Abstract
The human microbiome, consisting of microorganisms that coexist symbiotically with the body, impacts health from birth. Alterations in gut microbiota driven by factors such as diet and medication can contribute to diseases beyond the gut. Synthetic biology has paved the way for engineered microbial therapeutics, presenting promising treatments for a variety of conditions. Using genetically encoded biosensors and dynamic regulatory tools, engineered microbes can produce and deliver therapeutic agents, detect biomarkers, and manage diseases. This review organizes engineered microbial therapeutics by disease type, emphasizing innovative strategies and recent advancements. The scope of diseases includes gastrointestinal disorders, cancers, metabolic diseases, infections, and other ailments. Synthetic biology facilitates precise targeting and regulation, improving the efficacy and safety of these therapies. With promising results in animal models, engineered microbial therapeutics provide a novel alternative to traditional treatments, heralding a transformative era in diagnostics and treatment for numerous diseases.
Keywords: Engineered microbial therapeutics, synthetic biology, GI disease, tumor, metabolic disease, infection
Introduction
The human microbiome, a community of microorganisms coexisting symbiotically with the human body, has been present since birth [1, 2]. Human microbiota colonizes all body sites, with the gastrointestinal (GI) tract harboring the highest microbial population [3]. Recently, it has been revealed that modifications to the microbial community within the gut caused by various factors such as diet, drugs, and environmental signals can lead to diseases in not only the intestine but also various organs [4]. Consequently, there is a diversification of efforts aimed at directly or indirectly restoring or maintaining a healthy gut microbiome to regulate human health [5].
With advancements in synthetic biology, the creation of microbes performing desired functions through the combination of bio-parts has become feasible. Particularly, research on engineered microbial therapeutics using probiotics or dominant strains as chassis is gaining attention as next-generation treatments [6-9]. Bacteria-based therapies offer advantages such as self-replication, the potential for diagnostic functions via genetic circuits, the ability for on-site production and delivery of therapeutic agents, omitting costly downstream processes, and reducing side effects [8]. Research in this area not only involves conferring roles as delivery vehicles for therapeutics or vaccines [10] but also encompasses more complex functions. These functions include detecting biomarkers of inflammation, disease, and pathogens based on genetically encoded biosensors, diagnosing [7, 9], and recording using memory devices [11, 12]. Additionally, to ensure the stability of these therapeutic agents, physical or biological containment (biocontainment) technologies are being developed to prevent exposure to bacteria after and during action [13, 14].
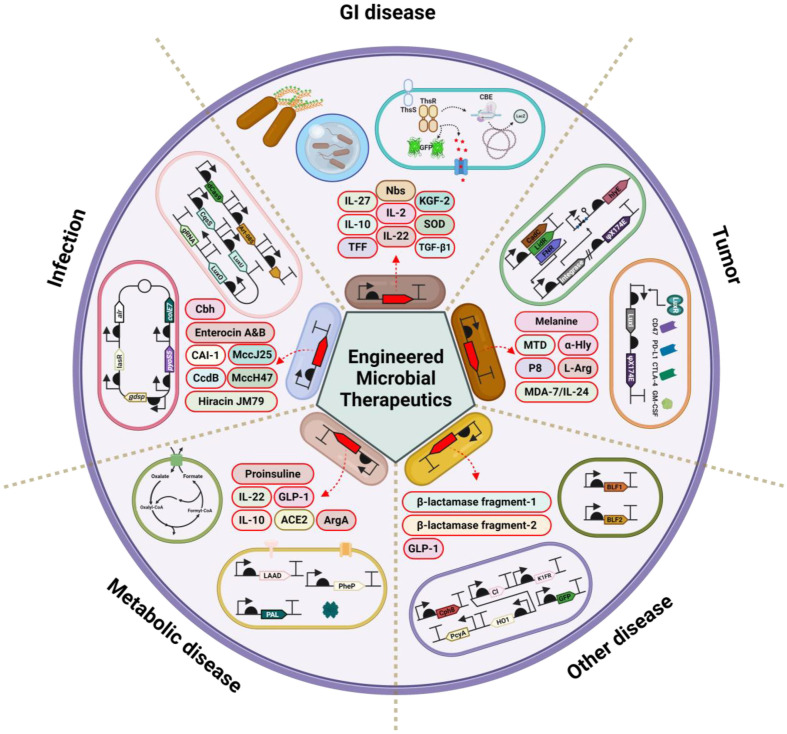
In this review, engineered microbial therapeutics based on synthetic biology are classified according to disease (Fig. 1). Furthermore, each study is summarized and introduced regarding the strategies used including bio-parts, actuators and genetic circuits (Tables 1-5). Five representative categories are identified as actively researched areas up to the present: GI tract disease, tumors, metabolic disease, infectious disease, and others. This information will provide resources for researchers involved in engineered microbial therapeutics.
Fig. 1. Schematic diagram of the utilization for engineered microbial therapeutics.
Engineered microbial therapeutics, developed through synthetic biology, are applied to various conditions including gastrointestinal disease, tumor, pathogen infection, metabolic disease, and others. These applications utilize strategies that include not only simple production and delivery of therapeutic substances but also sophisticated regulatory mechanisms using genetic circuits for precise diagnosis, monitoring, and treatment.
Table 1.
Reports on engineered bacterial therapeutics for GI disease.
| Chassis | Target disease | Genetically encoded biosensor | Actuator | Model | Ref. |
|---|---|---|---|---|---|
| E. coli Nissle 1917 | IBD | - | Encapsulated microbe | Sprague-Dawley rat | [14] |
| E. coli Nissle 1917 | Colitis | - | Curli-fused trefoil factor | C57BL/6NCrl mice | [18] |
| E. coli Nissle 1917 | Intestinal ulcerative disease | - | Epidermal growth factor | C57BL/6 mice | [19] |
|
E. coli Nissle 1917 ΔthyA, alr |
IBD | - | Type3 secretion apparatus TNF-α neutralizing nanobody |
C57BL/6 mice BALB/c mice |
[20] |
|
Lactococcus lactis ΔthyA |
IBD | - | Interleukin-10 | Piétrain/Landrace crossbred pigs | [13] |
| L. lactis | Colitis | - | Interleukin-27 | C57BL/6 mice | [24] |
| Lactobacillus reuteri | Intestinal disease (GvHD, IBD) | - | Interleukin-22 | - | [25] |
| Bacteroides ovatus | Chronic gut disorder | - | Interleukin-2 | - | [27] |
| B. ovatus | Chronic gut disorder | - | Trefoil factor | - | [28] |
| B. ovatus | Colitis | - | Transforming growth factor-β1 | C57BL/6 mice | [29] |
| B. ovatus | Colitis | - | Keratinocyte growth factor-2 | C57BL/6 mice | [30] |
| Bifidobacterium longum | Colitis | - | PEP-1 fused manganese superoxide dismutase | Sprague-Dawley rat | [32] |
| E. coli Nissle 1917 | IBD | Thiosulfate-inducible sensor for the sensing of biomarker and regulation of genes expression | Cytosine base editor sgRNA sfGFP Hly fused AvCystatin |
C57BL/6J mice | [12] |
Table 2.
