Abstract
Increases in sequencing capacity, combined with rapid accumulation of publications and associated data resources, have increased the complexity of maintaining associations between literature and genomic data. As the volume of literature and data have exceeded the capacity of manual curation, automated approaches to maintaining and confirming associations among these resources have become necessary. Here we present the Data Citation Explorer (DCE), which discovers literature incorporating genomic data that was not formally cited. This service provides advantages over manual curation methods including consistent resource coverage, metadata enrichment, documentation of new use cases, and identification of conflicting metadata. The service reduces labor costs associated with manual review, improves the quality of genome metadata maintained by the U.S. Department of Energy Joint Genome Institute (JGI), and increases the number of known publications that incorporate its data products. The DCE facilitates an understanding of JGI impact, improves credit attribution for data generators, and can encourage data sharing by allowing scientists to see how reuse amplifies the impact of their original studies.
Subject terms: Research data, Genomics
Introduction
The Department of Energy’s (DOE) Joint Genome Institute (JGI, jgi.doe.gov) is a national User Facility that provides state-of-the-art environmental genomics capabilities to the scientific community. JGI has directly supported more than 4,000 researchers and generated over 15 petabytes of data in its 25-year history. Following delivery to the primary investigators (PIs)and a short embargo period, data produced through JGI proposals is made available for public use through a variety of external and JGI-maintained systems. These include Integrated Microbial Genomes & Microbiomes (IMG/M)1, Phytozome2, MycoCosm3, PhycoCosm4, and the JGI Data Portal (data.jgi.doe.gov), as well as the National Center for Biotechnology Information’s (NCBI) Sequence Read Archive (SRA)5 and GenBank6 databases. This data is made public with the understanding that it may have impact beyond the PIs’ original intended uses. A wider appreciation of this concept has led in recent years to greater community emphasis on the importance not just of initial publication and the findings of original data generators, but also of the downstream reuse of public scientific data. Many efforts are underway to make public scientific data more Findable, Accessible, Interoperable, and Reusable (FAIR)7, in order to encourage and facilitate downstream impact.
Motivation: Understanding institutional and individual impact
Organizations providing services or products to a specific community can benefit from a better understanding of their impact within that community. This understanding is essential for directing service improvements that can benefit the organization and the community it serves. JGI thus has strong motivations for capturing citations of its products. Doing so enables the organization to better serve its users by identifying which data and thematic areas are heavily cited as well as those that are underutilized. This can inform researchers as to which topics may be ripe for innovative analysis and uncover ways in which older data can be reused. Citation capture can also direct improvements in operational efficiency and inform policy decisions. Knowing how specific workflows and product offerings contribute to downstream publications facilitates resource allocation for activities that have the greatest scientific impact.
A more complete understanding of data use is essential for appreciating the extent to which the activities of JGI users align with its policies, initiatives, and strategic goals. JGI also has an interest in identifying researchers, regions, institutions, or specific research fields that make heavy use of its products but with which it has little direct engagement or towards which it has not yet made concerted outreach efforts. Extending searches to include data citations in patents and other intellectual property provides a window into potential commercialization of JGI products and can demonstrate contributions to economic growth and innovation. Building a comprehensive picture of product utilization across science and industry demonstrates to key stakeholders (i.e., taxpayers, elected officials, and the scientific community) that funds granted represent a worthwhile investment with compounding returns over time.
Many organizations, including nonprofit entities like Research Data Alliance8 and government entities like NSF, NIH, and DOE9, have long advocated for more transparent links among data producers and data consumers. By building connections between citations of JGI data and the datasets themselves, JGI can attribute credit to the primary investigators and JGI personnel who contributed to the creation of a given dataset. Doing so preserves the provenance of an individual’s work and demonstrates their impact on downstream research. Systematically preserving contributor relationships and roles can inform funders, reviewers, and institutional evaluators of significant contributions that are currently missed when using traditional metrics such as the h-index10 as indicators of research productivity.
New metrics can account for deficiencies in existing methods for research evaluation that rely heavily on publication authorship, the standards for which are uneven at best11,12. For example, an individual’s overall citation count or h-index may not be high, or their inclusion in authorship lists may not be extensive, but their contributions to high-value, frequently utilized data may be significant (i.e., workflow managers and personnel responsible for sample processing). A more granular methodology of research evaluation can also benefit researchers with publications that may not see heavy citation activity, but whose overall contributions as data producers have significant impact across the community. Adopting a contributor evaluation model based on data citations (for example, the “data-index” proposed by Hood et al.13) thus offers improvements in equity and transparency over more traditional metrics grounded in the authorship of scientific publications.
The Problem: Too much literature, too much data
Through anecdotal evidence, the U.S. Department of Energy Joint Genome Institute (JGI) recognizes that a significant body of literature exists that incorporates JGI data products but that does not include formal data citations that attribute credit to either JGI or the individual contributors who produced the data. ‘Formal data citations’ refer in this work to structured, machine-resolvable references that adhere to recent recommendations and are added to the bibliography of a given publication14,15. While a full discussion of community citation practices in the biosciences is out of scope for this work, preliminary works in the earth16, social17, and biomedical sciences18 have begun to explore the extent to which formal data citations fail to represent the full scope of data usage. Other recent domain-agnostic studies found that fewer than 10% of articles describing data usage included a formal data citation19,20. Because the most current recommendations for formal data citations do not accommodate for all means by which data is referenced in the literature, the term ‘citation’ is used here to refer to the broader set of all means by which researchers refer to data leveraged in their studies. According to this usage, the inclusion of a domain-specific identifier in the body text of a publication, for example, would fall under the umbrella of ‘citation’ along with more formal bibliographic references. A recent study mapping full-text mentions of genomic data identifiers by articles in Europe PubMed Central indicates the extent to which these types of citations occur in the literature21.
