Abstract
Microsatellite unstable colorectal cancer (MSI-CRC) can arise through germline mutations in mismatch repair (MMR) genes in individuals with Lynch syndrome (LS), or sporadically through promoter methylation of the MMR gene MLH1. Despite the different origins of hereditary and sporadic MSI tumours, their genomic features have not been extensively compared. A prominent feature of MMR-deficient genomes is the occurrence of many indels in short repeat sequences, an understudied mutation type due to the technical challenges of variant calling in these regions. In this study, we performed whole genome sequencing and RNA-sequencing on 29 sporadic and 14 hereditary MSI-CRCs. We compared the tumour groups by analysing genome-wide mutation densities, microsatellite repeat indels, recurrent protein-coding variants, signatures of single base, doublet base, and indel mutations, and changes in gene expression. We show that the mutational landscapes of hereditary and sporadic MSI-CRCs, including mutational signatures and mutation densities genome-wide and in microsatellites, are highly similar. Only a low number of differentially expressed genes were found, enriched to interferon-γ regulated immune response pathways. Analysis of the variance in allelic fractions of somatic variants in each tumour group revealed higher clonal heterogeneity in sporadic MSI-CRCs. Our results suggest that the differing molecular origins of MMR deficiency in hereditary and sporadic MSI-CRCs do not result in substantial differences in the mutational landscapes of these tumours. The divergent patterns of clonal evolution between the tumour groups may have clinical implications, as high clonal heterogeneity has been associated with decreased tumour immunosurveillance and reduced responsiveness to immunotherapy.
Keywords: colorectal cancer, microsatellite instability, Lynch syndrome, whole genome sequencing, RNA sequencing
Introduction
Mismatch repair (MMR) is a highly active DNA repair mechanism that is particularly crucial during DNA replication to ensure fidelity. The MMR proteins target for repair any base–base mismatches and insertion–deletion loops that persist after proofreading by DNA polymerases [1]. MMR plays an important role in protecting the cell from accumulating potentially cancer-causing mutations. Approximately 15% of colorectal cancers (CRC) are characterised by deficient MMR (dMMR). The majority of these dMMR tumours, about 80%, occur sporadically, while the remaining 20% are hereditary tumours found in Lynch Syndrome (LS) patients [2].
Deficient MMR in tumour cells manifests as an order of magnitude increase in the rate of somatic mutations compared to the vast majority of MMR-proficient tumours [3]. Microsatellite repeats are particularly prone to accumulating mutations in the form of insertions or deletions in these short repetitive tracts. Indeed, dMMR has traditionally been diagnosed by identifying mutations in a panel of microsatellite repeat markers [2]. Consequently, dMMR is often equated with microsatellite instability (MSI) as a distinction from microsatellite stable (MSS) tumours.
LS is an autosomal dominantly inherited disease manifesting in a high rate of epithelial tumorigenesis, especially in the gastrointestinal and urinary tract and, for women, in the endometrium and ovary [4, 5]. The cancer risk is caused by pathogenic germline mutations causing loss-of-function in one of the genes involved with DNA mismatch repair [6]. The key MMR genes mutated in LS patients are MLH1 and MSH2, followed by MSH6 and PMS2 [7]. The inherited mutations are heterozygous and thus a second hit inactivating the remaining allele, typically a somatic mutation or promoter hypermethylation, is required for the emergence of dMMR tumour cells [2, 8]. By contrast, in sporadic MSI-CRCs, MLH1 expression is epigenetically silenced via bi-allelic promoter hypermethylation. We hypothesised that the differing origins of the hereditary and sporadic MSI-CRCs could result in differences in molecular characteristics of the tumours.
The objective of this investigation was to perform a comprehensive genomic and transcriptomic characterisation of sporadic and LS-associated MSI-CRCs. To this end, we performed whole genome sequencing and RNA-sequencing of sporadic and LS-associated MSI cancers representing in total 43 tumours: 14 MLH1 germline mutation carriers and 29 sporadic CRC patients.
Results
Study outline
We performed whole genome sequencing (WGS) and RNA-sequencing of MSI tumours from 14 hereditary MLH1-defective LS and 29 sporadic CRC patients and, where available, paired non-neoplastic colorectal tissues. (Table 1, Fig. 1A). For tumours with paired normal tissue, somatic mutations were detected using two main approaches. First, a GATK4 variant calling pipeline with Mutect2 [9, 10] was utilised to produce a catalogue of somatic single nucleotide variants (SNVs) and small insertion–deletion mutations (indels) in the tumours for comparisons between the hereditary and sporadic cancer groups. These calls were utilised in the analysis of protein-coding gene mutations and somatic mutation signatures across the genome. Second, genotyping of tandem repeats, that are difficult to call using standard tools, was performed using the GangSTR tool [11], followed by quality filtering and calling of the somatic changes in tumours compared to the paired normal colorectal DNA. The indel counts from tandem repeat regions with different repeat unit lengths were compared to the Mutect2 calls and between the LS and sporadic cancer groups, as well as annotated according to their genomic region. For the latter, untranslated regions (UTRs) were subjected to a more detailed analysis due to observed differences in mutation counts between the tumour groups. Gene expression changes between the cancer groups were analysed from the RNA-sequencing data using principal component analysis, clustering, and differential gene expression analysis.
Table 1.
Clinical features of 14 LS and 29 sporadic MSI-CRCs.
| n (%) | |||
|---|---|---|---|
| Hereditary | Sporadic | ||
| Total | 14 | 29 | |
| Gender | Male | 6 (42.9%) | 8 (27.6%) |
| Female | 8 (57.1%) | 21 (72.4%) | |
| Dukes | A | 3 (21.4%) | 6 (20.7%) |
| B | 4 (28.6%) | 13 (44.8%) | |
| C | 6 (42.9%) | 9 (31%) | |
| D | 1 (7.1%) | 1 (3.4%) | |
| Grade | I | 0 | 5 (17.2%) |
| II | 11 (78.6%) | 18 (62.1%) | |
| III | 3 (21.4%) | 3 (10.3%) | |
| IV | 0 | 1 (3.4%) | |
| Primary location | Colon | 13 (92.9%) | 25 (86.2%) |
| Rectum | 1 (7.1%) | 4 (13.8%) | |
| Distal/Proximal | Distal | 6 (42.9%) | 7 (24.1%) |
| Proximal | 8 (57.1%) | 22 (75.9%) | |
| Median | |||
| Age | Median | 41.5 [27–75] | 74 [41–88] |
| RIN | Median | 6.65 [5.1–8.9] | 7.5 [5–9.6] |
Figure 1.
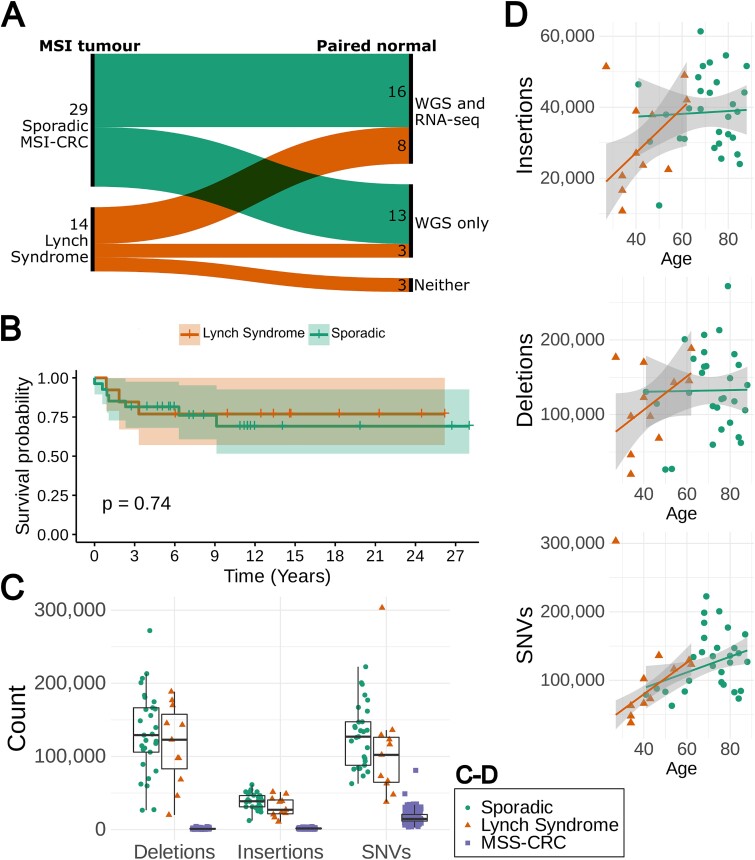
(A) Sankey diagram showing the 29 sporadic and 14 LS MSI-CRCs and the number of each that had paired normal data available for both WGS and RNA-seq, only for WGS, or for neither analysis. (B) Kaplan–Meier curve of CRC-specific survival in 27 sporadic MSI and 13 LS patients for whom up-to-date cause of death information was available or are still living. Time is shown in years. (C) The total numbers of deletions, insertions and SNVs in 11 LS and 29 sporadic MSI-CRCs compared to 221 MSS-CRCs [12]. (D) The number of variants from WGS data against the age of the patients in 11 LS and 29 sporadic MSI-CRCs with a robust linear regression model fitted.
Clinical characteristics
As has been previously observed for MSI-CRCs [13, 14], most patients in both tumour groups were female and most tumours were proximally located, particularly for the sporadic cases (Table 1). As expected, the 14 LS patients typically had a much younger age of diagnosis than the 29 sporadic CRC patients [15, 16] (Welch’s t-test P = 1.5 × 10−5, median 41.5 and 74 respectively). There was no significant difference between sporadic and hereditary tumours regarding the other available clinical characteristics including tumour location (colon vs rectum or proximal vs distal), histological grade, TNM stage, or sex (Table 1).
In this legacy series, the majority of LS tumours were discovered as a result of symptoms without prior diagnosis of the hereditary syndrome. In only three out of the thirteen LS patients with available records, was the tumour found during routine colonoscopy screening. Despite this, almost all LS patients had at least some family history of CRC, and for at least two patients, LS had been diagnosed in other family members. The known family history may have led to patients seeking treatment at a lower threshold than those with sporadic CRC.
There was no significant difference in the CRC-specific survival in the 27 sporadic CRC patients vs 14 LS patients for whom we had up-to-date cause of death information available or were still living (Fig. 1B; log-rank test P = 0.74). The LS patients showed a trend towards an increased overall survival, possibly explained by the typically younger age of diagnosis in this group of patients (Supplementary Fig. 1; log-rank test P = 0.068).