Reports on engineered bacterial therapeutics for tumor.
| Chassis | Target disease | Genetically encoded biosensor | Actuator | Model | Refs |
|---|---|---|---|---|---|
| E. coli χ6212 Δasd | Tumor | - | α-hemolysin | BALB/c mice | [36] |
| E. coli MG1655 | Tumor | - | Type 3 secretion system Mitochondrial targeting domain of Noxa |
BALB/c mice | [37] |
| E. coli Nissle 1917 ΔdapA, thyA | Tumor | - | Cyclic di-AMP | C57BL/6 mice BALB/c mice |
[38] |
| E. coli Nissle 1917 ΔargR, malEK::argAfbr | Tumor | - | L-Arginine | C57BL/6 mice | [39] |
| E. coli Nissle 1917 | Tumor | - | Melanine | BALB/c mice | [40] |
| Pediococcus pentosaceus SL4 Δalr | Colorectal cancer | - | P8 | BALB/c mice C57BL/6J mice |
[42] |
| E. coli Nissle 1917 | Colorectal cancer | Lactate, pH, Hypoxia-inducible sensor for the expression of serine integrase | Hemolysin | BALB/c mice C57BL/6J mice |
[43] |
| E. coli Nissle 1917 | Colorectal cancer | - | INP-tagged histone-like protein A YebF-fused I1 myrosinase |
BALB/c mice | [44] |
| Bifidobacterium breve | Head and neck squamous cell carcinoma | - | MDA-7/IL-24 | BALB/c mice | [49] |
| E. coli Pir1+ | Tumor | AHL-inducible sensor for the production of transcription factor and lysis protein | Phage-lysis protein (ϕX174E) HA-tagged CD47 antagonistic nanobody |
BALB/c mice C57BL/6 mice |
[50] |
| E. coli Nissle 1917 | Tumor | Phage-lysis protein (ϕX174E) HA-tagged PD-L1 blocking nanobody HA-tagged CTLA4 blocking nanobody Granulocyte-macrophage colony-stimulation factor |
BALB/c mice | [51] | |
| E. coli Nissle 1917 ΔclbA | Colorectal neoplasia | C57BL/6 mice | [52] |
Table 3.
Reports on engineered bacterial therapeutics for metabolic disorder.
| Chassis | Target disease | Genetically encoded biosensor | Actuator | Model | Refs |
|---|---|---|---|---|---|
| L. lactis | Obesity | - | GLP-1 | C57BL/6 mice | [54] |
| L. reuteri | Nonalcoholic fatty liver disease | - | Interleukin-22 | C57BL/6J mice | [60] |
| L. reuteri | Alcoholic liver disease | - | Interleukin-22 | C57BL/6 mice | [61] |
| L. lactis | Diabetes | - | Proinsulin Interleukin-10 |
NOD mice | [62] |
| Lactobacillus paracasei | Diabetic retinopathy | - | Angiotensin converting enzyme 2 | C57BL/6J mice | [63] |
| E. coli Nissle 1917 ΔthyA | Enteric hyperoxaluria | - | Oxalate/formate antiporter Oxalyl-CoA decarboxylase Formyl-CoA trasferase |
C57BL/6J mice Cynomolgus monkey |
[65] |
| E. coli Nissle 1917 ΔthyA, argR | Hepatic encephalopathy | - | N-acetylglutamate synthase | C57BL/6 mice Cynomolgus monkey Human (healthy) |
[66] |
| E. coli Nissle 1917 ΔdapA | Phenylketonuria | - | Phenylalanine transporter Phenylalanine ammonia lyase L-amino acid deaminase |
C57BL/6 mice Cynomolgus monkey |
[67] |
| Trans-cinnamate-inducible sensor for the screening of phenylalanine ammonia lyase activity | Phenylalanine transporter Phenylalanine ammonia lyase mutant L-amino acid deaminase |
Cynomolgus monkey | [68] | ||
| Small molecule-inducible sensor array for the regulation of enzyme expression | - | [69] | |||
Table 4.
Reports on engineered bacterial therapeutics for infection.
| Chassis | Pathogen | Genetically encoded biosensor | Actuator | Model | Refs |
|---|---|---|---|---|---|
| E. coli TOP10 | Pseudomonas aeruginosa | 3OC12HSL-inducible sensor for the production of cell lysis enzyme and bacteriocin | Pyocin S5 E7 lysis protein |
- | [72] |
| E. coli Nissle 1917 Δalr, dadX | 3OC12HSL-inducible sensor for the production of cell lysis enzyme, bacteriocin, and anti-biofilm enzyme | Pyocin S5 E7 lysis protein Dispersin B |
Caenorhabditis elegans ICR mice |
[74] | |
| E. coli UU2685 | 3OC12HSL-inducible sensor for the activation of motility and killing modules | Microcin S DNaseI CheZ fused degron |
- | [73] | |
| E. coli Nissle 1917 | Vibrio cholera | - | Cholera autoinducer 1 | CD-1 mice | [77] |
| E. coli XL2 Blue | - | Type II gyrase inhibiting toxin | - | [78] | |
| E. coli MG1655 | Cholera autoinducer 1-inducible sensor for expression of lysis enzyme | Artilysin YebF fused Artilysin |
Zebrafish larvae Crustacean larvae |
[79] | |
| E. coli Nissle 1917 | Salmonella enterica | - | Microcin J25 | Turkey | [82] |
| E. coli Nissle 1917 | Tetrathionate-inducible sensor for the production of antimicrobial peptide and competition of resource | Microcin H47 | - | [83] | |
| E. coli Nissle 1917 Δalr, dadX | Clostiridium difficile | Sialic acid-inducible sensor for diagnosis of antibiotic-induced dysbiosis | Bile salt hydrolase | C57BL/6 mice | [85] |
| E. coli Nissle 1917 |
Enterococcus faecium Enterococcus faecalis |
- | Enterocin A Enterocin B Hiracin JM79 |
Balb/cJ mice | [86] |
| E. coli NGF-1 | Candida albicans | Hydroxyphenylacetic acid-inducible sensor for the sensing of fungus | Cis-2-dodecenoic acid | - | [87] |
Table 5.
Reports on engineered bacterial therapeutics for other diseases.
| Chassis | Target disease | Genetically encoded biosensor | Actuator | Model | Refs |
|---|---|---|---|---|---|
| L. lactis | Parkinson’s disease | - | GLP-1 | C57BL/6 mice | [91] |
| E. coli Nissle 1917 | Parkinson’s disease | Red-light inducible chimeric light sensor for production of cell lysis enzyme | Exendin-4 fused anti-neonatal FC receptor affibody | C57BL/6 mice | [92] |
| L. lactis | Antibiotic-induced dysbiosis | - | SpyTag fused β-lactamase fragment 1 SpyCatcher fused β-lactamase fragment 2 |
C57BL/6 mice | [93] |
Gastrointestinal Disease
Synthetic biology strategies for treating GI diseases have become increasingly diverse (Table 1). The GI tract is the primary focus for the application of engineered microbial therapeutics developed through synthetic biology, due to the feasibility of localized treatment. The prevalence of GI diseases is increasing, affecting nearly 2 million individuals in North America and Europe with chronic inflammatory disorders such as inflammatory bowel disease (IBD), which includes ulcerative colitis and Crohn’s disease [15-17].
Encapsulation and Biocontainment
One notable approach involves the physical encapsulation of Escherichia coli Nissle 1917 (EcN), a potential therapeutic agent for intestinal diseases, within a chitosan-alginate matrix using layer-by-layer assembly and CaCl2 cross-linking. This method aims to ensure biocontainment and enhance survivability in the harsh environment of oral administration [14].
Genetic Modifications for Enhanced Therapeutic Action
Several strategies involve direct modifications to EcN. For example, therapeutic curli hybrids have been designed by expressing trefoil factors fused to curli-forming proteins, promoting mucosal healing through matrix-tethered therapeutic domains [18]. Another approach targets ulcerative diseases by delivering epidermal growth factor using EcN, facilitating cell migration and extracellular matrix formation for intestinal damage repair [19]. Moreover, strategies to inhibit tumor necrosis factor-α (TNF-α) crucial target in IBD treatment - have been explored, including the use of nanobodies to neutralize TNF-α, with EcN’s type III secretion system enhancing nanobody delivery due to its gram-negative nature [20].
Lactic Acid Bacteria (LAB) as Therapeutic Chassis
In addition to EcN, various strategies employ LAB as therapeutic chassis. These strategies primarily focus on protein secretion-based approaches, influencing the immune system directly or indirectly. Many target therapeutic molecules are interleukin (IL) family members, which have shown therapeutic efficacy in conditions like colitis [21-23]. For example, one study utilized Lactococcus lactis as a chassis to secrete IL-10 while knocking out the essential gene thyA to achieve auxotrophic biocontainment [13]. Other studies have enhanced wild-type L. lactis to secrete IL-27 to treat immune colitis [24], and genetically modified Lactobacillus reuteri to secrete IL-22 for conditions such as IBD and Graft versus Host Disease (GvHD) [25]. Furthermore, engineering efforts have increased IL-22 secretion by replacing the 2nd and 17th proline residues with glycine to counteract high proteolytic activity in Lactobacillus, preventing cleavage and enhancing IL-22 expression [26].
