Abstract
Colorectal cancer (CRC) is a multifactorial disease driven by genetic and epigenetic alterations that modulate specific metabolic pathways. Despite the availability of effective treatments like 5-fluorouracil (5-FU), pharmacological therapy for CRC still faces significant challenges, including drug resistance, toxicity, and limited specificity. Therefore, discovering new compounds remains critical to overcoming these barriers and expanding treatment options. This study evaluated the cytotoxicity of fluorinated quaternary ammonium salts (FQAS) library in CRC-derived cell lines with premetastatic and metastatic phenotypes. The genetic and epigenetic background of the CRC cell lines and the selectivity of cytotoxicity compared to nontumor cells and between different CRC stages were also assessed. Additionally, the in silico pharmacological properties of these FQASs were analyzed. Results showed that FQASs 9–14 exhibited significant cytotoxic activity against both premetastatic and metastatic CRC cell lines, with FQASs 9, 13, and 14 displaying selective toxicity toward CRC cells over normal murine colorectal cells. However, in silico studies indicated poor oral bioavailability for these compounds, suggesting that an injection-based delivery route may be more effective for targeting CRC cells. In conclusion, CF3-containing FQASs are promising therapeutic candidates for CRC treatment.
Keywords: biopharmaceutical properties, colon rectal cancer cell lines, epigenetic analysis, genetic background, physicochemical properties
1. Introduction
Colorectal cancer (CRC) is a multifactorial and heterogeneous disease that involves genetic and epigenetic alterations that modulate specific metabolic pathways [1, 2]. This neoplasm is the third most prevalent type of cancer and the second leading cause of cancer-related deaths [3]. In Colombia, CRC ranks fourth in prevalence and incidence and third in mortality rates [3], constituting a public health problem. Factors contributing to the development of CRC include age, family history, chemical exposure, western diet, smoking, mutations in tumor suppressor genes and oncogenes, microsatellites alterations, chromosomal instability (CIN), and epigenetic background [4, 5].
Oncogenic transformation in CRC is primarily driven by gene mutations such as APC, KRAS, SMAD4, and TP53, collectively known as the adenoma–carcinoma sequence (ACS) [6, 7]. Additionally, epigenetic changes, particularly DNA methylation and histone modifications, play crucial pathophysiological roles in CRC initiation and progression [6, 8]. On the other hand, hypermethylation of CpG islands in promoter regions of tumor-suppressor genes placed by DNA methyltransferases (DNMTs) inhibits gene expression, favoring tumorigenesis, while hypomethylation of promoters or distant super-enhancers can enhance the expression of proto-oncogenes [7–9].
CRC management relies on various systemic interventions, including resection surgery, chemotherapy, biotherapy, and immunotherapy [10–12]. Despite the availability of effective treatments like 5-fluorouracil (5-FU), a drug widely used in the pharmacological therapy for CRC, still faces significant challenges, including drug resistance, toxicity, limited specificity, and high cost [11–13]; in addition, genetic background is also a determinant factor for the therapeutic outcomes [11, 13]. Therefore, discovering new compounds remains critical to overcoming these barriers and expanding treatment options.
Quaternary ammonium salts (QASs) are a large family of organic salts characterized by at least one quaternary ammonium cation, where a nitrogen atom is covalently bonded to four carbon atoms [14]. These compounds have demonstrated a range of biological activities, including antiviral [15, 16], antibacterial [17], antifungal [18], and antitumoral effects [14, 19, 20]. Additionally, some QASs feature fluorine atoms or trifluoromethyl groups attached to the phenyl rings of their vinyl moieties, which have been associated with leishmanicidal [21] and anti-trypanosomal activities [22].
Fluorinated moieties, in general, are widely used in medicinal compounds due to the unique properties fluorine atoms confer. With a size like hydrogen but significantly higher electronegativity, fluorine's nonpaired electrons and strong C–F bonds often enhance the physicochemical properties of molecules, potentially improving biological activity or slowing metabolic degradation [23–25]. Trifluoromethyl groups (–CF3) can alter important drug characteristics such as steric effects, lipophilicity, pKa, and the capacity to form hydrogen bonds, all of which can significantly influence pharmacokinetic parameters like absorption and metabolism [26]. Similarly, recent studies suggest that the biological activity of QASs is shaped not only by their chemical structure but also by their hydrophilic and hydrophobic properties, pointing to specific biological mechanisms underlying their cytotoxic effects [27–29]. Therefore, by highlighting the influence of both fluorinated groups and the structural properties of QASs, these findings underscore the potential for designing more effective and targeted pharmacological agents.
The present work aims to design and synthesize a series of novel fluorinated QASs (FQASs) of general structure {[(Y-C6H5)2C=CH-(CH2)n]N(CH3)2CH2X}+ I− with different lengths in the carbon chain and variations in the fluorinated position, either meta or para positions and the subsequent determination of the cytotoxicity effects of each compound over three CRC-derived cell lines with different metastatic phenotypes, according to its genetic and epigenetic molecular phenotypes. The potential pharmacology of fluorinated and nonfluorinated QASs was also assessed using in silico approaches to identify promising hit molecules for developing specific antitumoral therapies for CRC.
2. Results and Discussion
2.1. Synthesis and Characterization of FQASs and QASs
The 14 FQASs synthesized were: C5CH3mF2 (1), C5IpF2 (2), C6ClpF2 (3), C6IpF2 (4), C5ClmCF3 (5), C5ImCF3 (6), C5CH3mCF3 (7), C5ClpCF3 (8), C5IpCF3 (9), C5CH3pCF3 (10), C6ImCF3 (11) C6ClpCF3 (12), C6IpCF3 (13), and C6CH3pCF3 (14). The two synthesized QASs corresponded to C5I (15) and C6I (16) (Figure 1).
Figure 1.
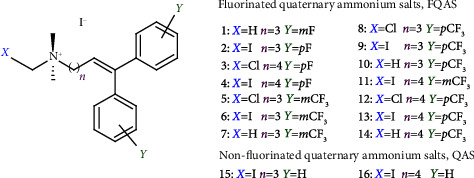
Generic structure of fluorinated and nonfluorinated quaternary ammonium salts.
2.2. Genetic and Epigenetic Analysis of CRC Cell Features
Understanding the genetic and epigenetic landscape of CRC cell lines with premetastatic, metastatic in situ, and metastatic invasive phenotypes is essential for identifying novel therapeutic targets, predicting drug efficacy, and developing more effective treatments for CRC. In the CRC cell lines studied (SW948, SW48, and SW620), eight mutated genes were identified (Table 1), all of which play crucial roles in gene regulation, cell proliferation, repair, angiogenesis, and metastasis. Any mutation in these genes may disrupt normal cell cycle control and proliferation processes.
Table 1.
Genetic alterations status of colorectal cancer cell lines SW620, SW48, and SW948.
| Cell line | Mutant gene | Encoded protein | Protein function | Zygosity | Mutation | Amino acid change | Platform | |
|---|---|---|---|---|---|---|---|---|
| ATCC | IGRhCellID | |||||||
| SW948 | KRAS | K-Ras | Cell proliferation and differentiation | Heterozygous | 182A > T | Q61L | x | x |
| PIK3CA | p110 alpha (p110α) protein | Cell proliferation and migration | Heterozygous | 1624G > A | E542K | x | x | |
| APC | APC protein | Tumor suppression | Heterozygous | 3340C > T | R1114∗ | x | x | |
| Heterozygous | 4285C > T | Q1429∗ | x | x | ||||
| TP53 | Tumor protein 53 | Tumor suppression | ND | 412 stop at 122 | Gly117FS | — | x | |
|
| ||||||||
| SW620 | KRAS | K-Ras | Cell proliferation and differentiation | Homozygous | 35G > T | G12V | — | x |
| Heterozygous | 182A > T | Q61L | — | x | ||||
| TP53 | Tumor protein 53 | Tumor suppression | Homozygous | 818G > A | R273H | — | x | |
| Homozygous | 925C > T | P309S | — | x | ||||
| ND | 881G > A | Arg273His | — | x | ||||
| ND | 988C > T | Pro309Ser | — | x | ||||
| APC | APC protein | Tumor suppression | Homozygous | 4012C > T | Q1338 | — | x | |
| Heterozygous | 3340C > T | p.R1114∗ | — | x | ||||
| Heterozygous | 4285C > T | p.Q1429∗ | — | x | ||||
| SMAD4 | TGFB | Tumor suppression | Homozygous | 5G > C | — | — | x | |
| PIK3CA | p110 alpha (p110α) protein | Cell proliferation and migration | Heterozygous | 1624G > A | p.E542K | — | x | |
|
| ||||||||
| SW48 | CTNNB1 | β-Catenin | Cell proliferation and differentiation | Heterozygous | 98C > A | S33Y | x | x |
| EGFR | Epidermal growth factor receptor | Cell proliferation and cell survival | Heterozygous | 2155G > A | G719S | x | x | |
| FBXW7 | F-box and WD repeat domain-containing 7 | F-box protein family | Heterozygous | 2001delG | S668fs∗39 | x | — | |
| TP53 | Tumor protein 53 | Tumor suppression | ND | 805C > T | Arg248Trp | — | x | |
Note: Data extracted from The Cancer Genome Atlas (TCGA) [30] at the NIH database (https://www.portal.gdc.cancer.gov), the American Type Culture Collection—ATCC (https://www.atcc.org/), and the IGRhCellID (https://igrcid.ibms.sinica.edu.tw).
