Abstract
Purpose
Tripartite motif-containing protein 13 (TRIM13) directly or indirectly participates in autophagy and apoptosis. However, it remains unclear whether TRIM13 participates in chronic obstructive pulmonary disease (COPD) progression. This study aimed to reveal the molecular mechanisms through which TRIM13 regulates alveolar epithelial cell injury in COPD to provide new molecular targets for COPD treatment.
Methods
The TRIM13 expression levels were determined in clinical COPD patients and a rat emphysema model. A cigarette smoke-induced model of endoplasmic reticulum stress (ERS) and endoplasmic reticulum autophagy (ER-phagy) was developed using A549 cells, and the effects of TRIM13 gene overexpression/knockdown on ERS, ER-phagy, and cell apoptosis were assessed in these cells.
Results
TRIM13 expression was significantly decreased in the lung tissues of COPD patients and rats with emphysema. Moreover, the apoptosis level was significantly increased in the lung tissues of rats with emphysema. TRIM13 gene overexpression reduced the expression levels of ERS-related molecules (GRP78, GRP94, XBP-1, and eIF2a) in the COPD model; it also lowered the ER-phagy level, as evidenced by decreased number of autolysosomes observed by transmission electron microscopy, improved endoplasmic reticulum structure, reduced LC3-II/LC3-I and Beclin1 expression levels, and increased expression level of the autophagy inhibitory molecule Bcl-2. TRIM13 gene knockdown, however, led to opposite results.
Conclusion
TRIM13 expression attenuated alveolar epithelial cell injury in COPD by inhibiting ERS-induced ER-phagy.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00408-024-00753-8.
Keywords: Chronic obstructive pulmonary disease, TRIM13, ERS, ER-phagy, Cell apoptosis
Introduction
Chronic obstructive pulmonary disease (COPD) is an irreversible condition caused by damage to the airways or alveoli by exposure to harmful particles or gases, resulting in persistent respiratory symptoms and airflow obstruction. Smoking is an important risk factor of COPD development [1–3]. Cigarette smoke (CS) can induce the production of reactive oxygen species in cells, thereby triggering endoplasmic reticulum stress (ERS)-mediated cell death [4, 5]. In the ERS process, various signaling pathways are activated in the cells, such as unfolded protein response, endoplasmic reticulum (ER) overload response, and apoptosis pathway, in response to conditions, such as protein misfolding, aggregation of unfolded proteins, and calcium ion imbalance within the ER. ERS can induce the expression of ER-related molecular chaperones, such as the glucose regulatory proteins GRP78 and GRP94, resulting in protective effects against ERS. It can also independently induce the apoptosis of endogenous cells, ultimately influencing how cells respond to stress through adaptation, injury, or apoptosis [6]. ERS can also promote ER autophagy (ER-phagy). ER-phagy is the process of regulating ER fragments and delivering them to lysosomes for degradation; it is used to degrade excess ER membranes, thereby maintaining the steady state of ER mass and capacity [7]. Several recent studies have indicated that COPD patients have ERS [4, 8, 9]. ERS and ER-phagy may be involved in COPD progression by inducing excessive ER pressure, oxidative stress, and inflammatory response in cells [10].
Tripartite motif-containing protein 13 (TRIM13) is a novel transmembrane E3 ubiquitin ligase located on the ER. It is a nonclassical ER phage receptor involved in protein degradation and ER homeostasis [11]. TRIM13 contains a RING domain that mediates ubiquitin transfer by binding to proteins or substrates [12, 13]. Recent studies have shown that the ER-phagy degradation process is initiated following the self-ubiquitination of TRIM13 through the lysine 63 (K63) chain [14]. The onset of COPD is associated with cellular autophagy and apoptosis. COPD patients with emphysema exhibit enhanced ubiquitination. Moreover, skeletal muscle atrophy, one of the main skeletal muscle/extrapulmonary complications of COPD, is also associated with ubiquitination. TRIM13 is directly or indirectly involved in cellular autophagy, apoptosis, and ubiquitination. However, there is currently no relevant research on the relationship between TRIM13 and COPD.
Therefore, in the present study, we aimed to elucidate the role and mechanism of TRIM13 in the occurrence and development of COPD to provide new targets for COPD treatment. We performed gene chip sequencing and differential expression analysis in the early stage to identify and compare differentially expressed genes in COPD patients and the control group. By conducting Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, we found that TRIM13 participates in multiple pathways involved in the pathogenesis of COPD [15]. Based on these findings, we speculate that TRIM13 may regulate alveolar epithelial cell damage in COPD through ERS-induced ER-phagy.
Materials and Methods
Collection of Clinical Samples
Noncancerous tissues adjacent to the edge of the excised tissue specimen and located at > 5 cm from the lesion were collected from lung cancer patients who were admitted to the Department of Thoracic Surgery of the First Affiliated Hospital of Kunming Medical University from April 2023 to October 2023 and underwent lung resection due to the indeterminate nature of their lung masses. Based on the presence of COPD, the patients were assigned to the COPD group (n = 9) and the control group (n = 6). This study was approved by the Ethics Committee of the First Affiliated Hospital of Kunming Medical University [Approval Number: (2022) Lun Shen L No. 196]. This study informed consent was obtained from the patients for participation in the study.
