Abstract
In the yeast Saccharomyces cerevisiae, 2 types of trehalase activities have been described. Neutral trehalases (Nth1 and Nth2) are considered to be the main proteins that catalyze intracellular trehalose mobilization. In addition to Nth1 and Nth2, studies have shown that acid trehalase Ath1 is required for extracellular trehalose degradation. Although both neutral and acid-type trehalases have been predominantly investigated in laboratory strains of S. cerevisiae, we sought to examine the phenotypic consequences of disrupting these genes in wild strains. In this study, we constructed mutants of the trehalose degradation pathway (NTH1, NTH2, and ATH1) in 5 diverse S. cerevisiae strains to examine whether published lab strain phenotypes are also exhibited by wild strains. For each mutant, we assessed a number of phenotypes for comparison to trehalose biosynthesis mutants, including trehalose production, glycogen production, cell size, acute thermotolerance, high-temperature growth, sporulation efficiency, and growth on a variety of carbon sources in rich and minimal medium. We found that all trehalase mutants including single deletion nth1Δ, nth2Δ, and ath1Δ, as well as double deletion nth1nth2Δ, accumulated higher intracellular trehalose levels compared to their isogenic wild-type cells. Also, nth1Δ and nth1Δnth2Δ mutants exhibited mild thermal sensitivity, suggesting a potential minor role for trehalose mobilization when cells recover from stress. In addition, we evaluated phenotypes more directly relevant to trehalose degradation, including both extracellular and intracellular trehalose utilization. We discovered that intracellular trehalose hydrolysis is critical for typical spore germination progression, highlighting a role for trehalose in cell cycle regulation, likely as a storage carbohydrate providing glycolytic fuel. Additionally, our work provides further evidence suggesting Ath1 is indispensable for extracellular trehalose utilization as a carbon source, even in the presence of AGT1.
Keywords: trehalose degradation, trehalase, Saccharomyces cerevisiae, genetic heterogeneity, NTH1, NTH2, ATH1
Introduction
Saccharomyces cerevisiae, the budding yeast, is a model organism for investigating fundamental aspects of eukaryotic cell biology (Duina et al. 2014). Among S. cerevisiae research studies, many utilize a small number of strains adapted to laboratory growth and manipulation, which has proven useful for comparing results between studies in identical strains. One common strain lineage started with the S288C strain, which was isolated by Robert Mortimer, and its derivatives are among the most widely used laboratory strains of S. cerevisiae (Mortimer and Johnston 1986; Engel et al. 2014). This strain background has been useful for studying mutation effects and gene functions, it was the first eukaryotic organism to have its genome completely sequenced, and it has been the source of multiple strain collections such as overexpression and deletion mutant libraries (Mortimer and Johnston 1986; Goffeau et al. 1996; Giaever et al. 2002; Huh et al. 2003; Sopko et al. 2006). However, this reference strain, like any genetic background, only represents a subset of the many aspects of natural yeast biology, highlighting the risk of extrapolating gene–trait correlations observed in lab-domesticated lineages to species as a whole (Warringer et al. 2011; Su et al. 2021; Scott et al. 2023). Although laboratory strains such as the S288C derivatives remain important for studying fundamental elements of eukaryotic biology, investigating the phenotypic variation of S. cerevisiae wild isolates can also be useful to identify both the phenotypic variability within strains of the same species and the genetic basis underlying observed phenotypic variance. As an example, we recently evaluated phenotypic consequences of trehalose biosynthesis mutants in a number of wild yeast strains compared to a laboratory yeast strain, which yielded multiple instances of strain-to-strain variation that enable evaluation of the genetic basis of respective phenotypes (Chen et al. 2022).
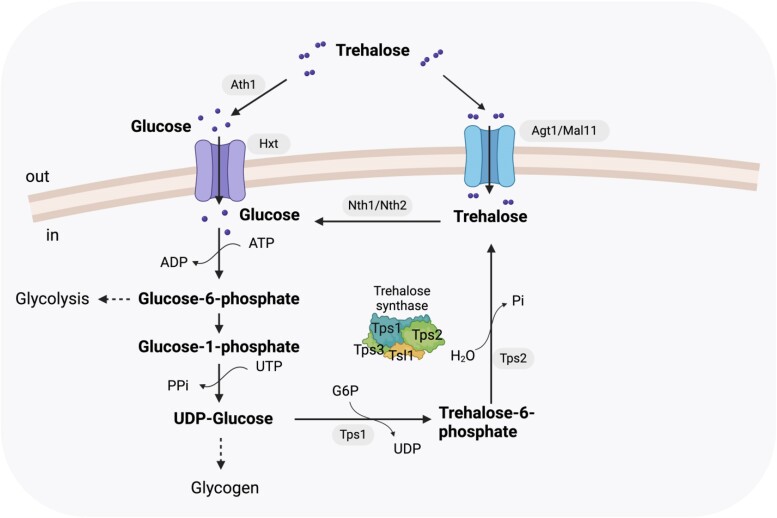
Trehalose is a nonreducing disaccharide in which 2 glucose molecules are connected with a α-1,1-glycosidic linkage. Trehalose metabolism is widely distributed in various organisms, including bacteria, fungi, plants, insects, and invertebrates (Elbein et al. 2003). In the yeast S. cerevisiae, trehalose may constitute as much as 15–20% of its dry weight when undergoing environmental stress (Van Dijck et al. 1995). While yeast cells actively growing on rich carbon sources contain very low levels of trehalose, as they enter the stationary phase when nutrients are exhausted or during growth on nonfermentable carbon sources, the amount of intracellular trehalose substantially increases (Yi et al. 2016). However, when nutrients are replenished, trehalose is rapidly converted to glucose (Nwaka and Holzer 1997). In S. cerevisiae, trehalose hydrolysis to glucose depends on 2 types of hydrolase enzymes: the neutral trehalases (Nth1 and Nth2) and acid trehalase (Ath1), named due to optimal pH for enzymatic activity (Fig. 1) (Jules et al. 2008). The trehalose degradation pathway has been examined in multiple laboratory strains, including S288C, CEN.PK, and SEY6210, among others, and this work has characterized the major genes and proteins involved in yeast trehalose degradation (Nwaka, Mechler, et al. 1995; Jules et al. 2004; Gibney et al. 2015). A wealth of information exists on the neutral trehalase enzymes Nth1 and Nth2, including their role in regulating intracellular trehalose levels within S. cerevisiae as well as characterization in other fungi (Thevelein 1984; Nwaka and Holzer 1997). Nth1 and Nth2 localize to the cytosol and have maximal activity at pH 6.8–7; Nth2 shares 77% identity at the amino acid level with Nth1 (Nwaka, Kopp, et al. 1995; Nwaka, Mechler, et al. 1995). Deletion of NTH1 prevents intracellular trehalose hydrolysis and a loss of measurable trehalase activity (Nwaka and Holzer 1997). NTH2 was found to be expressed at low levels in exponentially growing cells on glucose and at high levels in stationary phase after glucose exhaustion (Nwaka, Kopp, et al. 1995).
Fig. 1.
Schematic of trehalose metabolism in S. cerevisiae. Major metabolites (bold), enzymes/proteins (highlighted in gray), and other reactants/products are indicated. Additionally, branching pathways for glycolysis and glycogen synthesis are indicated. Tps3 and/or Tsl1 have proposed roles in supporting Tps1 and/or Tps2 function, though the mechanism is not clearly understood.
ATH1 encodes an acid trehalase enzyme with an optimal activity at pH 4.5–5 (Destruelle et al. 1995). Saccharomyces cerevisiaeAth1 includes an N-terminal signal peptide and transmembrane domain (Parrou et al. 2005). Deletion of ATH1 prevents cells from utilizing extracellular trehalose as a carbon source (Nwaka et al. 1996; Gibney et al. 2015). Acid trehalase activity has been detected in glucose-grown stationary phase cells and on respiratory substrates, suggesting its activity may be subject to glucose repression, though its association with the general stress response is unlikely because no stress response element (STRE) sequences have been observed in the ATH1 promoter (San Miguel and Argüelles 1994). Divergent observations regarding Ath1 localization have led to multiple models for extracellular trehalose consumption: one model proposes that extracellular Ath1 hydrolyzes trehalose into glucose before glucose import, while another model proposes that trehalose is first imported into the cell and then the vacuole where it is hydrolyzed to glucose by vacuolar-localized Ath1. Despite open questions related to the localization of trehalose catabolism by Ath1, there is a clear consensus that Ath1 is required for utilization of extracellular trehalose (Nwaka et al. 1996; Nwaka and Holzer 1997; Huang et al. 2007; He et al. 2009).
The main goal of this work was to evaluate phenotypic diversity of trehalose degradation mutants in wild strains compared to a lab strain and then use this information to better understand the physiological roles associated with trehalose degradation enzymes. To accomplish this goal, we separately deleted each of the 3 genes encoding proteins for trehalose mobilization (NTH1, NTH2, and ATH1), including a double gene deletion mutant of the neutral trehalase genes and a triple gene deletion mutant of all 3 trehalase genes. We constructed these deletion mutants in 2 commercial wine strains (Simi White and CSM), 1 vineyard isolate [Bb32(3)], 1 oak tree isolate (YPS1000), and an S288C derivative. With regard to phenotypic characterization, we employed 2 strategies. First, we evaluated a series of phenotypes that were previously evaluated with a panel of trehalose biosynthesis mutants made in the same strains; these phenotypes included stationary phase intracellular trehalose and glycogen concentrations, carbon source utilization, high-temperature survival and growth, and sporulation efficiency (Chen et al. 2022). This phenotypic characterization allows for comparing effects of failing to produce intracellular trehalose in biosynthesis mutants with the effects of failing to degrade intracellular trehalose in trehalose degradation mutants. Such comparisons can be helpful to identify phenotypes that are directly related to trehalose levels. Second, we evaluated a series of phenotypes that are more directly relevant to trehalose degradation mutants, including extracellular trehalose utilization and intracellular trehalose utilization during both lag phase and spore germination.
Phenotypic characterization of these trehalose degradation mutants has led to a number of insights, including confirmation of lab strain phenotypes in wild strains, identification of novel phenotypes, and conceptual models for the roles of trehalose and trehalases in cellular physiology. For example, spores from nth1Δnth2Δ strains exhibited a germination defect, indicating the importance of intracellular trehalose utilization to fuel exit from the arrested cell cycle associated with the spore state. Further, mutant strains lacking NTH1 exhibited mildly compromised thermotolerance, suggesting the importance of intracellular trehalose degradation when cells are recovering from thermal stress. As the localization and role of Ath1 in intracellular trehalose degradation have been controversial, we also present data exploring the function of Ath1. We confirmed that Ath1 is indispensable to utilizing extracellular trehalose, even in a strain that encodes AGT1, a disaccharide transporter. These results suggest that Agt1 may not be a typical route for natural trehalose import. In addition, slightly increased intracellular trehalose was found in many ath1Δ mutants, even with cytoplasmic trehalases present, illustrating the potential role of Ath1 in vacuolar degradation of trehalose. However, whether vacuolar trehalose or its metabolism has any physiological consequences remains unclear. We demonstrate that Ath1 is not involved in cytosolic trehalose degradation, as accumulated intracellular trehalose in nth1Δ nth2Δ mutants is not degraded in strains with wild-type or overexpressed ATH1. Altogether, these results compare trehalose degradation mutant phenotypes across diverse genetic backgrounds, highlight an important role for trehalose and trehalose degradation in spore germination, and clarify the role of Ath1 in extracellular trehalose utilization. This work provides a well-characterized panel of trehalose degradation mutants and expands the knowledge on how systems regulating intracellular and extracellular trehalose mobilization are connected.
Materials and methods
Yeast growth media
Yeast cell growth and genetic manipulations were performed using standard approaches (Guthrie and Fink 1991). All media used were either minimal (YNB: 0.67% w/v yeast nitrogen base without amino acids plus 2% w/v indicated carbon sources) or rich (YP: 2% w/v bactopeptone, 1% w/v yeast extract, 2% w/v indicated carbon sources). Exceptions are YPGE and SGE media, rich and minimal formulations, respectively, containing both 3% w/v glycerol and 2% w/v ethanol as respiratory carbon sources. YPD indicates rich media containing 2% glucose (dextrose). Solid media formulations included 2% w/v agar and were poured into standard round 10 cm plates or rectangular plates (Petri 1887).