Gene expression
The global gene expression profiles were highly similar in hereditary and sporadic MSI-CRCs. Both hierarchical clustering and a PCA plot of the most variable genes showed a separation between tumour and normal samples as expected, but the germline mutation status did not have an effect on global differential expression (Fig. 2A and B).
Figure 2.
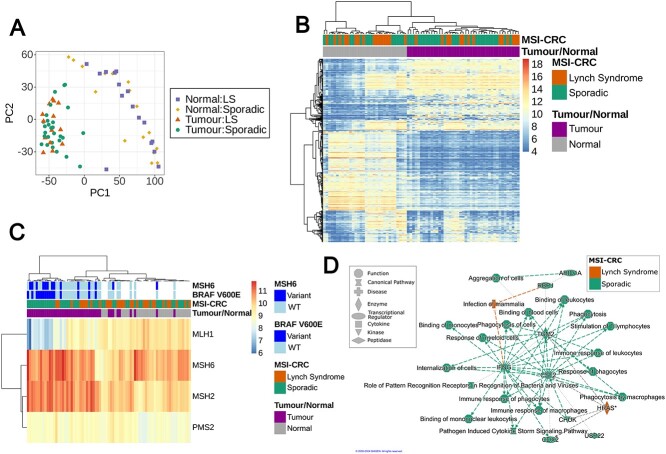
Global gene expression in 29 sporadic and 14 hereditary MSI-CRCs along with 16 sporadic and 16 hereditary normal tissues from CRC patients. (A) PCA plot of the 2000 genes with the highest variance in RNA-seq expression data normalised by variance stabilising transformation. (B) Hierarchical clustering with the top 500 most variable autosomal genes following normalisation by variance stabilising transformation. (C) Hierarchical clustering of the expression of four key MMR genes following normalisation by variance stabilising transformation. The annotation indicates the tumour/normal and sporadic/hereditary status of each sample, and whether the tumours carry a somatic BRAF V600E or non-synonymous MSH6 mutation. (D) Graphical summary of the main functions and proteins affected by the 200 differentially expressed genes from a gene ontology analysis by QIAGEN ingenuity pathway analysis software [17]. Green and orange represent a predicted increase in activation in sporadic and LS tumours, respectively.
Immunohistochemical staining of MMR proteins in tumour tissue to detect expression changes is often used in LS diagnostics. Hierarchical clustering of the RNA-sequenced samples based on the expression levels of the four major MMR genes mutated in LS showed separate clustering of the tumour and normal tissue samples. However, there was no clear separation between the sporadic and LS samples, either in the tumours or normal tissue (Fig. 2C). MLH1 was not differentially expressed between LS and sporadic MSI-CRCs.
Interestingly, a subset of sporadic tumours clustered together, displaying low MLH1 expression and a high frequency of somatic BRAF V600E mutation (Fig. 2C). The MLH1-low BRAF-mutated sporadic tumour cluster also displayed a high rate of somatic mutations in MSH6, another MMR gene more rarely affected by germline mutations in LS [18] (Fig. 2C). Sporadic MSI-CRCs carrying the BRAF V600E mutation consistently displayed low MLH1 expression while more variation in MLH1 expression is seen in BRAF wildtype tumours (WT; Supplementary Fig. 2). The low MLH1 expression in BRAF mutant tumours was also replicated in MSI-CRCs from The Cancer Genome Atlas (TCGA) [19–21] (Welch two sample t-test of log-transformed MLH1 RPKM values, P = 0.013, 14 BRAF-mutated and 13 WT samples; Supplementary Fig. 3) and iCAN dataset (Deseq2, P = 0.10, 8 BRAF-mutated and 3 WT samples).
A differential expression analysis of the sporadic vs hereditary MSI tumours found only 200 differentially expressed genes of which 136 had higher expression in sporadic tumours as compared to 64 genes with elevated expression in hereditary tumours (FDR 10%, |LFC| > 0.6; Supplementary Table 1). Eleven of the 200 genes are well-established cancer genes in the COSMIC catalogue, one of which (PTPRK) is a CRC-related gene [22]. None of the genes overlapped with those we identified as being most commonly differentially mutated between the tumour groups. Gene ontology analyses of the 200 differentially expressed genes using ToppGene and Ingenuity Pathway Analysis (IPA) tools independently identified increased activation of pathways related to the immune response, both innate and adaptive immunity, in sporadic compared to hereditary MSI-CRCs (Fig. 2D, Supplementary Table 2). Phagocytosis and phagocytic immune cells including macrophages were highlighted in the results by IPA, along with cytokines interferon-γ (IFNG) and colony stimulating factor 2 (CSF2; also known as granulocyte-macrophage colony stimulating factor) identified as key regulators of the differentially expressed immune pathways (Fig. 2D). Additionally, many gene ontology terms in the ToppGene analysis were related to developmental processes, particularly to angiogenesis, and the MAPK cascade (Supplementary Table 2). Genes more highly expressed in LS tumours were more often connected to terms relating to cell migration or morphogenesis.
Very few genes were differentially expressed between the normal colon tissues of 16 sporadic MSI and 16 LS patients. Only 11 genes were overexpressed in sporadic samples and 3 genes in hereditary samples (FDR 10%, |LFC| > 0.6; Supplementary Table 3). The most differentially expressed gene with higher expression in normal tissue from sporadic tumour patients was LRP2.
Deconvolution of the RNA-sequencing data was performed with CIBERSORTx [23] to estimate the immune cell contexture of the MSI-CRCs and normal colon tissues (Supplementary Fig. 4). The previously validated leukocyte gene signature matrix LM22 was applied in deconvolution to distinguish 22 human hematopoietic cell phenotypes [24]. The tumour and normal samples clustered separately based on the estimated cell type proportions, while sporadic MSI and LS samples did not. Six of the 22 immune cell types had significantly different proportions in sporadic and LS tumours (Supplementary Fig. 5, Supplementary Table 4). Plasma cells, CD4+ naive T-cells, and activated and resting dendritic cells had higher proportions in LS tumours (Mann Whitney U test, P = 4.4 × 10−3, 0.013, 0.017 and 0.018, respectively), while neutrophils and M0 macrophages had higher proportions in sporadic tumours (Mann Whitney U test, P = 0.031 and 0.041, respectively).
Mutation densities
As expected, the MSI-CRCs displayed a consistently higher number of somatic indels when compared to MSS-CRCs [12]. The same was true for somatic single nucleotide variants (SNVs; Fig. 1C; Welch two sample t-test P < 2.2 × 10−16, < 2.2 × 10−16 and = 1.54 × 10−15 for insertions, deletions and SNVs, respectively).
The genome-wide counts of insertions, deletions, and single nucleotide variants (SNVs), were similar in sporadic and LS MSI-CRCs, except for a trend towards elevated insertion counts in sporadic tumours (Fig. 1C). Sporadic tumours had 24%, 14% and 17% higher mean counts of insertions, deletions and SNVs, respectively, compared to LS tumours. Sporadic tumours also had a 17% higher mean count of coding mutations and a 33% higher mean count of gene truncating mutations. None of these differences were statistically significant. The SNV counts showed a weak correlation with age while the insertion and deletion counts did not (Spearman correlation coefficients of 0.36, 0.25 and 0.1 and p-values 0.02, 0.12 and 0.52 for SNVs, insertions and deletions, respectively; Fig. 1D).
In the regions accessible with short read sequencing, the mutation density of the sporadic tumours was on average 106 mutations/Mbp (range: 43–178) across all samples, slightly higher than the 90 mutations/Mbp (range: 24–189) in LS tumours. This trend was seen for insertions, deletions and SNVs (insertions: 14 and 11 mutations/Mbp, deletions: 47 and 41 mutations/Mbp, SNVs: 45 and 39 mutations/Mbp, in sporadic and LS tumours, respectively).
The mean transition-to-transversion rate was 21% higher in the LS tumours, although this was not significant in a Welch two sample t-test (Supplementary Fig. 6A; P = 0.0643, median 2.48 and 2.71, and ranges 1.49–4.41 and 2.11–4.92 in sporadic and LS tumours respectively). Of the individual single base substitution types, the only significant difference between LS and sporadic tumours was seen for the C > A transversions (Supplementary Fig. 6B; Welch two sample t-test P = 2.473 × 10−3).
Indel landscape in microsatellite repeats
As MSI-CRCs are characterised by the instability of microsatellite repeats whose mutations are poorly called by standard short read WGS variant calling pipelines, we looked specifically at mutations in short repeat sequences using the GangSTR tool [11]. The bulk of these mutations also measure time since loss of MMR.
Along with the 40 MSI-CRC tumour-normal pairs from our sample set, the indels from an additional 245 CRCs previously sequenced in-house were also identified to allow comparison. The additional CRCs consisted of 226 MSS-CRCs and 19 sporadic MSI-CRC tumour-normal pairs (Supplementary Fig. 7). In total, 1 916 246 loci were genotyped and 12 070 762 mutations at 972 173 different loci were identified across the 285 CRCs following Q score filtering (Supplementary Fig. 8).
As expected, the MSI-CRCs had a consistently higher number of indels than MSS-CRCs for repeats with short motifs (Fig. 3A). The difference between MSI- and MSS-CRC was significant in motifs of 1–5 bp in length (Wilcoxon test P = 1.7 × 10−12, 3 × 10−12, 1.15 × 10−10, 5.64 × 10−8 and 1.5 × 10−4, respectively, for each motif length, controlled for sequencing batch, Supplementary Fig. 7). In repeats with a motif length between 6 and 20 bp, only a very small proportion of loci were mutated and the mutation rate was similar in MSS- and MSI-CRCs (Supplementary Fig. 9). A small number of tumours were slight outliers with a low number of indels, particularly visible in regards to the dinucleotide repeats. We were not able to identify any clinical features that these tumours had in common.
