Abstract
Orchidaceae is one of the most prominent flowering plant families, with many species exhibiting highly specialized reproductive and ecological adaptations. An estimated 10% of orchid species in the American tropics are pollinated by scent-collecting male euglossine bees; however, to date, there are no published genomes of species within this pollination syndrome. In this study, we present the first draft genome of an epiphytic orchid from the genus Gongora, a representative of the male euglossine bee–pollinated subtribe Stanhopeinae. The 1.83-Gb de novo genome with a scaffold N50 of 1.7 Mb was assembled using short- and long-read sequencing and chromosome capture (Hi-C) information. Over 17,000 genes were annotated, and 82.95% of the genome was identified as repetitive content. Furthermore, we identified and manually annotated 26 terpene synthase genes linked to floral scent biosynthesis and performed a phylogenetic analysis with other published orchid terpene synthase genes. The Gongora gibba genome assembly will serve as the foundation for future research to understand the genetic basis of floral scent biosynthesis and diversification in orchids.
Keywords: genome assembly, Gongora gibba, chemotype A, orchid, floral scent, terpene synthesis
Introduction
Plant–pollinator interactions are thought to have played a key role in the diversification of flowering plants (Van der Niet et al. 2014). Orchidaceae is a particularly species-rich group, consisting of over 30,000 species and 880 genera (Dressler 2005; Chase et al. 2015; Christenhusz and Byng 2016; Govaerts et al. 2021; Pérez-Escobar et al. 2024). Orchids exhibit a wide range of highly specialized ecological and reproductive strategies, some of which are thought to contribute to their elevated diversification rates (Cozzolino and Widmer 2005; Xu et al. 2012). Multiple factors related to pollination, such as the evolution of deceptive pollination strategies and pollinator specialization, have been suggested to play a role in enhancing diversification rates (Givnish et al. 2015; Ackerman et al. 2023).
A striking example of a reproductive strategy linked to higher diversification rates is pollination by euglossine bees (or orchid bees; Apidae: Euglossini; Dressler 1968; Gerlach and Schill 1991; Ramírez et al. 2011; Givnish et al. 2015). Fragrance is thought to be the second most common reward among orchids, predominantly associated with euglossine bee pollination (Ackerman et al. 2023). In this pollination system, male bees pollinate plants while visiting inflorescences to collect chemical compounds, which they store in hind-leg pockets for later use acting as a pheromone analog during courtship display (Allen 1954; Eltz et al. 2005; Henske et al. 2023). The production of floral scent not only attracts pollinators but is itself the reward. Therefore, all Gongora species lack additional floral rewards, such as nectar.
Gongora, one of at least 22 genera that exhibit euglossine bee pollination, contains 60–70 recognized species. However, the taxonomy and the identification of species in the genus Gongora are notoriously difficult because multiple cryptic species, with little morphological variation, can coexist, and are only discernible by their floral scent (Dressler 1966; Whitten 1985; Jenny 1993; Hetherington-Rauth and Ramírez 2016; Guizar Amador 2022). Gongora emit species-specific floral scents, typically consisting of a few main compounds alongside minor compounds. Differences in the floral scent between chemotypes, or distinct chemical groups, lead to the attraction of different pollinators, which is thought to maintain reproductive isolation barriers (Guizar Amador 2022). Pollinator-driven diversification is hypothesized to have played a major role in the evolutionary history of Gongora (Dressler 1968; Williams and Dodson 1972; Ramírez et al. 2011); however, the molecular and genetic mechanisms underlying the origin and maintenance of reproductive barriers remain largely unexplored.
Generating genomic resources for Gongora is needed to elucidate the genetic basis of floral scent production and reveals how divergent floral scent phenotypes evolve and lead to reproductive isolation. To date, no reference genomes are available for any euglossine bee–pollinated orchids, a major obstacle toward studying the diversification of this group. In this study, we report the genome of Gongora gibba, a member of the Gongora subgenus previously described as chemotype A (Supplementary Fig. 1;Hetherington-Rauth and Ramírez 2016; Guizar Amador 2022). We report the assembly size, the repeat sequences detected, as well as an annotation. We confirm the identity of G. gibba by placing this species in a plastid phylogeny. We also conducted a high-quality annotation of terpene synthase (TPS) genes, laying the foundation for further research on floral scent biosynthesis.
Materials and methods
Sample preparation and sequencing
All plant materials used for the genome assembly were obtained from a mature G. gibba plant collected from the surroundings of the La Gamba Tropenstation in the province of Puntarenas, located in southwestern Costa Rica (BioSample no. SAMN37328957, BioProject no. PRJNA1014482). The sample was imported to the United States under the CITES Certificate of Scientific Exchange permit no. 14US51372B/9 and is currently located in the Botanical Conservatory at the University of California, Davis (ID: G-10, Supplementary Fig. 1b). For genome sequencing, we collected fresh leaves and the leaves were flash-frozen in liquid nitrogen. Two tissue samples were shipped to Dovetail Genomics (Santa Cruz, CA, USA) for the construction and sequencing of 2 Illumina libraries (Illumina HiSeq 2500 and NextSeq 2000, insert sizes 402 and 523 bp, respectively), 1 PacBio Sequel library (5 SMRT cells, 7,876 bp average read length), 1 Hi-C library (Illumina HiSeq X), and 1 Chicago library (Illumina HiSeq X). An additional sample was used for DNA extraction with a DNEasy Plant Mini Kit from Qiagen, followed by library construction and sequencing using an Illumina HiSeq 4000 platform. In total, we generated 412 Gb of raw reads that were then filtered based on sequencing quality and adapter contamination.
Genome size estimation
To estimate a genome size and a heterozygosity of G. gibba plant, we analyzed the k-mer frequency distribution from the 402-bp insert size Illumina library with Jellyfish (Marçais and Kingsford 2011). We also estimated the genome size of the plant with flow cytometry. Briefly, a 1.5-cm2 fresh orchid leaf was chopped with a fresh single-edge razor blade in a cold Galbraith buffer along with a similar-sized fresh leaf from either tomato (Solanum lycopersicum, 1C = 1,320.3) or pea (Pisum sativum, 1C = 4,591.71). The released nuclei were then filtered and stained with a cold solution of 25 mg/mL propidium iodide for 30 min in the dark. We quantified the relative fluorescence of 2C orchid and 2C standard nuclei using a Beckman Coulter CytoFLEX flow cytometer. A ploidy level was determined by the relative position of the 2C orchid and 2C standard peaks and by the estimated genome size based on the ratio of the 2C peak positions of the sample and standard times the amount of DNA in the standard.