Commensal Bacteria as Therapeutic Tools
Another research group utilized Bacteroides ovatus, a predominant gut commensal, as a chassis. They first engineered a system employing a xylan-inducible gene expression mechanism from the xylanase operon and a secretion signal sequence from the enterotoxin of Bacteroides fragilis to produce IL-2 [27]. They then modified this system to secrete trefoil factor-3 for validation purposes [28]. Further engineering enabled the secretion of transforming growth factor-β1 to evaluate therapeutic effects in a DSS-induced mouse model [29]. This was followed using the system to secrete keratinocyte growth factor 2, demonstrating its versatility [30]. The recent study showed improved secretion of heterologous therapeutic proteins in engineered Bacteroides species, particularly Bacteroides thetaiotaomicron, which is a significant component of the human gut microbiome [31]. The researchers identified novel signal peptides from B. thetaiotaomicron and Akkermansia muciniphila that enhance protein transport across cellular membranes. Additionally, they developed an episomal plasmid system that outperforms traditional chromosomal integration plasmids in protein secretion efficiency [31]. This plasmid system incorporates an essential gene (thyA)-based selection method to maintain plasmid stability without antibiotics, which is crucial for clinical applications. The study demonstrates the applicability of these advancements across multiple Bacteroides species, setting a new standard for developing live biotherapeutics aimed at treating gut diseases. Additionally, Bifidobacterium longum, known for its probiotic properties, was used as a chassis. In this study, a fusion protein of PEP-1 and manganese superoxide dismutase was expressed. This fusion protein was designed to convert reactive oxygen species, which increase during colitis onset, thus reducing intestinal inflammation [32].
Intelligent Whole-cell Systems
In addition, there have been reports on systems designed not only for inflammation treatment but also for detecting inflammation biomarkers and releasing drugs only when these biomarkers are detected. In one such study, a genetic circuit was engineered into EcN to regulate the expression of several components: the expression of superfolder green fluorescent protein (sfGFP) for real-time signal confirmation through a bacterial two-component system capable of detecting thiosulfate as the selected biomarker, the induction of cytosine base editor along with sgRNA for inducing inheritable signals through base editing, and the expression of AvCystatin, an immunomodulatory protein. This intelligent whole-cell engineered bacteria system was designed to alleviate inflammation, demonstrating its potential effectiveness in inflammation relief [12]. A whole-cell biosensor using engineered EcN was reported to diagnose gut inflammation by sensing nitrate levels, a biomarker of inflammation [33]. The study employed the NarX-NarL two-component regulatory system to create a nitrate-responsive genetic circuit. This biosensor was optimized for sensitivity and specificity and successfully detected elevated nitrate levels in a mouse model of colitis. Additionally, they introduced a Boolean AND logic gate combining nitrate and thiosulfate sensing, enhancing the biosensor's specificity for gut inflammation [33].
Chronic Inflammation Management
Chronic inflammation of the GI tract, the focus of these studies, is marked by recurrent cycles of symptom exacerbation and remission, with a multifactorial etiology. The complexity of its onset and progression often makes specific therapeutic interventions challenging, leading to the frequent use of systemic immunosuppressants, which carry significant risks of adverse effects [34]. The described research efforts continue to propose effective, less risky treatment modalities for managing the complexities of GI tract inflammation.
Tumor
The application of engineered microbial therapeutics, enhanced through synthetic biology, is increasingly prominent in cancer treatment. Bacteria-based therapies for tumors offer several advantages, including their ability to colonize tumors and deliver therapeutic molecules directly to the site. This capability facilitates the direct killing of tumor cells and the induction of antitumor immune responses [35]. Several therapeutic strategies have emerged based on these advantages (Table 2).
Facultative Anaerobes for Selective Colonization
Certain facultative anaerobes, known for their ability to selectively colonize tumors, have been utilized. One study modified E. coli χ6212 into an aspartate auxotroph by knocking out aspartate semialdehyde dehydrogenase (asd), allowing it to deliver pore-forming proteins from S. aureus to treat tumors [36]. Another strategy employed E. coli MG1655 as a chassis to secrete anti-tumor proteins via the type III secretion system from Salmonella [37].
Immune System Modulation
EcN was engineered to produce cyclic di-AMP, a stimulator of interferon genes agonist, to induce type 1 interferon production and enhance anticancer effects [38]. This research also included strategies involving the knockout of essential genes for biocontainment and plasmid selection [38]. Indirect therapeutic enhancements have also been explored. For instance, EcN was engineered to overproduce L-arginine, which enhances the effectiveness of programmed death-ligand 1 (PD-L1) blocking antibodies, key immune checkpoint inhibitors [39]. Another approach used EcN to produce melanin, enhancing tumor photothermal and immunotherapy, and delivered via outer membrane vesicles coated with calcium phosphate for increased delivery efficiency [40].
Targeted Tumor Recognition
Unique strategies for specific cancer types have been reported as well. For colorectal cancer (CRC), Pediococcus pentosaceus SL4 was engineered to produce P8, a protein from Lactobacillus rhamnosus CBT LR5, effectively reducing tumor volume and inhibiting growth [41, 42]. EcN was also designed to recognize the tumor microenvironment (TME) by incorporating sensors for lactate, pH, and hypoxia, leading to the secretion of hemolysin based on TME recognition [43]. Furthermore, a synthetic consortium of three bacterial strains was applied to treat CRC, reducing metabolic load and enhancing treatment efficacy [43].
Enzyme Expression for Anticancer Compounds
Enzyme expression for converting dietary substances into therapeutic agents has also been investigated. In one study, EcN was engineered to express histone-like protein A on its surface to bind specifically to CRC cells and secrete an enzyme that converts glucosinolate from cruciferous plants into the anticancer compound sulforaphane near the tumor site [44]. For head and neck squamous cell carcinoma, Bifidobacterium breve was engineered to secrete melanoma differentiation-associated gene-7/interleukin-24 (MDA-7/IL-24), which induces apoptosis in various cancer cells [45-49].
Innovative Bacterial Systems
Acyl homoserine lactone (AHL)-inducible sensors have been used to induce the production of therapeutic agents and cell lysis, facilitating therapeutic delivery. This system was initially used to design a strategy with CD47 antagonistic nanobodies, enhancing anti-tumor T cell priming by promoting phagocytosis of cancer cells and cross-presentation of tumor antigens [50]. Subsequently, therapies incorporating PD-L1 and CTLA-4 blocking nanobodies with granulocyte macrophage colony-stimulating factor production were developed [51]. This system was also applied to colorectal neoplasia, demonstrating therapeutic effects upon oral administration of engineered bacteria [52].
Challenges and Future Directions
Bacterial cancer therapies represent a significant segment of the therapeutic market utilizing engineered microbes. Ongoing research continues to refine the strengths and address the weaknesses of commonly used bacterial strains, expand therapeutic strategies, and improve methods for delivering treatments [35].
Metabolic Disease
Numerous studies have explored the treatment of metabolic diseases using engineered microbial therapeutics (Table 3). Metabolic diseases are conditions that disrupt the metabolic process, such as the conversion of food into energy at the cellular level. Diagnostic markers for these diseases involve the cellular ability to perform key biochemical reactions related to proteins (amino acids), carbohydrates (sugars and starches), or lipids (fatty acids)[53].
Obesity
Obesity, a significant factor in metabolic diseases, often results from the excessive accumulation of triglycerides in adipose tissue [54]. Gut microbial diversity and composition also play a crucial role [55]. In this context, one strategy for treating obesity involves engineered L. lactis strains that secrete glucagon-like peptide-1 (GLP-1), a hormone that stimulates insulin secretion in a glucose-dependent manner. This approach enhances fatty acid oxidation, reduces blood triglyceride levels, and activates peroxisome proliferator-activated receptors α (PPARα) to improve liver tissue, thus alleviating obesity [54].
