Abstract
Plant evolution is driven by key innovations of functional traits that enables their survivals in diverse ecological environments. However, plant adaptive evolution from land to atmospheric niches remains poorly understood. In this study, we use the epiphytic Tillandsioideae subfamily of Bromeliaceae as model plants to explore their origin, evolution and diversification. We provide a comprehensive phylogenetic tree based on nuclear transcriptomic sequences, indicating that core tillandsioids originated approximately 11.3 million years ago in the Andes. The geological uplift of the Andes drives the divergence of tillandsioids into tank-forming and atmospheric types. Our genomic and transcriptomic analyses reveal gene variations and losses associated with adaptive traits such as impounding tanks and absorptive trichomes. Furthermore, we uncover specific nitrogen-fixing bacterial communities in the phyllosphere of tillandsioids as potential source of nitrogen acquisition. Collectively, our study provides integrative multi-omics insights into the adaptive evolution of tillandsioids in response to elevated aerial habitats.
Subject terms: Genome, Plant evolution, Evolutionary genetics, Evolutionary ecology
The mechanisms by which tillandsioids adapt to elevated aerial habitats remain largely unexplored. Here, the authors report their evolution and link life history, diversification, comparative genomic, and functional changes to processes underlying the unique biology of air plants.
Introduction
Land plants evolved more than 500 million years ago (Mya) from green algae1,2. During the evolutionary process, terrestrial plants became less dependent on water resources compared to their aquatic ancestors. One of the major evolutionary innovations that enabled this transition was the acquisition of water-absorbing and -transporting tissues2–4. The emergence of root and vascular system solved the biggest challenge in successful land colonization of plants2–4. The spread of land plants on earth has accelerated the competition of living space and promoted speciation as they occupied new ecological niches.
Epiphytes are plants that grow on rocks or the surface of other plants without taking their nourishments5. Just like the challenge for the conquest of land of aquatic plants, undoubtedly, epiphytes were confronted with the acquisition of water and nutrition to adapt to the harsher atmospheric space. Evolutionary innovations in adaptive traits burst in many epiphytic lineages allowing plants to survive in the aerial niches. Epiphytic non-vascular plants are poikilohydrous, capable of drying out temporarily, thus avoiding drought stress rather than resisting it, until liquid water is available again6. Epiphytic vascular plants conquer the atmospheric space through the innovation of special tissues, such as the ‘nest’ leaves of epiphytic ferns7, the aerial roots of epiphytic orchids8 and the absorptive trichomes of epiphytic bromeliads9. Epiphytes have evolved among all major lineages of land plants and vascular epiphytes accounting for 9% of all vascular plants, involving 912 genera in 73 families5. Unexpectedly, epiphytes have evolved to be dominant in a few large vascular plant families after recent massive expansions10–12.
Tillandsioideae, the largest subfamily of Bromeliaceae, encompasses about two-thirds of all epiphytic species5,10,13,14. It exhibits a remarkable diversity of vegetative forms and habitats, spanning from arid deserts to perpetually moist rainforests15,16. This diversity arises from specialized traits such as impounding tanks, absorptive trichomes, and reduced root systems, which have facilitated their evolutionary success. Tillandsioids are typically classified into two types based on their life forms: tank-forming species, characterized by cup-shaped leaves that collect rainwater and relatively sparse absorptive trichomes on leaves (Supplementary Fig. 1a-h); and atmospheric species, which lack tanks but possess dense absorptive trichomes for extracting water from air moisture or fog, often with poorly developed or absent roots (Supplementary Fig. 1i-p). The atmospheric ones are commonly known as air plants due to their extreme adaptation, allowing them to thrive even on surfaces like telephone wires, giving up any contact with water or nutrient-supplying substrates. Givnish et al. 11 showed that tank formation evolved first in Tillandsioideae with atmospheric species later evolving in Tillandsia. The distinctive lineages of bromeliads exhibit several key innovations, such as the tank habit, absorptive trichomes, CAM photosynthesis, and avian pollination, which have enabled them to colonize various adaptive zones11. This may make bromeliads an excellent model for studying plant evolution.
Evolutionarily, the imposed environmental stress factors derived the rapid evolution of tillandsioids, which in turn, influenced our terrestrial ecosystem in water and mineral recycling17,18. However, several questions remain to be addressed: What are key environmental factors for driving epiphyte evolution? When and how epiphytes migrated from land to atmospheric niches? What is the molecular genetic basis for their adaptive evolution? And importantly, how they forage the nutrients in their aerial habitat?
In this work, we characterize the tempo of species evolution and divergence of epiphytes in Tillandsioideae by constructing a comprehensive and well resolved phylogenetic tree using transcriptomes of 143 species (representing 78% genera-richness of this subfamily). We also assemble the genomes of two typical types of tillandsioids and employ various omics approaches, including genomics, spatial transcriptomics, full-length transcriptomics, single-cell transcriptomics, and microbiomics, to uncover the genomic basis of their featured adaptive traits and potential nutrient sources. This study sheds lights on the implications of evolutionary trade-offs and adaptive evolution in ecological niches occupation.
Results
Core tillandsioids originated from Andes and diverged rapidly from mid-Pliocene period
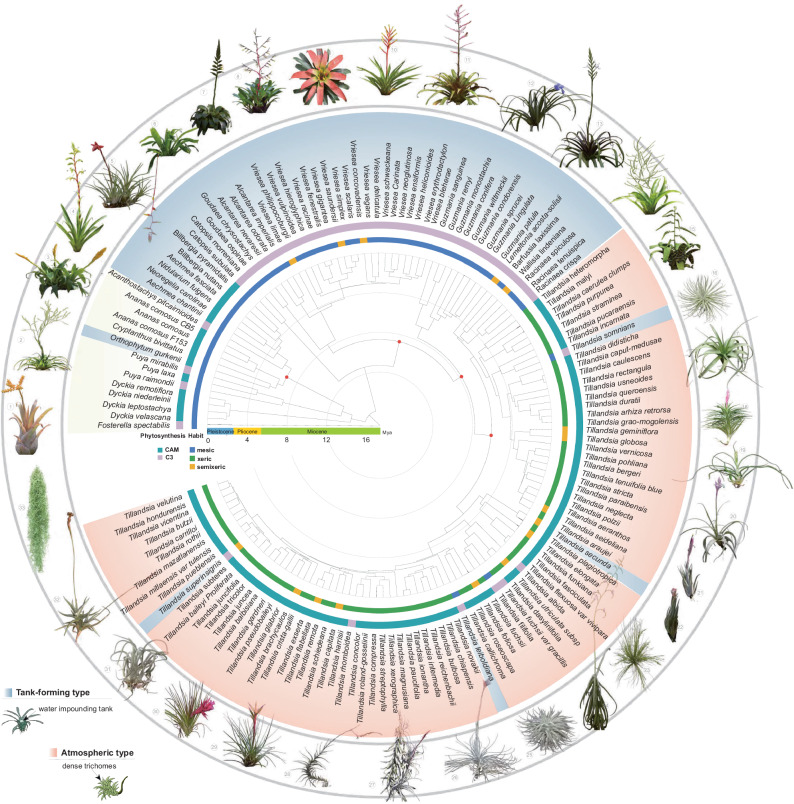
While previous phylogenetic trees of Tillandsioideae relied on plastid markers13,14, nuclear genes can help to resolve ambiguous relationships19–22. To achieve a comprehensive phylogeny reconstruction, we sequenced the transcriptomes of 143 species and obtained three genome sequences from public resources, totaling 146 bromeliad datasets (Fig. 1 and Supplementary Table 1). Nearly 78% of the genera richness of Tillandsioideae was covered by the samplings, which included 13 out-group species from Pitcairnioideae and Bromelioideae. Subsequently, largely based on nuclear transcriptomic sequences, five different sets of low-copy nuclear genes were used to construct phylogenetic trees, all of which exhibited similar tree topologies (Fig. 1 and Supplementary Fig. 2). In order to determine the evolutionary timeframe of Tillandsioideae lineages, we utilized the tree generated from the 91-gene sets with age calibrations derived from five fossils to estimate divergence times. Our chronogram reveals that Tillandsioideae is a relatively young and monophyletic subfamily, originating approximately 11.3 Mya (Fig. 1 and Supplementary Fig. 3). Tillandsiodeae is often seen as consisting of the core tillandsioids14 and non-core tillandsioids, with the latter consisting of Catopsis and Glomeropitcairnia11,13. Our phylogenetic analysis did not include Glomeropitcairnia, but it positions Catopsis as a sister group to the core Tillandsioideae (Fig. 1 and Supplementary Fig. 3). Shortly after the origin of core Tillandsioideae ( ~ 8.7 Mya), the first clade of Clade I emerged ( ~ 7.6 Mya), along with the divergence of Clade II ( ~ 7.7 Mya) from the rest of the Tillandsioideae species. Both Clade I and Clade II consist of C3 plants with well-developed water-impounding tanks, while the later-evolved atmospheric Clade IV ( ~ 5.6 Mya) and Clade V ( ~ 5.1 Mya) are CAM plants lacking tanks. Given that Tillandsioideae is a monophyletic group, it is likely that the ancestor of Tillandsioideae belonged to tank-forming plants using the C3 photosynthetic pathway. These ancestral traits have been retained in Clades I to III. Conversely, the absence of tanks and the adoption of the CAM photosynthetic pathway originated from more recent atmospheric ancestors in Clades IV, becoming the prevailing life form in these younger clades (Fig. 1 and Supplementary Fig. 3). These findings indicate that within a relatively short period of 2 million years, the tillandsioids underwent a significant divergence, resulting in two distinct life forms. This rapid divergence suggests that there were likely dramatic climatic changes or geological events in their habitats between 7.6 Mya and 5.6 Mya, which served as catalysts for the diversification of the tillandsioids. In addition, we observed four tank-forming plants within the atmospheric clades (Fig. 1). This may be due to either a single gain of the atmospheric habit at the base of Tillandsia and five subsequent losses within the genus, or two independent gains within Tillandsia and four losses.
Fig. 1. Phylogenetic relationships, geological time and ecological habitats of Tillandsioideae.
The ML tree based on sequence data from five gene sets is presented. Red dots on nodes indicate the presence of fossil evidence supporting the estimated time. The chronogram was estimated using the ML tree derived from the 91 gene sets tree using MEGA 11. Habitats, photosynthetic types, and species names are displayed around the tree. Mesic refers to plants with ligulate leaves featuring few, inconspicuous, appressed scales or trichomes, which have a tank without water-storing tissue; Semixeric refers to plants with poorly developed tanks or no tanks, narrow leaf blades with inconspicuous, appressed scales, and little or no water-storing tissue; Xeric refers to plants with conspicuous, spreading trichomes, narrow leaf blades, water-storing tissue, and no tank. Tank-forming plants are highlighted with a light blue background, atmospheric plants with a light pink background, and terrestrial plants with a yellow background. Images of two life forms from different tillandsioid genera are featured outside the tree. Numbers 1-15 represent tank-forming tillandsioids, while numbers 15-33 depict atmospheric tillandsioids. Detailed taxonomic names can be found in Supplementary Table 1.
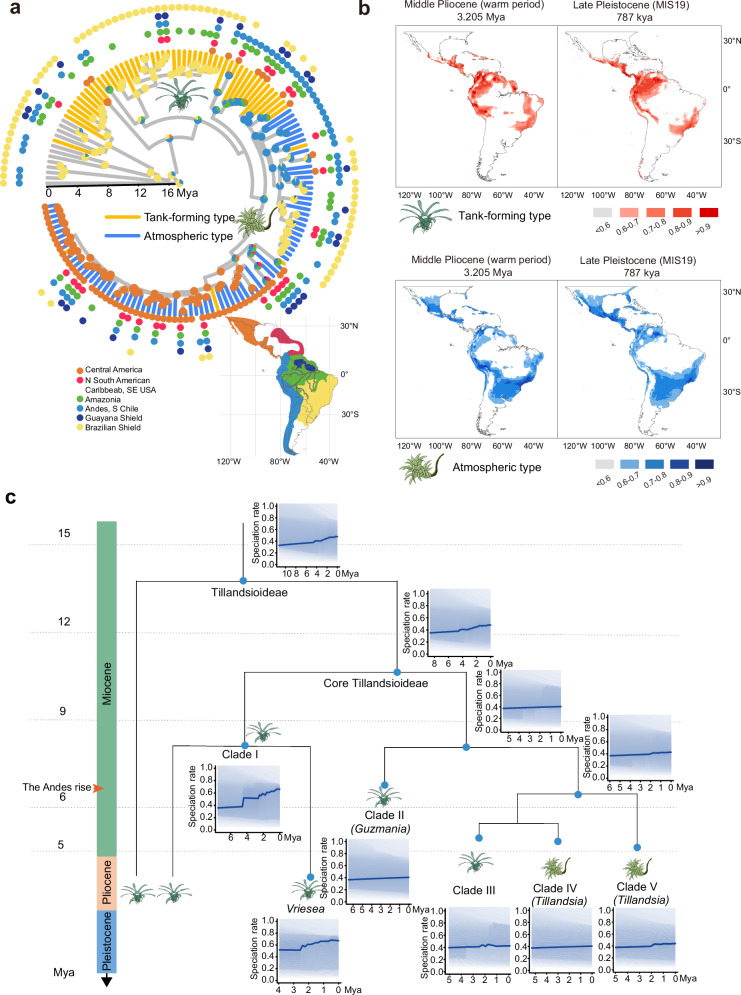
All Tillandsioideae species are indigenous to the tropics and subtropics of the New World, where the climate is significantly influenced by the Andes range. The two life forms of tillandsioids are found in habitats with differing water availability. In order to investigate the geographic origins and factors contributing to the rapid divergence of Tillandsioideae, we reconstructed the ancestral area of tillandsioid occurrences. Based on the chronogram and biogeographic reconstruction using Maximum Likelihood, we inferred that the ancestor of core tillandsioids most likely originated in the Andes around 11.3 Mya (Fig. 2a). This result aligns with the studies by Givnish et al. on the historical biogeography of Bromeliaceae14. The first emerged tank-forming tillandsioids ( ~ 7.6 Mya) spread and diversified primarily in the Brazilian shield, Amazonia, and the Andes (Fig. 2a). The significant shift in life form (atmospheric tillandsioids arose) occurred in the Andes around 5.6 Mya (Fig. 2a), coinciding with a period of rapid uplift of the Andes23.
Fig. 2. The geographic evolution, divergence dynamics and peripheral migration maps of Tillandsioideae.
a Temporal calibration of the geographic evolution of Tillandsioideae. The current distribution of individual species is represented by colored dots. Pie charts at nodes illustrate the ancestral distributions inferred through Bayesian inference (BI), with wedge width indicating the likelihood of alternative inferences. b Ecological niche modeling showcasing suitable habitats for two types of tillandsioids. The color scale reflects the suitability score. c Analysis of diversification dynamics within Tillandsioideae. Rates-through-time plots for all species within Tillandsioideae and distinct main clades are presented separately.