Abbreviations: APC gene, adenomatous polyposis coli; CTNNB1 gene, catenin beta-1; EGFR gene, epidermal growth factor receptor; FBXW7 gene, F-Box and WD Repeat Domain Containing 7; KRAS gene, Kirsten rat sarcoma viral oncogene; PIK3CA gene, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; SMAD4 gene, mothers against decapentaplegic member 4; TP53 gene, tumoral protein p53.
The TP53 gene was mutated in all three cell lines (premetastatic SW948, metastatic in situ SW48, and metastatic invasive SW620), underscoring their fundamental role in cell cycle control, senescence, apoptosis, and DNA repair. The common R273H mutation in TP53 was observed across these lines, a mutation known to maintain the regular conformation of the TP53 protein while inducing apoptosis [7]. However, this mutation also contributes to cell cycle dysregulation, affecting normal proliferation and compromising DNA repair mechanisms [31, 32].
The KRAS, PIK3CA, and APC genes were mutated in both premetastatic SW948 and metastatic invasive SW620 cell lines. These genes are involved in pathways that regulate the cell cycle, proliferation, and angiogenesis [9]. Specifically, mutations in the APC gene are linked to enhanced cell survival by modulating proteins such as BCL2, IAP, PCNA, eNOS, and A20, promoting abnormal cell proliferation [33, 34].
Uniquely, SW620 cells (metastatic invasive) also exhibited mutations in the SMAD4 gene, which is associated with promoting metastasis [35]. This finding suggests that the SMAD4 mutation could play a critical role in the progression from localized to invasive metastatic phenotypes. In contrast, the metastatic in situ SW48 cells showed distinct genetic mutations. Alongside TP53, SW48 cells presented mutations in CTNNB1, EGFR, and FBXW7, genes known for their involvement in oncogenesis and tumor progression [30, 36, 37]. These mutations could influence cell signaling pathways that drive tumor growth and potentially contribute to the early stages of metastasis.
Overall, the differences in molecular phenotypes and allelic expression of mutated genes between these CRC cell lines reflect the varying stages of metastatic potential. Mutations in TP53, KRAS, PIK3CA, and APC are shared by premetastatic and metastatic invasive cells, while SMAD4 mutations are specific to invasive cells, suggesting its role in metastasis. The distinct mutation profile of SW48 highlights the importance of CTNNB1, EGFR, and FBXW7 in metastatic in situ phenotypes.
On the other hand, understanding the molecular characteristics of CRC cell lines, including their microsatellite instability (MSI), microsatellite stability (MSS), CIN, and CpG island methylator phenotype (CIMP) status, is crucial for studying not only their biology and disease progression but also therapeutic susceptibility. Our analysis showed that SW48 cells are MSI-positive, while SW620 and SW948 are MSS (Table 2).
Table 2.
Epigenetic status of SW48, SW620, and SW948 cell linesa.
| Cell line | Disease | Stage | MSI/MSS status | CIMP Panel 1 | CIMP Panel 2 | CIN |
|---|---|---|---|---|---|---|
| SW948 | Adenocarcinoma | Premetastatic | MSS+ | ND | ND | ND |
| SW48 | Adenocarcinoma | Metastatic in situ | MSI+ | + | + | − |
| SW620 | Adenocarcinoma | Metastatic invasive | MSS+ | + | + | − |
Note: The microsatellite instability/stability status in CRC SW948, SW48, and SW620 cell lines. Panel 1 (CDKN2A(p16), MINT1, MINT2, MINT31, and MLH1); Panel 2 (CACNA1G, IGF2, NEUROG1, RUNX3 and SOCS1).
Abbreviations: CIMP, CpG island methylator phenotype; CIN, chromosomal instability pathway; MSI, microsatellite instability; MSS, microsatellite stability; ND, no data available.
aInformation was taken from the Cellosaurus platform (https://www.expasy.org) and [30].
The MSI-positive status in SW48 has important biological implications, influencing processes such as tumor development and progression, response to therapy, molecular subtyping, and genomic stability. MSI is typically associated with defects in the DNA mismatch repair system, leading to the accumulation of mutations in microsatellite regions, which contributes to cancer initiation and progression. As a result, SW48 cells, with their MSI-positive status, may exhibit enhanced tumor development and progression compared to MSS cells like SW620 and SW948. Furthermore, MSI-positive tumors, such as those derived from SW48 cells, tend to be more responsive to immunotherapy, particularly immune checkpoint inhibitors, making this cell line a valuable model for exploring therapeutic responses.
The increased genomic instability seen in MSI-positive cells like SW48 results from the accumulation of mutations, affecting crucial cellular processes such as proliferation, differentiation, and survival. In contrast, SW620 and SW948, being MSS, exhibit a more stable genomic environment. The MSS phenotype may correlate with lower mutation rates in microsatellite regions, and consequently, these cell lines might demonstrate different responses to therapeutic agents, particularly those targeting DNA repair pathways.
Regarding the CIMP profile, both SW48 and SW620 cells are CIMP-positive (Panels 1 and 2), indicating the presence of aberrant DNA methylation patterns commonly associated with altered gene expression and tumor behavior. CIMP-positive status can influence epigenetic regulation of oncogenes and tumor suppressor genes, contributing to tumor aggressiveness and resistance to some therapies. The presence of aberrant methylation in these cell lines suggests they may have distinct epigenetic landscapes, making them valuable models for exploring therapies targeting DNA methylation pathways.
On the other hand, SW948 is CIMP-negative but CIN-positive, reflecting increased chromosomal aberrations and aneuploidy, which contribute to tumor heterogeneity and progression. CIN-positive tumors are often characterized by high CIN, which may lead to resistance to certain treatments but could also provide opportunities for targeted therapies that exploit this instability. Notably, SW48 and SW620 cells are CIN-negative, signifying a more chromosomally stable profile. The lack of CIN in these cell lines might suggest that their tumorigenic potential is more strongly driven by epigenetic changes (CIMP) and mutations in key oncogenes or tumor suppressor genes rather than CIN.
2.3. Cytotoxicity of FQASs and QASs
The cytotoxicity of FQASs (compounds 1–14) and QASs (compounds 15-16) was evaluated in CRC cell lines SW48, SW620, and SW948, along with healthy murine colorectal tissue (MCRT) and peripheral blood mononuclear cells (PBMCs), using the half-maximal cytotoxic concentration (CC50) values (μM) and the cytopathic effect (CPE). Results are summarized in Table 3.
Table 3.
Cytotoxicity and cytopathic effect of FQASs and QASs in different cell lines.
| Compound | CC50 in μM (CPE)a | ||||
|---|---|---|---|---|---|
| SW948 | SW48 | SW948 | MCRT | PBMC | |
| 1 | 211.2 ± 7.4 (+) | 105.0 ± 12.0 (+) | 63.7 ± 15.3 (+) | 212.0 ± 4.0 (NA) | 29.8 ± 2.4 (NA) |
| 2 | 77.9 ± 9.1 (+) | 75.7 ± 14.2 (+) | 28.1 ± 6,1 (+) | 204.7 ± 14.5 (NA) | 35.6 ± 6.6 (NA) |
| 3 | 52.9 ± 13.8 (+) | 125.3 ± 5.1 (+) | 64.5 ± 8.1 (+) | 129.7 ± 20.0 (NA) | 39.7 ± 7.7 (NA) |
| 4 | 72.8 ± 12.8 (+) | 100.4 ± 3.0 (+) | 52.5 ± 5.6 (+) | 106.8 ± 4.5 (NA) | 45.1 ± 7.3 (NA) |
| 5 | 43.5 ± 4.0 (+++) | 52.0 ± 9.2 (+++) | 24.5 ± 2.3 (+++) | 121.0 ± 5.4 (NA) | 34.8 ± 6 (NA) |
| 6 | 45.6 ± 3.6 (++) | 55.5 ± 6.4 (++) | 38.8 ± 2.1 (++) | 129.0 ± 11.3 (NA) | 44.8 ± 6.4 (NA) |
| 7 | 20.0 ± 0.8 (++) | 81.4 ± 9.4 (++) | 31.1 ± 0.2 (++) | 123.2 ± 9.2 (NA) | 46.3 ± 7.5 (NA) |
| 8 | 20.4 ± 6.2 (++) | 50.8 ± 4.0 (++) | 19.2 ± 3.5 (++) | 102.0 ± 5.0 (NA) | 42.3 ± 9.3 (NA) |
| 9 | 20.0 ± 4.7 (++) | 40.2 ± 6.0 (++) | 32.2 ± 1.0 (++) | 206.1 ± 19.4 (NA) | 30.6 ± 2.1 (NA) |
| 10 | 13.4 ± 3.9 (++) | 44.0 ± 6.0 (++) | 18.8 ± 4.7 (++) | 119.4 ± 8.7 (NA) | 51.4 ± 5.3 (NA) |
| 11 | 21.5 ± 4.5 (+++) | 22.1 ± 4.0 (+++) | 18.1 ± 3.7 (+++) | 93.5 ± 13.9 (NA) | 33.4 ± 2.0 (NA) |
| 12 | 25.0 ± 4.8 (+++) | 48.0 ± 5.1 (+++) | 15.1 ± 1.5 (+++) | 110.6 ± 16.7 (NA) | 40.5 ± 9.5 (NA) |
| 13 | 26.9 ± 4.0 (+++) | 16.3 ± 2.0 (+++) | 16.0 ± 1.5 (+++) | > 292.7 (NA) | 53.9 ± 3.0 (NA) |
| 14 | 49.0 ± 11.1 (+++) | 18.5 ± 3.6 (+++) | 14.5 ± 2.9 (+++) | 419.4 ± 15.0 (NA) | 93.2 ± 13.1 (NA) |
| 15 | 98.2 ± 4.3 (+) | 123.7 ± 2.3 (+) | 157.5 ± 2.3 (+) | 255.9 ± 14.0 (NA) | 29.3 ± 1.4 (NA) |
| 16 | 71.0 ± 12.4 (+) | 65.2 ± 3.7 (+) | 72.9 ± 4.2 (+) | 134.9 ± 11.8 (NA) | 38.8 ± 5.6 (NA) |
| 5-FU | 30.3 ± 5.2 (+++) | 27.9 ± 9.6 (+++) | 30.3 ± 6.0 (+++) | 944.7 ± 43.7 (NA) | 38.7 ± 7.0 (NA) |
Note: Data represent the mean ± SD of three independent trials with two technical replicates of the CC50 values that provide a quantitative measure of cytotoxicity, with lower values indicating higher toxicity. The CPE describes the qualitative effects on cell viability, with categories ranging from mild to strong cytotoxic effects of the median cytotoxic concentration (CC50), where the higher the CC50 value is, the less cytotoxic the compound is. SW620: Dukes' type C, invasive metastatic colorectal adenocarcinoma; SW48: Dukes' type C, grade IV noninvasive (in situ) metastatic colorectal adenocarcinoma; SW948: Dukes' type C, grade III, pre-metastatic colorectal adenocarcinoma; +: Mild effect (10–30% reduction in viability); ++: Moderate effect (30–60% reduction in viability); +++: Strong effect (60–90% reduction in viability); (NA): not applicable in non-adherent MCRT and PBMC.