Yeast growth evaluation
Cell concentration estimation was evaluated by measuring transmittance of 600 nm light (OD600) through a cell suspension using a Gensys 6 UV-Vis spectrophotometer (Thermo Fisher) or using a Synergy H1 Hybrid reader (BioTek). For plate reader growth, 200 µL cultures were prepared in a clear, flat-bottom, Corning Costar 96-well plate; plates were sealed with a Breathe-Easy gas-permeable membrane from Research Products International Corporation. The 96-well plate was incubated at 30 °C with a double orbital shaking at a speed of 559 cpm. Measurements of cell density and cell size were performed using a Coulter Z2 Particle Count and Size Analyzer (Beckman Coulter) with a 100-µm aperture. Cell size measurements were converted from a spherical diameter to volume for reporting. For comparative growth assays, cell dilutions were spotted onto relevant solid growth media. Cell spotting was performed by dilution of a stationary phase culture to an initial OD600 of 1.0, followed by 10-fold serial dilutions. All dilutions were then spotted onto solid media using a replica plater for 96-well plate, either the 8 × 6 or 12 × 8 array as needed (Sigma-Aldrich). Plates were incubated at indicated temperatures for times noted in respective figures and captions. For growth in minimal media containing trehalose or maltose, cells were first grown to stationary phase in YNB + 2% glucose before being diluted to OD600 of 0.05 in minimal trehalose/maltose media. OD600 measurements were then taken every day for a total of 12 days. At least 3 independent biological replicates were performed on different days for spotting assays shown in figures, and a representative image is shown.
Yeast strain construction
The strains used in this study are listed in Supplementary Table 1. Gene deletions were constructed by transforming PCR products amplified from plasmids containing different deletion cassettes: pFA6a-kanMX for kanMX, pAG32 for hphMX, and pAC372 for natAC (Supplementary Table 2). Primers were designed with 40 flanking base pairs identical to the upstream and downstream of genes to be deleted by homologous recombination. All gene deletions in the S288C background (GAL+, HAP1-repaired, prototrophic derivatives) were made by transformation into a diploid to yield a heterozygous gene deletion, which was confirmed by PCR then dissected to get MATa and MATα segregants before mating together obtain a homozygous diploid. Similarly, mutant strains constructed from non-S288C strains (Simi White, YPS1000, CSM, and Bb32) were made in the same way, although homozygous diploids were obtained directly after tetrad dissection (the wild strains are homothallic; thus, after spores germinate, cells can switch mating types and mate to produce colonies that are essentially all diploid cells). All gene deletions were confirmed by PCR (Supplementary Fig. 1). All combinatorial gene deletion/insertion strains were made by mating, sporulating, and tetrad dissection, followed by selective plating. Sporulation was performed by growing cells to log phase in rich media, collecting cells by centrifugation, washing once in 1% w/v potassium acetate, and then resuspending in 1% w/v potassium acetate. Cells were then incubated at room temperature on a roller wheel for at least 6 days before tetrad dissection.
Plasmid construction
The plasmids used in this study are listed in Supplementary Table 2. Plasmids were built using pRS series shuttle vector backbones containing the THD3 promoter and CYC1 3′UTR (Sikorski and Hieter 1989; Mumberg et al. 1995). All inserts were amplified using primers from Integrated DNA Technologies containing 5′end SpeI and 3′end XhoI sites to use for restriction enzyme-based confirmation of inserts during cloning. ATH1 expression plasmids were cloned using Gibson assembly: plasmids p416GPD and p426GPD were linearized using SpeI and XhoI, then incubated with PCR inserts containing ATH1 amplified from DBY12007 genomic DNA along with the Gibson Assembly mixture at 50 °C for 1 h before transforming into TOP10 Escherichia coli (Gibson et al. 2009). After transformation, individual colonies were screened for correct insertion using restriction digest. All gene insertion and allele-specific mutations were confirmed by Sanger sequencing at the Cornell Institute of Biotechnology sequencing core facility.
Assessment of thermotolerance
To assess thermotolerance, minimal media cultures were inoculated with a single colony and grown overnight to stationary phase. To standardize the growth regime leading to a stationary phase culture, cells were then diluted into fresh minimal medium to an OD600 = 0.05 and grown another 24 h to stationary phase. Two aliquots of 0.8 mL cell culture were then removed into microcentrifuge tubes. For the heat shock, one of the aliquots was incubated in a 42 °C thermomixer for 2 h. Both pre- and postheat shocked cell dilutions were plated on rich media (YPD) and incubated at 30 °C for 2–3 days to measure viability by counting colony forming units. At least 3 independent biological replicates were performed for each thermotolerance assay.
Measurement of sporulation efficiency
Sporulation was performed by growing cells to log phase in rich media (except plasmid-containing cells, which were grown in minimal media for plasmid maintenance), collecting cells by centrifugation, washing twice in 1% w/v potassium acetate, and then resuspending in 1% w/v potassium acetate. Cells were then incubated at room temperature on a roller wheel for at least 6 days. Sporulated cultures were evaluated by counting tetrads, or asci containing 4 spores; 2-spore- and 3-spore-containing asci, dyads or triads, respectively, were not observed in any significant fraction. Sporulation efficiency was calculated as the proportion of observed tetrads over the total number of observed cells, with a minimum of 300 total cells counted. At least 3 independent biological replicates were performed for each sporulation efficiency assay.
Measurement of trehalose and glycogen
Trehalose and glycogen levels were measured essentially as described (Parrou and François 1997). Briefly, 10 OD600 units of stationary cells were harvested, washed in cold water, and resuspended in 250 µL of 0.25 M sodium carbonate. Cell mixtures were stored at −80 °C until the assay was performed. To begin the assay, cells were boiled at 95 °C for 4 h with occasional agitation—this step extracts the trehalose and glycogen, as both are highly stable and not heat-degraded (any residual glucose, however, is degraded under these alkaline, high-temperature conditions). Next, 150 µL of acetic acid was added to the sample, followed by 600 µL of 0.2 M sodium acetate. After mixing, two 350 µL aliquots were removed to fresh tubes, and either 5 µL of 70 U/mL trehalase (Megazyme) or 70 U/mL amyloglucosidase (Sigma-Aldrich) was added. These reactions were incubated overnight at 37 or 57 °C, respectively, in a thermomixer set at 550 rpm (Eppendorf). Next, the sample was clarified by centrifugation at maximum speed for 3 min, and 200 µL of each sample was used to measure the amount of glucose liberated from trehalose or glycogen using the Glucose (GO) Assay Kit (Sigma-Aldrich).
Evaluation of germination
Germination was initially evaluated from sporulated cultures after being incubated in sporulation medium (1% w/v potassium acetate) at room temperature for 6 days. Six tetrads from each culture were dissected on YPD plates (24 total spores per culture), which were incubated at 30 °C for 8 h before the number of resulting cells from each germinated spore was counted by visual observation using the tetrad dissection microscope. To evaluate the time required for bud emergence from spores between strains and their respective trehalose mutants, 2 tetrads (8 spores) from the sporulated cultures were dissected on YPD plates and incubated at 30 °C. Through visual inspection each hour, we evaluated the number of hours required for bud emergence. Finally, time required for bud emergence of S288C-derived strains containing ATH1 expression plasmids were similarly investigated to evaluate potential suppression activity of overexpressed ATH1 on nth1Δ nth2Δ delayed bud emergence. Plasmid-containing strains were sporulated, and for each, 2 tetrads were dissected from 3 independent cultures, resulting in 24 spores per tested strain. Bud emergence time was evaluated as described above. A number of data points were not included in the resulting figure as some spores either failed to germinate within the 10-h assay window or failed to germinate after 3 days growth at 30 °C (0/24 spores removed from ura3Δ0 + p416GPD, 1/24 removed from ura3Δ0 + p416GPD-ATH1, 9/24 spores removed from nth1Δ nth2Δ ura3Δ0 + p416GPD; 9/24 spores were removed from nth1Δ nth2Δ ura3Δ0 + p416GPD-ATH1).
Statistical analysis
All experiments were conducted using at least 3 independent biological replicates. Mutant phenotypes were evaluated for statistical significance compared to their isogenic wild-type strains using a 2-tailed paired t-test. Asterisks (*) in figures indicate the mutant phenotype showed a difference (P < 0.05) compared to its isogenic wild type (P-values were not corrected for multiple hypothesis testing). Separately, to evaluate statistically significant phenotypic differences between the wild strains examined, or also between different strains with identical gene deletions, 1-way ANOVA with post hoc Tukey honest significant difference (HSD) tests were performed (Supplementary Tables 3–7). Corresponding to the F-statistic of 1-way ANOVA, indicated P-values lower than 0.05 suggest that one or more evaluated treatments were significantly different.
Results and discussion
Trehalose mutant phenotypes under consideration
Trehalose is a widely distributed carbohydrate in nature that is synthesized in S. cerevisiae in response to nutrient limitation and during certain environmental stress (Lillie and Pringle 1980; Crowe et al. 1984; Thevelein 1984; Tapia et al. 2015). To examine the phenotypic diversity of mutants in the trehalose mobilization pathway, we constructed single deletion mutants (nth1Δ, nth2Δ, and ath1Δ), 1 double deletion mutant (nth1Δnth2Δ), and 1 triple deletion mutant (nth1Δnth2Δath1Δ) in 5 different strains of S. cerevisiae (Supplementary Table 1). These strains included 2 commercial wine strains (Simi White and CSM), 1 vineyard isolate [Bb32(3)], 1 oak tree isolate (YPS1000), and an S288C derivative for comparison. We initially focused on a core set of relevant phenotypes that could be easily compared to phenotypes of trehalose biosynthesis mutants in the same strain backgrounds (Chen et al. 2022). By evaluating these phenotypes, we can compare the consequences of failing to hydrolyze trehalose (excess intracellular trehalose) to the consequences of failing to produce trehalose (absence of intracellular trehalose). These comparisons can then be used to evaluate hypotheses about the role of intracellular trehalose on different cellular processes. We first quantified both intracellular trehalose and glycogen levels in single and double deletion trehalase mutants. Further, as stress tolerance has been correlated to intracellular trehalose concentration, we assessed thermotolerance (42 °C for 2 h) and ability to grow at an elevated temperature (growth at 37 °C). We also examined sporulation efficiency, cell size, and growth on multiple carbon sources in both rich and minimal media as trehalose biosynthesis mutants exhibit phenotypes in these conditions (Neves et al. 1995; de Silva-Udawatta and Cannon 2001; Gancedo and Flores 2004; Walther et al. 2013). We additionally evaluated trehalase roles in hydrolysis of both extracellular and intracellular trehalose, as these phenotypes are particularly relevant to the function of the Nth1, Nth2, and Ath1 enzymes.
