Abstract
Background and Introduction
Over the last decade, treatment of patients with advanced non-small cell lung cancer (NSCLC) has become dependent on tissue and/or blood biomarkers to guide treatment decisions. Timely access to comprehensive biomarker and tumor signature information is crucial for diagnostic testing. With the rapid development and implementation of complex biomarker testing, comprehensive molecular profiling with in-depth analysis of DNA, RNA, and proteins can be easily performed. Initial data from the MedStar Health system showed considerable disparities in the use of next generation sequencing (NGS) between hospitals, and there is a clear need to improve education and understanding regarding which cases are appropriate for NGS, as well as the use of protocols to make those decisions in an expeditious manner.
Materials and Methods
Clinical pathways are systems-based tools that aim to create greater transparency around care decision making, therapeutic selection, and care delivery. They enhance quality and efficiency by reducing non-value-added intra-provider variability in care. We aimed to create a comprehensive clinical pathway system, the Targetable Molecular Algorithm (TMA), to increase the understanding and use of NGS in NSCLC by physicians in training, oncology nurse navigators, nurse practitioners, and general oncologists.
Discussion
We provide an overview of the implementation of the platform along with navigation guide. A realistic case study—a typical clinical workflow for a patient requiring NGS testing with an EGFR mutation—is also reviewed, demonstrating how the TMA platform can be applied. Additionally, we highlight the importance of the resource, and discuss its strengths, weaknesses, and potential future applications.
Results and Conclusion
This new and innovative pathway system will make decision-making easier for clinicians trying to understand the appropriate tests and treatment algorithms for their patients. Our aim is to increase the appropriate and timely use of NGS among health-system providers with the hope that this system will empower physicians to provide better care by providing a quick, simple, user-friendly tool for comprehensive patient care.
Keywords: NSCLC, clinical pathways, biomarkers, targetable molecular algorithm, next generation sequencing, precision oncology, electronic learning system
Background and introduction
The delivery of precision care represents a significant paradigm in the field of oncology.1 Cancer diagnostics have progressed considerably in the last decade and includes the use of advanced molecular diagnostic technologies, such as Next-Generation Sequencing (NGS) to select targeted therapies in patients with advanced cancers. Such precision oncology studies have shown improvement in treatment outcomes for refractory cancer patients with advanced cancers while lowering average per-week healthcare costs, resource utilization, and end-of-life costs.2
Treatment decisions for advanced non-small cell lung cancer (NSCLC) has become increasingly reliant on tissue and/or blood biomarkers, but implementing molecular diagnostics remains a challenge. Studies have shown the superiority of a targeted therapy approach for patients with advanced NSCLC, but unfortunately, the proportion of patients who undergo tumor genetic testing remains low.3–7 Despite the development of targeted agents, the general medical and oncology communities still lack in-depth knowledge of these complex molecular pathways and more importantly, the knowledge of when and how to order the appropriate tests. It is therefore crucial to raise awareness of potentially targetable genetic alterations for improving patient prognosis.
Initial data from our institution at the MedStar Health system, along with published research,8–10 showed considerable disparities in the use of NGS. An analysis of tumor registry data from January 2017 to December 2019 at MedStar Washington Hospital Center (WHC) revealed that among the 47 stage III and 125 stage IV NSCLC patients who met the criteria for NGS, only 20 cases (11.6%) underwent NGS. Comparatively, practice at MedStar Georgetown University Hospital (GUH) during the same period had 100% of qualified cases sent for NGS. Over three and a half years, GUH saw 62% of patients with genomic alterations identified via NGS.
While official guidelines for molecular and biomarker testing exist, such as the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines https://www.nccn.org), a more streamlined algorithmic approach is needed to guide care providers efficiently and clearly. It is important to increase awareness about potentially targetable genetic alterations, and improve the understanding about which NSCLC cases are appropriate for NGS testing, and the use of protocols to make those decisions in an expeditious manner. There is hence a pressing need for a usable and practical solution that can empower clinicians to handle complex patient cases.
