Abstract
Fragile X syndrome (FXS) is a neurodevelopmental disorder caused by hypermethylation of expanded CGG repeats (>200) in the FMR1 gene leading to gene silencing and loss of Fragile X Messenger Ribonucleoprotein (FMRP) expression. FMRP plays important roles in neuronal function, and loss of FMRP in mouse and human FXS cell models leads to aberrant synaptic signaling and hyperexcitability. Multiple drug candidates have advanced into clinical trials for FXS, but no efficacious treatment has been identified to date, possibly as a consequence of poor translation from pre-clinical animal models to human. Here, we use a high resolution all-optical electrophysiology platform applied to multiple FXS patient-derived and CRISPR/Cas9-generated isogenic neuronal cell lines to develop a multi-parametric FXS disease phenotype. This neurophysiological phenotype was optimized and validated into a high throughput assay based on the amount of FMRP re-expression and the number of healthy neurons in a mosaic network necessary for functional rescue. The resulting highly sensitive and multiparameter functional assay can now be applied as a discovery platform to explore new therapeutic approaches for the treatment of FXS.
Subject terms: Excitability, Autism spectrum disorders
Deep functional characterization of Fragile X syndrome patient and isogenic neurons using all-optical electrophysiology and machine learning identifies a validated, FMR1-dependent cellular phenotype compatible with high throughput drug screening.
Introduction
Fragile X Syndrome (FXS) is the most common inherited form of intellectual disability and the leading monogenic cause of autism1–3. FXS is caused by transcriptional silencing of the FMR1 gene as a result of DNA methylation driven by a tri-nucleotide CGG repeat expansion (>200) in the 5’ UTR of the FMR1 gene1–4. Fragile X Messenger Ribonucleoprotein (FMRP), the product of the FMR1 gene, is an RNA-binding protein that regulates a large number of mRNAs in the brain, including transcripts encoding chromatin and synaptic activity associated proteins5–9. The loss of FMRP leads to elevated levels of basal protein synthesis particularly affecting the synapse5,10–15.
Much of the understanding of the functional impact of loss of FMRP has been gained from analyses of Fmr1 knockout mice16. Two core domains of neuronal dysfunction associated with FMRP deficiency revealed by these studies are altered synaptic plasticity6,17 and neuronal and network hyperexcitability18. A key study found that mGluR-dependent long-term depression, a form of synaptic plasticity, is increased in the hippocampus of Fmr1 knockout mice19, leading to the theory that protein-synthesis-dependent functions of mGluRs are exaggerated in the absence of FMRP, comprising a core pathogenic mechanism in FXS12. Further studies have found additional alterations in synaptic transmission and plasticity in Fmr1 knockout mice17,20–28. Observations of altered dendritic spine morphology in post-mortem brain tissue from FXS patients and in Fmr1 knockout mice are consistent with a pathogenic role of synaptic dysfunction in FXS28–31.
In addition to synaptic dysfunction, there are multiple reports of neuronal and circuit hyperexcitability in Fmr1 knockout mice18,32–34, consistent with the increased seizure susceptibility in FXS patients35,36 and in these mice37,38. Furthermore, altered patterns of sensory-evoked and resting EEG activity in FXS patients39,40 and Fmr1 knockout mice41 are indicative of network hyperexcitability in FXS. FMRP deficiency has been reported to impact multiple ion channels that may contribute to altered excitability, including large conductance calcium-activated (BK) channels, small conductance calcium-activated (SK) channels, sodium-activated potassium (Slack) channels, hyperpolarization-activated cyclic nucleotide-gated (HCN) channels, A-type potassium channels, sodium channels and L-type calcium channels42. Despite the substantial insights derived from analyses of Fmr1 knockout mice and the areas of alignment with observations in FXS patients, Fmr1 knockout mice do not fully recapitulate the epigenetic silencing-based pathophysiology found in FXS patients, creating a need to extend understanding of FXS pathogenic mechanisms to a human neuronal context. Human induced pluripotent stem cell (iPS cell)-derived models provide a powerful basis for mechanistic analyses of neurological diseases in a disease-relevant, human context with potential to be leveraged in phenotypic rescue applications in ways that cannot be readily achieved with in vivo models43,44.
Several studies have applied human iPS cell-derived neuronal models to characterize different aspects of FXS biology, including mechanisms of FMR1 gene silencing and reactivation45–48, effects of FMRP loss on neuronal growth and differentiation49–52 as well as impact of FMRP loss on neuronal activity53–57. While early studies reported that iPS cell-derived neurons lacking FMRP were deficient in action potential firing58, several recent studies have reported hyperexcitability in human FXS neuronal models, including increased neuronal firing rate55,59 and increased burst frequency53,54, as well as altered homeostatic plasticity56, consistent with altered regulation of excitability34. Recent studies have used human iPS cell-derived neuronal models to show rescue of a hyperexcitability phenotype upon reactivation of the silenced FMR1 gene in neurons59 and to examine the degree of FMRP expression and proportion of FMRP expressing cells required to normalize excitability55. While providing valuable insights into human neuronal FXS disease biology, the patch clamp and MEA techniques used in these studies have limitations in cellular throughput or resolution, constraining the ability to perform deep electrophysiological phenotyping across multiple independent iPS cell-derived neuronal lines with sufficient scale and sensitivity to capture potentially subtle, disease-relevant alterations in neuronal activity. There is a need to develop sensitive, multiparameter read-outs of the impact of FMRP loss on human neuronal activity within a format that is readily scalable to drug discovery applications.
To address this need, we have developed an integrated platform that combines human iPS cell-derived neuronal disease models, high-throughput all-optical electrophysiological characterization with single cell and millisecond temporal resolution, and machine-learning analytics for disease phenotype discovery60–63. Here we applied the platform to comprehensive electrophysiological phenotyping of multiple human isogenic and patient/control iPS cell-derived neuronal models of FXS. We developed an optimized multiparameter, neurophysiological disease phenotypic read-out consistent across multiple independent lines and rounds of analysis. We applied the assay in a mosaic configuration to determine the proportion of unaffected neurons necessary to rescue the disease phenotype and showed that the presence of 50% unaffected neurons rescues the disease phenotype within FMRP-deficient cells, highlighting the value of the scale of functional measurements at the cellular resolution achieved with our assay. We also utilized the assay system combined with high-content imaging of FMRP expression to quantitatively characterize the rescue profile for viral gene replacement, showing dose-dependent rescue of the identified multiparameter phenotype attributable to FMR1 restoration. Lastly, we screened a library of pharmacologically defined reference compounds and showed the degree of rescue achieved for different mechanisms with relevance to FXS. The platform provides a powerful tool for examining underlying neuronal disease biology of FXS. Collectively, we demonstrate the applicability of the platform for high-throughput screening and deeper understanding of FXS disease biology.
Results
Characterization of FXS and control cell lines
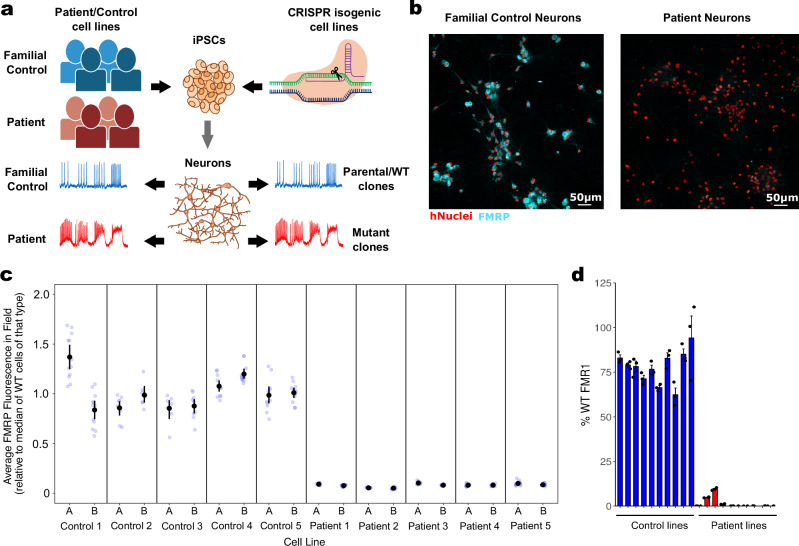
To better understand FXS disease biology in human neurons and to assess convergent, robust patterns of behavior, we developed a large and diverse set of FXS-relevant human cell models. Human iPS cell lines were differentiated into excitatory glutamatergic neurons via the overexpression of the transcription factor NGN264 combined with small molecule-mediated patterning to drive uniform differentiation65. To provide multiple convergent lines of evidence, we took a dual approach by differentiating both familial patient/control iPS cell lines and CRISPR/Cas9-edited cell lines in which the FMR1 gene was knocked out (KO), (Fig. 1a). We generated iPS cell lines and neuronal cell lines from five FXS patients, and from a healthy male family-member of each patient as a genetically-related matched control. Two clones from each of these 10 donors were generated to help account for clonal variability, for a total of 20 cell lines comprising our “patient/control” cell model of FXS. In parallel, we used CRISPR/Cas9 editing of a wild type (WT) cell line (28-year-old male donor) to generate two FMR1−/y (KO) clones. These KO lines paired with the WT parental cell line composed our “isogenic” cell model of FXS. All cell lines were karyotyped, tested for mycoplasma, and sequenced to confirm CGG repeat length (Supplementary Table 1), which ranged from 22 to 36 repeats for controls and 174–912 repeats for diagnosed FXS patients. FMR1 transcript levels were evaluated using qPCR and FMRP protein levels were evaluated via immunocytochemistry using single-cell analysis of high-content imaging scans (Fig. 1b–d). As expected, cells from FXS patients and FMR1−/y CRISPR KO clones lacked FMRP protein and FMR1 transcript, but not familial controls or from the isogenic WT line.
Fig. 1. Production and characterization of control cell lines and disease model cell lines lacking FMRP.
a Graphical representation of neuronal production from patient/control subjects and isogenic iPS cell lines to electrophysiologically-active neurons. b Representative high-content confocal image of control-derived neurons showing FMRP in cytoplasm (cyan) compared to patient-derived neurons that show no FMRP. hNuclei: staining for human-specific nuclear antigen (red). c Quantitative ICC showing levels of FMRP across all 10 control clones (from 5 control donors) and all 10 patient clones (from 5 patient donors). Fluorescence in the proximal cytoplasm was estimated for each cell (identified via nuclei) and aggregated to a field-level average, then rescaled such that the median control field = 1. Mean and 95% confidence intervals are shown in black. d qPCR showing average FMR1 expression in each of the lines. Some of the patient lines expressed breakthrough FMR1. The qICC data for these lines indicated that some individual neurons (about 0.5–3%) expressed FMRP while the rest did not.
