Abstract
Objectives
To develop and validate a real-time PCR assay detecting the sequence bridging Tn1549 and the Enterococcus faecium chromosome in the emerging vanB vancomycin-resistant E. faecium (VREfm) clone (ST80/CT2406).
Methods
The Tn1549 insertion site was determined on routinely sequenced VREfm isolates. The outer boundaries of Tn1549 and adjoining host bacterial sequences were determined using a BLAST search in the silent information regulator gene sir2. Next, the primers and probe were developed, targeting the sequence bridging Tn1549 and the E. faecium chromosome. Finally, the PCR assay was validated on well-characterized strains and prospectively performed on rectal screening samples submitted to our laboratory.
Results and conclusions
The PCR assay proved to be accurate and provide rapid diagnosis of the emerging vanB VREfm in rectal screening samples.
Introduction
Vancomycin-resistant Enterococcus faecium (VREfm) is a major cause of nosocomial infections. The vanA and vanB genotypes are the most prevalent among clinical isolates of E. faecium.1 The vanB gene cluster is carried by a 34 kb conjugative Tn1549 transposon and most often integrated into the chromosome.1 vanA/vanB PCR performed on rectal screening samples is accurate in detection of vanA VREfm; however, the positive predictive value for vanB in faecal screening samples is low, because of the presence of vanB in non-enterococcal gut bacteria. Therefore, vanB VREfm diagnosis on rectal screening samples is challenging as definite identification of vanB VREfm depends on culture (2–3 days) compared with PCR (6 h). vanA was initially dominant in the Capital Region of Denmark (2012–19); however, in 2018 a vanB VREfm clone [ST117, cgMLST complex type (CT) 36] was introduced to the region.2,3 Tn1549 was found to be inserted in an L-arabinose isomerase (araA2) gene.3,4 A specific PCR was developed to detect the sequence bridging Tn1549 and araA2 (araA2/Tn1549 PCR) allowing rapid and accurate detection of this vanB VREfm clone.4 In 2020–22, a shift in the dominating vanB clone was observed. The emerging vanB clone was ST80, cgMLST CT2406, which has a different insertion site for the Tn1549.3
The aim of the present study was to develop a real-time PCR assay detecting the sequence bridging Tn1549 and the E. faecium chromosome in the emerging vanB VREfm clone (ST80/CT2406). In addition, the PCR assay was to be validated on well-characterized strains and prospectively on rectal screening samples. VREfm are routinely whole-genome sequenced and used for validation.
Materials and methods
Determination of Tn1549 insertion site in the emerging vanB VREfm clone
VREfm is routinely sequenced at the Department of Clinical Microbiology (DCM), Hvidovre Hospital and the Tn1549 insertion site in the emerging vanB VREfm cluster (ST80/CT2406) was determined in contigs encoding Tn1549. A BLAST search of the Tn1549 complete sequence (AF_192329.1) was used to identify contigs. Most often, the Tn1549 sequence was identified in one contig. The outer boundaries of Tn1549 were identified and adjoining host bacterial sequences were determined. The adjoining host bacterial sequences were used for a BLAST search against the NCBI nr/nt database to determine the gene in which Tn1549 was inserted. BLAST revealed this to be a silent information regulator gene (sir2).
Primer and probe design
The target sequence for the sir2/Tn1549 PCR assay was selected with a forward primer in sir2 (TGTCAGTTCATAATTATATCTCTCTTGATCA), a reverse primer in Tn1549 (GGCTATACCGACATTCAAGAACTTC) and a TaqMan probe (LC640- TCCTCAGAATCGACAAAATTTTCCT-BBQ), amplifying a 121 bp fragment (Table S1, available as Supplementary data at JAC-AMR Online). The sir2/Tn1549 PCR assay was multiplexed to the existing araA2/Tn1549 TaqMan assay on the Roche FLOW System (Roche Diagnostics, Basel, Switzerland) that identified the ST117/CT36 clone.4
To determine the analytical specificity, a BLAST search against the NCBI nr/nt database (highly similar sequences) was performed for the 121 bp amplicon to ensure that this sequence was only found in E. faecium.
Validation of the PCR assay
The PCR assay was validated on 75 E. faecium isolates. Five isolates were vancomycin-susceptible E. faecium from clinical samples, and 70 isolates were VREfm well characterized by WGS. They either had Tn1549 integrated into sir2 or an intact sir2, as predicted by genome analysis. The validation of the sir2/Tn1549 PCR is summarized in Table 1. We found no false-positive and no false-negative isolates.