Liver Disease
The liver is vital in lipid and glucose metabolism in obese individuals. Liver diseases, which result from abnormalities in these metabolic processes, are categorized based on alcohol influence. Non-alcoholic fatty liver disease occurs when fat comprises more than 5% of liver weight due to non-alcoholic factors [56], while alcoholic liver disease results from excessive ethanol intake, leading to hepatic steatosis, alcoholic steatohepatitis, and hepatocellular carcinoma [57]. IL-20 family cytokines, particularly IL-22, have shown potential in reducing liver injury through the activation of the STAT3 signaling pathway [58, 59]. Engineered L. reuteri strains secreting IL-22 have demonstrated therapeutic effects in both alcoholic and non-alcoholic fatty liver diseases [60, 61].
Diabetes
Engineered microbes have also been utilized to treat diabetes, a prevalent metabolic disease. For autoimmune diabetes, L. lactis has been engineered to secrete human proinsulin and IL-10, which, when administered with low-dose anti-CD3 antibodies, reversed hyperglycemia and reduced insulin autoantibody levels, effectively reversing diabetes [62]. Additionally, in treating diabetic retinopathy, engineered Lactobacillus paracasei secreting angiotensin-converting enzyme 2 reduced inflammation and oxidative stress within the renin-angiotensin system, improving diabetic retinopathy outcomes [63].
Addressing Genetic Metabolic Disorders
Therapeutic interventions have also targeted metabolic diseases caused by the loss of genes encoding metabolic enzymes. For example, enteric hyperoxaluria (EH) results from the accumulation and absorption of dietary oxalate in the intestines [64]. A strategy to address EH involves engineering EcN to express oxalate transporters and enzymes that convert oxalate into fumarate, reducing intestinal oxalate levels [65]. Similarly, hyperammonemia, a condition causing hepatic encephalopathy due to ammonia accumulation, has been addressed by altering the inhibitory feedback mechanism in EcN. By knocking out the repressor ArgR and expressing a feedback-resistant version of N-acetylglutamate synthase, this modification demonstrated therapeutic efficacy in suppressing hyperammonemia [66]. For phenylketonuria, characterized by neurotoxicity due to phenylalanine accumulation, engineered EcN was used to convert phenylalanine into non-toxic trans-cinnamate (TCA) while also expressing membrane-anchored L-amino acid deaminase to convert phenylalanine into phenyl pyruvate extracellularly [67]. Subsequent improvements involved selecting a more efficient phenylalanine ammonia lyase mutant using a TCA biosensor, resulting in a microbial therapy twice as effective as the previous version [68]. Further enhancements included creating genomic landing pads on EcN chromosomes for gene insertion, utilizing transcription factor-based sensors with small molecule ligands to improve phenylalanine conversion by 50% compared to existing methods [69]. Continuous advancements in using engineered microbes to express enzymes or substances that activate desired metabolic pathways highlight the potential for treating metabolic disorders.
Infection
Infectious diseases remain a leading cause of mortality in clinical settings [70]. While antibiotic administration is a common treatment, it can eliminate beneficial microbes and promote antibiotic-resistant bacteria. Consequently, research on engineered microbes for treating infections is actively progressing (Table 4).
Pseudomonas aeruginosa
To combat P. aeruginosa infections in the GI tract [71], researchers have explored two main strategies across three studies. The first strategy utilized sensing, lysing, and killing devices. Two studies employed P. aeruginosa's type I quorum sensing mechanism to detect 3OC12HSL using a transcription factor-based sensor. Upon detection, these sensor cells expressed enzymes that caused self-lysis and released bacteriocins, resulting in the pathogen's destruction [72, 73]. The second strategy used the same biosensor but with a different output: secretion of DNaseI to break down biofilms, attraction of the pathogen through chemotaxis, and secretion of bacteriocins to kill the pathogen [74].
Vibrio cholerae
For V. cholerae, which causes 21,000 to 143,000 deaths annually [75], treatment strategies vary based on the use of biosensors. Without biosensors, one approach involved engineering EcN to express both CAI-1 and AI-2 to suppress virulence gene expression in V. cholerae [76, 77]. Another strategy targeted antibiotic-resistant V. cholerae by using a type II bacterial toxin-antitoxin system as an actuator, which selectively killed the pathogen upon horizontal gene transfer of the plasmid [78]. With biosensors, a sensing, lysing, and killing device was developed. This device used a CAI-1 sensor to trigger cell lysis and release lysing enzymes to kill the pathogen [79].
Salmonella enterica
To combat S. enterica, which causes significant foodborne illness [80, 81], strategies without biosensors involved engineering EcN strains to secrete antimicrobial peptides, achieving a 25-fold higher clearance rate in turkeys compared to controls [82]. Another approach used a tetrathionate sensor to regulate antimicrobial peptide expression, providing both bactericidal effects and resource competition [83].
Clostridium difficile
For C. difficile, which thrives due to gut dysbiosis, researchers engineered a sensor for sialic acid, which increases during dysbiosis [84]. This sensor regulated enzymes converting taurocholate, a C. difficile germination inducer, into cholate, thus preventing infection by modulating germination [85].
Enterococci and Candida albicans
Additionally, strategies to inhibit Enterococci growth, which causes nosocomial infections and shows antibiotic resistance, involved using three antimicrobial peptides simultaneously [86]. Research targeting C. albicans, a fungus causing opportunistic infections, focused on identifying and detecting secreted substances using biosensors to inhibit hypha formation, a virulence factor [87].
Ongoing Research and Future Directions
Various therapeutic strategies are being explored to replace antibiotics with engineered bacterial therapeutics, aiming to combat infections caused by bacteria and fungi more safely and effectively.
Other Diseases
In addition to GI diseases, tumor, and metabolic diseases, synthetic biology and engineered microbial therapeutics offer promising solutions for a range of other diseases. These approaches leverage the precision of genetic circuits and the versatility of microbial systems to address complex health challenges (Table 5).
Neurodegenerative Diseases
One significant area of application is neurodegenerative diseases, particularly through the regulation of the gut-brain axis. Two notable studies focus on treating Parkinson’s disease (PD), characterized by the loss of dopaminergic neurons in the brain. Both studies utilized GLP-1 and its analog Exendin-4 as therapeutic agents due to their known beneficial effects on PD [88]. GLP-1 can cross the blood-brain barrier, protect neurons from oxidative stress-induced apoptosis, and promote neuronal proliferation, making it a potential treatment [89, 90]. In one study, L. lactis was engineered to continuously express and secrete GLP-1 [91]. Another study, engineered a strain to constitutively express Exendin-4 and incorporated a red-light inducible chimeric light sensor to regulate the expression of a cell lysis enzyme, allowing controlled drug release through red-light stimulation with high skin penetrability [92]. These studies highlight the potential for treating diseases in organs beyond the gut through diverse therapeutic production and delivery methods.
Antibiotic-induced Dysbiosis
Another area of research targets antibiotic-induced dysbiosis. One study focused on β-lactamase and employed L. lactis as a chassis, splitting β-lactamase into two fragments fused with SpyTag/Catcher for extracellular secretion [93]. Unlike β-lactamase in Gram-negative bacteria, which is located in the periplasm and confers antibiotic resistance to single cells [94], this study used Gram-positive bacteria, making survival dependent on cell density. When the engineered microbes and antibiotics were administered, the gut microbiota exhibited less disruption compared to the control group, demonstrating an inhibitory effect on pathogenic strains such as C. difficile [93]. This research exemplifies the broader therapeutic applications of engineered microbes.
Indeed, synthetic biology-driven engineered bacterial therapeutics hold significant potential for targeting various organs and disease targets, contingent upon the identification and combination of appropriate bio-parts as demonstrated in these studies.