FXS-associated electrophysiological phenotypes
Both neuronal models of FXS were evaluated for a functional disease phenotype using an all-optical electrophysiology platform. The platform enabled simultaneous optical recordings of cellular voltage changes in many hundreds of cells through expression of a blue light-activated channelrhodopsin, CheRiff, to stimulate cells with different paradigms (steps, ramps, etc.), and a red light excited archaerhodopsin, QuasAr, to measure the resulting voltage changes60,62,63. Custom engineered instrumentation allowed for measurements in multi-well plate formats, providing a throughput of hundreds of thousands of neurons per day with near patch-clamp resolution. Fluorescence traces of action potential firing in response to blue light stimulation are shown for three example control lines and three example patient lines (Supplementary Fig. 1a). All iPS cell lines used in this study produced functional neurons. An automated analytics pipeline processed the raw movies to extract single-cell recordings. From each cellular recording, a set of 500 features was extracted describing spike shape and spike timing characteristics as a function of different stimuli as a way of quantitatively probing diverse neurophysiological response.
To identify FXS-associated functional phenotypes, we plated neurons from all 23 cell lines onto 96-well plates for measurements of intrinsic excitability and synaptic function in at least three independent replicate rounds. Phenotype exploration was performed using a machine-learning-based analytical approach that uses parameterized (e.g. average spike shape, spiking dynamics, etc.) neuronal fluorescence traces for each blue stimulation pattern to interrogate a diverse set of cellular functions (see Methods for description). Patient phenotypes confirmed in the isogenic cell model were carried forward into rescue, mosaicism, and screening experiments.
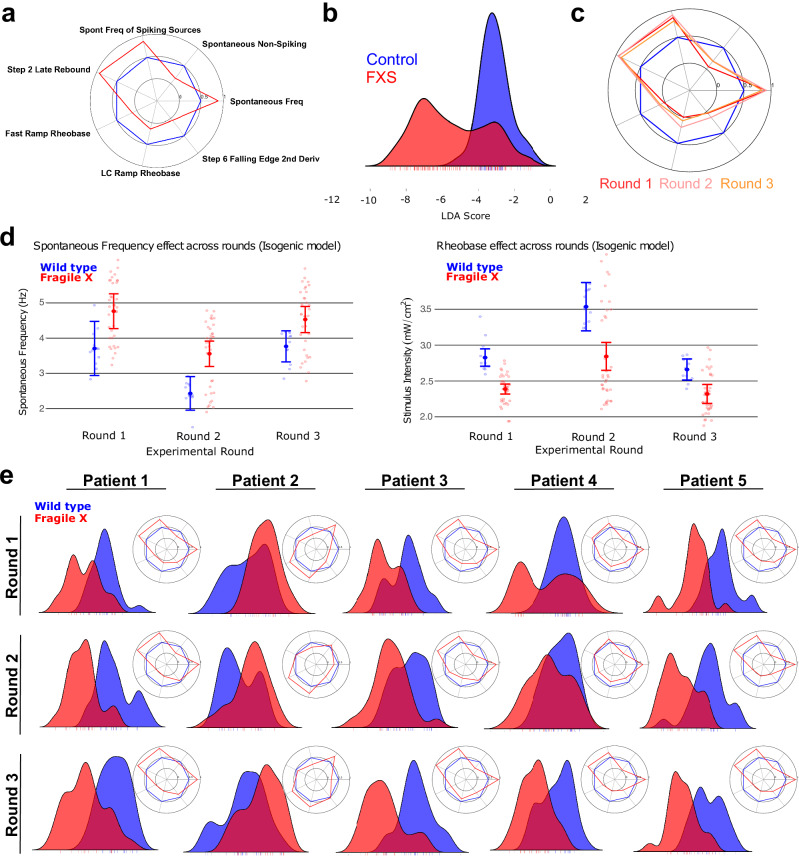
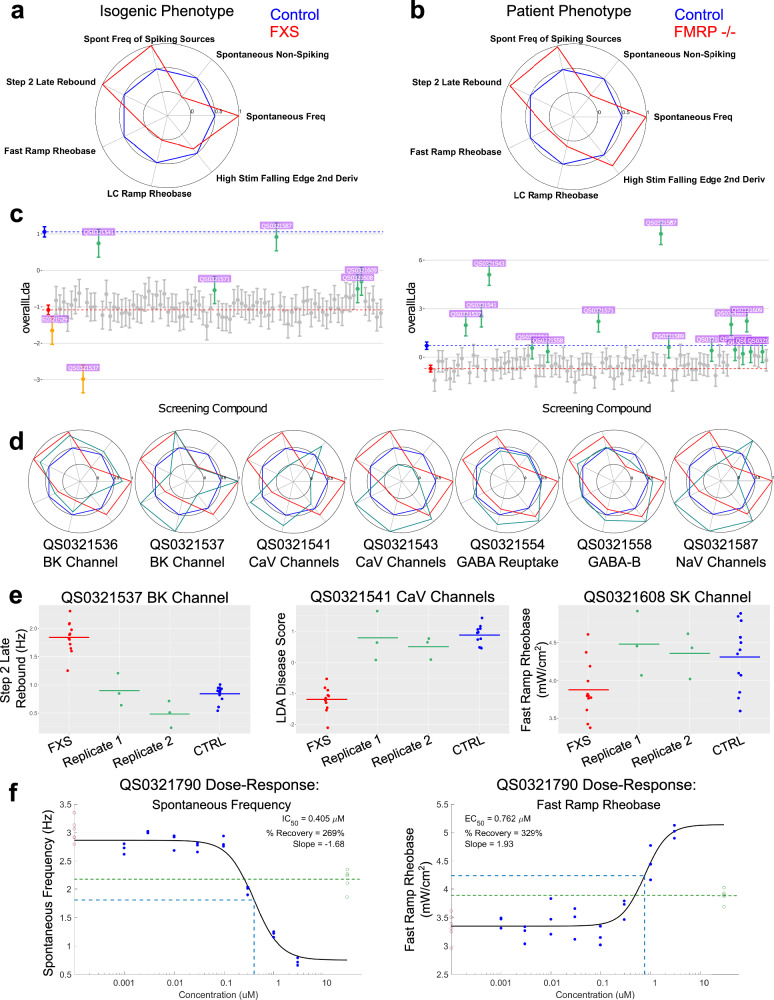
The resulting multiparameter phenotype had seven features, depicted in the form of radar plots (Fig. 2a; radar plot approach explained in Supplementary Fig. 2 for illustration purposes), including an increase in spontaneous frequency, a reduction in the number of cells that do not fire action potentials when unstimulated, a lower rheobase, and a higher rate of “rebound” action potential firing after a blue light stimulus has been turned off, all indicating a more granular view of hyperexcitability which is consistent with the FXS literature18. To help characterize overall phenotypic differences across these parameters, a composite disease score was created using a linear discriminant analysis, which maximizes separability when projecting these seven dimensions onto a single axis of difference (Fig. 2b). We found that the FXS phenotype was consistent across all three rounds of confirmatory measurements (Fig. 2c, d and Supplementary Fig. 3). The same 7-feature phenotype was apparent in 4 out of the 5 patient/control cell line comparisons, even when considering both clones for each line (Fig. 2e). The difference in the 5th pair (‘Patient/control pair 2’) cannot be explained by repeat number, FMRP expression, or a single functionally aberrant clone, but may be indicative of the role of background genetics in the biology of the familial controls and highlights the need for evaluating multiple patient-control pairs. The patient/control phenotype was also replicated across all 3 rounds of measurements (Fig. 2e). Our FXS fingerprint was also present at longer 45-day in vitro cultures with minimal change (Supplementary Fig. 4).
Fig. 2. FMRP-dependent electrophysiological FXS phenotypes in iPSC-derived neurons.
a Machine learning-identified intrinsic excitability FXS fingerprint in the isogenic reagents expressed as a radar plot of control (blue) vs. FXS (red). Values reflect the “common language effect size”, a parametric estimate of the probability that a random FXS well exceeds the value of a random CTRL well (where probability = 0.5 is a null effect). b LDA score for control (blue) vs. FXS (red) in the isogenic reagents, see methods for detail. Three FXS clones are represented – two show a distinct phenotype while the third (the peak inside the CTRL distribution) does not. c Radar plots showing the phenotype in isogenic reagents across 3 replicate rounds show consistency across fresh-from-thaw platings of neurons. d Difference in spontaneous frequency (left) and rheobase (right) of control (blue) vs. FXS (red) for each of three rounds. Error bars represent 95% confidence intervals around the well-level estimates. e LDA scores for family-matched controls vs. patient neurons from 5 different patient/control cell lines (labeled Pair 1 to Pair 5). Each Pair includes the average of 2 control clones and 2 patient clones. Radar plots for the FXS fingerprints are depicted in the inset. Data for 3 rounds is shown.
Lentiviral re-delivery of FMRP rescues FXS functional phenotype
To further validate the FMRP-dependence of the functional human neuronal phenotype, we designed FMR1 lentiviral constructs for re-expression of FMRP in the FXS neuronal models. We applied this approach to find (1) what is the minimum amount of FMRP needed to show a significant rescue effect towards control behavior? and (2) what is the approximate amount of FMRP required to show a near-complete reversal of the phenotype? These lentiviruses contained a wild-type full-length copy of FMR1 driven by a human Synapsin 1 promoter to elicit neuronal-specific expression of FMRP in transduced cells. We also designed one of the lentiviruses to include a translational attenuator to reduce lentivirus-attributable FMRP protein expression by 10-20x given an equivalent dose of the lentivirus66. Finally, we constructed a lentivirus driving expression of the fluorescent protein mOrange with the same promoter for use as a general transduction control.
As expected, delivery of increasing volumes of FMR1 lentivirus to FMR1−/y neurons resulted in a dose-dependent increase in FMRP as measured by immunocytochemistry using high content imaging (see Methods; Fig. 3a, b). Lentivirus delivered at 0.2% volume yielded FMRP expression around WT levels (Fig. 3b). Volumes of virus beyond this resulted in supraphysiological expression of FMRP. The translational attenuator was highly effective, yielding a >10x reduction in FMRP expression given equivalent volumes of virus (Fig. 3e).