Table 1.
WGS results and PCR results for all isolates used in the validation of the multiplex PCR
| WGS |
sir2/Tn1549 PCR n detected/ n not detected |
|---|---|
| Vancomycin susceptible | 0/5 |
| vanA | 0/33 |
| vanB (non-sir2) | 0/4 |
| vanA + vanB | 0/1 |
| vanB + araA2/Tn1549 | 0/6 |
| vanB + sir2/Tn1549 | 26/0 |
In silico PCRs were performed for the following targets: vanA, vanB, araA2/Tn1549 and sir2/Tn1549; in silico PCR detected targets are listed in the first column.
In addition, the PCR assay was performed on 1393 rectal screening samples submitted for VREfm screening to the DCM, Hvidovre Hospital, from 10 March 2022 to 24 October 2022, from the Southern part of the Capital Region of Denmark (approximately 1.0 million inhabitants). When receiving VREfm rectal screening samples, PCR assays for vanA/vanB in addition to sir2/Tn1549 and araA2/Tn1549 were performed. All positive samples (vanA, vanB, araA2/Tn1549 or sir2/Tn1549) were cultured and compared with WGS data. Routinely, a minimum of one VREfm isolate per patient per year was whole-genome sequenced. In silico PCR for vanB, araA2/Tn1549 and sir2/Tn1549 was performed on WGS data for the validation.
Culture and identification of VREfm
First, the sample was inoculated into a selective brain heart infusion (BHI) broth, containing 4 mg/L vancomycin and 60 mg/L aztreonam. On the next day, 10 µL of the enrichment broth was inoculated on a selective chromogenic agar (bioMérieux, Marcy-L’Étoile, France) for 48 h. If E. faecium grew on the agar (purple colonies) the species identity was confirmed using MALDI-TOF.
WGS and in silico PCR
WGS was performed locally at the DCM, Hvidovre Hospital. DNA was purified using the Nucleic Acid Extraction Kit For Bacteria Genomic DNA Extraction (#738, PentaBase, Denmark). DNA libraries were prepared on a Biomek 4000 (Bechman Coulter, IN, USA) with the Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA, USA). Shotgun sequencing was performed on a MiSeq instrument (Illumina Inc.) using MiSeq® Reagent Kit v2 (300 cycle) (Illumina, cat. no. 15033412), generating 2 × 150 bp paired-end reads. 5,6 Sequencing reads were de novo assembled using SKESA v. 2.2 with default settings except inclusion of the parameter –allow_snps.7 MLST and cgMLST were performed in SeqSphere v.8.2.0 (Ridom GmbH, Münster, Germany).8 All sequences were visualized on a minimum spanning tree (MST) with the setting of a maximum of 20 alleles between clusters.
In silico PCR was performed to detect the PCR amplicon in WGS data. A script performed a BLAST search of the genome with the sequences of the primers and probe with a perfect (100%) match. The script identified all matches where the forward and reverse primers were on the same contig with the direction pointing towards each other, and where the probe matched a sequence between the two primers. The script only identified amplicons with a perfect match (100%) to both primers and probe. Finally, the size of the amplicon was determined.
Ethics
This project was approved by the Danish Data Protection Agency (P-2022-653) and the Danish Health and Medicines Authority (3-3013-1118/1).
Results
Primer and probe design
A sequence of 151 bp bridging Tn1549 and the E. faecium chromosome in isolates belonging to the dominant vanB VREfm cluster was identified for primer and probe design. The PCR design resulted in a 121 bp amplicon (Figure S1). The analytical specificity of the sir2/Tn1549 PCR assay was examined. The 121 bp amplicon was identified in 10 E. faecium isolates (100% query cover and 100% identity). The remaining sequences identified by BLAST had a query cover of ≤66%, showing that the sir2/Tn1549 PCR assay did not have any significant hits in gut microbes.