Genome assembly
Given the high levels of heterozygosity and repetitive content in the G. gibba genome, we decided to use a hybrid strategy for the assembly. Long reads can improve the contiguity of an assembly; however, this technology is associated with high error rates (Zhang et al. 2020). We used FMLRC (Wang et al. 2018) with one of our Illumina libraries (SRCD2 S1 L001) to leverage the higher accuracy of the short reads to perform long-read error correction on the PacBio library. Before proceeding with the assembly, we used BLAST (Altschul et al. 1990) to identify long reads not belonging to the nuclear genome by comparing them against the Oncidium plastid (GQ324949.1) and mitochondrial (KJ501920.1) sequences downloaded from the NCBI (Wheeler et al. 2007). The resulting 85,553 reads were removed from the nuclear genome assembly and used separately to assemble the organelle genomes.
With the corrected and filtered PacBio reads as input, we used WTDBG2 (Ruan and Li 2020) for the de novo assembly. One of the short-read libraries (SRCD2 S1 L00) was then mapped to the contigs using BWA (Li 2013; Supplementary Table 1). Contigs with different levels of coverage were searched using the BLAST against the NCBI's nucleotide library to determine whether they belong to exogenous DNA. Contigs with >70% of their length are not covered by any Illumina reads that matched bacterial DNA, so 70% was established as a cutoff point to remove exogenous contigs from the assembly.
SSPACE v3.0 (Boetzer et al. 2011) and the 523-bp insert size Illumina library were used for a preliminary scaffolding step. To improve the accuracy of the assembly, pilon (Walker et al. 2014) and the 402-bp insert size library were then used to polish the assembly. This preliminary assembly was sent to Dovetail Genomics for further scaffolding with the Chicago and Hi-C libraries, which were used as input data for HiRise, a software pipeline designed for utilizing proximity ligation data to scaffold genome assemblies (Putnam et al. 2016). Completeness of the genome assembly was assessed using Benchmarking Universal Single-Copy Orthologs (BUSCO; Simão et al. 2015) with default parameters and the embryophyta dataset.
All raw sequence data and the final genome assembly were deposited under the BioProject accession no. PRJNA1014482. All details of parameters used in the genome assembly are given in the supplementary file Genome_Assembly_Report.ipynb available at OSF (https://osf.io/hqav3/?view_only=5d65957cc6474488b53847219a7b6ac4).
Plastid sequencing, assembly, and annotation of other Gongora species
Using the BLAST, we identified G. gibba scaffolds that matched the Oncidium organelle genomes. These scaffolds, and the previously identified chloroplast and mitochondrial PacBio reads, were used as an input for Canu (Koren et al. 2017) to perform de novo assemblies. SSPACE v3.0 (Boetzer et al. 2011), pilon (Walker et al. 2014), and GetOrganelle (https://github.com/Kinggerm/GetOrganelle) were used to improve the contiguity and accuracy of the assemblies.
The plastid genomes of 10 additional Gongora samples from Costa Rica and Panama were assembled to determine the phylogenetic position of the sequenced G. gibba (A-chemotype). Total DNA was extracted from the fresh leaves dried in silica gel using the CTAB method (Doyle and Doyle 1987). Library preparation and sequencing were performed at Genewiz GmbH (Leipzig, Germany) using the NovaSeq 6000 sequencing system. Paired-end reads of 150 bp were obtained for fragments with an insert size of 300–600 bp. Raw sequences were quality-filtered using Trim Galore v.0.6.5 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with a Phred quality threshold of 30 (-q 30 –paired) and a minimum read length of 20 (−length). De novo assembly of the plastid genomes was performed for each sample using GetOrganelle (https://github.com/Kinggerm/GetOrganelle; Jin et al. 2020).
The assembled plastid genomes were annotated with the GeSeq application (Tillich et al. 2017) in MPI-MP CHLOROBOX (https://chlorobox.mpimp-golm.mpg.de/index.html). Multigene alignments of all plastid genomes were constructed using the HomBlocks pipeline (Bi et al. 2018). Phylogenetic trees were reconstructed using maximum likelihood with RAxML-8.2.4 (Stamatakis 2014). Bootstrap percentages were calculated using 1,000 replicates to assess node support. The Erycina pusilla (L.) N.H. Williams and M.W. Chase (JF746994) plastid genome was selected as the outgroup, retrieved from the NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/). The resulting trees were visualized in FIGTREE v 1.4.4 software. The plastid raw reads were deposited under BioProject no. PRJNA1146482.
RNA-seq for gene annotation
To generate RNA-seq data for G. gibba, we collected 1 root tip, 1 young pseudobulb, and 1 young inflorescence in the day (between 8 and 9 Am), in addition to 8 floral samples, consisting of 3 hypochiles (a basal portion of labellum) and 3 epichiles (an apical portion of labellum) sampled during the day and 2 hypochiles sampled at night (9 Pm). The tissue samples were placed in 2-mL centrifuge tubes filled with two 3-mm glass beads and immediately flash-frozen in liquid nitrogen. The tissue was disrupted using a Mixer Mill MM400 (Retsch), using 3 rounds of 30-s homogenization at 30 Hz, with cooling in liquid nitrogen between runs. RNA was then extracted using a standard RNeasy Mini protocol with QIAshredder following manufacturer's instructions, using a final elution with 30 μL of water. RNA was freeze-dried in GenTegra tubes and rehydrated just before sending for sequencing, following the manufacturer's instructions. The samples were sent to Novogene for quality control, library preparation, and sequencing. The samples were sequenced using 150-bp paired-end reads on a NovaSeq 6000. This generated ∼47 million reads per library (mean = 47.17 million, SD = 4.29 million, n = 11). The raw sequence data were deposited in the SRA under the BioProject accession no. PRJNA1027883. We trimmed the reads using Trim Galore! (Martin 2011). We then mapped the reads to the G. gibba genome assembly using 2-pass mapping in STAR (Dobin et al. 2013). We concatenated the BAM file to use in the annotation as described below.
Repeat annotation
Tandem repeats and transposable elements were identified and annotated using the Extensive de novo TE Annotator (EDTA) v1.9.8 pipeline (Ou et al. 2019), RepeatModeler v.2.0.1 (Flynn et al. 2020), and RepeatMasker v.4.1.2 (Tarailo-Graovac and Chen 2009). Briefly, the EDTA pipeline and RepeatModeler were used for both ab initio and homology-based identification of TEs and tandem repeats, producing G. gibba repeat libraries. These libraries were combined using USEARCH to cluster sequences with >80% identity and remove all but 1 sequence from each cluster (Edgar 2010). We then used RepeatMasker with the custom library (Gongorav1_lib1.fa) to generate a masked genome (Gongorav1.fa.masked). Custom library and masked genome files can be found at the OSF (https://osf.io/hqav3/?view_only=5d65957cc6474488b53847219a7b6ac4).