Abbreviations: MCRT, healthy murine colorectal tissue; PBMC, peripheral blood mononuclear cells.
aCPE, cytopathic effect.
FQASs 5–14 displayed significantly stronger cytotoxicity across CRC cell lines, with CC50 values ranging from 14.5 to 49 μM and strong CPE (+++), indicating a pronounced reduction in cell viability (60%–90%). Notably, compound 5 had CC50 values of 43.5 μM in SW48, 52.0 μM in SW620, and 24.5 μM in SW948, all with strong cytotoxic effects (+++). The reference compound, 5-FU, showed comparable cytotoxicity with CC50 values of 30.3 μM in SW48, 27.9 μM in SW620, and 30.3 μM in SW948, along with strong CPE (+++). In contrast, the FQASs and QASs had a significantly reduced cytotoxic effect on MCRT and PBMC cell lines, where CC50 values were substantially higher, indicating lower toxicity. For example, compound 5 showed a CC50 of 121 μM in MCRT and 34.8 μM in PBMC. This trend was consistent across most compounds, including the reference 5-FU, which had a CC50 value of 944.7 μM in MCRT and 38.7 μM in PBMC (Figure 2).
Figure 2.
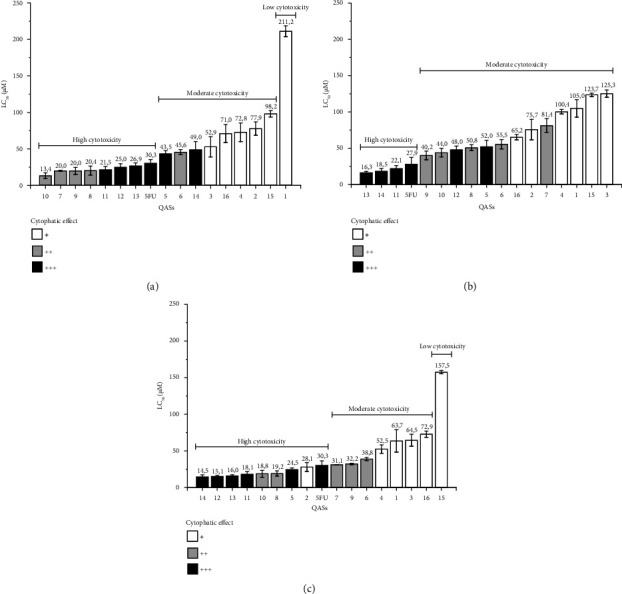
Cytotoxicity and cytopathic effect relationship of FQASs and QASs. The figure shows the cytotoxicity level of FQASs and QASs according to the median cytotoxic concentration (CC50) and the cytopathic effect in (a) SW948 Dukes' Type C, Grade III premetastatic colorectal cancer line; (b) SW48 Dukes' Type C, Grade IV in situ colorectal cancer line; (c) SW620 Dukes' Type C, Grade IV metastatic colorectal line. Cytopathic effect: mild (+), moderate (++), strong (+++).
The cytotoxicity of FQASs over the premetastatic CRC cell line SW948 (Dukes' Type C, Grade III) was moderate to high (Figure 2(a)). CC50 values for FQASs ranged between 211.2 μM (compound 1) and 13.4 μM (compound 10). Compounds 7–13 showed a cluster of similar CC50 values, exhibiting a slightly higher cytotoxicity than the reference drug 5-FU, which had a CC50 of 30.3 μM. A notable observation was that compounds with strong cytotoxicity also exhibited a comparable CPE to 5-FU. For example, compounds 7–14 were marked as (+++) for strong cytotoxicity, suggesting a potential shared mechanism of action with 5-FU. Interestingly, the premetastatic CRC cell line SW948 appeared to be more susceptible to QASs 15 and 16, which displayed moderate CC50 values. This suggests that QASs may have different mechanisms of action when compared to FQASs.
For the metastatic in situ CRC cell line SW48 (Dukes' Type C, Grade IV), FQASs demonstrated a wide range of cytotoxic effects, with CC50 values between 125.3 μM (compound 3) and 16.3 μM (compound 13) (Figure 2(b)). FQASs 11, 13, and 14 showed stronger cytotoxic effects than 5-FU (which had a CC50 of 27.9 μM) with respective CC50 values of 22.1, 18.5, and 16.3 μM. These three compounds also demonstrated strong CPEs (+++). Additionally, compounds 5 and 12 were also labeled as (+++) for strong CPE, correlating with their high cytotoxicity. In contrast, QAS 15 and 16 demonstrated only moderate cytotoxicity for SW48 cells.
The data revealed that FQASs showed a range of cytotoxic effects on the metastatic CRC cell line SW620 (Dukes' Type C, Grade IV) (Figure 2(c)). For FQASs, CC50 values spanned from 63.7 to 14.5 μM, with compounds 13 and 14 again standing out for their low CC50 values, indicating higher cytotoxicity. The strong CPE (+++) observed in the more cytotoxic FQASs further reinforces the relationship between cytotoxicity and cytopathic damage. QASs 15 and 16, while still effective, were less potent compared to FQASs, exhibiting higher CC50 values and only mild to moderate CPEs. Table 3 (second column) and Figure 2(b) also show that FQASs have moderate to highly cytotoxic effects for the metastatic in situ CRC cell line SW48, with CC50 values ranging from 125.3 to 16.3 mM. FQASs 11, 13, and 14 were slightly more cytotoxic than the reference 5-FU (CC50 values of 27.9 mM for 5-FU, 22.1, 18.5, and 16.3 mM for compounds 11, 14, and 13, respectively). Again, the top more cytotoxic FQASs (11, 13, and 14) had a robust CPE and FQASs 5 and 12 (labeled as +++). QAS 15 and 16 had moderate cytotoxicity in these CRC cells.
Structurally, the most cytotoxic FQASs for SW948 cells share certain features: (i) a random distribution of cytotoxicity attributes for the C3-tethered and C4-tethered salts, with similar CC50 values; (ii) the presence of a trifluoromethyl substituent attached to either the para-position or meta-position of the aromatic rings, although six out of seven FQASs are para-substituted; (iii) the presence or absence of a halogen atom attached to one of the methyl groups of the quaternary ammonium head, with a random distribution of cytotoxicity patterns for chlorinated, iodinated, or nonhalogenated salts. FQAS 1 shows low cytotoxicity (CC50 value of 211.2 μM), followed by 2, which are (i) salts with three methylenes, (ii) with or without C-halogenate in the quaternary head, and (iii) nonfluorinated or monofluorinated in the aromatic rings. In agreement with this observation, for premetastatic SW948 cells, the carbon bond length is not highly relevant. Moreover, for the SW948 line, the presence or absence of a halogen atom attached to one of the quaternary head's methyl groups does not seem relevant, as cytotoxicity results from a complex combination of structural variables in the organic cation.
In the case of SW48 CRC cells, the most cytotoxic FQASs share certain features: (i) a four-methyl bond between the quaternary nitrogen atom and the vinyl moiety; (ii) a trifluoromethyl substituent on the aromatic rings, with para-substituted molecules slightly more cytotoxic than meta-substituted ones; (iii) the presence or absence of an iodine atom attached to one of the methyl groups of the quaternary ammonium head (CH2I vs. CH3).
A random distribution of cytotoxic behavior of the salts studied suggests that for this type of CRC cell line, the length of the bound carbon is not highly relevant. However, qualitatively, the salts bound to four methyls tend to be the most cytotoxic [38, 39], in addition to the experimental results shown in Table 3. The presence or absence of a halogen atom bound to one of the methyl groups of the quaternary head is not significantly relevant for cytotoxicity, suggesting that cytotoxicity results from a complex combination of structural variables in the organic cation. As mentioned, the cytotoxic FQASs 5 and 11–14 showed strong (+++) CPE. Both salts carry a chlorine atom (C-Cl pattern) on the quaternary ammonium head and an aromatic CF3 substituent (meta and para, respectively). It is important to note that the epigenetic characteristics of SW48 cells mean that they do not respond to a defined pathway to 5-FU [40] and would be expected to behave similarly in the presence of FQASs 11, 13, and 14, despite being the most cytotoxic 5-FU [40]. Additionally, although the structure of 5-FU is dissimilar to that of FQASs, it is known to be an effective alternative for treating CRC and other cancers. As observed in our analyses, the behavior of these fluorinated salts is like that of 5-FU, suggesting that these molecules are optimal for developing an antitumoral activity. In practice, CRC patients with mutated KRAS and positive methylation status (epigenetic profile) do not benefit from 5-FU monotherapy because 5-FU is ineffective in CRC cells with this genotype, as in the SW48 cell line (Table 3).