Clinical pathway tools such as Wolters Kluwer’s UpToDate pathways guide physicians in making optimal screening, diagnosis, and treatment of patients in a time-efficient manner using evidence-based medicine. These pathways provide a question-and-answer based approach to a clinical question in a diagrammatic form, and then lead the clinician to choose the optimal treatment or screening based on their patient’s specific case.11,12
Objectives
We have developed Targetable Molecular Algorithm (TMA) as the first interactive online platform to train and assist physicians in making decisions on treating patients with actionable driver positive NSCLC.
The goals were 2-fold (1) to increase the appropriate and timely use of NGS among health-system providers so that patients receive the best treatment in an expeditious fashion and (2) to train, assist, and empower decision making in order to guide and expedite the treatment of NSCLC. The TMA platform is a collection of clinical algorithms specific to NSCLC that offers step-by-step, question-and-answer based algorithmic approach to navigate the complexities of diagnosis/treatment based on the type of mutation.
In this article, we provide an overview of how this online resource was constructed and demonstrate how such a tool can be used to assist physicians. We describe how the clinical pathways were generated using expert and best practices from clinical knowledge in conjunction with information from published literature. We also demonstrate the practical usability of this tool by describing a clinical case study. Lastly, we discuss the importance of this work and showcase its applications and future possibilities.
Methods
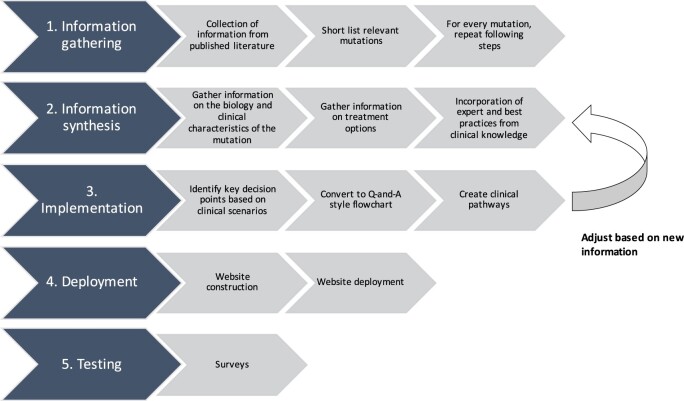
The TMA is an online platform that offers a comprehensive collection of clinical algorithms with step-by-step workflow instructions to help train and assist in making decisions on treating patients in NSCLC. This methodology has been structured into 5 parts, ie, Part 1: Information gathering, Part 2: Information synthesis, Part 3: Implementation, Part 4: Deployment, and Part 5: Testing.
Part 1: Information gathering
The clinical experts in our team systematically reviewed recent publications from Pubmed and Cochrane library in NSCLC. Genomic alterations that have been identified in lung cancer as targetable mutations were shortlisted. The frequencies of actionable molecular alterations were reviewed using publicly available datasets in NSCLC, and the clinical implications of the genomic alterations were studied. Based on this information gathered, the major targetable alterations in 7 genes in NSCLC including EGFR, ALK, ROS1, BRAF, NTRK, RET, and MET were selected for the TMA clinical pathway platform.
Part 2: Information synthesis
For each targetable mutation, our team gathered information on the incidence, biology, and clinical characteristics of the mutation. We also summarized the FDA approved current treatment options for each mutation, dosage, landmark clinical trials, publications, and known adverse events. Current treatment guidelines are available in FDA drug label information. Table 1 contains a summary of the key mutations in NSCLC along with the FDA drug information.
Table 1.
Summary of key genetic mutations in NSCLC.
NSCLC = Non small cell lung cancer; TMA = Tumor Molecular algorithm.
Once all the information was gathered, we incorporated best practices from clinical knowledge to identify various clinical scenarios and treatment paths for each genomic alteration. These clinical scenarios were the starting point that led to the implementation of this information in the form of flowcharts and pathways.
Part 3: Implementation
Flowcharts and pathways corresponding to each targetable alteration were created. Every mutation had a separate pathway in the TMA pathway system. The most established targetable mutation EGFR was chosen as a template. Key decision points such as whether a patient was new to therapy, presence of brain metastasis, progression on drug, etc., were first identified. These decision points were set up as a “yes/no” question and optimal management solutions for each possible answer were identified. Each algorithm included questions to help narrow down to the correct testing and treatment option for the patient, based on the targetable mutation. Following this process, a step-by-step flow chart was created with algorithms leading to the optimal management of a specific patient based on specific criteria. A general workflow diagram on the creation of these clinical pathways is shown in Figure 1.