Development of a Rat Model of Emphysema by Exposure to Cigarette Smoke
The animal experiments were approved by the animal ethics committee of our hospital (Approval Number: BST-RAT-20230121-01). A rat emphysema model was established by referring to previously reported methods [16]. Ten Sprague–Dawley rats (5 males and 5 females, specific pathogen-free grade, weight: 130–160 g) were randomly divided into a control group (n = 5) and an emphysema model group (n = 5). The rats were provided standard chow and water ad libitum. They were housed in a controlled experimental environment (temperature: 22 ± 2 °C; light hours: 6:00–18:00). For generating the emphysema model, the rats were placed in a PAB-S200 animal passive smoking exposure system comprising five components: a passive smoking smoke poisoning box, a cigarette smoke (CS) generator, a gas sensor, a signal conditioner, and a real-time data recording and analysis software. The rats were exposed to CS once per day using 5 sets of 3 cigarettes each/15 min with a 3-min rest between each set of cigarettes; each cigarette contained 1.0 mg of nicotine and 11 mg of tar. This exposure treatment was administered 6 days/week for 4 months. Rats in the control group were exposed to room air. All rats were sacrificed after the last CS exposure session.
Construction of CS-Induced ERS and ER-Phagy Cell Models
Preparation of CS Extract (CSE)
A vacuum pump was used to extract the smoke generated by the combustion of two cigarettes, and the extracted smoke particles were passed through and dissolved in 25 mL of serum-free Ham’s F-12 K medium to prepare a CSE. This extract was filtered and sterilized by passing a 0.22-µm filter and stored at 4 °C for subsequent use.
Cell Treatment
The CSE was added to Ham’s F-12 K complete medium to achieve working concentrations of 0% (control group) and 0.5%, 1.0%, 2.5%, 5.0%, and 10.0% (treatment groups).
Construction and Transfection of Lentiviral Vectors
Lentiviral vectors for OE-NC, OE-TRIM13, sh-NC, and sh-TRIM13 (OE-NC: Overexpression Control; OE-TRIM13: Stable TRIM13 Overexpression Cell Line; sh-NC: Interference Control; sh-TRIM13: Stable TRIM13 Interference Cell Line) were constructed by Heyuan Biotech (Shanghai, China). Lentivirus transfection was performed in accordance with a previously reported method [16]. After 72 h of transfection, the transfected cells were screened with 2-µg/mL puromycin, and transfection efficiency was confirmed by inverted fluorescence microscopy. The TRIM13 expression level was estimated by quantitative real-time PCR (RT-qPCR). The primer sequences are provided in Supplementary Material 1.
Pathological Testing
The expression and distribution of TRIM13 in lung tissues were determined by immunohistochemistry [16]. The paraffin-embedded tissue sections were dewaxed, hydrated, and washed with PBS. The sections were then incubated overnight at 4 °C with anti-TRIM13 antibodies (1:150, Abcam), followed by incubation with HRP-conjugated goat anti rabbit IgG secondary antibodies at 37 °C for 30 min. The sections were then stained with diaminobenzidine and observed and photographed under a microscope (Nikon). Quantitative analysis of tissue staining was performed using ImageJ Plus software.
Hematoxylin–Eosin (HE) Staining
The lung tissues were stained in accordance with the instructions of the HE Staining Kit and observed and photographed under a microscope.
qRT-PCR
Total RNA was extracted from tissues and cells using the TriQuick reagent, chloroform, isopropanol, and 75% ethanol. Next, the RT First-Strand cDNA Synthesis Kit (Servicebio®) was used to reverse transcribe the extracted total RNA into cDNA. RT-qPCR was performed using a 2× SYBR Green qPCR Master Mix Kit and the StepOnePlus Real-Time PCR system (Applied Biosystems). The expression level of the target genes relative to that of the GAPDH gene was estimated using the 2−∆∆Ct method [17]. The primer sequences are provided in Supplementary Material 2.
Western Blotting (WB) Assay
The tissues and cells were lysed in RIPA lysis buffer containing PMSF for extracting the total protein. The protein concentration was measured and homogenized using the BCA Protein Assay Kit. SDS-PAGE gel electrophoresis was then performed to separate the proteins, and the separated proteins were transferred to the polyvinylidene fluoride membrane. The membrane was then blocked at room temperature for 1 h and subsequently incubated overnight at 4 °C with antibodies against the target protein. The membrane was then incubated with HRP-conjugated secondary antibodies at room temperature for 1 h. Protein bands were visualized using the ECL reagent kit, and the target bands were visualized using a chemiluminescent imaging system. The scanned images were quantified with ImageJ software.
TUNEL Staining
TUNEL staining of tissues and cells was performed in accordance with the instructions of the TUNEL staining kit. The stained tissues and cells were observed and photographed under a fluorescence microscope. ImageJ software was used to quantify positive DAPI- and TUNEL-stained cells.