Similarly, for the SW948 line, the cytotoxic FQASs share certain features: (i) a random distribution of cytotoxicity patterns for the length of the tether, but the four top-cytotoxic FQASs 11–14 are four-methylene tethered ones; (ii) the presence of fluorine or a CF3 substituent attached to the aromatic rings, either to the para- or meta-position, although five out of eight FQASs are trifluoromethyl-substituted salts, mainly in the para-position; (iii) whether or not a halogen atom, attached to one of the methyl groups of the quaternary ammonium head, with a random distribution of cytotoxicity patterns for chlorinated, iodinated, or nonhalogenated salts, suggesting that this halogen atom is not significantly relevant for cytotoxicity against SW620 CRC cells. Although there is no clear profile, it is strongly suggested that metastatic CRC cells respond more to (both) four-methylene tethered and para-CF3-substituted FQASs.
MCRT and PBMCs control cell lines have differential susceptibilities to FQASs and QASs compounds compared to 5-FU susceptibility profiles (Table 3, fifth and sixth columns). FQASs were reported to be 50%–90% more cytotoxic than 5-FU against TCRM (CC50 for 5-FU 944.7 mM; meanwhile, FQASs CC50 ranges from 419.4 mM to 93.5 mM). However, PBMC cells showed a susceptibility profile in the same order of magnitude between most FQASs and the reference compound 5-FU (CC50 38.7 mM for 5-FU vs. CC50 ranging from 29.3 to 46.3 mM for FQASs). The observations regarding the QAS's cytotoxic effects against PBMC cells are comparable to the ones observed for FQASs in the same cell line, where QASs showed CC50 values comparable to the ones observed for 5-FU (CC50 values of 51.4, 53.9, and 93.2 mM for FQASs 10, 13, and 14, respectively). Both QASs 15 and 16 were highly cytotoxic for PBMCs; meanwhile, the MCRT FQASs 15 had moderate cytotoxicity, while compound 16 was noncytotoxic.
When comparing the CC50 values obtained for nontumoral MCRT and the CC50 obtained for CRC, it can be noticed that the values are much lower for CRC cells than MCRT, which means that the toxicity is higher in CRC cells (Table 3). This selectivity in cytotoxicity for some cells or others is reflected in IS higher than one, which is calculated by dividing the CC50 value obtained in nontumoral cells (MCRT or PBMC) by the CC50 value obtained for each CRC cell.
All the FQASs exhibited more cytotoxicity than 5-FU against TCRM mice, with a random distribution of cytotoxicity patterns compared to the structural features approached. Although this is not a standout result, it is noteworthy that several FQASs exhibit lower cytotoxicity against healthy PBMC cells. Ten FQASs 3, 4, 6, 7, 8, 10, and 12–14 were less cytotoxic (or comparable) than 5-FU against healthy PBMC cells (Table 3). While FQASs 13 and 14 tend to be cytotoxic molecules against the three CRC cells SW48, SW948, and SW620, these salts resulted in the least cytotoxic for healthy control PBMC cells, which means that at least eight salts (7–14) exhibit better IS than 5-FU (Table 4). Furthermore, a fundamental level of selectivity was observed for all the FQASs when comparing their cytotoxicity on healthy TCRM cells versus cytotoxicity on the three CRC cells studied. Indeed, according to Table 3, FQASs 13 and 14 (mainly the latter) exhibited IS values comparable to 5-FU (fourth, fifth, and sixth columns, entries 13 and 14). The best IS value was found for FQASs 14, which equals IS of 5-FU within experimental error (about the same cell lines). Although all FQASs and QASs also showed high cytotoxicity to PBMCs, the concentrations at which the mean toxic concentration is reached are higher than that observed for CRC cell lines. The cytotoxicity FQASs could be associated with mitochondrial damage because the MTT detects the activity of the mitochondrial enzyme succinate dehydrogenase; if this enzyme is altered, it is most likely that the mitochondria fail and generate cell death signals too early; nonetheless, this damage could be corrected, and cell recovery would occur. On the other hand, because these compounds are highly toxic, it is most likely that the damage is more significant than the repair events in the mitochondria. There is most likely no cell death due exclusively to mitochondrial damage but with additional adverse effects such as deterioration in other cellular organelles such as the membrane plasmatic [41].
Table 4.
Selectivity of FQASs and QASs for CRC cells vs. nontumoral cells MCRT and PBMC.
| Compound | Index of selectivity | |||||
|---|---|---|---|---|---|---|
| MCRT | PBMC | |||||
| SW948 | SW48 | SW620 | SW948 | SW48 | SW620 | |
| 1 | 1.0 | 2.0 | 3.3 | 0.1 | 0.3 | 0.5 |
| 2 | 2.6 | 2.7 | 7.3 | 0.5 | 0.47 | 1.3 |
| 3 | 2.5 | 1.0 | 2.0 | 0.8 | 0.32 | 0.6 |
| 4 | 1.5 | 1.1 | 2.0 | 0.6 | 0.45 | 0.9 |
| 5 | 2.8 | 2.3 | 4.9 | 0.8 | 0.66 | 1.4 |
| 6 | 2.8 | 2.3 | 3.3 | 1.0 | 0.80 | 1.2 |
| 7 | 6.2 | 1.5 | 4.0 | 2.3 | 0.57 | 1.5 |
| 8 | 5.0 | 2.0 | 5.3 | 2.1 | 0.8 | 2.2 |
| 9 | 10.3 | 5.1 | 6.4 | 1.5 | 0.8 | 1.0 |
| 10 | 8.9 | 2.7 | 6.4 | 3.8 | 1.2 | 2.7 |
| 11 | 4.4 | 4.2 | 5.2 | 1.6 | 1.5 | 1.9 |
| 12 | 4.4 | 2.3 | 7.3 | 1.6 | 0.8 | 2.7 |
| 13 | > 10.9 | > 18.0 | > 18.3 | 2.0 | 3.3 | 3.4 |
| 14 | 8.6 | 22.7 | 28.9 | 1.9 | 5.0 | 6.4 |
| 15 | 2.6 | 2.1 | 1.6 | 0.3 | 0.2 | 0.2 |
| 16 | 1.9 | 2.1 | 1.9 | 0.6 | 0.6 | 0.5 |
| 5-FU | 31.2 | 33.9 | 31.2 | 1.3 | 1.4 | 1.3 |
Note: Data correspond to the ratio between the CC50 for cells with the normal phenotype (MCRT and PBMC) and the CC50 for CRC cells (SW948, SW48, and SW620). SW948: Dukes' type C, grade III, pre-metastatic colorectal adenocarcinoma; SW48: Dukes' type C, grade IV noninvasive metastatic colorectal adenocarcinoma; SW620: Dukes' type C, invasive metastatic colorectal adenocarcinoma. IS = CC50 (MCRT or PBMC) ÷ CC50 (SW948, SW48 or SW620).
Abbreviations: 5-FU, 5-fluorouracil; MCRT, healthy murine colorectal tissue; PBMC, peripheral blood mononuclear cells.
The selectivity of FQAS and QAS for tumor cells over nontumor cells was calculated by comparing CRC cell lines to nontumor PBMC and MCRT cells. The calculated indexes of selectivity (IS) are summarized in Table 4. Notably, FQAS 13 exhibited significant selectivity for CRC cells, being up to 10 times more cytotoxic for SW948 cells and 18 times more cytotoxic for SW48 and SW620 cells. Similarly, FQAS 14 demonstrated high selectivity, with SI values of 22.7 for SW48 cells and 28.9 for SW620 cells, which correspond to noninvasive and invasive metastatic colorectal adenocarcinoma, respectively.
The majority of FQASs' selectivity index greater than 2 indicates that these compounds are more toxic to tumor cells than to nontumor cells, suggesting that they may be a good candidate in cancer therapy by minimizing damage to healthy cells.
As expected, 5-FU's antitumoral activity was selective for CRC cells instead of healthy MCRT, with IS values ranging between 31.2 and 33.9 (Table 4). Interestingly, no differences were observed in the compounds' selectivity for CRC cells and PBMCs, with IS values between 1.3 and 1.4.
The selectivity of FQASs 1–14 and QASs 15 and 16 between premetastatic (SW948), in situ metastatic SW48 cells, and invasive metastatic SW620 cells was also calculated by dividing the CC50 value obtained in noninvasive metastatic SW948 cells by the CC50 value obtained for in situ metastatic SW48 or invasive metastatic SW620 cells and, between in situ metastatic SW48 and invasive metastatic SW620 cells. The IS obtained are summarized in Table 5.
Table 5.