Figure 1.
Workflow process diagram.
Part 4: Deployment
Using the workflow process outlined in Figure 1, clinical pathways were created for every mutation. Once the pathway collection was developed, they were incorporated into a dynamic workflow in LucidChart (www.lucidchart.com) and embedded into an online platform built on the Google site platform (https://sites.google.com/). The website was developed as an online platform for clinical oncology training pathways and was called the TMA (https://tma.georgetown.edu). The online platform is hosted at Georgetown University Innovation Center for Biomedical Informatics (Georgetown-ICBI).
The website also leads users to additional resources and recommendations. It includes an overview of the mutations, possible treatments, and a quiz for users to test the knowledge gained. The pathways are also annotated with link outs to relevant references such as appropriate trials, FDA drug inserts for the drugs and articles used to support the algorithm development. The website also includes references, financial support information, and relevant clinical trials.
Such a visual algorithmic presentation enables the information to be presented as an interactive visual aid for clinicians to promptly search for an answer while being time efficient.
Part 5: Website testing—pilot phase
A soft launch of the website along was done in the spring of 2022. During the pilot phase, the primary audience for the platform were general oncologists, thoracic oncologists, the nurse navigators who worked with them, and pathologists from within the MedStar Health and Hackensack Meridian Health oncology networks. We also designed a short pre- and post-assessment survey to accompany the TMA during the pilot phase of implementation.
The TMA platform was promoted internally throughout MedStar Health and across participating oncology networks. This was done through a planned reach-out program to different medicine services, by setting up online meetings to introduce their departments to our project and pathways. Our aim was to evaluate 2 areas: knowledge gained by the participants and how that knowledge affected practices.
Overview of the resource and navigation guide
The TMA pathway system was constructed to help guide clinicians in selecting the appropriate test, and then managing a patient with a specific mutation. Our team aimed to create a comprehensive clinical pathway system to assist in making decisions on patient treatment for major targetable alterations in 7 genes in NSCLC: EGFR, ALK, ROS1, BRAF, NTRAK, RET, and MET.
The platform is available to the public using this link: https://tma.georgetown.edu. The front page has a brief summary of the advantages and applications of clinical pathway systems; and a background about NSCLC. Users are then encouraged to complete the Entry survey before they start exploring the clinical pathways. The left side of the page has a collapsible panel that lists the names of each clinical pathway—each one is designed like a dropdown that can be expanded to obtain an overview of the pathway, the pathway diagrams and a quiz. Users can explore each of the clinical pathways in any order they wish. The platform also provides information on resources that offer free drug assistance, information on patient advocacy for physicians, patients or their families who may be looking for this information. At the end, we ask users to complete an optional exit survey to obtain feedback on their experience and knowledge gained from this platform.
Case study
A typical clinical workflow for a patient that needs NGS testing, and has an EGFR mutation
Clinical summary: Patient is 55-year-old presents with shortness of breath and cough to his primary care physician. He has no significant past medical history. He was light smoker in the past but quit 20 years ago. His primary care sent patient for chest X-ray that was noted to have a possible mass on the right. CT scan was ordered and noted to have mass 6.5×7 cm in the right lower lobe, pleural effusion, mediastinal lymph nodes. Fluid was drained and send for the cytology that came back positive for adenocarcinoma lung primary. Patient was referred to medical oncology. He had PET/CT done and MRI of the brain. PET/CT demonstrated uptake with SUV 20 in the lung mass, 18 in the mediastinal lymph nodes, and also noted to have lesion on the 7th rib and iliac bone. MRI of the brain is positive for intracranial metastatic disease.