Gene prediction
We annotated the G. gibba genome using BRAKER3 (braker.pl v3.0.2) which combines RNA-seq and protein data in an automated pipeline (Lomsadze et al. 2005, 2014; Stanke et al. 2006, 2008; Gotoh 2008; Iwata and Gotoh 2012; Buchfink et al. 2015; Hoff et al. 2016, 2019; Kovaka et al. 2019; Brůna et al. 2020, 2021; Pertea and Pertea 2020; Gabriel et al. 2023). Along with the masked genome, we provided BRAKER with the RNA-seq data generated as described above and plant proteins downloaded from OrthoDB (odb10_plants; Zdobnov et al. 2021). In this pipeline, GeneMark-ETP was trained using the RNA-seq and protein hints, and then AUGUSTUS (Stanke et al. 2006, 2008) was run on the GeneMark-ETP prediction and also predicts a set of genes. TSEBRA (Gabriel et al. 2021) was then used to combine the predictions of AUGUSTUS and GeneMark-ETP to preserve only the genes with the highest evidence. The BUSCO (Simão et al. 2015) and OMArk (Nevers et al. 2022) were used to evaluate the completeness and consistency of the final set of gene models. We transferred the annotation to the final genome version after contaminant removal by the NCBI using Liftoff (Shumate and Salzberg 2021).
Annotation of TPS genes
To improve the annotation of TPS genes, we used 2 different pipelines. First, we used bitacora (Vizueta et al. 2020), a pipeline that curates an existing annotation and finds additional gene family members. To do this, BLASTP and hmmer were used to search the existing annotation for gene family members of interest (Altschul et al. 1990; Eddy 2011). Then, additional regions of the genome were searched using TBLASTN, annotated using GeMoMa (Keilwagen et al. 2019), and validated with hmmer to create a new set of genes to add to the originally annotated set. As input, we used 2 HMM profiles: Terpene_syth_C (PF03936) and TPS N-terminal domain (PF01397) downloaded from Pfam. In addition, we provided previously annotated genes from Arabidopsis thaliana and Oryza sativa (Chen et al. 2011; Yu et al. 2020; Jia et al. 2022). In addition, we searched the G. gibba genome with the same set of protein sequences using TBLASTN (Altschul et al. 1990) and then annotated proteins on these scaffolds using exonerate (Slater and Birney 2005). Gene models from both bitacora and exonerate, and genome-wide annotations from BRAKER, were compared and TPS genes were manually curated using IGV (Robinson et al. 2011). We also created an annotation based on the concatenated RNA-seq bam file using StringTie (Pertea et al. 2016) to compare with our other annotations in IGV (Robinson et al. 2011). To check the final set of TPS genes for complete protein domains, we used the NCBI conserved domain search (Marchler-Bauer et al. 2015). All TPS gene annotations were then merged with the genome-wide annotation file (Gongora_annotation.gff3). Sequences for all TPS genes are also available (TPS_dna.fa, TPS_aa.fa). Those sequences considered pseudogenes were not included in the following phylogenetic analyses (TPS_psuedo.fa). All files are available at the OSF (https://osf.io/hqav3/?view_only=5d65957cc6474488b53847219a7b6ac4).
The predicted G. gibba TPS protein sequences, along with those from A. thaliana and O. sativa, were combined with TPS sequences from other orchids (Apostasia shenzhenica, Dendrobium catenatum, Phalaenopsis aphrodite, Phalaenopsis equestris, and Vanilla planifolia; Yu et al. 2020; Huang, Huang, et al. 2021) and aligned with MAFFT v7.508 (-maxiterate 1000, using L-INS-I algorithm; Katoh and Standley 2013). We then trimmed the alignment using trimAl (-gt0.6, sites only included when present in 60% of sequences; Capella-Gutierrez et al. 2009). Based on this alignment, we reconstructed a gene tree using 198 sequences in IQ-TREE with the ModelFinder function to determine the best-fit model (1,000 bootstraps, model JTT + F + G4; Nguyen et al. 2015; Kalyaanamoorthy et al. 2017; Hoang et al. 2018). We rooted the tree using the midpoint.root function in the phytools package in R (Revell 2012; R Core Team 2023). This midrooted version was then plotted using the following packages in R: ape (Paradis and Schliep 2018), evobiR (Blackmon and Adams 2015), ggtree (Yu et al. 2017), and ggtreeExtra (Xu et al. 2021). All files for analysis are available at the OSF (https://osf.io/hqav3/?view_only=5d65957cc6474488b53847219a7b6ac4).
Results
Genome assembly
In this study, we reported the first draft of the G. gibba genome assembly (Supplementary Fig. 1). To overcome the high repeat content and heterozygosity, our assembly strategy consisted of combining short- and long-read sequencing with chromosome conformation capture (Hi-C) technologies. Based on a k-mer analysis, the final genome size was estimated to be 2.228 Gb (2.6 Gb with flow cytometry) with a heterozygosity of 5.9%. A total of 71 Gb of SMRT sequences were corrected with small reads and used for the initial contig assembly. After scaffolding and polishing, the total length of the assembly was 1.831 Gb, with a corresponding contig N50 value of 0.382 Mb (Supplementary Table 2). To further improve the assembly, 35.1 Gb of Chicago and 32 Gb of Hi-C library reads were used to anchor, order, and orient the contigs. The final assembly contains 9,019 scaffolds, with a total length of 1.832 Gb and an N50 value of 1.756 Mb (Table 1). About 50% of the total assembled genome was contained in the 262 longest scaffolds. Genome assembly completeness was assessed using the BUSCO with the embryophyta dataset. Of the 1,375 conserved core embryophyta genes used to assess genome completeness, 1,190 (86.6%) of core genes were represented in our genome assembly, compared with other published orchid genomes (Table 1, Supplementary Table 4).
Table 1.