Construction and phenotypic characterization of nth1Δ, nth2Δ, and nth1Δnth2Δ
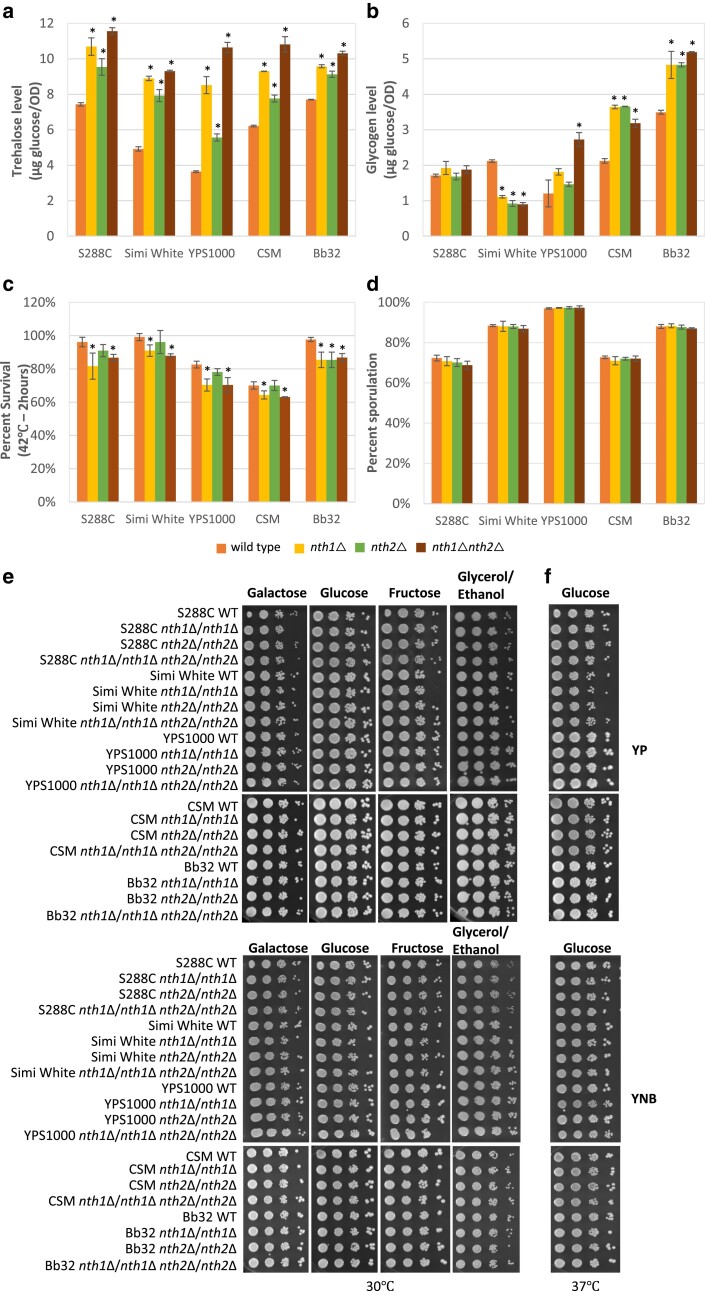
In S. cerevisiae, degradation of cytoplasmic trehalose is mainly catalyzed by neutral trehalase enzymes, encoded by NTH1 and NTH2 (Fig. 1) (Nwaka, Mechler, et al. 1995). Transcriptional activation of NTH1 depends on the general stress response mediated by the Msn2/Msn4 transcription factors through the STRE elements present in its promoter (Thevelein 1984; Zähringer et al. 2000; Panni et al. 2008). The Nth enzymes are also subject to activation through posttranslational modification (Alblova et al. 2019). In the present study, all nth1Δ and nth2Δ mutants exhibited increased trehalose levels compared to their isogenic wild-type strains (Fig. 2a). All 5 nth2Δ mutants accumulated trehalose to a lower extent than nth1Δ mutants, though still higher than their isogenic wild-type strains, highlighting that Nth2 plays a minor but significant role in intracellular trehalose degradation (Fig. 2a). All nth1Δnth2Δ double deletion mutants exhibited additive effects from deleting both neutral trehalases as they accumulated the highest level of intracellular trehalose (Fig. 2a). This result agrees with multiple previous studies demonstrating that the absence of Nth1 results in yeast cells unable to breakdown intracellular trehalose (Thevelein 1984; Nwaka, Kopp, et al. 1995; Gibney et al. 2015). Also, increased trehalose levels in nth2Δ mutants align well with the previous finding that overexpression of NTH2 in an nth1 mutant resulted in an approximately 10-fold increase of trehalose-hydrolyzing activity, suggesting Nth2 is a functional trehalase (Jules et al. 2008). Previous work using S288C-derived lab strains demonstrated a negative correlation between intracellular trehalose and glycogen levels, suggesting that when trehalose biosynthesis is disrupted, UDP-glucose gets shunted to glycogen production (François et al. 1991; de Silva-Udawatta and Cannon 2001). However, here we observed that intracellular glycogen levels were less consistent: mutants in different genetic backgrounds exhibited less, equivalent, or more glycogen than their isogenic wild-type strains, suggesting glycogen levels are either unrelated to flux through trehalose metabolism or the relationship is more complex than overflow metabolism (Fig. 2b). It is not clear why trehalase degradation mutants resulted in variation in the glycogen levels, but this further suggests that intracellular trehalose and glycogen levels are not necessarily anticorrelated, as also observed in trehalose biosynthesis mutants (Chen et al. 2022). We did not observe any significant cell size variations with any of these mutants (Supplementary Fig. 2a).
Fig. 2.
nth1Δ, nth2Δ, nth1Δ nth2Δ mutant phenotypes. a) Intracellular trehalose levels. b) Intracellular glycogen levels. c) Thermotolerance (heat shocked at 42 °C for 2 h). d) Sporulation efficiency. e and f) Growth at 30 and 37 °C on minimal (bottom panels) and rich (top panels) media. Indicated strains were grown overnight in YNB + 2% glucose liquid before 10-fold serial dilutions were prepared and spotted onto the indicated media. The initial dilution had an OD600 of 1.0. Plates were incubated at 30 °C for 2 days on rich media and 3 days for minimal media. Three biological replicates were performed for all tested phenotypes. Asterisks represent statistically significant differences (P < 0.05) between the mutants and their isogenic wild-type strains.
Trehalose has been proposed to be a protective agent against a variety of abiotic stresses (Hottiger et al. 1987; Hounsa et al. 1998; Singer and Lindquist 1998a; Elbein et al. 2003). Multiple studies revealed the ability of trehalose to stabilize proteins in vitro by retaining their native conformation at elevated temperatures and suppressing aggregation of denatured proteins (Singer and Lindquist 1998a, 1998b). However, recent work using the Agt1 overexpression system demonstrated that increased intracellular trehalose did not correlate with increased heat resistance (Gibney et al. 2015). Therefore, to further investigate the role of trehalose in heat stress protection in vivo, we tested thermotolerance in nth1Δ, nth2Δ, and nth1Δnth2Δ mutants by exposing the cells to 42 °C for 2 h. We found that none of the tested mutants had increased thermotolerance, despite higher intracellular trehalose levels. In contrast, all nth1Δ and nth1Δ nth2Δ mutants in all 5 strains exhibited mildly compromised thermotolerance when exposed to 42 °C for 2 h, ranging from 85.0 to 91.9% survival compared to their isogenic wild type (Fig. 2c). Four out of 5 nth2Δ mutants exhibited the same level of heat resistance as their isogenic wild types, except the Bb32 nth2Δ that had a survival rate similar to nth1Δ. Comparing results between Fig. 2a and c suggests that trehalose accumulation and increased thermotolerance do not correlate, further indicating that intracellular trehalose may not have a significant role in heat protection in vivo. In addition, mild thermosensitivity associated with these mutants suggests that trehalose degradation activity may play a minor role in cellular protection against heat, likely associated with recovery. Our data align with previous work demonstrating that nth1Δnth2Δ cells were found to be more thermosensitive than wild-type cells when exposed to 48 °C for 60 min and 50 °C for 20 min (Nwaka, Mechler, et al. 1995; Gibney et al. 2015). A possible explanation for this phenotype is based on the observation that trehalose can interfere with protein folding in vitro, which is also used to explain the rapid hydrolysis of trehalose in wild-type cells following heat shock as a mechanism to allow cells to more rapidly refold proteins and recover from heat shock (Singer and Lindquist 1998a). In addition to examining acute thermotolerance, we evaluated the ability of these mutants to grow at an elevated temperature. None of the deletion mutants exhibited any growth defect when incubated at 37 °C (Fig. 2e and f; Supplementary Fig. 3). In addition to evaluating heat stress response, we observed that sporulation efficiency was unaffected in nth1Δ, nth2Δ, and nth1Δnth2Δ mutants in any of the 5 tested genetic backgrounds (Fig. 2d).
We also examined whether the absence of neutral trehalase enzymes affects utilization of different carbon sources, which is a common phenotype associated with deletion of trehalose biosynthesis genes but found no noticeable growth defects with trehalase mutants (Fig. 2e). Trehalose biosynthesis mutants tps1Δ and tps2Δ exhibit defective growth on different carbon sources (highly fermentative carbon source growth defects for tps1Δ and respiratory carbon source growth defects for tps2Δ), and the severity of the growth defect is highly variable between strains (Navon et al. 1979; González et al. 1992; Stucka and Blázquez 1993; Walther et al. 2013; Gibney et al. 2015; Chen et al. 2022). The corresponding lack of carbon source utilization defects for the mutants tested here suggests that intracellular trehalose concentration is not relevant for carbon source utilization signaling. The observed phenotypes in trehalose biosynthesis mutants are likely more related to other hypotheses, including independent signaling roles for either Tps1 or trehalose-6-phosphate, among others (Chen et al. 2022). The absence of growth defects observed here is in contrast to a report using laboratory strain SEY6210, where nth1Δ grew poorly on rich media containing glycerol compared to its isogenic wild type (Nwaka, Mechler, et al. 1995). Notably, previous work has highlighted that multiple laboratory strains (CEN.PK, W303, and S288C) of S. cerevisiae are unable to use glycerol as a sole carbon source in a minimal medium, suggesting growth on rich media containing glycerol may involve utilization of additional carbon sources other than glycerol (Swinnen et al. 2013).
Construction and phenotypic characterization of ath1Δ
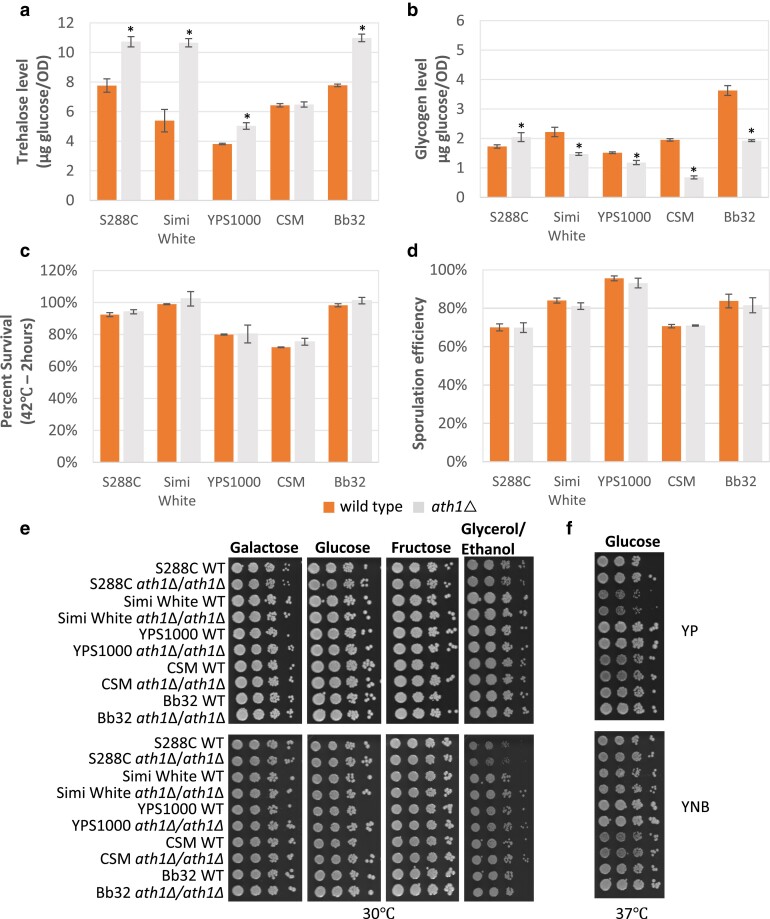
ATH1 encodes the acid trehalase enzyme with optimal activity at an acidic pH range (4.5–5) (Destruelle et al. 1995). We constructed ath1Δ mutants in all 5 genetic backgrounds and performed phenotype tests identical to those for the neutral trehalase mutants. Stationary phase intracellular trehalose levels in 4 of the 5 tested ath1Δ strains were significantly higher than their isogenic wild-type cells, ranging from 132 to 198%; CSM ath1Δ alone maintained wild-type levels of trehalose (Fig. 3a). Similarly, a previous study reported that ath1Δ in the S288C genetic background maintained higher intracellular trehalose accumulation than wild-type cells under both a saline-stress condition and in the postdiauxic phase during growth in 2% glucose; it was suggested that acid trehalase activities might be involved in intracellular trehalose metabolism under these stress conditions (Garre et al. 2009). Therefore, it is possible that Ath1 possesses dual roles in trehalose mobilization, both intracellular and extracellular, although if accurate, the functional role of intracellular trehalose degradation by Ath1 remains unclear. Glycogen levels of ath1Δ were slightly higher in the S288C strain, but lower in the 4 wild strains (Fig. 3b). We did not observe any significant cell size variations with any of the ath1Δ mutants (Supplementary Fig. 2b).