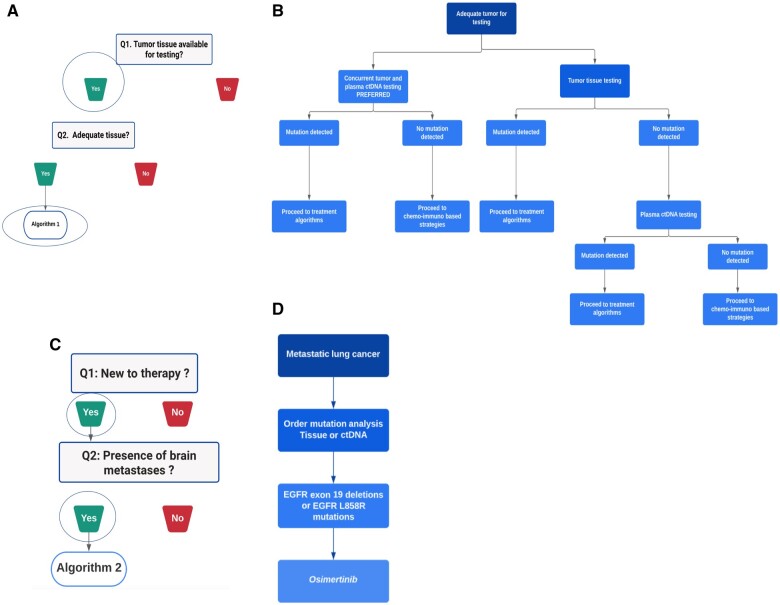
Solution: A physician when presented with such a clinical summary can navigate to the TMA platform and select the NGS testing tab on the left side panel. After reading the overview, the user would proceed to the clinical algorithm. Figure 2A shows the NGS testing clinical pathway. It showcases 2 key decision points: whether or not there is tumor tissue available for testing, and whether tissue quantity is available adequately. In this clinical question, if there is tumor tissue available adequately, the physician would click on Algorithm 1 (Figure 2B).
Figure 2.
(A) Two key decision points in the NGS testing clinical pathway. (B) Algorithm 1 of the NGS testing clinical pathway—a situation when tumor tissue is available adequately. (C) Key decision points in the EGFR pathway. (D) Algorithm 2 of the EGFR pathway.
Algorithm 1 clinical pathway provides a step-by-step guidance on which tests to perform, and the treatment strategies when a mutation is detected. In this algorithm, concurrent tumor and plasma ctDNA tests are recommended, and if a mutation is detected, mutation-based treatment strategies are suggested as a next step. If a mutation is not detected, traditional chemo-immuno based treatment strategies are recommended. Hence, a clinical question is “triaged” based on the medical evidence using a series of simple question-and-answer style flow charts in order to get the correct clinical guidance.
In this clinical scenario, the physician would understand that the recommendation is that whenever possible, tissue NGS testing and blood circulating tumor DNA (ctDNA) testing be done concurrently.
Let us assume that in this case, the result of the NGS testing indicated an EGFR mutation
The guideline on the TMA platform is to proceed to the correspond treatment algorithm for EGFR.
The user would navigate to the EGFR tab on the left side, read the overview and get to the clinical pathway section. According to the clinical summary, the patient was newly diagnosed with lung cancer and MRI indicated presence of brain metastasis. Following the flow chart presented on the website, the user would click on Algorithm 2 (Figure 2C). The tissue NGS test previously done would confirm the type of EGFR mutation. According to the flowchart, the physician would start the patient on Osimertinib (Figure 2D).
Pilot phase survey results
A soft launch of the website along was done in the spring of 2022 and the test users were asked to complete pre and post user surveys, and the feedback was collected. Target audience were Oncology, Internal medicine, and Pulmonology departments within MedStar Georgetown and Washington Hospital Center. The test users were asked to complete pre and post user surveys, and the feedback was collected. As of June 2023, the entry survey was completed by 57 participants. About 80% (46 count) of the participants identified themselves as Physician in training, 7% (4 count) identified as Oncology Physicians, 3 participants were Advanced practice providers (APP). As of June 2023, the exit survey was completed by 10 participants.
Summary of entry survey: The mode (the most common value) of entry survey data was the rating of 1 out of 10 indicating that participants had nearly no initial knowledge in NGS and targeted agents used to treat metastatic lung cancer.