Since the emergence of atmospheric tillandsioids around 5.6 Mya, multiple dispersal and extinction peaks have been observed, often accompanied by geographical isolations (Supplementary Fig. 4a). The most significant dispersal peak occurred during the middle Pleistocene, specifically between 2.0 and 1.0 Mya (Supplementary Fig. 4a). This period was characterized by repeated glacial and interglacial cycles, which brought about dramatic temperature changes. These fluctuations likely acted as another important factor contributing to the rapid extinction and spread of species. In order to investigate the primary environmental factors influencing the distribution of two types of tillandsioids, we conducted jackknife analysis on the regularized training gain, test gain, and AUC gain in MaxEnt models. The findings from all three models were consistent. The distribution of tank-forming tillandsioids is predominantly influenced by the dramatic change (increase or decrease) of temperature seasonality (Bio_4), whereas the distribution of atmospheric tillandsioids is primarily shaped by the mean temperature sharp change (increase or decrease) of the coldest quarter (Bio_11) variable (Supplementary Fig. 4). Ecological niche modeling reveals that regions with suitable environmental conditions for both tank-forming and atmospheric tillandsioids (suitability > 0.6) have remained interconnected from the mid-Pliocene to the present day. However, compared to the mid-Pliocene era, atmospheric tillandsioids have adapted to a broader range and displayed a noticeable trend of peripheral diffusion (Fig. 2b). This suggests that atmospheric tillandsioids evolved more rapidly and demonstrated a stronger ability to thrive in diverse environments.
To gain further insight into the dynamics of species diversification in Tillandsioideae, we conducted diversification rate and shift estimations using our time-calibrated phylogeny (Fig. 2c). Within the entire Tillandsioideae or core Tillandsioideae subfamily, we identified two notable shifts in diversification rate acceleration, occurring at approximately 4.3 Mya and 1.8 Mya, respectively. These shifts coincided with transitions in climate periods (Fig. 2c). Specifically, the diversification rate of tank-forming types exhibited a sharp increase around 4.3 Mya, approximately one million years after the onset of the Pliocene period. The Pliocene was characterized by a warmer and wetter climate, following a gradual cooling trend from the Miocene in South America. On the other hand, the speciation rate of atmospheric types experienced acceleration at 1.8 Mya, soon after the onset of the Pleistocene epoch, which was marked by repeated glacial and interglacial cycles, leading to significant climate fluctuations (Fig. 2c). These findings align with the primary environmental factors that influence their distribution (Supplementary Fig. 4b-c). Additionally, the ongoing uplift of the Andes throughout this period contributed to changes in atmospheric circulation patterns, further influencing climate change in South America, the habitat of tillandsioids, and consequently accelerated species formation.
LTRs expansions shape the genomes and drive adaptive evolution
The lack of genomic information in Tillandsioids has constrained both genome evolution and functional genomic studies in this group. To further explore the adaptive evolution of tillandsioids, we assembled the genome sequences of two representative types of tillandsioids, namely the tank-forming species Vriesea erythrodactylon and the atmospheric species Tillandsia duratii, utilizing PacBio RSII single-molecule real-time (SMRT) sequencing and Hi-C sequencing technology (Supplementary Table 2).
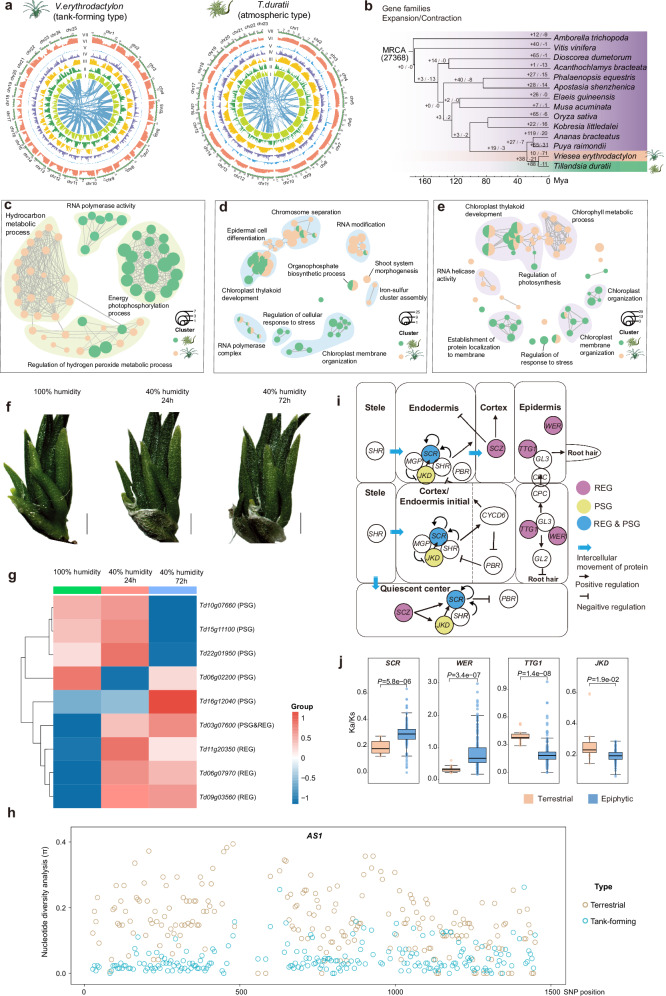
The total length of V. erythrodactylon genome assembly was 417.89 Mb with a contig N50 of 1.32 Mb (Supplementary Table 2). According to the BUSCO results, genome sequence completeness was estimated to be 98.41% anchoring onto 25 pseudochromosomes (Fig. 3a and Supplementary Fig. 5a), and a total of 20, 415 protein-coding genes were predicted and repetitive sequences accounts for 62.56% of the V. erythrodactylon genome (Supplementary Table 3). The genome size of V. erythrodactylon is comparable to the reported terrestrial bromeliads pineapple (Ananas comosus, 513 Mb) and Puya (Puya raimondii, 314 Mb), and the number of chromosomes is also equivalent to pineapple24,25. While, we assembled the atmospheric T. duratii genome sequence and obtained 1,030.11 Mb in size with a contig N50 of 1.92 Mb, which is about twice the size of these genomes, and was anchored onto 23 pseudochromosomes (Fig. 3a, Supplementary Fig. 5a and Supplementary Table 2). The chromosomes 11 and 16 were formed by the head-to-head fusion of two ancestral chromosomes each (Supplementary Fig. 5b). The genome of T. duratii is approximately twice the size of the other genomes, suggesting a potential whole-genome duplication (WGD) event following divergence. However, aside from the two ancient WGD events shared by all bromeliads, we did not identify any additional WGD event in the T. duratii genome (Supplementary Fig. 5c). The expansion in genome size is often driven by an increased abundance of repetitive sequences26. In our analysis, we annotated 26,185 protein-coding genes in T. duratii. The number of genes in T. duratii is similar to that of the Bromelioideae plants Ananas comosus CB5 (27,024 genes)24 and Puyoideae Puya raimondii (26,696 genes)27, but higher than the Tillandsioideae plant V. erythrodactylon (20,415 genes), and lower than Tillandsioideae plants T. fasciculata (34,886 genes) and T. leiboldiana (38,180 genes)28. In the genome of T. duratii, 76.93% consists of repetitive sequences (Supplementary Table 3). Among these, transposable elements (TEs) were the most common, with long-terminal repeat retrotransposons (LTRs) being the most abundant (Supplementary Fig. 6a). This dominance of LTR-RTs was also observed in the V. erythrodactylon (Supplementary Fig. 6a) and pineapple genomes24. Specifically, Gypsy and Copia were the predominant LTR types (Supplementary Fig. 6b), and while the average lengths of LTRs in these genomes were similar, their numbers and total lengths varied significantly. The T. duratii genome had a total length of LTRs reaching 318.23 Mb, which is much greater than the 82.18 Mb in V. erythrodactylon and 184.68 Mb in the pineapple genome (Supplementary Fig. 6c). These variations in LTR lengths correspond to the differences in genome size, indicating that the larger genome size of T. duratii primarily stems from the expansion of LTRs.
Fig. 3. High-quality reference genomes and comparative analysis of two types of tillandsioids.
a Genomic landscapes of the genomes of atmospheric tillandsioid T. duratii and tank-forming tillandisioid V. erythrodactylon. The rings indicate the pseudo-chromosomes (I), TE density (II), gene density (III), Gypsy retrotransposons density (IV), Copia retrotransposons density (V), Long interspersed nuclear elements density (VI), GC contents (VII), and inner lines indicate syntenic blocks. b Phylogenetic tree depicting 14 plant species and the evolution of gene families, with numbers next to each node indicating the counts of expanded and contracted gene families. c–e Gene Ontology terms associated with expanded gene families c, positively selected genes (PSGs) d, and rapidly evolving genes (REGs) e within the genomes of V. erythrodactylon and T. duratii. f Visual representation of T. duratii plants under different humidity conditions: >96% air humidity as control, and under 40-45% air humidity for 24 hours and 72 hours as drought stress treatment. g Expression patterns of PSGs and REGs enriched in the “response to stress” Gene Ontology terms under drought stress in T. duratii. Scale bars, 1 cm. h Nucleotide diversity analysis of AS1 coding sequences in 13 terrestrial and 49 tank-forming bromeliads. i Alterations in the root development pathway of tillandsioids. Genes highlighted in purple are rapidly evolving genes (REGs), those in yellow are under positive selection (PSGs), and genes in blue signify REGs that are also under positive selection. j Evaluation of evolutionary changes in five root development-related genes. Ka/Ks ratios illustrate the selective pressure on these genes in epiphytic (n = 137) and terrestrial (n = 17) bromeliads. Data are presented as mean values +/- SD. The box plots show the mean (black square), median (center line), quartiles (box limits) and 1.5× interquartile range (whiskers). Two-tailed Wilcoxon test was performed for P values. h Genetic variations across the AS1 coding region in 18 terrestrial and 50 tank-forming epiphytic bromeliads. Source data are provided as a Source Data file.
Furthermore, we observed that the proportion of solo LTRs in V. erythrodactylon is higher than that in T. duratii (Supplementary Fig. 6d). Solo-LTRs are generated when non-homologous recombination occurs between two LTR sequences within LTRs or between LTRs, representing a significant mechanism for genome clearance of LTRs29. The lower proportion of solo-LTRs in T. duratii suggests a slower clearance of LTRs, potentially contributing to the higher number of LTRs in T. duratii genomes compared to others. The integration of LTR retrotransposons serves as a major driver of genetic diversity26. Our analysis unveiled a recent upsurge in LTR insertions in both tillandsioid species, commencing approximately 6.0 Mya (Supplementary Fig. 6e), coinciding with the onset of the significant uplift of the Andes23. This discovery underscores the profound impact of the Andean uplift on the evolutionary trajectory of tillandsioids. Moreover, between 5.5 Mya and 2.2 Mya, the density of LTR insertions was higher in the tank-forming tillandsioid V. erythrodactylon than in the atmospheric tillandsioid T. duratii; however, post 2.2 Mya, the density of LTR insertions increased in T. duratii. These temporal trends align with the accelerated pace of species divergence in tank-forming and atmospheric tillandsioids (Fig. 2c), indicating that the expansion of LTR retrotransposons played a pivotal role in driving species diversification in tillandsioids. Furthermore, we observed that T. duratii had a greater number of genes affected by LTRs (4970 genes) compared to V. erythrodactylon (1255 genes). These LTRs were located in the gene coding region or within 3 kb upstream or downstream of genes. Gene functional enrichment analysis revealed that these genes were associated with various plant development processes (Supplementary Data 1-2) and exhibited significant responses to both cold stress and ultraviolet (UV) in T. duratii (Supplementary Data 1). Collectively, LTRs play a crucial role in the evolutionary adaptation of tillandsioid genomes.
Genomic selection signatures associated with adaptive evolution
As tillandsioids further adapted to aerial environments, they faced new selection pressures necessitating adjustments to their environmental adaptation. In order to delve deeper into the adaptive evolution of tillandsioids, we conducted a comparative genomic analysis involving 14 plant species (Fig. 3b). The functional enrichments of expanded gene families in T. duratii and V. erythrodactylon exhibit distinct patterns (Fig. 3c). The expanded gene families in the atmospheric tillandsioid T. duratii are primarily associated with energy metabolism (Fig. 3c), like the oxidoreductase activity acting on NAD(P)H. Many of these genes are likely involved as components of NAD(P)H dehydrogenase, facilitating cyclic electron flow and maintaining the effective operation of primary antioxidative systems30. The NAD(P)H dehydrogenase component also represents a pivotal factor in the adaptive evolution of early plants transitioning from aquatic to terrestrial habitats. Environmental stresses such as high light intensity and drought can trigger cyclic electron flow, leading to increased production of reactive oxygen and nitrogen species, thereby inducing nitro-oxidative stress31. Given that atmospheric tillandsioids typically inhabit exposed mountain rocks or arid deserts, with substantial exposure to UV radiation, the expansion of gene families related to NAD(P)H dehydrogenase activity could serve as a protective mechanism against nitro-oxidative stress arising from challenging environmental conditions.
Besides, the enriched gene families in the tank-forming tillandsioid V. erythrodactylon are also closely associated with its adaptation to a challenging aerial habitat, particularly in the regulation of hydrogen peroxide metabolic processes and terpene metabolism (Fig. 3c). Hydrogen peroxide plays a vital role in regulating plant metabolism and cellular signaling in response to environmental stresses32. Terpenoids encompass tens of thousands of small-molecule natural products found widely across all life domains. Driven by selective pressures to adapt to both biotic and abiotic environments within individual plant species’ ecological niches, specialized terpenoid metabolism has undergone extensive evolutionary divergence, resulting in lineage-specific pathways and products33. The biosynthesis and accumulation of these compounds may be tightly regulated by internal or external stimuli, enabling plants to finely adjust the deployment of terpenoids to mediate dynamic interactions with the environment26,34,35. As a result, these genes related to terpenoid metabolism likely play a critical role in the adaptability of tillandsioids to occupy new ecological niches.
As tillandsioids continued to adapt to aerial environments, they encountered new selection pressures that required modifications to their environmental adaptations. This included adjustments in molecular cell responses to stresses and changes in plant life forms. We subsequently investigated if these phenotypic and intracellular changes were driven by the evolution of the underlying protein-coding genes by identifying positively selected genes (PSGs) and rapidly evolving genes (REGs) within the entire genomes. These genes exhibit differences either among monocots compared to other monocots, within the Bromeliaceae family compared to other bromeliads, or in response to distinct selective pressures within tillandsioids. Our current research revealed that the genes responsible for chloroplast thylakoid development were evolving rapidly and experiencing positive selection in both tank-forming and atmospheric tillandsioids (Fig. 3d-e). Stress conditions can lead to irreversible damage to the chloroplast thylakoid membrane system, which is crucial for photosynthesis36. This finding suggests that structural alterations in the chloroplast thylakoid membranes of tillandsioids may have occurred during evolution to thrive in challenging aerial environments. The chloroplast thylakoids in tillandsioids demonstrate enhanced resistance to environmental stress, enabling plants to sustain photosynthesis and survival. Furthermore, we have pinpointed specific genes that directly react to stress in atmospheric tillandsioids (Fig. 3d-e). These tillandsioids typically thrive in drier habitats in contrast to tank-forming tillandsioids. To validate their role in responding to drought stress in atmospheric tillandsioids, we subjected T. duratii growth to two different environments: a moist tube culture with air humidity exceeding 96%, and a dry environment with an air humidity of 40-45% for drought treatment (Fig. 3f). We then observed the expressions of these stress response enrichment PSGs and REGs (Fig. 3e). As anticipated, the genes showed significant up-regulation under lower-humidity conditions (Fig. 3g), indicating their potential significance in adapting to arid habitats.
Moreover, we have identified several PSGs that are significantly enriched in epidermal cell differential differentiation in tank-forming tillandsioids (Fig. 3d). Among these genes, ROOT HAIRLESS 2 (RH2) and TRANSPARENT TESTA GLABRA 1 (TTG1) are known to play pivotal roles in the patterning and shape of leaf trichomes37,38. TTG1, in conjunction with the bHLH protein GLABRA3 (GL3) and the R2R3-MYB protein GLABRA1 (GL1) MYB, forms a TTG1-bHLH-MYB complex that directly regulates trichome cell fate and patterning38. The genetic variations and selection pressures acting on these two genes likely contributed significantly to the unique morphogenesis of trichomes in tillandsioids.