Fig. 3.
ath1 Δ mutant phenotypes. a) Intracellular trehalose levels. b) Intracellular glycogen levels. c) Thermotolerance (heat shocked at 42 °C for 2 h). d) Sporulation efficiency. e and f) Growth at 30 and 37 °C on minimal and rich media. Indicated strains were grown overnight in YNB + 2% glucose liquid before 10-fold serial dilutions were prepared and spotted onto the indicated media. The initial dilution had an OD600 of 1.0. Plates were incubated at 30 °C for 2 days on rich media and 3 days for minimal media. Three biological replicates were used for all tested phenotypes. Asterisks represent statistically significant differences (P < 0.05) between the mutants and their isogenic wild-type strains.
Deleting ATH1 did not significantly compromise thermotolerance of any tested strain (Fig. 3c). Several previous studies investigated the effect of Ath1 absence on stress resistance, though some of them were difficult to interpret because they were affected by a confounding difference in an auxotrophic nutrient requirement associated with a ura3Δ mutation between mutant and reference strains (Nwaka, Mechler, et al. 1995; Kim et al. 1996; Chopra et al. 1999). Nwaka, Mechler, et al. (1995) detected no significant Ath1 activity from cells recovered from thermal stress, suggesting that unlike NTH1, the ATH1 gene is not involved in the recovery of cells after heat shock. Notably ATH1 transcription is not activated by stress likely due to lack of a STRE in its promoter, which mediates general stress-induced transcriptional activity by binding the Msn2/4 transcription factor (Destruelle et al. 1995; Nwaka et al. 1996). As with thermotolerance, none of the ath1Δ strains exhibited a growth defect when incubated at high temperature, 37 °C (Fig. 3f; Supplementary Fig. 4). Sporulation efficiency and carbon source utilization were similarly unaffected by the absence of Ath1 in any of the 5 tested genetic backgrounds (Fig. 3d and e).
Comparing trehalose degradation phenotypes to trehalose biosynthesis phenotypes
It is noteworthy that many of the phenotypes evident in trehalose biosynthesis mutants are not evident in trehalose degradation mutants (Chen et al. 2022). Trehalose biosynthesis mutants evaluated with a similar panel of phenotypic tests exhibited heat survival defects, temperature-sensitive growth defects, sporulation defects, and carbon source utilization defects. Other than a mild defect in heat survival associated with nth1Δ, trehalose degradation mutants did not exhibit variability compared to wild-type cells for these phenotypes. Comparison of these phenotypes suggests that lack of trehalose itself is not responsible for the phenotypes described above associated with biosynthesis mutants. For example, accumulation of intracellular trehalose either by disruption of NTH genes or via import using constitutively expressed AGT1 is sufficient to induce both desiccation tolerance and freeze–thaw tolerance (Tapia et al. 2015; Chen and Gibney 2022). Together, these results suggest that trehalose itself acts as a molecular chaperone under desiccation or freeze–thaw conditions, both conditions with low water activity that may align with the notion that trehalose can act as a molecular chaperone by replacing the hydrogen-bonding activity associated with water molecules (Olsson et al. 2016; Shao et al. 2019). In contrast, accumulation of trehalose does not appear to enhance heat survival, suggesting that trehalose itself may not act as a molecular chaperone for heat shocked cells in conditions with high intracellular water activity (Gibney et al. 2015). The absence of phenotypes observed for trehalose degradation mutants compared to biosynthesis mutants, along with repeated observation in multiple strains, provides a useful comparison to evaluate direct effects of trehalose. Multiple studies have sought to further distinguish the trehalose-independent roles of trehalose metabolism in carbon source utilization, heat survival, and sporulation, though detailed molecular mechanisms that incorporate known phenotypic data remain elusive (Gibney et al. 2015; Chen et al. 2022). Another contrast to the phenotypic characterization of trehalose biosynthesis mutants is strain-to-strain phenotypic variability. For most of the biosynthesis mutants, at least one strain failed to exhibit the phenotype associated with the other strains. This was not the case with trehalose degradation mutants, which exhibited more uniformity among phenotypes tested here. One notable exception, similar to trehalose biosynthesis mutants, was the variability in stationary phase intracellular glycogen concentrations between strains. It is possible that these variations are related to variations in stationary phase between strains, such as differential timing for stationary phase initiation or differential expression levels of glycogen metabolism enzymes.
Extracellular trehalose utilization by Ath1
Beyond phenotypes useful for comparison with trehalose biosynthesis mutants, we sought to evaluate the consequences of trehalase gene deletion on a number of phenotypes directly relevant to conditions of enzymatic trehalose hydrolysis, including intracellular and extracellular trehalose utilization. Yeast growth using extracellular trehalose is relatively slow in a number of laboratory strain backgrounds (CEN.PK, S288C, and JF657). The assimilation of this disaccharide is purely oxidative, and in an S288C derivative background, it yields a doubling time of approximately 17 h (Nwaka et al. 1996; Jules et al. 2004; Gibney et al. 2015). Conflicting evidence regarding Ath1 localization and activity has resulted in multiple conceptual models for extracellular trehalose degradation. To investigate the cellular localization of Ath1, Huang et al. (2007) expressed a C-terminal, GFP-tagged Ath1 in cells grown in minimal glucose media to log phase, then used trafficking mutants to illustrate Ath1 reaches the vacuole via the multivesicular body pathway. They suggest a conceptual model wherein vacuolar Ath1 consumes extracellular trehalose in the vacuole. However, the publication also reports only roughly 50% of the acid trehalase activity is vacuolar, but did not specify localization of remaining activity (Huang et al. 2007). Furthermore, no evidence was presented regarding how extracellular trehalose is transported both into the cell and then into the vacuole. In contrast, He et al. (2009) used mCherry fused to Ath1 to demonstrate both vacuolar and plasma membrane localization. By disrupting delivery of Ath1 to the vacuole, this study demonstrated no effect on either Ath1 enzymatic activity or growth rate of cells on extracellular trehalose, indicating that vacuolar-localized Ath1 is not important for extracellular trehalose utilization (He et al. 2009). He et al. (2009) also demonstrated that fusing the signal peptide of invertase to an N-terminally truncated Ath1 missing its native signal sequence was sufficient to allow ath1Δ growth on trehalose, suggesting that functional localization of Ath1 at the cell periphery is required for extracellular trehalose mobilization. Additionally, fusion of the Ath1 signal sequence and transmembrane domain to an invertase allele lacking its own signal sequence and transmembrane domain is required to recover invertase activity and growth on sucrose in a suc2Δ mutant (He et al. 2009). This study supports the conceptual model that extracellular trehalose is hydrolyzed to glucose outside of the cell by extracellular Ath1. The 2 models are not mutually exclusive, as both mechanisms could be applicable. It is also possible that plasma membrane localization represents functional expression of Ath1, while vacuolar-localized Ath1 is merely a consequence of membrane recycling and is not connected to Ath1 function (Harris and Cotter 1988; Alizadeh and Klionsky 1996; Jules et al. 2004; Parrou et al. 2005; Huang et al. 2007). It is noteworthy that both studies employed fluorescent protein fusions to Ath1, as these strategies can lead to potential confounding artifacts. For example, GFP protein fusions causing mislocalization to the vacuolar lumen or YFP protein fusions forming nonnative structures (though the same study demonstrated that this was not an issue with an mCherry protein fusion) (Roberts et al. 1992; Swulius and Jensen 2012). These observations suggest caution should be applied to protein localization or quantification of localization based only on 1 type of data. Further, both Ath1 localization studies localized Ath1 during growth on glucose, a condition in which proper Ath1 localization for extracellular trehalose mobilization is likely unimportant (Huang et al. 2007; He et al. 2009).
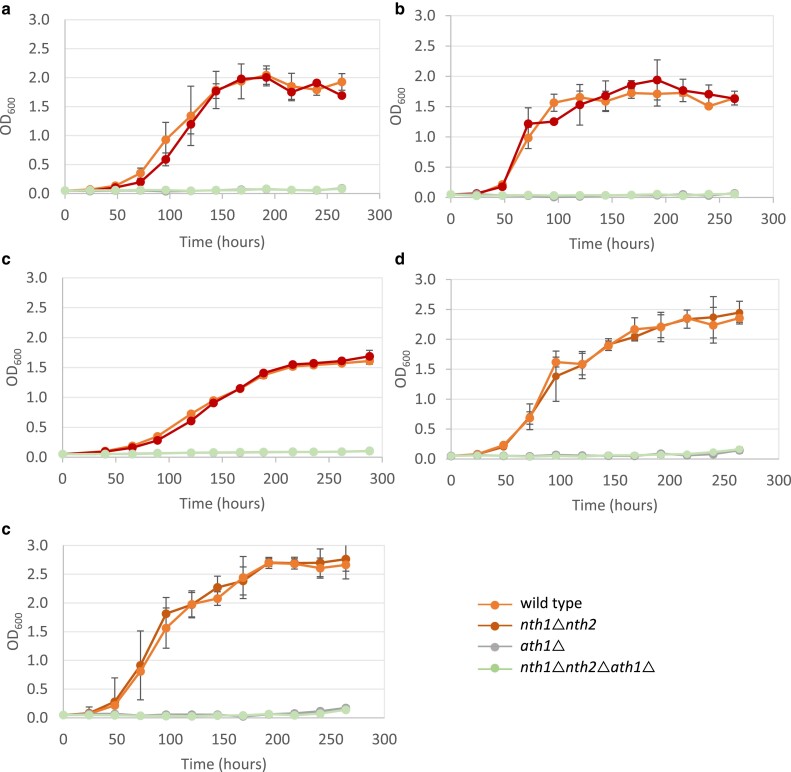
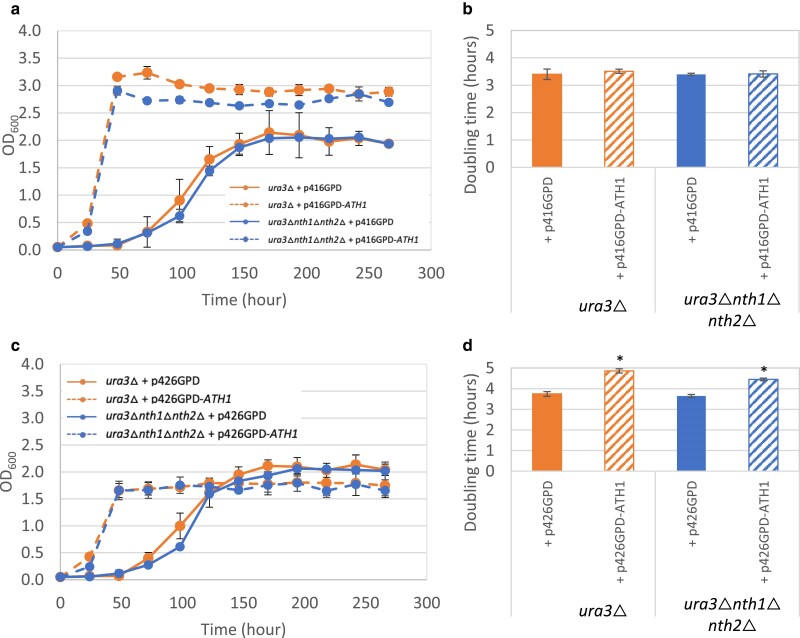
As multiple previous studies have demonstrated that Ath1 is required for extracellular trehalose degradation, we first sought to evaluate whether this holds true for the nonlaboratory strains tested here (Alizadeh and Klionsky 1996; Nwaka et al. 1996; Jules et al. 2004). We included deletion of the NTH genes, nth1Δ nth2Δ, with or without deletion of ATH1 for comparison with strains lacking either all cellular trehalase enzyme activity or only lacking cytosolic trehalase activity, respectively. Both ath1Δ single mutants and nth1Δnth2Δath1Δ triple mutants failed to grow on trehalose in all 5 genetic backgrounds, whereas nth1Δnth2Δ mutants exhibited similar growth rates compared to their isogenic wild-type strains in trehalose-containing media (Fig. 4). To evaluate effects of ATH1 overexpression on extracellular trehalose utilization, we used the S288C laboratory strain background due to an available auxotrophic mutation that prevents uracil biosynthesis, ura3Δ0, which was subsequently used in all experiments requiring plasmid selection. We also examined yeast cells overexpressing ATH1 from a low- or high-copy plasmid under control of the strong, constitutive TDH3 promoter (Mumberg et al. 1994). We found that low-copy overexpression of ATH1 (p416GPD-ATH1) in both wild type and nth1Δ nth2Δ decreased lag phase and increased growth rate in a minimal medium containing trehalose as the sole carbon source, highlighting the sufficiency of Ath1 for extracellular trehalose mobilization (Fig. 5a). This result also demonstrates that the presence or absence of Nth1/2 activity had no effect on extracellular trehalose utilization. Expression of ATH1 from a low-copy plasmid (p416GPD-ATH1) did not have any effects on growth rate in minimal glucose medium (Fig. 5b). These observations align with a previous study that observed constitutive overexpression of ATH1 under the control of the TDH3 promoter increased acid trehalase activity 4-fold in intact cells, with a 3-fold reduction of time in lag phase (Jules et al. 2004). ATH1 overexpression from a high-copy plasmid (p426GPD-ATH1) resulted in similar lag phase reduction and increased growth rate, though expression at high-copy levels also had negative consequences for cellular physiology, including lower stationary phase cell concentrations and a slower growth rate in standard minimal glucose medium (Fig. 5c and d). One potential interpretation of this phenomenon is protein burden, which has been observed in overexpression systems of multiple glycolytic proteins (Vind et al. 1993; Gong et al. 2006). It is also possible that as a membrane-localized protein, excessive Ath1 expression levels could decrease the efficiency or function of the protein secretion machinery or membrane functionality, ultimately leading to a growth defect. Together, these results corroborate the necessary and sufficient role of Ath1 in extracellular trehalose utilization, along with increased Ath1 activity associated with its overexpression.