Summary of exit survey: The mode (the most common value) of the exit survey was the rating of 7 out of 10 for indicating that participants improved their knowledge in NGS after using the TMA platform. In addition, 90% of the exit survey participants gave a high score between 6 and 10 to indicate their understating of the use of targeted therapy for treatment of the metastatic lung cancer after using the TMA platform. Similarly, 80% of the participants gave a high rating between 6 and 10 to indicate the ease of use of the TMA platform; and the mode (the most common value) of this survey question was 9. All the participants in exit survey said that (1) would consider using the TMA platform as a learning platform (2) would consider using an electronic algorithm platform in their clinical practice.
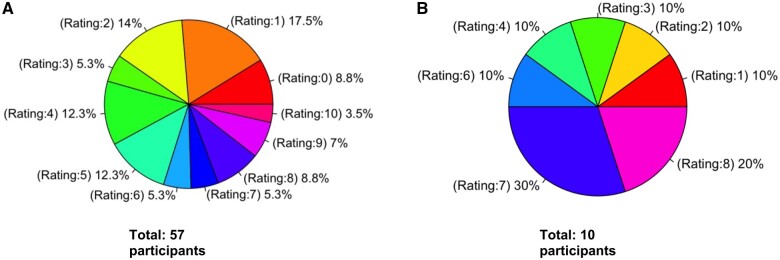
The participants’ knowledge of NGS testing before and after using the TMA platform was represented in the form of pie chart. Figure 3A shows initial knowledge of NGS which indicates that 70% of the participants rated their initial knowledge of NGS as between 0 and 5. Figure 3B shows the knowledge in NGS after using the TMA platform which shows that 60% of participants rated knowledge of NGS as between 6 and 10. These summary statistics show a clear trend from lower knowledge in the entry survey to higher knowledge in the exit survey demonstrating a positive impact and use of the platform.
Figure 3.
(A) Pie chart showing initial knowledge of NGS obtained from the entry survey (total 57 participants). It uses a rating scale from 0 to 10. (B) Pie chart showing knowledge in NGS after using the TMA platform obtained from the exit survey10 (total 10 participants). It uses a rating scale from 0 to 10.
Due to the low number participants in the exit survey, it was challenging to get significance from the statistical analyses. Fisher exact test results was performed to test the null hypothesis that the results from the entry survey and exit survey are the same meaning no knowledge was obtained from the system. The result obtained was close to statistical significance with a P-value of .08 (Tables 2 and 3).
Table 2.
Survey data binned to 2 rating groups—low scoring group that included ratings from 0 to 5 and a high scoring group that included ratings from 6 to 10.
| Rating group | Entry survey | Exit survey |
|---|---|---|
| Low score (0-5) | 40 | 4 |
| High score (6-10) | 17 | 6 |
| Total participants | 57 | 10 |
Table 3.
Results of Fishers exact test on the data from Table 2.
| Fishers exact test for count data: test results | |
|---|---|
| Alternative hypothesis | True odds ratio is not equal to 1 |
| 95% confidence interval | (0.7155281, 18.9113351) |
| Sample estimates: odds ratio | 3.455926 |
| P-value | .08038 |
Discussion
Importance of this resource
The TMA platform we developed contains a schematic approach to NGS and mutation testing for NSCLC. Such a system allows a user to walk through a series of key decision nodes and is then guided through a pathway leading to the optimal management of a specific patient. When a physician has a case study with a particular targetable mutation of interest, the algorithm questions help narrow down to the correct testing and treatment option for the patient.
We hope such an algorithmic approach can help increase the understanding and use of NGS in NSCLC by physicians in training, oncology nurse navigators, nurse practitioners, and general oncologists. This new and innovative pathway training system will make decision-making easier for clinicians who are trying to understand the appropriate tests and treatment algorithms for their patients. Such a tool allows for creating greater transparency around care decision making, therapeutic selection, and care delivery.
Strengths and weaknesses
Based on the pilot phase survey results, it is clear that such an electronic learning system can have positive impact on users of the platform. It can greatly improve not only their knowledge of NGS, but also offer assistance in their clinical practice. We recommend users to view this website on regular laptops and desktop computers, instead of mobile devices.
One of the challenges we faced was the low number participants who completed the exit survey, making it challenging to get significance from the statistical analyses. It is also important to keep the resource updated as new therapies and are developed, and clinical guidelines are updated.
