Abstract
The WHO guidelines for classifying central nervous system (CNS) tumours are changing considerably with each release. The classification of CNS tumours is uniquely complex among most other solid tumours as it incorporates not just morphology, but also genetic and epigenetic features. Keeping current with these changes across medical fields can be challenging, even for clinical specialists. Large language models (LLMs) have demonstrated their ability to parse and process complex medical text, but their utility in neuro‐oncology has not been systematically tested. We hypothesised that LLMs can effectively diagnose neuro‐oncology cases from free‐text histopathology reports according to the latest WHO guidelines. To test this hypothesis, we evaluated the performance of ChatGPT‐4o, Claude‐3.5‐sonnet, and Llama3 across 30 challenging neuropathology cases, which each presented a complex mix of morphological and genetic information relevant to the diagnosis. Furthermore, we integrated these models with the latest WHO guidelines through Retrieval‐Augmented Generation (RAG) and again assessed their diagnostic accuracy. Our data show that LLMs equipped with RAG, but not without RAG, can accurately diagnose the neuropathological tumour subtype in 90% of the tested cases. This study lays the groundwork for a new generation of computational tools that can assist neuropathologists in their daily reporting practice.
Keywords: large language models, neuropathology, adult‐type diffuse gliomas, decision support tools
Introduction
Large language models (LLMs) have shown promising performance on several healthcare‐related tasks including medical education [1, 2], and administrative tasks such as writing hospital discharge summaries [3] and clinic letters [4] for cancer patients in a variety of scenarios. While LLMs have also been applied to patient‐facing tasks, such as medical chatbots [5], concerns remain regarding hallucinations (where a generative response contains false or misleading information), their inability to provide references for factual information, and the lack of transparency in decision‐making [6].
One potential application of LLMs is as decision support tools to aid practitioners in the interpretation of clinical guidelines [7]. However, to our knowledge this has not been trialled in a neuropathology setting. We performed a PubMed search and found one study that assessed the diagnostic accuracy of vision‐LLMs on neuropathology images of neurodegenerative diseases [8]. A further seven studies examining the utility of LLMs for diagnostic support tasks within the field of neurology were identified, however none were applied in a neuropathology setting. Our search criteria and a summary of our findings can be found in supplementary material, Table S1.
Given the challenges presented by neuropathology diagnostics due to recent changes in the diagnostic approach and the multitude of potential diagnoses [9], we hypothesised that LLMs could potentially benefit neuropathology practitioners.
This work aims to assess the ability of three leading LLMs, one open‐source (Llama3‐70b from Meta) and two proprietary (ChatGPT‐4o from OpenAI, Claude‐3.5‐sonnet from Anthropic), to provide accurate diagnoses in neuropathology. Since adult‐type diffuse gliomas represent a significant proportion of the diagnostic work in adult neuro‐oncology practice [10], we created 30 realistic free‐text neuropathology reports and asked the models to make a diagnosis based on the histopathological description.
Additionally, we evaluated the standard ‘zero‐shot’ responses against Retrieval‐Augmented Generation (RAG) responses, where the models were provided with the latest WHO guidelines. RAG is a framework which limits the LLM to utilising data provided by the user, such as diagnostic guidelines, rather than relying on data acquired during training or from the internet. Moreover, this approach has been shown to mitigate some limitations of standard LLMs, including hallucinations [11].
Both responses were compared to an expert‐generated ground truth and for strict concordance with the latest WHO guidelines. Thereby, we hypothesise that the RAG responses will outperform the zero‐shot responses.
Materials and methods
Cohort description
We generated 30 artificial neuropathology cases: 10 each for astrocytoma, oligodendroglioma, and glioblastoma, with varying grade, morphological, and molecular features. For accuracy, these cases were based on real data, chosen semi‐randomly from the pseudonimised University College London Hospitals (UCLH) dataset. Cases were selected to represent a full range of features that a neuropathologist might encounter when reporting these entities. Only the diagnostic details necessary for reaching a diagnosis were kept, such as morphological features (e.g. gemistocytes, presence or absence of necrosis and/or mitoses, microvascular proliferations) and the results of further testing (e.g. immunohistochemistry and/or molecular analysis). Tumour descriptions were otherwise rephrased while trying to preserve different writing styles, including any grammar or spelling errors (e.g. ‘mitoticly’ and ‘cells positive’ instead of ‘cells are positive’). This experiment was conducted in concordance with the Declaration of Helsinki. No patient‐identifiable data were accessed or used during this project.