Publications that cite data informally or implicitly are difficult for JGI to identify for two reasons. First, as these data are hosted by JGI as well as the US National Institutes for Health (NIH) National Center for Biological Information (NCBI), researchers frequently cite NCBI5,6 and other external identifiers (e.g., Human Oral Microbiome Database, Human Microbiome Project) for a given dataset rather than JGI identifiers. The relationship between these external metadata and JGI data to which they refer are often complex and difficult to traverse. Second, the context within individual publications in which citations of JGI-linked identifiers occur can often be ambiguous or left unindexed by full-text search tools. For example, a JGI-linked identifier could be mistaken for some other identifier by naíve text matching, or could be buried within the supplemental materials of a publication beyond the reach of most searches.
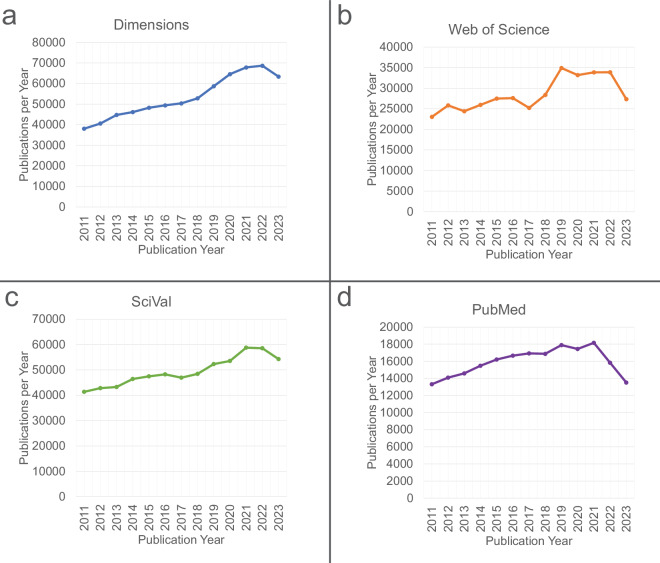
With the rates at which new literature is published and the scale of data production by JGI, it is not feasible for humans to identify all citations of JGI data without automated assistance. Publications associated with the field of “Genetics”’ published from 2011 to 2023 were found to number between 206,974 and 693,491 using research area or MeSH term queries in Web of Science (Clarivate), PubMed (National Library of Medicine)22, Dimensions (Digital Science)23,24, and SciVal (Elsevier). Though growth in the yearly volume of Genetics publications during this time has slowed in recent years, all four sources show steady increases throughout much of this period (Fig. 1). During the same period, the yearly JGI output of raw sequence data increased from just over 30,000 gigabases per year to over 700,000 gigabases per year (Fig. 2).
Fig. 1.
Yearly genetics-associated publications from 2011-2023 in Dimensions (a), Web of Science (b), SciVal (c), and PubMed (d). The net increases in yearly publications were as follows: Dimensions, 66.4% (693,491 total publications). Web of Science, 18.65% (371,079 total publications). SciVal, 31.24% (642,528 total publications), PubMed, 1.48% (206,974 total publications). The research area or MeSH terms used to generate each publication set were “Genetics" (PubMed, SciVal, Dimensions) and “Genetics & Heredity" (Web of Science).
Fig. 2.
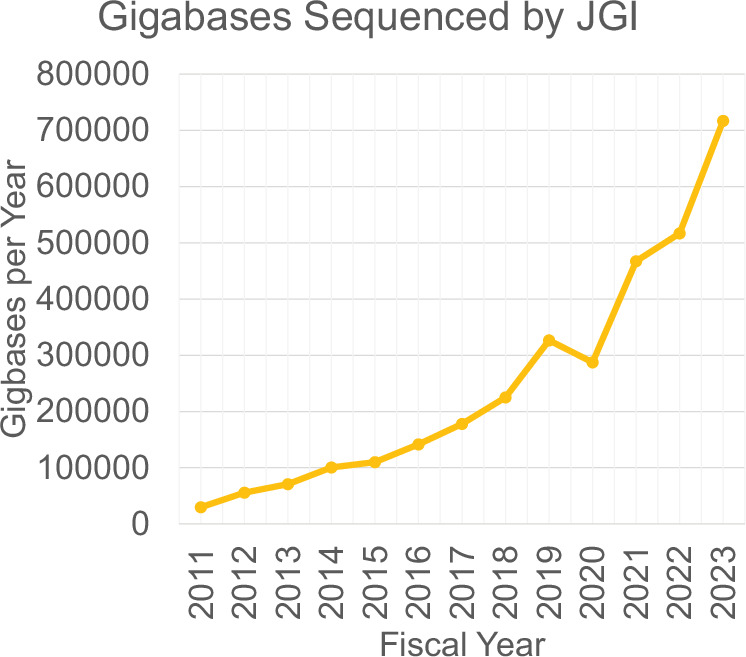
Yearly sequencing production at the DOE Joint Genome Institute (JGI). Note that the fiscal year begins on October 1st of the preceding calendar year. The decrease in FY20 sequencing output was due to the COVID-19 pandemic.
Positive identification of literature citing JGI products is a labor-intensive process when performed manually. This manual process relies on an individual’s expert knowledge within a specific field, which may have specialized literature, subject language terminology, and data resources (see Lafia et al.17 for an example from the social sciences). This process often involves repetitive searches across numerous sources using distinct queries intended to capture a limited set of JGI products. The timeliness, scope, comprehensiveness, and repeatability of the manual search process is limited by the availability of skilled data curators. Consistency and accuracy of manually compiled citation data varies not only among those performing the searches but also individually over time. The accuracy of even well-trained curators may decline over the course of a single work day. Thus, at present, verification and validation of highly accurate citation data requires a duplication of effort among multiple reviewers in order to identify and correct errors. Even with redundant human reviewers, this form of precise manual tracking of information sources is impractical if not impossible when branching paths are discovered through multiple online resources. Nonetheless, manual literature searches are important for collecting initial data and verifying citations, but there are many repetitive components of this process that are amenable to automation. The DCE automates those repetitive tasks, freeing skilled personnel to focus their attention on tasks that resist automation.