Utilization and applications of this work
While the preliminary data from our study show good utility in NGS use and education that TMA platform can offer, we believe that including clinicians beyond the initial target audience can further our knowledge on key elements that impact adoption of NGS and understanding of the benefits of such a system. We plan to continue expanding the tool’s accessibility to a wider audience. With the pilot phase now complete, the TMA platform is being advertised to local regional conferences and the molecular tumor boards, as well as conference presentations to expand its visibility.
Future work
Incorporating real world evidence from sources such as oncoKB (https://www.oncokb.org), Clingen (https://clinicalgenome.org) and other evidence obtained using natural language processing (NLP) tools, can improve the knowledge offered in this platform. Incorporation of real-world evidence into the TMA platform is currently being implemented as the next phase of this platform.
Clinical pathways have been implemented in clinical care in multiple countries. The TMA platform is currently optimized to work on laptop and desktop computers. Offering such a solution as a mobile application would even more help apply evidence-based medicine to routine clinical care in an even efficient and timely manner. Future work is to expand number of pathways in lung cancer and other cancers type, create mobile phone application for daily use, and provide real-time updates on the platform as new treatment pathways emerge. We can envision this platform being implemented as a decision support tool, where information about enrolled patients is entered into a model built using data from this platform, predicting the best next steps for a patient along with reliability statistics to validate the model’s accuracy.
Conclusion
In the past decade, the number of successful targeted therapies with attendant mutational biomarkers has steadily increased. While oncologists have access to guidelines and up-to-date literature on advancements in oncology, navigating them can be challenging and time consuming.
In this article, we report the successful development and implementation of an interactive online decision-tool to educate and inform providers on the importance of biomarker testing and clinical consequences of these results. The TMA platform is a collection of clinical algorithms specific to NSCLC that offers step-by-step, question-and-answer based algorithmic approach to navigate the complexities of diagnosis/treatment based on the type of mutation and assist the clinician in making decisions on patient treatment. The goal of this pilot phase of our TMA platform was to increase the appropriate and timely use of NGS among health-system providers, so that patients receive the best treatment or therapy in an expeditious fashion.
As we bring the TMA platform to a wider audience, we foresee such a solution being offered for more cancers with such predictive markers. Such a tool allows for creating greater transparency around care decision making, therapeutic selection and care delivery; and improve quality and efficiency by reducing non-value-added intra-provider variability in care.
Supplementary Material
Contributor Information
Krithika Bhuvaneshwar, Georgetown University Innovation Center for Biomedical Informatics (Georgetown-ICBI), Georgetown University Medical Center, Washington, DC 20007, United States.
Chul Kim, Oncology, Medstar Georgetown Cancer Center, Washington, DC 20007, United States.
Kaushal Parikh, Oncology, Hackensack Meridian Health, Neptune City, NJ 07753, United States.
Joshua E Reuss, Oncology, Medstar Georgetown Cancer Center, Washington, DC 20007, United States.
Camelia Bencheqroun, Georgetown University Innovation Center for Biomedical Informatics (Georgetown-ICBI), Georgetown University Medical Center, Washington, DC 20007, United States.
Anvitha G Agraharam, Georgetown University Innovation Center for Biomedical Informatics (Georgetown-ICBI), Georgetown University Medical Center, Washington, DC 20007, United States.
Ayesha Munir, Oncology, Medstar Georgetown Cancer Center, Washington, DC 20007, United States.
Adil Alaoui, Georgetown University Innovation Center for Biomedical Informatics (Georgetown-ICBI), Georgetown University Medical Center, Washington, DC 20007, United States.
Yuriy Gusev, Georgetown University Innovation Center for Biomedical Informatics (Georgetown-ICBI), Georgetown University Medical Center, Washington, DC 20007, United States.
Irina G Veytsman, Oncology, Medstar Georgetown Cancer Center, Washington, DC 20007, United States.
Author contributions
Irina G. Veytsman designed the study. Chul Kim, Kaushal Parikh, Joshua E Reuss, and Ayesha Munir created the content for the clinical pathways. Yuriy Gusev, Anvitha G. Agraharam, Krithika Bhuvaneshwar, Camelia Bencheqroun, and Anvitha G. Agraharam developed the platform. Krithika Bhuvaneshwar drafted the paper. All authors reviewed and edited the paper.