Validation and application of the specific sir2/Tn1549 PCR assay
Twenty-six isolates with Tn1549 integrated into the sir2 gene were correctly identified. In addition, 49 isolates carrying an intact sir2 gene were negative in the PCR assay. Next, the PCR was performed prospectively on 1393 rectal screening samples. Of these samples, 141 (10.1%) were sir2/Tn1549 positive. All positive samples were cultured, and 129 (91.5%) were culturable. Of the non-culturable samples (n = 12), five patients had a culturable sir2/Tn1549 VREfm sample within 1 week, and two patients had a culturable sir2/Tn1549 VREfm sample 2 months prior to the present sample. Three samples had a high Cq value (the number of PCR cycles until threshold) of 37–39, and two isolates had Cq values of 30 for both vanB and sir2/Tn1549. Next, we looked at the WGS results and identified 91 of the 129 cultured sir2/Tn1549 isolates (70.5%). In all these sequences, in silico PCR identified the 121 bp amplicon of the sir2/Tn1549 PCR assay. The results from PCR and in silico PCR for each WGS isolate are shown in Table S2.
Finally, we searched through our collection of WGS isolates to identify vanB VREfm from rectal screening samples during the study period that were negative in the sir2/Tn1549 PCR assay. Sixteen isolates were identified, of which 15 had an intact sir2 gene (true negatives). One isolate had Tn1549 integrated in sir2 (6.3% of negatives). This isolate had a high Cq value of 40 for vanB, indicating a low number of VREfm in the sample.
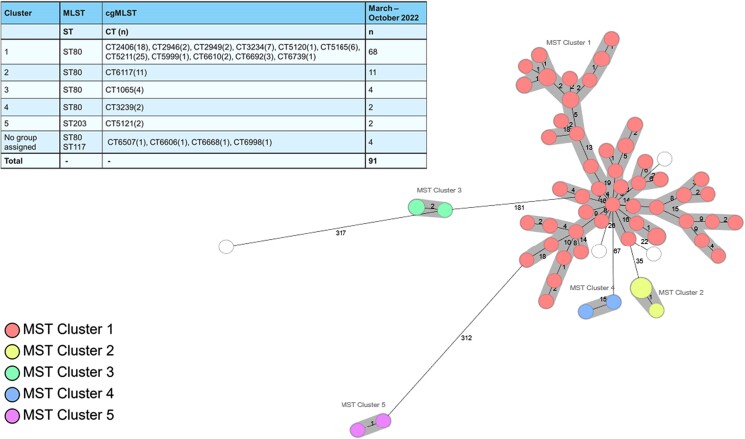
As shown on the MST, most of the isolates belonged to the ST80/CT2406 cluster (74.7%) (Figure 1). Four additional clusters and four singletons (ST80, ST203 and ST117) were identified, indicating that the PCR assay also identified vanB clusters other than the dominant one.
Figure 1.
MST of 91 isolates made with SeqSphere v.8.2.0 (Ridom GmbH, Münster, Germany). The table shows the MLST and the cgMLST for each cluster.
Discussion
The sir2/Tn1549 PCR assay was developed, validated and implemented in a multiplex PCR assay combined with the araA2/Tn1549 PCR assay in the routine laboratory at the DCM, Hvidovre Hospital.4 Compared with WGS of all vanB VREfm identified during the study period, all sir2/Tn1549 isolates were identified by the PCR assay with the exception of one isolate. This isolate had the Tn1549 in the sir2 gene on WGS results but had a high Cq value of 40 for the vanB gene, which can explain the negative outcome of the sir2/Tn1549 PCR. Twelve patients had a sir2/Tn1549-positive sample that could not be cultured; however, the majority (n = 7; 58%) of these patients had a culturable VREfm sample prior to this sample. The remaining five samples showed high Cq values. Most likely, this represents higher sensitivity of the PCR assay compared with culture rather than false-positive results of the sir2/Tn1549 PCR assay.
The PCR assay provides rapid and accurate vanB VREfm diagnosis with direct patient-related consequences. Isolation precautions can be implemented or withdrawn in less than 12 hours after admission to hospital with our multiplex PCR. If conventional culture-based methods are used instead, the diagnosis takes 2–3 days. In the Capital Region of Denmark, VREfm clonal shifts have been observed multiple times.2–6 It was also demonstrated by Zhou and colleagues in 20189 that Tn1549 can be located in different insertion sites. Therefore, it is important to perform and survey VREfm through WGS data to ensure that new emerging vanB clones do not escape the very specific PCR assays.