RAG approach
Relevant diagnostic criteria are based on the CNS WHO 2021 fifth edition [9]. The chapters on adult‐type diffuse gliomas, along with paragraphs from the foreword and introduction discussing changes to the diagnostic approach from the previous edition, were collated into a Microsoft Word document. Relevant tables were converted to plain text to make the information accessible to the LLMs.
Large language models
We compared ChatGPT‐4o, Claude‐3.5‐sonnet, and Llama3‐70b‐groq. The Llama3‐70b model with the groq extension was selected for its RAG capabilities, and was used for both the zero‐shot and RAG experiments. All models were accessed via a web interface: ChatGPT through chatgpt.com, Claude through claude.ai, and Llama3 through poe.com. Further details of the prompts used for both the zero‐shot and the RAG experiments are provided in supplementary material, Table S2. The experiments were conducted between 8 May and 26 June 2024.
Analysis
The full neuropathological diagnosis consists of a histopathological diagnosis, a molecular diagnosis, and a grade [9] (e.g. astrocytoma, IDH‐mutant, CNS WHO grade 3). Each response generated by the network was reviewed for these three components, and was deemed correct only if all three completely matched the WHO guidelines. The diagnostic criteria for adult‐type diffuse gliomas can be found in Table 1.
Table 1.
Diagnostic criteria
| Essential criteria | Additional information | |
|---|---|---|
|
Astrocytoma, IDH‐mutant |
Diffusely infiltrating glioma AND IDH‐mutation AND ATRX mutation OR exclusion of whole arm deletions of 1p and 19q |
ATRX mutation can be demonstrated as loss of nuclear ATRX expression on immunohistochemistry or ATRX mutation on molecular testing. |
|
Oligodendroglioma, IDH‐mutant 1p/19q‐codeleted |
Diffusely infiltrating glioma AND IDH‐mutation AND Whole arm deletions of 1p and 19q |
Recommended that 1p/19q assays be able to detect whole‐arm chromosomal losses, such as FISH or molecular genetic testing. |
|
Glioblastoma, IDH‐wildtype |
Diffuse astrocytic glioma AND IDH‐wildtype AND H3‐wildtype |
Must additionally demonstrate one or more of: microvascular proliferation, necrosis, TERT promoter mutation, EGFR gene amplification, chromosome +7/−10 copy number alterations. |
This table summarises the diagnostic criteria for the neuropathological diagnosis of adult‐type diffuse gliomas according to WHO CNS fifth edition [9]. Once a diffusely infiltrating glioma has been identified on histopathology, these criteria must be demonstrated either by immunohistochemistry or DNA sequencing. Alternatively, DNA methylation profiling can also be used. WHO CNS5 also recommends that grade is designated in Arabic numerals, rather than Roman numerals, as in previous editions.
Results
Zero‐shot LLMs are ineffective at neuropathology diagnosis
We compared the ‘zero‐shot’ (standard) performance of all three models (zGPT, zLlama, and zClaude) in diagnosing adult‐type diffuse gliomas from neuropathological descriptions. Although the vast majority of LLM‐generated diagnoses were close to the ground truth, because we held the LLM to clinical standards many responses were classified as ‘incorrect’ by human experts (Table 2; Figure 1).
Table 2.
