum could be a biomarker in ccRCC. Whether sB7-H3 plays a potential role in the metastatic niche formation in ccRCC deserves exploration.
Gene expression analysis from a sub-group of patients showed a similar correlation between B7-H3 protein expression evaluated from TMA sections and the B7-H3 mRNA expression from matching whole tissue sections. We also observed a good correlation of top differentially expressed genes in our pilot experiment with bigger datasets, such as the TCGA dataset. Of the top differentially expressed genes in B7-H3 high tumors, we observed higher expression of JAK3 and IRS1 genes involved in signaling pathways JAK/STAT and PI3K/AKT, respectively. We and others have previously linked these two pathways to the pro-oncogenic role of B7-H3.38,44 We also observed increased expression of MMP14, which is also associated with tumor prognosis and immune invasion.45 This is in consistence with other reports associating the expression of B7-H3 and MMP14 in other cancer forms.45,46 CSF1R, also known as the macrophage colony-stimulating factor receptor (M-CSFR), was found highly expressed in the B7-H3 high tumor group. M-CSFR is a tyrosine kinase and has previously been linked to ccRCC progression, and it is considered a potential therapeutic target.47,48 Whether B7-H3 recruits TAMs to the tumor microenvironment in ccRCC by upregulation of CSF1R, deserves dedicated studies. The biological relevance of downregulation of GK and ALDH1A1 metabolic enzymes, BMP2, and HAVCR2/TIM3 in B7-H3 high tumors also needs further elucidation. In this regard, B7-H3 has been linked with the glycolytic capacity of tumor cells.38 HAVCR2/TIM3 is an immune checkpoint protein whose low expression has been found associated with poor outcome of ccRCC,49 which is in line with the B7-H3 pro-oncogenic role.
Pathway analysis showed an increased myeloid immune evasion score and macrophage cell type score in B7-H3 high tumors. This is in consistence with recently published results from CRISPR/Cas knockout of B7-H3 in colorectal cancer spheroids that resulted in lower macrophage recruitment into the tumor spheroid.50 We also observed a significant correlation between high protein levels of B7-H3 and a T cell exhaustion score and higher exhausted CD8 cells. Advanced ccRCC was recently linked to progressive immune dysfunction, with increased populations of terminally exhausted CD8 T cells and M2-like macrophages.51 Spatial expression analysis of different immune cell populations would provide additional insights into the immune–tumor cell interactions in ccRCC.52 The immune evasive role of B7-H3 in ccRCC deserves further studies.
Our in vitro results suggest that inhibition of the immune checkpoint protein B7-H3 could have therapeutic benefits in ccRCC in combination with tyrosine kinase inhibitors, which deserves further investigation. Additional studies in cocultures or immune competent mouse models would be necessary to assess the potential of B7-H3 inhibition to promote both the anti-tumor immune response and sensitivity to VEGF inhibition in ccRCC.
Together, our findings highlight B7-H3 as an actionable novel immune checkpoint protein in advanced and metastatic ccRCC and suggest the involvement of B7-H3 in JAK/STAT and PI3K/AKT signaling pathways, as well as in T cell and myeloid immune evasion in ccRCC.
Supplementary Material
Acknowledgments
We thank all technical and administrative personnel from the Biobizkaia Health Research Institute for their expert assistance. We would also like to thank Arantza Perez Dobaran (University of the Basque Country, Leioa, Spain) and the Genomics Core Facility and the Flow Cytometry Core Facility (Oslo University Hospital, Oslo, Norway) for their expert technical support. We are grateful to NanoString for the Science Never Stops in the Nordics Grant to CENX. We would also like to thank the Asociación de Mujeres de Leioa for their support.
Funding Statement
This work of CENX is funded by Instituto de Salud Carlos III (ISCIII) through the projects [CP20/00008, PI22/00386 and MV22/00019] (Spain and co-funded by European Union), Biobizkaia Health Research Institute (IISB, Ayudas para el fortalecimiento de grupos emergentes; [grant number BB/I/EMERG/24/001], Spain), and Stiftelsen til fremme av forskning innen nyresykdommer (Unifor, Norway). M.E. is the recipient of a Fellowship 2023/2024 from Biobizkaia Health Research Institute [grant number BCB/I/Fellowship/23/002], and fellowships from the Jesus Gangoiti Barrera Foundation [grant numbers FJGB21/006, FJGB22/006]. E.R-I. is the recipient of a predoctoral fellowship from Asociación Española Contra el Cáncer (AECC), [grant number PRDVZ222375REY, Junta Provincial de Bizkaia, Spain].