A comparison between orchid genome assemblies.
| Phalaenopsis equestris | Dendrobium catenatum | Phalaenopsis aphrodite | Cymbidium sinensea | Cymbidium goeringiia | Gongora gibba | |
|---|---|---|---|---|---|---|
| Year | 2015 | 2016 | 2018 | 2021 | 2021 | 2022 |
| Est. size (Gb) | 1.6 | 1.11 | 1.2 | 3.52 | 4.0 | 2.23 |
| Assembled size (Gb) | 1.09 | 1.01 | 1.02 | 3.45 | 3.99 | 1.83 |
| Scaffold N50 (Mb) | 0.36 | 0.39 | 0.95 | NA | NA | 1.756 |
| Contig N50 (kb) | 20.55 | 33.09 | 18.81 | 1,110 | 377.6 | 382.58 |
| Longest scaffold (Mb) | 81.76 | 2.59 | 10.39 | NA | NA | 17.67 |
| Repeat content (%) | 62 | 78.1 | 60.3 | 77.78 | 88.87 | 82.95 |
| BUSCO (%) | 91 | 92.46 | 95 | 91 | 87.8 | 86.6 |
| No. of genes | 29,431 | 28,910 | 28,902 | 29,638 | 29,556 | 17,374 |
| No. of scaffolds | 236,185 | 72,901 | 13,732 | 20 | 20 | 9,019 |
| Reference | Cai et al. 2015 | Zhang et al. 2016 | Chao et al. 2018 | Yang et al. 2021 | Chung et al. 2022 | This study |
a Chromosome-level assembly.
The mitochondrial genome was assembled into 13 scaffolds with a total length of 462,164 bp. The size of the plastid genome was 156,794 bp in length including a pair of inverted repeats named IRa and IRb of 26,677 bp that divide the plastid genome into a large single-copy (84,915 bp) and a small single-copy region (18,525 bp). We identified and annotated a total of 114 unique genes, 80 coding sequences, including 21 genes duplicated in the IR region, 30 distinct tRNAs, and 4 distinct rRNA genes.
We confirmed the identity of chemotype A as G. gibba by creating a plastid phylogeny with 10 additional Gongora samples from Panama. G. gibba from Panama and chemotype A from La Gamba, Costa Rica, formed a well-supported clade, and we therefore named chemotype A as G. gibba (Supplementary Fig. 2).
Gene prediction
Using both de novo and library-based repetitive sequence annotation, we annotated 82.95% of the G. gibba genome as repeat elements. The repetitive content of G. gibba was higher than the most of the other sequenced orchids except for Cymbidium goeringii (88.87%; Chung et al. 2022). Retrotransposable elements, known to be the dominant form of repeats in angiosperm genomes, constitute a large part of the genome and include the most abundant subtypes, such as LTR/Copia (15.82%), LTR/Ty3 (10.99%; Wei et al. 2022), LINE/L1 (0.83%), and LINE/RTE-BovB (1.13%), among others (Supplementary Table 3). Of the repetitive elements, 30.88% could not be classified into any known families, consistent with previous reports from other orchid genomes, suggesting that there may be new repetitive or transposable elements unique to the family Orchidaceae (Zhang et al. 2016; Yu et al. 2020; Yang et al. 2021).
Protein-coding gene models were constructed using a pipeline combining de novo prediction and homology-based prediction methods. In total, 17,374 protein-coding genes were annotated in G. gibba. Using the BUSCO to assess the completeness of genic regions using the embryophyta database, we found that 83.6% (1,150/1,375) of orthologous groups were present in annotation (Supplementary Table 4). Furthermore, we assessed annotation quality using the OMArk which assesses not only completeness but also consistency of the annotation compared with the most closely related species available. The OMArk used the Magnoliopsida database to calculate the BUSCO scores, with 88.5% (6,915/7,818) orthologous groups present in the G. gibba annotation. Furthermore, 92.0% of the proteome was assessed as having consistent lineage placement, with no contamination (genes whose closest gene families is from another lineage and likely come from contamination), no partial mapping (genes that have <80% of the sequence with shared k-mer content from its closest gene family) or fragments (genes with a length less than half the median gene content of its closest gene family) detected. This suggested that both the assembly and annotation quality of the G. gibba genome were high.
Annotation of TPS genes
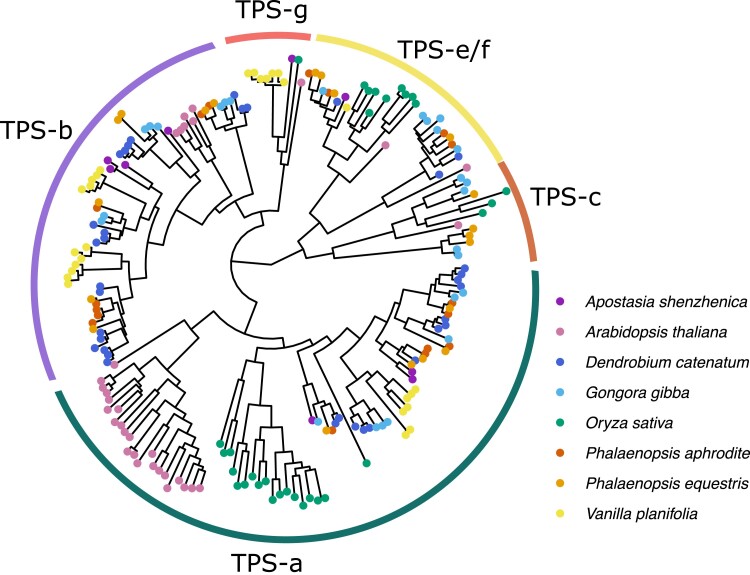
We annotated 26 TPS genes in G. gibba, similar to the previously described orchid TPS families. To resolve the phylogenetic relationship of the TPS genes and those of other orchids, we constructed a phylogenetic tree based on their amino acid sequences and included TPS gene sequences derived from A. thaliana and O. sativa, as well as the orchids Ap. shenzhenica, D. catenatum, D. officinale, P. equestris, and V. planifolia. The 26 putative TPS genes in G. gibba were ascribed to the previously recognized TPS subfamilies in angiosperms: TPS-a, TPS-b, TPS-c, and TPS-e/f (Fig. 1). No members of TPS-g were found.
Fig. 1.
A phylogenetic analysis of flowering plant TPSs. TPS family classifications are illustrated.
Discussion
Some orchid species, such as those belonging to the Catasetinae and Stanhopeinae, exhibit highly specialized pollination associations with fragrance-collecting male euglossine bees. In this system, differences in the floral scent profile of closely related lineages can mediate reproductive isolation because floral scent regulates pollinator attraction and specificity (Dressler 1968; Williams and Dodson 1972; Williams and Whitten 1983; Hetherington-Rauth and Ramírez 2016; Guizar Amador 2022). Therefore, the study of speciation in these orchids requires understanding the molecular basis of floral scent emission and the evolutionary forces promoting its differentiation. So far, research in this area has been limited by a lack of genomic resources. In this study, we construct a genome assembly of G. gibba, an orchid from the subtribe Stanhopeinae, with an assembled genome size of 1.83 Gb. Alongside the assembly, we present a genome-wide annotation and a high-quality annotation of TPS genes involved in floral scent production. These new resources will facilitate further investigation into the genetic architecture of floral scent and its role in reproductive isolation.