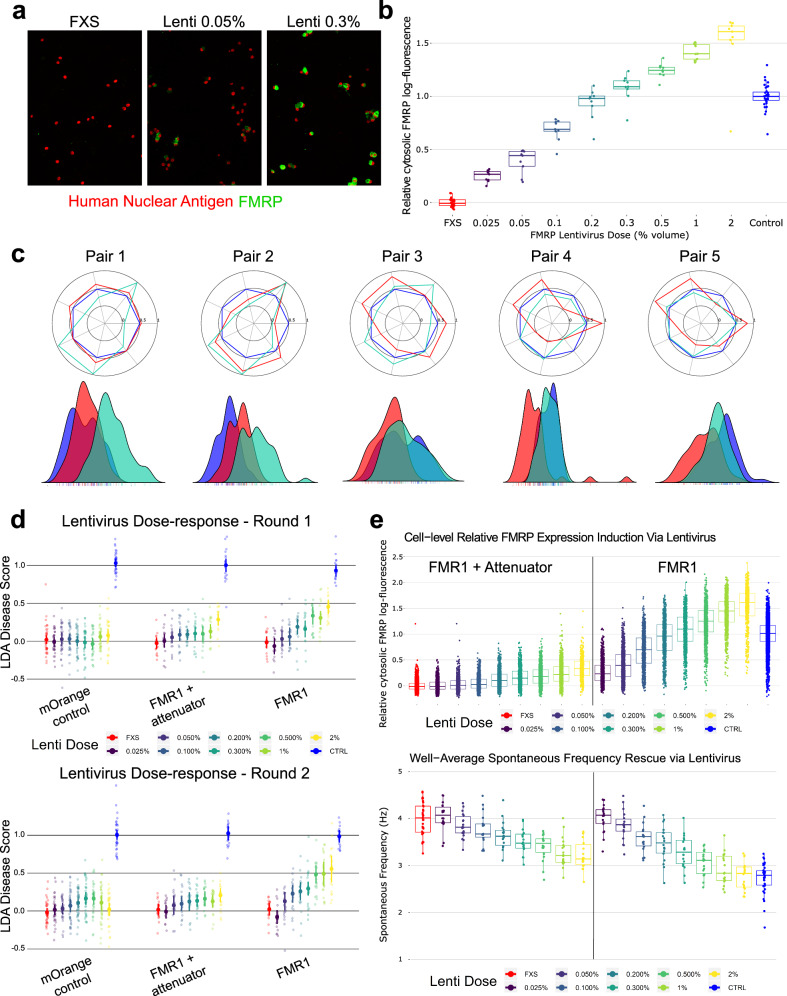
Fig. 3. Use of FMR1 lentiviruses to establish FMRP levels needed to rescue FXS phenotypes.
a Representative high-content confocal images showing dose-sensitive restoration of FMRP expression in FXS neurons. At low doses (e.g., 0.05% volume), only some neurons express FMRP after transduction. At higher doses, nearly every neuron expresses FMRP, but increased lentivirus doses increase average per-cell expression. b Quantitative ICC showing amount of FMRP introduced via the lentivirus across a range of doses. Fluorescence in the proximal cytoplasm was estimated for each cell (identified via nuclei) and aggregated to a field-level average, then log-transformed and rescaled such that the on-plate control group = 1, after which data from all plates were combined. c Radar plots showing the FXS phenotype (red) vs CTRL wells (blue) alongside FXS + lentivirus at a single 1% by-volume dose (teal) for each of the five patient-control pairs (both clones for each donor). Values reflect the common language effect size. Beneath is the LDA scores for each group, fit over the isogenic phenotype and applied to each patient-control pair. d LDA score for isogenic FMR1−/y neurons treated with 8 increasing doses (represented as % volume of lentivirus) of either an mOrange fluorescent tag, an attenuated form of FMR1, or a full-strength FMR1, for each of two replicate rounds (round 1 top, round 2 bottom) plated fresh-from-thaw. For each lentivirus condition, lentivirus treated neurons were compared to undosed FXS and CTRL neurons from the same row on the 96-well plate. e Top: Quantitative ICC of FMRP fluorescence of control neurons (blue) and FXS neurons (red) treated with 8 increasing doses (represented as % volume of lentivirus) of either an FMR1 lentivirus (right) or attenuated FMR1 lentivirus (left). Each dot is a neuron. Fluorescence in the proximal cytoplasm was estimated for each cell (identified via nuclei) and aggregated to a field-level average, then log-transformed and rescaled such that the on-plate control group = 1, after which data from all plates were combined. Bottom: Rescue of Spontaneous Frequency phenotype in optical electrophysiology analysis from the same round of data, with the same conditions. Each dot is a well (to better see the change in a subtle phenotype).
To evaluate FMRP-mediated changes to the functional phenotype, FMR1 lentivirus, attenuated FMR1 lentivirus, and the mOrange fluorescent tag control lentivirus were tested in 8-point dose-response in FXS neurons. For the isogenic line, we observed phenotype rescue in the full-strength FMR1 lentivirus in the composite linear discriminant analysis (LDA) score and each major domain of behavior for both replicate rounds (Fig. 3d). The lowest significant rescue dose was often observed at ~0.2% FMR1 lentivirus volume (Round 1 LDA disease-score difference from mOrange = 0.50 [95% CI 0.112–0.887], t(14.81) = 2.53, p = 0.023), which achieved ~75% of WT FMRP. Full rescue showed for most features at 1% and 2% FMR1 lentivirus (e.g. Round 1 Spontaneous Frequency at 2% lentivirus differed from the corresponding mOrange control dose by −0.559 Hz [95% CI −0.303–−0.815], t(6.77) = 4.27, p = 0.004), approximately matching control cell behavior, as shown in Fig. 3c–e; Round 1 fast-ramp rheobase at 1% lentivirus differed from the corresponding mOrange control dose by 0.338 mW/cm2 [95% CI 0.263–0.413], t(17.57) = 8.79, p < 0.001), nominally exceeding the 0.301 mW/cm2 phenotype effect of control vs FXS, though this tended to be >1.5× WT FMRP levels (Fig. 3b). The mOrange negative control showed little effect on the FXS phenotype (Round 1 LDA disease-score vs un-dosed FXS p-values > 0.2 for all lentivirus doses). Interestingly, the attenuated FMR1 lentivirus often showed phenotype rescue effects at ~0.3% volume of lentivirus (Round 1 LDA disease-score difference from mOrange = 0.469 [95% CI 0.128–0.811], t(21.41) = 2.69, p = 0.013), where FMRP expression was closer to ~20% WT FMRP; (Fig. 3d, e). The attenuated lentivirus may show signs of rescue at lower average FMRP expression levels than full-strength FMR1 lentivirus because fewer cells remain FMRP-deficient when the well was dosed with more lentivirus particles (see images in Fig. 3a and single-cell quantifications in Fig. 3e), but additional experiments would be required to better understand these differences. FMR1 lentivirus rescue in a representative patient/control line manifests the same pattern (Supplementary Fig. 5a, b).
To evaluate whether restoration of FMRP generally rescues the FXS phenotype across most patient genotypes, we ran a single-dose study that transduced all 10 patient cell lines (two clones from each of five donors) with 1% full-strength FMR1 lentivirus. The lentivirus had a similar effect on all patient lines (Fig. 3c), though two patients did not show sufficient difference in the un-treated FXS and matched control lines to properly assess rescue. Similar rescue was observed whether FMR1 was delivered ~1 month before imaging (DIV9/10; Fig. 3) or 2 weeks before imaging (DIV30) (Supplementary Fig. 5c).
Neuronal network and glial effects of the presence or absence of FMRP
Fragile X syndrome disease severity is known to be modified by mosaicism67. In this context, we explored such effects by co-culturing FXS and control neurons in different proportions. One recent multi-electrode array study suggests that 20–50% of neurons in a network must be corrected to move the entire network to WT excitability levels55. We expanded on this work by evaluating spiking behavior while under synaptic blockade at the time of imaging, where the surrounding WT cells might freely alter the maturation and development of FMRP-deficient neurons during the preceding incubation but would not propagate normal functional signatures through the network during recording. Additionally, because our optogenetic system provides single-cell resolution and compatibility with other fluorophores, we were able to fluorescently tag WT and FXS neurons (Fig. 4a; see Methods for additional details) and assess phenotype rescue specifically in FXS neurons that have been co-cultured with different proportions of WT neurons. While the recent study provided evidence that the network as a whole can be rescued by some proportion of FMRP-expressing neurons55, we evaluated whether co-cultured networks of different proportions of WT neurons can rescue the single cell-resolved, cell autonomous excitability phenotypes in FXS neurons.
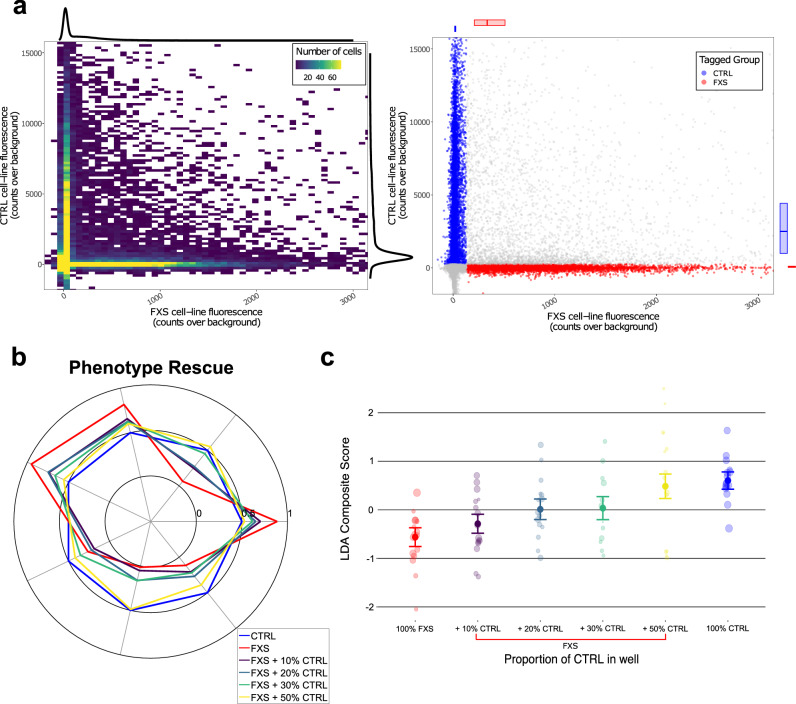
Fig. 4. Modulation of electrophysiological FXS phenotypes via mosaic co-cultures.
a Left: 2D histogram showing the number of cells fluorescing in each of the tag channels (where the orthogonal expression is apparent in the vertical and horizontal ridges) with colors aliased at a max of 75 cells per bin to prevent washout by the high-density peak of non-fluorescers, with 1D kernel density estimates over cells for each channel along the margins. Right: every individual cell in the same data set, color-coded to indicate the assigned tag label, with FXS labels in red and CTRL labels in blue, and ambiguous or non-fluorescing cells in gray. b Radar plot showing the dose-dependent rescue of the phenotype in isogenic reagents via increasing proportions of CTRL neurons in well. Values reflect the common language effect size. Only FXS neurons are reflected in the data from the co-cultured wells (meaning that these data show rescue specifically in cells that do not express FMRP, rather than simply of the overall network which includes CTRL neurons). Neurons are under synaptic blockade at the time of imaging but spent the prior >29 days in vitro without synaptic blockers. c LDA score for 100% control (blue) vs. 100% FMR1−/y (red) and increasing amounts of WT neurons in the network (purple, dark green, light green, yellow). Error bars indicate standard error. Points are well-averages. Point size reflects sample weight (square root of number of quality FXS neurons identified via fluorescent tag in that well). Only FXS neurons are reflected in the data from the co-cultured wells.
Co-culturing FMR1−/y neurons with increasing percentages of WT neighbors in the neuronal network monotonically corrected the FXS neuron intrinsic excitability behavior for each of the individual phenotypic parameters (Fig. 4b) and the composite disease score (Fig. 4c). Functional rescue was seen by just 20–30% network rescue (LDA disease-score difference between 20% co-culture and monoculture FXS = 0.649 [95% CI 0.167–1.131], t(81.1) = 2.637, p = 0.01, percent recovery = 52.7%), and 50% network rescue approximately restored WT behavior (LDA disease-score difference between 50% co-culture and monoculture FXS = 1.223 [95% CI 0.731–1.715], t(81.1) = 4.88, p < 0.001, percent rescue = 99.3%), despite the absence of FMRP in the cells being measured. This finding suggests that only part of the network needs restoration of FMRP expression to provide a therapeutic benefit to FMRP-lacking neurons within the network.