Challenges and Future Directions
While engineered microbial therapeutics hold significant promise, several challenges must be addressed to bring these treatments from the lab to the clinic. For GI diseases, one major hurdle is ensuring the stability and survivability of therapeutic microbes in the harsh conditions of the GI tract. Approaches like encapsulation [14] and biocontainment [13, 95, 96] are being developed to overcome these challenges, but further research is needed to ensure consistent efficacy in human trials. In the case of cancers, the primary challenge lies in the selective targeting of tumors [44] and minimizing off-target effects [44, 97]. Strategies such as engineering microbes to respond to tumor-specific biomarkers are promising but require rigorous validation. Metabolic diseases present a unique challenge in achieving sustained therapeutic effects without disrupting the body's metabolic balance [98]. Finally, for infectious diseases, the main obstacle is developing therapies that are potent enough to eradicate pathogens without promoting antibiotic resistance or disrupting the native microbiota [73].
To gain approval as new drugs, engineered microbial therapeutics must overcome significant regulatory hurdles. These include demonstrating safety and efficacy through preclinical studies and clinical trials. Additionally, regulatory agencies require comprehensive data on the genetic stability and biocontainment of these organisms to prevent unintended environmental release [95]. Establishing standardized protocols for the production and quality control of these biotherapeutics is also crucial. Collaborative efforts between researchers, regulatory bodies, and industry stakeholders will be essential to streamline this process. To create safe and effective engineered microbial therapeutics, several standards and genetic methods should be applied. These include genetic stability, rigorous testing, standardization, and regulatory compliance. To ensure the safety and efficacy of engineered microbial therapeutics, several critical measures must be taken. First, incorporating genetic safeguards such as kill switches and biocontainment mechanisms is essential to prevent horizontal gene transfer and environmental persistence [14, 92, 99, 100], thereby ensuring genetic stability. Rigorous testing, including extensive in vitro and in vivo studies, is necessary to evaluate the long-term safety and therapeutic efficacy of these microbes. Developing standardized protocols for genetic modifications and therapeutic production is crucial to maintain consistency across different batches and studies. Finally, adherence to regulatory guidelines set by agencies such as the FDA and EMA is imperative. This includes submitting detailed data on genetic constructs, production methods, and safety evaluations to comply with regulatory requirements. By implementing these measures, the development of safe and effective microbial therapeutics can be achieved.
The use of live genetically modified microorganisms presents several challenges, including environmental exposure, phenotypic changes, and non-specific mutations. To mitigate these issues, several strategies must be employed. First, physical and genetic biocontainment strategies are crucial to prevent the release and spread of modified organisms in the environment [96]. Ensuring the phenotypic stability of therapeutic strains through rigorous genetic and phenotypic screening is essential. Advanced gene editing techniques should be used to minimize off-target effects and non-specific mutations. Additionally, designing therapeutics that do not rely on antibiotic resistance markers and developing alternative selection methods can help manage antibiotic resistance [13, 31]. Finally, conducting detailed pharmacokinetic studies is important to understand the distribution, persistence, and clearance of these therapeutics in the human body. By addressing these challenges, the safe and effective use of live genetically modified microorganisms can be achieved.
Conclusion
Looking ahead, several key areas should be prioritized to further advance the field of synthetic biology and engineered microbial therapeutics. First, there is a need for continued development and refinement of genetic circuits that can provide more precise and robust control over microbial functions. This includes the creation of more sophisticated biosensors and actuators that can respond to a wider range of biological signals and environmental conditions. Additionally, enhancing the stability and safety of these engineered microbes is crucial, particularly through the implementation of biocontainment strategies that prevent unintended spread and ensure that therapeutic functions are restricted to the target site. Another important direction is the integration of engineered microbial therapeutics with other treatment modalities, such as conventional drugs, immunotherapies, and personalized medicine approaches. Combining these therapies can enhance their effectiveness and reduce the likelihood of resistance. For example, engineered bacteria that deliver checkpoint inhibitors directly to tumor sites could complement systemic immunotherapies, providing a more targeted and potent anti-cancer strategy. Moreover, advancements in genome editing technologies, such as CRISPR-Cas systems, can facilitate the development of more precise and efficient microbial therapeutics, enabling the correction of genetic disorders and the modulation of complex metabolic pathways [101, 102]. Finally, rigorous clinical evaluation and regulatory frameworks are essential to translate the promising research findings into safe and effective therapies for patients. This includes conducting well-designed clinical trials to assess the efficacy, safety, and long-term effects of engineered microbial therapeutics in diverse patient populations. Collaboration between researchers, clinicians, regulatory agencies, and industry stakeholders will be critical to overcoming the challenges and accelerating the adoption of these innovative therapies in clinical practice. In conclusion, advancements in synthetic biology and engineered microbial therapeutics hold great promise for revolutionizing the treatment of a wide array of diseases. By continuing to innovate and address the remaining challenges, we can unlock the full potential of these cutting-edge therapies and improve the health and well-being of patients worldwide.
Acknowledgements
This work was supported by the Bio & Medical Technology Development Program [grant number 2018M3A9H3024746] of the National Research Foundation funded by the Ministry of Science and ICT of the Republic of Korea and the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program (KGM5402423).
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
References
- 1.Korpela K, de Vos WM. Early life colonization of the human gut: microbes matter everywhere. Curr. Opin. Microbiol. 2018;44:70–78. doi: 10.1016/j.mib.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Korpela K, Helve O, Kolho KL, Saisto T, Skogberg K, Dikareva E, et al. Maternal fecal microbiota transplantation in cesareanborn infants rapidly restores normal gut microbial development: a proof-of-concept study. Cell. 2020;183:324–334.:e325. doi: 10.1016/j.cell.2020.08.047. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, et al. Strains, functions and dynamics in the expanded human microbiome project. Nature. 2017;550:61–66. doi: 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N. Engl. J. Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 5.Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021;19:55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 6.Selvakumar R, Kumar I, Onajobi GJ, Yu Y, Wilson CJ. Engineering living therapeutics and diagnostics: a new frontier in human health. Curr. Opin. Syst. Biol. 2024;37:100484. doi: 10.1016/j.coisb.2023.100484. [DOI] [Google Scholar]