Species identification within the genus Gongora poses significant challenges due to little genetic and morphological variation, with cryptic species differentiated by their floral scent (Dressler 1966; Whitten 1985; Jenny 1993; Hetherington-Rauth and Ramírez 2016; Guizar Amador 2022). The genome assembly generated belongs to a group previously treated as chemotype A from La Gamba, Costa Rica (Hetherington-Rauth and Ramírez 2016; Guizar Amador 2022). In this study, we name chemotype A as G. gibba. The type specimen of G. gibba is from Colon, Panama, a geographically distant location from La Gamba. This species is characterized by specific scent compounds, mainly trans-elemicin and trans-methyl cinnamate (Whitten 1985). In contrast, chemotype A produces trans-methyl-methoxy-cinnamate, estragole, cis-methyl-methoxy-cinnamate, and chavicol and attracts different bee pollinators (Hetherington-Rauth and Ramírez 2016). However, plants of G. gibba from Panama are morphologically indistinguishable from plants treated as chemotype A from La Gamba, which is confirmed by their close phylogenetic relationship. This chemotype, along with others, is being actively investigated for the presence of cryptic species (Hetherington-Rauth and Ramírez 2016; Guizar Amador 2022). This highlights the need for comprehensive investigations to delineate species boundaries and relationships within Gongora chemotypes. However, we emphasize the utility of the genome reported here for bioinformatic studies at the genus level due to the potentially high genetic similarity among Gongora species.
A nearly 3,000-fold range of genome sizes has been described in land plants, with larger genomes tending to have higher percentages of transposable elements (Kress et al. 2022). Within the flowering plants, Orchidaceae has the most variation in genome size (Leitch et al. 2009). The size of the G. gibba genome, at 1.83 Gb, is comparable with other sequenced orchids from the subfamily Epidendroideae. The G. gibba genome is around half the size of previously sequenced Cymbidium genomes and around double that of previously sequenced Dendrobium and Phalaenopsis genomes. Repeat content is high, 83%, in the G. gibba genome, with only C. goeringii described as having a higher percentage of repeats at 89%. Given the smaller genome size in G. gibba compared with C. goeringii, this high percentage of repetitive content is perhaps surprising. Fewer genes were annotated in G. gibba compared with other orchids. This could be due to the high ratio of repeats to genome size or due to the stringent criteria used when combining evidence from both RNA-seq and protein homology during annotation.
For decades, biologists have studied plant metabolism, trying to elucidate both the evolutionary function and the underlying genetic basis of plant chemical diversity (Firn and Jones 2003). The plants also exhibit variation in chemical production within species as well as variation between different families or species (van Leur et al. 2006; Dussarrat et al. 2023). In some species, this variation is so large that chemotypes can be described based on the presence of certain compounds or their ratios. In many cases, the ecological function of these chemotypes and their genetic basis remain unknown. Gongora orchids produce a floral scent that attracts their pollinators: male euglossine bees (Allen 1954; Eltz et al. 2005). The scent compounds are then collected by the bees, and they use them as a perfume during courtship displays. Different Gongora chemotypes produce distinct floral scents, which attract nonoverlapping sets of pollinator species, providing an excellent system to study the evolution of plant chemical diversity (Hetherington-Rauth and Ramírez 2016; Guizar Amador 2022). This study provides the genomic resources needed to make this possible.
Many volatile compounds emitted by Gongora flowers are terpenes, a large and structurally diverse class of compounds (Hetherington-Rauth and Ramírez 2016). The diversity of terpenes found in nature is mainly due to the diversification of the TPS gene family that carries out key steps in terpene formation (Tholl 2006). This family of enzymes is present in all land plants and has evolved rapidly through gene duplication and sequence divergence, resulting in an astounding diversity of often lineage-specific terpene compounds (Chen et al. 2011). The total number of TPS genes in a genome differs between species, and in orchids, they have been found to range from 14 in Ap. shenzhenica (Yu et al. 2020) to 48 in Dendrobium chrysotoxum (Zhang et al. 2021). TPS genes are classified into 7 subfamilies: TPS-a, TPS-b, TPS-c, TPS-d, TPS-e/f, TPS-g, and TPS-h (Chen et al. 2011). In the G. gibba genome, we identified and annotated 26 different TPS genes belonging to multiple subfamilies. Based on the previously identified chemotypes (Hetherington-Rauth and Ramírez 2016), we expect G. gibba (chemotype A) to be able to produce both monoterpene and sesquiterpenes (terpenes of different sizes). We find that G. gibba has 6 TPSs from the TPS-e/f subfamily and 8 from the TPS-b subfamily, both shown to be involved in monoterpene biosynthesis in the floral tissue of Phalaenopsis bellina (Huang, Kuo, et al. 2021). Furthermore, we identified 7 TPSs belonging to the TPS-a family, most of which have been described as sesquiterpene synthases (Chen et al. 2011). Lineage-specific expansions are present in all 3 subfamilies (TPS-a, TPS-b, and TPS-e/f), highlighting the dynamic nature of the evolution of this gene family.
Changes in the number of TPS genes, or in their coding sequences, are not the only mechanisms for scent differentiation. In fact, especially among more closely related lineages, we expect differences in floral scent to be driven by differential expression and regulatory mechanisms upstream of biosynthetic pathways underlying the production of volatile organic compounds. We propose that Gongora is an excellent system for studying the rapid evolution of floral scent, particularly those changes involving different biosynthetic pathways. For example, G. gibba (chemotype A) and chemotype M from La Gamba are closely related to each other, occur sympatrically and have overlapping flowering phenologies. Through pollinator network reconstruction, we have previously shown that each chemotype attracts a unique set of pollinator species, but reproductive isolation is not complete (Guizar Amador 2022). Despite the occurrence of gene flow, no plants with intermediate floral phenotypes have been observed so far, suggesting that hybridization is rare or that selection against intermediate phenotypes is strong. Not only are the floral scents different between the 2 chemotypes, but the volatile compounds emitted by these 2 lineages are the products of 2 unrelated biosynthetic pathways: G. gibba (chemotype A) produces mainly aromatic compounds and chemotype M emits monoterpenoids (Hetherington-Rauth and Ramírez 2016; Guizar Amador 2022). Due to the close evolutionary relationship between the 2 chemotypes, we expect that identifying differentially expressed genes in the labellum of these orchids will shed light on the regulatory networks involved in differential floral scent biosynthesis.