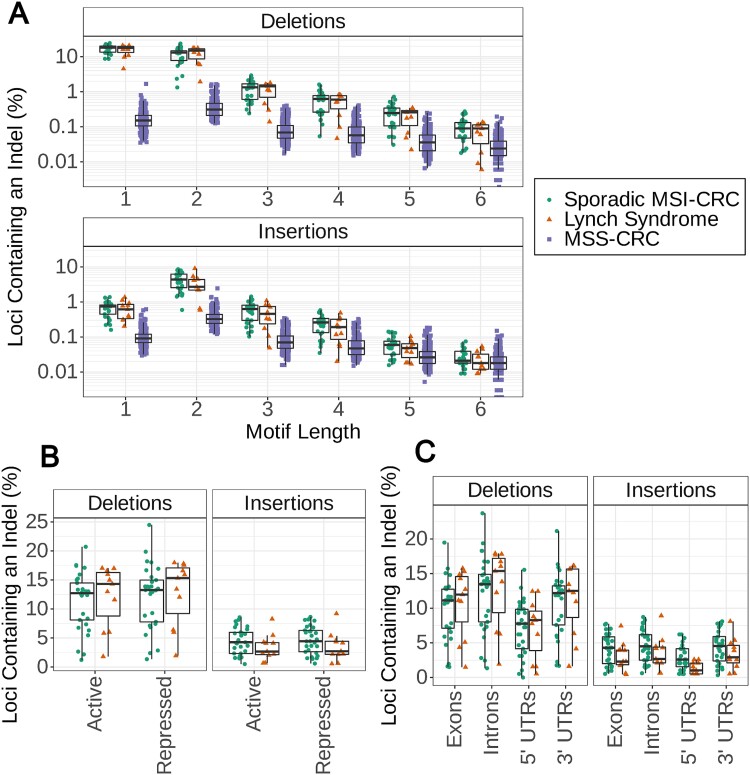
Figure 3.
Somatic indels in microsatellite repeats, determined from GangSTR genotype calls in 29 sporadic MSI and 11 LS CRCs. (A) The number of microsatellite repeat loci with insertions or deletions normalised by the total number of loci of each motif length. The x-axis separates microsatellite repeats with motif lengths from 1 to 6 in 226 MSS-CRCs as well as the MSI-CRCs. The y-axis is on a log scale. (B) The number of loci in dinucleotide repeats in active and repressed genomic regions with insertions or deletions. Counts normalised by the total number of loci in dinucleotide repeats for each annotation and shown as a percentage. Annotations are based on annotations from the roadmap epigenomics project. (C) The number of loci in dinucleotide repeats in protein-coding genes with insertions or deletions. Counts normalised by the total number of loci in dinucleotide repeats for each annotation and shown as a percentage. Annotations are based on GENCODE biotype annotations.
Overall, the microsatellite repeat mutation patterns were highly similar between the hereditary and sporadic MSI tumours. Sporadic MSI-CRCs showed a trend of having a higher number of insertions than hereditary tumours, particularly in dinucleotide repeats, while the number of deletions was more similar between the two sample groups (Fig. 3A, Supplementary Fig. 10). Conversely, hereditary tumours showed a tendency towards having more dinucleotide repeat deletions than sporadic tumours (Supplementary Fig. 10). These differences were not significant, however (Mann–Whitney U test).
Chromatin state annotation from the roadmap epigenomics project gave insight into the indels in active regions of the genome as compared to repressed regions [25]. In both active and repressed regions, the number of indels in sporadic and hereditary tumours in relation to each other followed similar trends as we saw overall and there was no significant difference between them for any motif length (Fig. 3B; Supplementary Fig. 11). When considering all 40 MSI-CRCs together, a higher proportion of mononucleotide repeats in active regions contained deletions than those in repressed regions (Mann–Whitney U test, P = 3.52 × 10−6) but no difference was seen in repeats with longer motif lengths. No difference was observed in the insertion counts between active and repressed regions in microsatellite repeats with motifs of any length. Additionally, no difference was observed between sporadic and LS tumours for any of the individual chromatin state annotations (Supplementary Fig. 12).
To further compare the indels in different regions of the genome, the mutated loci were annotated to identify loci located in protein-coding exons, 5′ UTRs, 3′ UTRs and protein-coding introns based on feature annotations from the GENCODE project [26]. Only the number of insertions in dinucleotide repeats in the 5′ UTR was significantly different between sporadic and hereditary tumours; fewer such insertions were observed in LS tumours (Fig. 3C; Mann–Whitney U test, P = 0.0175). In tri- and tetranucleotide repeats, fewer deletions were also observed in 5′ UTRs in LS tumours, despite LS tumours otherwise tending to have more deletions than sporadic tumours in microsatellite repeats (Supplementary Fig. 13).
To look more closely at the indels in the 5′ UTR regions and to further explore the difference between the variant landscapes of sporadic and LS MSI-CRCs, the variants in 5′ UTRs were annotated with the UTRannotator tool [27]. UTRannotator annotates SNVs and small indels up to 5 bp which includes the vast majority, 88%, of the loci across the 40 MSI-CRCs in which we detected indels. UTRannotator identified 22 loci which were mutated in at least one of our forty MSI-CRCs with the potential for upstream AUG (uAUG) gain, and 9, 20 and 425 mutated loci across the tumours with potential for uAUG loss, upstream STOP (uSTOP) loss, and predicted upstream open reading frame (uORF) frameshift indels, respectively. No 5′ UTR loci were identified that were convincingly differentially mutated between the two MSI-CRC groups. Additionally, no connections between the mutated 5′ UTR loci and differential gene expression between sporadic MSI and LS tumours were identified.
Interestingly, we didn’t observe a correlation between the number of indels and the age of the patient or the TNM stage. The number of indels was slightly, but insignificantly, different in proximal and distal tumours. In LS tumours, there were 21% more insertions and 11% more deletions in distal tumours as opposed to proximal tumours. While in sporadic tumours, the opposite trend was seen with 17% more deletions in proximal tumours than distal tumours. Proximal tumours were more often observed in female patients (23/30 of proximal tumours and 6/13 distal tumours from our tumour set were in female patients). There was no significant difference between the number of indels in male and female patients.
Protein-coding gene mutations
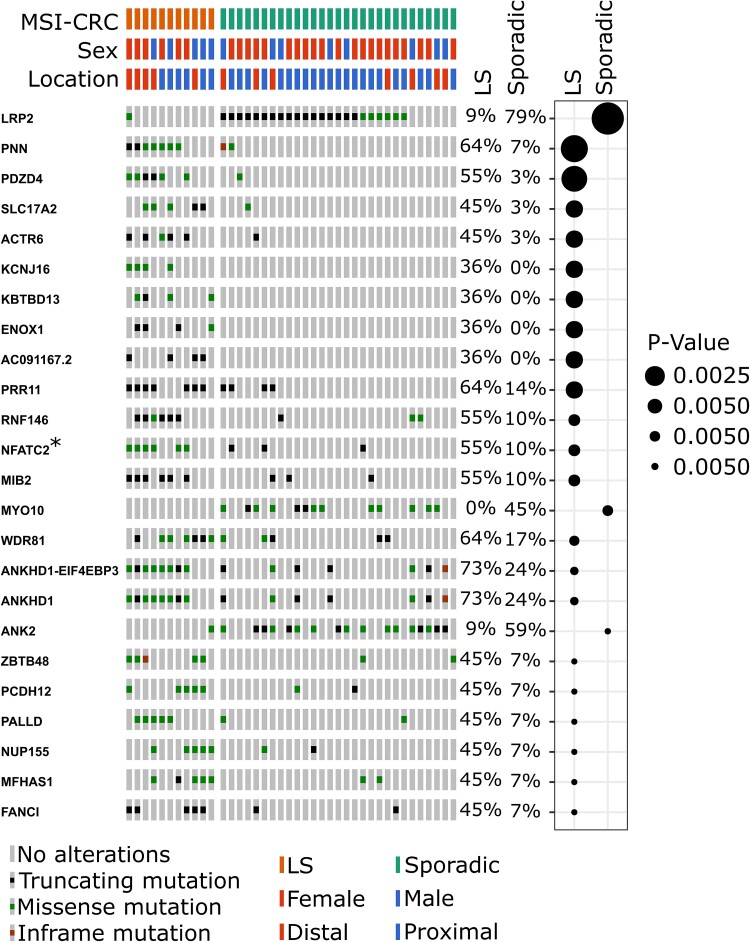
As expected, a large number of genes were recurrently mutated across the 40 MSI-CRCs for which paired WGS data was available, including many non-synonymous SNVs and indels. There was some variation in the genes that were most frequently mutated in sporadic MSI and LS tumours. A comparison of non-synonymous mutation counts in LS and sporadic MSI tumours based on a fisher’s test indicated that 178 genes were differentially mutated between the two groups (unadjusted P-value < 0.05; Supplementary Table 5). A gene ontology analysis with the ToppGene tool [28] suggested that genes involved in potassium ion transport were more often mutated in sporadic MSI than LS tumours. The top genes are shown in Fig. 4. LRP2 was the most differentially mutated gene and more often mutated in sporadic MSI-CRCs. LRP2 was also one of the genes identified as being more highly expressed in normal tissue from sporadic MSI-CRC patients. (Supplementary Table 3). PNN and PDZD4 followed and were more often mutated in LS tumours. Seventeen of the top 20 differentially mutated genes were more frequently mutated in LS tumours, among them were also MIB2 and ANKHD1. Along with LRP2, the two genes more commonly mutated in sporadic MSI tumours were MYO10 and ANK2.
Figure 4.
The top twenty genes with the most differential number of samples carrying non-synonymous mutations between sporadic and hereditary tumours, as ranked by Fisher’s test p-values. The COSMIC cancer gene is indicated by an asterisk. On the right-side, the percentages indicate the proportion of samples carrying mutations, and the plot indicates whether sporadic MSI or LS tumours more frequently carry mutations in each gene and the Fisher’s test p-value. Plots produced by Oncoprinter.
Twelve of the 178 significantly differentially mutated genes are included in the COSMIC cancer gene census as cancer genes. NFATC2, PIK3CA, NAB2, MUC1 and EPHA3 which were more frequently mutated in LS CRCs, and BRAF, HNF1A, CHD4, BAZ1A, MECOM, SPECC1 and BRIP1 which were more frequently mutated in sporadic MSI-CRCs. Three of these genes, BRAF, BAZ1A and PIK3CA, have been recognised as CRC-related genes (Supplementary Fig. 14). Along with BRAF, BAZ1A was more frequently mutated in sporadic MSI-CRCs. BRAF mutations in particular are well-known to be frequently present in sporadic MSI tumours while absent in LS tumours; this was also evident in our tumour set [29]. PIK3CA, on the other hand, was more frequently mutated in LS CRCs.