Immunofluorescence Assay
Immunofluorescence detection of LC3 expression in A549 cells was performed as follows. Following lentiviral transfection, the cells were treated with 10% CSE for 24 h. After cell fixation and permeabilization, PBS containing bovine serum albumin was added to reduce nonspecific fluorescence. The cells were initially incubated overnight with anti-LC3 antibodies (dilution: 1∶500), followed by incubation with secondary antibodies (dilution: 1:2000) for 30 min. The cell nuclei were stained with DAPI for 2 min, and fluorescence imaging was performed after the slides were blocked with a blocking agent.
Transmission Electron Microscopy
Cells were fixed in glutaraldehyde for performing transmission electron microscopy. The samples were observed with the JEM-1400Plus transmission electron microscope.
Monodansylcadaverine (MDC) Staining
MDC is a fluorescent probe used to detect autophagy in cells. Lentivirus-transfected A549 cells were uniformly seeded onto a cell culture plate and treated with 10% CSE for 24 h. The fluorescence was detected using a Cell Autophagy Staining Kit (MDC method) in according with the manufacturer’s instructions. Autophagy was observed under a fluorescence microscope.
Flow Cytometry Analysis
Cell apoptosis was detected using the Annexin V-Fluor 647/PI Apoptosis Detection Kit. After cell resuspension, 5-µL Annexin V-Fluor 647 and 5-µL propidium iodide were added to the cells for staining for 10 min. The BD Accuri™ C6 Plus Flow Cytometer was used to detect the number of apoptotic cells within 1 h (based on analysis of 10,000 cells per sample). FlowJo software was utilized to analyze the data.
Statistical Analysis
SPSS 27.0 and GraphPad Prism 7.0 were used to analyze the statistical data. The data are expressed as mean ± standard deviation (SD). Unpaired t test and one-way analysis of variance were used to evaluate significant differences between two and multiple groups, respectively. p < 0.05 was considered to indicate a significant difference.
Results
TRIM13 is Expressed at a Low Level in COPD Patients’ Lung Tissues with Alveolar Cell Apoptosis
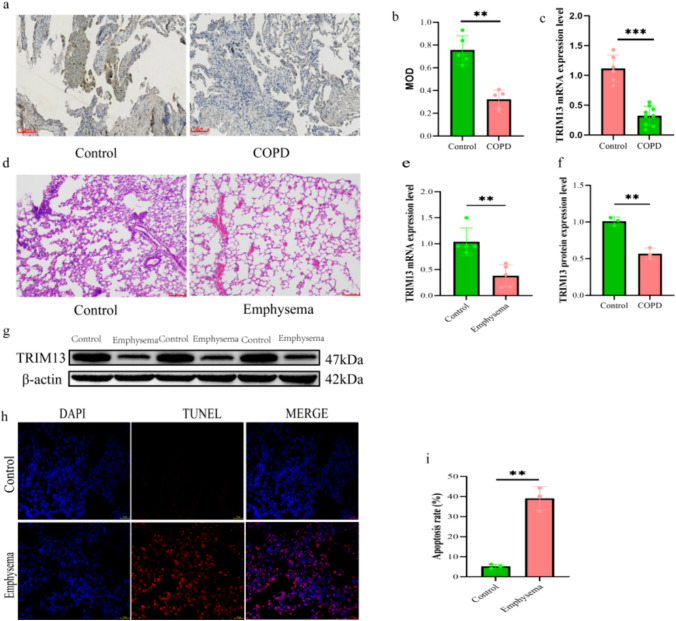
The TRIM13 expression level was significantly reduced in the lung tissues of COPD patients (p < 0.01; Fig. 1a–c). HE staining of the lung tissues of the rat emphysema model showed apparent disorganization of the tissue, dilated alveoli, and thin or even broken alveolar septa; the dilated alveoli fused to form larger cystic cavities, together with the presence of degenerated and necrotic bronchial epithelial cells. These findings indicated that the rat emphysema model was successfully constructed (Fig. 1d). The TRIM13 expression level was significantly decreased in the lung tissues of the emphysema model group (p < 0.01; Fig. 1e–g), and the apoptosis rate was also significantly increased (p < 0.01; Fig. 1h, i).
Fig. 1.