Selectivity of FQASs and QASs for premetastatic SW948 CRC cells vs. in situ metastatic SW48 and invasive metastatic SW620 CRC cells.
| Compound | Index of selectivity | ||
|---|---|---|---|
| SW948 vs. SW48a | SW948 vs. SW620b | SW48 vs. SW620c | |
| 1 | 2.0 | 3.3 | 1.7 |
| 2 | 1.0 | 2.8 | 2.7 |
| 3 | 0.4 | 0.8 | 1.9 |
| 4 | 0.7 | 1.4 | 1.9 |
| 5 | 0.8 | 1.8 | 2.1 |
| 6 | 0.8 | 0.8 | 1.4 |
| 7 | 0.2 | 1.6 | 2.6 |
| 8 | 0.4 | 1.1 | 2.6 |
| 9 | 0.5 | 0.6 | 1.2 |
| 10 | 0.3 | 0.7 | 2.3 |
| 11 | 1.0 | 1.2 | 1.2 |
| 12 | 0.5 | 1.7 | 3.2 |
| 13 | 1.7 | 1.7 | 1.0 |
| 14 | 2.6 | 3.4 | 1.3 |
| 15 | 0.8 | 0.6 | 0.8 |
| 16 | 1.1 | 1.0 | 0.9 |
| 5-FU | 1.1 | 1.0 | 0.9 |
Note: Data correspond to the ratio between the CC50 for CRC cells SW948 and SW48, SW948 and SW620, and SW48 and SW620. SW948: Dukes' type C, grade III, pre-metastatic colorectal adenocarcinoma; SW48: Dukes' type C, grade IV noninvasive metastatic colorectal adenocarcinoma; SW620: Dukes' type C, invasive metastatic colorectal adenocarcinoma.
Abbreviation: 5-FU, 5-fluorouracil.
aIS = CC50 SW948 ÷ CC50 SW48.
bIS = CC50 SW948 ÷ CC50 SW620.
cIS = CC50 SW48 ÷ CC50 SW620.
The IS values of most FQASs were < 1.0. Only FQASs 14, 1, and 13 had IS values of 2.6, 2.0, and 1.7, respectively (Table 5). Similarly, the comparison of the selectivity of FQASs and QASs between premetastatic versus invasive CRC cells showed that FQASs 14, 1, 2, 5, 13, 12, 7, 4, and 11 were selective for premetastatic cells with IS values of 3.4, 3.3, 2.8, 1.8, 1.7, 1.7, 1.6, 1.4, and 1.2, respectively. As expected, compound 5-FU showed no selectivity between CRC cells with values close to 1.0 (Table 5).
2.4. In Silico Analysis of Physicochemical Properties and Pharmacology of FQASs 1–14 and QASs 15 and 16
Simplified Molecular Input Line Entry Specification formulas (SMILES) were generated for each FQAS and QAS and subsequently loaded to the ADMETLab2.0 platform to calculate the physicochemical and pharmacodynamic parameters and distribution. Calculated parameters are summarized in Table 6. The molar mass of FQASs and QASs ranged from 443.30 to 683.30 g/mol, with larger molecular weights being a common feature across the compounds. All FQASs had 1 hydrogen bond acceptor, while the number of hydrogen bond donors was zero for all compounds, indicating limited potential for hydrogen bonding with biological targets. The number of bond rotations varied slightly: 9 bond rotations for compound 5 and 10 bond rotations for compounds 11 and 13. The variability in bond rotations may affect the molecular flexibility and interaction with biological systems.
Table 6.
Physicochemical parameters and medicinal chemistry parameters in silico analysis of the FQAS and QAS.
| Compound | Physicochemical parameters | Medicinal chemistry | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MW | nHA | nHD | nRot | TPSA (Å2) | LogS | LogP | LogD | Lipinsky rule | Veber rule | PAINS alert | BBB-penetrance | PPB % | |
| 1 | 457.3 | 1 | 0 | 7 | 0 | −0.033 | 3.894 | 3.003 | A | A | No | Yes | 87.43 |
| 2 | 569.2 | 1 | 0 | 7 | 0 | −0.596 | 3.847 | 3.217 | R | R | No | Yes | 87.32 |
| 3 | 491.8 | 1 | 0 | 8 | 0 | −1.47 | 4.204 | 3.470 | R | R | No | Yes | 90.89 |
| 4 | 583.2 | 1 | 0 | 8 | 0 | 0.1.048 | 4.221 | 3.295 | R | R | No | Yes | 89.80 |
| 5 | 577.7 | 1 | 0 | 9 | 0 | −3.711 | 4.870 | 4.307 | R | R | No | No | 95.72 |
| 6 | 669.2 | 1 | 0 | 9 | 0 | −3.413 | 4.899 | 4.177 | R | R | No | No | 94.91 |
| 7 | 557.3 | 1 | 0 | 9 | 0 | −2.618 | 4.931 | 4.141 | R | R | No | No | 5.32 |
| 8 | 577.8 | 1 | 0 | 9 | 0 | −4.142 | 5.024 | 4.385 | R | R | No | No | 95.88 |
| 9 | 669.2 | 1 | 0 | 9 | 0 | −3.965 | 5.049 | 4.242 | R | R | No | No | 95.07 |
| 10 | 557.3 | 1 | 0 | 9 | 0 | −3.632 | 5.073 | 4.141 | R | R | No | No | 95.45 |
| 11 | 683.3 | 1 | 0 | 10 | 0 | −3.827 | 5.277 | 4.156 | R | R | No | No | 95.45 |
| 12 | 591.8 | 1 | 0 | 10 | 0 | −4.373 | 5.415 | 4.344 | R | R | No | No | 96.37 |
| 13 | 683.2 | 1 | 0 | 10 | 0 | −4.221 | 5.424 | 4.220 | R | R | No | No | 95.63 |
| 14 | 571.3 | 1 | 0 | 10 | 0 | −4.054 | 5.445 | 4.160 | R | R | No | No | 96.02 |
| 15 | 406,1 | 1 | 0 | 7 | 0 | −0.027 | 3647 | 2935 | A | R | No | Yes | 85.23 |
| 16 | 420,1 | 1 | 0 | 8 | 0 | −0.248 | 3982 | 2992 | A | R | No | Yes | 88.09 |
| Accepted values | < 400 | 0–12 | 0–7 | 0–11 | 0–140 | −4.0 to 0.5 | 0–3 | 1–3 | — | — | No | No | < 90 |
Note: Lipinsky's and Veber's rules accepted values.
Abbreviations: A, accepted; BBB, blood–brain barrier; LogD, LogP at physiological pH 7,4; LogP, logarithm of the octanol/water partition coefficient; LogS, logarithm of the aqueous solubility; MW, molar weight; nHA, number of acceptor hydrogens; nHD, number of donor hydrogens; nRot, number of bonds; P, probably; PAINS, Pan-Assay INterference Substances; PPB, plasma protein binding; R, rejected; TSPA, topological polar surface area.
The calculated polar topological surface area (TPSA) for all compounds was 0.000 Å2, indicating that these molecules have no polar surface area, which may hinder interactions with polar solvents and reduce the likelihood of crossing biological barriers like the blood–brain barrier (BBB). The LogS values, which reflect the aqueous solubility of the compounds, ranged from −4.373 to 0.033, indicating that all FQASs and QASs are poorly soluble in polar solvents. The LogP values, representing the partition coefficient between octanol and water (log octanol/water), indicate that FQASs and QASs are highly apolar molecules. Their high solubility in organic solvents suggests poor solubility in aqueous environments. The values for LogD and LogP at pH 7.4 were above optimal levels, ranging from 3.003 to 4.385, suggesting that these compounds are more lipophilic than recommended for drug-like properties.
The pharmacological properties of FQASs 2-16 were evaluated based on Lipinski's Rule of Five and Veber's criteria. All compounds were rejected due to their molar mass exceeding 400 g/mol and LogP values greater than 4, indicating they may have poor bioavailability and permeability. QASs 9–14 were predicted not to cross the BBB, which suggests limited utility in targeting central nervous system (CNS) disorders. High plasma protein binding (PPB) values (indicating strong binding to plasma proteins) were reported for most compounds, except for FQASs 1 and 2, which had optimal PPB values < 90%, suggesting these two compounds may be better candidates for bioavailability.
The FQASs and QASs studied display high molecular weight (MW), poor aqueous solubility, and high lipophilicity, which leads to their rejection based on Lipinski's and Veber's rules for drug-likeness. While FQASs 9–14 cannot cross the BBB, they display high PPB values, whereas FQASs 1 and 2 have more favorable PPB properties. Despite their pharmacokinetic challenges, the lack of PAINS alerts suggests these compounds could be refined for specific, targeted applications.
Fluorine-containing compounds have emerged as a robust and practical tool in rational drug design, leveraging their advantages in potency and absorption, distribution, metabolism, excretion, and toxicity (ADMET) considerations [42]. Fluorine substitution is renowned for conferring exceptional electronic, physical, and biological attributes to organic molecules, making it a cornerstone in modern pharmaceuticals. This substitution often elevates pharmacological properties, including metabolic stability, membrane permeability, and receptor binding affinity. Incorporating fluorine into QASs holds promise for enhancing the potency and bioavailability of these compounds. Notably, approximately half of the top-selling drugs, or blockbuster drugs, incorporate fluorine-containing compounds. As a result, approximately half of the top-selling drugs, also known as blockbuster drugs, incorporate fluorine-containing compounds.