Highlights
B7-H3 expression associates with synchronous metastasis and poor outcomes in clear cell renal cell carcinoma (ccRCC).
Molecular studies suggest B7-H3 involvement in JAK/STAT and PI3K/AKT pro-oncogenic pathways, T cell exhaustion and myeloid immune evasion in ccRCC.
Our findings highlight B7-H3 as an actionable novel immune checkpoint protein in ccRCC in combination with the tyrosine kinase inhibitor axitinib.
Credit author statement
M.E., E.R-I., Á.M., L.M., D.L., T.Ø., N.R., G.M.M., J.A., abd P.E.: investigation, visualization, analysis, and validation; G.L., R.P., J.I.L.: investigation, conceptualization, visualization, analysis validation, and writing – review and editing; C.E.N-X.: investigation, conceptualization, visualization, analysis, validation, supervision, funding acquisition, project administration, and writing – review and editing.
Data availability statement
All data generated in this study are included in the manuscript or in the supplementary data. The full nCounter analysis generated datasets are available from the corresponding authors on reasonable request.
Ethical statement
The study was performed in accordance with the Declaration of Helsinki. Ethical approval was obtained from Comité de Ética de la Investigación con medicamentos de Euskadi (CEIm-E), Spain.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/2162402X.2024.2419686
Abbreviations
- ccRCC
clear cell renal cell carcinomas
- chRCC
chromophobe renal cell carcinomas
- CPI
immune checkpoint inhibitors
- IHC
immunohistochemistry
- OS
overall survival
- PD-L1
programmed death ligand 1
- PD-1
programmed cell death protein 1
- PRCC
papillary renal cell carcinomas
- TIL
tumor infiltrating lymphocytes
- TMA
tissue microarray
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A.. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–15. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Xu W, Atkins MB, McDermott DF.. Checkpoint inhibitor immunotherapy in kidney cancer. Nat Rev Urol. 2020;17(3):137–150. doi: 10.1038/s41585-020-0282-3. [DOI] [PubMed] [Google Scholar]
- 3.de Velasco G, Miao D, Voss MH, Hakimi AA, Hsieh JJ, Tannir NM, de Velasco G, Tamboli P, Appleman LJ, Rathmell WK, et al. Tumor mutational load and immune parameters across metastatic renal cell carcinoma risk groups. Cancer Immunol Res. 2016;4(10):820–822. doi: 10.1158/2326-6066.CIR-16-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun DA, Hou Y, Bakouny Z, Ficial M, Sant’ Angelo M, Forman J, Ross-Macdonald P, Berger AC, Jegede OA, Elagina L, et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat Med. 2020;26(6):909–918. doi: 10.1038/s41591-020-0839-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angulo JC, Shapiro O. The changing therapeutic landscape of metastatic renal cancer. Cancers (Basel). 2019;11(9):1227. doi: 10.3390/cancers11091227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun DA, Bakouny Z, Hirsch L, Flippot R, Van Allen EM, Wu CJ, Choueiri TK. Beyond conventional immune-checkpoint inhibition — novel immunotherapies for renal cell carcinoma. Nat Rev Clin Oncol. 2021;18(4):199–214. doi: 10.1038/s41571-020-00455-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373(19):1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motzer RJ, Tannir NM, McDermott DF, Aren Frontera O, Melichar B, Choueiri TK, Plimack ER, Barthélémy P, Porta C, George S, et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N Engl J Med. 2018;378(14):1277–1290. doi: 10.1056/NEJMoa1712126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDermott DF, Huseni MA, Atkins MB, Motzer RJ, Rini BI, Escudier B, Fong L, Joseph RW, Pal SK, Reeves JA, et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med. 2018;24(6):749–757. doi: 10.1038/s41591-018-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rini BI, Powles T, Atkins MB, Escudier B, McDermott DF, Suarez C, Bracarda S, Stadler WM, Donskov F, Lee JL, et al. Atezolizumab plus bevacizumab versus sunitinib in patients with previously untreated metastatic renal cell carcinoma (IMmotion151): a multicentre, open-label, phase 3, randomised controlled trial. Lancet. 2019;393(10189):2404–2415. doi: 10.1016/S0140-6736(19)30723-8. [DOI] [PubMed] [Google Scholar]