Fig. 4.
Extracellular trehalose utilization via Ath1. Growth of wild type, nth1Δ nth2Δ, ath1Δ, and nth1Δ nth2Δ ath1Δ mutants in trehalose: S288C a), Simi White b), YPS1000 c), CSM d), and Bb32 e). Cells were first inoculated into YNB + 2% glucose, incubated overnight, washed once with water, and subcultured to OD600 of 0.05 into YNB + 1% trehalose. OD600 was recorded every day for 12 days. Three biological replicates were performed, and the values are presented as the mean ± SD.
Fig. 5.
Plasmid-based ATH1 overexpression increases the rate of extracellular trehalose utilization. Growth of wild-type and trehalase mutants with and without ATH1 overexpressed from a low-copy a and b) or high-copy c and d) plasmid. a and c) Cells were grown to stationary phase in YNB + 2% glucose, washed once with sterile water, and subcultured to OD600 of 0.05 in YNB + 1% trehalose followed by daily measurement of cell concentration. b and d) Cells were grown to stationary phase in YNB + 2% glucose before subcultured to OD600 of 0.05 in YPD in a 96-well plate, which was then incubated at 30 °C for 48 h. Growth curves were used to calculate doubling time during exponential growth. Three biological replicates were performed for each experiment, and the values are presented as the mean ± SD. Asterisks represent statistically significant differences (P < 0.05) between the mutants and their isogenic control strains.
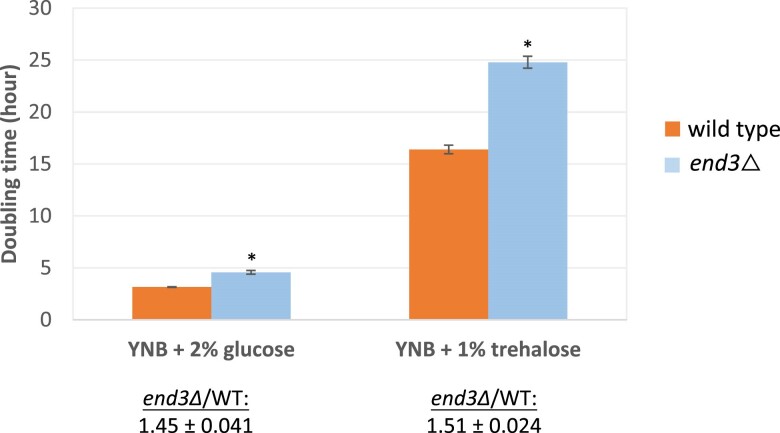
However, these results do not inform whether or not trehalose is consumed by extracellular hydrolysis to glucose or import and hydrolysis in the vacuole. If consumption of extracellular trehalose occurs through import of trehalose into cells and then into the vacuole for subsequent hydrolysis to glucose by Ath1, then one possibility for trehalose import and transport to the vacuole is endocytosis. One study mentioned that an endocytosis mutant failed to grow on trehalose, potentially supporting this model, though no data were shown (Nwaka et al. 1996). Therefore, to test this hypothesis, we constructed an end3Δ mutant, which is reportedly defective in receptor-mediated and fluid-phase endocytosis (Raths et al. 1993). Despite its decreased endocytic activity, end3Δ had a similar growth defect on trehalose compared to its growth defect on glucose: roughly 50% slower than wild type on either carbon source (Fig. 6). This suggests that endocytosis is not a major mechanism of trehalose import, and no alternative model for extracellular trehalose transport to the vacuole has been proposed. This result supports the alternative model described previously in which extracellular trehalose is metabolized to glucose by extracellular Ath1 tethered to the plasma membrane (Jules et al. 2004; He et al. 2009). However, we and others have observed an increase in intracellular trehalose in an ath1Δ strain, which may represent accumulated trehalose in the vacuole, though the cellular role or significance of vacuolar-localized trehalose is unclear (Fig. 2) (Garre et al. 2009).
Fig. 6.
Comparison of doubling times between wild type and end3Δ mutant in glucose and trehalose. Cells were grown to stationary phase in YNB + 2% glucose before being diluted into fresh YNB + 2% glucose or YNB + 1% trehalose liquid media to an initial OD600 of 0.05 in 96-well microplates. The plates were then shaken at 30 °C, and the absorbance of each well was measured every 15 min (48 h in total) in double orbital shaking mode at a speed of 559 cpm. Three biological replicates were performed to calculate exponential doubling times, and the values are presented as the mean ± SD. Asterisks represent statistically significant differences (P < 0.05) between the mutants and their isogenic control strains. Note that the average doubling time ratio (end3Δ/WT) from the 3 replicates is reported below each carbon source; the 2 doubling time ratios are not significantly different based on a t-test with a significance threshold of P < 0.05.
The role of Agt1 in extracellular trehalose utilization
In addition to Ath1, some S. cerevisiae strains possess an alternate path for consuming extracellular trehalose: Agt1-mediated trehalose transport into the cytoplasm followed by intracellular Nth1/2-mediated hydrolysis (Jules et al. 2004; Gibney et al. 2015). The Agt1 transporter is a proton-disaccharide symporter also known as Mal11. Agt1 has accumulated mutations conferring a broader substrate range than its ancestral maltose transporter and can transport trehalose among other disaccharides (Fig. 1) (Han et al. 1995; Brown et al. 2010; Gibney et al. 2015). The observation that deletion of ATH1 abrogates growth on trehalose in all tested strains implies that trehalose import through the Atg1 transport protein may not be a common, natural alternate route for trehalose import. To further investigate this possibility, we first designed primers to selectively amplify a fragment AGT1 (Supplementary Fig. 5). PCR amplification and gel electrophoresis revealed that S288C, YPS1000, and Bb32 contain AGT1, while Simi White and CSM do not (Supplementary Fig. 5). As AGT1 belongs to the MAL gene family, we hypothesized that preculturing on maltose might induce native Agt1 expression, enabling growth on trehalose despite the absence of ATH1; pregrowth in maltose to increase trehalose utilization has been previously described (François and Parrou 2001). Therefore, we first evaluated whether these strains contain a maltose transport gene and the ability to grow in maltose. MAL31, another gene in the MAL family, encodes a transport protein specific to maltose. While PCR confirmed that all 5 tested strains contain MAL31, S288C and YPS1000 did not grow in maltose (Supplementary Fig. 5). S288C-derived strains encode a mutated, inactive maltose regulator protein, resulting in failure to express any MAL gene and concomitant inability to grow on maltose (Supplementary Fig. 5) (Brown et al. 2010). Consequently, we were only able to pregrow Simi White, Bb32, and CSM in maltose (though Bb32 is the only 1 of these 3 containing AGT1), while S288C and YPS1000 were pregrown in glucose. While all wild-type strains were able to grow in minimal trehalose medium, none of the ath1Δ mutants exhibited growth under the same conditions, regardless of pregrowth regime (Supplementary Fig. 5e). As a positive control, we compared the growth of ath1Δ strains with an ath1Δ mutant strain overexpressing Agt1 from the S288C genetic background (Gibney et al. 2015). As expected, based on previous literature, overexpressed Agt1 restored growth on trehalose, recapitulating the observation that the Agt1 transporter allows growth in trehalose when ATH1 is absent (Supplementary Fig. 5e) (Jules et al. 2004; Gibney et al. 2015). In our experiment, the only tested strain able to pregrow on maltose that also encodes AGT1 was Bb32. As Bb32 ath1Δ was also unable to grow using extracellular trehalose regardless of pregrowth regime, it is possible that either Bb32's AGT1 allele contains loss-of-function mutations, or that it is not expressed in the conditions tested potentially due to promoter mutation or because it is part of a silenced copy of the MAL regulon in the subtelomeric region of the chromosome (Brown et al. 2010). Based on these results, while AGT1 can clearly be used to transport extracellular trehalose when constitutively overexpressed, it is unclear how often native expression of AGT1 supports growth on extracellular trehalose. Evaluation of further strains will be required to demonstrate a role for native expression of AGT1 in extracellular trehalose utilization.