An automated solution: The Data Citation Explorer
The overarching goal of the DCE is to systematically and reproducibly identify literature that relies on data stored in JGI repositories but that does not formally cite said data. The system was designed to encapsulate the expertise of data curators into the business logic of a web service. Using a selected subset of metadata fields from JGI genome projects as input, this web service can consistently apply curatorial methods to incrementally discover and traverse additional resources and metadata that are directly associated with a specific genome. Starting with a limited set of validated genome metadata stored in JGI systems, the service automatically performs an exhaustive search of targeted literature and online resources to discover any uses of that associated data. The service provides users with an audit trail that provides a precise explanation of how each additional resource was discovered. In testing, the DCE has been shown to uncover multiple paths to new information about a genome. The service can also re-process genomes at any time to discover new uses and citations, whether or not the data was formally cited. Briefly, the process occurs in two phases:
Crawl genomic data repositories to accumulate new metadata that has been produced in downstream resources
Search for in-text occurrences of unique identifiers in publicly available literature.
Results
During the initial trial of the DCE, 238,994 metadata records from the JGI Archive and Metadata Organizer (JAMO) were fed into the system. This resulted in hits linking 30,641 publications to 78,104 JAMO records. These publications were automatically identified by the DCE via full-text searches in publicly available sources like PubMed Central (see Methods). From this larger set, 998 audit trails linking 576 unique publications to 282 individual JAMO records were sampled and manually evaluated. Evaluation was accomplished by checking the nature of the hits in the citing publication as well as by verifying the feasibility of relationship between keys in the larger audit trail. Briefly, audit trails refer to multi-step linkages established between publications and data that are determined by the DCE. Stratified random sampling of the larger full set of results was used to select a more manageable set for manual evaluation (refer to the Methods section for a full description of the sampling process). Only 10 of the 998 audit trails accounting for 10 out of 576 publications led to irrelevant results (false positives). Examples of true and false positive hits can be seen in Figs. 3 and 4. From the perspective of individual publications, the precision value for connections between publications and JAMO records in this sample was 0.983 (Table 1).
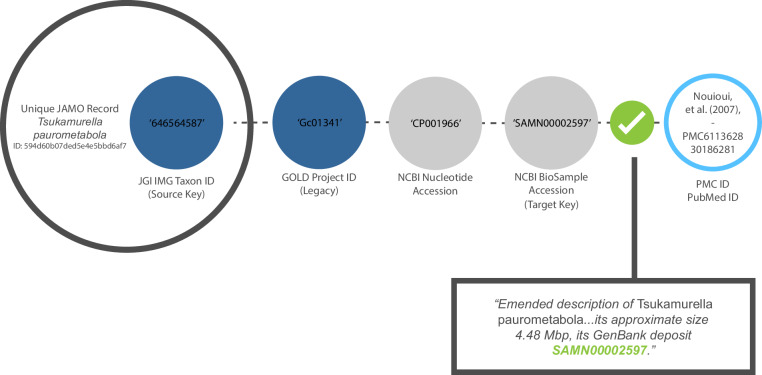
Fig. 3.
Example of an audit trail returning a relevant publication. The target key refers unambiguously to the data in the original JAMO record.
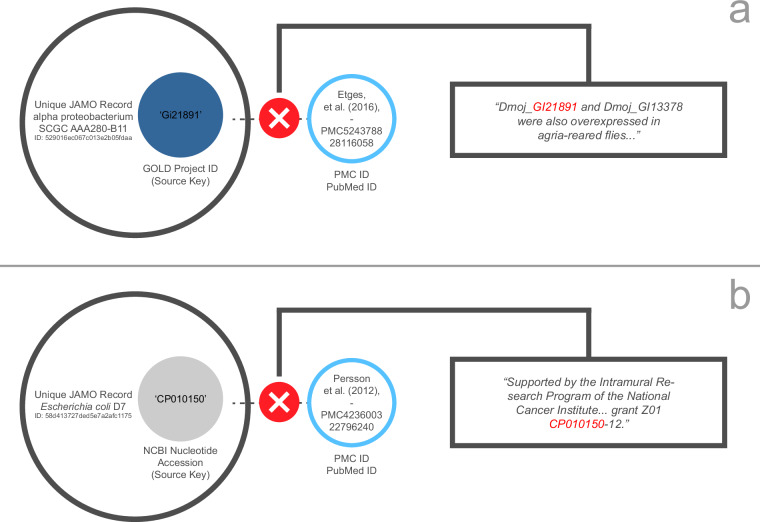
Fig. 4.
Examples of audit trails returning irrelevant publications. The error in example (a) results from a namespace collision with a grant number, while the error in example (b) results from a namespace collision between a GOLD Project ID and a FlyBase gene identifier. Some namespace collisions could only be resolved by retrieving and parsing full GenBank records, which degrades performance but improves accuracy.
Table 1.
Manual evaluation results.
| Indexer | Unique Keys | Total Publications | Relevant Publications | Irrelevant Publications | Precision |
|---|---|---|---|---|---|
| Public | 489 | 576 | 566 | 10 | .983 |
| Dimensions | 489 | 1,027 | 1,018 | 9 | .991 |
| Both | 489 | 1,234 | 1,218 | 16 | .987 |
The ‘Public’ row includes sampled results using just what the DCE returned via searches in public sources. The ‘Dimensions’ row indicates results returned by manually using the keys that generated hits in the ‘Public’ row to search Dimensions full-text articles. The ‘Both’ row combines all unique publications from each of the other two rows. Precision values are calculated using the values in the ‘Relevant Publications’ and ‘Total Publications’ Columns.