- 11.Motzer RJ, Penkov K, Haanen J, Rini B, Albiges L, Campbell MT, Venugopal B, Kollmannsberger C, Negrier S, Uemura M, et al. Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med. 2019;380(12):1103–1115. doi: 10.1056/NEJMoa1816047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choueiri TK, Motzer RJ, Rini BI, Haanen J, Campbell MT, Venugopal B, Kollmannsberger C, Gravis-Mescam G, Uemura M, Lee JL, et al. Updated efficacy results from the JAVELIN renal 101 trial: first-line avelumab plus axitinib versus sunitinib in patients with advanced renal cell carcinoma. Ann Oncol. 2020;31(8):1030–1039. doi: 10.1016/j.annonc.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nunes-Xavier CE, Angulo JC, Pulido R, Lopez JI. A critical insight into the clinical translation of PD-1/PD-L1 blockade therapy in clear cell renal cell carcinoma. Curr Urol Rep. 2019;20(1):1. doi: 10.1007/s11934-019-0866-8. [DOI] [PubMed] [Google Scholar]
- 14.Flem-Karlsen K, Fodstad O, Nunes-Xavier CE. B7-H3 immune checkpoint protein in human cancer. Curr Med Chem. 2020;27(24):4062–4086. doi: 10.2174/0929867326666190517115515. [DOI] [PubMed] [Google Scholar]
- 15.Emaldi M, Nunes-Xavier CE. B7-H4 immune checkpoint protein affects viability and targeted therapy of renal cancer cells. Cells. 2022;11(9):1448. doi: 10.3390/cells11091448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou QH, Li KW, Chen X, He HX, Peng SM, Peng SR, Wang Q, Li Z-A, Tao Y-R, Cai W-L, et al. HHLA2 and PD-L1 co-expression predicts poor prognosis in patients with clear cell renal cell carcinoma. J Immunother Cancer. 2020;8(1):e000157. doi: 10.1136/jitc-2019-000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin X, Zhang H, Ye D, Dai B, Zhu Y, Shi G. B7-H3 is a new cancer-specific endothelial marker in clear cell renal cell carcinoma. Onco Targets Ther. 2013;6:1667–1673. doi: 10.2147/OTT.S53565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emaldi M, Alamillo-Maeso P, Rey-Iborra E, Mosteiro L, Lecumberri D, Pulido R, López JI, Nunes-Xavier CE. A functional role for glycosylated B7-H5/VISTA immune checkpoint protein in metastatic clear cell renal cell carcinoma. iScience. 2024;27(9):110587. doi: 10.1016/j.isci.2024.110587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flem-Karlsen K, Fodstad O, Tan M, Nunes-Xavier CE. B7-H3 in cancer - beyond immune regulation. Trends Cancer. 2018;4(6):401–404. doi: 10.1016/j.trecan.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Zhou C, Zhang D, Huang Z, Zhang G. The anti-apoptotic effect on cancer-associated fibroblasts of B7-H3 molecule enhancing the cell invasion and metastasis in renal cancer. Onco Targets Ther. 2019;12:4119–4127. doi: 10.2147/OTT.S201121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Zhang G, Zhang X, Lv G, Wei X, Yuan H, Hou J. Overexpression of B7-H3 in CD14+ monocytes is associated with renal cell carcinoma progression. Med Oncol. 2014;31(12):349. doi: 10.1007/s12032-014-0349-1. [DOI] [PubMed] [Google Scholar]
- 22.Zhao J, Lei T, Xu C, Li H, Ma W, Yang Y, Fan S, Liu Y. MicroRNA-187, down-regulated in clear cell renal cell carcinoma and associated with lower survival, inhibits cell growth and migration though targeting B7-H3. Biochem Biophys Res Commun. 2013;438(2):439–444. doi: 10.1016/j.bbrc.2013.07.095. [DOI] [PubMed] [Google Scholar]