- 7.Kim K, Kang M, Cho BK. Systems and synthetic biology-driven engineering of live bacterial therapeutics. Front. Bioeng. Biotechnol. 2023;11:1267378. doi: 10.3389/fbioe.2023.1267378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pedrolli DB, Ribeiro NV, Squizato PN, de Jesus VN, Cozetto DA Team AQAUai, author. Engineering microbial living therapeutics: the synthetic biology toolbox. Trends Biotechnol. 2019;37:100–115. doi: 10.1016/j.tibtech.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Zhao N, Song Y, Xie X, Zhu Z, Duan C, Nong C, et al. Synthetic biology-inspired cell engineering in diagnosis, treatment, and drug development. Signal. Transduct. Target. Ther. 2023;8:112. doi: 10.1038/s41392-023-01375-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexander LM, van Pijkeren JP. Modes of therapeutic delivery in synthetic microbiology. Trends Microbiol. 2023;31:197–211. doi: 10.1016/j.tim.2022.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mimee M, Tucker AC, Voigt CA, Lu TK. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst. 2015;1:62–71. doi: 10.1016/j.cels.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou ZP, Du Y, Fang TT, Zhou Y, Ye BC. Biomarker-responsive engineered probiotic diagnoses, records, and ameliorates inflammatory bowel disease in mice. Cell Host Microbe. 2023;31:199–212.:e195. doi: 10.1016/j.chom.2022.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Steidler L, Neirynck S, Huyghebaert N, Snoeck V, Vermeire A, Goddeeris B, et al. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat. Biotechnol. 2003;21:785–789. doi: 10.1038/nbt840. [DOI] [PubMed] [Google Scholar]
- 14.Luo X, Song H, Yang J, Han B, Feng Y, Leng Y, Chen Z. Encapsulation of Escherichia coli strain Nissle 1917 in a chitosanalginate matrix by combining layer-by-layer assembly with CaCl(2) cross-linking for an effective treatment of inflammatory bowel diseases. Colloids Surf. B Biointerfaces. 2020;189:110818. doi: 10.1016/j.colsurfb.2020.110818. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 2015;12:720–727. doi: 10.1038/nrgastro.2015.150. [DOI] [PubMed] [Google Scholar]
- 16.Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390:2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 17.Ng SC. Emerging trends of inflammatory bowel disease in Asia. Gastroenterol. Hepatol. 2016;12:193–196. [PMC free article] [PubMed] [Google Scholar]
- 18.Praveschotinunt P, Duraj-Thatte AM, Gelfat I, Bahl F, Chou DB, Joshi NS. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat Commun. 2019;10:5580. doi: 10.1038/s41467-019-13336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu M, Kim J, Ahn JH, Moon Y. Nononcogenic restoration of the intestinal barrier by E. coli-delivered human EGF. JCI Insight. 2019;4:e125166. doi: 10.1172/jci.insight.125166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch JP, Gonzalez-Prieto C, Reeves AZ, Bae S, Powale U, Godbole NP, et al. Engineered Escherichia coli for the in situ secretion of therapeutic nanobodies in the gut. Cell Host Microbe. 2023;31:634–649.:e638. doi: 10.1016/j.chom.2023.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, Falk W, et al. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289:1352–1355. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 22.Sasaoka T, Ito M, Yamashita J, Nakajima K, Tanaka I, Narita M, et al. Treatment with IL-27 attenuates experimental colitis through the suppression of the development of IL-17-producing T helper cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;300:G568–576. doi: 10.1152/ajpgi.00329.2010. [DOI] [PubMed] [Google Scholar]
- 23.Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29:947–957. doi: 10.1016/j.immuni.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanson ML, Hixon JA, Li W, Felber BK, Anver MR, Stewart CA, et al. Oral delivery of IL-27 recombinant bacteria attenuates immune colitis in mice. Gastroenterology. 2014;146:210–221.:e213. doi: 10.1053/j.gastro.2013.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortiz-Velez L, Goodwin A, Schaefer L, Britton RA. Challenges and pitfalls in the engineering of human lnterleukin 22 (hIL-22) secreting Lactobacillus reuteri. Front. Bioeng. Biotechnol. 2020;8:543. doi: 10.3389/fbioe.2020.00543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duar RM, Clark KJ, Patil PB, Hernandez C, Bruning S, Burkey TE, et al. Identification and characterization of intestinal lactobacilli strains capable of degrading immunotoxic peptides present in gluten. J. Appl. Microbiol. 2015;118:515–527. doi: 10.1111/jam.12687. [DOI] [PubMed] [Google Scholar]
- 27.Farrar MD, Whitehead TR, Lan J, Dilger P, Thorpe R, Holland KT, Carding SR. Engineering of the gut commensal bacterium Bacteroides ovatus to produce and secrete biologically active murine interleukin-2 in response to xylan. J. Appl. Microbiol. 2005;98:1191–1197. doi: 10.1111/j.1365-2672.2005.02565.x. [DOI] [PubMed] [Google Scholar]
- 28.Hamady ZZR, Farrar MD, Whitehead TR, Holland KT, Lodge JPA, Carding SR. Identification and use of the putative Bacteroides ovatus xylanase promoter for the inducible production of recombinant human proteins. Microbiology. 2008;154:3165–3174. doi: 10.1099/mic.0.2008/019109-0. [DOI] [PubMed] [Google Scholar]
- 29.Hamady ZZ, Scott N, Farrar MD, Wadhwa M, Dilger P, Whitehead TR, et al. Treatment of colitis with a commensal gut bacterium engineered to secrete human TGF-beta1 under the control of dietary xylan 1. Inflamm. Bowel Dis. 2011;17:1925–1935. doi: 10.1002/ibd.21565. [DOI] [PubMed] [Google Scholar]
- 30.Hamady ZZ, Scott N, Farrar MD, Lodge JP, Holland KT, Whitehead T, et al. Xylan-regulated delivery of human keratinocyte growth factor-2 to the inflamed colon by the human anaerobic commensal bacterium Bacteroides ovatus. Gut. 2010;59:461–469. doi: 10.1136/gut.2008.176131. [DOI] [PubMed] [Google Scholar]
- 31.Kim TH, Ju K, Kim SK, Woo S-G, Lee J-S, Lee C-H, et al. Novel signal peptides and episomal plasmid system for enhanced protein secretion in engineered Bacteroides species. ACS Synth. Biol. 2024;13:648–657. doi: 10.1021/acssynbio.3c00649. [DOI] [PubMed] [Google Scholar]
- 32.Liu M, Li S, Zhang Q, Xu Z, Wang J, Sun H. Oral engineered Bifidobacterium longum expressing rhMnSOD to suppress experimental colitis. Int. Immunopharmacol. 2018;57:25–32. doi: 10.1016/j.intimp.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Woo S-G, Moon S-J, Kim SK, Kim TH, Lim HS, Yeon G-H, et al. A designed whole-cell biosensor for live diagnosis of gut inflammation through nitrate sensing. Biosens. Bioelectron. 2020;168:112523. doi: 10.1016/j.bios.2020.112523. [DOI] [PubMed] [Google Scholar]
- 34.Sandborn WJ, Loftus EV. Balancing the risks and benefits of infliximab in the treatment of inflammatory bowel disease. Gut. 2004;53:780–782. doi: 10.1136/gut.2003.020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raman V, Deshpande CP, Khanduja S, Howell LM, Van Dessel N, Forbes NS. Build-a-bug workshop: using microbial-host interactions and synthetic biology tools to create cancer therapies. Cell Host Microbe. 2023;31:1574–1592. doi: 10.1016/j.chom.2023.09.006. [DOI] [PubMed] [Google Scholar]
- 36.St Jean AT, Swofford CA, Panteli JT, Brentzel ZJ, Forbes NS. Bacterial delivery of Staphylococcus aureus alpha-hemolysin causes regression and necrosis in murine tumors. Mol. Ther. 2014;22:1266–1274. doi: 10.1038/mt.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim D, Jung WC, Jeong JH, Song M. Targeted delivery of the mitochondrial target domain of noxa to tumor rissue via synthetic secretion system in E. coli. Front. Bioeng. Biotechnol. 2020;8:840. doi: 10.3389/fbioe.2020.00840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leventhal DS, Sokolovska A, Li N, Plescia C, Kolodziej SA, Gallant CW, et al. Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat. Commun. 2020;11:2739. doi: 10.1038/s41467-020-16602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canale FP, Basso C, Antonini G, Perotti M, Li N, Sokolovska A, et al. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature. 2021;598:662–666. doi: 10.1038/s41586-021-04003-2. [DOI] [PubMed] [Google Scholar]