Briefly, manual searches using the proprietary Dimensions database were used to investigate the extent to which keys in our sample would generate hits using a larger corpus of full-text article content than is available through public sources like those maintained by NCBI. Of the 489 keys used for full-text searches in Dimensions, 341 (69.7%) resulted in hits. The total number of individual publications returned was 1,027, substantially greater than that returned through NCBI (Fig. 5). The precision value for individual publications returned via Dimensions was evaluated using the same methods as with the initial sample and was determined to be 0.991. Combining these results with those from public indexing sources, the test set of keys generated hits on 1,234 unique publications (Table 1). Over half of these were only identifiable through the proprietary Dimensions database.
Fig. 5.
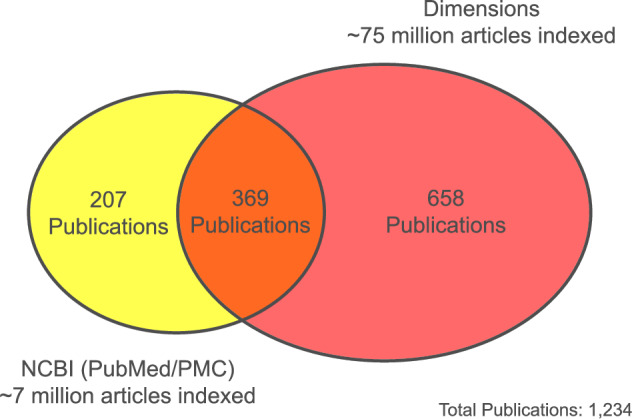
Comparison of publication results returned through the Data Citation Explorer using two distinct publication indexes and a sample of 282 JAMO records. The precision value for both sets of results was over 0.98.
Though precision values are often used in concert with recall and F-scores to determine the efficacy of retrieval systems, the latter two metrics were unobtainable in this context. The wide range of means used to indicate data usage and the inexactitude that characterizes many of these means25 implies that one cannot confidently gather the entire set of articles citing or using a given dataset. There is simply too much variation for each dataset to be fully anticipated. As a result, the variable that is used as the denominator when calculating recall (all results relevant to the original query), which is itself used to calculate F-scores, cannot be determined. However, in this instance precision remains a useful evaluation metric in itself because it allows one to gauge the relevance of those results that are returned by a system.
Discussion
Corpus expansion and context analysis
Comparing results between the two publication data sources indicates the degree to which access to a larger corpus of full-text literature could increase the number of hits returned by the Data Citation Explorer’s search feature, particularly for disciplines not indexed by PubMed or PubMed Central. As all of the 207 publications returned by searching NCBI sources and not returned by Dimensions were later found to be indexed by Dimensions, it is likely that the differences in results returned by each were due to variations in search functionalities and full-text document indexing between the two providers. Investigation of these differences is beyond the scope of this work but represents an opportunity for future work. In addition to expanding the body of literature available to the DCE, further exploration of strategies for parsing non-standard supplemental material files included with scientific articles may increase this number still further. Internally, JGI can improve the comprehensiveness and quality of citable metadata indexed by JAMO for any given dataset to account for a wider range of possible citation methods.
Though the results of this evaluation indicate high levels of reliability, the system currently does not feature means for distinguishing the significance of one positively identified citation from another. Previous work has shown the possibility of applying natural language processing (NLP) techniques to the textual context surrounding individual citations26,27. Similar techniques could be applied to evaluate DCE results beyond a simple relevance assessment. Furthermore, a rigorous investigation of the relative frequencies of ‘informal’ and ‘formal’ citations of data in the genomics field could provide another avenue for future work.
Generalization
Expanding the service model embodied by the DCE to other disciplines (e.g., physics, earth sciences) is the next goal for future development and collaboration. Generalization of the service for use by other public data resources could maximize its impact and greatly improve wider knowledge of how public data is being used and by whom, supporting a Data Ecosystem that encourages connections among cross-organizational resources. Admittedly, the DCE was designed with biosciences literature and metadata in mind, a field that is dominated by relatively unique identifier strings. Other fields (i.e. the social sciences) in which plain-language mentions of dataset names are more prevalent could require additional NLP-based features to ensure relevance of search results17,27. While these results validate the conceptual underpinnings of the service’s architecture, much work remains to maximize the usefulness of the DCE’s service model to users beyond JGI and the biosciences field.
Accessibility and collaboration
Much development work remains to be done to determine how best to make the results accessible to our user community. The citations and audit paths are back-propagated into the JAMO database, which could serve as the integration point for making this data available via the JGI Data Portal (Fig. 6) for a more user-friendly interface.
Fig. 6.
A mockup of how data citations discovered by the DCE might be presented to a user via the JGI Data Portal.
Some examples of potentially desirable features could include research-facilitating search tools for the Joint Genome Institute and associated DOE organizations like the National Microbiome Data Collaborative (NMDC)28 and the Systems Biology Knowledgebase (KBase)29. Generally available reports could describe who is using the work products produced by a given researcher and provide public-facing views that increase the visibility of JGI work products. From a user’s perspective, such features can illustrate previous use cases (or lack thereof) of any dataset of interest. The lessons learned from this project could be applied in other organizations that have an interest in mining highly focused genomic literature.
FAIRness
The metadata and literature connections established by the DCE enable JGI to more equitably attribute credit to individual contributors for downstream outcomes of individual work products, though the specific metrics and methodology for doing so remain undetermined. Automated and standardized means for determining the extent of the service’s contributions to the findability, accessibility, interoperability, and reusability (FAIR) of public genomic data, similar to the framework developed by Wilkinson et al.30, would allow for continuous reassessment of the DCE for potential future updates.
Methods
JGI source data: JAMO
The data source for the DCE is the JGI Archive and Metadata Organizer (JAMO), which is JGI’s primary data management system. It manages most of the data assets the organization produces and caches associated metadata from the other data support systems, for example the Genomes On-Line Database (GOLD)31. JAMO also archives data to several geographically dispersed high-performance tape systems, manages the restore and purge policies of files on spinning disk, provides publish/subscribe services to internal pipelines, and creates a single connection point for all internal data systems to communicate with each other with regard to metadata services.