In conclusion, we here report a vanB sir2/Tn1549 PCR assay that has been thoroughly validated and is currently part of our routine VREfm diagnostic pathway to ensure rapid and accurate VREfm diagnostic on rectal screening samples. We continuously survey our VREfm WGS data for new emergent VREfm and are ready to develop new specific vanB PCR assays.
Supplementary Material
Acknowledgements
Part of the data was presented at the 33rd European Congress of Clinical Microbiology and Infectious Diseases (Poster 737). We thank the WGS staff at the DCM, Hvidovre for excellent technical assistance.
Contributor Information
Maja Johanne Søndergaard Knudsen, Department of Clinical Microbiology, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark.
Christel Barker Jensen, Department of Clinical Microbiology, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark.
Rikke Lind Jørgensen, Department of Clinical Microbiology, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark.
Andreas Munk Petersen, Department of Clinical Microbiology, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark; Gastrounit, Medical Division, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark; Department of Clinical Medicine, University of Copenhagen, Copenhagen, Denmark.
Gitte Qvist Kristiansen, Department of Clinical Microbiology, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark.
Jan Gorm Lisby, Department of Clinical Microbiology, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark.
Peder Worning, Department of Clinical Microbiology, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark.
Henrik Westh, Department of Clinical Microbiology, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark; Department of Clinical Medicine, University of Copenhagen, Copenhagen, Denmark.
Mette Pinholt, Department of Clinical Microbiology, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark.
Funding
This work was supported by the Scandinavian Society for Antimicrobial Chemotherapy (SSAC) Foundation, the Regionernes Medicin- og Behandlingspulje, the Danmarks Frie Forskningsfond and internal funding.
Transparency declarations
None to declare.
Data availability
Data are uploaded in BioProject PRJNA1023182.
Supplementary data
Figure S1 and Tables S1 and S2 are available as Supplementary data at JAC-AMR Online.
References
- 1. Werner G, Coque TM, Hammerum AM et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill 2008; 13: 19046. 10.2807/ese.13.47.19046-en [DOI] [PubMed] [Google Scholar]
- 2. Hammerum AM, Baig S, Kamel Y et al. Emergence of vanA Enterococcus faecium in Denmark, 2005–15. J Antimicrob Chemother 2017; 72: 2184–90. 10.1093/jac/dkx138 [DOI] [PubMed] [Google Scholar]
- 3. DANMAP . DANMAP 2021. Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark. 2021. https://www.danmap.org/reports/2021
- 4. Pinholt M, Mollerup S, Boye K et al. Investigation of the introduction and dissemination of vanB Enterococcus faecium in the Capital Region of Denmark and development of a rapid and accurate clone-specific vanB E. faecium PCR. J Antimicrob Chemother 2021; 76: 2260–7. 10.1093/jac/dkab198 [DOI] [PubMed] [Google Scholar]
- 5. Pinholt M, Gumpert H, Bayliss S et al. Genomic analysis of 495 vancomycin-resistant Enterococcus faecium reveals broad dissemination of a vanA plasmid in more than 19 clones from Copenhagen, Denmark. J Antimicrob Chemother 2017; 72: 40–7. 10.1093/jac/dkw360 [DOI] [PubMed] [Google Scholar]
- 6. Pinholt M, Bayliss SC, Gumpert H et al. WGS of 1058 Enterococcus faecium from Copenhagen, Denmark, reveals rapid clonal expansion of vancomycin-resistant clone ST80 combined with widespread dissemination of a vanA-containing plasmid and acquisition of a heterogeneous accessory genome. J Antimicrob Chemother 2019; 74: 1776–85. 10.1093/jac/dkz118 [DOI] [PubMed] [Google Scholar]
- 7. Souvorov A, Agarwala R, Lipman DJ. SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol 2018; 19: 153. 10.1186/s13059-018-1540-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Been M, Pinholt M, Top J et al. Core genome multilocus sequence typing scheme for high-resolution typing of Enterococcus faecium. J Clin Microbiol 2015; 53: 3788–97. 10.1128/JCM.01946-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou X, Chlebowicz MA, Bathoorn E et al. Elucidating vancomycin-resistant Enterococcus faecium outbreaks: the role of clonal spread and movement of mobile genetic elements. J Antimicrob Chemother 2018; 73: 3259–67. 10.1093/jac/dky349 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are uploaded in BioProject PRJNA1023182.