Conclusion
We generated a high-quality reference genome for G. gibba which will serve as a crucial resource for understanding the evolution and maintenance of reproductive barriers in euglossine bee–pollinated orchids. Future studies may focus on elucidating the molecular mechanisms that control pollinator specialization in this group by investigating the expression and regulation of biosynthetic pathways involved in floral scent production.
Supplementary Material
Acknowledgments
The authors thank the Ramírez laboratory for their support and feedback throughout the project. The authors also thank the Tina Honfmann from CIMi (Center for Mycological Research) for her help with the DNA extraction for the Gongora plastome sequencing. The authors thank MiAmbiente of Panama and SINAC of Costa Rica (R-SINAC-SE-DT-PI-006-2023) for the collection permits and the Comisión Institucional de Biodiversidad of the Universidad de Costa Rica for the permit to access the genetic and biochemical elements of Costa Rica's biodiversity under the project C4074. We thank the staff at La Gamba Tropenstation for support during fieldwork.
Contributor Information
Maria Fernanda Guizar Amador, Department of Evolution and Ecology, University of California, Davis, Davis, CA 95616, USA.
Kathy Darragh, Department of Evolution and Ecology, University of California, Davis, Davis, CA 95616, USA.
Jasen W Liu, Department of Evolution and Ecology, University of California, Davis, Davis, CA 95616, USA.
Cheryl Dean, Department of Evolution and Ecology, University of California, Davis, Davis, CA 95616, USA.
Diego Bogarín, Lankester Botanical Garden, University of Costa Rica, P.O. Box 302-7050, Cartago 30109, Costa Rica; Evolutionary Ecology Group, Naturalis Biodiversity Center, 2333 CR Leiden, The Netherlands.
Oscar A Pérez-Escobar, Lankester Botanical Garden, University of Costa Rica, P.O. Box 302-7050, Cartago 30109, Costa Rica; Royal Botanic Gardens, Kew, Richmond, Surrey TW9 3AE, UK.
Zuleika Serracín, Herbario UCH, Universidad Autónoma de Chiriquí, P.O. Box 0427, David, Chiriquí 0427, Panamá.
Franco Pupulin, Lankester Botanical Garden, University of Costa Rica, P.O. Box 302-7050, Cartago 30109, Costa Rica.
Santiago R Ramírez, Department of Evolution and Ecology, University of California, Davis, Davis, CA 95616, USA; Lankester Botanical Garden, University of Costa Rica, P.O. Box 302-7050, Cartago 30109, Costa Rica.
Data availability
All raw sequence data and the genome assembly were deposited to the NCBI under the BioProject accession no. PRJNA1014482. The raw RNA-seq data used for genome annotation were deposited in the SRA under the BioProject accession no. PRJNA1027883 with accession nos. SAMN37807278–SAMN37807288. The plastid sequences were deposited under the BioProject no. PRJNA1146482. Notebooks with parameter details, scripts for phylogenetic analyses, and the genome annotation are available at the OSF (DOI 10.17605/OSF.IO/HQAV3).
Supplemental material available at G3 online.
Funding
S.R.R. received support from the David and Lucile Packard Foundation. Z.S. received funding from the Senacyt (APY-NI-2017-05). D.B. and F.P. received support from the Universidad de Costa Rica (project no. C4074).
Literature cited
- Ackerman JD, Phillips RD, Tremblay RL, Karremans A, Reiter N, Peter CI, Bogarín D, Pérez-Escobar OA, Liu H. 2023. Beyond the various contrivances by which orchids are pollinated: global patterns in orchid pollination biology. Bot J Linn Soc. 202(3):295–324. doi: 10.1093/botlinnean/boac082. [DOI] [Google Scholar]
- Allen PH. 1954. Pollination in Gongora maculata. Ceiba. 4:121–124. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22. doi: 10.1016/j.ygeno.2017.08.001. [DOI] [PubMed] [Google Scholar]
- Blackmon H, Adams RA. 2015. EvobiR: Tools for comparative analyses and teaching evolutionary biology. Zenodo. doi: 10.5281/zenodo.30938. [DOI]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 27(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- Brůna T, Hoff KJ, Lomsadze A, Stanke M, Borodovsky M. 2021. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genomics Bioinforma. 3(1):lqaa108. doi: 10.1093/nargab/lqaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brůna T, Lomsadze A, Borodovsky M. 2020. GeneMark-EP+: eukaryotic gene prediction with self-training in the space of genes and proteins. NAR Genomics Bioinforma. 2(2):lqaa026. doi: 10.1093/nargab/lqaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Cai J, Liu X, Vanneste K, Proost S, Tsai WC, Liu KW, Chen LJ, He Y, Xu Q, Bian C, et al. 2015. The genome sequence of the orchid Phalaenopsis equestris. Nat Genet. 47(1):65–72. doi: 10.1038/ng.3149. [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao YT, Chen WC, Chen CY, Ho HY, Yeh CH, Kuo YT, Su CL, Yen SH, Hsueh HY, Yeh JH. 2018. Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol J. 16(12):2027–2041. doi: 10.1111/pbi.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase MW, Cameron KM, Freudenstein JV, Pridgeon AM, Salazar G, Van den Berg C, Schuiteman A. 2015. An updated classification of Orchidaceae. Bot J Linn Soc. 177(2):151–174. doi: 10.1111/boj.12234. [DOI] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E. 2011. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J Cell Mol Biol. 66(1):212–229. doi: 10.1111/j.1365-313X.2011.04520.x. [DOI] [PubMed] [Google Scholar]
- Christenhusz MJM, Byng JW. 2016. The number of known plants species in the world and its annual increase. Phytotaxa. 261(3):201–217. doi: 10.11646/phytotaxa.261.3.1. [DOI] [Google Scholar]
- Chung O, Kim J, Bolser D, Kim H-M, Jun JH, Choi J-P, Jang H-D, Cho YS, Bhak J, Kwak M. 2022. A chromosome-scale genome assembly and annotation of the spring orchid (Cymbidium goeringii). Mol Ecol Resour. 22(3):1168–1177. doi: 10.1111/1755-0998.13537. [DOI] [PubMed] [Google Scholar]
- Cozzolino S, Widmer A. 2005. Orchid diversity: an evolutionary consequence of deception? Trends Ecol Evol. 20(9):487–494. doi: 10.1016/j.tree.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J, Doyle J. 1987. Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem Bull. 19:11. [Google Scholar]
- Dressler R. 1966. Some observations on Gongora. Orchid Dig. 30:220–223. [Google Scholar]
- Dressler R. 1968. Observations on orchids and euglossine bees in Panama and Costa Rica. Rev Biol Trop. [Google Scholar]
- Dressler RL. 2005. How many orchid species? Selbyana. 26:155–158. [Google Scholar]