Metadata in JAMO are organized by JGI sequencing product/pipeline and file type, each of which has a defined dictionary of required and optional metadata. There are over 3,000 distinct metadata fields in JAMO across approximately 400 product, pipeline, and file types. The metadata in JAMO can be broken down into several classes: operational data, project and proposal information, genomic classifications, data ownership, data usability, internally produced public identifiers, and external public identifiers. Data provenance is also captured in the operational and project metadata.
The DCE pilot project focused on the metadata likely to be present in publications: NCBI GenBank Accessions, NCBI BioProject and BioSample Accessions, references to the NCBI taxonomy (from GOLD), IMG Taxon Object IDs (from JGI’s Integrated Microbial Genomes & Microbiomes system), SRA IDs (from NCBI’s Sequence Read Archive), and contact information (from JGI’s proposal system). After evaluation and testing, the initial production run of the DCE included 1.7 million selected JAMO records. These records were those that either were published at NCBI’s SRA or were downloaded by two or more distinct non-JGI users from JGI systems between February 22, 2019 and August 30, 2022.
Citation discovery process
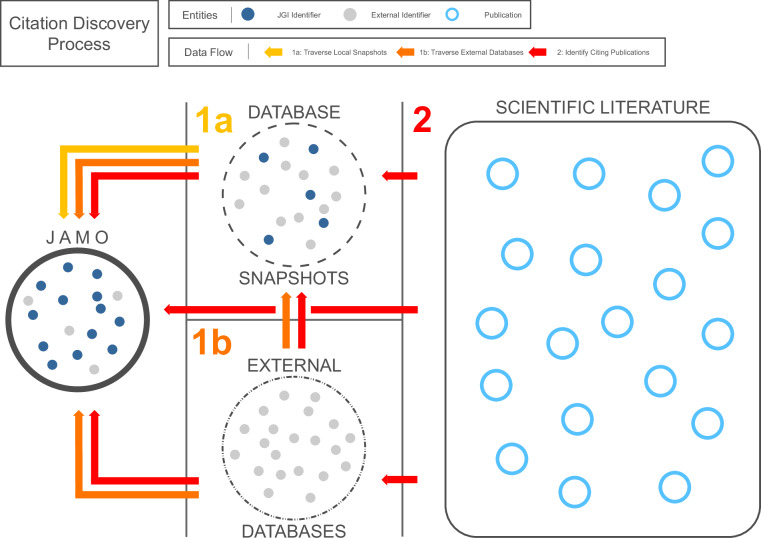
The citation discovery process runs in two phases: a metadata collection phase and a citation search phase (Fig. 7). The metadata collection phase crawls genomic data repositories to accumulate new metadata that has been produced in downstream resources. This phase starts with an initial metadata registry of selected fields from JGI’s JAMO database, then incrementally adds newly discovered metadata to the registry by crawling other data repositories via unique identifiers (e.g., genome assemblies, sequence data, sequence reads, bioprojects, biosamples) while respecting identifier hierarchies and cardinality. Specifically, the DCE avoids traversal of accession relations that have one-to-many cardinality (e.g., BioProject to BioSample) that could result in connections to non-targeted datasets. This process continues iteratively until no additional resources are discovered. As a new accession is discovered, its source is stored with the accession in a relational database in a form that preserves the graph representation of the DCE’s traversal through all discovered resources. The resulting graph may be used for evaluating the correctness of the system. Note that while some classes of identifier are ambiguous, these cases are generally mitigated by the addition of identifier namespaces or prefixes that are respected by the relational database schema and associated business logic (Fig. 8). Although this ambiguity issue is not completely solved for protein identifiers and grant numbers (Fig. 4), it is partially addressed with special handling of some document sections (i.e., “front matter" and “back matter" in the NLM XML schema), and by retrieving and parsing full GenBank records to confirm or negate matching strings. This issue remains an area of active development.
Fig. 7.
The citation discovery process for an individual JAMO record (the target genome) happens in two phases: (1) crawl genomic data repositories to accumulate new metadata that has been produced in downstream resources, and (2) search for occurrences of unique identifiers in publicly available literature.
Fig. 8.
A sample audit trail for a set of connected genomic data resources as they are stored in the underlying namespace-aware relational database, retrieved as a set of triples via a SQL view, and reconstructed as a directed, acyclic graph. These audit trails support the validation of discovered citations. A depth-first traversal of the graph can identify all known paths between any two identifiers in the audit trail, as well as the shortest path between any two identifiers.
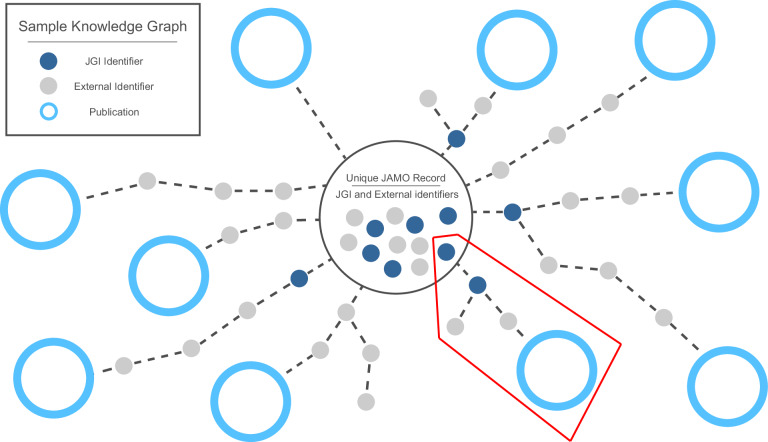
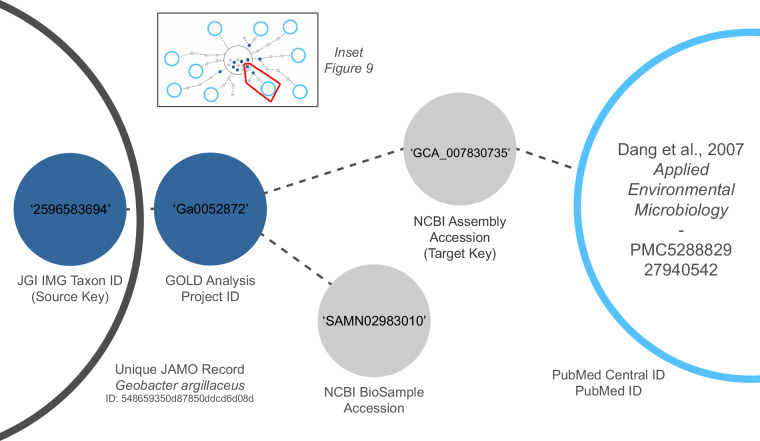
In the second phase of the process, the DCE searches the Open Access (OA) literature for occurrences of any of the collected identifiers that identify the project of interest. Similar to methods employed recently to explore full-text hits on identifiers by articles in Europe PubMed Central21, the DCE gathers a portion of its search results using pattern matching on the content of articles indexed by PubMed Central. Additional results were added via full-text pattern searches for known accession formats on a locally compiled corpus of OA taxonomic literature for prokaryotes (see below). Further, any full-text content discovered is downloaded and processed in a similar method to the taxonomic corpus. The results of each search are added to the metadata registry and stored in the database. At this point, the citation discovery process is complete (Figs. 9 and 10). Please refer to the DCE source data32 for all materials used to create the full knowledge graph, including descriptions of the JAMO fields used to source initial metadata.