FMRP is not expressed exclusively in neurons and has been detected in astrocytes. The influence of FMRP-negative astrocytes on WT neurons has been explored using mosaic cultures from Fmr1 knockout and WT mice (WT and mutant neurons plated with WT and mutant glia) and suggests that FMRP-deficient glia may play an important role in FXS pathophysiology68,69. Given that our phenotyping experiments were measured in the presence of WT mouse astrocytes, we aimed to better understand the effect of mutant astrocytes on our FXS neuronal hyperexcitability phenotype. To this end, we separately cultured WT and FMR1−/y human neurons on Fmr1+/y and on Fmr1−/y mouse glia. We observed that culture of WT neurons on Fmr1−/y rodent glia significantly moves neuronal behavior towards the FXS state and culture of FMR1−/y human neurons on Fmr1−/y mouse glia results in a more pronounced disease phenotype (Supplementary Fig. 6). Correspondingly, the largest phenotypic window is observed when WT neurons are plated with WT glia and KO neurons are plated with KO glia.
Assay optimization and validation
To further validate and test the performance of our human neuronal FXS assay, we performed a pilot screen of a set of pharmacologically defined reference compounds that modulate targets of past FXS drug candidates, potential FXS therapeutic targets described in the literature or targets deemed relevant to the multiparameter FXS functional phenotype. Several experiments around assay optimization were performed prior to screening (see Methods). Plate layouts and experiment design for the compounds screen are shown in Supplementary Fig. 7. Screening was performed in both the isogenic pair and a single FXS patient/control pair.
Clear phenotypes were observed on the sentinel plate and on-plate controls of the screening plates for both sets of cell lines (Fig. 5a, b, median Z’ of screening plate composite LDA scores = −0.016 for isogenic and −1.77 for patient/control screens). Ten clear hits were identified in the isogenic screen, while five additional molecules were flagged as “borderline/partial hits”, and one molecule showed “anti-hit” behavior (Supplementary Table 2). Screening of the same compound set in the patient/control line identified the same ten clear hits, and in this experiment the five “borderline” molecules also appeared as hits (Supplementary Table 2). Several well-validated FXS targets from the literature yielded hits (BK channels, GABA receptors, Slack channels), and we were also able to identify multiple hits for the same target, lending further evidence to the relevance of these targets (Supplementary Table 2). Hits identified as correcting the LDA score are shown in Fig. 5c. Radar plots depicting the FXS fingerprint with compound treatment for 7 example hits are also shown (Fig. 5d). Correction of the raw values of example features from a subset of hits is shown in Fig. 5e. Targets that were identified to correct at least some parameters of the phenotype include: BK channels, SK channels, GABA receptors and GABA transporter, Rac1, NaV channels, and CaV channels.
Fig. 5. Small molecule screen identifies multiple hits and targets for therapeutic development.
a Recapitulation of the FXS electrophysiological fingerprint in isogenic neurons measured in a sentinel plate on the day of screening. Values on the radar plot reflect the common language effect size. b Recapitulation of the FXS electrophysiological fingerprint in patient/control neurons measured in a sentinel plate on the day of screening. Values on the radar plot reflect the common language effect size. c The LDA score for on-plate control (blue), on-plate FXS (red), and FXS neurons treated with 86 unique small molecule compounds for the isogenic reagents (left) and a representative patient/control pair (right). All hits identified as green on the graph. Anti-hits marked in yellow-orange. Bars are 95% confidence intervals. d Radar plots for 7 example small molecule compound hits represented as green on each radar plot compared to the control (blue) vs. FMR1−/y (red) FXS fingerprint. For each example hit, the compound ID and molecular target are listed. e Rebound spike rate (left), LDA score (middle) and rheobase (right) for different example compound hits plotted per well and separated by replicate plates. f Dose-response graphs for spontaneous frequency (left) and rheobase (right) for an example compound on the isogenic cell lines. On-plate controls shown in red (FXS, left) and green (CTRL, right), with the dose and the response of the EC50/IC50 marked with dotted blue line. Dotted green line is the mean of the controls.
We next performed hit confirmation that included re-screening the 16 hits, including the anti-hit, from the primary screen plus an additional 8 compounds for a subset of targets to build additional confidence in the specificity of select target mechanisms (See Supplementary Table 2 for information on compound mechanism of action). These 24 compounds were screened in 8-point concentration-response format ranging from 0.001 μM to 3 μM. Out of the 24 compounds screened in dose-response, seven of the ten strongest hits from the primary screen showed clear dose-response in this range, building support for these targets. Example concentration-response curves for a single hit for two example features are shown (Fig. 5f). Additional compound concentration-response data can be found in Supplementary Fig. 8. Overall, this pilot screen demonstrates the applicability of the assay to compound screening and the examination of molecular underpinnings of FXS disease mechanisms.
Discussion
Gaining a deeper understanding of the neurophysiological impact of loss of FMRP in a human neuronal context is important to enable identification and qualification of disease modifying therapies for FXS. We have applied our CNS discovery platform of high-throughput all-optical electrophysiology with single cell and millisecond temporal resolution and machine learning analytics, to comprehensive neurophysiological phenotyping of human iPS cell-derived models of FXS. From this analysis, we have developed an optimized multiparameter phenotype that captures a broad set of intrinsic neurophysiological features that are perturbed in the absence of FMRP. The identification of these altered neuronal properties and the optimization to an assay configuration suitable for drug discovery applications were uniquely enabled by the high throughput and automated characteristics of the all-optical electrophysiology and deep analytics, which permit synthesis of information recorded from vast numbers of individual neurons derived from multiple cell lines and clones. The assay provides a tool for probing neurophysiological disease mechanisms underlying FXS and a basis for therapeutic approaches that address this core pathogenic domain.
The multiparameter functional phenotype we identified includes increases in (1) spontaneous action potential frequency, (2) the average firing frequency within the population of spontaneously active neurons, (3) the proportion of neurons in a well that don’t spontaneously spike, (4) increased rebound firing after stimulus termination, as well as decreased rheobase, during the (5) fast ramp and (6) linear conductivity ramp (a blue stimulus stimulation that linearly increases the proportion of open CheRiff channels across 2.5 s). Additionally we identified changes in (7) how quickly the downstroke of the action potential is elicited by the highest stimulation. Many of these features indicate a hyperexcitable state in FXS neurons relative to control neurons. This hyperexcitability is generally in line with previous studies of Fmr1 KO mice and human iPS cell-derived neurons18,53–56,59. Multiple previous electrophysiological studies of human iPS cell-derived neuronal models of FXS using either MEA or traditional patch clamp methodology have reported increased neuronal firing rate or bursting frequency, consistent with our results. However, our study extends on previous findings in several key aspects. The high throughput nature of our optical electrophysiology platform enabled analysis of a large number of cell lines with multiple clones per line and multiparametric electrophysiological analyses of many thousands of neurons per line. This scale yielded identification of a consensus, multi-feature phenotype that includes discrete active membrane properties as well as more complex action potential firing properties. We propose that this higher resolution phenotype, which can probe modulation of multiple underlying components, is essential for identifying targets and compounds that specifically reverse, rather than simply mask, underlying disease biology. Second, the scale and format of our FXS functional assay is directly amenable to higher throughput drug discovery applications, providing a significantly improved basis for incorporating read-outs of fundamental neurophysiological aspects of FXS human disease biology into target identification and therapeutic screening and qualification efforts.
Though FXS has a number of core, commonly observed features, there is a significant degree of phenotypic and genetic heterogeneity in FXS70. Mosaicism in CGG repeat length or degree of FMR1 gene methylation are key direct genetic factors that can influence disease phenotype70. In addition, variation in genetic background likely contributes to disease heterogeneity70. This variability could present a challenge for development of a generally disease-relevant neuronal model of FXS. We took several approaches to address this variability. First, we evaluated functional phenotype in neurons derived from five unrelated FXS patient/matched control pairs to ensure that detected phenotypes are representative across individual patients. Second, we used matched controls to limit the impact of background genetic variability in each patient/control comparison to focus analysis on disease mechanisms directly related to FMRP deficiency. Third, we selected patients that scored within a defined range in relevant cognitive tests to ensure a degree of uniformity among patients in disease severity for the intellectual disability domain. Fourth, we confirmed the lack of FMRP expression in all patient cellular reagents. These elements of our approach likely contributed to our ability to identify a consensus, multiparameter phenotype that was present in the majority of case/control pairs. The deviation from this phenotype in 1/5 pairs could result from background genetic variation as we confirmed that it did not correlate with repeat length or symptom severity. Extending the analysis to a larger number of case/control pairs may be useful to determine whether there are any additional recurrent multiparameter phenotypes that may be relevant to subsets of patients across the spectrum of genetic diversity.
We validated the relevance of the composite phenotype to FXS disease by using lentiviral-mediated re-expression of FMRP to confirm the dependence of the phenotype on FMRP deficiency. By using two lentiviral FMRP expression vectors, a full-strength vector, and a translation attenuated form, we were able to generate a high-resolution titration of FMRP re-expression in FMRP-deficient neuronal cultures. This approach revealed FMRP-dependent rescue of the composite functional phenotype, with a clear relationship between FMRP expression level and degree of rescue for each vector. The results indicated that statistically significant rescue of FMRP-deficient neurons towards control behavior occurs at expression levels as low as ~20% of WT FMRP expression. Full rescue of most of the individual components of the disease phenotype was achieved with FMRP re-expression, albeit at levels ~1.5-fold the endogenous level. Not all phenotype parameters achieved full rescue by the maximum dose (e.g. rebound firing), and correspondingly, the LDA composite phenotype score also only showed partial rescue.
A second approach we applied to validate FXS disease relevance of the assay was to examine modulation of the functional phenotype by mosaicism achieved through co-culturing FMRP-deficient and control neurons in different proportions in the assay cultures. Cellular mosaicism is a known modulator of FXS disease severity67, providing a means to test whether the assay is sensitive to a disease-relevant modulatory factor. We found that presence of ~50% control neurons in the cultures was sufficient to fully rescue the functional disease phenotype in the FMRP-deficient cell population. Importantly, the cellular resolution of the assay system enabled demonstration that presence of this percentage of control neurons in the cultures fully rescued the intrinsic disease phenotype in FMRP-deficient neurons, indicating complete rescue throughout the neuronal network. It is intriguing that maturation within a network containing control neurons can correct intrinsic deficits in FMRP-deficient neurons, indicating potential for non-cell autonomous therapeutic mechanisms.