- 23.Inamura K, Amori G, Yuasa T, Yamamoto S, Yonese J, Ishikawa Y. Relationship of B7-H3 expression in tumor cells and tumor vasculature with FOXP3+ regulatory T cells in renal cell carcinoma. Cancer Manag Res. 2019;11:7021–7030. doi: 10.2147/CMAR.S209205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JH, Kim YJ, Ryu HW, Shin SW, Kim EJ, Shin SH, Park JY, Kim SY, Hwang CS, Na J-Y, et al. B7-H3 expression is associated with high PD-L1 expression in clear cell renal cell carcinoma and predicts poor prognosis. Diagn Pathol. 2023;18(1):36. doi: 10.1186/s13000-023-01320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crispen PL, Sheinin Y, Roth TJ, Lohse CM, Kuntz SM, Frigola X, Thompson RH, Boorjian SA, Dong H, Leibovich BC, et al. Tumor cell and tumor vasculature expression of B7-H3 predict survival in clear cell renal cell carcinoma. Clin Cancer Res. 2008;14(16):5150–5157. doi: 10.1158/1078-0432.CCR-08-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saeednejad Zanjani L, Madjd Z, Axcrona U, Abolhasani M, Rasti A, Asgari M, Fodstad Ø, Andersson Y. Cytoplasmic expression of B7-H3 and membranous EpCAM expression are associated with higher grade and survival outcomes in patients with clear cell renal cell carcinoma. Ann Diagn Pathol. 2020;46:151483. doi: 10.1016/j.anndiagpath.2020.151483. [DOI] [PubMed] [Google Scholar]
- 27.Mischinger J, Frohlich E, Mannweiler S, Meindl C, Absenger-Novak M, Hutterer GC, Seles M, Augustin H, Chromecki TF, Jesche-Chromecki J, et al. Prognostic value of B7-H1, B7-H3 and the stage, size, grade and necrosis (SSIGN) score in metastatic clear cell renal cell carcinoma. Cent Eur J Urol. 2019;72(1):23–31. doi: 10.5173/ceju.2018.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loo D, Alderson RF, Chen FZ, Huang L, Zhang W, Gorlatov S, Burke S, Ciccarone V, Li H, Yang Y, et al. Development of an Fc-enhanced anti–B7-H3 monoclonal antibody with potent antitumor activity. Clin Cancer Res. 2012;18(14):3834–3845. doi: 10.1158/1078-0432.CCR-12-0715. [DOI] [PubMed] [Google Scholar]
- 29.Wang G, Wu Z, Wang Y, Li X, Zhang G, Hou J. Therapy to target renal cell carcinoma using 131I-labeled B7-H3 monoclonal antibody. Oncotarget. 2016;7(17):24888–24898. doi: 10.18632/oncotarget.8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352):357(6352. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research N . Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nunes-Xavier CE, Kildal W, Kleppe A, Danielsen HE, Waehre H, Llarena R, Mælandsmo GM, Fodstad Ø, Pulido R, López JI, et al. Immune checkpoint B7-H3 protein expression is associated with poor outcome and androgen receptor status in prostate cancer. Prostate. 2021;81(12):838–848. doi: 10.1002/pros.24180. [DOI] [PubMed] [Google Scholar]
- 35.Errarte P, Guarch R, Pulido R, Blanco L, Nunes-Xavier CE, Beitia M, Gil J, Angulo JC, López JI, Larrinaga G, et al. The expression of fibroblast activation protein in clear cell renal cell carcinomas is associated with synchronous lymph node metastases. PLOS ONE. 2016;11(12):e0169105. doi: 10.1371/journal.pone.0169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez JI, Errarte P, Erramuzpe A, Guarch R, Cortes JM, Angulo JC, Pulido R, Irazusta J, Llarena R, Larrinaga G, et al. Fibroblast activation protein predicts prognosis in clear cell renal cell carcinoma. Hum Pathol. 2016;54:100–105. doi: 10.1016/j.humpath.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Nunes-Xavier CE, Emaldi M, Mingo J, Oyjord T, Maelandsmo GM, Fodstad O, Errarte P, Larrinaga G, Llarena R, López JI, et al. The expression pattern of pyruvate dehydrogenase kinases predicts prognosis and correlates with immune exhaustion in clear cell renal cell carcinoma. Sci Rep. 2023;13(1):7339. doi: 10.1038/s41598-023-34087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nunes-Xavier CE, Karlsen KF, Tekle C, Pedersen C, Oyjord T, Hongisto V, Nesland JM, Tan M, Sahlberg KK, Fodstad Ø, et al. Decreased expression of B7-H3 reduces the glycolytic capacity and sensitizes breast cancer cells to AKT/mTOR inhibitors. Oncotarget. 2016;7(6):6891–6901. doi: 10.18632/oncotarget.6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jimenez C, Jones DR, Rodriguez-Viciana P, Gonzalez-Garcia A, Leonardo E, Wennstrom S, von Kobbe C, Toran JL, Luis R, Calvo V, et al. Identification and characterization of a new oncogene derived from the regulatory subunit of phosphoinositide 3-kinase. Embo J. 1998;17(3):743–753. doi: 10.1093/emboj/17.