Fig. 9.
High-level depiction of a DCE knowledge graph as it branches out from a single JAMO record. Some data citations are connected via a single identifier, but others have more complex paths, often branching or forming redundant connections to publications. In this simplified image, for example, the highlighted branching path indicates that two separate JGI-external identifiers are both linked via a common JGI-internal identifier. One of these links to a publication, while the other does not.
Fig. 10.
Sample audit trail for a single path to a publication that relies on a genome of interest (see inset). In this example, a JGI-internal identifier from the source JAMO record links to another JGI-internal identifier from a second resource. This second identifier, in turn, is linked through an external NCBI identifier to the citing publication, represented here by a PubMed Central identifier. The source and target resource from which each accession is discovered is stored as a triple, providing a traceable path that can explain how each resource was discovered. This is an important improvement over manual searches, as tracking the source of each individual metadata entry would be burdensome and error-prone for a human data curator. Note that a linkage to a separate external NCBI identifier forms a branch of this audit trail but does not link to any publications. These identifiers are also stored, as additional connections may be discovered at a later time.
Evaluation procedures
In order to evaluate the system’s performance prior to large-scale production batches, an initial trial was performed by processing 238,994 JAMO metadata records with the Data Citation Explorer. The major goal of the evaluation process was to determine the extent to which in-text citations returned by the DCE truly referenced the data represented by each JAMO record. Records were selected if they represented genomic data downloaded by external users of JGI’s Genome Portal33 four or more times between the dates of February 22, 2019 and May 31, 2020. These dates span the period between when internal file request information became available and the start of the DCE trial. These metadata records describe publicly available genomic data that was produced by JGI or uploaded to its systems between January 2009 and May 2020. Of these 238,994 records, approximately 78,104 were linked to publications during the initial DCE trial. 30,641 unique publications were retrieved in this step.
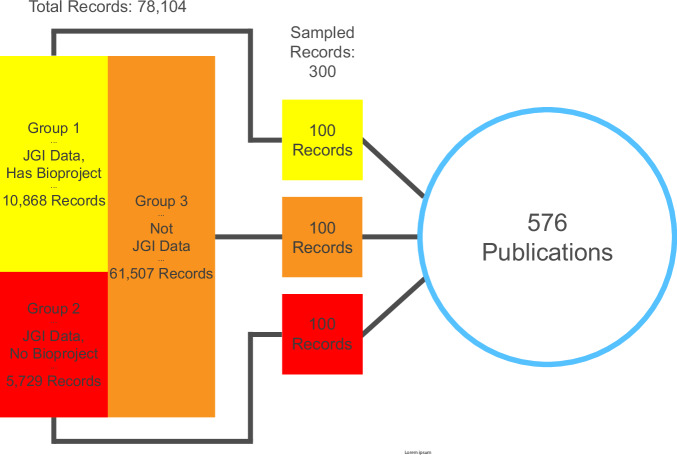
As manual evaluation of connections between tens of thousands of records and publications would be impossible, a subset of records with linked publications from the initial test run was selected. The subset was produced via stratified random sampling to avoid overrepresenting JGI projects and data that have disproportionately high numbers of associated records. This would ensure that a diverse set of citations was available for evaluation. The stratified sample consisted of three groups of 100 records from larger, mutually-exclusive sets distinguished by particular characteristics of interest:
All records for data that were associated with a NCBI BioProject ID in the GOLD system. Because GOLD indexes genome announcement publications using a semi-manual system that predates the DCE34, these records and their pre-existing publication linkages could be used in the future for comparative purposes. Total records: 5,729
All records generated from JGI-sequenced data that were not registered at NCBI. The inclusion of this group would determine whether the DCE could find citations where external metadata IDs were limited or non-existent in the JAMO records. Total records: 10,868
All JAMO records generated from sequence data that were not produced by JGI, primarily gene annotations and similar data from external sources uploaded to JGI’s IMG/M system. The goal for this group was similar to Group 2 with the added benefit of determining whether or not the DCE would reject citations from upstream data sources (i.e., publications from the original sequence data). Total records: 61,507
Of the 300 sampled records, 18 could not be evaluated because of a technical error that prevented the audit trails from resolving. Though the bug causing this error was fixed prior to deployment, it was decided at this point to leave these records out and proceed with the evaluation. The 282 remaining records in the resulting sample were linked to 576 unique publications via 998 audit trails by the DCE (Fig. 11).
Fig. 11.
