Abstract
Diffuse large B-cell lymphoma (DLBCL) is an aggressive B-cell lymphoma characterized by distinct subtypes and heterogeneous treatment outcomes. Oxidative stress and the dysregulation of related regulatory genes are prevalent in DLBCL, prompting an investigation into the nuclear factor erythroid 2-related factor 2 (Nrf2)-kelch-like ECH-associated protein 1 (Keap1) signaling pathway and associated genes. The present study assessed pathological specimens and clinical data from 43 newly diagnosed patients with DLBCL, comparing the associations and correlations between the expression of Nrf2, Keap1, microtubule-associated protein 1 light chain 3β (LC3B) and nitrotyrosine and the activated B-cell (ABC) and germinal center B-cell (GCB) subtypes of DLBCL using immunohistochemistry and digital image analysis software. Nuclear Nrf2 activation was observed in 33.3% of patients with DLBCL ABC, demonstrating a higher prevalence of hepatitis B surface antigen positivity, calcium ions and significant body weight loss (P<0.05). Total Nrf2 expression was associated with the DLBCL GCB subtype and inversely correlated with Keap1 expression in the DLBCL ABC subtype. Furthermore, a positive correlation was demonstrated between Nrf2 and LC3, indicating that total Nrf2 is inhibited by Keap1 and regulates LC3 expression. The ABC subtype was also associated with lower white blood cell counts and more frequent chemotherapy courses than the GCB subtype. These findings suggest that nuclear Nrf2 could be a biomarker for DLBCL clinical diagnosis.
Keywords: diffuse large B-cell lymphoma, nuclear factor erythroid 2-related factor 2, activated B-cell, germinal center B-cell, kelch-like ECH-associated protein 1, nitrotyrosine, microtubule-associated protein 1 light chain 3β
Introduction
Diffuse large B-cell lymphoma (DLBCL) is an aggressive malignant lymphoma and despite significant therapeutic advances in recent years, relapsed/refractory DLBCL occurs in 30–40% of patients due to DLBCL morphological and molecular heterogeneity (1–4). According to the World Health Organization, DLBCL subgroups are classified based on gene expression profiling or cell of origin (5) and primarily stratified into germinal center B-cell (GCB) and activated B-cell (ABC) subtypes. In comparison with the GCB subtype of DLBCL, the ABC subtype is associated with a poor prognosis, with a high expression of BCL2, cMYC and BCL6, and a typical progression of clinical features (6,7). Despite the effectiveness of lenalidomide, BTK and PI3K inhibitors (8–13), ABC-type DLBCL still displays an inadequate response to standard immunotherapy (6). Furthermore, although the pathogenesis of these two DLBCL subtypes involves different mutated genes and activated signaling pathways, with corresponding drugs already available, the mechanisms remain unclear and require investigation.
Oxidative stress and related regulatory genes are considered hallmarks of cancer progression and therefore can be used to assess disease course and prognosis. The Kelch-like ECH-associated protein 1 (Keap1)-nuclear factor erythroid 2-related factor 2 (Nrf2) signaling pathway is a typical antioxidant stress pathway that exhibits abnormalities in several human malignant tumors, such as breast cancer, lung cancer, liver cancer, thyroid cancer, ovarian cancer and gastric cancer (14–17). Nrf2 is a transcriptional regulatory factor for antioxidant stress and is induced by oxidative stress to enter the nucleus to activate downstream antioxidant genes to protect cells from oxidative and electrophilic stress. Conversely, upregulation of Nrf2 activity within cancer cells suppresses drug-induced reactive oxygen species (ROS) production, leading to the development of drug resistance, thereby facilitating cancer cell survival and proliferation (18). Recent studies have reported that Nrf2 is overexpressed in cancer cells, indicating that it may serve an oncogenic role in carcinogenesis (15). Moreover, due to its dual role, Nrf2 and its antagonist Keap1 have become the subject of debate regarding their specific roles in preventing or promoting tumor progression (18). Exploring the related gene expression of this pathway is of great significance for cancer prevention and treatment. In addition, previous studies have also reported the association between Nrf2 and autophagy. The autophagy receptor p62 is also a target of Nrf2, whereby induction of autophagy leads to increased p62 levels. Consequently, p62 interacts with Keap1, activating Nrf2, further promoting cancer cell survival (19–22).