The in silico analyses provide information on physicochemical parameters, allowing an approach to some properties related to the potential pharmacology of the FQASs under study. Firstly, it was observed that all the FQASs evaluated are highly lipophilic and poorly soluble in polar solvents since, according to the maximum levels of bioavailability, they are out of the standard parameters that a drug should have to be administered orally. This observation is congruent with their chemical structure. Despite being QASs, their tail contains a long carbon chain and two aromatic rings, which make them more lipophilic. Furthermore, the trifluoromethyl (p-CF3 or m-CF3) or fluorine (m-F or p-F) groups located on both aromatic rings at the end of the chain are not sufficient to decrease these liposolubility values. It was observed that the MWs of these compounds range from 330 to almost 556.09 g/mol, indicating that they are heavy molecules compared to most of the compounds used orally. The TPSA value of 0.000 Å2 suggests minimal polar surface area, while the LogS values of −4.373 and 0.033 indicate poor aqueous solubility. These findings support the conclusion that FQASs are poorly soluble molecules in polar-type solvents. The LogP partition coefficient indicates that FQASs are highly apolar molecules with higher solubility in organic solvents. These align with the observed poor aqueous solubility and support the conclusion that FQASs are more soluble in nonpolar solvents. The values of LogD and LogP above the optimum for this parameter (ranging from 3.003 to 4.385) suggest that FQASs may have challenges in terms of distribution and permeability. The rejection of FQASs 2–16 based on their molar mass (> 400) and LogP (> 4) aligns with the Lipinski and Veber rules, which are commonly used guidelines for assessing the drug-likeness of compounds. FQASs 9–14 are not crossing the BBB but have high PPB values, which indicates potential challenges in terms of CNS penetration and distribution in the bloodstream. Lastly, the absence of alerts for PAINS suggests that FQASs do not exhibit problematic chemical interactions with various biological targets, which is a positive finding for their potential as drug candidates. These properties have proven to be a valid descriptor in many models and rules to quickly estimate absorption, distribution, metabolism, and excretion (ADME) properties, especially for crossing biological barriers such as absorption and accessing the brain or BBB [43].
The in vitro experiments provide a controlled environment to evaluate the efficacy and toxicity profiles of FQAS candidates, laying the foundation for selecting lead compounds for further preclinical and in vivo assessments. Moreover, in vitro assays enable screening a broad range of FQAS derivatives to identify potent candidates with promising antitumor properties, enhancing the efficiency of subsequent in vivo studies. Additionally, in vitro studies contribute to establishing structure–activity relationships and identifying lead compounds with favorable pharmacokinetic and pharmacodynamic profiles for subsequent in vivo efficacy and safety evaluations. Therefore, in vitro cytotoxicity and pharmacology data of FQAS against CRC cell lines presented in this work are promising.
Nonetheless, it is crucial to recognize that the study lacks comprehensive in vivo validation that would complement the existing in vitro findings and provide a more holistic understanding of the therapeutic potential of FQAS. The current study's limitation of in vivo data is mainly due to the initial approach: identifying lead molecules among a battery of compounds to preselect the most promising ones. This limitation will be addressed in future studies, where priority will be given to in vivo experiments to bridge the gap between preclinical findings and potential clinical applications of FQAS in treating CRC. The absence of comprehensive in vivo data currently limits the extrapolation of our findings to clinical settings, underscoring the preliminary nature of this study. Addressing this limitation through future research initiatives will strengthen the scientific basis of our work and provide a more complete understanding of the therapeutic mechanisms of FQAS.
3. Conclusions
The synthesized FQASs 9–14, particularly those containing CF3, show promising potential as antitumor agents for advanced stages of CRC, such as Dukes' Type C and Grade III. The selectivity of FQASs toward CRC tumor cells, without affecting PBMC, makes them attractive candidates for further development. Even though 5-FU is a widely used and effective drug for treating CRC, its long-term efficacy diminishes due to several limitations, mainly associated to drug resistance, variable patient response, and side effects. These limitations highlight the need for new compounds to overcome drug resistance, reduce toxicity, and improve treatment outcomes for CRC patients.
The FQASs 9–14 containing CF3 represent potential candidates for antitumor agents against CRC, particularly in advanced stages like Dukes' Type C and Grade III. The observed selectivity of FQASs toward tumor cells over normal tissue, without affecting PBMC, suggests the need for strategies facilitating direct tissue delivery while circumventing systemic circulation. Addressing potential challenges associated with utilizing FQASs as drug candidates may involve employing specific release formulations, such as enterically coated tablets or capsules resistant to gastric juices. Additionally, utilizing prodrugs activated by colonic enzymes or administering drugs directly into the colon via techniques like enemas could enhance efficacy. Targeted delivery systems, such as nanoparticles or functionalized microspheres, tailored to release the drug selectively in the colon while minimizing systemic absorption, present further avenues for exploration.
The synthesized FQASs demonstrate promising cytotoxicity and selectivity against CRC cells, positioning them as potential candidates for therapeutic development. The observed correlations between genotype and cellular phenotype suggest that FQASs could be tailored to create personalized treatments for specific subgroups of CRC patients. However, further studies are needed to optimize the structure of FQASs to enhance their pharmacological activity, improve their anticancer efficacy, and minimize side effects linked to chemical groups not involved in antitumor mechanisms. Additionally, advancing preclinical studies using relevant CRC models is crucial to comprehensively assess the efficacy and safety of FQASs in CRC treatment. Moreover, metabolomic research should focus on exploring the metabolic pathways modulated by FQASs in CRC cell lines (SW620, SW48, and SW948). This will provide deeper insights into the mechanisms of action and the accessory pathways contributing to the antitumor response of these compounds. Such studies hold promise for developing new treatment modalities, especially for CRC patients resistant to current therapies. The ability to tailor treatments based on individual cancer characteristics highlights the growing importance of precision medicine in improving patient outcomes.
4. Experimental
4.1. Chemistry
4.1.1. General Procedures for the Synthesis of FQASs and QASs
The QASs and FQASs were synthesized and prepared according to the well-known four-step synthetic sequence, which involves (a) in situ generation of trifluoromethyl- or fluorine-containing aryl magnesium Grignard reagents (either meta- or para-substituted) and their reaction with ethyl ω-bromoesters; (b) acid-catalyzed dehydration of the resulting tertiary α,α-bis-aryl-ω-bromoalcohols, giving rise to the respective ω-bromoolefins; (c) classical SN2 displacement of the bromide group by aqueous dimethylamine, to generate the respective tertiary amines; and (d) finally, reaction with either CH2I2 or ICH2Cl or CH3I in acetonitrile to produce iodide salts with -CH2I, -CH2Cl, or -CH3 groups respectively bound to the quaternary nitrogen [22, 38, 45].
The salts were crystallized from isopropanol–water binary mixtures and characterized by MS, 1H, and 13C NMR. Crystallographic analysis of representative QA's salts was performed, and signals are in accordance with our previous results reported elsewhere [22, 45].
The compounds were solubilized in dimethyl sulfoxide (DMSO, Fischer Chemical, NJ, USA) at 50,000 μg/mL and stored at −20°C until use. From the stock solution, six serial 1:2 double dilutions of each compound were prepared to start with concentrations of 100, 50, 25, 12.5, 6.25, and 3.125 μg/mL, respectively. 5-FU, one of the conventional drugs to treat CRC [34], was used as an antitumor compound control at concentrations ranging between 800 and 0.625 μM.
4.2. Pharmacological/Biological Assays
4.2.1. Cell Lines Phenotype and Culture Conditions
SW948 (Dukes' Type C, Grade III, premetastatic colorectal adenocarcinoma—CCL-237), SW48 (Dukes' Type C, Grade IV, noninvasive colorectal adenocarcinoma—CCL231), and SW620 (Dukes' Type C, Grade IV invasive colorectal adenocarcinoma—CCL227) (American Type Culture Collection Manassas, VA, USA) were maintained in standard conditions at 37°C, without a CO2 atmosphere. PBMCs and MCRT were used as nontumoral control cells. PBMCs were obtained by healthy donors, separated using Histopaque-1077 (Sigma-Aldrich) and cultured in RPMI-1640 medium (Sigma-Aldrich) supplemented with 10% SFB and 1% penicillin/streptomycin [46]. In turn, MCRT were obtained from healthy BALB/c mice; for this, mice were euthanized with CO2 after previous sedation, the colorectal tissue removed, cleaned up, and cut into 2 × 2 mm pieces, suspended in supplemented DMEM, and used immediately after collection. Both PBMC and MCRT were used as nontumoral control cells to assess the selective toxicity of FQASs. Moreover, the inclusion of PBMC also served to assess the systemic toxicity of FQAS.
4.2.2. Analysis of Genetic and Epigenetic Profiling of CRC Cell Lines
The genetic features of the CRC cell lines were analyzed using the information available in the ATCC database (https://www.atcc.org/products/tcp-1007) and the Taiwanese Igrcid cell bank (https://igrcid.ibms.sinica.edu.tw/cgi-bin/cell_line_view.cgi). The epigenetic profile in these cell lines was verified according to parameters related to MSS or MSI, CIN, and CIMP reported in the scientific literature [39, 47–49] and the Cellosaurus database (https://www.expasy.org).