TRIM13 expression in the lung tissues of COPD patients and rats with emphysema. a Immunohistochemical staining results of TRIM13 expression in the lung tissues of COPD patients (scale bar = 100 µm). b Quantitative analysis of immunohistochemical staining of TRIM13. c qRT-PCR analysis for detecting the TRIM13 expression level in the lung tissues of COPD patients. d HE staining of rat lung tissues (scale bar = 100 µm). e qRT-PCR analysis for detecting the TRIM13 expression level in rat lung tissues. f, g Western blotting assay for detecting the TRIM13 expression level in rat lung tissues. h Determination of cell apoptosis in rat lung tissues (scale bar = 25 µm). i Apoptosis rate of cells in rat lung tissues. **p < 0.01 vs. control, ***p < 0.001 vs. control
CSE Induces ERS and ER-Phagy in A549 Cells
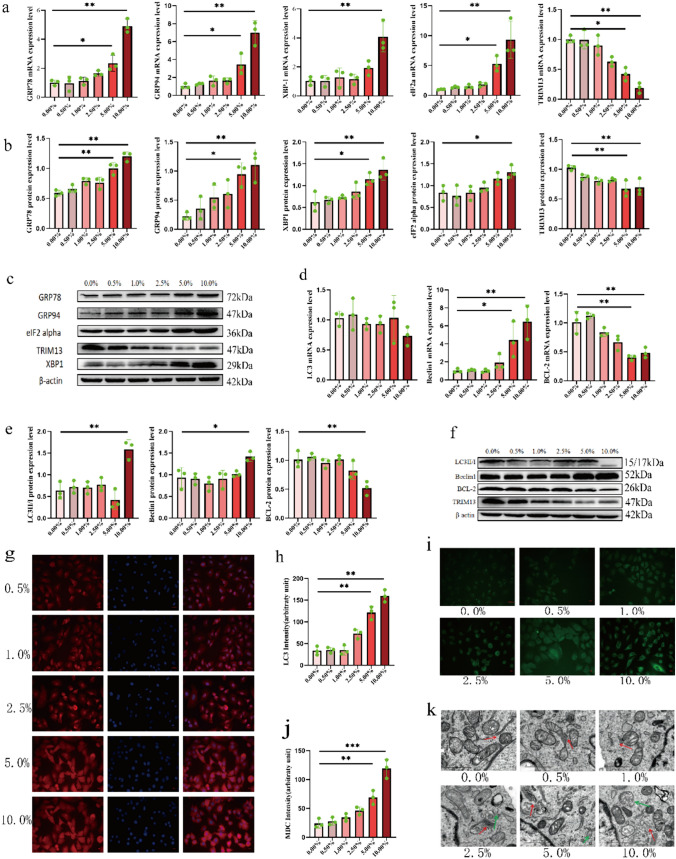
A549 cells were treated with medium containing different concentrations of CSE for 24 h, and the expression of ERS-related molecules and ER-phagy-related molecules was detected in alveolar cells. Autophagy was observed by transmission electron microscopy and MDC staining. qRT-PCR analysis, WB assay, and immunofluorescence assay showed a significant increase in the expression levels of ERS-related molecules (GRP78, GRP94, XBP-1, and eIF2a) and ER-phagy-related molecules (Beclin1 and LC3-II/LC3-I) in cells treated with 10% CSE; however, the expression levels of TRIM13 and Bcl-2 were significantly reduced (Fig. 2a–h). MDC staining also showed that the number of autophagosomes was significantly increased in 10% CSE-treated A549 cells (Fig. 2i, j). Transmission electron microscopy revealed autophagic vesicles with crescent-shaped or cup-shaped, bilayered, or multilayered membranes with an increase in encapsulated periplasmic components around the ER; moreover, the swelling of the ER increased as CSE concentration increased (Fig. 2k). These results indicated that 10% CSE could significantly induce ERS and ER-phagy in A549 cells. Hence, 10% CSE was identified as the concentration for developing the ERS and ER-phagy cell models and then used in subsequent experiments.
Fig. 2.
Construction of the ERS and ER-phagy cell models. a mRNA expression levels of ERS-related molecules after CSE treatment. b, c Protein expression levels of ER chaperone-related molecules after CSE treatment. d mRNA expression levels of ER-phagy-related molecules after CSE treatment. e, f Protein expression levels of ER-phagy-related molecules after CSE treatment. g, h Immunofluorescence detection of LC3 expression after CSE treatment (scale bar = 25 µm). i, j MDC staining for detecting cellular autophagosomes after CSE treatment (scale bar = 25 µm). k Transmission electron microscopy for detecting intracellular autophagosomes and ultrastructural changes after CSE treatment (scale bar = 1 µm, red arrowheads represent ER, green arrowheads represent autophagic vesicles). *p < 0.05 vs. control, **p < 0.01 vs. control, ***p < 0.001 vs. control
TRIM13 Inhibits CSE-Induced ER-Phagy in A549 Cells
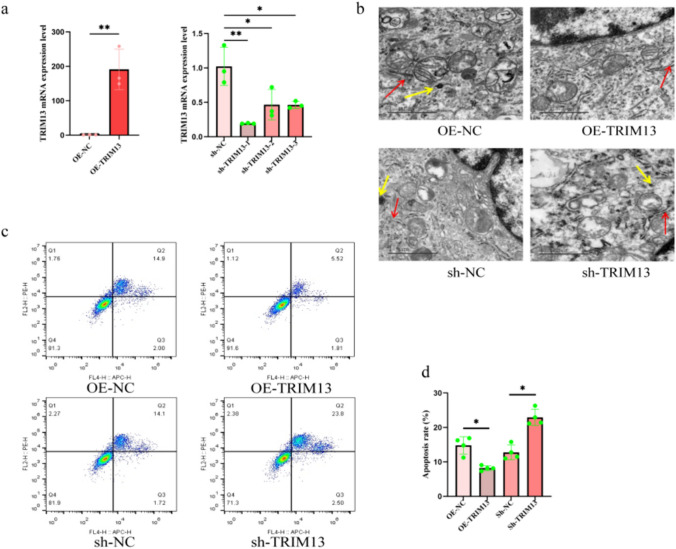
Following lentiviral transfection of A549 cells for 72 h, the TRIM13 mRNA expression levels showed significant changes. The TRIM13 gene was successfully overexpressed or knocked down in A549 cells (Fig. 3a). The number of autophagosomes decreased in the OE-TRIM13 group after the treatment of cells with 10% CSE; moreover, the number of structurally intact apoptotic cells in the ER-phagy group was significantly reduced. In contrast, in the sh-TRIM13 group, the number of autophagosomes increased, the endoplasmic reticulum was severely swollen, and the number of apoptotic cells increased significantly (Fig. 3b–d). Therefore, we concluded that TRIM13 overexpression enabled to improve ER-phagy and attenuate apoptosis, which is a potential COPD-targeted therapeutic approach.