In addition to trichome traits, shoot morphogenesis leading to the formation of impounding tanks is another critical innovation that enables tillandsioids to thrive in aerial habitats. The tank formation arises from the overlapping of wide leaves, a feature absents in bromeliads with slender and narrow leaves incapable of forming tanks. Hence, the presence of broad leaf bases is essential for tank formation39. Through our analysis of positively selected genes, we have identified nine genes associated with shoot system morphogenesis in tank-forming tillandsioids (Fig. 3d). Among these genes, ASYMMETRIC LEAVES 1 (AS1) stands out as the best candidate gene. In Arabidopsis, AS1 exhibits specific expression in the petiole, and loss of function in this gene leads to wider petioles40. Furthermore, AS1 is the homolog of ROUGH SHEATH2 (RS2) in maize, which plays a crucial role in maize leaf sheath formation41. Therefore, we hypothesize that in tank-forming tillandsioids, the genetic variations and selection pressures acting on the AS1 gene may symbolize molecular evolutionary events in innovating tank-forming phenotypes.
Rapid evolution and positive selections of essential root development genes of tillandsioids
Tillandsioids, particularly atmospheric tillandsioids, are known for their limited or entirely absent root systems, with the poorly developed roots serving primarily as a mechanical “holdfast“15. To investigate whether the evolution of this root development trait in tillandsioids is driven by selection acting on genes controlling root development, we assessed the rates of molecular evolution of key genes within the root development network42,43 (Fig. 3h). As anticipated, five out of ten genes in tillandsioids exhibited positive selection or rapid evolution (Fig. 3h). Specifically, we identified two positively selected loci, SCARECROW (SCR) and JACKDAW (JKD), which are involved in root endodermis and quiescent center development (Fig. 3h). Additionally, four loci—WEREWOLF (WER), TRANSPARENT TESTA GLABRA (TTG1), SCHIZORHIZA (SCZ), and SCR— associated with root endodermis, quiescent center, cortex, or epidermis displayed rapid evolution (Fig. 3h), indicating that genes related to the development of nearly all root tissue components are subject to selection pressure. To further assess whether this variation and selection is a common feature across all tillandsioids, we compared the extent of evolutionary change in these five genes between epiphytic tillandsioids and terrestrial bromeliads using transcriptome or genome data from 143 bromeliads. Our analysis revealed significant differences in the Ka/Ks ratios of four genes (WER, TTG1, SCZ, and SCR) between epiphytic tillandsioids and terrestrial bromeliads (Fig. 3i). The Ka/Ks ratios of SCR and WER were significantly higher in epiphytic tillandsioids, suggesting stronger selection pressure on these two genes in all tillandsioids. This genetic variation in key root development genes may play a role in the limited root development observed in tillandsioids.
The poorly developed root system in epiphytic tillandsioids primarily functions as holdfasts, lacking typical root characteristics such as geotropism and hydrotropism (Supplementary Fig. 7). ALTERED RESPONSE TO GRAVITY 1 (ARG1) serves as a crucial positive regulator of plant responses to gravity44. Our investigation revealed that the roots of T. duratii are insensitive to gravity and lose their gravitropic response immediately upon germination (Supplementary Fig. 7d). We observed the loss of the ARG1 gene in both T. duratii and V. erythrodactylon genomes, along with another significant gene involved in regulating plant hydrotropic response, ALTERED HYDROTROPIC RESPONSE 1 (AHR1)45, during evolution. Furthermore, our analysis identified two genes related to root gravitropism and haptotropism, WAVY GROWTH 2 (Td20g02820) and ENDOBINDING 1 (Td07g02930), are under positive selection. WAVY GROWTH 2 has been shown to negatively regulate gravity-induced root bending46, while ENDOBINDING1 positively regulates plant responses to touch47. We hypothesize that the loss or potential functional alteration of genes controlling root growth direction frees tillandsioids roots from external constraints, enabling them to better adapt to atmospheric ecological niches and effectively serve as secure holdfasts for anchoring.
Furthermore, we observed that tillandsioid roots, particularly the atmospheric types, either lack or minimally possess lateral roots (Supplementary Fig. 7). Consequently, we speculated that the genes responsible for lateral root development in tillandsioids may have undergone evolutionary changes. The Arabidopsis MADS-box gene ANR1 is a crucial regulator of lateral root development in response to external nitrate supply48. We found that the ortholog of ANR1 was lost in both T. duratii and V. erythrodactylon. The MADS-box gene subfamily ANR1 was also reduced to only two members in both T. duratii and V. erythrodactylon, compared with three members in Puya (Puya raimondii) and five members in pineapple (Ananas comosus, CB5) (Supplementary Fig. 8). The contraction of subfamily ANR1 may affect the lateral roots development in tillandsioids.
Key genes involved in root lignification contribute to the holdfast role in tillandsioids
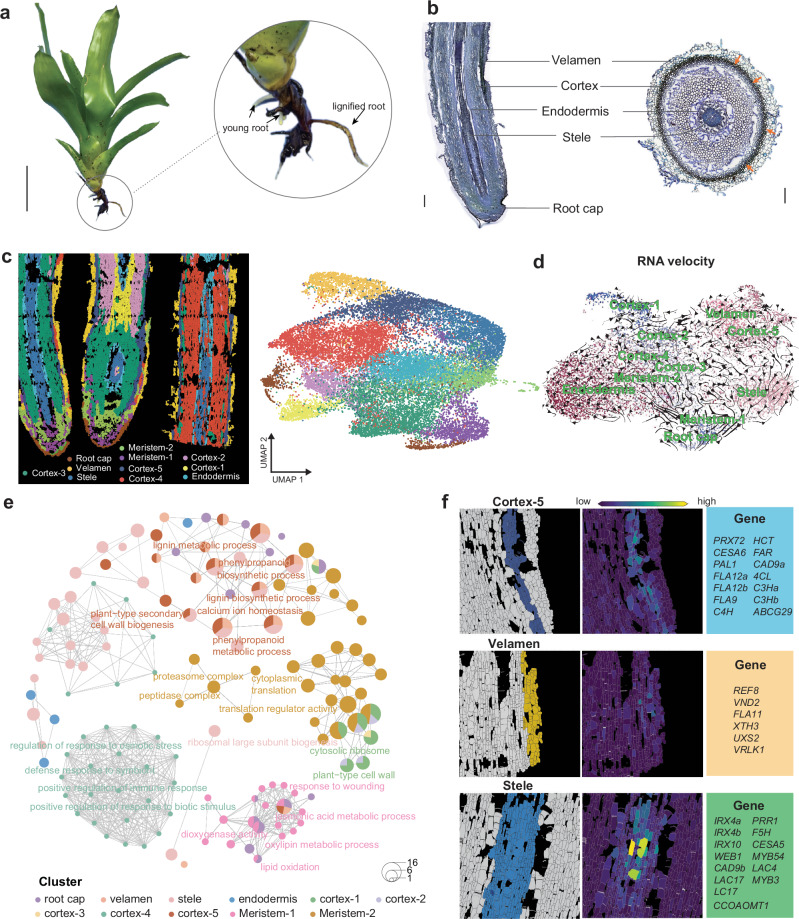
The roots of tillandsioids exhibit significant lignification that contributes to their exceptional durability and mechanical strength (Supplementary Fig. 7). It was observed that these roots underwent rapid lignification upon emergence, forming distinct structures (Fig. 4a and Supplementary Fig. 7d). These roots retain initial root tissues, with all components (stele, endodermis, cortex) present along with a root cap (Fig. 4b). The outer part of the roots is covered by a multilayered rhizodermis, resembling the velamen that covers the roots of epiphytic Orchidaceae8. The cortex typically exhibits differentiation, with the interzone constituting the majority of the root volume. While the endodermis has thick walls, the cortex primarily provides the root’s strength49. Cell walls of the outer cortex display centripetal sclerification/lignification49 (Fig. 4b). To delve into the detailed cellular and molecular mechanisms underlying the development of mechanical support in tillandsioid roots, we conducted spatial transcriptomic analysis on roots undergoing lignification (Fig. 4c). Longitudinal sections of these roots were utilized for spatial transcriptomic analysis on BMKMANU S1000 (Fig. 4c).
Fig. 4. Spatial gene expression atlas of root organ development using spatial RNA-seq.
a Root structure of V. erythrodactylon. Root growth was stimulated by with abundant water. The new roots are white and quickly lignified. Scale bar, 10 cm. b Longitudinal and cross-sectional views of V. erythrodactylon root. Scale bar, 500 µm. c Spatial transcriptomics-based cell clustering and cell-type identification in V. erythrodactylon roots. Visualization and annotation of identified cell types, with corresponding markers listed in Supplementary Data 3. Different cell types are color-coded. d RNA velocity analysis of root cells, indicating direction and speed of cellular movement. e Network plots displaying enriched GO terms and pathways of marker genes in various root cell types from spatial RNA-seq. f Spatial expression patterns of marker gene modules specific to cortex-5, stele, and velamen in V. erythrodactylon roots. Detailed gene names for each module are provided in the right border.
We captured 11,173 spots at a resolution of 50 μm, detecting a total of 18,130 genes. Uniform manifold approximation and projection (UMAP) method was used to visualize the data, with spots effectively clustered based on the marker genes (Fig. 4c, Supplementary Fig. 9 and Supplementary Data 3). The cortex cells were divided into 5 clusters, indicating the functional differentiation of cortex. RNA velocity analysis depicted a trajectory from cortex-1 to cortex-2, cortex-3, and cortex-4, with no trajectory leading to cortex-5 from other cortex clusters, which represents the outer layer of the cortex (Fig. 4d). Cortex-5 and its adjacent cells displayed mutual differentiation, with toluidine blue-stained root sections indicating that cortex-5 cells possess thick walls (Fig. 4B). Functional enrichment analysis of specifically expressed marker genes in these cell clusters unveiled unique molecular characteristics for each cell type (Fig. 4e). Gene Ontology (GO) term analysis demonstrated enrichment for processes such as lignin biosynthesis, phenylpropanoid biosynthesis, and secondary cell walls SCWs biogenesis in the cortex-5, as well as in the stele and endodermis clusters (Fig. 4e). SCWs are crucial for providing robust mechanical support50, and their deposition in these three root cell clusters enhances the mechanical role of the root as a ‘holdfast’. The abundance of cortex cells with thick walls exceeded that of the other two clusters (Fig. 4b), underscoring the importance of genes governing SCWs biosynthesis in the cortex for the root’s function as a ‘holdfast’.
Although there are three types of cells with SCWs deposition, the genes involved in this process are not identical. These genes consist of some well-known regulators and enzymes responsible for SCWs biosynthesis (Fig. 4f)50. They play respective roles in depositing SCWs in specific cell types within tillandsioid roots. For example, PEROXIDASE 72 (PRX72), CELLULOSE SYNTHASE 6 (CESA6), PHE AMMONIA LYASE 1 (PAL1), and others are specifically involved in cortex SCWs biosynthesis; CESA5, MYB54, IRX10, and others are specifically involved in steles SCWs biosynthesis; and VASCULAR-RELATED NAC-DOMAIN 2 (VND2), REDUCED EPIDERMAL FLUORESCENCE 8 (REF8) and others are specifically involved in velamen SCWs biosynthesis (Fig. 4f). To further validate the vital and specific role of these genes in SCWs biosynthesis of different cell types in tillandsioid roots, we also conducted the cross-section of the newly emerged and lignifying roots (Supplementary Fig. 10-11). The spatial visualization of the expression of these gene modules specific to cortex, velamen or stele, further proved their specific roles acting in different cell types. As the roots develop, the genes specifically expressed in the cortex show an expression trend from outer to inner cells, which is consistent with the cortex cell lignification pattern, further validating the crucial role of these genes in root lignification (Supplementary Fig. 10-11). The lignification of roots is the key process enabling them to act as holdfast in tillandsioids.
To explore the conservation of the root lignification network across different type tillandsioids, we conducted SMART RNA-seq analysis on the root cell division zone (newly initiated) and the elongation/lignification zone of both tank-forming tillandsioid V. erythrodactylon and atmospheric tillandsioid T. duratii (Fig. 4a and Supplementary Fig. 7d). Given the challenging germination of T. duratii seeds, we cultivated them on MS medium (Supplementary Fig. 7d). Functional enrichment analysis of the up-regulated genes in the root elongation/lignification zone of both tillandsioids revealed significant enrichment in the phenylpropanoid biosynthetic pathway (Supplementary Fig. 12a-b). The phenylpropanoid pathway is known for providing essential metabolites in plants and serving as a precursor for lignin biosynthesis, preceding lignin accumulation51. Additionally, through collinearity analysis, we identified that the majority of enriched genes involved in phenylpropanoid biosynthesis are orthologs in both types of tillandsioids (Supplementary Fig. 12c). These findings indicate that the genetic function related to root lignification is conserved across all tillandsioids, playing a crucial role in promoting root lignification for structural support.
Tandem duplicates of a cytochrome P450 gene associated with trichome formation in tillandsioids
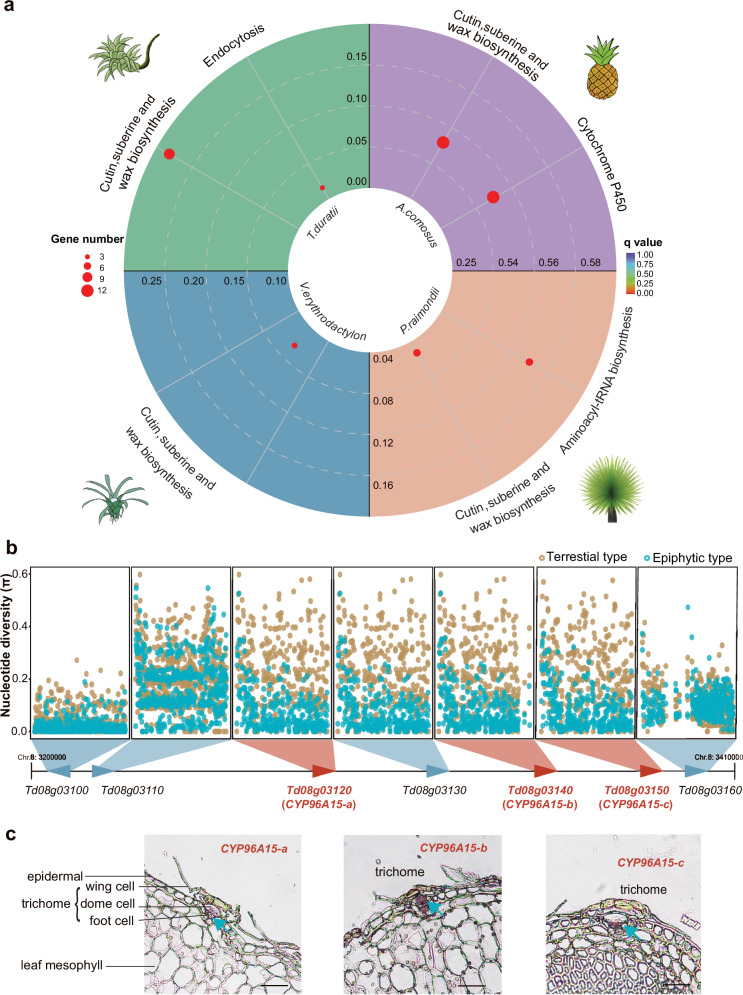
As the roots lose their absorbing function, specialized trichomes replace the roots and help to acquire water and nutrients for the epiphyte bromeliads. These trichomes are multicellular structures in the leaf epidermis consisting of a living stalk and a dead shield with absorbing function9. Besides, they are also important parts of plants to prevent evaporation, resist ultraviolet rays and catch moisture when exposed to the arid conditions. In fact, almost all bromeliads possess multicellular trichomes on leaf surface, and even the simplest, and unbranched appendages consist of adjoined cells15. Trichomes are an extension of the above-ground epidermal cells and defined as highly specialized epidermal cells in plants52. Like the normal epidermal cells, they are usually covering cuticle to protect plants against biotic and abiotic stress53–55. Raux et al.9 found that while there is no cuticle covering the wing cells, a thin cuticle layer lines the lateral walls of the dome cells and trichome stalks in Tillandsia species. This cuticle prevents capillary flow within the wall space connecting the outer trichome to the mesophyll, which is crucial for forming the trichome structure necessary for its absorption function. When we performed the functional enrichment analysis of the co-expanding gene families shared by both terrestrial and epiphytic bromeliads, we found that the function of co-expanding gene families is mainly associated with cutin, suberine and wax biosynthesis (Fig. 5a). These genes are tandemly repeated in T. duratii genome, and encode CYP96A15 duplications (Fig. 5b). CYP96A15 is a cytochrome P450 enzyme known for its involvement in stem cuticular wax biosynthesis in Arabidopsis56. Through sliding window analysis of the coding sequences of these genes and their neighboring genes within a 1 M region, using transcriptomic data from 143 terrestrial and epiphytic bromeliads, we discovered conserved and fixed sequence variations within the CYP96A15 duplications specific to epiphytic bromeliads (Fig. 5b). To further explore the gene expression patterns in trichomes, we conducted in situ hybridization experiments on T. duratii leaves. The results demonstrated the specific expressions of the three CYP96A15 duplicates in trichome dome and foot cells (Fig. 5c), providing additional evidence for their role in trichome development in tillandsioids.