Intracellular trehalose utilization during lag phase
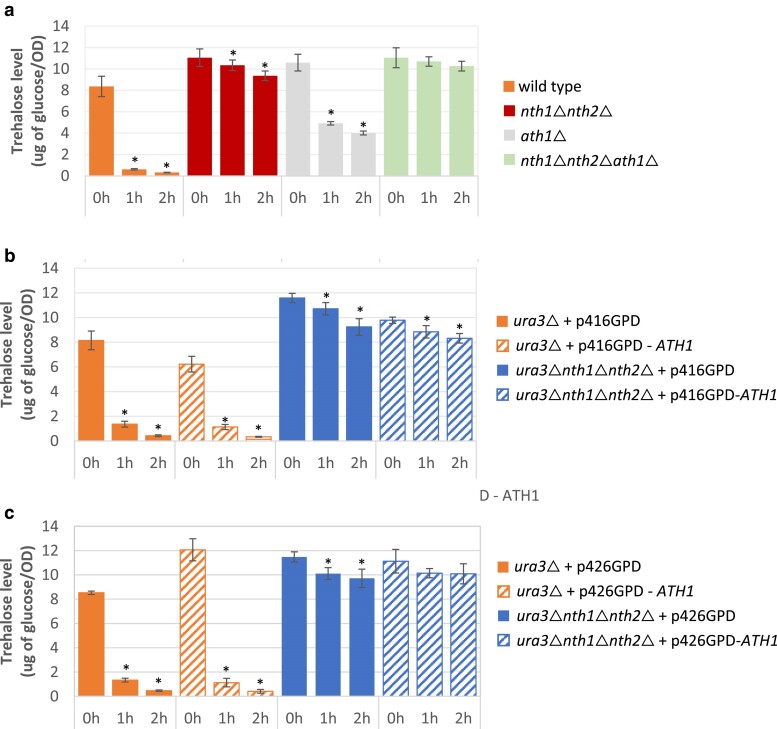
Slow growing or nongrowing cells produce and accumulate high levels of intracellular trehalose, up to 15–20% of dry cell weight, which is quickly mobilized after addition of sufficient nutrients (Silljé et al. 1999). To further examine the role of trehalase enzymes in intracellular trehalose degradation, we monitored the change in intracellular trehalose levels during lag phase in wild-type cells and multiple trehalase mutants from the S288C genetic background. When we transferred stationary phase cells to fresh growth media, we observed that most of the intracellular trehalose in wild-type cells was hydrolyzed within 1–2 h (Fig. 7a). By taking time points at 1 and 2 h after exposure to fresh media, we expect that reduction in trehalose levels represents intracellular enzymatic trehalose degradation rather than dilution by cell division. As anticipated, when both NTH1 and NTH2 were deleted (nth1Δnth2Δ), the initial concentration of intracellular trehalose and rate of trehalose degradation was much slower compared to wild type. In the ath1Δ mutant, intracellular trehalose was still consumed, though not as completely as observed for wild-type cells. In the triple trehalase gene deletion strain (nth1Δnth2Δath1Δ), intracellular trehalose levels did not change significantly between tested time points. This is consistent with previous findings by Parrou et al. (2005) that demonstrated that an nth1nth2 mutant could still mobilize trehalose during prolonged incubation in the stationary phase, but further deletion of ATH1 completely abolished this mobilization (Parrou et al. 2005). These results align with the notion that the majority of intracellular trehalose degradation relies on the Nth1/2 enzymes, with Ath1 also playing some role in intracellular trehalose utilization, potentially related to vacuolar localization.
Fig. 7.
Intracellular trehalose utilization during lag phase. a–c) Stationary phase cells (0 h) from indicated strains pregrown overnight in YNB + 2% glucose were diluted into fresh YNB +2% glucose liquid media with an OD600 of 0.5 and then incubated at 30 °C for 1 hour (1 h) and 2 hours (2 h) before samples were taken for trehalose content measurement. Three biological replicates were performed, and the values are presented as the mean with SD. Asterisks represent statistically significant differences (P < 0.05) between an individual time point compared to its respective time 0.
We also evaluated the effect of ATH1 overexpression on the degradation rate of intracellular trehalose during lag phase. Plasmid-based ATH1 expression was insufficient to degrade intracellular trehalose to wild-type levels when the Nth1/2 enzymes were absent (Fig. 7b and c). This suggests that increased Ath1 activity associated with overexpression is unable to hydrolyze bulk cytoplasmic trehalose, a function performed by Nth1/2 enzymes, which also aligns with their pH optima. However, unconsumed trehalose in ath1Δ mutants during lag phase indicates that a fraction of Ath1-susceptible intracellular trehalose produced in the cytoplasm by glucose-grown stationary phase cells can accumulate in a cellular compartment not accessible to or functionally compatible with Nth1/2, likely the vacuole. While there is no evidence that extracellular trehalose is transported into the vacuole as a utilization route, these results suggest that intracellular trehalose can enter the vacuole, though the mechanism of transport is not characterized. Together, these results suggest that Ath1 degrades vacuolar-localized trehalose, though the biological significance of vacuolar trehalose accumulation/degradation remains unclear and unrelated to the role of Ath1 in extracellular trehalose utilization. Overall, these results indicate that intracellular cytoplasmic trehalose is primarily hydrolyzed by Nth1/2, though Ath1 also has a minor role in intracellular trehalose hydrolysis, likely inside the vacuole.
Intracellular trehalose utilization during spore germination
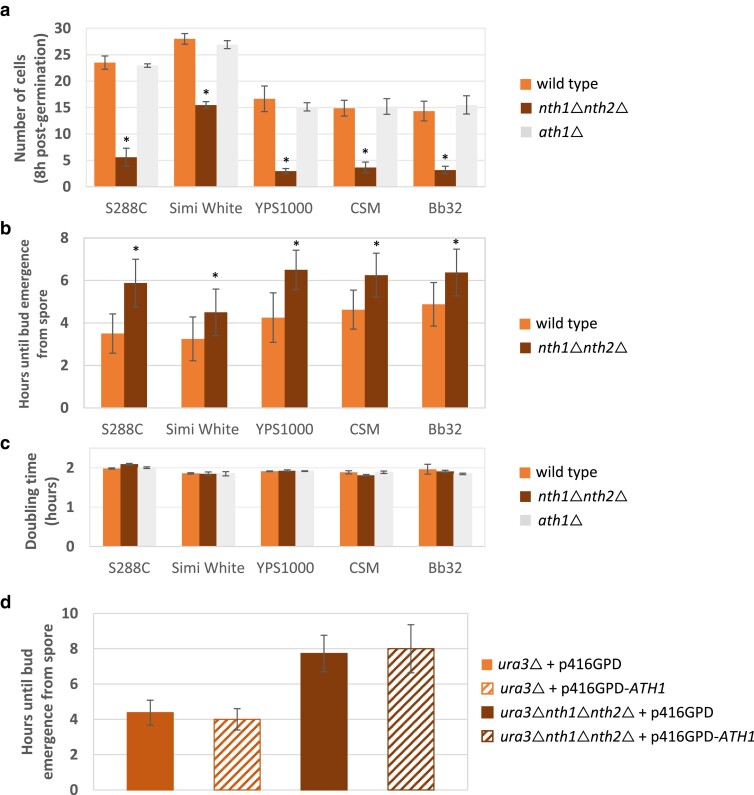
Sporulation in the budding yeast S. cerevisiae involves meiotic cell division followed by formation of 4 haploid spores containing high levels of trehalose (Geijer et al. 2012). Upon exposure to glucose and other nutrients, spores exit dormancy through germination and resume growth. Spore germination of S. cerevisiae is a multistep developmental path in which trehalose is rapidly mobilized (Rousseau et al. 1972). Therefore, we sought to evaluate the role of intracellular trehalose in spore germination. Germination was evaluated after tetrads were dissected on glucose-containing rich media. All 5 tested nth1Δnth2Δ mutants exhibited significantly slower germination than their isogenic parent and respective ath1Δ mutant (Fig. 8a). This germination defect was associated with the first cell division, as all evaluated nth1Δnth2Δ mutants displayed significantly slower emergence of the first cell compared to their isogenic wild-type parent (Fig. 8b). There was no effect on subsequent exponential growth rate, or, presumably, cell cycle progression, as all the trehalase mutants had statistically indistinguishable doubling times compared to their isogenic wild-type parent (Fig. 8c). Similar results were observed in the fission yeast Schizosaccharomyces pombe when spores from strains lacking neutral trehalase (ntp1-) were induced to germinate: the onset of this process was markedly delayed and germination rate was reduced (Beltran et al. 2000). To determine whether this was a germination-specific defect or a general lag phase defect associated with dormant cells, we also examined single-cell lag phase times for quiescent trehalase mutants (nth1Δnth2Δ and ath1Δ) and observed no lag phase extension (Supplementary Fig. 6). These results indicate that intracellular cytoplasmic trehalose hydrolysis has an important role in promoting exit from spore dormancy, though not in exiting the quiescent state. None of the ath1Δ spores or quiescent cells exhibited slower cell division upon exposure to nutrients, and all exhibited the same logarithmic growth rate as their isogenic wild types in glucose media, indicating that failure to degrade vacuolar trehalose, if present, has no effect on spore germination or postquiescent lag phase (Fig. 8a and c). It seems likely that the germination defect in neutral trehalase mutants results from decreased available trehalose-derived glucose, suggesting an important role for trehalose as a storage carbohydrate specifically during sporulation, though not during quiescence. This result contrasts with that described by Shi et al. (2010), where wild-type CEN.PK cells were able to recover from quiescence faster if they contained more intracellular trehalose (pregrowth in rich medium) compared to cells with less intracellular trehalose (pregrowth in minimal medium). While Shi et al. indicates a correlation between intracellular trehalase content and lag phase duration postquiescence, a number of other explanations could also explain the result, including that the laboratory CEN.PK strain lineage contains a mutation in adenylate cyclase (cyr1-K1867M) that affects nutrient sensing and signaling, or variable survivability in rich vs minimal medium (Vanhalewyn et al. 1999; Dumortier et al. 2000).
Fig. 8.
Absence of neutral trehalases results in a spore germination defect. a) Cells were grown overnight in YPD before being sporulated for 6 days at room temperature. Six tetrads for each strain were dissected onto YPD plates, and the number of cells grown from each germinated spore was counted after an 8-h incubation at 30 °C. b) Two tetrads (8 spores) from each sporulated cultures were dissected on YPD plates and incubated at 30 °C. Through visual inspection each hour, the number of hours required until bud emergence was recorded. c) Doubling times for indicated strains in YPD were calculated from plate reader-based growth curves as described in Materials and methods. d) Six tetrads (24 spores) from 3 independent sporulated cultures were dissected on YPD plates and incubated at 30 °C. Bud emergence time was evaluated as above. Values are presented as the mean with SD. Asterisks represent statistically significant differences (P < 0.05) between the mutants and their isogenic wild-type strains.
Previous studies have suggested the possibility that intracellular trehalose accumulation may be important for cell cycle regulation. As G1/G0 cells accumulate intracellular trehalose and hydrolyze it upon cell cycle start, it has been proposed that trehalose is important for entering the cell cycle, though no cell cycle defects have been observed for trehalose metabolism mutants growing in standard growth media conditions (Küenzi and Fiechter 1969; Silljé et al. 1999; Futcher 2006; Chen et al. 2007; Shi et al. 2010). In this conceptual model, cells require glucose from hydrolyzed trehalose as the glycolytic fuel to initiate the cell cycle. In support of this notion, trehalose and glycogen hydrolyzing enzymes are regulated by cell cycle regulators, and cells lacking both neutral trehalase and glycogen phosphorylase activity (nth1Δnth2Δgph1Δ) exhibited significant changes in the dynamics of central carbon metabolites and amino acid metabolism during cell cycle progression, alongside a complete loss of sporulation ability (Ewald et al. 2016; Zhao et al. 2016). In the same studies, Nth1 was activated at the G1/S transition, which would funnel trehalose-derived glucose into glycolysis during S phase; this activation was proposed to support cell cycle progression in poor nutrient environments. In accordance with this observation, Ewald et al. (2016) demonstrated that cells unable to hydrolyze trehalose and glycogen (nth1Δnth2Δgph1Δ) had a lower probability of completing a cell cycle after shifting from 0.005% w/v glucose to 0% glucose in a microfluidic device. This phenotype was reversed when Nth1 function was restored (Ewald et al. 2016). The spore germination defect observed associated with all nth1Δnth2Δ strains tested in this study demonstrates another cell cycle progression defect associated with failure to hydrolyze trehalose.
The germination defect associated with nth1Δnth2Δ also provides an opportunity to further test whether Ath1 has a role in liberating glucose from intracellular trehalose to support cellular growth. If ATH1 overexpression is able to suppress the germination defect associated with nth1Δnth2Δ, this would be evidence for a functional role associated with Ath1-based hydrolysis of intracellular trehalose. However, overexpression of ATH1 did not decrease the amount of time required for bud emergence from nth1Δnth2Δ spores (Fig. 8d). This result further suggests that Ath1 does not appear to have a meaningful intracellular role in producing glucose from intracellular trehalose. It is noteworthy that in this experiment, 8 of the 48 tested nth1Δnth2Δ spores completely failed to germinate, compared with 1 of the 48 tested wild-type spores (Supplementary Fig. 7). This suggests that the increased germination lag phase observed in these mutants can sometimes result in a complete germination failure, though the mechanism determining these variable outcomes remains elusive. Similarly, Ewald et al. (2016) demonstrated that mutants unable to hydrolyze trehalose and glycogen (nth1Δnth2Δgph1Δ) had a significantly higher rate of cell divisions resulting in nonviable cells during starvation compared to wild type, ∼15% vs ∼3%.