Example results
| Case number: ground truth | Experimental approach | Model and responses | ||
|---|---|---|---|---|
| ChatGPT | Llama | Claude | ||
|
Case A2: Astrocytoma IDH‐mutant Grade 2 |
Zero‐shot |
Astrocytoma IDH‐mutant Grade II |
Astrocytoma IDH‐mutant Grade II |
Astrocytoma IDH‐mutant Grade 2 |
| RAG |
Astrocytoma IDH‐mutant Grade 2 |
Astrocytoma IDH‐mutant Grade 2 |
Astrocytoma IDH‐mutant Grade 2 |
|
|
Case G10: Glioblastoma IDH‐wildtype Grade 4 |
Zero‐shot |
Glioblastoma IDH‐wildtype Grade 4 |
Anaplastic astrocytoma Grade III |
Astrocytoma IDH‐wildtype Grade 4 |
| RAG |
Glioblastoma IDH‐wildtype Grade 4 |
Glioblastoma IDH‐wildtype Grade 4 |
Glioblastoma IDH‐wildtype Grade 4 |
|
|
Case O9: Oligodendroglioma IDH‐mutant, 1p/19q co‐deleted Grade 3 |
Zero‐shot |
Anaplastic oligodendrogl. IDH‐mutant, 1p/19q co‐del. Grade III |
Glioblastoma IDH‐mutant Grade IV |
Anaplastic oligodendrogl. IDH‐mutant, 1p/19q co‐del. Grade II |
| RAG |
Oligodendrogl. IDH‐mutant, 1p/19q co‐del. Grade 3 |
Astrocytoma IDH‐mutant Grade 3 |
Oligodendrogl. IDH‐mutant, 1p/19q co‐del. Grade 3 |
|
This table provides examples of the diagnoses given by each model, for each experimental approach and tumour type. One case for each tumour type was chosen and the diagnoses given by each model for both experimental approaches are provided, to illustrate how the models performed. Responses that were classified as incorrect are highlighted in red. In most instances, the diagnosis provided by the model was close to the ground truth, but were deemed incorrect because they failed to meet the current WHO guidance [9]. For example, the ChatGPT response in the zero‐shot experiment for case A2 was deemed incorrect because the grade was given in Roman, rather than in Arabic, numerals. This change to how grade should be noted was implemented by the most recent edition of the WHO CNS 5 guidelines. Many of the diagnostics errors made by the models in the zero‐shot experiments related to changes that were introduced in the WHO CNS 5 guidelines, such as astrocytoma and glioblastoma being exclusively IDH‐mutant and IDH‐wildtype respectively, and the term anaplastic no longer being used. Full responses are provided in supplementary material, Full responses by case.
Figure 1.
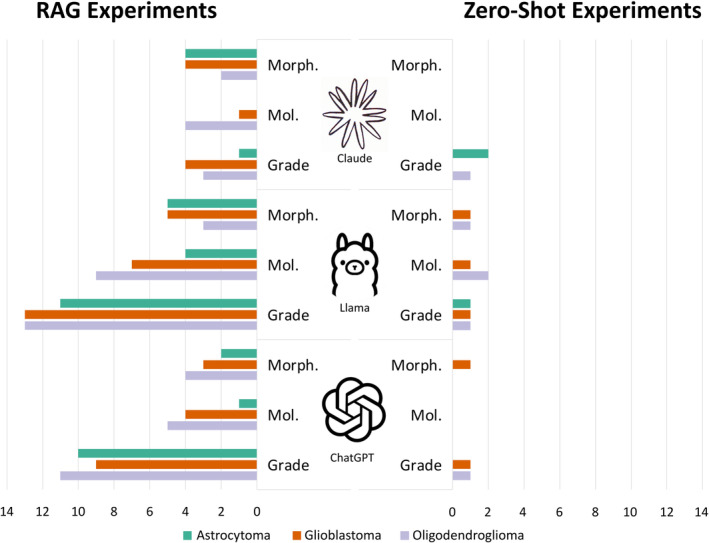
Mistakes by model and tumour type. This bar chart shows the total number of mistakes made by each model for each tumour type, where green represents astrocytoma, orange represents glioblastoma and grey oligodendroglioma. The bars are absent for some of the RAG experiments because no incorrect diagnoses were generated for these cases. The diagnostic accuracy of the models was judged according to three components: the morphological diagnosis, e.g. astrocytoma; the molecular diagnosis, e.g. IDH‐mutant; and the grade, e.g. grade 3. Errors related to grade were either that the incorrect grade was given by a model or the grade was given in Roman rather than Arabic numerals. Errors relating to morphology were either that the morphological diagnosis itself was incorrect, or an outdated term such as ‘anaplastic’ was used. Errors relating to molecular status were either that the molecular diagnosis provided was wrong, or a molecular status was not provided at all. In the instances where an outdated term or a grade in Roman numerals was provided, the response was deemed incorrect, regardless of whether the morphological diagnosis or the grade itself was correct. This level of accuracy was chosen because we wanted to hold the models to clinical standards. This figure shows that mistakes of all types were made much more frequently in the zero‐shot experiments than the RAG experiments.
In the astrocytoma cases, zClaude provided the correct diagnosis in 6/10 cases and zGPT in 1/10, whereas zLlama was unable to provide any correct diagnoses. For the glioblastoma cases, zClaude provided 5/10 correct diagnoses, zGPT provided 3/10, and zLlama provided no correct diagnoses. As for the oligodendroglioma cases, only zClaude provided correct diagnoses, doing so in 5/10 cases. Neither zGPT nor zLlama provided any correct responses.