Fig. 3.
Effect of TRIM13 overexpression/knockdown on ER-phagy and apoptosis in CSE-treated A549 cells. a Relative expression levels of TRIM13 after lentiviral transfection. b Ultrastructural changes in autophagosomes after the overexpression/knockdown of TRIM13 (scale bar = 1 µm, red arrowheads represent ER, yellow arrowheads represent autophagosomes). c, d Apoptosis after overexpression/knockdown of TRIM13. *p < 0.05, **p < 0.01, ***p < 0.001 vs. the OE-NC and sh-NC groups. OE overexpression, sh knockdown, NC negative control
Molecular Mechanism of TRIM13 Inhibition of CSE-Induced ER-Phagy
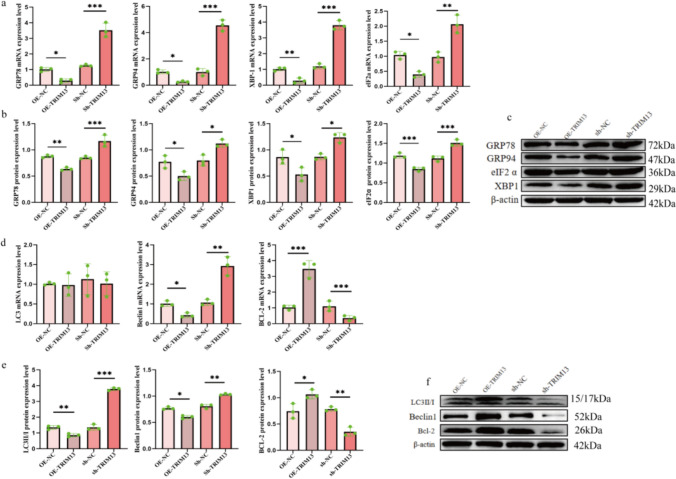
TRIM13 overexpression in 10% CSE-treated A549 cells decreased the expression levels of GRP78, GRP94, XBP-1, and eIF2a as compared to those in the OE-NC group. In contrast, TRIM13 knockdown elevated the expression levels of GRP78, GRP94, XBP-1, and eIF2a, thus suggesting that TRIM13 overexpression alleviated the status of CSE-induced ERS by downregulating the expression of ERS-associated molecules (Fig. 4a–c). In the ER-phagy cell model, we found that Beclin1 and LC3-II/ LC3-I expression levels were significantly reduced, and Bcl-2 expression level was significantly elevated following TRIM13 overexpression. A contrasting result was observed following TRIM13 knockdown, which suggested that TRIM13 overexpression suppressed ER-phagy (Fig. 4d–f). ERS and ER-phagy induced disorganization of the structure and function of the ER, which may trigger apoptosis. Combined with the previous experimental results, TRIM13 overexpression attenuated apoptosis in A549 cells by inhibiting ERS-induced ER-phagy; this might be the molecular mechanism underlying the protective effect of TRIM13 against COPD.
Fig. 4.
Effect of TRIM13 on the expression of CSE-induced ER-phagy-related molecules in A549 cells. a–c Effect of TRIM13 overexpression/knockdown on the expression level of ERS-related molecules in 10% CSE-treated alveolar epithelial cells. d–f Effect of TRIM13 overexpression/knockdown on the expression level of autophagy- and apoptosis-related molecules in 10% CSE-treated alveolar epithelial cells. *p < 0.05, **p < 0.01, ***p < 0.001 vs. the OE-NC group and the sh-NC group
Discussion
CS contains many toxic substances, such as tar, nicotine, and carbon monoxide. The stimulation of lung cells by CS and other toxic substances triggers the ERS process. Moreover, if ERS persists or is too strong, it will lead to cellular dysfunction and even apoptosis, thereby promoting COPD development [18]. Several researchers have previously constructed COPD models using the CS-exposed mice and cells and evaluated the expression profiles of ERS-related molecules. Jorgensen et al. performed high-density microarray and protein blotting analysis and reported that the expression levels of several ERS regulators, such as eIF2α, and ERS marker genes, such as XBP1, are upregulated in the CS-exposed human bronchial epithelioid-like cells. Moreover, the expression levels of ERS-related proteins were gradually increased [19]. Kenche et al. found that eIF2α and XBP1 expression levels were increased in mice exposed to CS [20]. Another study also reported an increase in the GRP78 expression level in the bronchoalveolar lavage fluid of smokers [21]. In the present study, we successfully constructed COPD rat and cell models using a CS exposure system and a growth medium containing CSE, respectively. Furthermore, ERS and ER-phagy were successfully induced by treating A549 cells with 10% CSE for 24 h. These findings indicated that CS can trigger ERS and ER-phagy in alveolar cells. Dithiothreitol-induced ERS increases ER-phagy-related protein levels in hepatocytes [22]. Furthermore, ERS can promote structural and functional changes in the ER [23]. Additionally, an excessive level of ER-phagy caused increased apoptosis in HeLa cells and human embryonic kidney (HEK) cells [24, 25]. These findings suggest that ERS induces ER-phagy and therefore promotes apoptosis.