4.2.3. In Vitro and Ex Vivo Cytotoxicity of FQASs and QASs
The cytotoxic activity of synthesized FQASs and QASs was studied by measuring cell viability/mortality, determined with the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazole bromide) method (VWR Chemicals and Reagents Avantor), upon exposure of each cell line to six serial dilutions of the different compounds independently [50]. Unexposed cells were included as viability controls, and cells exposed to 5-FU were included as cytotoxicity controls. Briefly, cells were adjusted to 200,000 cells/mL in the corresponding supplemented culture medium, and 100 μL (20,000 cells/well) were dispensed into each 96-well cell culture plate (Nunc, Thermo Fisher Scientific). Twelve hours after seeding the monolayer, 100 μL dilution of each compound was added to each well; the plates containing CRC were incubated at 37°C in the absence of CO2, while plates with PBMCs and MCRT were incubated at 37°C and 5% CO2 atmosphere. For MCRT, each piece was placed into each well of a 96-well plate and exposed to each dilution of the FQASs and QASs. After 48 h of incubation, 30 μL/well of MTT (5 mg/mL) was added, and plates were left at 37°C for 3 h, protected from light. After this time, 100 μL/well of DMSO was added to solubilize the formazan crystals formed. Plates were read at 570 nm on a Varioskan plate reader (Thermo Fisher), and nonspecific absorbance was corrected by subtracting the optical densities (OD) of the blank solutions corresponding to the supplemented culture media (RPMI 1640, DMEM, or L-15 media) and 0.5% DMSO. Each concentration of the FQASs, QASs, and controls was evaluated in duplicate in three independent experiments.
The percentages of viability (equation (1)) and mortality (equation (2)) were calculated with the net OD:
| (1) |
| (2) |
The %mortality data for each compound concentration was recorded, and the median cytotoxic concentration (CC50), corresponding to the concentration at which 50% of cell death occurs, was calculated using GraphPad Prism 8 software (GraphPad, Software, Inc., La Jolla, CA, USA). The cytotoxic activity of the evaluated FQASs and QASs was classified as high, moderate, or low according to the CC50 values obtained for compound 5-FU as follows: high cytotoxicity for CC50 values < 50 μM, moderate for CC50 values > 50 and < 150 μM, and low for values > 150 μM. Lastly, the index of selectivity (IS) was calculated by dividing the CC50 value obtained for control cells (MCRT and PBMCs) by the CC50 value obtained for each CRC cell phenotype using the formula: IS = CC50 (MCRT or PBMC) ÷ CC50 (SW948, SW48 or SW620).
4.2.4. In Silico Analysis of Physicochemical Properties and Pharmacology
The open-access multiparameter platform ADMETLab2.0 (https://admetmesh.scbdd.com) [51] was utilized to analyze the physicochemical and distribution parameters of FQASs and QASs, as well as their pharmacological properties according to the Lipinski et al. [52] and Veber et al. [53] rules. The physicochemical properties analysis included parameters such as MW, number of hydrogen donors (nHD), number of hydrogen acceptors (nHA), number of rotations (nRot), TPSA, logarithm of aqueous solubility (LogS), log octanol/water partition coefficient (LogP), and LogP at physiological pH 7.4 (LogD). Distribution parameters encompassed the assessment of BBB permeability and PPB. Lastly, the oral bioavailability of compounds was evaluated based on the compliance with Lipinski's rule of five (MW < 500, log p < 5, hydrogen bond donors < 5, and hydrogen bond acceptors < 10) [52] and Veber's rule (rotatable bonds [RTB] < 10 and TPSA < 140 Å2) [53].
4.2.5. Statistical Analysis
All experimental assays were conducted in duplicate across a minimum of three independent experiments. For comparisons between treatment groups, one- or two-way ANOVA with Tukey's or Dunnett's post hoc test, involving multiple treatments, was performed after confirming the normal distribution of the data calculated using GraphPad Prism 8 software (GraphPad, Software, Inc., La Jolla, CA, USA). Statistical significance was determined with a p value < 0.05 (∗), < 0.01 (∗∗), and < 0.001 (∗∗∗) considered as statistically significant.
Contributor Information
Luz Amalia Ríos-Vásquez, Email: amalia.rios@ucaldas.edu.co.
Sara M. Robledo, Email: sara.robledo@udea.edu.co.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Statement
The procedure for collecting venous blood from healthy donors was endorsed by the Bioethical Committee of the Universidad de Antioquia (Act 16-05-727), and the subjects provided informed consent. In addition, the obtention of MCRT was approved by the Animal Experimentation Ethics Committee of the University of Antioquia (Act 124, May 14, 2019), and all international animal welfare standards for animal experimentation were followed.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding
This work was supported by the Colombian Ministry of Science, Technology, and Innovation (Code 112777757135), the Caldas University (Grants 0421722 and 00H342), the Antioquia University (RR 51054, February 29, 2024), and the Innovation Corporation CIDEPRO.
References
- 1.Nguyen H., Duong H., Duong H., Duong H H. The Molecular Characteristics of Colorectal Cancer: Implications for Diagnosis and Therapy. Oncology Letters . 2018;16:p. 9. doi: 10.3892/ol.2018.8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huan L., Guo T., Wu Y., et al. Hypoxia Induced LUCAT1/PTBP1 Axis Modulates Cancer Cell Viability and Chemotherapy Response. Molecular Cancer . 2020;19:p. 1. doi: 10.1186/s12943-019-1122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Global Cancer Observatory. Colombia: GLOBOCAN 2020. International Agency for Research on Cancer . 2021;509:1–2. [Google Scholar]
- 4.Berretta M., Alessandrini L., De Divitiis C., et al. Serum and Tissue Markers in Colorectal Cancer: State of Art. Critical Reviews in Oncology . 2017;111:103–116. doi: 10.1016/j.critrevonc.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Inamura K. Colorectal Cancers: An Update on Their Molecular Pathology. Cancers . 2018;10(1):p. 26. doi: 10.3390/cancers10010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roa I., Sanchez T., Majlis A., Schalper K. KRAS Gene Mutation in Colorectal Cancer. Revista Medica de Chile . 2013;141(9):1166–1172. doi: 10.4067/S0034-98872013000900009. [DOI] [PubMed] [Google Scholar]
- 7.Soussi T., Wiman K. G. Shaping Genetic Alterations in Human Cancer: The P53 Mutation Paradigm. Cancer Cell . 2007;12(4):303–312. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Afanador C. H., Palacio K. A., Isaza L. F., Ahumada E., Ocampo C. M., Muñetón C. M. Caracterización Molecular de Pacientes con Cáncer Colorrectal. Biomedica . 2022;42:154–171. doi: 10.7705/biomedica.5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aldecoa F., Ávila J J. La vía Canónica PI3K/AKT/mTOR y sus Alteraciones en Cáncer. Horizons Medicaux . 2021;21:p. e1547. [Google Scholar]
- 10.Biller L. H., Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA . 2021;325(7):p. 669. doi: 10.1001/jama.2021.0106. [DOI] [PubMed] [Google Scholar]
- 11.Matsuda T., Yamashita K., Hasegawa H., et al. Recent Updates in the Surgical Treatment of Colorectal Cancer. Annals of Gastroenterological Surgery . 2018;2:129–136. doi: 10.1002/ags3.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf A. M. D., Fontham E. T. H., Church T. R., et al. Colorectal Cancer Screening for Average‐Risk Adults: 2018 Guideline Update From the American Cancer Society. CA: A Cancer Journal for Clinicians . 2018;68(4):250–281. doi: 10.3322/caac.21457. [DOI] [PubMed] [Google Scholar]
- 13.Granados-Romero J. J., Valderrama-Treviño A. I., Contreras-Flores E. H., et al. Colorectal Cancer: A Review. International Journal of Research in Medical Sciences . 2017;5(11):p. 4667. doi: 10.18203/2320-6012.ijrms20174914. [DOI] [Google Scholar]
- 14.Egorova K. S., Gordeev E. G., Ananikov V. P. Biological Activity of Ionic Liquids and Their Application in Pharmaceutics and Medicine. Chemical Reviews . 2017;117(10):7132–7189. doi: 10.1021/acs.chemrev.6b00562. [DOI] [PubMed] [Google Scholar]
- 15.Mavromoustakos T., Papadopoulos A., Theodoropoulou E., Dimitriou C., Antoniadou-Vyza E. Thermal Properties of Adamantanol Derivatives and Their β-Cyclodextrin Complexes in Phosphatidylcholine Bilayers. Life Sciences . 1998;62(20):1901–1910. doi: 10.1016/S0024-3205(98)00155-6. [DOI] [PubMed] [Google Scholar]
- 16.Shiraishi M., Aramaki Y., Seto M., et al. Discovery of Novel, Potent, and Selective Small-Molecule CCR5 Antagonists as Anti-HIV-1 Agents: Synthesis and Biological Evaluation of Anilide Derivatives With a Quaternary Ammonium Moiety. Journal of Medicinal Chemistry . 2000;43(10):2049–2063. doi: 10.1021/jm9906264. [DOI] [PubMed] [Google Scholar]
- 17.Rego P., Casas O., Sieiro C. M., Rodríguez L. A., Pombar Gómez A. Evaluación de la Eficacia de Detergentes a Base de Amonio Cuaternario Frente a Cepas de “Salmonella“ y “E. coli“ Aisladas de Alimentos. Revista de Tecnología e Higiene de los Alimentos . 2005;364:p. 114. [Google Scholar]