Across all zero‐shot experiments, the correct diagnosis was given in just 20/90 cases (22.2%); 13.3% (n = 4) by zGPT and 53.3% (n = 16) by zClaude. zLlama provided no correct responses. The most common reason for responses being deemed incorrect was that the grade was given in Roman, rather than Arabic numerals. This was the case in 18/30 astrocytoma cases (60%), 18/30 glioblastoma cases (60%), and 21/30 oligodendroglioma cases (70%). Figures 1 and 2 provide an overview of the mistakes and correct diagnoses made by each model.
Figure 2.
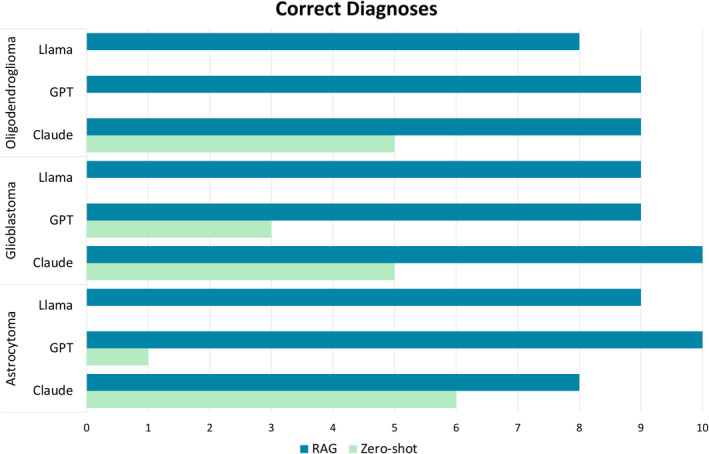
Correct diagnoses by tumour type. This bar chart presents the total number of correct diagnoses made by each model for each tumour type, where dark blue represents zero‐shot correct response and light green RAG correct responses. The bar is absent for some of the zero‐shot experiments because no correct diagnoses were generated for these cases. This figure demonstrates that correct diagnoses were made much more frequently by all models in the RAG experiments than the zero‐shot experiments.
RAG can provide an accurate neuropathology diagnosis
Next, we compared performance of all three models when utilising a RAG approach (rGPT, rLlama, and rClaude).
In the astrocytoma cases, rClaude provided the correct diagnosis in 8/10 cases, rLlama in 9/10 cases, and rGPT in 10/10 cases. For the glioblastoma cases, rClaude provided the correct diagnosis in 10/10 cases, while rGPT and rLlama both provided the correct diagnosis in 9/10 cases. As for the oligodendroglioma cases, both rGPT and rClaude correctly diagnosed 9/10 cases, whereas rLlama correctly diagnosed 8/10 cases.
Across all RAG experiments, the correct diagnosis was provided in 81/90 cases (90%); 93.3% (n = 28) by rGPT, 86.7% (n = 26) by rLlama, and 90% (n = 27) by rClaude. A results summary can be found in Figures 1 and 2.
Summaries of the results given by all three models across both the zero‐shot and RAG experiments for the astrocytoma, glioblastoma and oligodendrogioma case experiments are provided in supplementary material, Tables S3, S4 and S5 respectively.
Discussion
Our study has evaluated the performance of LLMs on neuropathology cases, providing evidence against the use of zero‐shot LLMs within this field. Despite all three models being capable of providing neuropathology diagnoses based on histological descriptions of brain tumours, the diagnoses frequently used outdated terminology and often failed to meet current diagnostic standards. Conversely, the RAG approach yielded higher‐quality results, providing an accurate diagnosis in 90% of cases. ChatGPT with RAG was the most accurate among the models, with only two misdiagnoses. One misdiagnosis consisted of an incorrect grade in an oligodendroglioma case, which is arguably subjective and prone to inter‐observer variability, even among experts. However the other misdiagnosis was more consequential, as it was a glioblastoma case misdiagnosed as a low‐grade astrocytoma. This has significant impact for both management [12] and outcome [10], but as this case was an ‘early’ glioblastoma, it also presents a diagnostic challenge.
Our findings support RAG's potential to improve the diagnostic capabilities of LLMs. By constraining the natural language processing power of LLMs within clinical guidelines, RAG can mitigate some of the primary concerns associated with LLMs, such as lack of transparency and fabricated results. Through additional prompt engineering, LLMs with RAG could benefit clinicians working in neuropathology.