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mingo J, Luna S, Gaafar A, Nunes-Xavier CE, Torices L, Mosteiro L, Ruiz R, Guerra I, Llarena R, Angulo JC, et al. Precise definition of PTEN C-terminal epitopes and its implications in clinical oncology. npj Precis Onc. 2019;3(1):11. doi: 10.1038/s41698-019-0083-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nunes-Xavier CE, Aurtenetxe O, Zaldumbide L, Lopez-Almaraz R, Erramuzpe A, Cortes JM, López JI, Pulido R. Protein tyrosine phosphatase PTPN1 modulates cell growth and associates with poor outcome in human neuroblastoma. Diagn Pathol. 2019;14(1):134. doi: 10.1186/s13000-019-0919-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muglia VF, Prando A. Renal cell carcinoma: histological classification and correlation with imaging findings. Radiol Bras. 2015;48(3):166–174. doi: 10.1590/0100-3984.2013.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masuda A, Arai K, Nishihara D, Mizuno T, Yuki H, Kambara T, Betsunoh H, Abe H, Yashi M, Fukabori Y, et al. Clinical significance of serum soluble T cell regulatory molecules in clear cell renal cell carcinoma. Biomed Res Int. 2014;2014:1–6. doi: 10.1155/2014/396064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Tekle C, Chen YW, Kristian A, Zhao Y, Zhou M, Liu Z, Ding Y, Wang B, Mælandsmo GM, et al. B7-H3 silencing increases paclitaxel sensitivity by abrogating Jak2/Stat3 phosphorylation. Mol Cancer Ther. 2011;10(6):960–971. doi: 10.1158/1535-7163.MCT-11-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, Li S, Zhou L, Yang L, Wu X, Tang B, Xie S, Fang L, Zheng S, Hong T, et al. Immune infiltration of MMP14 in pan cancer and its prognostic effect on tumors. Front Oncol. 2021;11:717606. doi: 10.3389/fonc.2021.717606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng R, Wang B, Cai XR, Chen ZS, Du Q, Zhou LY, Ye J-M, Chen Y-L. CD276 promotes vasculogenic mimicry formation in hepatocellular carcinoma via the PI3K/AKT/MMPs pathway. Onco Targets Ther. 2020;13:11485–11498. doi: 10.2147/OTT.S271891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soares MJ, Pinto M, Henrique R, Vieira J, Cerveira N, Peixoto A, Martins AT, Oliveira J, Jerónimo C, Teixeira MR, et al. CSF1R copy number changes, point mutations, and RNA and protein overexpression in renal cell carcinomas. Mod Pathol. 2009;22(6):744–752. doi: 10.1038/modpathol.2009.43. [DOI] [PubMed] [Google Scholar]
- 48.Menke J, Kriegsmann J, Schimanski CC, Schwartz MM, Schwarting A, Kelley VR. Autocrine CSF-1 and CSF-1 receptor coexpression promotes renal cell carcinoma growth. Cancer Res. 2012;72(1):187–200. doi: 10.1158/0008-5472.CAN-11-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao G, Wang P, Wang Y. Identification of the prognosis value and potential mechanism of immune checkpoints in renal clear cell carcinoma microenvironment. Front Oncol. 2021;11:720125. doi: 10.3389/fonc.2021.720125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durlanik S, Fundel-Clemens K, Viollet C, Huber HJ, Lenter M, Kitt K, Pflanz S. CD276 is an important player in macrophage recruitment into the tumor and an upstream regulator for PAI-1. Sci Rep. 2021;11(1):14849. doi: 10.1038/s41598-021-94360-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braun DA, Street K, Burke KP, Cookmeyer DL, Denize T, Pedersen CB, Gohil SH, Schindler N, Pomerance L, Hirsch L, et al. Progressive immune dysfunction with advancing disease stage in renal cell carcinoma. Cancer Cell. 2021;39(5):632–648.e8. doi: 10.1016/j.ccell.2021.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez JI, Hogan MF, Sutton B, Church SE, Angulo JC, Nunes-Xavier CE. Distinct spatial landscapes in clear-cell renal cell carcinoma as revealed by whole transcriptome analysis. Immunooncol Technol. 2024;21:100690. doi: 10.1016/j.iotech.2023.100690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are included in the manuscript or in the supplementary data. The full nCounter analysis generated datasets are available from the corresponding authors on reasonable request.