Implementation of gene-based therapeutic approaches to FXS involving FMR1 gene replacement or reactivation requires understanding of the level of FMRP expression and the proportion of FMRP expressing cells required for therapeutic benefit. The lentiviral expression and mosaicism experiments, in addition to validating disease-relevance of the assay, indicate the utility of the assay to address these important questions for enablement of gene-based therapeutic strategies. A previous study identified a network hyperexcitability phenotype in a human iPS cell-derived neuronal FXS model and applied the phenotype to address these similar questions55. While general conclusions were similar to those of our study, there are quantitative differences between the findings, with lower mean FMRP expression and a lower degree of mosaicism found to normalize the phenotype in the prior study55. There are several key differences between the two studies that may explain this discrepancy. The previous study used MEA recordings from spontaneously active networks in the absence of synaptic blockade and measured a weighted firing average while our study focused on a multiparameter intrinsic activity phenotype. Additionally, the FMRP titration in the previous study was achieved by titrating an FMR1 antisense oligonucleotide in control neurons to knock down FMRP to different degrees, while our titration was achieved via lentiviral delivery to neurons lacking FMRP in a genetic rescue context. Finally, the mosaicism analysis in the prior study did not specifically measure phenotype rescue within FMRP-deficient neurons but rather measured the entire network, including the control cells. Further work, including evaluation of rescue by different FMRP isoforms and varying kinetics of FMRP introduction during neuronal differentiation and maturation should help to more definitively address these questions to enable gene-based therapeutic approaches.
To validate assay performance and assess the multiparametric read-out in relation to targets previously associated with FXS disease biology, we screened a small library of pharmacologically defined reference compounds with reported relevance to FXS. A total of 86 compounds were tested and 10 hits were identified in the screen on the isogenic pair, all of which also appeared as hits in the patient/control screen. There were an additional 5 borderline compounds in the isogenic screen that were detected as true hits in the patient/control screen. Compounds targeting several different ion channels previously implicated in FXS pathogenesis were identified as hits in the screen, and the individual hits rescued at least some parameters of the phenotype. We anticipate that compounds with ability to fully rescue all parameters would be more likely to address and correct underlying FXS disease biology and thereby have greater potential to yield therapeutic efficacy, representing the benefit of a high-throughput, disease-focused, multiparametric neurophysiological screening assay.
In addition to excitatory neurons, there is evidence for FXS pathogenic mechanisms in other cell types, including inhibitory neurons and astrocytes27,68,71,72. Our preliminary work with Fmr1 knockout mouse astrocytes indicated that FMRP-deficient astrocytes can exacerbate the composite disease phenotype, consistent with pathogenic contributions from multiple cell types. Future work applying our all-optical electrophysiology platform to examine human iPS cell derived FXS co-culture models including excitatory and inhibitory neurons and glial cells will be an important extension of the current study.
In summary, we have developed a novel assay of altered neurophysiology in human neuronal models of FXS. Our results highlight the value of scalable, multiparameter read-outs that can broadly capture neurophysiological effects stemming from disease-relevant perturbations in a human neuronal context. The assay system provides a tool for further studies of FXS disease biology. The platform can be similarly applied to other neurological disorders with a genetic basis and is applicable across multiple therapeutic modalities. Given the pleiotropic nature of FMRP, which is known to regulate many targets in the brain relevant to neuronal and synaptic activity, comprehensive assays of this type are essential for identifying and qualifying candidate targets and therapeutics to address core disease biology.
Methods
Human iPS cell line culture and NGN2 neuronal production
For the generation of FXS patient-derived iPSC cell models, we used sample collection protocol and consent forms approved by RUSH University Medical Center’s IRB to obtain peripheral blood samples (2–4 mL) from 5 male Fragile X Syndrome patients and from 5 healthy control male relatives (brother or father). Peripheral blood mononuclear cells (PBMCs) were then isolated from these samples by density gradient centrifugation with Ficoll. PBMCs were reprogrammed into induced pluripotent stem cells (iPSCs) using non-integrating transient overexpression of the pluripotency factors OCT4, KLF4, SOX2 and c-MYC, via Sendai virus transduction (Cytotune 2.0 kit, Life Technologies). At least four to five independent reprogrammed clones per donor were established into iPSC lines, and these cells were routinely expanded in culture using mTeSR1TM medium (STEMCELL Technologies) and 6-well cell culture dishes precoated with MatrigelTM (Corning). At least two iPSC lines per donor were used for subsequent QC assays, neuronal production and phenotyping. For the generation of FMR1+/y and FMR1−/y isogenic iPSC lines, we used CRISPR/Cas9 to target the disruption of the FMR1 gene in the control iPSC line (FMR1+/y) M28, which was initially derived from a neurologically healthy male donor. Cas9 and gRNAs targeting an early FMR1 exon were used to induce insertion/deletion mutations resulting in early truncations via frameshift of the FMR1 mRNA. CRISPR/Cas9 edited clones were screened via Sanger sequencing and lack of FMRP expression as a result of FMR1 genetic disruption was confirmed in the selected iPSC clones using a Simple WesternTM assay. At least two edited iPSC lines were used for subsequent QC assays, neuronal production and phenotyping. Neuronal production was carried out as previously described60. iPS cell lines were differentiated into cortical excitatory “NGN2” neurons using a transcriptional programming approach mediated by doxycycline induction of the pro-neuronal transcription factor NEUROGENIN-260,64, but combined with small molecule modulation of key developmental pathways (DUAL SMAD “D.S.” inhibition) as previously reported65. These neurons were referred to as NGN2 + D.S. To initiate neuronal cultures for optical physiology and immunocytochemistry assays, differentiated neurons were plated at 70,000 cells +20,000 mouse glia per well of a 96-well plate (IbidiTM) and maintained for 30 days (days in vitro 30 or DIV30) or 45 days (DIV45) in neuronal maturation medium as previously reported60.
Delivery of all-optical physiology components
Lentiviral transduction of cultured neurons with actuator and reporter components was performed 2 weeks before imaging for the intrinsic excitability measurements. Neurons were transduced with lentiviral particles encoding the blue light-activated channelrhodopsin CheRiff-BFP and the voltage reporter QuasAr-Citrine60. All lentiviral particles were produced at Q-State and all expression constructs utilized a human synapsin 1 (hSyn1) promoter for neuronal-specific expression.
Neuronal all-optical physiology measurements and quality criteria
Custom blue stimulus protocols were used to evoke a range of firing behaviors. QuasAr fluorescence was captured using a custom high-throughput microscope73 at ~1 kHz frame rate. The movies were then segmented and voltage signals from individual cells extracted74,75, yielding a throughput of about 60,000 cells per hour, with patch clamp-like resolution. Intrinsic excitability measurements, with cells under total synaptic blockade at the time of imaging, were carried out at both 30 and 45 DIV. Patient/control data were utilized for data mining, as the five donor families (two clones each from a patient and a familial control) were convenient and appropriate holdout folds for cross-validation (where holdout data shares no confounding dependencies with training data). Candidate parameters from this data mining step were then confirmed with inferential statistics tests using the isogenic cell model data, with adjustments for multiple comparisons (see Phenotyping section). All inferential tests reported in this manuscript are two-sided, with alpha = 0.05.
Movies of the QuasAr fluorescence channel were typically recorded at ~1 kHz and functionally segmented to find spatially coherent pixels with correlated signals serving as neuronal candidates, called “sources”. All experiments were recorded in a custom in-house BrainPhys imaging buffer. The BrainPhys medium used for neuronal imaging is prepared with the following components, all purchased from Sigma-Aldrich: 115 mM NaCl (S5886), 18 mM Sodium gluconate (G9005), 4 mM Glucose (G8270), 3 mM KCl (P5405), 1.1 mM CaCl2 (C5670), 1 mM MgSO4 (M7506), 0.5 mM Na2HPO4 (S5136), 0.45 mM NaH2PO4 (S5011), and 0.15 mM Sodium Pyruvate (P2256). To prepare the medium, powders for each component were weighed out and prepared as 50X stock solutions in deionized water (dH2O) for each component. 50× stock solutions were then added to a volumetric flask to achieve final concentrations specified above. 1 M HEPES (Gibco, 15630-080) was added to achieve a final concentration of 10 mM. Deionized water (dH2O) was then added to bring the medium to the final volume. The pH of the BrainPhys medium was adjusted to 7.4, and the solution was filter-sterilized prior to use. For measurements of intrinsic excitability, D-AP5 (25 µM), GABAzine (20 µM) and NBQX (10 µM) were included in the buffer to block most relevant synaptic transmission. The average fluorescence trace for each source was then corrected for photobleaching and optical crosstalk and run through a spike detection algorithm. The trace was split into epochs at changes in stimulation. Parameters of the spike shapes (e.g. height, width, afterhyperpolarization, 1st and 2nd derivatives of the rising and falling edge), spike timings (e.g. time to first spike, time of last spike, inter-spike intervals), spiking patterns (e.g. frequency, bursting, onset frequency at the start of stimulation, stable “plateau” frequency during stimulation, rebound firing after stimulation), and changes in spiking behavior (e.g. adaptation in firing rate over constant stimulation) were then quantified for each stimulation epoch of each trace, with some stimulation-specific measures (e.g. rheobase during a ramp, frequency of bursts during long spontaneous periods)61. Source morphology was also parameterized, and source candidates were automatically rejected if they did not meet standard quality criteria for size (minor width of an ellipse fit to the estimated cell body >10 microns), clarity (cells for which the ratio of the brightness of the source mask and the brightness of an equivalent image with no speckling is greater than 10 are discarded), brightness (average fluorescence over background), and signal-to-noise ratio (where the spikes are the signal and the high-frequency shot noise in the trace is the noise, requiring a ratio greater than 3). We also rejected the nearly-non-spiking sources (a unique second population of sources, distinct from the primary distribution, that only spike ~1–3 times total during an entire protocol, typically comprising 5–20% of the otherwise “quality” sources in any given experiment), as we expected a subtle phenotype and anticipated that these low-information sources would decrease the sensitivity of our assay. The quality sources were then aggregated to the well level with an algorithm that estimates the centrality of each parameter using the statistic (e.g. mean, median, trimmed mean) most appropriate for that parameter class (mostly considering skew, outliers, discretization, and the information of greatest biologic interest). Finally, additional derived features were constructed using a sparse PCA over the training data. The output was a table with wells down rows and measurements (called “features”) across columns.
Phenotyping
Phenotyping was conducted primarily on the large intrinsic excitability data set shown in Fig. 2, spanning five patient/control pairs as well as CRISPR/Cas9 iPSC isogenic models (all with multiple clones). Neurons were evaluated while under synaptic blockade after 30 DIV, recording 3 movies per well. The ultimate goal of phenotyping was to infer FMRP-dependent changes in cell behavior given the data collected across cell lines, rounds, plates, and cell models (patient vs CRISPR/Cas9-isogenic). With hundreds of measured outcomes, we used the patient/control reagents as a training set for feature selection using machine-learning, and the CRISPR lines as a confirmatory holdout using inferential statistics. Three rounds of data were used for both training (patient/control reagents) and for validation with inferential methods (CRISPR/Cas9-isogenic reagents), with 12 wells plated per cell line per round, split across different plates at counterbalanced plate locations. With 2 FXS and 2 control cell lines for each of the five patient/control pairs, this yielded 240 wells per round for the feature search, for a total of 720 balanced training samples available across the three rounds. The isogenic reagents only had a single control line plated at the same 12 wells per round, leaving the validation set imbalanced (though note that we are validating the individual features, not a predictive model).