JAMO record sampling for manual evaluation. 300 JAMO records were selected from three stratified groups. After filtering for records with unresolved audit trails, 282 total records with 998 unique audit trails connecting to 576 unique publications were evaluated.
Following sample generation, the validity of each audit trail between a JAMO record and a publication was evaluated manually (see Data Availability statement for evaluation files). An audit trail was considered “valid" only if the following conditions are met:
The key tied directly to or found within a target publication is an actual data identifier and not a false positive
The key tied directly to or found within a target publication unambiguously refers to the JGI data entity represented by the source JAMO record.
Only results of ‘valid’ audit trails would be considered relevant in the later precision calculations.
The initial proof of concept for the Data Citation Explorer included searches over a core full-text corpus composed of the primary taxonomic literature of prokaryotes (primarily the International Journal of Systematic and Evolutionary Microbiology from 2005 through 2018 and Standards in Genomic Sciences v1-9). The corpus was expanded prior to the initial evaluation to include literature searches in two publicly available indexing services, NCBI’s PubMed and PubMed Central.
To understand how the Data Citation Explorer performs with access to a larger corpus of publications, search results from the original corpus were compared with results from Digital Science’s Dimensions platform. Dimensions was used because it is at the time of this writing the only subscription service that provides full-text search functionality. This sets the service apart from tools like Web of Science or Scopus, which only index articles at the citation level. Because the DCE attempts to identify informal citations that are not indexed as formal references, the citation-level metadata provided by these other services do not provide an adequate point of comparison. This step was accomplished manually using Dimensions’ public search interface to run individual full-text searches for each of the 489 unique keys that led to hits in public sources from the initial sample (see Lafia et al.17 for a similar use case). The process returned hits for a total of 1,027 publications.
Acknowledgements
The authors would like to thank Tatyana Smirnova at JGI for contributing JGI Data Portal mockup images. The work conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under Contract No. DE-AC02-05CH11231. Certain data included herein are derived from Clarivate Web of Science. Copyright Clarivate 2021. All rights reserved. This paper was written using data obtained on September 8, 2021 (DCE results) and January 22, 2024 (Genetics publications) from Digital Science’s Dimensions platform, available at https://app.dimensions.ai.
Author contributions
N.B. designed and conducted DCE evaluation experiments. C.P. designed and developed the DCE. C.B. generated the evaluation sample and supervised DCE integration with JAMO. T.B.K.R. advised on JGI metadata usage, facilitated access to GOLD metadata, and provided guidance on NCBI and PubMed integration. H.S. supervised initial JAMO integration activities. G.G. and K.F. conceived of the presented idea and supervised the project. N.B., C.P., C.B., G.G. and K.F. contributed to the writing and revision of the manuscript.
Data availability
Materials used for manual evaluation of DCE results as well as all source data for the initial trial run can be found at the following Zenodo repository: 10.5281/zenodo.13830817. Genomic metadata can be found at the Genomes OnLine Database (gold.jgi.doe.gov), the JGI Genome Portal (genome.jgi.doe.gov), the JGI Data Portal (data.jgi.doe.gov), Integrated Microbial Genomes & Microbiomes (img.jgi.doe.gov), Phytozome (phytozome.jgi.doe.gov), PhycoCosm (phycocosm.jgi.doe.gov), MyCosm (mycocosm.jgi.doe.gov), GenBank (ncbi.nlm.nih.gov/genbank), and the Sequence Read Archive (ncbi.nlm.nih.gov/sra). Publication data can be found at PubMed (pubmed.ncbi.nlm.nih.gov) and PubMed Central (www.ncbi.nlm.nih.gov/pmc). Additionally, the analyses include results returned via searches over proprietary full-text data contained within the Dimensions database (app.dimensions.ai) that was not directly accessed and cannot be exposed publicly.
Code availability
The source code for the Data Citation Explorer is hosted in a GitLab repository at https://code.jgi.doe.gov/data-citation-explorer/. The authors will assist with any reasonable replication attempts for two years following publication. At the time of submission, JGI’s hosted Data Citation Explorer web application is on a private network, but is planned to be made publicly accessible at https://dce.jgi.doe.gov.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Neil Byers, Charles Parker.
References
- 1.Chen, I.-M. A. et al. The img/m data management and analysis system v.7: content updates and new features. Nucleic acids research51, d723–d732, 10.1093/nar/gkac976 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research40, d1178–d1186, 10.1093/nar/gkr944 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grigoriev, I. V. et al. Mycocosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Research42, d699–d704, 10.1093/nar/gkt1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grigoriev, I. V. et al. Phycocosm, a comparative algal genomics resource. Nucleic Acids Research49, d1004–d1011, 10.1093/nar/gkaa898 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katz, K. et al. The sequence read archive: a decade more of explosive growth. Nucleic Acids Research50, gkab1053–, 10.1093/nar/gkab1053 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, D. A. et al. Genbank. Nucleic Acids Research41, d36–d42, 10.1093/nar/gks1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson, M. D. et al. The fair guiding principles for scientific data management and stewardship. Scientific Data3, 160018, 10.1038/sdata.2016.18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cousijn, H., Feeney, P., Lowenberg, D., Presani, E. & Simons, N. Bringing citations and usage metrics together to make data count. Data Science Journal18, 10.5334/dsj-2019-009 (2019).
- 9.Wood-Charlson, E. M., Crockett, Z., Erdmann, C., Arkin, A. P. & Robinson, C. B. Ten simple rules for getting and giving credit for data. PLoS Computational Biology18, e1010476, 10.1371/journal.pcbi.1010476 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsch, J. E. An index to quantify an individual’s scientific research output. Proceedings of the National Academy of Sciences102, 16569–16572, 10.1073/pnas.0507655102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross, M. B. et al. Women are credited less in science than men. Nature608, 135–145, 10.1038/s41586-022-04966-w (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Why we’re removing the rg score (and what’s next), https://www.researchgate.net/researchgate-updates/removing-the-rg-score. Accessed: 2023-03-08 (2022).