TRIM13 is a nonclassical ER-phagy receptor, and among more than 80 TRIM family members, TRIM13 is the only protein localized to the ER membrane [26]. TRIM13 is involved in a variety of ER and protein degradation processes, and it is an important member of the ER to maintain homeostasis [27, 28]. A previous study revealed that the TRIM family members are involved in ER-phagy activities [26]. The primary mechanism of action involves the interaction of the TRIM protein with p62, utilizations of the LIR structural domain of the p62 protein to complete LC3 recruitment, and promotion of the formation of the isolation membrane and autophagosomes [11]. Tomar et al. observed that TRIM13 regulates the initiation of autophagy during ERS in HEK293 cells [29]. The downregulation of the expression of the TRIM family members, including TRIM13, TRIM32, TRIM44, and TRIM59, induced cell cycle arrest and increased cellular apoptosis in T-cell acute lymphoblastic leukemia. Based on this finding, it was inferred that the upregulation of TRIM13 expression could decrease cell death [30]. The results of the present study demonstrated that TRIM13 overexpression inhibits alveolar epithelial cell apoptosis by alleviating ERS and ER-phagy, which could be the underlying molecular mechanism through which TRIM13 protects against COPD.
To date, studies on the association between TRIM13 and COPD have not yet been reported. However, previous studies have shown that TRIM13 deficiency can lead to age-related autoinflammatory diseases. In one study, 5-month-old TRIM13-deficient mice showed no apparent signs of inflammation; however, 10-month-old TRIM13-deficient mice showed extensive inflammatory cell infiltration in the lungs [31]. COPD is a typical age-related disease and its pathogenesis involves lung inflammation; furthermore, TRIM13 may be directly or indirectly involved in the pathogenesis of COPD. The present study observed for the first time that the downregulation of TRIM13 is involved in the pathogenesis of COPD in both COPD patients and mouse lung tissues. The regulation of the TRIM13 expression level can effectively control ERS and ER-phagy levels in COPD patients, thereby reducing damage to alveolar epithelial cells. This may provide new directions and targets for developing drugs for COPD treatment. The upregulation of the TRIM13 protein expression level has also been reported in macrophages after stimulation with the TLR2 ligand [32]. This encouraging result suggests that patients with COPD may benefit from similar treatments.
The present study has a limitation. Although this study showed that TRIM13 is closely related to ER-phagy and changes in the ER were observed before and after overexpression/knockdown of TRIM13, it was not possible to determine whether these changes represent the occurrence of ER-phagy; this aspect will be investigated in future studies.
Conclusion
The present study confirmed that TRIM13 reduces alveolar epithelial cell damage in COPD by inhibiting ER-phagy induced by ERS. In conclusion, the results of this study provide a theoretical basis for using TRIM13 as a target for ERS and ER-phagy therapy in COPD.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank TopEdit for its linguistic assistance during the preparation of this manuscript.
Author Contributions
All authors significantly contributed to the present work. Jianqing Zhang supervised and guided the study and was involved in study concept and design. Yaling Xiang performed the experiments, analyzed the data, and wrote the manuscript. Chuntao Li, Zhiyuan Wang, Jiagang Feng, and Jiaqiang Zhang revised and critical reviewed the manuscript. Yue Yang and Jinbiao Zhou assisted in completing the experiments. All authors have approved the final manuscript version submitted for publication and agree to take responsibility for all aspects of the work.
Funding
This work for supported by the National Natural Science Foundation of China (Grant Number: 8206010196).
Data Availability
No datasets were generated or analyzed during the current study. The copyright of the images used in this article belongs to the author and can be copied, edited, or used by the journal.
Declarations
Conflict of interest
The authors declare that they have no potential conflicts of interest related to this work.
Consent to Participate
This study informed consent was obtained from the patients for participation in the study.