A separate look at the most frequently mutated genes in the two MSI-CRC groups also reveals some similarities between the tumour types. Power to detect the differences depend on the likelihood of mutation in individual genes which depends on many factors such as the gene length, the number of repeats and the lengths of the repeats within the coding sequence. To account for differing gene lengths, the counts were normalised by their coding sequence (CDS) length (Supplementary Fig. 15). The most frequently mutated genes in sporadic MSI-CRCs were RPL22, C12orf76 and SUMO1. RPL22 and C12orf76 are similarly among the top 20 frequently mutated genes in LS. The most frequently mutated genes in LS were SMKR1, DIABLO and AC107959.5. None of these three were among the top 20 genes in sporadic MSI-CRCs.
RPL22 is a COSMIC cancer gene, as are B2M and ACVR2A which were also among the top 20 genes in sporadic tumours. B2M and KRAS, meanwhile, were among the top 20 in LS tumours.
Mutational signatures
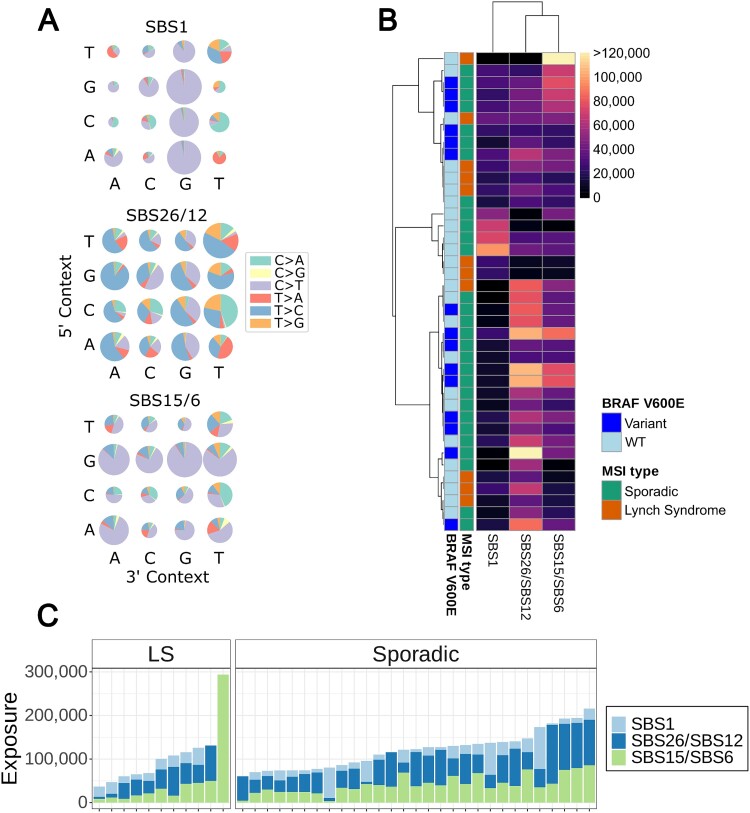
We identified the genome-wide somatic mutational signatures most prevalent in the tumours [30]. Single base substitution (SBS) signatures were extracted from 40 MSI-CRCs that had paired normal WGS data available from the adjacent colon. The signatures were extracted with all tumours together and sporadic and hereditary tumours were then compared.
Three distinct SBS signatures were identified from the tumours. As expected, one of the signatures corresponded to the dMMR-related signatures, SBS15 and SBS6, listed in the COSMIC database [31]. The second signature corresponded to the age-related signature SBS1, while the third most closely resembled the COSMIC signatures SBS26 and SBS12 (Fig. 5A). SBS26 is another dMMR-related signature, while SBS12 has an unknown aetiology, but may be related to transcription-coupled nucleotide excision repair [31].
Figure 5.
Mutational signature analysis of single base pair substitutions of 11 hereditary and 29 sporadic MSI-CRCs. (A) The three signatures extracted, similar to cosmic SBS1, SBS26/SBS12, and SBS15/SBS6. (B) Hierarchical clustering of MSI-CRCs for the three signatures with cosine distance and average linkage. Colour scale indicates the mutation counts. The BRAF V600E mutation status of the tumours is annotated. (C) The number of mutations corresponding to the three signatures in each tumour.
For the two MSI-related signatures, a higher overall SNV count in the tumours correlated to a higher number of mutations contributing to the signatures, particularly for SBS15/SBS6 (linear regression, R2 = 0.6636 and 0.1916, and P = 1.594 × 10−10 and 0.00471 for SBS15/SBS6 and SBS26/SBS12, respectively). The same was true for insertions (R2 = 0.2161 and 0.1786, and P = 0.002511 and 0.006596, respectively) and deletions (R2 = 0.1078 and 0.1858, and P = 0.02856 and 0.005488, respectively). This correlation was not observed for SBS1.
Sporadic and hereditary MSI-CRCs did not form distinctly separate groups when hierarchical clustering was performed based on the tumours’ mutational signatures. Tumours carrying a BRAF V600E mutation likewise did not cluster separately from the wildtype tumours (Fig. 5B).
The vast majority of tumours displayed all three signatures at varying exposures. Four outliers, two sporadic and two hereditary tumours, showed only one or two of the signatures. One hereditary tumour in particular had a high count of the MSI-related SBS15/SBS6 mutations but had an exposure of 0 for the other two signatures. Sporadic and hereditary tumours did not have significantly different exposures to the observed mutational signatures (Mann–Whitney U test P-values of 0.1567, 0.8084 and 0.1162 for SBS15/SBS6, SBS1 and SBS26/SBS12 respectively; Fig. 5C and Supplementary Fig. 16A).
One of the MSI-related signatures, SBS15/6, showed a weak correlation with the age of the patient, unlike the SBS1 previously associated with age or the MSI-related SBS26/12 signature (Spearman correlation coefficients of 0.36, 0.3 and 0.1, and p-values 0.022, 0.058 and 0.54, for SBS15/6, SBS26/12 and SBS1 respectively; Supplementary Fig. 16B). These unexpected results could be explained by high variability in mutation counts in the MSI tumours resulting in high variance in the SBS exposures. Also, the MSI-related DNA methylation defects could contribute to the extra variation in SBS1, characterised by deamination of methylated CpGs.
One LS tumour, had a particularly high number of MSI-related SBS15/SBS6 mutations (Fig. 5C). This patient was of a young age, 27 years at the time of diagnosis, and was an outlier with a very high number of somatic SNVs, including R1858C in the Domain of Unknown Function (DUF1744) of POLE. This tumour did not, however, have more indels than other tumours in our sample set (Fig. 1C). The sporadic tumours with a particularly high number of SBS1 mutations were three of the five rectal tumours, all of whom were from male patients, and were low stage (TNM I/II) and low grade (Grade I/II) tumours.
To complement the SBS signatures, doublet-base substitution (DBS) signatures and indel (ID) signatures were extracted with the SigProfiler tool [32] (Supplementary Fig. 17). Indel signatures ID1, ID2 and ID7 were identified from the Mutect2 indel calls; ID2, ID12, and the de novo signatures dnID-A and dnID-B were identified from the gangSTR indel calls (Supplementary Fig. 18). ID1, ID2 and ID7 have all been recognised as prevalent in dMMR tumours. ID1 and ID2 represent insertions and deletions in homopolymers, respectively, and are thought to be due to strand slippage in DNA replication [31], while ID7 is less common and is additionally characterised by deletions in dinucleotide repeats. The additional ID signatures identified in the gangSTR calls, ID12 and the two de novo signatures, do not have known aetiologies. The de novo ID signatures are characterised by deletions of a length of 2 bp or longer in microsatellite repeats of varying lengths. The deletions include those in microsatellite repeats of a longer length than the deletions characterised by ID12 (Supplementary Fig. 18). DBS signatures DBS8, DBS14 and de-novo signature dnDBS-A were identified (Supplementary Fig. 19). dnDBS-A was the most prevalent DBS signature and characterised by thymidine dimers mutating to a different homodimer (i.e. TT > CC;AA;GG; Supplementary Fig. 19). Similar to the SBS signatures, none of the DBS or ID signatures significantly differed between sporadic MSI and LS tumours.
Tumour Clonality
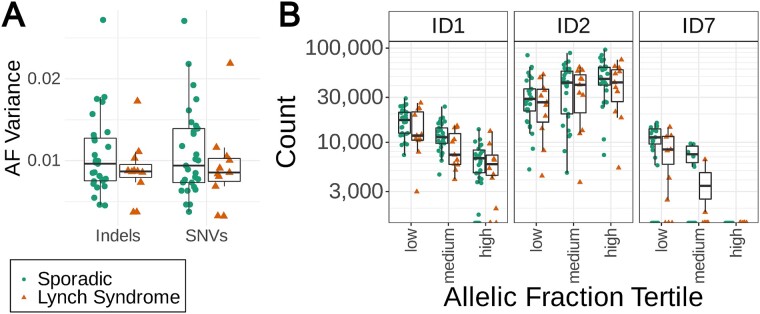
We compared the allelic fractions (AF) of the somatic variants as an indicator of clonality and growth patterns within the tumours. Higher variance of the AF indicates more diverse clonal structure suggesting slower tumour growth, while smaller variance can be considered a marker for a rapidly growing tumour [33]. Overall, sporadic tumours showed wider distribution of AF variances for both SNVs and indels (Fig. 6A, Supplementary Fig. 20). In particular, the absence of high AF variances among LS tumours suggests a high selection pressure against emerging immunogenic tumour cell clones in these patients that have a history of frequent exposure to dMMR cell clones due to their genetic background. The immune cell infiltration estimates from deconvolution of RNA-sequencing data were compared to the AF variances and two immune cell types showed a suggestive positive correlation (Supplementary Fig. 21, Supplementary Table 6). These were resting CD4+ memory T cells (Spearman’s test, rho = 0.34 and 0.41, and P = 0.03 and 8.9 × 10−3, to the indel and SNV AF variances, respectively) and ɣδ T cells (Spearman’s test, rho = 0.41 and 0.42, and P = 9.1 × 10−3 and 6.4 × 10−3, to indel and SNV AF variances, respectively), yet the latter cell type was identified only in three tumours. The AF variances did not appear to explain the clustering of sporadic MSI and LS tumours based on the overall pattern of estimated immune cell infiltration for 22 cell phenotypes (Supplementary Fig. 4).
Figure 6.
(A) Variance of allelic fractions of biallelic indel and SNV loci in sporadic and LS tumours. (B) Indel signatures by sample in low, medium and high allelic fraction tertiles.