Fig. 5. Genomic selection signatures of co-expanded genes involved in trichome development in epiphytic bromeliads.
a KEGG pathway enrichment analysis of co-expanded gene families in T. duratii, V. erythrodactylon, A. comosus, and P. raimondii. Bonferroni multiple comparison Hypergeometric Test was performed for p values. b Genetic variations across the region containing the three expanded genes in 17 terrestrial and 127 epiphytic bromeliads. c In situ detection of three expanded genes. Spatial expression of CYP96A15 duplications within trichome cells, with blue arrows indicating specific expression in the pitted trichome cells. Scale bars, 100 µm.
Spatial expression atlas of key genes regulating specialized trichomes development in tillandsioids
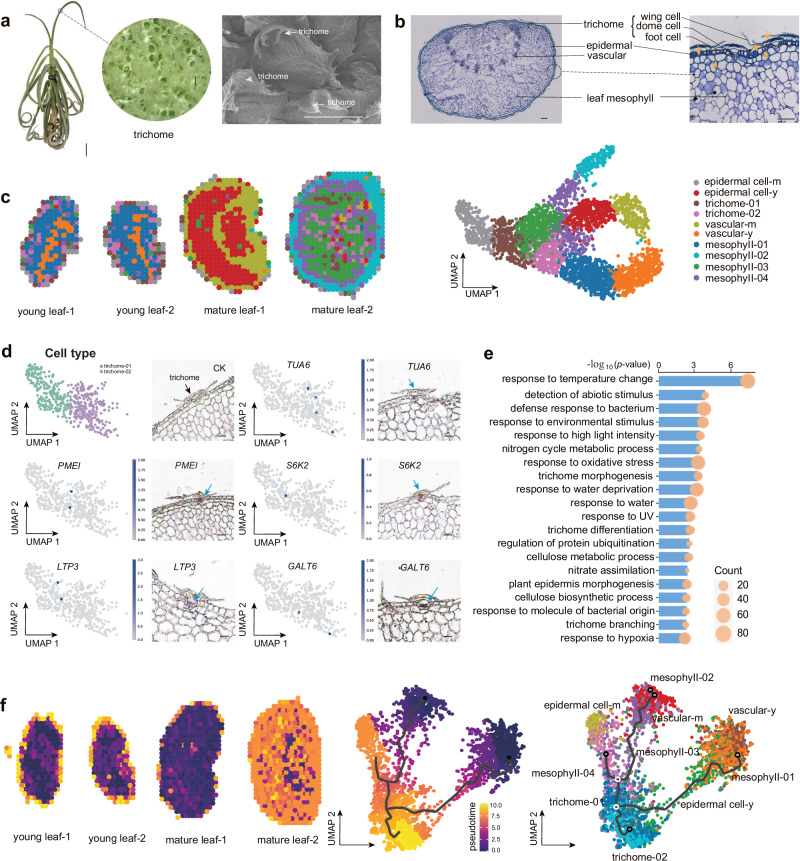
To comprehensively investigate the spatial expression patterns and some key genes involved in trichome development in tillandsioids, we collected the leaves of T. duratii with dense trichomes in young and adult stages for cryo sections and performed in-situ omics sequencing with greatly improved resolutions (Stereo-seq)57 (Fig. 6a-b). The high-quality newly assembled reference genome of T. duratii were subsequently employed for raw reads blasting and generated a total of 17,158 genes. To further provide an overall spatial information, 100 × 100 spots (calculated as a 50 µm2 area) on the Stereo-chip were used as a bin, a total of 3015 spots were finally obtained. Unsupervised clustering analysis of these spots revealed 10 clusters, each spatially distributed within the samples (Fig. 6c). The clusters, which indicate the trichome structures consistently matched well for both stage trichomes while the trichomes were separated well into two sub-groups. To compare the cell clusters in spatial positions in the leaves, the clusters correspond to the actual trichomes arrangements (Fig. 6c). To identify the specific genes involved in trichome development, we first scrutinized the marker genes across various cell types and classified the cell types based on these markers (Fig. 6c, Supplementary Fig. 13a and Supplementary Data 4). To confirm the spatial expression patterns of these trichome-related marker genes, we conducted in situ hybridization for five such markers. Among these markers, TUBULIN ALPHA-6 (TUA6), GALT6, and SERINE/THREONINE PROTEIN KINASE 2 (S6K2) have been implicated in the trichome branching process, while LIPID TRANSFER PROTEIN 3 (LTP3) harbors a trichome-specific promoter encoding a putative lipid transfer protein58–60. The outcomes were consistent with the spatial data (Fig. 6d), underscoring the reliability of these trichome markers. These marker genes likely play pivotal roles in the development of distinct and innovative trichomes in tillandsioids.
Fig. 6. Spatial gene expression profilings of leaf trichome of T. duratii.
a Morphology of T. duratii showcasing dense trichomes on the surface. Scale bars, 5 cm (whole plant), 100 µm (trichome). b Cross-section of a T. duratii leaf illustrating the structure of leaf and trichomes with different cell types. Scale bars, 100 µm. c Cell clustering and identification of cell types in T. duratii leaf based on spatial transcriptomics, with visualization and annotation of identified cell types. Corresponding markers listed in Supplementary Data 4. Cell types distinguished by different colors. d Validation of trichome-specific marker genes using RNA in situ hybridization. Scale bars, 100 µm. e Enriched GO terms and pathways of DEGs between trichome 01 and trichome 02 clusters identified from spatial RNA-seq. Bonferroni multiple comparison hypergeometric test was performed for P values. f Monocle 3 analysis depicting the trajectory of leaf cells in T. duratii, highlighting trichome cells as the final cells to differentiate.
Through functional enrichment analysis of specifically expressed marker genes in different cell clusters, we identified distinct molecular characteristics associated with each cell type. Genes related to the “response to water deprivation” Gene Ontology (GO) term were found in both epidermal cells and outer layer mesophyll cells of mature leaves (Supplementary Fig. 13b). This suggests their potential involvement in regulating responses to the relatively dry aerial environment. Marker genes expressed in trichomes displayed significant enrichment in GO terms related to the “protein polyubiquitination process” (Supplementary Fig. 13b), which aligns with previous reports linking ubiquitination to trichome development61. Furthermore, we observed functional enrichment of certain trichome markers, along with markers for mature epidermal cells, in lignin biosynthesis (Supplementary Fig. 13b). Lignin serves as both a mechanical support and a natural UV-blocking material for plant protection62. This finding underscores the similarity between these two cell types in terms of their protective functions. Additionally, the presence of thick SCWs in mature epidermal and trichome cells, as revealed by toluidine blue staining (Fig. 6b), further supports this observation.
The enrichment of GO terms among the Differentially Expressed Genes (DEGs) between the two trichome clusters (trichome-01 and trichome-02) further supports the protective role of trichomes in tillandsioids. These GO terms include responses to extreme temperatures, water deprivation, high light intensity, UV radiation, and other environmental stimuli, as well as defense responses to bacteria (Fig. 6e). Furthermore, we identified genes involved in trichome morphogenesis and differentiation, such as homologs of known genes associated with trichome development. For instance, PERMEABLE LEAVES3 (PEL3) mutations lead to altered trichome phenotypes with tangled trichomes during leaf expansion63; DISTORTED TRICHOMES 2 (DIS2) plays a role in trichome maturation, with mutants exhibiting enlarged trichomes64; JMJ29, encoding an H3K9me2 demethylase, regulates trichome morphogenesis through the modulation of GL365; and UBIQUITIN-PROTEIN LIGASE 3 (UPL3) is involved in trichome cell morphogenesis, with mutants showing supernumerary trichome branches66. These genes likely contribute to the distinct shield-like morphology of trichomes in tillandsioids. Furthermore, the three tandem repeat CYP96A15 genes identified through comparative genomics (Fig. 5) were further validated for their potential involvement in trichome development through spatial transcriptome analysis, predominantly showing expression in trichome cells (Supplementary Fig. 14 and Fig. 6b). To investigate the differentiation and development processes of trichomes, pseudo time analysis was conducted on all clusters generated by Monocle3. The results indicated that trichome cells differentiated from young epidermal or mesophyll cells near the epidermis (Fig. 6b, f), representing the final stage of cell differentiation in tillandsioid leaves (Fig. 6b, f). In conclusion, the genes highlighted in the spatial expression atlas of trichome development likely play crucial roles in shaping trichomes into the distinctive shield-like structures found in tillandsioids.
Phyllospheric nitrogen-fixing bacteria as potential source of nitrogen nutrition for tillandsioid
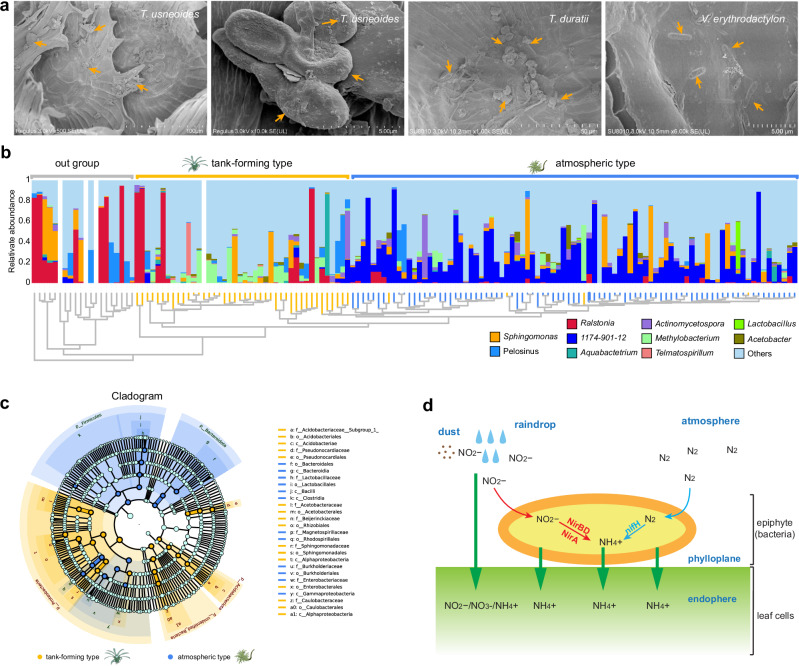
In addition to water acquisition, nutrient acquisition poses the other big challenge for tillandsioids, which do not rely on soil. The nutrients available through rainwater and airborne dust are relatively scarce. To meet their nutritional needs and support optimal growth, epiphytic bromeliads must develop innovative mechanisms to obtain nutrients, with a specific emphasis on nitrogen (N). N2-fixation by bacteria is an important source of N nutrition for plants, like the classical leguminous plants67. Phyllospheres have been shown to provide appropriate conditions for colonization by microorganisms, including N2-fixing bacteria that are able to fix atmospheric N268–71. Incremental evidence shows that phyllosphere microorganisms control plant nutrient availability and acquisition by free-living N2-fixation or associative N2- fixation with plants, influencing plant growth67–70. Obtaining clean DNA from the leaves of atmospheric tillandsioids presents a significant challenge due to frequent contamination by microbial DNA. This led us to speculate about potential microbial symbiosis with tillandsioids. Brighigna et al.72 and Givnish et al.39 have also discovered nitrogen-fixing bacteria in tanks and foliar trichomes of bromeliads. Despite not detecting any endophytes in the leaves of tillandsioids using transmission electron microscopes (TEM), we observed numerous bacteria on the trichomes located on the leaf surface through scanning electron microscope (SEM) analysis (Fig. 7a). To further identify these bacteria species and their potential host specificity, we conducted an analysis of phyllospheric bacteria (bacteria from leaf interior and surface) from 129 tillandsioids using 16S rRNA sequencing.
Fig. 7. Identification and phylogeny of nitrogen-fixing bacteria in the tillandsioid phyllosphere.
a SEM revealing microorganisms on the leaf surfaces of T. duratii and V. erythrodactylon. The orange arrows indicate bacteria or their excretions. b Top-10 phyllospheric bacterial communities of 127 tillandsioids and 17 out-group bromeliads with genera divergence. The phylogenetic tree constructed by bacterial communities mirrors the phylogenetic tree based on tillandsioids genera divergence as shown in Fig. 1. The relative abundance of phyllospheric bacterial communities is presented at the genus level, with different genera depicted in different colors. c Cladogram generated from the LEfSe analysis indicating the phylogenetic distribution from phylum to genus of the microbiota of two types of tillandsioids. In the phylogenetic tree, the concentric circles radiating outward represent taxonomic levels from phylum to genus (or species). Each small circle at different taxonomic levels represents a classification at that level, with the diameter of each circle proportional to its relative abundance. Species with no significant differences are uniformly colored light blue. Species (or genera, or families, or orders, or phylum) with differences are colored according to their groups; orange nodes represent bacterial groups that play important roles in tan-forming plants, while dark blue nodes represent those in atmospheric-type plants. Species (or genera, or families, or orders, or phylum) names represented by letters in the figure are explained in the legend at the bottom. d Nitrogen metabolism pathways in T. usneoides phyllospheric bacterial communities. Two ammonification pathways were identified in the phyllospheric bacterial metagenome, including the N2-fixation and Nitrate reduction pathways.