Currently, there is limited data regarding the impact of Nrf2 expression in different DLBCL subtypes on subsequent treatment or clinical outcomes. Therefore, the present study assessed the gene expression of NFE2L2 (Nrf2), KEAP1 (Keap1) and MAP1LC3B (LC3B), as well as ROS, in the cells of different subtypes (ABC and GCB) of newly diagnosed patients with DLBCL. Subsequently, the present study analyzed the correlation between these expression profiles and clinical pathological data, as well as the correlations among diverse genes to assess the impact of varying gene expression on DLBCL subtypes to identify potential prognostic biomarkers.
Materials and methods
Public databases for gene expression analysis
DLBCL samples from the Cancer Genome Atlas (TCGA) (https://www.cancer.gov/ccg/research/genome-sequencing/tcga) and the Genotype-Tissue Expression (GTEx) (https://gtexportal.org/home/) databases were analyzed using Gene Expression Profiling Interactive Analysis 2 (http://gepia2.cancer-pku.cn/) and the University of Alabama at Birmingham Cancer data analysis Portal (https://ualcan.path.uab.edu/) software (23,24). The gene expression data included NFE2L2, KEAP1 and MAP1LC3B and the value |log2FoldChange|>1 and q values <0.01 were considered to indicate differential expression. These databases provide tumor/normal differential expression analysis to aid in the analysis of RNA-sequencing data.
Study design
Following approval by the Institutional Review Board of Kaohsiung Medical University Chung-Ho Memorial Hospital [Kaohsiung, Taiwan; approval nos. KMUHIRB-E(I)-20210119 and KMUHIRB-E(I)-20220298], pathological specimens of patients diagnosed with DLBCL at Kaohsiung Medical University Chung-Ho Memorial Hospital between July 2015 and December 2021 were collected, along with clinical and biochemical laboratory data for subsequent analysis. A total of 43 specimens were obtained and DLBCL subtypes were classified as ABC or GCB subtypes using immunohistochemistry (IHC) analysis (25). The inclusion criteria included the following: i) Histopathologically-confirmed DLBCL; ii) age of ≥18 years; and iii) pathological tissue sections measuring ≥1×1×5 mm. The exclusion criteria included the following: i) Concurrent malignancies; ii) aged of <18 years; and iii) adequate pathological specimens were unavailable or too small for analysis.
IHC
The staining process was automated using a BOND-MAX Automated IHC Staining System (Leica Biosystems) following a standardized protocol (26). Briefly, pre-existing formalin-fixed paraffin-embedded blocks were sectioned to 4-µm thick, deparaffinized with xylene at 72°C and pre-treated for permeabilization using Epitope Retrieval Solution 1 (citrate, pH 6.0; Leica Biosystems) at 100°C for 20 min. Hydroperoxide blocking was performed for 5 min using the Bond Polymer Refine Detection Kit (Leica Biosystems Newcastle Ltd, United Kingdom), incubated with the following primary antibodies at room temperature for 30 min: Primary antibodies targeting Nrf2 (EP1808Y; monoclonal; 1:50; cat. no. ab62352; Abcam), Keap1 (lB4; monoclonal; 1:150; cat. no. ab119403; Abcam), LC3B (polyclonal; 1:100; cat. no. ab63817; Abcam) and nitrotyrosine (39B6; monoclonal; 1:100; cat. no. sc-32757; Santa Cruz Biotechnology, Inc.). Afterward, the tissue was incubated with the secondary antibody from the BOND Polymer Refine Detection Kit (Leica Biosystems) at 25°C for 15 min. Polymer incubation lasted for 15 min before development with 3,3′-diaminobenzidine tetrahydrochloride hydrate (DAB) chromogen for 10 min. The specimens were counterstained with hematoxylin at 25°C for 5 min. Positive and negative controls consisted of squamous cell carcinoma tissues. The staining of cytoplasmic Nrf2, Keap1, LC3B and nitrotyrosine was visualized using the TissueFAXS PLUS system (version 4.2; TissueGnostics GmbH) and the Zeiss Observer microscope with a 20X objective lens (Ziess GmbH). HistoQuest software (version 4.0; TissueGnostics GmbH) was used to quantify cytoplasmic Nrf2, Keap1, LC3B, nitrotyrosine and hematoxylin staining (Fig. S1). Regions of interest (ROIs) within each slide were selected for analysis, with ≥3 representative areas measured for consistency. Positive cell expression within the ROIs was quantified as %. Staining intensity was assessed by assigning arbitrary numbers based on grayscale pixel conversion, with the numbers of DAB-positive and hematoxylin-positive events used for optimization. This approach ensured robust analysis of immunohistochemically stained samples adhering to established protocols for accurate interpretation. Due to the limitations of imaging software in accurately distinguishing between nuclear and cytoplasmic Nrf2, pathologists qualitatively assessed the nuclear expression of Nrf2 in DLBCL using optical microscopy. A positivity threshold of ≥10% was used to define a sample as positive.