Three dMMR-related indel signatures ID1, ID2 and ID7 were identified among tumour somatic Mutect2 variants and these were divided into low, medium, and high allelic fractions. Homopolymer insertion-related ID1, and ID7, were more prevalent among variants with a low allelic fraction while the converse was true for homopolymer deletion-related ID2 (Fig. 6B, Supplementary Fig. 22). The signatures did not differ significantly between the two tumour groups in any of the low, medium or high allelic fraction subsets.
Discussion
The increasing accessibility and affordability of whole genome sequencing technologies has led to a substantial increase in research being undertaken at the genomic level in cancers and is no longer so limited to exonic regions of the genome. MSI-CRCs too have attracted a lot of attention as MSI is an important prognostic factor and can affect clinical treatment decisions including the suitability of chemotherapy or immunotherapy [34, 35]. However, MSI-CRCs have been disproportionately neglected in whole genome analyses given the additional complexities that exist due to their hypermutated phenotype at several stages of the analysis. Firstly, indels in microsatellite repeats, frequent in these tumours, are more prone to technical errors during sequencing. Secondly, longer microsatellite repeats can be longer than the WGS read length and so additional tools are required to estimate the length of the microsatellite repeat and indels within it. Thirdly, with such a high density of variants in the tumours, it is particularly challenging to identify which may be influential to tumour growth and behaviour, and which are simply passenger mutations.
The aim of this study was to comprehensively compare the characteristics of sporadic and hereditary MSI-CRCs at the genome and transcriptome level. Despite sporadic MSI and LS tumours having common driver genes, the MMR genes, their molecular origins differ. Sporadic MSI-CRCs most commonly lose MLH1 expression through promoter hypermethylation while heterozygous germline SNVs or indels are typically present in LS patients along with a somatic second hit leading to tumour development [3]. In a clinical setting differences are also apparent. MSI-CRCs have been observed to differ from MSS-CRCs in the patient’s prognosis and response to chemotherapy and immunotherapy [36]. Analyses from WGS and RNA-seq data, along with clinical information, were integrated for a set of 43 MSI-CRCs. In order to compare the 14 LS and 29 sporadic MSI-CRCs, we looked at the tumour clinicopathological parameters, mutation densities, microsatellite repeat variant landscape, mutational signatures, non-synonymous gene mutations, global gene expression and differential gene expression.
As expected, sporadic MSI patients were significantly older than LS patients, explaining their shorter overall survival [2, 37]. There was no difference in CRC-specific survival or in the other clinical characteristics compared, such as tumour differentiation, stage, or tumour location, although there was a trend towards a higher proportion of distal tumours in LS tumours as would be expected [37]. As is well known, most of the MSI tumour samples had a very stable karyotype compared to microsatellite stable tumours [38]: only one tumour displayed a chromosomal instability phenotype in addition to MSI.
Global gene expression profiles were similar across all tumours, consistent with previous findings [39]. Only 200 genes were differentially expressed, the majority, 136, of which were more highly expressed in sporadic MSI-CRCs. Many were related to the immune response, both innate and adaptive arms, which warrants further study. Immunotherapy is increasingly commonly used to treat MSI-CRC patients and a difference in the immune landscapes of these tumours could have clinical implications. Pathway analysis of the differentially expressed genes predicted reduced activity of multiple interferon-γ regulated immune response pathways in LS tumours compared to sporadic MSI-CRCs. Interferon-γ is a cytokine with key roles in the activation of anti-tumour immune responses and induction of antigen presentation in tumour cells; however, tumours may also develop interferon-γ resistance that remains a major obstacle for responsiveness to immunotherapy [40]. Sporadic MSI and LS tumours did not cluster separately based on the estimated proportions of 22 immune cell types from deconvolution of RNA-sequencing data. Six immune cell types had significantly different estimated proportions in sporadic and LS tumours. Compatible with the significantly higher estimated proportions of M0 macrophages and neutrophils in sporadic MSI vs LS tumours, pathway analysis of differentially expressed genes highlighted increased expression of multiple pathways related to phagocytic immune cells in sporadic MSI tumours. A suggestive positive correlation was also observed between resting CD4+ memory T cell proportions and tumour AF variances. These observations based on in silico estimates of immune cell proportions need confirmation in future studies using more direct methods. We also observed lower MLH1 gene expression in the sporadic tumours carrying a BRAF V600E mutation compared to the sporadic BRAF WT tumours. This is consistent with previous research that has recognised that BRAF V600E mutations are more frequent in sporadic MSI-CRCs with extensive MLH1 promoter methylation [41].
Taking into consideration variants genome-wide, without a focus on repeated genomic regions, there was no significant difference between the two MSI-CRC groups in the total number of insertions, deletions, or SNVs. However, sporadic MSI-CRCs showed a trend towards higher indel and SNV counts. This is consistent with Sato et al who identified a significantly higher number of indels in sporadic MSI than LS tumours from whole exome sequence data, but in contrast they observed no difference in the SNV counts [42]. The slightly higher mutation counts in sporadic MSI-CRCs may reflect the later age of diagnosis of the sporadic cases, although only the SNV counts showed a weak correlation with age.
MSI-CRCs are known to have a very high number of indels in microsatellite repeats genome-wide [43–45], many of which may disrupt crucial genomic elements, such as genes, regulatory elements [43, 46] or chromatin organisation. Additionally, the vast majority of the top 1% of conserved regions across the human genome have been identified as being non-coding regulatory regions [46]. This demonstrates the importance of considering the impact of indels in non-coding genomic regions in cancer. To derive a more comprehensive view of the mutation landscape in MSI-CRCs, we looked specifically at indels located in microsatellite repeats genome-wide. To our knowledge, this is the first time a genome-wide analysis of microsatellite indels comparing sporadic MSI to LS tumours has been performed. The number of insertions or deletions in LS and sporadic MSI tumours did not significantly differ in microsatellite repeats of any motif length.
LS tumours had a particularly low number of insertions in 5′ UTRs when compared to sporadic MSI-CRCs: in dinucleotide repeats this difference was significant. Despite LS tumours overall tending towards a higher number of microsatellite deletions than sporadic MSI-CRCs, the opposite trend was observed at 5′ UTR loci in tri- and tetranucleotide repeats. Despite the global difference in 5′ UTR indel rate, we were unable to identify any specific 5′ UTR loci where indels were significantly more frequent in sporadic tumours.
Microsatellite indels have previously been observed across many MSI cancer types to be enriched in actively-transcribed genomic regions, promoters and enhancers [43]. In our analysis, the number of indels identified in our tumour set was largely consistent across active and repressed regions of the genome. The only exception was that we observed that mononucleotide repeats more often carried deletions in active than repressed genomic regions. The chromatin state did not affect the indel counts in microsatellite repeats in a differential manner in sporadic MSI as compared to LS tumours.
Mutational signatures corresponding to SBS1, SBS15 and SBS26 have been previously identified in MSI-CRC exomes, and SBS26 has been associated with poor survival and immunotherapy response in dMMR CRC tumours [47–49]. Expanding to a genome-wide scale, we identified similar mutational signatures, corresponding to SBS1, SBS15/SBS6 and SBS26/SBS12. Also similar to Giner-Calabuig et al [47], we observed a subset of tumours with a comparatively high proportion of SBS1 mutations while fewer dMMR-related mutations. These may have lost MMR capacity at a relatively late stage of tumorigenesis.
In addition to single-base substitution signatures, we extracted doublet base substitution and indel signatures from the Mutect2 somatic calls and indel signatures from the gangSTR calls and compared these between the sporadic MSI and LS tumours. ID1, ID2 and ID7, identified in the Mutect2 calls, along with DBS7 and DBS10 have been recognised as related to dMMR [31]. Unexpectedly, the known MSI-related signatures DBS7 and DBS10 were not detected in our samples. This is consistent with recent studies where ID1, ID2 and a single de-novo DBS signature were identified in MSI-CRCs [45, 50]. We also identified the dMMR-related ID2 and ID7, and three DBS signatures DBS8, DBS14 and a novel signature which we designated dnDBS-A. DBS8 has been associated with hypermutated tumours, although not dMMR, and was only present in a small number of our tumour set with a strong overrepresentation in one tumour. The outlying sample did not otherwise have an unusually high number of somatic variants. The signatures extracted from the gangSTR indel calls reflect the higher representation of indels in microsatellites of longer motif lengths whereas the Mutect2 indel calls are dominated by mononucleotide repeats following the standard pipeline filters. ID12, of unknown aetiology, and the two de novo signatures all predominantly represent deletions in microsatellite repeats, typical of dMMR tumours. The de novo indel signatures identified may reflect the variant calling method which identifies longer indels than are typically recognised by standard Mutect2 calling methods. Mutect2 recognises short indels in complex areas and largely excludes repetitive areas, in contrast to the gangSTR indel calls which only includes repetitive genomic regions. The mutational signatures in sporadic MSI and LS tumours did not significantly differ from each other in any mutation type.
We further analysed somatic indel signatures in the tumours with respect to clonality of variants associated with each signature. Homopolymer insertions represented by ID1 are more frequently of low clonality which may be because they accumulate later in tumorigenesis or that insertions have more deleterious effects and so are more often selected against. Conversely, homopolymer deletions represented by ID2 are more frequent among the highly clonal variants, perhaps reflecting their accumulation throughout tumorigenesis and a lower negative selection pressure.
MSI-CRCs carry many non-synonymous mutations in coding regions of genes, especially in genes containing many long microsatellite repeats. However, this effect is expected to be the same when comparing the two MSI tumour groups. We identified 178 genes that were significantly differentially mutated between LS and sporadic MSI-CRCs. LRP2 was the most differentially mutated gene and more often mutated in sporadic MSI compared to LS tumours. LRP2, also known as megalin, a multi-ligand endocytic receptor, was the most differentially mutated gene. It was also slightly more highly expressed in normal tissue from sporadic MSI than LS patients; in the tumours LRP2 was not differentially expressed. LRP2 mutations have been associated with immune cell infiltration, immune-related gene expression, and in melanoma, increased OS following immunotherapy [51]. LRP2 hypermethylation, suggesting low expression, has also been associated with a reduced rate of recurrence following stage II CRC and an enrichment of activated B cell signatures, mTORC1 and DNA repair pathways [52]. PNN, on the other hand, was more often mutated in LS tumours. High PNN expression has previously been linked to poor progression free survival and overall survival along with a worse response to immunotherapy [53, 54].