The operational taxonomic units (OTUs) were clustered and then annotated based on 97% sequence similarities. The rarefaction curve of the OTU number against the sequences number indicated that the sequencing data covered nearly all of the diversity that expected to be found in these samples (Supplementary Fig. 15a). Based on Principal Coordinates Analysis (PCoA) and relative abundance analysis, significant differences in compositions of phyllospheric bacteria were observed among plants of different types (Supplementary Fig. 15b). However, there were no discernible differences in the relative abundances of phyllospheric bacteria among different types of tillandsioids (Supplementary Fig. 15c). The compositions of top-10 dominant bacteria at genera level are also similar in the same type of tillandsioid (Fig. 7B). According to phylogenetic relationships of species, phyllospheric bacteria of tank-forming tillandsioids mainly came from phyla Firmicutes and Actinobacteria, as well as two classes Gammaproteobacteria and Alphaproteobacteria, while atmospheric tillandsioids mainly distributed in Alphaproteobacteria class (Fig. 7b-c and Supplementary Fig. 15d). Bacteria of Methylorubrum and Telmatospirillum are two dominant genera in tank-forming tillandsioids phyllosphere (Fig. 7b-c). As a genus of G-bacteria with both methylotrophic and methanotrophic properties, Methylorubrum can use glucose or other complex nutrients as carbon and energy sources73,74. The abundance of Methylobacteria on the leaf surface of tank-forming tillandsioids may be from the rainwater, fallen leaves and other sediments collected in the tank, and provides a good habitat and carbon source for Methylobacteria, which are involved in many biological activities such as N2-fixation, phosphorus affluence, abiotic stress tolerance, promotion of plant growth, and biocontrol activity against plant pathogens69. Thus, a large number of Methylobacteria on leaf surface of tank-forming tillandsioids may play a favorable role in their growth and development. Same as Methylorubrum, the enrichment of Telmatospirillum in tank-forming tillandsioids phyllosphere may be due to the abundant organic sediment such as leaves in the tank. Telmatospirillum bacteria can help degrade organic matter, and at the same time can participate in the sulfur cycle, relieve the toxicity in the impounding tank, avoid the toxic substances produced through the degradation of organic matter in the impounding tank to damage the plant, and provide the plant with available nutrients75.
1174-901-12 is conspicuous among the phyllospheric bacteria in atmospheric tillandsioid, followed by Sphingomonas (Fig. 7b and Supplementary Fig. 15e). Many bacteria of Sphingomonas have been identified as N2-fixing bacteria, such as S. trueperi, S. azotifigens and S. paucimobilis76–78, which are usually associated with gramineous crops. 1174-901-12 were dominant in clustering phyllosphere bacterial community for almost all atmospheric type tillandsioids (Fig. 7b) and was predicted to be the most important genus for tillandsioids(Supplementary Fig. 16a). These findings implied selection bias of 1174-901-12 bacteria with atmospheric tillandsioids co-evolution. 1174-901-12 belongs to family Beijerinckiaceae, order Rhizobiales, and class Alphaproteobacteria. There are very few studies on the bacteria in the genus 1174-901-12, and all bacteria in this genus are uncultured. Some bacteria of 1174-901-12 have capacity to fix N, which enables them to inhabit environments with low nitrogen levels79. Hence, we speculate that bacteria in the genus 1174-901-12 must play important roles in the growth of atmospheric tillandsioids, especially in terms of nutrients provision.
Based on functional predictions, we failed to find the commonalities of phyllospheric bacteria among different types of bromeliads (Supplementary Fig. 16b) To further confirm the role of these phyllospheric bacteria, we generated a metagenome of the phyllospheric bacterial communities found on T. usneoides. Due to the dense trichomes covering the leaf surface of atmospheric tillandsioid plants, the wings of the trichomes adhere to the leaf surface when they come into contact with liquid, making it challenging to isolate pure microorganisms9. T. usneoides has a higher leaf area ratio compared to most tillandsioids, which helps lower the possibility of plant genome contamination when extracting the DNA of phyllospheric microorganism (Supplementary Fig. 1k). This makes it an excellent candidate for studying the metagenome of phyllospheric bacterial communities in atmospheric tillandsioids. We annotated 63185 genes in metagenome TU, and identified some genes encoding enzymes involved in nitrogen metabolism (Supplementary Fig. 17). Based on the positions of these enzymes in the nitrogen metabolism pathway, it can be seen that phyllosphere bacteria on atmospheric tillandsioids participate in multiple processes of reducing nitrite to ammonia (Supplementary Fig. 17). These nitrites may be obtained from rainwater and dust on the leaf surface (Fig. 7d). Phyllosphere bacteria reduce them to ammonia, which is more readily absorbed and utilized by plants. Moreover, we identified three nifH genes and three nifA genes in the TU metagenome (Fig. 7d, and Supplementary Fig. 16c and Supplementary Fig. 17). The nifH gene is the most widely sequenced marker gene used to identify nitrogen-fixing bacteria80, and nifA gene product can activate the expression of nitrogenase genes81. These intriguing results strongly suggest the important role of the phyllosphere microorganisms in providing the nutrition source for atmospheric tillandsioids, allowing them to survive in the air with poor nutrient access.
Discussion
Bromeliads are capable of inhabiting diverse ecological habitats owing to their strong adaptability. Redeployment of absorptive function from roots to trichomes has rendered these bromeliads as an extant model for studying Darwinian natural selection and adaptive evolution in the plant kingdom.
Utilizing a comprehensive dataset of multiple nuclear genes, we constructed a highly resolved Tillandsioideae phylogeny that captures 78% of the genera-richness within this subfamily by nuclear genes (Fig. 1). Barfuss et al. had constructed a phylogeny of Tillandsioideae by plastome markers13,82. Compared to plastome markers, nuclear genes have an overall faster rate of sequence evolution, and provide multiple independent (unlinked) loci and biparental inheritance information83. In some cases, the nuclear phylogenies are incongruent with plastome phylogenies84–86. Due to differences in the species sampled between our study and the Barfuss et al. research13,82, direct comparison of the specific positions of species in the phylogenetic tree is difficult. Nevertheless, we have observed incongruence in species locations, particularly among some species within the Tillandsia genus. But in terms of broader clades, our nuclear phylogeny is mostly consistent with the plastome phylogeny as reported by Barfuss et al. Both researches revealed that Tillandsioidese was divided into core and non-core groups13. Our molecular clock and biogeographic analyses revealed that core tillandsioids originated from the Andes at the mid-late Miocene (Fig. 1), a period characterized by significant global climatic shifts marked by abrupt cooling following distinct warming phases87. The first to evolve is the tank-forming type tillandsioid, and within a relatively short period of 2 million years ( ~ 7.6 Mya to ~5.6 Mya), tillandsioids underwent a significant divergence, evolving into two distinct types (Fig. 1). This rapid speciation is often closely associated with abrupt climate changes88. While the Earth did not undergo substantial geoclimatic shifts from ~7.6 Mya to ~5.6 Mya, the Andes experienced a dramatic uplift started from 6 Mya23. The elevation of mountains can impact local climate conditions, thereby influencing species evolution89. With the emergence of the atmospheric type of tillandsioid also originating from the Andes, it is plausible to attribute the origin of this type to the uplift of the Andes. Consequently, we speculate that the origin and evolutionary history of atmospheric tillandsioids unfolded as follows: The sharp uplift of the mountains altered the elevation of certain tank-forming tillandsioids’ habitats in the Andes, transitioning from regions with higher rainfall to areas with less precipitation. As a result of this environmental shift, tillandsioids were no longer able to rely on tanks to collect rainwater. However, species with an increased presence of trichomes were able to capture water from clouds or fog, thus ensuring their survival and subsequent evolution.
In the past 5 million years since the emergence of atmospheric tillandsioids, their species have spread to almost all regions of Latin America, ranging from moist rainforest areas to arid coastal desert regions (Fig. 2a). The tillandsioids plants are extraordinarily remarkable in both the speed of their species variation and dispersion, as well as in the diversity of their habitats11. Both De Candolle and Darwin have explicitly pointed out that plants with a broader distribution range generally tend to exhibit more variations90. Givnish et al. indicated that epiphytism accelerates species diversification11,12. The speciation rates of the two types of tillandsioids accelerated, occurring shortly after significant climate changes on earth during transitions between geological periods. This phenomenon aligns with the general trend observed in many plant species88. However, our research revealed a key distinction: while both types of tillandsioids experienced these shifts between geological periods, each type only exhibited one time of accelerated speciation. This suggests that the environmental factors influencing the diversification of these two types differ. Our subsequent MaxEnt analysis of environmental variables for these species revealed that temperature seasonality is the primary factor driving the evolution of tank-forming tillandsioids, while atmospheric tillandsioids are primarily affected by substantial changes in temperature during the coldest quarter (Supplementary Fig. 4b-c). Compared to the Middle Pliocene, when global seasonal temperature differences were lower, tank bromeliad distribution increased in the Amazon and Central America but decreased in the Brazilian Shield during the Late Pleistocene (Fig. 2b), a period marked by greater seasonal temperature fluctuations. The uplift of the Brazilian Shield during this time may have diminished local seasonality. Considering this factor, we propose that tank bromeliads thrive in regions with more pronounced seasonal variations. In contrast, atmospheric bromeliads saw an increase across Central America, the Amazon, and the Brazilian Shield, with only a minor decrease noted in a small section of the Andes, which experienced lower temperatures during the coldest quarter compared to the Middle Pliocene (Fig. 2b). Thus, we speculate that the rise in atmospheric bromeliads correlates with the drop in temperatures during the coldest quarter. These findings are in line with the environmental changes that occurred during the geological periods corresponding to the acceleration of their evolution rates: one transitioned from the gradual cooling Miocene to the warm Pliocene, while the other shifted from the warm Pliocene to the Pleistocene characterized by repeated glacial and interglacial cycles (Fig. 2c). Furthermore, while tank-forming tillandsioids emerged earlier than atmospheric tillandsioids, the diverse distribution habitats of atmospheric tillandsioids are much broader than those of tank-forming tillandsioids. This demonstrates that atmospheric tillandsioids are more prone to variation and possess stronger environmental adaptability. In our subsequent genome characteristic studies of the two types of tillandsioids, we discovered that the rapid diversification rates of atmospheric tillandsioids may be attributed to a significantly higher number of LTR insertions in their genomes. The abundance of LTRs enables atmospheric tillandsioids to vary easily, aiding them in conquering diverse habitats.
Two types of tillandsioids occupy diverse habitats, associated with several key innovations of traits, including photosynthetic types, degree of root degradation, presence of tanks, and density of trichomes. Particularly, the photosynthetic types are strongly correlated with the dryness of their habitats (Fig. 1). Through comparative genomic and transcriptomic analysis, Crego et al. found that the rewiring of photosynthetic metabolism is mainly obtained through regulatory elements rather than coding sequence evolution and CAM-related gene families manifesting differential expression underwent accelerated gene family expansion in the constitutive CAM species28. Although photosynthetic types influence plant habitats, they are not the decisive factor in the evolution of Tillandsioids from terrestrial to epiphytic forms. The impounding tank, absorptive trichome and degraded root system with only mechanical role are the three key functional innovations of tillandsioids, facilitating them to adapt the aerial niches of diverse habitats8,15,26,91–93. Through genome assembly of two different types of tillandsioids and comparative genomic analysis, we successfully identified the key gene AS1 that may regulate the formation of the impounding tank (Fig. 3h). In Arabidopsis, mutations in this gene resulted in the widening of petiole, and homolog of it in maize directly regulated sheath formation40,41. When the base of the leaf widens but the diameter of the shortened stem remains unchanged, the leaves fold over each other, eventually forming a tank structure. Additionally, we identified a P450 gene associated with trichome development that underwent duplication in all bromeliads (Fig. 5a). In contrast to terrestrial bromeliads, three tandemly duplicated genes acquired the same variations in epiphytic species (referred to here as tillandsioids), potentially linked to the development of specialized trichome structures for water absorption in tillandsioids. However, how water enters through the trichome cap cells, subsequently reaches the live stem cells, and supplies moisture to the leaves requires further investigation. Furthermore, through analysis of key genes involved in root development and morphology, we successfully identified the crucial genes leading to the characters of degeneration, lack of geotropism, and hydrotropism of roots in tillandsioids. Variations in root development key genes WER, TTG1, SCZ, and SCR42,43 resulted in abnormal or complete loss of root development in tillandsioids, while the gene loss of gravity-responsive ARG144 and water-responsive AHR145 leads to the absence of geotropism and hydrotropism in tillandsioid roots. Additionally, through spatial transcriptomics, we identified some potential genes that enable tillandsioid roots to possess stable mechanical fixation capabilities (Fig. 4). These genes are specifically expressed in the root cortex, allowing tillandsioid roots to quickly lignify after growth and adhere to surfaces. Just as illustrated in Darwin’s theory of evolution, when one organ of an organism acquires the function of the other organ and is easier to perfect, the function of the other organ will be changed or even completely eliminated. Tillandsioids have gained the ability to absorb water through their trichomes, and in the barren limestone mountains of the Andes, they can obtain water more effectively from the moist air than through their roots. As a multicellular organism with unique morphology, the crucial innovation of, and development of trichomes must involve a complex gene network. Utilizing spatial transcriptomic technologies, we identified numerous marker genes involved in tillandsioid trichome development, some responding to stress and some participating in trichome morphogenesis (Fig. 6). The discovery of these trichome marker genes provides a new avenue for understanding the formation of the unique morphology of tillandsioids in future studies. Collectively, the assembly of the tillandsioid genome sequences, along with the discovery of genes potentially controlling a series of innovative traits in tillandsioids, undoubtedly mark a milestone in tillandsioid research and will greatly propel further studies on these magical species.
Nutrition poses a significant challenge for tillandsioids to thrive in aerial environments. The nutrition of most soil-rooted plants is aided by microbial mutualists, like endomycorrhizal fungi and bacteria67,94. Reports of microbial mutualists in epiphyte roots are few and attempts at identification cursory. The well-known case is the epiphytic orchids roots that have symbiosis with endomycorrhizal fungi94. Apart from this symbiotic interaction, asymbiotic relationship between plants and their hosts has become a research hotspot in recent years, especially the asymbiotic nitrogen fixation. N2-fixing bacteria live in close association with the plants and fix N with a loose mutualism pattern95. This associative N2-fixation mode exists not only on the surface of the root system, but also on the leaves68–71. Although tillandsioids, especially atmospheric type ones, do not possess a well-developed root system, the dense peltate-shape trichomes on leaf surfaces provides good habitats for bacteria. The protective role of trichomes on the leaf surface shields the plant from environmental stresses while providing a conducive environment for bacterial growth. By employing 16S rRNA sequencing, we identified diverse nitrogen-fixing bacteria residing on tillandsioids’ phyllosphere, with distinct microbial communities based on host life forms (Fig. 7). The dominance of bacteria belonging to the 1174-901-12 genus in all atmospheric tillandsioids’ phyllosphere suggested a specific interaction between these bacteria and the atmospheric tillandsioids. A recent report found the function of tillandsioid trichomes, may not only serve as for protection, but also secretion96. Polysaccharides were found on the surface of the trichomes96, which might be the carbon sources attracting those bacteria. However, research on the bacteria of genus 1174-901-12 is limited, possibly due to difficulties in artificial culturing. Their exact effects on the growth and development is unknown. In addition to the conspicuous 1174-901-12, we did find plenty of bacteria of Sphingomonas on the leaf surface of tillandsioids, in which many bacteria have been identified as N2-fixing bacteria76–78. Our subsequent identification of nifH genes in the newly assembled metagenome of one atmospheric tillandsioid provides robust evidence of the presence of nitrogen-fixing bacteria on leaf surface, underscoring their importance in nutrient acquisition (Supplementary Fig. 16c). While previous literature has suggested rainwater and dust as potential nutrient sources for atmospheric tillandsioids10,15, our study offers a new perspective on the nutritional ecology of epiphytes.
In summary, our study presents a comprehensive analysis of the evolutionary dynamics and genetic basis of the rapid differentiation, speciation, dispersal, and adaptation to various habitats and aerial ecological niches of tillandsioids (Fig. 8). This study sheds new lights on the implications of evolutionary trade-offs of functional traits and adaptive evolution of epiphytes transition from land to air.