- 18.Ng C. K. L., Obando D., Widmer F., Wright L. C., Sorrell T. C., Jolliffe K. A. Correlation of Antifungal Activity With Fungal Phospholipase Inhibition Using a Series of Bisquaternary Ammonium Salts. Journal of Medicinal Chemistry . 2006;49(2):811–816. doi: 10.1021/jm0508843. [DOI] [PubMed] [Google Scholar]
- 19.Dias A. R., Costa-Rodrigues J., Fernandes M. H., Ferraz R., Prudêncio C. The Anticancer Potential of Ionic Liquids. ChemMedChem . 2017;12(1):11–18. doi: 10.1002/cmdc.201600480. [DOI] [PubMed] [Google Scholar]
- 20.Mavromoustakos T., Calogeropoulou T., Koufaki M., et al. Ether Phospholipid-AZT Conjugates Possessing Anti-HIV and Antitumor Cell Activity. Synthesis, Conformational Analysis, and Study of Their Thermal Effects on Membrane Bilayers. Journal of Medicinal Chemistry . 2001;44(11):1702–1709. doi: 10.1021/jm001121c. [DOI] [PubMed] [Google Scholar]
- 21.Duque-Benítez S. M., Ríos-Vásquez L. A., Ocampo-Cardona R., et al. Synthesis of Novel Quaternary Ammonium Salts and Their In Vitro Antileishmanial Activity and U-937 Cell Cytotoxicity. Molecules . 2016;21(4):p. 381. doi: 10.3390/molecules21040381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López-Muñoz M., Gomez-Peña J. J., Ríos-Vásquez L. A., et al. Novel Fluorinated Quaternary Ammonium Salts and Their In Vitro Activity as Trypanocidal Agents. Medicinal Chemistry Research . 2019;28(3):300–319. doi: 10.1007/s00044-018-02285-2. [DOI] [Google Scholar]
- 23.Smart B. E., Fluor J. Fluorine Substituent Effects (on Bioactivity) Chem . 2001;109:p. 3. doi: 10.1016/S0022-1139(01)00375-X. [DOI] [Google Scholar]
- 24.López S. E. Grupo Pentafluorosulfanilo (-SF5): ¿Un Nuevo Bioisóstero del Grupo Trifluorometilo (-CF3)? Revista Facultad de Farmacia . 2014;77:p. 54. [Google Scholar]
- 25.Gillis E. P., Eastman K. J., Hill M. D., Donnelly D. J., Meanwell N. A. Applications of Fluorine in Medicinal Chemistry. Journal of Medicinal Chemistry . 2015;58(21):8315–8359. doi: 10.1021/acs.jmedchem.5b00258. [DOI] [PubMed] [Google Scholar]
- 26.López S. E., Romero A. Grupo Trifluorometilo: Un Sustituyente Importante en Química Medicinal. Revista Facultad de Farmacia . 2011;74:p. 40. [Google Scholar]
- 27.Li Q., Li Q., Tan W., Zhang J., Guo Z. Phenolic-Containing Chitosan Quaternary Ammonium Derivatives and Their Significantly Enhanced Antioxidant and Antitumor Properties. Carbohydrate Research . 2020;498:p. 108169. doi: 10.1016/j.carres.2020.108169. [DOI] [PubMed] [Google Scholar]
- 28.Kopiasz R. J., Tomaszewski W., Kuźmińska A., et al. Hydrophilic Quaternary Ammonium Ionenes—Is There an Influence of Backbone Flexibility and Topology on Antibacterial Properties? Macromolecular Bioscience . 2020;20(7):p. e2000063. doi: 10.1002/mabi.202000063. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L., Feng X. Z., Xiao Z. Q., et al. Design, Synthesis, Antibacterial, Antifungal and Anticancer Evaluations of Novel β-Pinene Quaternary Ammonium Salts. International Journal of Molecular Sciences . 2021;22(20):p. 11299. doi: 10.3390/ijms222011299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Chen H., Bao H., Zhang J., Wu R., Zhu L. Comprehensive Characterization of FBXW7 Mutational and Clinicopathological Profiles in Human Colorectal Cancers. Frontiers Oncology . 2023;13:p. 1154432. doi: 10.3389/fonc.2023.1154432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X. L., Zhou J., Chen Z. R., Chng W. J. P53 Mutations in Colorectal Cancer—Molecular Pathogenesis and Pharmacological Reactivation. World Journal of Gastroenterology . 2015;21(1):84–93. doi: 10.3748/wjg.v21.i1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivlin N., Brosh R., Oren M., Rotter V. Mutations in the P53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer . 2011;2(4):466–474. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L., Shay J. W. Multiple Roles of APC and its Therapeutic Implications in Colorectal Cancer. Journal of the National Cancer Institute . 2017;109(8):p. djw332. doi: 10.1093/jnci/djw332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longley D. B., Harkin D. P., Johnston P. G. 5-Fluorouracil: Mechanisms of Action and Clinical Strategies. Nature Reviews Cancer . 2003;3(5):330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 35.Zhang B., Halder S. K., Kashikar N. D., et al. Antimetastatic Role of Smad4 Signaling in Colorectal Cancer. Gastroenterology . 2010;138(3):969–980.e3. doi: 10.1053/j.gastro.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karczmarski J., Goryca K., Pachlewski J., et al. Mutation Profiling of Premalignant Colorectal Neoplasia. Gastroenterology Research and Practice . 2019:p. 2542640. doi: 10.1155/2019/2542640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawashita Y., Morine Y., Ikemoto T., et al. Loss of Fbxw7 Expression Is a Predictor of Recurrence in Colorectal Liver Metastasis. Journal of Hepato-Biliary-Pancreatic Sciences . 2017;24(10):576–583. doi: 10.1002/jhbp.500. [DOI] [PubMed] [Google Scholar]
- 38.Fernandez M., Murillo J., Ríos-Vásquez L. A., et al. In Vivo Studies of the Effectiveness of Novel N-Halomethylated and Non-Halomethylated Quaternary Ammonium Salts in the Topical Treatment of Cutaneous Leishmaniasis. Parasitology Research . 2018;117(1):273–286. doi: 10.1007/s00436-017-5702-9. [DOI] [PubMed] [Google Scholar]
- 39.Weinstein J. N., Collisson E. A., Mills G. B., et al. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nature Genetics . 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goyle S., Maraveyas A. Chemotherapy for Colorectal Cancer. Digestive Surgery . 2006;22(6):401–414. doi: 10.1159/000091441. [DOI] [PubMed] [Google Scholar]
- 41.Diaz-Vegas A., Sanchez-Aguilera P., Krycer J. R., et al. Is Mitochondrial Dysfunction a Common Root of Noncommunicable Chronic Diseases? Endocrine Reviews . 2020;41(3):p. bnaa005. doi: 10.1210/endrev/bnaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheikhi N., Bahraminejad M., Saeedi M., Mirfazli S. S. A Review: FDA-Approved Fluorine-Containing Small Molecules from 2015 to 2022. European Journal of Medicinal Chemistry . 2023;260:p. 115758. doi: 10.1016/j.ejmech.2023.115758. [DOI] [PubMed] [Google Scholar]
- 43.Zhu L., Zhao J., Zhang Y., et al. ADME Properties Evaluation in Drug Discovery: In Silico Prediction of Blood–Brain Partitioning. Molecular Diversity . 2018;22(4):979–990. doi: 10.1007/s11030-018-9866-8. [DOI] [PubMed] [Google Scholar]
- 44.Ahmed D., Eide P. W., Eilertsen I. A., et al. Epigenetic and Genetic Features of 24 Colon Cancer Cell Lines. Oncogenesis . 2013;2(9):p. e71. doi: 10.1038/oncsis.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molina D. A., Ríos-Vásquez L. A., Toscano R. A., Ocampo-Cardona R., Gomez-Peña J. J., Macías M. A. Synthesis and Crystallographic Study of Six Quaternary Salts of N-Halomethylated and Non-N-Halomethylated Ammonium: Importance of C-H…X (X = F, I) and I…I− Halogen Interactions in the Supramolecular Structures. Journal of Molecular Structure . 2022;1271:p. 133962. doi: 10.1016/j.molstruc.2022.133962. [DOI] [Google Scholar]
- 46.Montoya A., Daza A., Muñoz D., et al. Development of a Novel Formulation with Hypericin to Treat Cutaneous Leishmaniasis Based on Photodynamic Therapy in In Vitro and In Vivo Studies. Antimicrobial Agents and Chemotherapy . 2015;59(9):5804–5813. doi: 10.1128/AAC.00545-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Armaghany T., Wilson J. D., Chu Q., Mills G. Genetic Alterations in Colorectal Cancer. Gastrointestinal Cancer Research . 2012;5:19–27. [PMC free article] [PubMed] [Google Scholar]
- 48.Baharudin R., Ab Mutalib N. S., Othman S. N., et al. Identification of Predictive DNA Methylation Biomarkers for Chemotherapy Response in Colorectal Cancer. Frontiers in Pharmacology . 2017;8:p. 47. doi: 10.3389/fphar.2017.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Setaffy L., Langner C. Microsatellite Instability in Colorectal Cancer: Clinicopathological Significance. Polish Journal of Pathology . 2015;66:203–218. doi: 10.5114/pjp.2015.54953. [DOI] [PubMed] [Google Scholar]
- 50.Niks M., Otto M. Towards an Optimized MTT Assay. Journal of Immunological Methods . 1990;130(1):149–151. doi: 10.1016/0022-1759(90)90309-j. [DOI] [PubMed] [Google Scholar]
- 51.Xiong G., Wu Z., Yi J., et al. ADMETlab 2.0: An Integrated Online Platform for Accurate and Comprehensive Predictions of ADMET Properties. Nucleic Acids Research . 2021;49(W1):W5–W14. doi: 10.1093/nar/gkab255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings 1PII of Original Article: S0169-409X(96)00423-1. The Article Was Originally Published in Advanced Drug Delivery Reviews 23 (1997) 3–25. 1. Advanced Drug Delivery Reviews . 2001;46(1-3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 53.Veber D. F., Johnson S. R., Cheng H. Y., Smith B. R., Ward K. W., Kopple K. D. Molecular Properties that Influence the Oral Bioavailability of Drug Candidates. Journal of Medicinal Chemistry . 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