Of the twelve significantly differentially mutated genes that are considered cancer-related genes in the COSMIC database, three are CRC-related: BRAF, PIK3CA and BAZ1A. As expected, BRAF mutations were only present in the sporadic MSI tumours while absent in LS tumours [29]. BAZ1A is involved in the repair of DNA double-strand breaks by nonhomologous end-joining [55], while BRAF and PIK3CA are involved in the MAPK and PI3K signalling pathways, respectively. Mutations in all three genes have previously been associated with a poor prognosis in cancer, in CRC in the case of BRAF and PIK3CA, and in BAZ1A in breast cancer [55–57]. Although the prognostic significance of PIK3CA is not without dispute [58]. NFATC2 was the most differentially mutated gene among the COSMIC cancer genes with more LS tumours carrying mutations. It has been suggested to promote carcinogenesis and low NFATC2 expression has been suggested to reduce stem cell like properties of CRC stem cells [59, 60]. MUC1, also more often mutated in LS tumours, has been identified as having a role in immune suppression in the tumour microenvironment and in CRC its upregulation has been linked to a worse prognosis and metastasis [61].
LS tumours were more clonal compared to sporadic MSI tumours as indicated by the absence of high variances in the allelic fractions of somatic variants. This can also be considered a marker for a rapidly growing tumour [33, 62, 63]. In agreement, several studies have suggested accelerated progression from adenoma to carcinoma in LS patients compared to the general population [64], and based on pathology review, presence of tumour subclones is much more common in sporadic compared to LS-associated MSI-CRCs [37]. The low clonal diversity in LS tumours is also compatible with the notion that LS patients have been partially immunised against dMMR tumours as a consequence of repeated exposure to early-stage tumours that are eliminated by the immune system. The frequent mutations in coding microsatellites in MSI tumours generate frameshift peptides acting as neoantigens recognized by the immune system, and higher neoantigen burden has been reported in LS tumours compared to sporadic MSI [65, 66]. Although both sporadic and hereditary MSI tumours are highly infiltrated with T cells [37, 66–69], T cell reactivity against frameshift neoantigens is detectable already in healthy carriers of LS-associated germline mutations [70]. This suggests a vaccination-like effect and a high selection pressure against emerging new immunogenic dMMR tumour subclones. Interestingly, tumours with high clonal heterogeneity have been associated with decreased immunosurveillance and reduced responsiveness to immunotherapy across several cancer types [71]. However, it remains unclear whether a strong anti-tumour immune response restricts tumour clonality by pruning out subclones, or whether high clonal heterogeneity directly impairs the anti-tumour immune response [71]. Our results from the comparison of two highly similar subgroups of CRCs, differing mainly in their clonal evolution shaped by immunosurveillance, may lend support to the former hypothesis, providing evidence that the primed immune system in LS patients yields more clonal tumours. Yet further studies in larger numbers of cancers are warranted. Taken together, our results suggest different patterns of clonal evolution in LS tumours and sporadic MSI-CRCs that may arise in part due to differences in tumour immune surveillance. This could have clinical implications, in particular for the subgroup of sporadic MSI-CRCs showing high clonal heterogeneity, as MSI status alone is an insufficient predictor of immunotherapy response with response rates of 40%–50% in patients with dMMR/MSI-CRC [72]. Response rates have not been observed to differ between LS and MSI-CRC patients, but studies have largely been limited by small numbers of LS patients [73, 74].
This research was challenged by the small tumour sample size, limited by the rarity of LS cases and the challenge of extracting high quality RNA from old tumours. This was further affected by an incomplete data set of the paired normal tissue, particularly for the RNA-seq analysis. The single-region tumour sampling and the degree of somatic copy number alterations and tumour purity can affect the accuracy of estimates of clonal heterogeneity [71].
A deeper understanding of the biology of sporadic MSI and LS tumours has the potential to translate to clinical benefits for patients, such as having implications in CRC diagnosis or therapies. The genome-wide microsatellite indel landscape of MSI-CRC in particular is still a largely understudied aspect of these tumours and continuing research will be required to better understand the role that such variants play in CRC development and maintenance. The increasing quality and accessibility of long-read sequencing provides new opportunities to explore repetitive regions of the MSI-CRC genome more thoroughly along with the integration of further datasets, such as epigenetic data, as the quantity of data available continues to increase. In particular, this line of research could shed light on the role of microsatellite indel mutations in regulatory functions, genetic basis of cancer, as well as population-level effects of germline microsatellite variants in disease susceptibility.
Materials and methods
Study approval
This research was conducted in accordance with the Declaration of Helsinki and was reviewed and approved by the ethics committee of the hospital districts of Helsinki and Uusimaa. For all samples, either the patients provided their signed informed consent or authorisation was obtained from the National Supervisory Authority for Welfare and Health to allow the use of the samples in this study.
Patient material
Analysis was performed starting from fresh frozen adenocarcinoma tissue and, where possible, corresponding normal colorectal tissue collected from CRC patients in Finland between 1994 and 2017. The sample set consisted of 14 MLH1-defective tumour-normal pairs from 13 LS patients, and 29 tumour-normal pairs from sporadic MSI-CRC patients. In addition we analysed normal colorectal tissue from 9 unpaired MLH1-defective LS patients. To our knowledge, none of the LS patients are closely related to each other. We had access to the relevant detailed clinical information of the patient for all tumours. Radiation therapy prior to surgery was not performed for any of the five patients with rectal tumours that were included in this study.
The MSI status of the tumours had been determined prior to this study by radioactive labelling techniques, fluorescence-based PCR methods or fragment analysis as described in detail by Kondelin et al [75].
Survival curves were generated in R with the survival (v3.5.5) and survminer (v0.4.9) packages [76, 77].
RNA sequencing
RNA-seq was performed for tumours, and where possible, their normal pair, as indicated in Fig. 1A. Additionally, unmatched normal tissue from 9 LS patients was included in the normal tissue DE analysis and hierarchical clustering plots. RNA was extracted from frozen tissue with the Trizol method. Paired-end sequencing was performed as a Macrogen service using the Illumina TruSeq Stranded Total RNA with Ribo-Zero Human library construction kit and sequencing was performed by the Illumina NovaSeq6000 platform. The read lengths were 101 bp.
RNA-seq analysis
The GRCh38 reference genome was used for sequence alignment. Transcript counts were estimated from the raw data with Salmon (v0.12.0). Transcript analysis was performed with the DeSeq2 R package [78, 79] (v1.30.1). All genes with at least 10 transcripts across all 43 tumours were included and all transcripts were mapped to the primary ENSGs. Gene expression was compared between sporadic MSI and LS tumour samples with the tumour percentage and scaled RIN included in the model as covariates. When comparing tumour and normal samples, and sporadic MSI and LS normal samples, only the scaled RIN was included as a covariate.
In the differential gene expression analysis apeglm unadapted shrinkage was applied and genes with an adjusted P-value > 0.1 and |LFC| > 0.6 were retained [80] (v1.12.0).
Genes considered as COSMIC cancer genes were those in the COSMIC Cancer Gene Census [81] (v97). CRC genes were those indicated as being associated with colon or colorectal cancer.
PCA analysis and clustering were performed with counts normalised by the VST method with DeSeq2 [79]. The pheatmap R package was used to make the heat maps [82] (v1.0.12). Hierarchical clustering in the heat maps used cosine similarity and complete-linkage clustering.
Gene ontology analysis was performed with the ToppGene Suite [28] (date accessed 22.09.2022) and QIAGEN Ingenuity Pathway Analysis [17] (QIAGEN Inc., https://digitalinsights.qiagen.com/IPA; v81348237).
Validation of BRAF mutations against MLH1 expression was performed with TCGA data accessed through the cBioPortal [19–21] (v6.0.5) and with the iCAN—Digital Precision Cancer Medicine Flagship Discovery Platform (https://ican.fi/). With the iCAN data, a likelihood ratio test was performed with DESeq2 (v.1.40.1) including RNA-sequencing batch as a covariate in the model.
Immune cell deconvolution was performed with the CIBERSORTx tool [23], using the LM22 signature matrix to profile 22 immune cell subsets [24].
Whole genome sequencing
DNA was extracted from fresh-frozen tissue using standard methods. WGS was performed for tumours and, where possible, their normal pairs, as indicated in Fig. 1A. For the 43 MSI-CRCs sequenced for this project, libraries were prepared with the Illumina TrueSeq Nano DNA kit and paired-end sequencing was performed by the Illumina NovaSeq600 platform as a Macrogen service. The read lengths were 150 bp.
WGS data from 226 MSS-CRCs and 19 additional MSI-CRC tumour-normal pairs was already available in-house as described in detail previously [12] (Somatic data EGA database accession code EGAS00001004710). For these samples, paired end sequencing was performed with either Illumina HiSeq 2000 as an Illumina service, HiSeq X Ten as a SciLifeLab service, or HiSeq X Ten as a BGI service. Read lengths were 100, 151 bp and 150 bp respectively.
Variant analysis
Sequence data pre-processing and somatic variant calling was performed following a workflow similar to the GATK4 best practices for all tumour-normal pairs [9] (v4.0.4.0). Somatic variants which were designated as “PASS” by Mutect2 were included in later analyses (v4.0.4.0). The GRCh38 reference genome was used for sequence alignment and in all analyses.
The genes considered as COSMIC cancer genes were the same as described under “RNA seq analysis”.
Somatic variant counts and annotations were extracted with BasePlayer with no additional filters applied [83] (v1.0.2). To evaluate the mutation rate in the accessible genomic regions, the pilot style callability mask from the 1000 genomes project was applied [84] (phase 3).
Oncoprints were generated by the Oncoprinter tool in the cBioPortal [20, 21] (v5.3—v5.4). The alteration types provided to Oncoprinter were determined based on the somatic alterations called by BasePlayer [83].
Microsatellite repeat profiling
The tool GangSTR was used to extract indel calls in microsatellite repeats [11] (v2.5.0). A custom reference file was created merging mononucleotide repeats of at least 7 repeats with the reference file recommended for use with GangSTR which contained repeats with motifs of 2–20 bp (hg38 v13) [11]. In the recommended GangSTR reference file from Mousavi et al, microsatellite repeats of 2 and 3 bp were required to have a minimum of 5 and 4 copies, respectively, while a minimum of three copies were required for repeats with motif lengths of four or more base pairs. Additional filters they applied are detailed in Mousavi et al [11]. To avoid complex repeat regions, mononucleotide repeats that were positioned within 50 bp of another microsatellite repeat on the panel were excluded. This led to the exclusion of the bethesda panel markers.