In the patient/control data, we began by removing extremely colinear features. In these cells, it was found that similarly situated parameterizations of many spike shape characteristics (e.g. shape of the first spike vs average shape across all spikes within a stimulation) tended to be correlated. As adaptation changes have been observed in the literature, we elected to keep the shape parameters from the first spike, before significant adaptation could affect behavior (which can confound shape and activity signals). Features were then trimmed via an iterative algorithm that interrogates the correlation matrix and drops features with information already captured elsewhere. Then, we evaluated independent predictive value for the diagnostic labels (via logistic regression). For each feature, models were fit using leave-one-group-out cross validation over the five patient/control pairs. Feature importance was computed as the average of the regression coefficient over folds weighted by cross-validated F1 score. Measures of rebound firing, rheobase, and proportion spontaneously active cells dominated rankings of activity features, while high-stim spike width and mid-stim AHP time were prominent among shape features. The top feature was area (a morphology measure). Features were dropped from consideration in subsequent analyses if mean cross-validated F1 scores was less than 0.5 or had a feature importance more than an order of magnitude less informative than the best features. This approach biases our search towards independent phenotypes over which we can search for correction by compound or reintroduction of FMRP (particularly in the case of partial correction, where we want to clarify which features are corrected and to what extent), but has the consequence of missing decision boundaries that traverse feature combinations instead of any particular feature.
Next, additional data mining was performed to further reduce the feature set towards an optimal mix using embedded feature selection methods. As low-information features have already been removed, this can be interpreted as a search for a favorable set of features carrying some orthogonal signal. To this end, robust elastic-net models were fit using all the remaining features, again with a cross-validation rotation over patient-control pairs (centered to the control of each pair to remove pair effects), implemented by scikit-learn76. An iterated hyperparameter search optimized the regularization scale (“alpha”), and found the maximal elastic net mixing parameter (l1_ratio in scikit-learn) that kept validation loss of the whole model to within tolerance of the best-performing model, in order to encourage more aggressive feature trimming within a specified information loss (if a pure L1 model is in fact optimal in this regard, it should be approached). Composite feature importance values were calculated on average over the folds by weighting the individual coefficients from each model proportional to the validation performance of that model. Top activity features again included rheobase and rebound firing, but also included measures of adaptation, onset frequency, and proportion of stimulations with just 1 or 0 spikes. Top shape features included spike height, depth of the afterhyperpolarization, and the first and second derivates of the falling edge of the action potential under high stimulation. The top feature was again area. A final pass over the top features was then conducted to drop features that were not statistically robust in the training set between replicate rounds. These feature candidates, plus promising candidates from the independent phenotypes (e.g. spontaneous frequency) were then taken to the isogenic reagents for statistical inference, with a Bonferroni correction for multiple comparisons, and features that did not validate in the orthogonal cell model across multiple rounds were dropped. Many features (including area) only reached significance in one or two rounds. The final phenotype is shown in Fig. 2.
Composite metrics
A single, composite outcome was used in instances where the average extent of therapeutic rescue was of interest, such as for lentivirus experiments and compound screens. In such cases, we conducted a linear discriminant analysis (LDA) over phenotype features to find the linear projection that maximized group separability. The LDA scores shown in Figs. 2–5 used the features shown in the corresponding radar plots, but removed remaining sources of high collinearity (e.g. we only use two of the three spontaneous activity features) or instability (e.g. the shape feature, which proved unreliable). The screen and some lentiviral interventions also utilized an on-target/off-target vector decomposition method, which uses a numerically stable implementation of the cosine rule to project the intervention wells onto the axis defined by the positive and negative controls for the “on target” score and captured the orthogonal distance to the intervention well as the “off target” score. This used nearly all measurements as inputs and weighed them approximately equally but expressed all vector magnitudes via Mahalanobis distance to account for data correlations. In cases where a sentinel plate was available, these projection methods were fit using the sentinel plate and then applied to the experimental plates, to prevent overly optimistic assay quality scores for each plate (using the on-plate controls).
Lentivirus phenotype rescue
Several experiments were conducted to evaluate phenotype rescue following reintroduction of FMR1 via lentivirus. Three lentiviruses were used (1) healthy, full-length copy of FMR1 (2) an FMR1 engineered to reduce expression by 10–20×66, and (3) an fluorescent mOrange construct as a negative control. All 3 viruses were driven by the human synapsin1 (HSyn) promoter. Lentivirus was delivered to neurons on either DIV9/10 or DIV30 and then imaged on DIV45, as indicated. After titrating the approximate dose of lentivirus required to restore some FMRP to the large majority of cells using automated high-content imaging (see below), we conducted a single-dose rescue experiment over all 20 patient-control cell lines (every cell line received a dose) in addition to the isogenic model (Fig. 3c). The experiment was repeated two additional times using fresh-from-thaw platings of neurons. Each plate of cells only had up to 4 cell lines with several on-plate replicates. To handle this hierarchical data structure, statistical tests were conducted for each feature of the phenotype using linear mixed-effects models of disease state (FXS vs control) * lentivirus dose (0% vs 1%), with a by-plate random intercept and a by-plate random slope for disease state, lentivirus dose, their interaction, and all correlations to the random intercept. Models were fit with the lme477 R package. The marginal effect of lentivirus dose within the FXS cell lines was derived using the emmeans78 R package and evaluated for significance using a Satterthwaite approximation for degrees of freedom.
We then evaluated the concentration-response of phenotype rescue on select cell lines for both the FMR1 lentivirus and the FMR1+Attenuator lentivirus, versus equivalent doses of a control mOrange lentivirus with the same basic design and promotor (Fig. 3d) across a 8-point dose-response. Statistical tests were conducted for each feature of the phenotype and the LDA composite using linear mixed-effects models of lentivirus (mOrange, FMR1+Attenuator, FMR1) * dose, with a by-plates random intercept and a by-plates random slope for lentivirus, dose, their interaction, and all correlations to the random intercept. The mOrange control and 0% dose were set as reference levels in the dummy-coding, so effects of interest (marginal rescue above and beyond mOrange for each of the doses) were directly estimated by the model and did not require computation of marginal means for post-hoc contrasts. Models were again fit via lme4, and statistical significance was evaluated via the lmerTest79 R package. For all rounds, plates were fixed and stained immediately after functional imaging, and immunocytochemistry was used to quantify FMRP expression.
Fmr1−/y glia model
Because our human neuronal assay utilizes mouse glia to help support stable long-term cultures, and wild-type mouse glia express trace amounts of FMRP (several orders of magnitude lower than the neurons), we evaluated the impact of Fmr1−/y mouse glia on the FXS phenotype. Male Fmr1 knockout mice (Fmr1-KO) on the C57BL/6J background were used as a source of glial cells in all relevant experiments. To generate these animals, the breeding scheme crossed a wild-type male C57BL/6J mouse (Jackson Laboratory Stock Number: 000664) with a female Fmr1 heterozygous mouse (Jackson Laboratory Stock Number: 003025). Weened progeny were group housed with sex matched littermates on static racks and maintained on a 12:12 h light:dark cycle. All experimental techniques were approved by the Committee on Animal Care at MIT and all animals were handled in accordance with NIH and MIT guidelines. Males were genotyped to identify Fmr1−/y and Fmr1+/y mice. Glia were obtained from each group and frozen, pooled across individual mice. Concurrently, non-littermate male Fmr1−/y glia were obtained from a different mouse colony using a different breeding scheme, to protect against breeding-pair-specific idiosyncrasies. Glia from the wild-type and two Fmr1−/y groups were plated alongside wild-type and FMRP deficient human neurons, then allowed to grow for 30 days in vitro. Optical electrophysiology recordings were collected for all neurons (Supplemental Fig. 6). Statistical tests were conducted for each feature of the phenotype and the LDA composite using linear mixed-effects models of human neuron genotype (FMRP deficient vs wild-type) * mouse glia group (wild-type, littermate Fmr1−/y, alternate site Fmr1−/y), with the maximal by-plates random effect structure (random slopes and intercepts for both main effects and interactions with correlations). Models were fit with lme4, evaluated with the emmeans package, and plotted with the sjPlot80 R package.
Mosaicism model
FMR1−/y neurons were plated at various densities and co-cultured with complementary fractions of WT neurons, with each genotypic group being differentially labeled with red (for FMR1−/y) or green (WT) fluorescent protein (RFP and EGFP, respectively) reporters via Cre-mediated recombination. Supplemental images acquired following all-optical physiology measurements were used to quantify the RFP and EGFP fluorescence for each of the neurons detected in the functional recordings. Figure 4a (left) shows the distribution of cells expressing the two different fluorescent reporters, showing efficient orthogonal expression of the Cre-dependent constructs. Cells unambiguously expressing only one tag were then labeled as FXS or control neurons (Fig. 4a, right). More specifically, an allotment of FXS neurons were plated on the first day of the experiment and dosed with Cre. The Cre virus was washed out the next day, and a complementary amount of control neurons was added without Cre. Later, a Cre-on fluorescent tag was added (which should fluoresce in the FXS neurons but not in the wild type neurons) along with a Cre-off fluorescent tag in a different channel (which should fluoresce in the control neurons). After collecting optical electrophysiology measurements from each movie, still images of the two fluorescence channels were collected and the local mean fluorescence for each source over the local background in that channel was computed (note that this occasionally returns a negative value when a source is dimmer on average than the local surroundings). Because the fluorescent tags were orthogonal, each channel had sources distributed in a zero-inflated right-skewed distribution: a significant proportion of cells didn’t express that tag, and the sources that expressed the tag did so to varying degrees. We identified the hinge in the zero-inflated portion of each distribution using estimates of the post-peak second derivative and used these values as thresholds in the two channels. Cells were tagged as being FXS in genotype if they expressed the Cre-on tag at levels outside the zero-inflated peak, but inside the zero-inflated Cre-off peak. Similarly, cells were tagged as being control if they expressed the Cre-on tag at levels inside the zero-inflated peak, but outside the zero-inflated Cre-off peak (Fig. 4a). All other sources (which expressed both tags or neither tag) were labeled as “ambiguous” and removed from further analysis. The tagged FXS sources were evaluated using the identified FXS phenotype, with groups separated by the proportion of control cells with which they were initially co-plated. Importantly, these functional readouts were collected while under synaptic blockade, and reflect the consequence of long-term exposure to control neurons on isolated intrinsic excitability. Statistical significance of rescue was evaluated using a linear mixed-effects model with the maximal by-plates random effects structure, with a Satterthwaite approximation to degrees of freedom.
Primary screen
We performed several experiments including varying the blue stimulus protocol, imaging parameters, and cell density to optimize the assay window to fully enable large scale studies of FXS and screening applications. These individual changes resulted in phenotype window improvements as indicated. The final, fully optimized screening assay resulted in a single-well Z’ of −0.168 between wells of WT and FXS neurons across 12 plates; groups of 3 wells were averaged to further improve the screening window. The compatibility of our assay with detecting “hits” from screening was also assessed by discriminating blinded delivery of an FMR1 lentivirus vs. a fluorescent tag.