Ethical Approval
This study was approved by the Ethics Committee of the First Affiliated Hospital of Kunming Medical University [Approval Number: (2022) Lun Shen L No. 196]. The animal experiments were approved by the animal ethics committee of our hospital (Approval Number: BST-RAT-20230121-01).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Adeloye D, Song P, Zhu Y, Campbell H, Sheikh A, Rudan I (2022) Global, regional, and national prevalence of, and risk factors for, chronic obstructive pulmonary disease (COPD) in 2019: a systematic review and modelling analysis. Lancet Respir Med 10(5):447–458. 10.1016/s2213-2600(21)00511-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Safiri S, Carson-Chahhoud K, Noori M, Nejadghaderi SA, Sullman MJM, Ahmadian Heris J, Ansarin K, Mansournia MA, Collins GS, Kolahi AA, Kaufman JS (2022) Burden of chronic obstructive pulmonary disease and its attributable risk factors in 204 countries and territories, 1990–2019: results from the Global Burden of Disease Study 2019. BMJ 378:e069679. 10.1136/bmj-2021-069679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sevilla-Montero J, Labrousse-Arias D, Fernández-Pérez C, Fernández-Blanco L, Barreira B, Mondéjar-Parreño G, Alfaro-Arnedo E, López IP, Pérez-Rial S, Peces-Barba G, Pichel JG, Peinado VI, Cogolludo Á, Calzada MJ (2021) Cigarette smoke directly promotes pulmonary arterial remodeling and Kv7.4 channel dysfunction. Am J Respir Crit Care Med 203(10):1290–1305. 10.1164/rccm.201911-2238OC [DOI] [PubMed] [Google Scholar]
- 4.Fan L, Li L, Yu X, Liang Z, Cai T, Chen Y, Xu Y, Hu T, Wu L, Lin L (2020) Jianpiyifei II granules suppress apoptosis of bronchial epithelial cells in chronic obstructive pulmonary disease via inhibition of the reactive oxygen species-endoplasmic reticulum stress-Ca(2+) signaling pathway. Front Pharmacol 11:581. 10.3389/fphar.2020.00581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King AP, Wilson JJ (2020) Endoplasmic reticulum stress: an arising target for metal-based anticancer agents. Chem Soc Rev 49(22):8113–8136. 10.1039/d0cs00259c [DOI] [PubMed] [Google Scholar]
- 6.Hetz C, Zhang K, Kaufman RJ (2020) Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol 21(8):421–438. 10.1038/s41580-020-0250-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei F, Xie Q, Huang Z, Yang A, Duan Y (2022) Induction of autophagy and endoplasmic reticulum autophagy caused by cadmium telluride quantum dots are protective mechanisms of yeast cell. J Appl Toxicol 42(7):1146–1158. 10.1002/jat.4282 [DOI] [PubMed] [Google Scholar]
- 8.Chen R, Michaeloudes C, Liang Y, Bhavsar PK, Chung KF, Ip MSM, Mak JCW (2022) ORMDL3 regulates cigarette smoke-induced endoplasmic reticulum stress in airway smooth muscle cells. J Allergy Clin Immunol 149(4):1445-1457.e5. 10.1016/j.jaci.2021.09.028 [DOI] [PubMed] [Google Scholar]
- 9.Wang HL, Chen FQ, Wu LJ (2022) Ephedrine ameliorates chronic obstructive pulmonary disease (COPD) through restraining endoplasmic reticulum (ER) stress in vitro and in vivo. Int Immunopharmacol 103:107842. 10.1016/j.intimp.2021.107842 [DOI] [PubMed] [Google Scholar]
- 10.Li X, Jiang X, Zeng R, Lai X, Wang J, Liu H, Wu H, He J, Liu L, Zhu Z, Li J, Liang X (2024) Formononetin attenuates cigarette smoke-induced COPD in mice by suppressing inflammation, endoplasmic reticulum stress, and apoptosis in bronchial epithelial cells via AhR/CYP1A1 and AKT/mTOR signaling pathways. Phytother Res. 10.1002/ptr.8104 [DOI] [PubMed] [Google Scholar]
- 11.Ji CH, Kim HY, Heo AJ, Lee SH, Lee MJ, Kim SB, Srinivasrao G, Mun SR, Cha-Molstad H, Ciechanover A, Choi CY, Lee HG, Kim BY, Kwon YT (2019) The N-degron pathway mediates ER-phagy. Mol Cell 75(5):1058-1072.e9. 10.1016/j.molcel.2019.06.028 [DOI] [PubMed] [Google Scholar]
- 12.Ikeda K, Inoue S (2012) TRIM proteins as RING finger E3 ubiquitin ligases. Adv Exp Med Biol 770:27–37. 10.1007/978-1-4614-5398-7_3 [DOI] [PubMed] [Google Scholar]
- 13.Ozato K, Shin DM, Chang TH, Morse III HC (2008) TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol 8(11):849–860. 10.1038/nri2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji CH, Kim HY, Heo AJ, Lee MJ, Park DY, Kim DH, Kim BY, Kwon YT (2020) Regulation of reticulophagy by the N-degron pathway. Autophagy 16(2):373–375. 10.1080/15548627.2019.1695402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C, Liu H, Zhang J, Zhang J, Feng J (2020) LncRNA BMF-AS1 exerts anti-apoptosis function in COPD by regulating BMF expression. Pak J Zool. 10.17582/journal.pjz/20190909090912 [Google Scholar]