Following the GangSTR run for all tumour and normal samples, the VCF files were filtered to contain only the variants that were present in all 48 normal samples, and only variants in autosomes, the X chromosome, and in samples from male patients, the Y chromosome.
The genotype of each tumour-normal pair was compared and the tumour VCF files annotated with the somatic mutations. To determine the somatic indel length, the shorter normal and tumour allele were paired to each other, as were the longer alleles. The resulting indels were filtered to exclude those with a Q score below 0.98: this amounted to the exclusion of 79.3% of the calls.
The biotype annotation was made with v39 GFF3 files from the GENCODE project [26]. The core 15 state model from the roadmap epigenomics project was used to annotate the epigenome using the hg38 lift mnemonics BED file from the colonic mucosa epigenome [25] (E075; release date 30.3.18). The consequences of indels in the 5′ and 3′ UTRs were analysed with the Ensembl Variant Effect Predictor (VEP) plugin UTRAnnotator [27, 85] (VEP v106.1).
Mutational signatures
Somatic mutation signatures from single base substitutions (SBSs) were extracted from the WGS data of 11 hereditary and 29 sporadic tumours based on the method from Alexandrov et al [30] The 96 SBS mutation context counts for each tumour were obtained using BasePlayer [83] and based on the silhouette score and frobenius error, three signatures were extracted.
Indel and doublet base substitutions (DBSs) were extracted with the SigProfiler tool (SigProfilerExtracter v1.1.23, SigProfilerMatrixGenerator v1.2.23 and SigProfilerAssignment v0.1.1) [32]. Indel signatures were extracted separately for the Mutect2 and GangSTR somatic indel calls.
Tumour Clonality
The allelic fraction tertiles were determined for each tumour separately with cut offs selected to divide the indels into three equal groups by their allelic fraction. Indels from Mutect2 were used in this analysis. Indel mutational signatures were calculated with the SigProfiler tool as described under “Mutational signatures”.
Data plotting and statistics
Unless otherwise mentioned, statistical analyses were performed with R, data was plotted with ggplot2 [86] (v3.3.5) and p-values were not corrected for multiple testing. The sankey diagram was produced with RAWgraphs [87]. Scripts are available on Zenodo under the DOI 10.5281/zenodo.10887161.
Supplementary Material
Acknowledgements
The authors would like to thank Inga-Lill Åberg, Justyna Kolakowska, Alison London, Sini Marttinen, Heikki Metsola, Marjo Rajalaakso, Sirpa Soisalo and Iina Vuoristo for their technical assistance.
The authors would like to acknowledge the computational resources provided by the ELIXIR node, hosted at the CSC-IT Center for Science, Finland. Part of this research has been conducted using the iCAN—Digital Precision Cancer Medicine Flagship Discovery Platform.
Contributor Information
Samantha Martin, Medicum/Department of Medical and Clinical Genetics, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland; Applied Tumor Genomics Research Program, Research Programs Unit, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland.
Riku Katainen, Medicum/Department of Medical and Clinical Genetics, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland; Applied Tumor Genomics Research Program, Research Programs Unit, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland.
Aurora Taira, Medicum/Department of Medical and Clinical Genetics, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland; Applied Tumor Genomics Research Program, Research Programs Unit, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland.
Niko Välimäki, Medicum/Department of Medical and Clinical Genetics, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland; Applied Tumor Genomics Research Program, Research Programs Unit, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland.
Ari Ristimäki, Applied Tumor Genomics Research Program, Research Programs Unit, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland; Department of Pathology, HUSLAB, HUS Diagnostic Center, University of Helsinki and Helsinki University Hospital, Haartmaninkatu 3, 00290 Helsinki, Finland.
Toni Seppälä, Applied Tumor Genomics Research Program, Research Programs Unit, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland; Department of Surgery, Helsinki University Central Hospital, Hospital District of Helsinki and Uusimaa, Haartmaninkatu 4, 00290 Helsinki, Finland; Department of Gastroenterology and Alimentary Tract Surgery, Tampere University Hospital and TAYS Cancer Centre, Kuntokatu 2, 33520 Tampere, Finland; Faculty of Medicine and Health Technology, Tampere University, Kalevantie 4, 33100 Tampere, Finland; iCAN Digital Precision Cancer Medicine Flagship, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland.
Laura Renkonen-Sinisalo, Applied Tumor Genomics Research Program, Research Programs Unit, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland; Department of Surgery, Helsinki University Central Hospital, Hospital District of Helsinki and Uusimaa, Haartmaninkatu 4, 00290 Helsinki, Finland.
Anna Lepistö, Applied Tumor Genomics Research Program, Research Programs Unit, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland; Department of Surgery, Helsinki University Central Hospital, Hospital District of Helsinki and Uusimaa, Haartmaninkatu 4, 00290 Helsinki, Finland.
Kyösti Tahkola, Faculty of Medicine and Health Technology, Tampere University, Kalevantie 4, 33100 Tampere, Finland; Department of Surgery, Central Finland Health Care District, Keskussairaalantie 19, 40620 Jyväskylä, Finland.
Anne Mattila, Department of Surgery, Central Finland Health Care District, Keskussairaalantie 19, 40620 Jyväskylä, Finland.
Selja Koskensalo, The HUCH Gastrointestinal Clinic, Helsinki University Central Hospital, Stenbäckinkatu 9A, 00029 Helsinki, Finland.
Jukka-Pekka Mecklin, Department of Education and Research, The Wellbeing Services of Central Finland, Hoitajatie 1, 40620 Jyväskylä, Finland; Department of Sport and Health Sciences, University of Jyväskylä, Seminaarinkatu 15, 40014 Jyväskylä, Finland.
Kristiina Rajamäki, Medicum/Department of Medical and Clinical Genetics, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland; Applied Tumor Genomics Research Program, Research Programs Unit, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland.
Kimmo Palin, Medicum/Department of Medical and Clinical Genetics, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland; Applied Tumor Genomics Research Program, Research Programs Unit, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland; iCAN Digital Precision Cancer Medicine Flagship, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland.
Lauri A Aaltonen, Medicum/Department of Medical and Clinical Genetics, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland; Applied Tumor Genomics Research Program, Research Programs Unit, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland; iCAN Digital Precision Cancer Medicine Flagship, University of Helsinki, Haartmaninkatu 8, 00014 Helsinki, Finland.
Conflict of interest statement: T.T.S. discloses the following potential conflicts of interest: consultation fees from Amgen Finland, Nouscom and Tillots Pharma. T.T.S. is the CEO and shareholder of Healthfund Finland and Clinical Advisory board member of LS Cancer Diag.
Funding
This study was supported by grants from the Research council of Finland Finnish Center of Excellence Program 2018–2025 (No. 352814), Academy Professor grants (No. 319083, 320149), iCAN Digital Precision Cancer Medicine Flagship (320185), Cancer Foundation Finland, Sigrid Juselius Foundation (230002), Jane and Aatos Erkko Foundation, Relander Foundation, HiLIFE Fellows 2023–2025, and State Research Funding (VTR) by HUS.
References
- 1. Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Ann Rev Microbiol 2003;57:579–608. [DOI] [PubMed] [Google Scholar]
- 2. Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology 2010;138:2073–2087.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim TM, Laird PW, Park PJ. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell 2013;155:858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meyer LA, Broaddus RR, Lu KH. Endometrial cancer and Lynch syndrome: clinical and pathologic considerations. Cancer Control 2009;16:14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aarnio M, Mecklin JP, Aaltonen LA. et al. Life-time risk of different cancers in hereditary non-polyposis colorectal cancer (HNPCC) syndrome. Int J Cancer 1995;64:430–433. [DOI] [PubMed] [Google Scholar]
- 6. Peltomäki P. DNA mismatch repair and cancer. Mutat Res 2001;488:77–85. [DOI] [PubMed] [Google Scholar]
- 7. Peltomäki P. Update on Lynch syndrome genomics. Fam Cancer 2016;15:385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moreira L, Muñoz J, Cuatrecasas M. et al. Prevalence of somatic mutl homolog 1 promoter hypermethylation in Lynch syndrome colorectal cancer. Cancer 2015;121:1395–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van der Auwera GA, Carneiro MO, Hartl C. et al. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Benjamin D, Sato T, Cibulskis K. et al. Calling somatic SNVs and Indels with Mutect2. Calling somatic SNVs and Indels with Mutect2. bioRxiv 2019;861054. 10.1101/861054. [DOI] [Google Scholar]
- 11. Mousavi N, Shleizer-Burko S, Yanicky R. et al. Profiling the genome-wide landscape of tandem repeat expansions. Nucleic Acids Res 2019;47:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Katainen R, Dave K, Pitkänen E. et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet 2015;47:818–821. [DOI] [PubMed] [Google Scholar]
- 13. Ward R, Meagher A, Tomlinson I. et al. Microsatellite instability and the clinicopathological features of sporadic colorectal cancer. Gut 2001;48:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Microsatellite instability in cancer of the proximal colon. Science 1993;260:816–819. [DOI] [PubMed] [Google Scholar]
- 15. Lynch HT, Lynch PM, Lanspa SJ. et al. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet 2009;76:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baretti M, Le DT. DNA mismatch repair in cancer. Pharmacol Ther 2018;189:45–62. [DOI] [PubMed] [Google Scholar]
- 17. Krämer A, Green J, Pollard J Jr. et al. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014;30:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Olkinuora AP, Peltomäki PT, Aaltonen LA. et al. From APC to the genetics of hereditary and familial colon cancer syndromes. Hum Mol Genet 2021;30:R206–R224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cancer Genome Atlas Network . Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cerami E, Gao J, Dogrusoz U. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao J, Aksoy BA, Dogrusoz U. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:l1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tate JG, Bamford S, Jubb HC. et al. COSMIC: the catalogue of somatic mutations In cancer. Nucleic Acids Res 2019;47:D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Newman AM, Steen CB, Liu CL. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37:773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Newman AM, Liu CL, Green MR. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roadmap Epigenomics Consortium, Kundaje A, Meuleman W. et al. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frankish A, Diekhans M, Ferreira AM. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 2019;47:D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang X, Wakeling M, Ware J. et al. Annotating high-impact 5′untranslated region variants with the UTRannotator. Bioinformatics 2021;37:1171–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J, Bardes EE, Aronow BJ. et al. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009;37:W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deng G, Bell I, Crawley S. et al. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res 2004;10:191–195. [DOI] [PubMed] [Google Scholar]