- 40.Chen X, Li P, Luo B, Song C, Wu M, Yao Y, et al. Surface mineralization of engineered bacterial outer membrane vesicles to enhance tumor photothermal/immunotherapy. ACS Nano. 2024;18:1357–1370. doi: 10.1021/acsnano.3c05714. [DOI] [PubMed] [Google Scholar]
- 41.An BC, Ryu Y, Yoon YS, Choi O, Park HJ, Kim TY, et al. Colorectal cancer therapy using a Pediococcus pentosaceus SL4 drug delivery system secreting lactic acid bacteria-derived protein p8. Mol. Cells. 2019;42:755–762. doi: 10.14348/molcells.2019.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chung Y, Ryu Y, An BC, Yoon YS, Choi O, Kim TY, et al. A synthetic probiotic engineered for colorectal cancer therapy modulates gut microbiota. Microbiome. 2021;9:122. doi: 10.1186/s40168-021-01071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou T, Wu J, Tang H, Liu D, Jeon BH, Jin W, et al. Enhancing tumor-specific recognition of programmable synthetic bacterial consortium for precision therapy of colorectal cancer. NPJ Biofilms Microbiome. 2024;10:6. doi: 10.1038/s41522-024-00479-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho CL, Tan HQ, Chua KJ, Kang A, Lim KH, Ling KL, et al. Engineered commensal microbes for diet-mediated colorectalcancer chemoprevention. Nat. Biomed Eng. 2018;2:27–37. doi: 10.1038/s41551-017-0181-y. [DOI] [PubMed] [Google Scholar]
- 45.Gopalan B, Shanker M, Chada S, Ramesh R. MDA-7/IL-24 suppresses human ovarian carcinoma growth in vitro and in vivo. Mol. Cancer. 2007;6:11. doi: 10.1186/1476-4598-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhutia SK, Das SK, Kegelman TP, Azab B, Dash R, Su ZZ, et al. mda-7/IL-24 differentially regulates soluble and nuclear clusterin in prostate cancer. J. Cell. Physiol. 2012;227:1805–1813. doi: 10.1002/jcp.22904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhutia SK, Das SK, Azab B, Menezes ME, Dent P, Wang XY, et al. Targeting breast cancer-initiating/stem cells with melanoma differentiation-associated gene-7/interleukin-24. Int. J. Cancer. 2013;133:2726–2736. doi: 10.1002/ijc.28289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dash R, Bhoopathi P, Das SK, Sarkar S, Emdad L, Dasgupta S, et al. Novel mechanism of MDA-7/IL-24 cancer-specific apoptosis through SARI induction. Cancer Res. 2014;74:563–574. doi: 10.1158/0008-5472.CAN-13-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L, Vuletic I, Deng D, Crielaard W, Xie Z, Zhou K, et al. Bifidobacterium breve as a delivery vector of IL-24 gene therapy for head and neck squamous cell carcinoma in vivo. Gene Ther. 2017;24:699–705. doi: 10.1038/gt.2017.74. [DOI] [PubMed] [Google Scholar]
- 50.Chowdhury S, Castro S, Coker C, Hinchliffe TE, Arpaia N, Danino T. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat. Med. 2019;25:1057–1063. doi: 10.1038/s41591-019-0498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gurbatri CR, Lia I, Vincent R, Coker C, Castro S, Treuting PM, et al. Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci. Transl. Med. 2020;12:eaax0876. doi: 10.1126/scitranslmed.aax0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gurbatri CR, Radford GA, Vrbanac L, Im J, Thomas EM, Coker C, et al. Engineering tumor-colonizing E. coli Nissle 1917 for detection and treatment of colorectal neoplasia. Nat. Commun. 2024;15:646. doi: 10.1038/s41467-024-44776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grundy SM, Brewer HB, Jr., Cleeman JI, Smith SC, Jr., Lenfant C, American Heart A, et al. Definition of metabolic syndrome: report of the national heart, lung, and blood institute/American heart association conference on scientific issues related to definition. Circ. 2004;109:433–438. doi: 10.1161/01.CIR.0000111245.75752.C6. [DOI] [PubMed] [Google Scholar]
- 54.Wang L, Chen T, Wang H, Wu X, Cao Q, Wen K, et al. Engineered Bacteria of MG1363-pMG36e-GLP-1 attenuated obesityinduced by high fat diet in mice. Front. Cell. Infect. Microbiol. 2021;11:595575. doi: 10.3389/fcimb.2021.595575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torres-Fuentes C, Schellekens H, Dinan TG, Cryan JF. The microbiota-gut-brain axis in obesity. Lancet Gastroenterol. Hepatol. 2017;2:747–756. doi: 10.1016/S2468-1253(17)30147-4. [DOI] [PubMed] [Google Scholar]
- 56.European Association for the Study of the L, European Association for the Study of D, European Association for the Study of O, author. EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J Hepatol. 2016;64:1388–1402. doi: 10.1016/j.jhep.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 57.Rehm J, Samokhvalov AV, Shield KD. Global burden of alcoholic liver diseases. J. Hepatol. 2013;59:160–168. doi: 10.1016/j.jhep.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 58.Wang X, Ota N, Manzanillo P, Kates L, Zavala-Solorio J, Eidenschenk C, et al. Interleukin-22 alleviates metabolic disorders and restores mucosal immunity in diabetes. Nature. 2014;514:237–241. doi: 10.1038/nature13564. [DOI] [PubMed] [Google Scholar]
- 59.Pan CX, Tang J, Wang XY, Wu FR, Ge JF, Chen FH. Role of interleukin-22 in liver diseases. Inflamm. Res. 2014;63:519–525. doi: 10.1007/s00011-014-0727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oh JH, Schueler KL, Stapleton DS, Alexander LM, Yen CE, Keller MP, et al. Secretion of recombinant interleukin-22 by engineered Lactobacillus reuteri reduces fatty liver disease in a mouse model of diet-induced obesity. mSphere. 2020;5:e0018320. doi: 10.1128/mSphere.00183-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hendrikx T, Duan Y, Wang Y, Oh JH, Alexander LM, Huang W, et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut. 2019;68:1504–1515. doi: 10.1136/gutjnl-2018-317232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takiishi T, Cook DP, Korf H, Sebastiani G, Mancarella F, Cunha JP, et al. Reversal of diabetes in NOD mice by clinical-grade proinsulin and IL-10-secreting Lactococcus lactis in combination with low-dose anti-CD3 depends on the induction of Foxp3-positive T cells. Diabetes. 2017;66:448–459. doi: 10.2337/db15-1625. [DOI] [PubMed] [Google Scholar]
- 63.Verma A, Xu K, Du T, Zhu P, Liang Z, Liao S, et al. Expression of human ACE2 in Lactobacillus and beneficial effects in diabetic retinopathy in mice. Mol. Ther. Methods Clin. Dev. 2019;14:161–170. doi: 10.1016/j.omtm.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Attalla K, De S, Monga M. Oxalate content of food: a tangled web. Urology. 2014;84:555–560. doi: 10.1016/j.urology.2014.03.053. [DOI] [PubMed] [Google Scholar]
- 65.Lubkowicz D, Horvath NG, James MJ, Cantarella P, Renaud L, Bergeron CG, et al. An engineered bacterial therapeutic lowers urinary oxalate in preclinical models and in silico simulations of enteric hyperoxaluria. Mol. Syst. Biol. 2022;18:e10539. doi: 10.15252/msb.202110539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kurtz CB, Millet YA, Puurunen MK, Perreault M, Charbonneau MR, Isabella VM, et al. An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci. Transl. Med. 2019;11:eaau7975. doi: 10.1126/scitranslmed.aau7975. [DOI] [PubMed] [Google Scholar]
- 67.Isabella VM, Ha BN, Castillo MJ, Lubkowicz DJ, Rowe SE, Millet YA, et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 2018;36:857–864. doi: 10.1038/nbt.4222. [DOI] [PubMed] [Google Scholar]
- 68.Adolfsen KJ, Callihan I, Monahan CE, Greisen PJ, Spoonamore J, Momin M, et al. Improvement of a synthetic live bacterial therapeutic for phenylketonuria with biosensor-enabled enzyme engineering. Nat. Commun. 2021;12:6215. doi: 10.1038/s41467-021-26524-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Triassi AJ, Fields BD, Monahan CE, Means JM, Park Y, Doosthosseini H, et al. Redesign of an Escherichia coli Nissle treatment for phenylketonuria using insulated genomic landing pads and genetic circuits to reduce burden. Cell Syst. 2023;14:512–524.:e512. doi: 10.1016/j.cels.2023.05.004. [DOI] [PubMed] [Google Scholar]