For the compound screen, we compiled a set of 86 compounds. As this purpose of the screen was to validate our FXS functional phenotype and assay, these compounds were tool compounds selected to modulate a variety of targets previously shown to be implicated in FXS models (see Supplemental Table 2). These compounds were each screened in two 3-well replicates at single dose (1 µM), with each 3-well replicate located across 2 different plates in different plate locations. This allowed us to approximately de-confound imaging-order and plate location effects and provided a within-experiment off-plate replication of hits to reduce false positives. Cells were treated 24 h before imaging and again ~30 min before imaging while in the imaging buffer, such that the compounds were present during the imaging session. Responses to the compounds were measured in triplicate on each of two replicate plates (for a total of 6 wells per compound) at a single dose of 1 µM. A “zebra stripe” sentinel plate was used to fit the weights used for LDA and on-target/off-target composite scores, then applied to all screening plates. Hit selection was conducted via statistical inference, which allowed us to incorporate information about compound variability and pool the information pertinent to estimating and correcting plate effects via linear mixed effects regression. The 95% confidence intervals of all compounds are shown in Fig. 5c, with the compounds statistically significantly different from the un-dosed FXS wells highlighted. Hits were prioritized if they corrected more than one component of the phenotype. This procedure was conducted independently for a CRISPR cell model screen and a patient-control cell model screen, chosen for the largest stable screening window representative of the phenotype also observed in other lines from that cell model, which allowed for a validation of compound effects. Though dozens of compounds showed some signs of activity along the FXS phenotype detectable via the all-optical excitability assay with this acute dosing regime, 10 compounds were clear multi-feature hits in both screens, which formed the basis for dose-response hit confirmation (along with other compounds of interest intended to further interrogate the most promising targets).
Dose-response hit confirmation
24 compounds were run in 8-point dose-response, in triplicate. The sentinel plate was again used to fit LDA and on-target/off-target projections, which were then applied to all screening plates. Each compound was subject to a 4-parameter log-logistic sigmoid model that freely varied the lower asymptote, upper asymptote, slope, and EC50. All models were fit via Levenberg-Marquardt algorithm, but each well was weighted proportional to the square root of the sample size of sources in that well (primarily to accommodate cases where a toxic dose killed most of the cells). Fits were rejected if the slope was higher than biochemically plausible (defined as traversing >55% of the response range in <5% of the dose range), if the nominal EC50 was not captured within the dose range (an unreliable basis for estimation), or if the nominal high-dose asymptote was less than 0.5 standard deviations away from the negative control mean (indicating an insufficient maximal response for a meaningful CRC function). We performed the hit confirmation in the same original cell lines and added an additional patient/control pair to confirm phenotype rescue in a holdout cell model.
Automated quantitative immunocytochemistry
Immunocytochemistry assays were carried out as previously described (Williams et al.60). In brief, cultured cells were fixed using 4% (v/v) paraformaldehyde for 20 min, permeabilized with 0.2% (v/v) Triton X-100, blocked with 10% donkey serum (Jackson ImmunoResearch) in 0.2% Tween-PBS and treated with primary antibodies overnight at 4 °C. Primary antibodies used were human nuclear-specific antibody (EMD Millipore; MAB1281, 1:1000), mature pan-neuronal marker MAP2 antibody (Novus Biological; NB300-213, 1:3000) and FMRP antibody (Cell Signaling, 7104S, 1:100). After five washes with 0.1% Tween-PBS, cells were treated with secondary (Alexa®-conjugated) antibodies for 1 h at room temperature. Cell nuclei (DNA) were stained with DAPI. Images were acquired as Maximum Intensity Z-projections using a GE IN Cell Analyzer 6000 with 20X (0.75NA) objective. Single-cell FMRP expression was estimated using a custom automated microscopy pipeline. For each field, a stack of ~20× confocal images was collected in fluorescence channels for primary antibodies for FMRP, MAP2, human nuclear antigen (“HuNu”; MAB1281 above), and for DAPI. All z-stacks for each channel were consolidated via a max projection. The HuNu channel was first segmented using custom histogram-splitting algorithms and the proximal cytoplasm for each nucleus was then found using the MAP2 channel. The centrality of the FMRP fluorescence for the cytoplasmic pixels was then estimated for each cell using the mean of the middle 80% of the data. In multi-plate experiments with on-plate controls, data from each plate was typically log-transformed then rescaled such that the mean of the negative controls was at “0” and the mean of the positive controls was at “1” prior to being combined.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We would like to thank the anonymous reviewers and the editor for their contributions to the revisions of this manuscript. M.F.B. and D.S. are supported by NIH grant R21NS123499. Quiver Bioscience would like to acknowledge NIH, in particular NIMH for funding the development of the high throughput FireflyTM instruments for all-optical electrophysiology recordings (grant R44MH112474) used in this study for phenotype discovery and compound screening.
Author contributions
J.J.F., N.D.B., R.D., E.N., C.F., C.H., M.F.B., L.A.W., D.G., M.B., B.F., and G.T.D. designed the study. E.B.K. recruited and collected patient/control samples for stem cell differentiation. J.F., K.H., H.U., T.N., H.S., J.J., and J.G. acquired the data. D.S. and M.F.B. provided mouse tissue for glia co-culture experiments. N.D.B., S.J.R., and J.J.F. analyzed the data. J.J.F., N.D.B., R.D., E.N., C.F., M.F.B., L.A.W., D.G., M.B., B.F., and G.T.D. interpreted the results. J.J.F., N.D.B., R.D., E.N., C.F., D.G., B.F., and G.T.D. prepared the manuscript. All authors contributed to manuscript revision and approved the submitted version.
Peer review
Peer review information
Communications Biology thanks Karo Talvio and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Christian Wozny and Joao Valente.
Data availability
The data that support the findings of this study are available as Supplementary Data in the form of excel worksheets.
Material availability
Patient-derived stem cell lines were generated as described (see Methods). Patient-derived iPS lines generated as part of this study can be requested to RUSH University Medical Center or Quiver Bioscience.
Code availability
The code81 used to perform the machine learning-based phenotype analysis is available: 10.5281/zenodo.13936561. Link to Zenodo: https://zenodo.org/records/13936561. Link to GitHub: https://github.com/q-state-biosciences/fragile-x-manuscript-2024.
Competing interests
J.J.F., K.H., S.J.R., J.J., J.G., L.A.W., and G.T.D. are current employees of Quiver Bioscience and may hold stock options in Quiver Bioscience. D.G. was an employee of Q-State Bioscience, is currently a consultant for Quiver Bioscience and has received stock options in Quiver Bioscience. E.N., C.F., M.B., B.F. are current employees of Vertex Pharmaceuticals Inc. and hold stock in Vertex Pharmaceuticals Inc. N.D.B. is a former employee of Q-State Biosciences and may be an inventor on relevant patents, but holds no financial stake, stock, or options. M.F.B. is a co-founder of Allos Pharma, and a consultant and owner of stock options for Addex Therapeutics and Luminopia. H.U. and H.S. are current employees of Wave Life Sciences and report stock ownership in Wave Life Sciences. R.P.D. was employed at Vertex Pharmaceuticals at the time of the study, is a current employee of Intellia Therapeutics, and reports stock ownership in Intellia Therapeutics. Elizabeth Berry-Kravis has received funding from Acadia, Alcobra, AMO, Asuragen, Avexis, Biogen, BioMarin, Cydan, Engrail, Erydel, Fulcrum, GeneTx, GW, Healx, Ionis, Jaguar, Kisbee, Lumos, Marinus, Moment Biosciences, Neuren, Neurogene, Neurotrope, Novartis, Orphazyme/Kempharm, Ovid, PTC Therapeutics, Retrophin, Roche, Seaside Therapeutics, Taysha, Tetra, Ultragenyx, Yamo, Zynerba, and Vtesse/Sucampo/Mallinckrodt Pharmaceuticals, to consult on trial design or run clinical or lab validation trials in genetic neurodevelopmental or neurodegenerative disorders, all of which is directed to RUMC in support of rare disease programs; B.-K. receives no personal funds and RUMC has no relevant financial interest in any of the commercial entities listed.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: James J. Fink, Nathaniel Delaney-Busch, Ryan Dawes, Evanthia Nanou.
These authors jointly supervised this work: Brinley Furey, Graham T. Dempsey.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07120-6.