Limitations of our study include the scope. Although adult‐type diffuse gliomas are a fundamental part of neuropathology, numerous other entities are routinely encountered by neuropathologists on a daily basis. Further work to evaluate LLMs on a wider variety of entities and differentials, obtained from a variety of different data sources, is necessary. This should include rare cases and cases where the diagnosis is uncertain, such as entities which are a diagnosis of exclusion, to assess how the LLMs deal with ambiguity.
Additionally, before any AI model can be used clinically, the issue of safeguarding confidential patient data needs to be addressed. Many approaches for this exist, including computational privacy‐preserving techniques [13] and locally run LLM anonymisation pipelines [14]. Regardless of the approach, the underlying principle remains the same: sensitive data should not be shared with cloud‐based services (including LLMs) unless the provider can ensure that the data is treated according to the relevant legislations, i.e. Health Insurance Portability and Accountability Act in the USA and General Data Protection Regulation in the European Union.
Author contributions statement
KJH is involved in the conception of the study; data acquisition and analysis; writing of first draft; review and critique; and final approval. ICW is involved in the conception of the study; review and critique; and final approval. ZIC is responsible for review and critique; and final approval. LB is involved in data acquisition; review and critique; and final approval. TOM is involved in the data acquisition; review and critique; and final approval. SB is involved in the data acquisition; review and critique; and final approval. JNK is involved in the conception of the study; review and critique; and final approval.
Supporting information
Table S1. Results of literature review
Table S2. Prompts
Table S3. Summary of astrocytoma case results
Table S4. Summary of glioblastoma case results
Table S5. Summary of oligodendroglioma case results
Full responses by case
Acknowledgements
The dataset on which the cases were based was obtained from UCLH via Brain UK (REF:22/011). The authors thank both Brain UK and UCLH for their roles in this work. JNK is supported by the German Cancer Aid (DECADE, 70115166), the German Federal Ministry of Education and Research (PEARL, 01KD2104C; CAMINO, 01EO2101; SWAG, 01KD2215A; TRANSFORM LIVER, 031L0312A; TANGERINE, 01KT2302 through ERA‐NET Transcan; Come2Data, 16DKZ2044A; DEEP‐HCC, 031L0315A), the German Academic Exchange Service (SECAI, 57616814), the German Joint Federal Committee (TransplantKI, 01VSF21048), the European Union's Horizon Europe and innovation programme (ODELIA, 101057091; GENIAL, 101096312), the European Research Council (NADIR, 101114631), the National Institutes of Health (EPICO, R01 CA263318), and the National Institute for Health and Care Research (NIHR; NIHR203331) Leeds Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care. This work was funded by the European Union. Views and opinions expressed are, however, those of the authors only and do not necessarily reflect those of the European Union. Neither the European Union nor the granting authority can be held responsible for them. Open Access funding enabled and organized by Projekt DEAL.
Conflict of interest statement: JNK declares consulting services for Bioptimus, Owkin, France; DoMore Diagnostics, Norway; Panakeia, UK; AstraZeneca, UK; Mindpeak, Germany; and MultiplexDx, Slovakia. Furthermore, he holds shares in StratifAI GmbH, Germany, Synagen GmbH, Germany, and has received a research grant by GSK, and has received honoraria by AstraZeneca, Bayer, Eisai, Janssen, Merck, MSD, BMS, Roche, Pfizer, and Fresenius.
Data availability statement
The UCLH dataset was acquired through Brain UK (REF: 22/011). Inquiries regarding access to this dataset should be directed to SB. Llama3‐70b‐groq is freely accessible via poe.com, whereas ChatGPT‐4o requires a paid subscription. Anthropic offers limited free access to Claude, but full access requires a paid subscription.
References
References 15, 16, 17, 18, 19, 20, 21 are cited only in the supplementary material.