- 70.Renwick MJ, Brogan DM, Mossialos E. A systematic review and critical assessment of incentive strategies for discovery and development of novel antibiotics. J. Antibiot. 2016;69:73–88. doi: 10.1038/ja.2015.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fujitani S, Sun HY, Yu VL, Weingarten JA. Pneumonia due to Pseudomonas aeruginosa: part I: epidemiology, clinical diagnosis, and source. Chest. 2011;139:909–919. doi: 10.1378/chest.10-0166. [DOI] [PubMed] [Google Scholar]
- 72.Saeidi N, Wong CK, Lo TM, Nguyen HX, Ling H, Leong SS, et al. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol. Syst. Biol. 2011;7:521. doi: 10.1038/msb.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hwang IY, Koh E, Wong A, March JC, Bentley WE, Lee YS, Chang MW. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun. 2017;8:15028. doi: 10.1038/ncomms15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hwang IY, Tan MH, Koh E, Ho CL, Poh CL, Chang MW. Reprogramming microbes to be pathogen-seeking killers. ACS Synth. Biol. 2014;3:228–237. doi: 10.1021/sb400077j. [DOI] [PubMed] [Google Scholar]
- 75.Ali M, Nelson AR, Lopez AL, Sack DA. Updated global burden of cholera in endemic countries. PLoS Negl. Trop. Dis. 2015;9:e0003832. doi: 10.1371/journal.pntd.0003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duan F, March JC. Interrupting Vibrio cholerae infection of human epithelial cells with engineered commensal bacterial signaling. Biotechnol. Bioeng. 2008;101:128–134. doi: 10.1002/bit.21897. [DOI] [PubMed] [Google Scholar]
- 77.Duan F, March JC. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc. Natl. Acad. Sci. USA. 2010;107:11260–11264. doi: 10.1073/pnas.1001294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lopez-Igual R, Bernal-Bayard J, Rodriguez-Paton A, Ghigo JM, Mazel D. Engineered toxin-intein antimicrobials can selectively target and kill antibiotic-resistant bacteria in mixed populations. Nat. Biotechnol. 2019;37:755–760. doi: 10.1038/s41587-019-0105-3. [DOI] [PubMed] [Google Scholar]
- 79.Jayaraman P, Holowko MB, Yeoh JW, Lim S, Poh CL. Repurposing a Two-Component System-Based Biosensor for the Killing of Vibrio cholerae. ACS Synth. Biol. 2017;6:1403–1415. doi: 10.1021/acssynbio.7b00058. [DOI] [PubMed] [Google Scholar]
- 80.Gould LH, Walsh KA, Vieira AR, Herman K, Williams IT, Hall AJ, et al. Surveillance for foodborne disease outbreaks - United States, 1998-2008. MMWR Surveill Summ. 2013;62:1–34. [PubMed] [Google Scholar]
- 81.Centers for Disease C, Prevention, author. Surveillance for foodborne disease outbreaks--United States, 2009-2010. MMWR Morb. Mortal. Wkly Rep. 2013;62:41–47. doi: 10.1016/j.annemergmed.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Forkus B, Ritter S, Vlysidis M, Geldart K, Kaznessis YN. Antimicrobial probiotics reduce Salmonella enterica in Turkey gastrointestinal tracts. Sci. Rep. 2017;7:40695. doi: 10.1038/srep40695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Palmer JD, Piattelli E, McCormick BA, Silby MW, Brigham CJ, Bucci V. Engineered probiotic for the inhibition of Salmonella via tetrathionate-induced production of microcin H47. ACS Infect. Dis. 2018;4:39–45. doi: 10.1021/acsinfecdis.7b00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koh E, Hwang IY, Lee HL, De Sotto R, Lee JWJ, Lee YS, et al. Engineering probiotics to inhibit Clostridioides difficile infection by dynamic regulation of intestinal metabolism. Nat. Commun. 2022;13:3834. doi: 10.1038/s41467-022-31334-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Geldart KG, Kommineni S, Forbes M, Hayward M, Dunny GM, Salzman NH, Kaznessis YN. Engineered E. coli Nissle 1917 for the reduction of vancomycin-resistant Enterococcus in the intestinal tract. Bioeng. Transl. Med. 2018;3:197–208. doi: 10.1002/btm2.10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tscherner M, Giessen TW, Markey L, Kumamoto CA, Silver PA. A Synthetic system that senses Candida albicans and inhibits virulence factors. ACS Synth. Biol. 2019;8:434–444. doi: 10.1021/acssynbio.8b00457. [DOI] [PubMed] [Google Scholar]
- 88.Ji C, Xue GF, Lijun C, Feng P, Li D, Li L, et al. A novel dual GLP-1 and GIP receptor agonist is neuroprotective in the MPTP mouse model of Parkinson's disease by increasing expression of BNDF. Brain Res. 2016;1634:1–11. doi: 10.1016/j.brainres.2015.09.035. [DOI] [PubMed] [Google Scholar]
- 89.Holscher C. Insulin, incretins and other growth factors as potential novel treatments for Alzheimer's and Parkinson's diseases. Biochem. Soc. Trans. 2014;42:593–599. doi: 10.1042/BST20140016. [DOI] [PubMed] [Google Scholar]
- 90.Ji C, Xue GF, Li G, Li D, Holscher C. Neuroprotective effects of glucose-dependent insulinotropic polypeptide in Alzheimer's disease. Rev Neurosci. 2016;27:61–70. doi: 10.1515/revneuro-2015-0021. [DOI] [PubMed] [Google Scholar]
- 91.Fang X, Tian P, Zhao X, Jiang C, Chen T. Neuroprotective effects of an engineered commensal bacterium in the 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine Parkinson disease mouse model via producing glucagon-like peptide-1. J. Neurochem. 2019;150:441–452. doi: 10.1111/jnc.14694. [DOI] [PubMed] [Google Scholar]
- 92.Zhang X, Pang G, Sun T, Liu X, Pan H, Zhang Y, et al. A red light-controlled probiotic bio-system for in-situ gut-brain axis regulation. Biomaterials. 2023;294:122005. doi: 10.1016/j.biomaterials.2023.122005. [DOI] [PubMed] [Google Scholar]
- 93.Cubillos-Ruiz A, Alcantar MA, Donghia NM, Cardenas P, Avila-Pacheco J, Collins JJ. An engineered live biotherapeutic for the prevention of antibiotic-induced dysbiosis. Nat. Biomed. Eng. 2022;6:910–921. doi: 10.1038/s41551-022-00871-9. [DOI] [PubMed] [Google Scholar]
- 94.Bush K, Bradford PA. Epidemiology of beta-Lactamase-producing pathogens. Clin. Microbiol. Rev. 2020;33:e00047–19. doi: 10.1128/CMR.00047-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pantoja Angles A, Valle-Perez AU, Hauser C, Mahfouz MM. Microbial biocontainment systems for clinical, agricultural, and industrial applications. Front. Bioeng. Biotechnol. 2022;10:830200. doi: 10.3389/fbioe.2022.830200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee JW, Chan CTY, Slomovic S, Collins JJ. Next-generation biocontainment systems for engineered organisms. Nat. Chem. Biol. 2018;14:530–537. doi: 10.1038/s41589-018-0056-x. [DOI] [PubMed] [Google Scholar]
- 97.Shen H, Zhang C, Li S, Liang Y, Lee LT, Aggarwal N, et al. Prodrug-conjugated tumor-seeking commensals for targeted cancer therapy. Nat. Commun. 2024;15:4343. doi: 10.1038/s41467-024-48661-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gao Y, Li W, Huang X, Lyu Y, Yue C. Advances in gut microbiota-targeted therapeutics for metabolic syndrome. Microorganisms. 2024;12:851. doi: 10.3390/microorganisms12050851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Inda-Webb ME, Jimenez M, Liu Q, Phan NV, Ahn J, Steiger C, et al. Sub-1.4 cm3 capsule for detecting labile inflammatory biomarkers in situ. Nature. 2023;620:386–392. doi: 10.1038/s41586-023-06369-x. [DOI] [PubMed] [Google Scholar]
- 100.Tang TC, Tham E, Liu X, Yehl K, Rovner AJ, Yuk H, et al. Hydrogel-based biocontainment of bacteria for continuous sensing and computation. Nat. Chem. Biol. 2021;17:724–731. doi: 10.1038/s41589-021-00779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jones TS, Oliveira SMD, Myers CJ, Voigt CA, Densmore D. Genetic circuit design automation with Cello 2.0. Nat. Protoc. 2022;17:1097–1113. doi: 10.1038/s41596-021-00675-2. [DOI] [PubMed] [Google Scholar]
- 102.Taketani M, Zhang J, Zhang S, Triassi AJ, Huang YJ, Griffith LG, et al. Genetic circuit design automation for the gut resident species Bacteroides thetaiotaomicron. Nat. Biotechnol. 2020;38:962–969. doi: 10.1038/s41587-020-0468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]