Conceptual model for trehalose degradation in S. cerevisiae
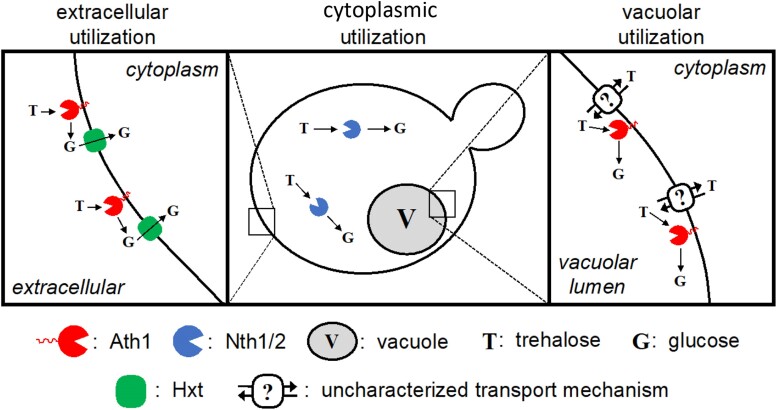
In Fig. 9, we have produced a conceptual model for intracellular and extracellular trehalose utilization in S. cerevisiae. Previously published information along with data presented here support the notion that extracellular trehalose is primarily utilized through the action of extracellular Ath1. This enzyme can hydrolyze trehalose into glucose, which is then imported and can be used for glycolysis. Further, while the Agt1 protein has the ability to transport trehalose when overexpressed, it remains unclear how often this route of trehalose import/utilization is used instead of Ath1-based hydrolysis, especially when AGT1 is under the regulation of its native promoter. Previously published information along with data presented here support the notion that cytosolic trehalose is primarily utilized through the action of Nth1/2. We also demonstrated that Nth1/2-based trehalose mobilization is required to support rapid germination from the spore state. Overexpression of ATH1 was unable to completely hydrolyze intracellular trehalose during lag phase and failed to suppress the nth1Δnth2Δ germination defect, further suggesting that Ath1 does not typically function to release glucose from intracellular trehalose. This is in line with its reported plasma membrane and vacuolar localization, in addition to its acidic pH optimum. Finally, we suggest that the fraction of intracellular trehalose present in ath1Δ mutants represents vacuolar trehalose. While it appears that vacuolar Ath1 can hydrolyze vacuolar trehalose, the functional consequences of this activity are an open question. Similarly, it remains to be seen whether Ath1 vacuolar localization is important for cellular physiology or if it is simply an artifact associated with plasma membrane recycling.
Fig. 9.
Conceptual model illustrating multiple modes of trehalose degradation in S. cerevisiae. Left: extracellular utilization of trehalose as a carbon source requires extracellular degradation of trehalose into glucose via the Ath1 trehalase enzyme. While the Agt1/Mal11 protein (not pictured) can also import extracellular trehalose when overexpressed, it remains unclear whether it is expressed/utilized for trehalose consumption in wild strains. Center: cytoplasmic trehalose is degraded into glucose via the Nth1 and Nth2 trehalase enzymes, an activity required for proper spore germination. Right: while of unclear functional significance, vacuolar trehalose enters the vacuole through an uncharacterized mechanism and is degraded by the Ath1 trehalase enzyme.
Supplementary Material
Acknowledgments
We would like to thank members of the Gibney lab for helpful discussions and critical reading of this manuscript. We would also like to thank the genomics facility (Research Resource Identifier: SCR_021727) of the Biotechnology Resource Center of Cornell Institute of Biotechnology for Sanger sequencing services.
Contributor Information
Anqi Chen, Department of Food Science, Cornell University, Ithaca, NY 14853, USA; Science Center for Future Foods, Jiangnan University, Wuxi 214122, China.
Sara E Stadulis, Department of Food Science, Cornell University, Ithaca, NY 14853, USA.
Kayla deLeuze, Department of Food Science, Cornell University, Ithaca, NY 14853, USA.
Patrick A Gibney, Department of Food Science, Cornell University, Ithaca, NY 14853, USA.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material available at G3 online.
Funding
This research was supported by startup funds provided to PAG by the Department of Food Science in the College of Agriculture and Life Sciences at Cornell University and also provided by E&J Gallo Winery .
Literature cited
- Alblova M, Smidova A, Kalabova D, Lentini Santo D, Obsil T, Obsilova V. 2019. Allosteric activation of yeast enzyme neutral trehalase by calcium and 14-3-3 protein. Physiol Res. 68(2):147–160. doi: 10.33549/physiolres.933950. [DOI] [PubMed] [Google Scholar]
- Alizadeh P, Klionsky DJ. 1996. Purification and biochemical characterization of the ATH1 gene product, vacuolar acid trehalase, from Saccharomyces cerevisiae. FEBS Lett. 391(3):273–278. doi: 10.1016/0014-5793(96)00751-X. [DOI] [PubMed] [Google Scholar]
- Beltran FF, Castillo R, Vicente-Soler J, Cansado J, Gacto M. 2000. Role for trehalase during germination of spores in the fission yeast Schizosaccharomyces pombe. FEMS Microbiol Lett. 193(1):117–121. doi: 10.1111/j.1574-6968.2000.tb09412.x. [DOI] [PubMed] [Google Scholar]
- Brown CA, Murray AW, Verstrepen KJ. 2010. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr Biol. 20(10):895–903. doi: 10.1016/j.cub.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Gibney PA. 2022. Intracellular trehalose accumulation via the Agt1 transporter promotes freeze–thaw tolerance in Saccharomyces cerevisiae. J Appl Microbiol. 133(4):2390–2402. doi: 10.1111/jam.15700. [DOI] [PubMed] [Google Scholar]
- Chen A, Vargas-Smith J, Tapia H, Gibney PA. 2022. Characterizing phenotypic diversity of trehalose biosynthesis mutants in multiple wild strains of Saccharomyces cerevisiae. G3 (Bethesda). 12(11):jkac196. doi: 10.1093/g3journal/jkac196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Odstrcil EA, Tu BP, McKnight SL. 2007. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science. 316(5833):1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- Chopra R, Sharma VM, Ganesan K. 1999. Elevated growth of Saccharomyces cerevisiae ATH1 null mutants on glucose is an artifact of nonmatching auxotrophies of mutant and reference strains. Appl Environ Microbiol. 65(5):2267–2268. doi: 10.1128/AEM.65.5.2267-2268.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JH, Crowe LM, Chapman D. 1984. Preservation of membranes in anhydrobiotic organisms: the role of trehalose. Science. 223(4637):701–703. doi: 10.1126/science.223.4637.701. [DOI] [PubMed] [Google Scholar]
- de Silva-Udawatta MN, Cannon JF. 2001. Roles of trehalose phosphate synthase in yeast glycogen metabolism and sporulation. Mol Microbiol. 40(6):1345–1356. doi: 10.1046/j.1365-2958.2001.02477.x. [DOI] [PubMed] [Google Scholar]
- Destruelle M, Holzer H, Klionsky DJ. 1995. Isolation and characterization of a novel yeast gene, ATH1, that is required for vacuolar acid trehalase activity. Yeast. 11(11):1015–1025. doi: 10.1002/yea.320111103. [DOI] [PubMed] [Google Scholar]
- Duina AA, Miller ME, Keeney JB. 2014. Budding yeast for budding geneticists: a primer on the Saccharomyces cerevisiae model system. Genetics. 197(1):33–48. doi: 10.1534/genetics.114.163188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumortier F, Vanhalewyn M, Debast G, Colombo S, Ma P, Winderickx J, Van Dijck P, Thevelein JM. 2000. A specific mutation in Saccharomyces cerevisiae adenylate cyclase, Cyr1(K1876M), eliminates glucose- and acidification-induced cAMP signalling and delays glucose-induced loss of stress resistance. Int J Food Microbiol. 55(1–3):103–107. doi: 10.1016/S0168-1605(00)00184-7. [DOI] [PubMed] [Google Scholar]
- Elbein AD, Pan YT, Pastuszak I, Carroll D. 2003. New insights on trehalose: a multifunctional molecule. Glycobiology. 13(4):17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- Engel SR, Dietrich FS, Fisk DG, Binkley G, Balakrishnan R, Costanzo MC, Dwight SS, Hitz BC, Karra K, Nash RS, et al. 2014. The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 (Bethesda). 4(3):389–398. doi: 10.1534/g3.113.008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald JC, Kuehne A, Zamboni N, Skotheim JM. 2016. The yeast cyclin-dependent kinase routes carbon fluxes to fuel cell cycle progression. Mol Cell. 62(4):532–545. doi: 10.1016/j.molcel.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François J, Neves MJ, Hers HG. 1991. The control of trehalose biosynthesis in Saccharomyces cerevisiae: evidence for a catabolite inactivation and repression of trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase. Yeast. 7(6):575–587. doi: 10.1002/yea.320070605. [DOI] [PubMed] [Google Scholar]
- François J, Parrou JL. 2001. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 25(1):125–145. doi: 10.1111/j.1574-6976.2001.tb00574.x. [DOI] [PubMed] [Google Scholar]
- Futcher B. 2006. Metabolic cycle, cell cycle, and the finishing kick to start. Genome Biol. 7(4):107. doi: 10.1186/gb-2006-7-4-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo C, Flores CL. 2004. The importance of a functional trehalose biosynthetic pathway for the life of yeasts and fungi. FEMS Yeast Res. 4(4–5):351–359. doi: 10.1016/S1567-1356(03)00222-8. [DOI] [PubMed] [Google Scholar]
- Garre E, Pérez-Torrado R, Gimeno-Alcañiz JV, Matallana E. 2009. Acid trehalase is involved in intracellular trehalose mobilization during postdiauxic growth and severe saline stress in Saccharomyces cerevisiae. FEMS Yeast Res. 9(1):52–62. doi: 10.1111/j.1567-1364.2008.00453.x. [DOI] [PubMed] [Google Scholar]
- Geijer C, Pirkov I, Vongsangnak W, Ericsson A, Nielsen J, Krantz M, Hohmann S. 2012. Time course gene expression profiling of yeast spore germination reveals a network of transcription factors orchestrating the global response. BMC Genomics. 13:554. doi: 10.1186/1471-2164-13-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418(6896):387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gibney PA, Schieler A, Chen JC, Rabinowitz JD, Botstein D. 2015. Characterizing the in vivo role of trehalose in Saccharomyces cerevisiae using the AGT1 transporter. Proc Natl Acad Sci U S A. 112(119):6116–6121. doi: 10.1073/pnas.1506289112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell G, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. 1996. Life with 6000 genes. Science. 274(5287):546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Gong M, Gong F, Yanofsky C. 2006. Overexpression of tnaC of Escherichia coli inhibits growth by depleting tRNA2Pro availability. J Bacteriol. 188(5):1892–1898. doi: 10.1128/JB.188.5.1892-1898.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González MI, Blázquez MA, Gancedo C, Stucka R, Feldmann H. 1992. Molecular cloning of CIF1, a yeast gene necessary for growth on glucose. Yeast. 8(3):183–192. doi: 10.1002/yea.320080304. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. 1991. Guide to Yeast Genetics and Molecular Biology. 1st ed.(Methods in Enzymology, vol. 194). Academic Press. p. 1–863. [Google Scholar]
- Han EK, Cotty F, Sottas C, Jiang H, Michels CA. 1995. Characterization of AGT1 encoding a general α-glucoside transporter from Saccharomyces. Mol Microbiol. 17(6):1093–1107. doi: 10.1111/j.1365-2958.1995.mmi_17061093.x. [DOI] [PubMed] [Google Scholar]
- Harris SD, Cotter DA. 1988. Transport of yeast vacuolar trehalase to the vacuole. Can J Microbiol. 34(7):835–838. doi: 10.1139/m88-143. [DOI] [PubMed] [Google Scholar]
- He S, Bystricky K, Leon S, François JM, Parrou JL. 2009. The Saccharomyces cerevisiae vacuolar acid trehalase is targeted at the cell surface for its physiological function. FEBS J. 276(19):5432–5446. doi: 10.1111/j.1742-4658.2009.07227.x. [DOI] [PubMed] [Google Scholar]
- Hottiger T, Boller T, Wiemken A. 1987. Rapid changes of heat and desiccation tolerance correlated with changes of trehalose content in Saccharomyces cerevisiae cells subjected to temperature shifts. FEBS Lett. 220(1):113–115. doi: 10.1016/0014-5793(87)80886-4. [DOI] [PubMed] [Google Scholar]