- 1. Singhal K, Tu T, Gottweis J, et al. Towards expert‐level medical question answering with large language models. arXiv 2023. 10.48550/arXiv.2305.09617. [DOI]
- 2. Kung TH, Cheatham M, Medenilla A, et al. Performance of ChatGPT on USMLE: potential for AI‐assisted medical education using large language models. PLoS Digit Health 2023; 2: e0000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Patel SB, Lam K. ChatGPT: the future of discharge summaries? Lancet Digit Health 2023; 5: e107–e108. [DOI] [PubMed] [Google Scholar]
- 4. Ali SR, Dobbs TD, Hutchings HA, et al. Using ChatGPT to write patient clinic letters. Lancet Digit Health 2023; 5: e179–e181. [DOI] [PubMed] [Google Scholar]
- 5. Ayers JW, Poliak A, Dredze M, et al. Comparing physician and artificial intelligence Chatbot responses to patient questions posted to a public social media forum. JAMA Intern Med 2023; 183: 589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rao A, Kim J, Kamineni M, et al. Evaluating ChatGPT as an adjunct for radiologic decision‐making. medRxiv 2023. 10.1101/2023.02.02.23285399. [DOI] [PMC free article] [PubMed]
- 7. Kresevic S, Giuffrè M, Ajcevic M, et al. Optimization of hepatological clinical guidelines interpretation by large language models: a retrieval augmented generation‐based framework. npj Digit Med 2024; 7: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ono D, Dickson DW, Koga S. Evaluating the efficacy of few‐shot learning for GPT‐4Vision in neurodegenerative disease histopathology: a comparative analysis with convolutional neural network model. Neuropathol Appl Neurobiol 2024; 50: e12997. [DOI] [PubMed] [Google Scholar]
- 9. WHO Classification of Tumours Editorial Board . Central Nervous System Tumours. WHO Classification of Tumours Series, Volume 6 (5th edn). International Agency for Research on Cancer: Lyon, 2021. [Google Scholar]
- 10. Ostrom QT, Price M, Neff C, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2016–2020. Neuro Oncol 2023; 25: iv1–iv99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiong G, Jin Q, Lu Z, et al. Benchmarking retrieval‐augmented generation for medicine. arXiv 2024. 10.48550/arXiv.2402.13178. [DOI]
- 12. Weller M, van den Bent M, Preusser M, et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol 2020; 18: 170–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khalid N, Qayyum A, Bilal M, et al. Privacy‐preserving artificial intelligence in healthcare: techniques and applications. Comput Biol Med 2023; 158: 106848. [DOI] [PubMed] [Google Scholar]
- 14. Wiest IC, Leßmann M‐E, Wolf F, et al. Anonymizing medical documents with local, privacy preserving large language models: the LLM‐anonymizer. bioRxiv 2024. 10.1101/2024.06.11.24308355. [DOI]
- 15. Wada A, Akashi T, Shih G, et al. Optimizing GPT‐4 turbo diagnostic accuracy in neuroradiology through prompt engineering and confidence thresholds. Diagnostics 2024; 14: 1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kumar RP, Sivan V, Bachir H, et al. Can artificial intelligence mitigate missed diagnoses by generating differential diagnoses for neurosurgeons? World Neurosurg 2024; 187: e1083–e1088. [DOI] [PubMed] [Google Scholar]
- 17. Zaki HA, Aoun A, Munshi S, et al. The application of large language models for radiologic decision making. J Am Coll Radiol 2024; 21: 1072–1078. [DOI] [PubMed] [Google Scholar]
- 18. Madadi Y, Delsoz M, Lao PA, et al. ChatGPT assisting diagnosis of neuro‐ophthalmology diseases based on case reports. medRxiv 2023. 10.1101/2023.09.13.23295508. [DOI]
- 19. Chen TC, Kaminski E, Koduri L, et al. Chat GPT as a neuro‐score calculator: analysis of a large language model's performance on various neurological exam grading scales. World Neurosurg 2023; 179: e342–e347. [DOI] [PubMed] [Google Scholar]
- 20. Koga S, Martin NB, Dickson DW. Evaluating the performance of large language models: ChatGPT and Google Bard in generating differential diagnoses in clinicopathological conferences of neurodegenerative disorders. Brain Pathol 2024; 34: e13207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nazario‐Johnson L, Zaki HA, Tung GA. Use of large language models to predict neuroimaging. J Am Coll Radiol 2023; 20: 1004–1009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Results of literature review
Table S2. Prompts
Table S3. Summary of astrocytoma case results
Table S4. Summary of glioblastoma case results
Table S5. Summary of oligodendroglioma case results
Full responses by case
Data Availability Statement
The UCLH dataset was acquired through Brain UK (REF: 22/011). Inquiries regarding access to this dataset should be directed to SB. Llama3‐70b‐groq is freely accessible via poe.com, whereas ChatGPT‐4o requires a paid subscription. Anthropic offers limited free access to Claude, but full access requires a paid subscription.