References
- 1.Lozano, R., Rosero, C. A. & Hagerman, R. J. Fragile X spectrum disorders. Intractable & Rare Diseases Research, 10.5582/irdr.2014.01022 (2014). [DOI] [PMC free article] [PubMed]
- 2.Penagarikano, O., Mulle, J. G. & Warren, S. T. The Pathophysiology of Fragile X Syndrome. Annual Review of Genomics and Human Genetics, 10.1146/annurev.genom.8.080706.092249 (2007). [DOI] [PubMed]
- 3.Hagerman, R. J. et al. Fragile X syndrome. Nat. Rev. Dis. Prim.3, 17065 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Verkerk, A. J. et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell65, 905–914 (1991). [DOI] [PubMed] [Google Scholar]
- 5.Darnell, J. C. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell146, 247–261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhakar, A. L., Dölen, G. & Bear, M. F. The pathophysiology of fragile X (and what it teaches us about synapses). Annu Rev. Neurosci.35, 417–443 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korb, E. et al. Excess Translation of Epigenetic Regulators Contributes to Fragile X Syndrome and Is Alleviated by Brd4 Inhibition. Cell170, 1209–1223.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richter, J. D., Bassell, G. J. & Klann, E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat. Rev. Neurosci.16, 595–605 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hale, C. R. et al. FMRP regulates mRNAs encoding distinct functions in the cell body and dendrites of CA1 pyramidal neurons. Elife10, e71892 (2021). [DOI] [PMC free article] [PubMed]
- 10.Greenough, W. T. et al. Synaptic regulation of protein synthesis and the fragile X protein. Proc. Natl Acad. Sci. USA98, 7101–7106 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zalfa, F. et al. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell112, 317–327 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Bear, M. F., Huber, K. M. & Warren, S. T. The mGluR theory of fragile X mental retardation. Trends Neurosci.10.1016/j.tins.2004.04.009 (2004). [DOI] [PubMed]
- 13.Dölen, G. et al. Correction of fragile X syndrome in mice. Neuron56, 955–962 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osterweil, E. K., Krueger, D. D., Reinhold, K. & Bear, M. F. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J. Neurosci.30, 15616–15627 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin, M., Kang, J., Burlin, T. V., Jiang, C. & Smith, C. B. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J. Neurosci.25, 5087–5095 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell78, 23–33 (1994). [PubMed]
- 17.Telias, M. Molecular Mechanisms of Synaptic Dysregulation in Fragile X Syndrome and Autism Spectrum Disorders. Front Mol. Neurosci.12, 51 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Contractor, A., Klyachko, V. A. & Portera-Cailliau, C. Altered Neuronal and Circuit Excitability in Fragile X Syndrome. Neuron10.1016/j.neuron.2015.06.017 (2015). [DOI] [PMC free article] [PubMed]
- 19.Huber, K. M., Gallagher, S. M., Warren, S. T. & Bear, M. F. Altered synaptic plasticity in a mouse model of fragile X mental retardation. PNAS99, 7746–7750 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bureau, I., Shepherd, G. M. G. & Svoboda, K. Circuit and plasticity defects in the developing somatosensory cortex of FMR1 knock-out mice. J. Neurosci.28, 5178–5188 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, J., Pelletier, M. R., Perez Velazquez, J.-L. & Carlen, P. L. Reduced cortical synaptic plasticity and GluR1 expression associated with fragile X mental retardation protein deficiency. Mol. Cell Neurosci.19, 138–151 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Paluszkiewicz, S. M., Olmos-Serrano, J. L., Corbin, J. G. & Huntsman, M. M. Impaired inhibitory control of cortical synchronization in fragile X syndrome. J. Neurophysiol.106, 2264–2272 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desai, N. S., Casimiro, T. M., Gruber, S. M. & Vanderklish, P. W. Early postnatal plasticity in neocortex of Fmr1 knockout mice. J. Neurophysiol.96, 1734–1745 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Harlow, E. G. et al. Critical period plasticity is disrupted in the barrel cortex of FMR1 knockout mice. Neuron65, 385–398 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domanski, A. P. F., Booker, S. A., Wyllie, D. J. A., Isaac, J. T. R. & Kind, P. C. Cellular and synaptic phenotypes lead to disrupted information processing in Fmr1-KO mouse layer 4 barrel cortex. Nat. Commun.10, 4814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bostrom, C. A. et al. Rescue of NMDAR-dependent synaptic plasticity in Fmr1 knock-out mice. Cereb. Cortex25, 271–279 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Cea-Del Rio, C. A. et al. Disrupted inhibitory plasticity and homeostasis in Fragile X syndrome. Neurobiol. Dis.142, 104959 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irwin, S. A., Galvez, R. & Greenough, W. T. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb. Cortex10, 1038–1044 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Irwin, S. A. et al. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am. J. Med. Genet.98, 161–167 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Nimchinsky, E. A., Oberlander, A. M. & Svoboda, K. Abnormal development of dendritic spines in FMR1 knock-out mice. J. Neurosci.21, 5139–5146 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He, C. X. & Portera-Cailliau, C. The trouble with spines in fragile X syndrome: density, maturity and plasticity. Neuroscience251, 120–128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson, J. R., Bartley, A. F., Hays, S. A. & Huber, K. M. Imbalance of neocortical excitation and inhibition and altered UP states reflect network hyperexcitability in the mouse model of fragile X syndrome. J. Neurophysiol.100, 2615–2626 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bülow, P., Segal, M. & Bassell, G. J. Mechanisms Driving the Emergence of Neuronal Hyperexcitability in Fragile X Syndrome. Int. J. Mol. Sci.23, 6315 (2022). [DOI] [PMC free article] [PubMed]
- 34.Liu, X., Kumar, V., Tsai, N.-P. & Auerbach, B. D. Hyperexcitability and Homeostasis in Fragile X Syndrome. Front. Mol. Neurosci.14, 805929 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berry-Kravis, E. Epilepsy in fragile X syndrome. Dev. Med. Child Neurol.44, 724–728 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Berry-Kravis, E. et al. Seizures in fragile X syndrome: characteristics and comorbid diagnoses. Am. J. Intellect. Dev. Disabil.115, 461–472 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Musumeci, S. A. et al. Audiogenic seizures susceptibility in transgenic mice with fragile X syndrome. Epilepsia41, 19–23 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Chen, L. & Toth, M. Fragile X mice develop sensory hyperreactivity to auditory stimuli. Neuroscience103, 1043–1050 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Wang, J. et al. A resting EEG study of neocortical hyperexcitability and altered functional connectivity in fragile X syndrome. J. Neurodev. Disord.9, 11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ethridge, L. E. et al. Auditory EEG Biomarkers in Fragile X Syndrome: Clinical Relevance. Front Integr. Neurosci.13, 60 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Razak, K. A., Binder, D. K. & Ethell, I. M. Neural Correlates of Auditory Hypersensitivity in Fragile X Syndrome. Front. Psychiatry12, 720752 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng, P.-Y. & Klyachko, V. A. Channelopathies in fragile X syndrome. Nat. Rev. Neurosci.22, 275–289 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vadodaria, K. C., Jones, J. R., Linker, S. & Gage, F. H. Modeling Brain Disorders Using Induced Pluripotent Stem Cells. Cold Spring Harb. Perspect. Biol.12, a035659 (2020). [DOI] [PMC free article] [PubMed]
- 44.Barral, S. & Kurian, M. A. Utility of Induced Pluripotent Stem Cells for the Study and Treatment of Genetic Diseases: Focus on Childhood Neurological Disorders. Front Mol. Neurosci.9, 78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abu Diab, M. & Eiges, R. The Contribution of Pluripotent Stem Cell (PSC)-Based Models to the Study of Fragile X Syndrome (FXS). Brain Sci.9, 42 (2019). [DOI] [PMC free article] [PubMed]
- 46.Brykczynska, U. et al. CGG Repeat-Induced FMR1 Silencing Depends on the Expansion Size in Human iPSCs and Neurons Carrying Unmethylated Full Mutations. Stem Cell Rep.7, 1059–1071 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou, Y., Kumari, D., Sciascia, N. & Usdin, K. CGG-repeat dynamics and FMR1 gene silencing in fragile X syndrome stem cells and stem cell-derived neurons. Mol. Autism7, 42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vershkov, D. et al. FMR1 Reactivating Treatments in Fragile X iPSC-Derived Neural Progenitors In Vitro and In Vivo. Cell Rep.26, 2531–2539.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Sheridan, S. D. et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS ONE10.1371/journal.pone.0026203 (2011). [DOI] [PMC free article] [PubMed]
- 50.Doers, M. E. et al. iPSC-Derived Forebrain Neurons from FXS Individuals Show Defects in Initial Neurite Outgrowth. Stem Cells Dev.23, 1777–1787 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boland, M. J. et al. Molecular analyses of neurogenic defects in a human pluripotent stem cell model of fragile X syndrome. Brain140, 582–598 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sunamura, N., Iwashita, S., Enomoto, K., Kadoshima, T. & Isono, F. Loss of the fragile X mental retardation protein causes aberrant differentiation in human neural progenitor cells. Sci. Rep.8, 11585 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gildin, L. et al. Impaired Functional Connectivity Underlies Fragile X Syndrome. Int. J. Mol. Sci.23, 2048 (2022). [DOI] [PMC free article] [PubMed]
- 54.Das Sharma, S. et al. Cortical neurons derived from human pluripotent stem cells lacking FMRP display altered spontaneous firing patterns. Mol. Autism11, 52 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Graef, J. D. et al. Partial FMRP expression is sufficient to normalize neuronal hyperactivity in Fragile X neurons. Eur. J. Neurosci.51, 2143–2157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, Z. et al. The fragile X mutation impairs homeostatic plasticity in human neurons by blocking synaptic retinoic acid signaling. Sci. Transl. Med.10, eaar4338 (2018). [DOI] [PMC free article] [PubMed]
- 57.Susco, S. G. et al. FMR1 loss in a human stem cell model reveals early changes to intrinsic membrane excitability. Dev. Biol.468, 93–100 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Telias, M., Kuznitsov-Yanovsky, L., Segal, M. & Ben-Yosef, D. Functional Deficiencies in Fragile X Neurons Derived from Human Embryonic Stem Cells. J. Neurosci.35, 15295–15306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu, X. S. et al. Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell172, 979–992.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams, L. A. et al. Scalable Measurements of Intrinsic Excitability in Human iPS Cell-Derived Excitatory Neurons Using All-Optical Electrophysiology. Neurochem. Res.44, 714–725 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Werley, C. A. et al. All-optical electrophysiology for disease modeling and pharmacological characterization of neurons. Curr. Protoc. Pharmacol.2017, 11.20.1–11.20.24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu, P. W. et al. A phenotypic screening platform for chronic pain therapeutics using all-optical electrophysiology. Pain.165, 922-940 (2024). [DOI] [PMC free article] [PubMed]
- 63.Williams, L. A. et al. Discovery of novel compounds and target mechanisms using a high throughput, multiparametric phenotypic screen in a human neuronal model of Tuberous Sclerosis. preprint at bioRxiv.10.1101/2024.02.22.581652 (2024).
- 64.Zhang, Y. et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron78, 785–798 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nehme, R. et al. Combining NGN2 Programming with Developmental Patterning Generates Human Excitatory Neurons with NMDAR-Mediated Synaptic Transmission. Cell Rep.23, 2509–2523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferreira, J. P., Overton, K. W. & Wang, C. L. Tuning gene expression with synthetic upstream open reading frames. Proc. Natl Acad. Sci. USA110, 11284–11289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajaratnam, A. et al. Fragile X syndrome and fragile X-associated disorders. F1000Res.6, 2112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jacobs, S., Cheng, C. & Doering, L. C. Hippocampal neuronal subtypes develop abnormal dendritic arbors in the presence of Fragile X astrocytes. Neuroscience324, 202–217 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Jacobs, S. & Doering, L. C. Astrocytes Prevent Abnormal Neuronal Development in the Fragile X Mouse. J. Neurosci.10.1523/JNEUROSCI.5027-09.2010 (2010). [DOI] [PMC free article] [PubMed]
- 70.Verdura, E. et al. Heterogeneity in Fragile X Syndrome Highlights the Need for Precision Medicine-Based Treatments. Front. Psychiatry12, 722378 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cea-Del Rio, C. A. & Huntsman, M. M. The contribution of inhibitory interneurons to circuit dysfunction in Fragile X Syndrome. Front. Cell Neurosci.8, 245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng, C., Sourial, M. & Doering, L. C. Astrocytes and developmental plasticity in fragile X. Neural Plast.2012, 197491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Werley, C. A., Chien, M.-P. & Cohen, A. E. An ultrawidefield microscope for high-speed fluorescence imaging and targeted optogenetic stimulation. Biomed. Opt. Express8, 5794 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mukamel, E. A., Nimmerjahn, A. & Schnitzer, M. J. Automated Analysis of Cellular Signals from Large-Scale Calcium Imaging Data. Neuron63, 747–760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hochbaum, D. R. et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods11, 825–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pedregosa, F. et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res.12, 2825–2830 (2011). [Google Scholar]
- 77.Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw.67, 1–48 (2015). [Google Scholar]
- 78.Lenth, R. V. Emmeans: Estimated Marginal Means, Aka Least-Squares Meanshttps://CRAN.Rproject.org/package=emmeans (2021).
- 79.Kuznetsova, A., Brockhoff, P. B. & Christensen, R. H. B. lmerTest Package: Tests in Linear Mixed Effects Models. J. Stat. Softw.82, 1–26 (2017). [Google Scholar]
- 80.Lüdecke, D. sjPlot: Data Visualization for Statistics in Social Sciencehttps://CRAN.Rproject.org/package=sjPlot (2022).
- 81.Delaney-Busch, N. & Ryan, S. J. Fragile X Patient Phenotyping Code10.5281/zenodo.13936561 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data that support the findings of this study are available as Supplementary Data in the form of excel worksheets.
Patient-derived stem cell lines were generated as described (see Methods). Patient-derived iPS lines generated as part of this study can be requested to RUSH University Medical Center or Quiver Bioscience.
The code81 used to perform the machine learning-based phenotype analysis is available: 10.5281/zenodo.13936561. Link to Zenodo: https://zenodo.org/records/13936561. Link to GitHub: https://github.com/q-state-biosciences/fragile-x-manuscript-2024.