- 30. Alexandrov LB, Nik-Zainal S, Wedge DC. et al. Deciphering signatures of mutational processes operative in human cancer. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013;3:246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alexandrov LB, Kim J, Haradhvala NJ. et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Islam SMA, Díaz-Gay M, Wu Y. et al. Uncovering novel mutational signatures by extraction with SigProfilerExtractor. Cell Genom 2022;2:100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun R, Hu Z, Sottoriva A. et al. Between-region genetic divergence reflects the mode and tempo of tumor evolution. Nat Genet 2017;49:1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kavun A, Veselovsky E, Lebedeva A. et al. Microsatellite instability: a review of molecular epidemiology and implications for immune checkpoint inhibitor therapy. Cancers 2023;15:2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Battaglin F, Naseem M, Lenz HJ. et al. Microsatellite instability in colorectal cancer: overview of its clinical significance and novel perspectives. Clin Adv Hematol Oncol 2018;16:735–745. [PMC free article] [PubMed] [Google Scholar]
- 36. Gatalica Z, Vranic S, Xiu J. et al. High microsatellite instability (MSI-H) colorectal carcinoma: a brief review of predictive biomarkers in the era of personalized medicine. Fam Cancer 2016;15:405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Young J, Simms LA, Biden KG. et al. Features of colorectal cancers with high-level microsatellite instability occurring in familial and sporadic settings: parallel pathways of tumorigenesis. Am J Pathol 2001;159:2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Palin K, Pitkänen E, Turunen M. et al. Contribution of allelic imbalance to colorectal cancer. Nat Commun 2018;9:3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kruhøffer M, Jensen JL, Laiho P. et al. Gene expression signatures for colorectal cancer microsatellite status and HNPCC. Br J Cancer 2005;92:2240–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han J, Wu M, Liu Z. Dysregulation in IFN-γ signaling and response: the barricade to tumor immunotherapy. Front Immunol 2023;14:1190333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koinuma K, Shitoh K, Miyakura Y. et al. Mutations of BRAF are associated with extensive hMLH1 promoter methylation in sporadic colorectal carcinomas. Int J Cancer 2004;108:237–242. [DOI] [PubMed] [Google Scholar]
- 42. Sato K, Kawazu M, Yamamoto Y. et al. Fusion kinases identified by genomic analyses of sporadic microsatellite instability-high colorectal cancers. Clin Cancer Res 2019;25:378–389. [DOI] [PubMed] [Google Scholar]
- 43. Cortes-Ciriano I, Lee S, Park WY. et al. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun 2017;8:15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sonay TB, Koletou M, Wagner A. A survey of tandem repeat instabilities and associated gene expression changes in 35 colorectal cancers. BMC Genomics 2015;16:702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fujimoto A, Fujita M, Hasegawa T. et al. Comprehensive analysis of indels in whole-genome microsatellite regions and microsatellite instability across 21 cancer types. Genome Res 2020;30:334–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Halldorsson BV, Eggertsson HP, Moore KHS. et al. The sequences of 150,119 genomes in the UK biobank. Nature 2022;607:732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Giner-Calabuig M, De Leon S, Wang J. et al. Mutational signature profiling classifies subtypes of clinically different mismatch-repair-deficient tumours with a differential immunogenic response potential. Br J Cancer 2022;126:1595–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Meier B, Volkova NV, Hong Y. et al. Mutational signatures of DNA mismatch repair deficiency in and human cancers. Genome Res 2018;28:666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gulhan DC, Viswanadham V, Muyas F. et al. Predicting response to immune checkpoint blockade therapy among mismatch repair-deficient patients using mutational signatures. medRxiv 2024;24301236. 10.1101/2024.01.19.24301236. [DOI] [Google Scholar]
- 50. Farmanbar A, Kneller R, Firouzi S. Mutational signatures reveal mutual exclusivity of homologous recombination and mismatch repair deficiencies in colorectal and stomach tumors. Sci Data 2023;10:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li C, Ding Y, Zhang X. et al. Integrated in silico analysis of LRP2 mutations to immunotherapy efficacy in pan-cancer cohort. Discov Oncol 2022;13:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tournier B, Aucagne R, Truntzer C. et al. Integrative clinical and DNA methylation analyses in a population-based cohort identifies and as risk recurrence factors in stage II colon cancer. Cancers 2022;15:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wei Z, Ma W, Qi X. et al. Pinin facilitated proliferation and metastasis of colorectal cancer through activating EGFR/ERK signaling pathway. Oncotarget 2016;7:29429–29439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang H, Jin M, Ye M. et al. The prognostic effect of PNN in digestive tract cancers and its correlation with the tumor immune landscape in colon adenocarcinoma. J Clin Lab Anal 2022;36:e24327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li Y, Gong H, Wang P. et al. The emerging role of ISWI chromatin remodeling complexes in cancer. J Exp Clin Cancer Res 2021;40:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tan ES, Fan W, Knepper TC. et al. Prognostic and predictive value of PIK3CA mutations in metastatic colorectal cancer. Target Oncol 2022;17:483–492. [DOI] [PubMed] [Google Scholar]
- 57. Tan E, Whiting J, Xie H. et al. BRAF mutations are associated with poor survival outcomes in advanced-stage mismatch repair-deficient/microsatellite high colorectal cancer. Oncologist 2022;27:191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Voutsadakis IA. The landscape of PIK3CA mutations in colorectal cancer. Clin Colorectal Canc 2021;20:201–215. [DOI] [PubMed] [Google Scholar]
- 59. Gerlach K, Daniel C, Lehr HA. et al. Transcription factor NFATc2 controls the emergence of colon cancer associated with IL-6-dependent colitis. Cancer Res 2012;72:4340–4350. [DOI] [PubMed] [Google Scholar]
- 60. Lang T, Ding X, Kong L. et al. NFATC2 is a novel therapeutic target for colorectal cancer stem cells. Onco Targets Ther 2018;11:6911–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Guo M, You C, Dou J. Role of transmembrane glycoprotein mucin 1 (MUC1) in various types of colorectal cancer and therapies: current research status and updates. Biomed Pharmacother 2018;107:1318–1325. [DOI] [PubMed] [Google Scholar]
- 62. Noble R, Burley JT, Le Sueur C. et al. When, why and how tumour clonal diversity predicts survival. Evol Appl 2020;13:1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shibata D, Navidi W, Salovaara R. et al. Somatic microsatellite mutations as molecular tumor clocks. Nat Med 1996;2:676–681. [DOI] [PubMed] [Google Scholar]
- 64. Ahadova A, Seppälä TT, Engel C. et al. The “unnatural” history of colorectal cancer in Lynch syndrome: lessons from colonoscopy surveillance. Int J Cancer 2021;148:800–811. [DOI] [PubMed] [Google Scholar]
- 65. Saeterdal I, Bjørheim J, Lislerud K. et al. Frameshift-mutation-derived peptides as tumor-specific antigens in inherited and spontaneous colorectal cancer. Proc Natl Acad Sci USA 2001;98:13255–13260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liu GC, Liu RY, Yan JP. et al. The heterogeneity between Lynch-associated and sporadic MMR deficiency in colorectal cancers. J Natl Cancer Inst 2018;110:975–984. [DOI] [PubMed] [Google Scholar]
- 67. Mlecnik B, Bindea G, Angell HK. et al. Integrative analyses of colorectal cancer show Immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 2016;44:698–711. [DOI] [PubMed] [Google Scholar]
- 68. Tougeron D, Maby P, Elie N. et al. Regulatory T lymphocytes are associated with less aggressive histologic features in microsatellite-unstable colorectal cancers. PLoS One 2013;8:e61001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ahtiainen M, Wirta EV, Kuopio T. et al. Combined prognostic value of CD274 (PD-L1)/PDCDI (PD-1) expression and immune cell infiltration in colorectal cancer as per mismatch repair status. Mod Pathol 2019;32:866–883. [DOI] [PubMed] [Google Scholar]
- 70. Schwitalle Y, Kloor M, Eiermann S. et al. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology 2008;134:988–997. [DOI] [PubMed] [Google Scholar]
- 71. Dijkstra KK, Wu Y, Swanton C. The effects of clonal heterogeneity on cancer Immunosurveillance. Annu Rev Cancer Biol 2023;7:131–147. [Google Scholar]
- 72. Galbraith NJ, Wood C, Steele CW. Targeting metastatic colorectal cancer with immune oncological therapies. Cancers 2021;13:3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Williams MH, Hadjinicolaou AV, Norton BC. et al. Lynch syndrome: from detection to treatment. Front Oncol 2023;13:1166238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Therkildsen C, Jensen LH, Rasmussen M. et al. An update on immune checkpoint therapy for the treatment of Lynch syndrome. Clin Exp Gastroenterol 2021;14:181–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kondelin J, Martin S, Katainen R. et al. No evidence of EMAST in whole genome sequencing data from 248 colorectal cancers. Gene Chromosomes Cancer 2021;60:463–473. [DOI] [PubMed] [Google Scholar]
- 76. Kassambara A, Kosinski M, Biecek P. Drawing Survival Curves using “ggplot2”, 2021. https://CRAN.R-project.org/package=survminer.
- 77. Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. Berlin, Germany: Springer Science & Business Media, 2013. [Google Scholar]
- 78. Patro R, Duggal G, Love MI. et al. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017;14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhu A, Ibrahim JG, Love MI. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 2019;35:2084–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sondka Z, Bamford S, Cole CG. et al. The COSMIC cancer gene census: describing genetic dysfunction across all human cancers. Nat Rev Cancer 2018;18:696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kolde R. pheatmap: Pretty Heatmaps, 2019. https://CRAN.R-project.org/package=pheatmap.
- 83. Katainen R, Donner I, Cajuso T. et al. Discovery of potential causative mutations in human coding and noncoding genome with the interactive software BasePlayer. Nat Protoc 2018;13:2580–2600. [DOI] [PubMed] [Google Scholar]
- 84. 1000 Genomes Project Consortium, Auton A, Brooks LD. et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. McLaren W, Gil L, Hunt SE. et al. The Ensembl variant effect predictor. Genome Biol 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag, 2016. [Google Scholar]
- 87. Mauri M, Elli T, Caviglia G. et al. RAWGraphs: A Visualisation Platform to Create Open Outputs, Proceedings of the 12th Biannual Conference on Italian SIGCHI Chapter. New York, NY, USA: Association for Computing Machinery, 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.