- Dussarrat T, Schweiger R, Ziaja D, Nguyen TTN, Krause L, Jakobs R, Eilers EJ, Müller C. 2023. Influences of chemotype and parental genotype on metabolic fingerprints of tansy plants uncovered by predictive metabolomics. Sci Rep. 13(1):11645. doi: 10.1038/s41598-023-38790-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Eltz T, Sager A, Lunau K. 2005. Juggling with volatiles: exposure of perfumes by displaying male orchid bees. J Comp Physiol A. 191(7):575–581. doi: 10.1007/s00359-005-0603-2. [DOI] [PubMed] [Google Scholar]
- Firn RD, Jones CG. 2003. Natural products—a simple model to explain chemical diversity. Nat Prod Rep. 20(4):382–391. doi: 10.1039/b208815k. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 117(17):9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel L, Bruna T, Hoff KJ, Ebel M, Lomsadze A, Borodovsky M, Stanke M. 2023. BRAKER3: fully automated genome annotation using RNA-Seq and protein evidence with GeneMark-ETP, AUGUSTUS and TSEBRA. Genome Res. doi: 10.1101/gr.278090.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel L, Hoff KJ, Brůna T, Borodovsky M, Stanke M. 2021. TSEBRA: transcript selector for BRAKER. BMC Bioinformatics. 22(1):566. doi: 10.1186/s12859-021-04482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach G, Schill R. 1991. Composition of orchid scents attracting euglossine bees. Bot Acta. 104(5):379–384. doi: 10.1111/j.1438-8677.1991.tb00245.x. [DOI] [Google Scholar]
- Givnish TJ, Spalink D, Ames M, Lyon SP, Hunter SJ, Zuluaga A, Iles WJD, Clements MA, Arroyo MTK, Leebens-Mack J, et al. 2015. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc R Soc B Biol Sci. 282(1814):20151553. doi: 10.1098/rspb.2015.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh O. 2008. A space-efficient and accurate method for mapping and aligning cDNA sequences onto genomic sequence. Nucleic Acids Res. 36(8):2630–2638. doi: 10.1093/nar/gkn105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govaerts R, Nic Lughadha E, Black N, Turner R, Paton A. 2021. The world checklist of vascular plants, a continuously updated resource for exploring global plant diversity. Sci Data. 8(1):215. doi: 10.1038/s41597-021-00997-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizar Amador MF. 2022. The Ecological and Genetic Basis of Floral Scent Differentiation in the Orchid Genus Gongora. Ph.D. Dissertation. [Google Scholar]
- Henske J, Saleh NW, Chouvenc T, Ramírez SR, Eltz T. 2023. Function of environment-derived male perfumes in orchid bees. Curr Biol. 33(10):2075–2080.e3 doi: 10.1016/j.cub.2023.03.060. [DOI] [PubMed] [Google Scholar]
- Hetherington-Rauth MC, Ramírez SR. 2016. Evolutionary trends and specialization in the euglossine bee–pollinated orchid genus Gongora. Ann Missouri Bot Gard. 100(4):271–299. doi: 10.3417/2014035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. 2016. BRAKER1: unsupervised RNA-seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinforma Oxf Engl. 32(5):767–769. doi: 10.1093/bioinformatics/btv661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lomsadze A, Borodovsky M, Stanke M. 2019. Whole-genome annotation with BRAKER. Methods Mol Biol. 1962:65–95. doi: 10.1007/978-1-4939-9173-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Kuo Y-W, Chuang Y-C, Yang Y-P, Huang L-M, Jeng M-F, Chen W-H, Chen H-H. 2021. Terpene synthase-b and terpene synthase-e/f genes produce monoterpenes for Phalaenopsis bellina floral scent. Front Plant Sci. 12:700958. doi: 10.3389/fpls.2021.700958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L-M, Huang H, Chuang Y-C, Chen W-H, Wang C-N, Chen H-H. 2021. Evolution of terpene synthases in Orchidaceae. Int J Mol Sci. 22(13):6947. doi: 10.3390/ijms22136947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H, Gotoh O. 2012. Benchmarking spliced alignment programs including Spaln2, an extended version of Spaln that incorporates additional species-specific features. Nucleic Acids Res. 40(20):e161. doi: 10.1093/nar/gks708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny R. 1993. Monograph of the Genus Gongora Ruiz & Pavon. Champaign, IL: Koeltz Scientific Books. [Google Scholar]
- Jia Q, Brown R, Köllner TG, Fu J, Chen X, Wong GK-S, Gershenzon J, Peters RJ, Chen F. 2022. Origin and early evolution of the plant terpene synthase family. Proc Natl Acad Sci U S A. 119(15):e2100361119. doi: 10.1073/pnas.2100361119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilwagen J, Hartung F, Grau J. 2019. Gemoma: homology-based gene prediction utilizing intron position conservation and RNA-seq data. Methods Mol Biol. 1962:161–177. doi: 10.1007/978-1-4939-9173-0_9. [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, Pertea M. 2019. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20(1):278. doi: 10.1186/s13059-019-1910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress WJ, Soltis DE, Kersey PJ, Wegrzyn JL, Leebens-Mack JH, Gostel MR, Liu X, Soltis PS. 2022. Green plant genomes: what we know in an era of rapidly expanding opportunities. Proc Natl Acad Sci U S A. 119(4):e2115640118. doi: 10.1073/pnas.2115640118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Kahandawala I, Suda J, Hanson L, Ingrouille MJ, Chase MW, Fay MF. 2009. Genome size diversity in orchids: consequences and evolution. Ann Bot. 104(3):469–481. doi: 10.1093/aob/mcp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv 13033997. 10.48550/arXiv.1303.3997, preprint: not peer reviewed. [DOI]
- Lomsadze A, Burns PD, Borodovsky M. 2014. Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res. 42(15):e119. doi: 10.1093/nar/gku557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33(20):6494–6506. doi: 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 27(6):764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res. 43(D1):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Nevers Y, Rossier V, Train CM, Altenhoff A, Dessimoz C, Glover N. 2022. Multifaceted quality assessment of gene repertoire annotation with OMArk.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Su W, Liao Y, Chougule K, Agda JRA, Hellinga AJ, Lugo CSB, Elliott TA, Ware D, Peterson T, et al. 2019. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20(1):275. doi: 10.1186/s13059-019-1905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Schliep K. 2018. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 35(3):526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- Pérez-Escobar OA, Bogarín D, Przelomska NAS, Ackerman JD, Balbuena JA, Bellot S, Bühlmann RP, Cabrera B, Cano JA, Charitonidou M, et al. 2024. The origin and speciation of orchids. New Phytol. 242(2):700–716. doi: 10.1111/nph.19580. [DOI] [PubMed] [Google Scholar]