- Hounsa C-G, Brandt EV, Thevelein J, Hohmann S, Prior BA. 1998. Role of trehalose in survival of Saccharomyces cerevisiae under osmotic stress. Microbiology. 144:671–680. doi: 10.1099/00221287-144-3-671. [DOI] [PubMed] [Google Scholar]
- Huang J, Reggiori F, Klionsky DJ. 2007. The transmembrane domain of acid trehalase mediates ubiquitin-independent multivesicular body pathway sorting. Mol Biol Cell. 18(7):2511–2524. doi: 10.1091/mbc.e06-11-0995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature. 425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jules M, Beltran G, François J, Parrou JL. 2008. New insights into trehalose metabolism by Saccharomyces cerevisiae: NTH2 encodes a functional cytosolic trehalase, and deletion of TPS1 reveals Ath1p-dependent trehalose mobilization. Appl Environ Microbiol. 74(3):605–614. doi: 10.1128/AEM.00557-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jules M, Guillou V, François J, Parrou JL. 2004. Two distinct pathways for trehalose assimilation in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol. 70(5):2771–2778. doi: 10.1128/AEM.70.5.2771-2778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Alizadeh P, Harding T, Hefner-Gravink A, Klionsky DJ. 1996. Disruption of the yeast ATH1 gene confers better survival after dehydration, freezing, and ethanol shock: potential commercial applications. Appl Environ Microbiol. 62(5):1563–1569. doi: 10.1128/aem.62.5.1563-1569.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küenzi MT, Fiechter A. 1969. Changes in carbohydrate composition and trehalase-activity during the budding cycle of Saccharomyces cerevisiae. Archiv für Mikrobiologie. 64(4):396–407. doi: 10.1007/BF00417021. [DOI] [PubMed] [Google Scholar]
- Lillie SH, Pringle JR. 1980. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 143(3):1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. 1986. Genealogy of principal strains of the yeast genetic stock center. Genetics. 113(1):35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22(25):5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 156(1):119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Navon G, Shulman RG, Yamane T, Eccleshall TR, Lam K-B, Baronofsky Jerald J., Marmur Julius. 1979. Phosphorus-31 nuclear magnetic resonance studies of wild-type and glycolytic pathway mutants of Saccharomyces cerevisiae. Biochemistry. 18(21):4487–4499. doi: 10.1021/bi00588a006. [DOI] [PubMed] [Google Scholar]
- Neves MJ, Hohmann S, Bell W, Dumortier F, Luyten K, Ramos J, Cobbaert P, de Koning W, Kaneva Z, Thevelein JM. 1995. Control of glucose influx into glycolysis and pleiotropic effects studied in different isogenic sets of Saccharomyces cerevisiae mutants in trehalose biosynthesis. Curr Genet. 27(2):110–122. doi: 10.1007/BF00313424. [DOI] [PubMed] [Google Scholar]
- Nwaka S, Holzer H. 1997. Molecular biology of trehalose and the trehalases in the yeast Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 58:197–237. doi: 10.1016/S0079-6603(08)60037-9. [DOI] [PubMed] [Google Scholar]
- Nwaka S, Kopp M, Holzer H. 1995. Expression and function of the trehalase genes NTH1 and YBR0106 in Saccharomyces cerevisiae. J Biol Chem. 270(17):10193–10198. doi: 10.1074/jbc.270.17.10193. [DOI] [PubMed] [Google Scholar]
- Nwaka S, Mechler B, Destruelle M, Holzer H. 1995. Phenotypic features of trehalase mutants in Saccharomyces cerevisiae. FEBS Lett. 360(3):286–290. doi: 10.1016/0014-5793(95)00105-i. [DOI] [PubMed] [Google Scholar]
- Nwaka S, Mechler B, Holzer H. 1996. Deletion of the ATH1 gene in Saccharomyces cerevisiae prevents growth on trehalose. FEBS Lett. 386(2–3):235–238. doi: 10.1016/0014-5793(96)00450-4. [DOI] [PubMed] [Google Scholar]
- Olsson C, Jansson H, Swenson J. 2016. The role of trehalose for the stabilization of proteins. J Phys Chem B. 120(20):4723–4731. doi: 10.1021/acs.jpcb.6b02517. [DOI] [PubMed] [Google Scholar]
- Panni S, Landgraf C, Volkmer-Engert R, Cesareni G, Castagnoli L. 2008. Role of 14-3-3 proteins in the regulation of neutral trehalase in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 8(1):53–63. doi: 10.1111/j.1567-1364.2007.00312.x. [DOI] [PubMed] [Google Scholar]
- Parrou JL, François J. 1997. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal Biochem. 248(1):186–188. doi: 10.1006/abio.1997.2138. [DOI] [PubMed] [Google Scholar]
- Parrou JL, Jules M, Beltran G, François J. 2005. Acid trehalase in yeasts and filamentous fungi: localization, regulation and physiological function. FEMS Yeast Res. 5(6–7):503–511. doi: 10.1016/j.femsyr.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Petri JR. 1887. Eine kleine Modification des Koch'schen Plattenverfahrens (A minor modification of the pating technique of Koch). Centralblatt für Bacteriologie und Parasitenkunde. 1:279–280. [Google Scholar]
- Raths S, Rohrer J, Crausaz F, Riezman H. 1993. End3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 120(1):55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Nothwehr SF, Stevens TH. 1992. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J Cell Biol. 119(1):69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau P, Halvorson HO, Bulla LA, St Julian G. 1972. Germination and outgrowth of single spores of Saccharomyces cerevisiae viewed by scanning electron and phase-contrast microscopy. J Bacteriol. 109(3):1232–1238. doi: 10.1128/jb.109.3.1232-1238.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Miguel PF, Argüelles JC. 1994. Differential changes in the activity of cytosolic and vacuolar trehalases along the growth cycle of Saccharomyces cerevisiae. Biochim Biophys Acta. 1200(2):155–160. doi: 10.1016/0304-4165(94)90130-9. [DOI] [PubMed] [Google Scholar]
- Scott TD, Xu P, McClean MN. 2023. Strain-dependent differences in coordination of yeast signalling networks. FEBS J. 290(8):2097–2114. doi: 10.1111/febs.16689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Q, Wang J, Zhu W. 2019. Trehalose stabilizing protein in a water replacement scenario: insights from molecular dynamics simulation. bioRxiv. 10.1101/2019.12.27.889063, preprint: not peer reviewed. [DOI]
- Shi L, Sutter BM, Ye X, Tu BP. 2010. Trehalose is a key determinant of the quiescent metabolic state that fuels cell cycle progression upon return to growth. Mol Biol Cell. 21(12):1982–1990. doi: 10.1091/mbc.e10-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silljé HHW, Paalman JWG, Ter Schure EG, Olsthoorn SQB, Verkleij AJ, Boonstra J, Verrips CT. 1999. Function of trehalose and glycogen in cell cycle progression and cell viability in Saccharomyces cerevisiae. J Bacteriol. 181(2):396–400. doi: 10.1128/JB.181.2.396-400.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MA, Lindquist S. 1998a. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol Cell. 1(5):639–648. doi: 10.1016/S1097-2765(00)80064-7. [DOI] [PubMed] [Google Scholar]
- Singer MA, Lindquist S. 1998b. Thermotolerance in Saccharomyces cerevisiae: the Yin and Yang of trehalose. Trends Biotechnol. 16(11):460–468. doi: 10.1016/S0167-7799(98)01251-7. [DOI] [PubMed] [Google Scholar]
- Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, et al. 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 21(3):319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Stucka R, Blázquez MA. 1993. The fdp1 and cif1 mutations are caused by different single nucleotide changes in the yeast CIF1 gene. FEMS Microbiol Lett. 107(2–3):251–253. doi: 10.1111/j.1574-6968.1993.tb06038.x. [DOI] [PubMed] [Google Scholar]
- Su Y, Macías LG, Heras JM, Querol A, Guillamón JM. 2021. Phenotypic and genomic differences among S. cerevisiae strains in nitrogen requirements during wine fermentations. Food Microbiol. 96:103685. doi: 10.1016/j.fm.2020.103685. [DOI] [PubMed] [Google Scholar]
- Swinnen S, Klein M, Carrillo M, McInnes J, Nguyen HTT, Nevoigt E. 2013. Re-evaluation of glycerol utilization in Saccharomyces cerevisiae: characterization of an isolate that grows on glycerol without supporting supplements. Biotechnol Biofuels. 6(1):157. doi: 10.1186/1754-6834-6-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swulius MT, Jensen GJ. 2012. The helical mreb cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-terminal yellow fluorescent protein tag. J Bacteriol. 194(23):6382–6386. doi: 10.1128/JB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia H, Young L, Fox D, Bertozzi CR, Koshland D. 2015. Increasing intracellular trehalose is sufficient to confer desiccation tolerance to Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 112(19):6122–6127. doi: 10.1073/pnas.1506415112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM. 1984. Regulation of trehalose mobilization in fungi. Microbiol Rev. 48(1):42–59. doi: 10.1128/mr.48.1.42-59.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijck P, Colavizza D, Smet P, Thevelein JM. 1995. Differential importance of trehalose in stress resistance in fermenting and nonfermenting Saccharomyces cerevisiae cells. Appl Environ Microbiol. 61(1):109–115. doi: 10.1128/aem.61.1.109-115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhalewyn M, Dumortier F, Debast G, Colombo S, Ma P, Winderickx J, Van Dijck P, Thevelein JM. 1999. A mutation in Saccharomyces cerevisiae adenylate cyclase, Cyr1(K1876M), specifically affects glucose- and acidification-induced cAMP signalling and not the basal cAMP level. Mol Microbiol. 33(2):363–376. doi: 10.1046/j.1365-2958.1999.01479.x. [DOI] [PubMed] [Google Scholar]
- Vind J, Sørensen MA, Rasmussen MD, Pedersen S. 1993. Synthesis of proteins in Escherichia coli is limited by the concentration of free ribosomes: expression from reporter genes does not always reflect functional mRNA levels. J Mol Biol. 231(3):678–688. doi: 10.1006/jmbi.1993.1319. [DOI] [PubMed] [Google Scholar]
- Walther T, Mtimet N, Alkim C, Vax A, Loret M-O, Ullah A, Gancedo C, Smits GJ, François JM. 2013. Metabolic phenotypes of Saccharomyces cerevisiae mutants with altered trehalose 6-phosphate dynamics. Biochem J. 454(2):227–237. doi: 10.1042/BJ20130587. [DOI] [PubMed] [Google Scholar]
- Warringer J, Zörgö E, Cubillos FA, Zia A, Gjuvsland A, Simpson JT, Forsmark A, Durbin R, Omholt SW, Louis EJ, et al. 2011. Trait variation in yeast is defined by population history. PLoS Genet. 7(6):e1002111. doi: 10.1371/journal.pgen.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Wang F, Dong S, Li H. 2016. Changes of trehalose content and expression of relative genes during the bioethanol fermentation by Saccharomyces cerevisiae. Can J Microbiol. 62(10):827–835. doi: 10.1139/cjm-2015-0832. [DOI] [PubMed] [Google Scholar]
- Zähringer H, Thevelein JM, Nwaka S. 2000. Induction of neutral trehalase Nth1 by heat and osmotic stress is controlled by STRE elements and Msn2/Msn4 transcription factors: variations of PKA effect during stress and growth. Mol Microbiol. 35(2):397–406. doi: 10.1046/j.1365-2958.2000.01706.x. [DOI] [PubMed] [Google Scholar]
- Zhao G, Chen Y, Carey L, Futcher B. 2016. Cyclin-dependent kinase co-ordinates carbohydrate metabolism and cell cycle in S. cerevisiae. Mol Cell. 62(4):546–557. doi: 10.1016/j.molcel.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material available at G3 online.