- 13.Hood, A. S. C. & Sutherland, W. J. The data-index: An author-level metric that values impactful data and incentivizes data sharing. Ecology and Evolution11, 14344–14350, 10.1002/ece3.8126 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenner, M. et al. A data citation roadmap for scholarly data repositories. Scientific Data6, 28, 10.1038/s41597-019-0031-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stall, S. et al. Journal production guidance for software and data citations. Scientific Data10, 656, 10.1038/s41597-023-02491-7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vannan, S., Downs, R. R., Meier, W., Wilson, B. & Gerasimov, I. V. Data sets are foundational to research. why don’t we cite them? Eos101, 10.1029/2020EO151665 (2020).
- 17.Lafia, S., Thomer, A., Moss, E., Bleckley, D. & Hemphill, L. How and why do researchers reference data? a study of rhetorical features and functions of data references in academic articles. Data Science Journal22, 10, 10.5334/dsj-2023-010 (2023). [Google Scholar]
- 18.Park, H., You, S. & Wolfram, D. Informal data citation for data sharing and reuse is more common than formal data citation in biomedical fields. Journal of the Association for Information Science and Technology69, 1346–1354, 10.1002/asi.24049 (2018). [Google Scholar]
- 19.Mayo, C., Vision, T. J. & Hull, E. A. The location of the citation: changing practices in how publications cite original data in the dryad digital repository. International Journal of Digital Curation11, 150–155, 10.2218/ijdc.v11i1.400 (2016). [Google Scholar]
- 20.Zhao, M., Yan, E. & Li, K. Data set mentions and citations: A content analysis of full-text publications. Journal of the Association for Information Science and Technology69, 32–46, 10.1002/asi.23919 (2017). [Google Scholar]
- 21.Lange, M. et al. Quantitative monitoring of nucleotide sequence data from genetic resources in context of their citation in the scientific literature. GigaScience10, giab084, 10.1093/gigascience/giab084 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White, J. Pubmed 2.0. Medical Reference Services Quarterly39, 382–387, 10.1080/02763869.2020.1826228 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Herzog, C., Hook, D. & Konkiel, S. Dimensions: Bringing down barriers between scientometricians and data. Quantitative Science Studies1, 387–395, 10.1162/qss_a_00020 (2020). [Google Scholar]
- 24.Hook, D. W., Porter, S. J. & Herzog, C. Dimensions: Building context for search and evaluation. Frontiers in Research Metrics and Analytics3, 23 10.3389/frma.2018.00023 (2018). [Google Scholar]
- 25.Moss, E. & Lyle, J. Opaque data citation: Actual citation practice and its implication for tracking data use. https://hdl.handle.net/2027.42/142393 Accessed: 2024-07-25 (2018).
- 26.Nicholson, J. M. et al. scite: A smart citation index that displays the context of citations and classifies their intent using deep learning. Quantitative Science Studies2, 882–898, 10.1162/qss_a_00146 (2021). [Google Scholar]
- 27.Lane, J., Gimeno, E., Levitskaya, E., Zhang, Z. & Zigoni, A. Data inventories for the modern age? using data science to open government data. Harvard Data Science Review10.1162/99608f92.8a3f2336 (2022).
- 28.Wood-Charlson, E. M. et al. The national microbiome data collaborative: enabling microbiome science. Nature Reviews Microbiology18, 313–314, 10.1038/s41579-020-0377-0 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Arkin, A. P. et al. Kbase: The united states department of energy systems biology knowledgebase. Nature Biotechnology36, 566–569, 10.1038/nbt.4163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkinson, M. D. et al. Evaluating fair maturity through a scalable, automated, community-governed framework. Scientific Data6, 174, 10.1038/s41597-019-0184-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjee, S. et al. Twenty-five years of genomes online database (gold): data updates and new features in v.9. Nucleic acids research51, d957–d963, 10.1093/nar/gkac974 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker, C. et al. Source data for manuscript: Identifying genomic data use with the data citation explorer (1.1.0). 10.5281/zenodo.13830817 (2024). [DOI] [PMC free article] [PubMed]
- 33.Nordberg, H. et al. The genome portal of the department of energy joint genome institute: 2014 updates. Nucleic Acids Research42, d26–d31, 10.1093/nar/gkt1069 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liolios, K., Tavernarakis, N., Hugenholtz, P. & Kyrpides, N. C. The genomes on line database (gold) v.2: a monitor of genome projects worldwide. Nucleic Acids Research34, d332–d334, 10.1093/nar/gkj145 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Materials used for manual evaluation of DCE results as well as all source data for the initial trial run can be found at the following Zenodo repository: 10.5281/zenodo.13830817. Genomic metadata can be found at the Genomes OnLine Database (gold.jgi.doe.gov), the JGI Genome Portal (genome.jgi.doe.gov), the JGI Data Portal (data.jgi.doe.gov), Integrated Microbial Genomes & Microbiomes (img.jgi.doe.gov), Phytozome (phytozome.jgi.doe.gov), PhycoCosm (phycocosm.jgi.doe.gov), MyCosm (mycocosm.jgi.doe.gov), GenBank (ncbi.nlm.nih.gov/genbank), and the Sequence Read Archive (ncbi.nlm.nih.gov/sra). Publication data can be found at PubMed (pubmed.ncbi.nlm.nih.gov) and PubMed Central (www.ncbi.nlm.nih.gov/pmc). Additionally, the analyses include results returned via searches over proprietary full-text data contained within the Dimensions database (app.dimensions.ai) that was not directly accessed and cannot be exposed publicly.
The source code for the Data Citation Explorer is hosted in a GitLab repository at https://code.jgi.doe.gov/data-citation-explorer/. The authors will assist with any reasonable replication attempts for two years following publication. At the time of submission, JGI’s hosted Data Citation Explorer web application is on a private network, but is planned to be made publicly accessible at https://dce.jgi.doe.gov.