- Pertea G, Pertea M. 2020. GFF utilities: GffRead and GffCompare. F1000Res. 9:ISCB Comm J-304. doi: 10.12688/f1000research.23297.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, O’Connell BL, Stites JC, Rice BJ, Blanchette M, Calef R, Troll CJ, Fields A, Hartley PD, Sugnet CW, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26(3):342–350. doi: 10.1101/gr.193474.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2023. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Ramírez SR, Eltz T, Fujiwara MK, Gerlach G, Goldman-Huertas B, Tsutsui ND, Pierce NE. 2011. Asynchronous diversification in a specialized plant-pollinator mutualism. Science. 333(6050):1742–1746. doi: 10.1126/science.1209175. [DOI] [PubMed] [Google Scholar]
- Revell LJ. 2012. Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3(2):217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol. 29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H. 2020. Fast and accurate long-read assembly with wtdbg2. Nat Methods. 17(2):155–158. doi: 10.1038/s41592-019-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumate A, Salzberg SL. 2021. Liftoff: accurate mapping of gene annotations. Bioinformatics. 37(12):1639–1643. doi: 10.1093/bioinformatics/btaa1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Slater GSC, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 6(1):31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 24(5):637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Stanke M, Schöffmann O, Morgenstern B, Waack S. 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 7(1):62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinforma. Chapter 4:4.10.1–4.10.14. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- Tholl D. 2006. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr Opin Plant Biol. 9(3):297–304. doi: 10.1016/j.pbi.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. Geseq—versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Niet T, Peakall R, Johnson SD. 2014. Pollinator-driven ecological speciation in plants: new evidence and future perspectives. Ann Bot. 113(2):199–212. doi: 10.1093/aob/mct290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leur H, Raaijmakers CE, van Dam NM. 2006. A heritable glucosinolate polymorphism within natural populations of Barbarea vulgaris. Phytochemistry. 67(12):1214–1223. doi: 10.1016/j.phytochem.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Vizueta J, Sánchez-Gracia A, Rozas J. 2020. Bitacora: a comprehensive tool for the identification and annotation of gene families in genome assemblies. Mol Ecol Resour. 20(5):1445–1452. doi: 10.1111/1755-0998.13202. [DOI] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JR, Holt J, McMillan L, Jones CD. 2018. FMLRC: hybrid long read error correction using an FM-index. BMC Bioinformatics. 19(1):50. doi: 10.1186/s12859-018-2051-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K, Aldaimalani R, Mai D, Zinshteyn D, Prv S, Blumenstiel JP, Kelleher ES, Brooks E. 2022. Rethinking the “Gypsy” Retrotransposon: A Roadmap for Community-Driven Reconsideration of Problematic Gene Names. OSF. [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. 2007. Database resources of the national center for biotechnology information. Nucleic Acids Res. 35(Database):D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitten WM. 1985. Variation in floral fragrances and pollinators in the Gongora quinquenervis complex (Orchidaceae) in central Panama. University of Florida.
- Williams NH, Dodson CH. 1972. Selective attraction of male euglossine bees to orchid floral fragrances and its importance in long distance pollen flow. Evolution. 26(1):84–95. doi: 10.2307/2406985. [DOI] [PubMed] [Google Scholar]
- Williams NH, Whitten WM. 1983. Orchid floral fragrances and male euglossine bees: methods and advances in the last sesquidecade. Biol Bull. 164(3):355–395. doi: 10.2307/1541248. [DOI] [Google Scholar]
- Xu S, Dai Z, Guo P, Fu X, Liu S, Zhou L, Tang W, Feng T, Chen M, Zhan L, et al. 2021. ggtreeExtra: compact visualization of richly annotated phylogenetic data. Mol Biol Evol. 38(9):4039–4042. doi: 10.1093/molbev/msab166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Schlüter PM, Schiestl FP. 2012. Pollinator-driven speciation in sexually deceptive orchids. Int J Ecol. 2012:e285081. doi: 10.1155/2012/285081. [DOI] [Google Scholar]
- Yang F-X, Gao J, Wei Y-L, Ren R, Zhang G-Q, Lu C-Q, Jin J-P, Ai Y, Wang Y-Q, Chen L-J, et al. 2021. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol J. 19(12):2501–2516. doi: 10.1111/pbi.13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. 2017. Ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 8(1):28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- Yu Z, Zhao C, Zhang G, Teixeira da Silva JA, Duan J. 2020. Genome-wide identification and expression profile of TPS gene family in Dendrobium officinale and the role of DoTPS10 in linalool biosynthesis. Int J Mol Sci. 21(15):5419. doi: 10.3390/ijms21155419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Kuznetsov D, Tegenfeldt F, Manni M, Berkeley M, Kriventseva EV. 2021. OrthoDB in 2020: evolutionary and functional annotations of orthologs. Nucleic Acids Res. 49(D1):D389–D393. doi: 10.1093/nar/gkaa1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G-Q, Xu Q, Bian C, Tsai W-C, Yeh C-M, Liu K-W, Yoshida K, Zhang L-S, Chang S-B, Chen F, et al. 2016. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci Rep. 6(1):19029. doi: 10.1038/srep19029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Jain C, Aluru S. 2020. A comprehensive evaluation of long read error correction methods. BMC Genomics. 21(S6):889. doi: 10.1186/s12864-020-07227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang G-Q, Zhang D, Liu X-D, Xu X-Y, Sun W-H, Yu X, Zhu X, Wang Z-W, Zhao X, et al. 2021. Chromosome-scale assembly of the Dendrobium chrysotoxum genome enhances the understanding of orchid evolution. Hortic Res. 8(1):183. doi: 10.1038/s41438-021-00621-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Blackmon H, Adams RA. 2015. EvobiR: Tools for comparative analyses and teaching evolutionary biology. Zenodo. doi: 10.5281/zenodo.30938. [DOI]
Supplementary Materials
Data Availability Statement
All raw sequence data and the genome assembly were deposited to the NCBI under the BioProject accession no. PRJNA1014482. The raw RNA-seq data used for genome annotation were deposited in the SRA under the BioProject accession no. PRJNA1027883 with accession nos. SAMN37807278–SAMN37807288. The plastid sequences were deposited under the BioProject no. PRJNA1146482. Notebooks with parameter details, scripts for phylogenetic analyses, and the genome annotation are available at the OSF (DOI 10.17605/OSF.IO/HQAV3).
Supplemental material available at G3 online.