- 16.Zhong Y, Li C, Xiang Y, Zhou J, Zhang J (2023) LncRNA RP11–521C20.3 inhibits cigarette smoke extract-induced apoptosis in A549 cells by targeting BMF signaling. Int J Chron Obstruct Pulmon Dis 18:669–682. 10.2147/copd.S395568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD (2013) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 18.Malhotra D, Thimmulappa R, Vij N, Navas-Acien A, Sussan T, Merali S, Zhang L, Kelsen SG, Myers A, Wise R, Tuder R, Biswal S (2009) Heightened endoplasmic reticulum stress in the lungs of patients with chronic obstructive pulmonary disease: the role of Nrf2-regulated proteasomal activity. Am J Respir Crit Care Med 180(12):1196–1207. 10.1164/rccm.200903-0324OC [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Jorgensen E, Stinson A, Shan L, Yang J, Gietl D, Albino AP (2008) Cigarette smoke induces endoplasmic reticulum stress and the unfolded protein response in normal and malignant human lung cells. BMC Cancer 8:229. 10.1186/1471-2407-8-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenche H, Baty CJ, Vedagiri K, Shapiro SD, Blumental-Perry A (2013) Cigarette smoking affects oxidative protein folding in endoplasmic reticulum by modifying protein disulfide isomerase. FASEB J 27(3):965–977. 10.1096/fj.12-216234 [DOI] [PubMed] [Google Scholar]
- 21.Aksoy MO, Kim V, Cornwell WD, Rogers TJ, Kosmider B, Bahmed K, Barrero C, Merali S, Shetty N, Kelsen SG (2017) Secretion of the endoplasmic reticulum stress protein, GRP78, into the BALF is increased in cigarette smokers. Respir Res 18(1):78. 10.1186/s12931-017-0561-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo YX, Han B, Yang T, Chen YS, Yang Y, Li JY, Yang Q, Xie RJ (2022) Family with sequence similarity 134 member B-mediated reticulophagy ameliorates hepatocyte apoptosis induced by dithiothreitol. World J Gastroenterol 28(23):2569–2581. 10.3748/wjg.v28.i23.2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao T, Wang J, Wang X, Wang Y, Mao H, Liu X (2019) The PERK/Nrf2 pathway mediates endoplasmic reticulum stress-induced injury by upregulating endoplasmic reticulophagy in H9c2 cardiomyoblasts. Life Sci 237:116944. 10.1016/j.lfs.2019.116944 [DOI] [PubMed] [Google Scholar]
- 24.Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson ÅB (2012) Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem 287(23):19094–19104. 10.1074/jbc.M111.322933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang S, Lin Y, Yao H, Yang C, Zhang L, Luo B, Lei Z, Cao L, Lin N, Liu X, Lin Z, He C (2018) The role of unfolded protein response and ER-phagy in quantum dots-induced nephrotoxicity: an in vitro and in vivo study. Arch Toxicol 92(4):1421–1434. 10.1007/s00204-018-2169-0 [DOI] [PubMed] [Google Scholar]
- 26.Hatakeyama S (2017) TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci 42(4):297–311. 10.1016/j.tibs.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 27.Di Rienzo M, Romagnoli A, Antonioli M, Piacentini M, Fimia GM (2020) TRIM proteins in autophagy: selective sensors in cell damage and innate immune responses. Cell Death Differ 27(3):887–902. 10.1038/s41418-020-0495-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altier C, Garcia-Caballero A, Simms B, You H, Chen L, Walcher J, Tedford HW, Hermosilla T, Zamponi GW (2011) The Cavβ subunit prevents RFP2-mediated ubiquitination and proteasomal degradation of L-type channels. Nat Neurosci 14(2):173–180. 10.1038/nn.2712 [DOI] [PubMed] [Google Scholar]
- 29.Tomar D, Singh R, Singh AK, Pandya CD, Singh R (2012) TRIM13 regulates ER stress induced autophagy and clonogenic ability of the cells. Biochim Biophys Acta 1823(2):316–326. 10.1016/j.bbamcr.2011.11.015 [DOI] [PubMed] [Google Scholar]
- 30.Cheng H, Chen L, Hu X, Qiu H, Xu X, Gao L, Tang G, Zhang W, Wang J, Yang J, Huang C (2019) Knockdown of MAML1 inhibits proliferation and induces apoptosis of T-cell acute lymphoblastic leukemia cells through SP1-dependent inactivation of TRIM59. J Cell Physiol 234(4):5186–5195. 10.1002/jcp.27323 [DOI] [PubMed] [Google Scholar]
- 31.Li X, Yu Z, Fang Q, Yang M, Huang J, Li Z, Wang J, Chen T (2022) The transmembrane endoplasmic reticulum-associated E3 ubiquitin ligase TRIM13 restrains the pathogenic-DNA-triggered inflammatory response. Sci Adv 8(4):eabh0496. 10.1126/sciadv.abh0496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang B, Baek SH (2017) Trim13 potentiates toll-like receptor 2-mediated nuclear factor κB activation via K29-linked polyubiquitination of tumor necrosis factor receptor-associated factor 6. Mol Pharmacol 91(4):307–316. 10.1124/mol.116.106716 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analyzed during the current study. The copyright of the images used in this article belongs to the author and can be copied, edited, or used by the journal.