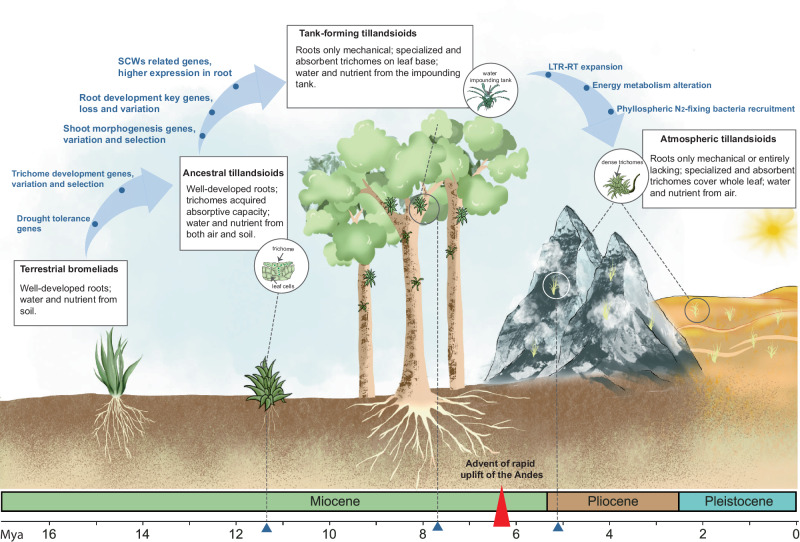
Fig. 8. The evolutionary dynamics of tillandsioids.
About 11.3 Mya in the Andes, the trichomes of the tillandsioid ancestor acquired the function of absorption and diverged from bromeliads with well-developed root systems. Since then, without needs of reliance on soil, tillandsioids could escape intense ground-level competition for space, allowing them to occupy new ecological niches. Approximately 2.6 million years later, tank-forming tillandsioids emerged with the tightly folded leaf rosettes forming tanks that collected and stored rainwater. The root absorption function became obsolete, leading to the gradual loss of genes controlling root hair formation,root geotropism and hydrotropism. Concurrently, many genes involved in SCWs synthesis were highly expressed in roots, enhancing their mechanical support as ‘holdfast’ structures. Around 6 million years ago, the Andes underwent rapid uplift, influencing regional climate and accelerating the genome expansion and evolution of tillandsioids. By approximately 5.1 Mya, another type of tillandsioid emerged with well adaptation to more arid habitats. These plants developed leaves densely covered with trichomes, protecting them from sunlight, reducing water evaporation, and mitigating high temperatures. They also formed a symbiotic relationship with phyllosphere bacteria, which provided nutrients, enabling them to thrive as true ‘air plants’ in harsh environments such as deserts.
Methods
Sample collection
A total of 143 species of Bromeliaceae were collected. These plants have been gradually collected over the past few decades from South America and botanical gardens worldwide. They are currently being cultivated at the Shanghai Chenshan Botanical Garden and the Bromeliads Research Center of Zhejiang Academy of Agricultural Sciences in China. Specific planting locations for each species are listed in Supplementary Data 5. Habits of all plants are classified according to the descriptions by Gilmartin97 and Barfuss et al.13 Briefly, plants with ligulate leaves that have few, inconspicuous, appressed scales or trichomes and lack a water-storing tank are classified as mesic; those with underdeveloped or absent tanks, narrow leaf blades featuring inconspicuous, appressed scales, and minimal or no water-storing tissue are considered as semixeric; and plants characterized by prominent, spreading trichomes, narrow leaf blades, the presence of water-storing tissue, and the absence of a tank are termed xeric. Types of photosynthesis of them were determined according to tissue carbon isotope ratio (δ13C)98.
De novo transcriptome assembly
Total RNA was extracted from fresh leaves using a Trizol kit (Invitrogen, USA), and 1 μg of RNA from each sample was utilized for sequencing on an Illumina HiSeq 2000 platform. Subsequently, Trinity v2.5.1 was employed for de novo transcriptome assembly99, and CD-HIT 4.6100 was used to eliminate redundant contigs (-c parameter: 0.98). The assembly completeness was assessed using BUSCO v3.0.2101. These de novo assembled transcriptomes were then utilized for constructing phylogenetic trees.
Phylogenetic tree construction
The newly denovo assembled 143 transcriptomes and the coding sequences derived from the genome sequences of A. comosus CB5 and F15325, and P. raimondii27 were used for the construction of phylogenetic tree. Python scripts developed for this analysis are available via GitHub (https://github.com/bpucker/Tillandsioideae). First, the identification of coding sequences in the transcriptome assemblies was performed based on integration of the results of ORFpredictor102, ORFfinder103, and TransDecoder104,105. The resulting coding sequences were in silico translated with transeq.py (https://github.com/bpucker/PBBtools/). BUSCO v3.0.2101 was run with eudicots_odb10 as reference data set on all species using wrapper script run_BUSCO_across_spec.py106. The Python script group_sequences.py (https://github.com/bpucker/Tillandsioideae) was applied to collect all sequences with orthologs in at least 140 species. Peptide sequences of each orthogroup were aligned with MAFFT v7.475107 using the auto function. RAxML-NG v.1.0.1108 was run using the parameters --bootstrap --model LG + G8 + F --bs-trees 1000 to construct preliminary trees based on aligned peptide sequences. Aligned peptide sequences were substituted with corresponding codons using pxaa2cdn as part of Phyx109 to increase the phylogenetic signal. Alignment columns with an occupancy below 10% were removed with the Python script algn_trim.py110. Phylogenetic trees were constructed with IQ-Tree v.1.6.12111 with -alrt 1000 -bb 1000. For ASTRAL analysis. 98 single copy genes were aligned with MAFFT v7.490 (https://mafft.cbrc.jp/alignment/software/)107, and the results were trimmed using trimAl v1.4. (https://vicfero.github.io/trimal/)112. Subsequently, the JTT model within MEGA v11113 was adopted to deduce the maximum likelihood tree for each individual single copy gene. We use the Weighted ASTRAL Hybrid method in ASTER to evaluate species trees based on each single copy gene tree, and calculate branch support using local posterior probabilities (LPP) support.
Divergence time estimation
The four fossil dates were collected from the TimeTree Resource (https://timetree.org/), and then were used to estimate divergence time by MEGA 11113. The input tree used for the analysis was derived from maximum likelihood reconstruction based on 91 concatenated orthologs, and the topology was designated according to Fig. 1. For the dating analyses, four fossils and a secondary calibration point were chosen, and the branch lengths were subsequently estimated.
Ecological niche modelling
We compiled 82 and 49 distinct geographic records of identified atmospheric and tank-forming tillandsioids for analysis. Using PaleoClim114, we obtained 19 bioclimatic variables at a 2.5-minute resolution for middle Pliocene paleoclimatic data (mid-Pliocene warm period, 3.205 Mya, v1.0*) and late Pleistocene data (MIS19, circa 787 ka, v1.0*). The original variables were retained during the analysis process, as removing collinear variables was found to have no significant impact on the maximum entropy model’s performance115. We exhaustively tested various settings using the R package ENMeval (v.0.3.1)116 and cross-validated the model evaluations. Different settings were chosen for different types to assess variable importance; for tank-forming types, LQ_0.5 and LQ_2 settings were used, while for atmospheric types, LQ_1.5 and LQH_2.5 settings were employed. Subsequently, MaxEnt (v.3.4.4)117 and ENMeval results were utilized to produce habitat suitability projections with 10 subsample repetitions and 25% random test samples.
Ancestral area reconstruction and speciation rate estimation
To evaluate evolutionary adaptation in different regions of South and Central America, and reconstruct the ancestral area, we employed the Bayesian Binary MCMC (BBM) method in RASP v. 4.2118. The input files for the BBM analysis in RASP comprise a geographical distribution statistics table of species (featuring two columns: species ID and geographical distribution) and the newly constructed time-calibrated tree was employed for this analysis constructed via RAxML v 8.2.1. Native distribution data for 150 species were sourced from Plants of the World Online records (https://powo.science.kew.org/). Six biogeographical regions were defined based on the current distribution of extant species, geological history, and modern climate14: (1) Guayana Shield; (2) Brazilian Shield (encompassing Serra do Mar, Serra da Mantiqueira, and adjacent Phanerozoic deposits); (3) Amazonia; (4) Caribbean; (5) Central America; and (6) the Andes (comprising all Andean regions, including the coastal desert). To perform the analysis, the tree file and species distribution files were loaded, the maximum number of areas any species could occupy was set to 6, and default parameters were used. Subsequently, RASP v. 4.2 generated results on evolutionary adaptation using the BBM method. The estimation of speciation rate for different plant genera/groups is carried out as descripted in Supplementary Method 1
DNA extraction and genome assembly
Genomic DNAs from tank-forming V. erythrodactylon and atmospheric T. duratii types were extracted from young leaves using the DNAsecure Plant Kit (Tiangen, Biotech, Beijing, China). PacBio sequencing libraries were prepared with high-quality DNA, fragmented to approximately 20 kb, repaired, ligated with adaptors, and sequenced on a Pacbio Sequel platform. For Illumina sequencing, DNA was fragmented using a Covaris ultrasonic crusher, followed by adaptor addition, PCR amplification, and sequencing on an Illumina platform. Hi-C libraries were prepared by fixing leaf samples, digesting chromatin with DPNII, biotin-labeling DNA ends, ligating with T4 DNA ligase, fragmenting DNA, repairing ends, isolating biotin-labeled fragments with Dynabeads®, and sequencing on an Illumina HiSeq X Ten sequencer after verification. Genome assembly, quality assessment and annotation were conducted as descripted in Supplementary Methods 2-4. Additionally, genome features, including the chromosomal rearrangements and LTR-retrotransposons, were analyzed in different bromeliad genomes following the methods outlined in Supplementary Methods 5-10.
Comparative genomic analysis
To explore the adaptive evolution of tillandsioids, a comparative genomic analysis was conducted involving 14 plant species from various plant lineages. The identification of one-to-one orthologous genes was performed as descripted as in Supplementary Method 11. Following the neutral theory of molecular evolution, the ratio of nonsynonymous substitution rate (Ka) to synonymous substitution rate (Ks) in protein-coding genes can pinpoint genes exhibiting signs of natural selection. To this end, we computed average Ka/Ks values and performed a two-ratio model analysis using Codeml from the PAML v4.9 package119 to detect rapidly evolving and positively selected genes within the monocot branch of six species, as illustrated in Fig. 3b. The branches of the two tillandsioids were designated as foreground branches, respectively. Genes with a ω value > 1 for the foreground branch and a P-value < 0.05 under the two-ratio model were classified as positively selected genes. Regarding rapidly evolving genes, those with a ω value for the background branch exceeding the ω value for the foreground branch ( < 1) and a P-value < 0.05 were identified as rapidly evolving genes. P-values were computed using chi-square statistics and adjusted for multiple testing via the FDR method. Additionally, the analysis of gene family expansion and contraction, as well as the identification of whole-genome duplication events, were conducted as described in Supplementary Method 12.
Nucleotide diversity analysis
The coding sequences of homologous genes derived from the recently assembled transcriptome and genome of 147 species, along with three published genomes, were employed for the analysis. Nucleotide diversity was assessed by calculating the π value for each population utilizing VCFtools v0.1.12b120. The transcriptome-assembled data by Trinity v2.5.199 were used to predict open reading frames (ORFs) using TransDecoder v5.7.1 (https://transdecoder.github.io/). Next, the target gene sequences were aligned with the 147 predicted transcripts using Diamond v2.0.15 (https://github.com/bbuchfink/diamond/releases/tag/v2.0.15). The best alignment results were retained, and the coding sequences (CDS) of all genes were aligned using Mafft v7.453 (http://mafft.cbrc.jp/alignment/software/). The multiple sequence alignment (MSA) results were then converted into a simplified VCF format. Nucleotide diversity (π) was estimated for each population using Python scripts (https://github.com/simonhmartin/genomics_general/blob/master/popgenWindows.py). The π value was measured using a 10 bp window with a step size of 5 bp. Finally, the results were visualized using ggplot2 in R.
UMI (Unique Molecular Identifier) RNA-seq
Total RNAs of T. duratii leaves under different conditions (under 100% humidity as control; under 40% for 24 and 72 hours for drought treatment) were extracted using the RNA Plant kit (TIANGEN, China) for RNA-seq. Each treatment was carried out with three biological replicates. Subsequent to fragmentation and reverse transcription, the ligation of adapters with UMI was performed. The resulting products were then sequenced using an Illumina HiSeq 6000 platform. The newly assembled T. duratii genome was used as a reference for reads alignment by HISAT2 v2.2.1 (https://daehwankimlab.github.io/hisat2/). The mapped reads of each sample were assembled using StringTie v2.2.1 with default parameters. HTSeq v0.6.1121 was used to estimate the expression levels of all transcripts and perform expression abundance for mRNAs by calculating FPKM (fragment per kilobase of transcript per million mapped reads) value122 and DESeq2 v1.12.3123 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) was used to perform differential gene expression.
SMART RNA-seq
Total RNAs from root samples at the early and late (lignifying) stages of T. duratii and V. erythrodactylon were extracted using the Trizol kit (Invitrogen, USA). Each stage was replicated three times for robustness. Total RNAs of picogram grade or from single/small cell populations were reverse-transcribed using SMARTScribe™ Reverse Transcriptase. The resulting double-stranded cDNAs were amplified via Polymerase Chain Reaction (PCR) until a sufficient quantity of cDNAs was obtained for sequencing on the Illumina Novaseq 6000 platform. Transcriptome analysis performed as the above section. The recently assembled genome sequences of T. duratii or V. erythrodactylon were used as reference for reads alignment.
Tissue cryosectioning
Leaf samples from the early and late stages of T. duratii and root samples from the early and late stages of V. erythrodactylon were each immersed in pre-chilled OCT solutions (Tissue-Tek® O.C.T.™ Compound, Sakura). Subsequently, they were promptly frozen on dry ice and preserved at −80 °C until required. The pre-frozen samples embedded in OCT were sliced into 10 μm sections for spatial transcriptome sequencing and histological examination. For spatial transcriptome sequencing, we utilized BMKMANU S1000 RNA-seq and BGI Spatial RNA-seq.