Supplementary material
Supplementary material is available at JAMIA Open online.
Research ethics and patient consent
N/A.
Funding
This work is funded by the Pfizer grant and Georgetown Lombardi Cancer Center Support Grant (CCSG).
Conflicts of interest
J.R. is Advisory Board/Consultant at Genentech/Roche, Sanofi/Genzyme, Personalis, Guardant, Astrazeneca, BMS, Arcus, Abbvie, Daiichi Sankyo, Catalym. He receives research funding (to institution) from: Genentech/Roche, Verastem, Nuvalent, Mesothelioma Applied Research Foundation, LUNGevity Foundation. He has received honoraria/Speaking Fees from Astrazeneca, Merck.
C.K. is Consultant/advisory board member at Novartis, Janssen, AstraZeneca, Sanofi, PierianDx, Diffuse pharmaceuticals, Mirati, Jazz Pharmaceuticals, Arcus Biosciences, Daiichi Sankyo, Eisai, Regeneron. He receives research funding (to institution) from: AstraZeneca, Bristol-Myers Squibb, Novartis, Genentech, Janssen, Regeneron, Debiopharm, Karyopharm, Daiichi-Sankyo, Lyell Immunopharma.
Data availability
The TMA platform can be accessed via any web browser at https://tma.georgetown.edu. We recommend users to view this website on regular laptops and desktop computers, instead of mobile devices.
References
- 1. Garraway LA, Verweij J, Ballman KV.. Precision oncology: an overview. JCO. 2013;31:1803-1805. [DOI] [PubMed] [Google Scholar]
- 2. Haslem DS, Chakravarty I, Fulde G, et al. Precision oncology in advanced cancer patients improves overall survival with lower weekly healthcare costs. Oncotarget. 2018;9:12316-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gutierrez ME, Choi K, Lanman RB, et al. Genomic profiling of advanced non-small cell lung cancer in community settings: gaps and opportunities. Clin Lung Cancer. 2017;18:651-659. [DOI] [PubMed] [Google Scholar]
- 4. MacLean E, Louder A, Saverno K, et al. Molecular testing patterns in metastatic non-small cell lung cancer. Am J Manag Care. 2016;22:e60-7. [PubMed] [Google Scholar]
- 5. Celine Audibert MS, Glass D, Kozak M, et al. Trends in the molecular diagnosis of lung cancer, results from an online market research survey. Friends Cancer Res. 2017. [Google Scholar]
- 6. Illei PB, Wong W, Wu N, et al. ALK testing trends and patterns among community practices in the United States. JCO Precis Oncol. 2018;2:1-11. [DOI] [PubMed] [Google Scholar]
- 7. Smeltzer MP, Wynes MW, Lantuejoul S, et al. The International Association for the Study of Lung Cancer Global Survey on molecular testing in lung cancer. J Thorac Oncol. 2020;15:1434-1448. [DOI] [PubMed] [Google Scholar]
- 8. Rivera MP, Charlot M, Durham DD, et al. Molecular biomarker and programmed death-ligand 1 expression testing in patients with advanced stage non-small cell lung cancer across North Carolina community hospitals. Chest. 2021;160:1121-1130. [DOI] [PubMed] [Google Scholar]
- 9. Enewold L, Thomas A.. Real-world patterns of EGFR testing and treatment with erlotinib for non-small cell lung cancer in the United States. PLoS One. 2016;11:e0156728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shen C, Kehl KL, Zhao B, et al. Utilization patterns and trends in epidermal growth factor receptor (EGFR) mutation testing among patients with newly diagnosed metastatic lung cancer. Clin Lung Cancer. 2017;18:e233-e241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barrett J, Turner B, Silva S, et al. Clinical pathways on a mobile device. BMJ EBM. 2020;25:131-137. [DOI] [PubMed] [Google Scholar]
- 12. Kluwer W. UpToDate Clinical Pathways. 2023. Accessed June 2, 2022. https://www.uptodate.com/contents/es/table-of-contents/pathways
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The TMA platform can be accessed via any web browser at https://tma.georgetown.edu. We recommend users to view this website on regular laptops and desktop computers, instead of mobile devices.