BMKMANU S1000 RNA-seq
The MKMANU S1000 RNA-seq analysis was performed as described by Song et al.124. In brief, root tissue sections were collected for RNA extraction for quality control, which required RIN value ≥ 7 to ensure no degradation of RNA during tissue freezing. For gene expression library construction and quality control, tissue permeabilization released mRNA and bound it to capture probes optimized through tissue experiments using the BMKMANU S1000 Tissue Optimization Kit (BMKMANU, ST03003). mRNA was reverse transcribed to obtain complete cDNA, followed by synthesis, denaturation, amplification, and purification. Purified cDNA concentration and peak shape were analyzed using Qubit and Agilent Bioanalyzer. A 10 µL cDNA sample underwent fragmentation, end repair, A tailing, junction ligation, and PCR indexing for library preparation. Libraries, typically 300-700 bp, were quantified with Qubit 4.0 and analyzed using Qseq400, and finally sequenced using the Illumina NavoSeq 6000 platform, and the sequencing strategy was PE150. Sequencing data quality is assessed using Clean Data post-initial quality control, focusing on key metrics like Q30 ≥ 80% for base quality, stable base content distribution (20%-60% GC), and tissue coverage (25%-90%) verified via toluidine blue staining on microarrays. BSTMatrix v2.0 and BSTMatrix-ONT v1.1employs image analysis algorithms to analyze tissue areas based on grayscale values, using the recently assembled genome of V. erythrodactylon as reference. A corresponding level 7 matrix (50 μm) was utilized for downstream analyses. UMAP and clustering details were generated using an R script (Bmk Space mapping.R, available at http://www.bmkmanu.com/archives/513), utilizing Seurat 4.3.0125 following the manufacturer protocols. The following trajectory and RNA velocity analysis were conducted as descripted in Supplementary Method 13
BGI spatial RNA-seq
The freshly prepared BGI spatial transcriptomic chips were affixed to a 10 µm slice exhibiting an optimal structure, following standard protocols outlined in the Stereo-seq transcriptome set (BGI, 111ST114) for cDNA preparation57. The amplified fragments underwent sequencing on the MGI DNBSEQ-T7 platform, and raw reads containing MIDs with N bases or >2 bases with low-quality scores ( < 10) were excluded. Subsequently, the filtered reads were aligned to the de novo high-quality genomes of T. duratii and V. erythrodactylon assembled in this study, utilizing the latest version of the SAW software (https://github.com/BGIResearch/SAW) with default parameters. Reads uniquely mapped to coding regions of the genomes were tallied. Original expression matrices were created with a spot resolution of 100 × 100 (equivalent to a bin size of 50 × 50 µm) and subsequently analyzed using Stereopy tools (https://github.com/BGIResearch/stereopy) for data quality assessment, spot clustering, UMAP visualization, identification of marker genes, and cell annotation. Differentially expressed marker genes, with the log2fc threshold parameter set to 0.25 in each cell cluster, were chosen for subsequent in situ validation experiments. The identification of differentially expressed genes (DEGs) was performed as descripted in Supplementary Method 14. Finally, the top 100 differentially expressed genes in each cell cluster were selected for Gene Ontology (GO) enrichment analysis, which carried out as descripted in Supplementary Method 15. Additionally, potential interactions between these genes and GO terms were investigated.
16S rRNA sequencing
We conducted 16S rRNA gene sequencing analysis following the method outlined by Peng et al.126. In brief, microbial DNA was extracted from 135 tillandsioids leaves (82 atmospheric and 49 tank-forming tillandsioids) using the E.Z.N.A.® Stool DNA Kit (Omega Bio-tek, Norcross, GA, USA) in accordance with the manufacturer’s protocols. Subsequently, PCR amplification of the V3-V4 hypervariable regions of the bacterial 16S rRNA gene was carried out using universal primers (338 F: 5′-ACTCCTACGGGAGGCAGCAG-3′, 806 R: 5′-GGACTACHVGGGTWTCTAAT-3′) incorporating the FLX Titanium adaptors and a barcode sequence. The resulting purified amplicons were pooled in equimolar amounts and subjected to paired-end sequencing on an Illumina NovaSeq. The following operational taxonomic units (OTUs) identification was conducted as descripted in Supplementary Method 16.
Bacterial diversity and abundance analysis
The diversity of 16S rRNA gene sequencing was analyzed using the Quantitative Insights Into Microbial Ecology platform and QIIME v1.4.0127. Principal coordinates analysis (PCoA) was conducted based on Binary jaccard distance, with the principal coordinate combination showing the highest contribution rate selected for visualization. PERMANOVA analysis (also known as ADONIS analysis) was performed using the ADONIS function from the R vegan package (https://cran.r-project.org/web/packages/vegan/index.html). QIIME v1.9.1 with default parameters were used to analyze rarefaction curves, and the vegan package of R v2.15.3 was used to plot. The two clustering ellipses were computed with the ggplot2 function stat_ellipse, applying a 95% confidence level. The linear discriminant analysis effect size (LEfSe) algorithm128 revealed statistically significant differences in the relative abundances of genera between GK rats and Wistar rats. Specifically, LDA values > 2.5 with a P value < 0.05 indicated significant enrichment.
Co-occurrence networks analysis
By calculating correlation indices (Spearman correlation coefficient SCC or Pearson correlation coefficient PCC) across all samples and obtaining a species correlation matrix, the following filtering criteria were applied: Firstly, remove connections with correlation coefficients <0.6; Secondly, filter out self-connections of nodes; Finally, exclude connections with node abundance <0.005%. Finally, use Graphviz v2.38.0 (https://graphviz.org/) for visualization and display, with the key parameters are as follows: Network diameter (ND) is 11, average degree (AD) is 17.79381, modularity is 0.5601838, clustering coefficient (CC) is 0.6945564, graph density (GD) is 0.09078477, and average path length (APL) is 17.79381.
Random forest analysis
The random forest model, a well-known machine learning approach based on the classification tree algorithm introduced by Breiman129, was utilized. Briefly, using the species abundance table at the genus level as the feature input data, we allocated 70% of the data to the training set and 30% to the testing set. An evaluation model was constructed using the randomForest package (https://cran.r-project.org/web/packages/randomForest/) in R. The ROC curve was plotted, and the AUC value was calculated using the pROC package (https://cran.r-project.org/web/packages/pROC/index.html) to assess the model’s performance. The top 30 genera were selected based on the mean decrease accuracy and mean decrease gin features, and bar charts were drawn for each.
DNA extraction and metagenome assembly
The phyllospheric microbiota of an atmospheric tillandsioid, T. usneoides, was further investigated through metagenomic sequencing following the method outlined by Dai et al.130. In brief, the total genomic DNA of the phyllospheric microbiota was extracted using the E.Z.N.A® Stool DNA kit (Omega Bio-Tek, USA) in accordance with the manufacturer’s instructions. Subsequently, the DNA was fragmented to an average size of approximately 300 bp using the TruSeq™ DNA Sample Prep Kit with Covaris M220 (Gene Company Limited, China) for the construction of paired-end libraries. The metagenomic sequencing process was carried out on an Illumina HiSeq4000 platform following the manufacturer’s protocols. Metagenome assembly and annotation were conducted as descripted in Supplementary Methods 17-19.
SEM observation
The leaves of V. erythrodactylon and T. duratii were collected and fixed overnight at 4 °C using a 100 mM sodium phosphate buffer (pH 7.0) containing 2.5% glutaraldehyde. Subsequently, they were treated with a 1% osmic acid solution for 2 hours, followed by dehydration using a series of graded aqueous ethanol solutions (50%, 70%, 80%, 90%, 95%) and two rounds of dehydration in 100% ethanol. The samples were then subjected to critical-point drying utilizing liquid CO2 (Hitachi HCP-2, Japan), mounted on metallic stubs using double-sided carbon adhesive tape, and coated with a layer of gold. Images were taken using a Hitachi SU-8010 (Japan) or Regulus 3.0kVx500 SE(UL) (Japan) after the sample preparation steps were completed.
Bright-field microscopic observation
We collected the leaves and roots of V. erythrodactylon and T. duratii, followed by fixation, embedding, and sectioning131. The sections were cut into slices of 2μm thickness and stained with 0.1% Toluidine Blue O for 1 minute. Subsequently, the sections were rinsed with distilled water (ddH2O) and covered with a cover glass. Finally, images were captured using a Nikon DS-Ri1 microscope (Nikon, Japan).
In situ hybridization histochemistry assay
The leaves of T. duratii were fixed in Formalin-Aceto-Alcohol (FAA), treated with diethypyrocarbonate (DEPC), and subsequently embedded in paraffin for sectioning131. The sections were then prepared for in situ hybridization132,133 utilizing digoxin-labeled ribo-probes (refer to Supplementary Table 4) for detection of the target RNA.
Statistics and reproducibility
Statistical analyses were conducted using R software (version 4.3.1). The Two-tailed Wilcoxon test was employed to assess statistical significance for comparisons between two groups. Data collection and analysis were not blinded with respect to the experimental conditions, and no data points were omitted from the analyses. The GO enrichment analysis was conducted using TBtools II, employing Hypergeometric testing and applying Bonferroni correction for P value adjustment. All statistical tests were conducted at a significance threshold of P < 0.05. Results are expressed as mean ± standard deviation (S.D.) or mean ± standard error of the mean (S.E.M.). The presented data originate from a minimum of three independent experiments, and all images displayed are representative of at least three such experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Dr. Xingxing Shen (Zhejiang University) and Dr. Chuyu Ye (Zhejiang University) for helpful discussions of the manuscript. We also thank Biomarker for the technical assistance of spatial transcriptome analysis. This work was supported by the Special Support Plan for High-level Talents of Zhejiang Province (2021R51007) and the Fundamental Research Funds for the Central Universities ( + 226-2022-00100).
Author contributions
M.Z., X.L. and J.Y. conceived and designed the project; X.L. conducted most of the experiments and analysis; P.L. and L.J. collected the plant materials; F.Y. contributed to the spatial transcriptome analysis; B.P. contributed to the phylogenetic tree construction; C.W. contributed to the statistical analysis; L.S. and M. Zhao contributed to the DNA and RNA extraction; L.L., Y.Z. and Q.Y. contributed to the figures’ modification; X. Li and K.X. contributed to microscopy observation. J. Yang and Z.H. contributed to data analysis. X.L. wrote the manuscript, and B.P., M.Z., and J.Y. revised it.
Peer review
Peer review information
Nature Communications thanks Thomas Givnish and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The genome sequencing and assembly for T. duratii and V. erythrodactylon have been deposited in the National Center for Biotechnology Information (NCBI) under BioProject PRJNA1090265 and the corresponding genome annotation files are available on Figshare [https://figshare.com/articles/dataset/_i_Vriesea_erythrodactylon_i_genome/25887199; https://figshare.com/articles/dataset/Tillandsia_duratii_genome/25887259]. The transcriptome and 16S sequencing data are available at NCBI under BioProjects accession PRJNA1090267 and PRJNA1090269, respectively. The metagenome sequencing and assembly data can be accessible at NCBI BioProject PRJNA1164103, and with annotation files deposited on Figshare [https://figshare.com/articles/dataset/hong_genome_of_Bromeliaceae/25887490]. Furthermore, the spatial transcriptomic data for tillandsioids are accessible in NCBI under BioProject PRJNA1163761 or in the CNGB Sequence Archive (CNSA) of the China National GeneBank DataBase (CNGBdb) with accession number CNP0005627. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiaolong Lyu, Ping Li, Liang Jin, Feng Yang.
Contributor Information
Jingquan Yu, Email: jqyu@zju.deu.cn.
Mingfang Zhang, Email: mfzhang@zju.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53756-7.
References
- 1.Banks, J. A. et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science332, 960–963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowles, A. M. C., Paps, J. & Bechtold, U. Water-related innovations in land plants evolved by different patterns of gene cooption and novelty. N. Phytol.235, 732–742 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu, B. et al. Contribution of NAC transcription factors to plant adaptation to land. Science343, 1505–1508 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Woudenberg, S., Renema, J., Tomescu, A. M. F., De Rybel, B. & Weijers, D. Deep origin and gradual evolution of transporting tissues: Perspectives from across the land plants. Plant Physiol.190, 85–99 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zotz, G. Plants on Plants – The Biology of Vascular Epiphytes, (Springer Cham, 2016).
- 6.VanBuren, R. et al. Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature527, 508–511 (2015). [DOI] [PubMed] [Google Scholar]
- 7.OLIWA, J. & SKOCZOWSKI, A. Different response of photosynthetic apparatus to high-light stress in sporotrophophyll and nest leaves of Platycerium bifurcatum. Photosynthetica57, 147–159 (2019). [Google Scholar]
- 8.Zhang, G. Q. et al. The Apostasia genome and the evolution of orchids. Nature549, 379–383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raux, P. S., Gravelle, S. & Dumais, J. Design of a unidirectional water valve in Tillandsia. Nat. Commun.11, 396 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benzing, D. H. The Evolution of Epiphytism. In Vascular Plants as Epiphytes: Evolution and Ecophysiology, U. Lüttge, ed., (Springer Berlin Heidelberg, 1989).
- 11.Givnish, T. J. et al. Adaptive radiation, correlated and contingent evolution, and net species diversification in Bromeliaceae. Mol. Phylogenet Evol.71, 55–78 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Givnish, T. J. et al. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. Biol. Sci.282 (2015). [DOI] [PMC free article] [PubMed]
- 13.Barfuss, M. H. J. et al. Taxonomic revision of Bromeliaceae subfam. Tillandsioideae based on a multi-locus DNA sequence phylogeny and morphology. Phytotaxa279, 1-+ (2016).
- 14.Givnish, T. J. et al. Phylogeny, adaptive radiation, and historical biogeography in Bromeliaceae: insights from an eight-locus plastid phylogeny. Am. J. Bot.98, 872–895 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Benzing, D. H. Bromeliaceae: Profile of An Adaptive Radiation, (2000).
- 16.Lüttge, U. Vascular Plants as Epiphytes, (Springer Berlin, Heidelberg, 2012).
- 17.Cavender-Bares, J., Kozak, K. H., Fine, P. V. & Kembel, S. W. The merging of community ecology and phylogenetic biology. Ecol. Lett.12, 693–715 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Van Stan, J. T. 2nd & Pypker, T. G. A review and evaluation of forest canopy epiphyte roles in the partitioning and chemical alteration of precipitation. Sci. Total Environ.536, 813–824 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Zeng, L. et al. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat. Commun.5, 4956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernández, R. et al. Phylogenomics, Diversification Dynamics, and Comparative Transcriptomics across the Spider Tree of Life. Curr. Biol.28, 1489–1497.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Chen, L. et al. Large-scale ruminant genome sequencing provides insights into their evolution and distinct traits. Science364 (2019). [DOI] [PubMed]
- 22.Guo, J. et al. Phylotranscriptomics in Cucurbitaceae Reveal Multiple Whole-Genome Duplications and Key Morphological and Molecular Innovations. Mol. Plant13, 1117–1133 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Garzione, C. N. et al. Rise of the Andes. Science320, 1304–1307 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Ming, R. et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet47, 1435–1442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, L. Y. et al. The bracteatus pineapple genome and domestication of clonally propagated crops. Nat. Genet51, 1549–1558 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Niu, S. et al. The Chinese pine genome and methylome unveil key features of conifer evolution. Cell185, 204–217.e14 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Liu, L. et al. Draft genome of Puya raimondii (Bromeliaceae), the Queen of the Andes. Genomics113, 2537–2546 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Groot Crego, C. et al. CAM evolution is associated with gene family expansion in an explosive bromeliad radiation. Plant Cell, (2024). [DOI] [PMC free article] [PubMed]
- 29.Hughes, J. F. & Coffin, J. M. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl Acad. Sci. USA101, 1668–1672 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller, P., Li, X. P. & Niyogi, K. K. Non-photochemical quenching. A response to excess light energy. Plant Physiol.125, 1558–1566 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corpas, F. J. & Barroso, J. B. NADPH-generating dehydrogenases: their role in the mechanism of protection against nitro-oxidative stress induced by adverse environmental conditions. Frontiers in Environmental Science2, 55-Article No.: 55 (2014).
- 32.Slesak, I., Libik, M., Karpinska, B., Karpinski, S. & Miszalski, Z. The role of hydrogen peroxide in regulation of plant metabolism and cellular signalling in response to environmental stresses. Acta Biochim Pol.54, 39–50 (2007). [PubMed] [Google Scholar]
- 33.Karunanithi, P. S. & Zerbe, P. Terpene Synthases as Metabolic Gatekeepers in the Evolution of Plant Terpenoid Chemical Diversity. Front Plant Sci.10, 1166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol.9, 297–304 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Schmelz, E. A. et al. Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J.79, 659–678 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Li, J., Liu, L. N., Meng, Q., Fan, H. & Sui, N. The roles of chloroplast membrane lipids in abiotic stress responses. Plant Signal Behav.15, 1807152 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugimoto-Shirasu, K., Stacey, N. J., Corsar, J., Roberts, K. & McCann, M. C. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr. Biol.12, 1782–1786 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Zhao, M., Morohashi, K., Hatlestad, G., Grotewold, E. & Lloyd, A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development135, 1991–1999 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Givnish, T. J. & Sytsma, K. J. Consistency, characters, and the likelihood of correct phylogenetic inference. Mol. Phylogenet Evol.7, 320–330 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Theodoris, G., Inada, N. & Freeling, M. Conservation and molecular dissection of ROUGH SHEATH2 and ASYMMETRIC LEAVES1 function in leaf development. Proc. Natl Acad. Sci. USA100, 6837–6842 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsiantis, M., Schneeberger, R., Golz, J. F., Freeling, M. & Langdale, J. A. The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science284, 154–156 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Ishida, T., Kurata, T., Okada, K. & Wada, T. A genetic regulatory network in the development of trichomes and root hairs. Annu Rev. Plant Biol.59, 365–386 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Slovak, R., Ogura, T., Satbhai, S. B., Ristova, D. & Busch, W. Genetic control of root growth: from genes to networks. Ann. Bot.117, 9–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou, N. et al. GSA-1/ARG1 protects root gravitropism in Arabidopsis under ammonium stress. N. Phytol.200, 97–111 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Saucedo, M. et al. An altered hydrotropic response (ahr1) mutant of Arabidopsis recovers root hydrotropism with cytokinin. J. Exp. Bot.63, 3587–3601 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mochizuki, S. et al. The Arabidopsis WAVY GROWTH 2 protein modulates root bending in response to environmental stimuli. Plant Cell17, 537–547 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gleeson, L., Squires, S. & Bisgrove, S. R. The microtubule associated protein END BINDING 1 represses root responses to mechanical cues. Plant Sci.187, 1–9 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Gan, Y., Filleur, S., Rahman, A., Gotensparre, S. & Forde, B. G. Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta222, 730–742 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Proença, S. L. & Sajo, M. D. G. Rhizome and root anatomy of 14 species of Bromeliaceae X1 - Anatomia de raízes e rizomas de 14 espécies de Bromeliaceae. Rodriguésia.59, 113–128 (2008). [Google Scholar]
- 50.Taylor-Teeples, M. et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature517, 571–575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fraser, C. M. & Chapple, C. The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book9, e0152 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, X., Shen, C., Meng, P., Tan, G. & Lv, L. Analysis and review of trichomes in plants. BMC Plant Biol.21, 70 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang, J. H. et al. The tomato odorless-2 mutant is defective in trichome-based production of diverse specialized metabolites and broad-spectrum resistance to insect herbivores. Plant Physiol.154, 262–272 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hegebarth, D., Buschhaus, C., Wu, M., Bird, D. & Jetter, R. The composition of surface wax on trichomes of Arabidopsis thaliana differs from wax on other epidermal cells. Plant J.88, 762–774 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Zhao, Q. & Chen, X. Y. Development: A new function of plant trichomes. Nat. Plants2, 16096 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Greer, S. et al. The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol.145, 653–667 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell185, 1777–1792.e21 (2022). [DOI] [PubMed] [Google Scholar]
- 58.Liu, H. C., Creech, R. G., Jenkins, J. N. & Ma, D. P. Cloning and promoter analysis of the cotton lipid transfer protein gene Ltp3(1). Biochim Biophys. Acta1487, 106–111 (2000). [PubMed] [Google Scholar]
- 59.Abe, T. & Hashimoto, T. Altered microtubule dynamics by expression of modified alpha-tubulin protein causes right-handed helical growth in transgenic Arabidopsis plants. Plant J.43, 191–204 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Henriques, R. et al. Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. Embo j.29, 2979–2993 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, Z., Yang, Z. & Li, F. Updates on molecular mechanisms in the development of branched trichome in Arabidopsis and nonbranched in cotton. Plant Biotechnol. J.17, 1706–1722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh, P., Singh, A. & Choudhary, K. K. Revisiting the role of phenylpropanoids in plant defense against UV-B stress. Plant Stress.7 (2023).
- 63.Tanaka, T., Tanaka, H., Machida, C., Watanabe, M. & Machida, Y. A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J.37, 139–146 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Saedler, R. et al. Actin control over microtubules suggested by DISTORTED2 encoding the Arabidopsis ARPC2 subunit homolog. Plant Cell Physiol.45, 813–822 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Hung, F. Y. et al. Arabidopsis JMJ29 is involved in trichome development by regulating the core trichome initiation gene GLABRA3. Plant J.103, 1735–1743 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Downes, B. P., Stupar, R. M., Gingerich, D. J. & Vierstra, R. D. The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J.35, 729–742 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Griesmann, M. et al. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science361 (2018). [DOI] [PubMed]
- 68.Fürnkranz, M. et al. Nitrogen fixation by phyllosphere bacteria associated with higher plants and their colonizing epiphytes of a tropical lowland rainforest of Costa Rica. Isme j.2, 561–570 (2008). [DOI] [PubMed] [Google Scholar]
- 69.Madhaiyan, M., Alex, T. H., Ngoh, S. T., Prithiviraj, B. & Ji, L. Leaf-residing Methylobacterium species fix nitrogen and promote biomass and seed production in Jatropha curcas. Biotechnol. Biofuels8, 222 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perreault, R. & Laforest-Lapointe, I. Plant-microbe interactions in the phyllosphere: facing challenges of the anthropocene. Isme j.16, 339–345 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Howe, A. et al. Seasonal activities of the phyllosphere microbiome of perennial crops. Nat. Commun.14, 1039 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brighigna, L. et al. Role of the nitrogen‐fixing bacterial microflora in the epiphytism of Tillandsia (Bromeliaceae). Am. J. Bot.79, 723–727 (1992). [Google Scholar]
- 73.Bijlani, S. et al. Methylobacterium ajmalii sp. nov., Isolated From the International Space Station. Front Microbiol12, 639396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Green, P. N. & Ardley, J. K. Review of the genus Methylobacterium and closely related organisms: a proposal that some Methylobacterium species be reclassified into a new genus, Methylorubrum gen. nov. Int J. Syst. Evol. Microbiol68, 2727–2748 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Hausmann, B. et al. Draft Genome Sequence of Telmatospirillum siberiense 26-4b1, an Acidotolerant Peatland Alphaproteobacterium Potentially Involved in Sulfur Cycling. Genome Announc6 (2018). [DOI] [PMC free article] [PubMed]
- 76.Xie, C. H. & Yokota, A. Sphingomonas azotifigens sp. nov., a nitrogen-fixing bacterium isolated from the roots of Oryza sativa. Int J. Syst. Evol. Microbiol56, 889–893 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Videira, S. S., de Araujo, J. L., Rodrigues Lda, S., Baldani, V. L. & Baldani, J. I. Occurrence and diversity of nitrogen-fixing Sphingomonas bacteria associated with rice plants grown in Brazil. FEMS Microbiol Lett.293, 11–19 (2009). [DOI] [PubMed] [Google Scholar]
- 78.Yang, S. et al. Growth-promoting Sphingomonas paucimobilis ZJSH1 associated with Dendrobium officinale through phytohormone production and nitrogen fixation. Micro. Biotechnol.7, 611–620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lopez-Echartea, E., Strejcek, M., Mukherjee, S., Uhlik, O. & Yrjälä, K. Bacterial succession in oil-contaminated soil under phytoremediation with poplars. Chemosphere243, 125242 (2020). [DOI] [PubMed] [Google Scholar]
- 80.Fischer, H. M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev.58, 352–386 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kennedy, C. & Robson, R. L. Activation of nif gene expression in Azotobacter by the nifA gene product of Klebsiella pneumoniae. Nature301, 626–628 (1983). [DOI] [PubMed] [Google Scholar]
- 82.Barfuss, M. et al. Taxonomic revision of Bromeliaceae subfam. Tillandsioideae based on a multi-locus DNA sequence phylogeny and morphology. Phytotaxa279, (2016).
- 83.SmallA, R., Cronn, R. & WendelC, J. L. A. S. Johnson Review No. 2. Use of nuclear genes for phylogeny reconstruction in plants. Australian Systematic Botany - AUST SYSTEMATIC BOTANY17 (2004).
- 84.Pelser, P. B. et al. Patterns and causes of incongruence between plastid and nuclear Senecioneae (Asteraceae) phylogenies. Am. J. Bot.97, 856–873 (2010). [DOI] [PubMed] [Google Scholar]
- 85.Galbany-Casals, M. et al. Phylogenetic relationships in Helichrysum (Compositae: Gnaphalieae) and related genera: Incongruence between nuclear and plastid phylogenies, biogeographic and morphological patterns, and implications for generic delimitation. TAXON63, 608–624 (2014). [Google Scholar]
- 86.Favre, A., Paule, J. & Ebersbach, J. Incongruences between nuclear and plastid phylogenies challenge the identification of correlates of diversification in Gentiana in the European Alpine System. Alp. Bot.132, 29–50 (2022). [Google Scholar]
- 87.Herbert, T. D. et al. Late Miocene global cooling and the rise of modern ecosystems. NATURE GEOSCIENCE9, 843–84 (2016). [Google Scholar]
- 88.Levin, D. A. Plant speciation in the age of climate change. Ann. Bot.124, 769–775 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu, R. et al. Adaptive evolution of the enigmatic Takakia now facing climate change in Tibet. Cell186, 3558–3576.e17 (2023). [DOI] [PubMed] [Google Scholar]
- 90.Darwin, C. On the origin of species. (1859).
- 91.Badouin, H. et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature546, 148–152 (2017). [DOI] [PubMed] [Google Scholar]
- 92.Zhang, L. et al. The water lily genome and the early evolution of flowering plants. Nature577, 79–84 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang, X. et al. Genomes of the Banyan Tree and Pollinator Wasp Provide Insights into Fig-Wasp Coevolution. Cell183, 875–889.e17 (2020). [DOI] [PubMed] [Google Scholar]
- 94.Johnson, L., Kane, M. E., Zettler, L. W. & Mueller, G. M. Diversity and specificity of orchid mycorrhizal fungi in a leafless epiphytic orchid, Dendrophylax lindenii and the potential role of fungi in shaping its fine-scale distribution. Front. Ecol Evol.11 (2023).
- 95.Haskett, T. L. et al. Engineered plant control of associative nitrogen fixation. Proc. Natl Acad. Sci. USA119, e2117465119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Papini, A., Tani, G., Falco, P. & Brighigna, L. The ultrastructure of the development of Tillandsia (Bromeliaceae) trichome. Flora - Morphology, Distribution. Funct. Ecol. Plants205, 94–100 (2010). [Google Scholar]
- 97.Gilmartin, A. J. Evolution of Mesic and Xeric Habits in Tillandsia and Vriesea (Bromeliaceae). Syst. Bot.8, 233–242 (1983). [Google Scholar]
- 98.Crayn, D. M., Winter, K., Schulte, K. & Smith, J. A. C. Photosynthetic pathways in Bromeliaceae: phylogenetic and ecological significance of CAM and C3 based on carbon isotope ratios for 1893 species. Botanical J. Linn. Soc.178, 169–221 (2015). [Google Scholar]
- 99.Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol.29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 102.Min, X. J., Butler, G., Storms, R. & Tsang, A. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res33, W677–W680 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wheeler, D. L. et al. Database resources of the National Center for Biotechnology. NUCLEIC ACIDS RESEARCH31, 28–33 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc.8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Haak, M. et al. High Quality de Novo Transcriptome Assembly of Croton tiglium. Front Mol. Biosci.5, 62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pucker, B. & Brockington, S. F. Genome-wide analyses supported by RNA-Seq reveal non-canonical splice sites in plant genomes. BMC Genom.19, 980 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol.30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kozlov, A. M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics35, 4453–4455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brown, J. W., Walker, J. F. & Smith, S. A. Phyx: phylogenetic tools for unix. Bioinformatics33, 1886–1888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pucker, B. & Iorizzo, M. Apiaceae FNS I originated from F3H through tandem gene duplication. PLoS One18, e0280155 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol.32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mello, B. Estimating TimeTrees with MEGA and the TimeTree Resource. Mol. Biol. Evol.35, 2334–2342 (2018). [DOI] [PubMed] [Google Scholar]
- 114.Brown, J. L., Hill, D. J., Dolan, A. M., Carnaval, A. C. & Haywood, A. M. PaleoClim, high spatial resolution paleoclimate surfaces for global land areas. Sci. Data5, 180254 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Feng, X., Park, D. S., Liang, Y., Pandey, R. & Papeş, M. Collinearity in ecological niche modeling: Confusions and challenges. Ecol. Evol.9, 10365–10376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Muscarella, R. et al. ENMeval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for MAXENT ecological niche models. Methods Ecol. Evol.5 (2014).
- 117.Phillips, S. J., Anderson, R. P., Dudík, M., Schapire, R. E. & Blair, M. E. Opening the black box: an open-source release of Maxent. ECOGRAPHY40, 887–893 (2017). [Google Scholar]
- 118.Yu, Y., Blair, C. & He, X. RASP 4: Ancestral State Reconstruction Tool for Multiple Genes and Characters. Mol. Biol. Evol.37, 604–606 (2020). [DOI] [PubMed] [Google Scholar]
- 119.Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol.24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 120.Danecek, P. et al. The variant call format and VCFtools. Bioinformatics27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol.28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Song, X. et al. Spatial transcriptomics reveals light-induced chlorenchyma cells involved in promoting shoot regeneration in tomato callus. Proc. Natl Acad. Sci. USA120, e2310163120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Peng, W. et al. Association of gut microbiota composition and function with a senescence-accelerated mouse model of Alzheimer’s Disease using 16S rRNA gene and metagenomic sequencing analysis. Aging (Albany NY)10, 4054–4065 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kuczynski, J. et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. MicrobiolChapter 1, Unit 1E.5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol.12, R60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Breiman, L. Random forests. Mach. Learn.45, 5–32 (2001). [Google Scholar]
- 130.Dai, H. L. et al. Metagenomic and metabolomic analysis reveals the effects of chemical phosphorus recovery on biological nutrient removal system. Chem. Engineer. J.328, 1087–1097 (2017). [Google Scholar]
- 131.Lyu, X. L. et al. Characterization of watermelon anther and its programmed cell death-associated events during dehiscence under cold stress. Plant Cell Reports38, 1551–1561 (2019). [DOI] [PubMed] [Google Scholar]
- 132.Nikovics, K. et al. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell18, 2929–2945 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lyu, X. et al. A natural mutation of the NST1 gene arrests secondary cell wall biosynthesis in the seed coat of a hull-less pumpkin accession. Hortic. Res9, uhac136 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The genome sequencing and assembly for T. duratii and V. erythrodactylon have been deposited in the National Center for Biotechnology Information (NCBI) under BioProject PRJNA1090265 and the corresponding genome annotation files are available on Figshare [https://figshare.com/articles/dataset/_i_Vriesea_erythrodactylon_i_genome/25887199; https://figshare.com/articles/dataset/Tillandsia_duratii_genome/25887259]. The transcriptome and 16S sequencing data are available at NCBI under BioProjects accession PRJNA1090267 and PRJNA1090269, respectively. The metagenome sequencing and assembly data can be accessible at NCBI BioProject PRJNA1164103, and with annotation files deposited on Figshare [https://figshare.com/articles/dataset/hong_genome_of_Bromeliaceae/25887490]. Furthermore, the spatial transcriptomic data for tillandsioids are accessible in NCBI under BioProject PRJNA1163761 or in the CNGB Sequence Archive (CNSA) of the China National GeneBank DataBase (CNGBdb) with accession number CNP0005627. Source data are provided with this paper.