Cell culture
Human DLBCL cancer cell lines, U2932 (Guangzhou Ubigene Biosciences Co., Ltd.) and HT (cat. no. 60486; Bioresource Collection and Research Center, Taiwan) were cultured in RPMI-1640 supplemented with 10% FBS (Merck KGaA), 2 mM L-glutamine (Gibco; Thermo Fisher Scientific, Inc.), 10 mM HEPES (BioConcept AG), 1 mM sodium pyruvate (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.) in 5% CO2 at 37°C. A total of 2×106 cells were initially seeded into a 75T flask with 10 ml medium. Every 3–4 days, the cells and medium were transferred to a 15 ml centrifuge tube and centrifuged at 150 × g for 5 min at 25°C. After centrifugation, the supernatant was carefully discarded and half of the cells were resuspended in fresh medium.
Reverse transcription-quantitative (q)PCR
Total RNA was isolated from the U2932 and HT cell lines using the TOOLSmart RNA Extractor reagent (cat. no. DPT-BD24; BIOTOOLS Co., Ltd.) and 1 µg RNA reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (cat. no. 4368814; Thermo Fisher Scientific, Inc.) at 25°C for 10 min, followed by 37°C for 120 min, 85°C for 5 min and maintenance at 4°C. Each 20 µl reaction contained 2× SYBR Green Master Mix (cat. no. A46012; Applied Biosystems; Thermo Fisher Scientific, Inc.), 1 µl forward and reverse primers (KEAP1 forward: 5′-CGTAGCCCCCATGAAGCA-3′ and reverse: 5′-ACTCCACACTGTCCAGGAACGT-3′; GAPDH forward: 5′-GCACCACCAACTGCTTAGCA-3′ and reverse: 5′-TCTTCTGGGTGGCAGTGATG-3′) and 2 µl cDNA. qPCR was performed using the QuantStudio real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) and the thermocycling conditions were as follows: 95°C for 1 min, followed by 40 cycles at 95°C for 10 sec and 60°C for 60 sec; 65°C for 10 sec for melting curves; and 40°C for 30 sec for cooling. Target gene expression was quantified using the 2−ΔΔCq method with GAPDH used as the internal control (27,28). The PCR reactions were performed in triplicate.
Statistical analysis
Statistical analyses were performed using SPSS software (version 19; IBM Corp.). Pearson's correlation coefficient was used to assess the correlation between Nrf2, Keap1, LC3B and nitrotyrosine. Continuous variables were compared between groups using an unpaired t-test. Fisher's exact test or the χ2 test were used to evaluate the association of categorical variables. Multiple logistic regression models were also used to assess the association between nuclear Nrf2 and clinical characteristics. Independent variables with P<0.05 in univariable analysis were selected in the multivariable analysis. A forward selection approach determined the final multivariable model. Continuous data are presented as mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
Differential distribution of Nrf2 in subtypes and stages of DLBCL
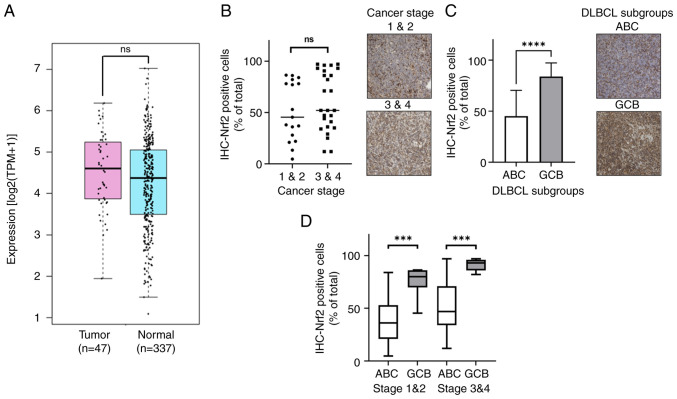
Nrf2 is a crucial gene involved in mitigating ROS and is present in several cancers (Fig. S2). In DLBCL, the NFE2L2 (Nrf2) mRNA level also demonstrated a significantly higher expression, with transcripts per million (TPM) of 23.29, compared with 19.69 TPM in normal tissues, as revealed by analysis of data from the TCGA and GTEx databases (Fig. 1A). Furthermore, a higher proportion of Nrf2-positive cancer cells was demonstrated in the advanced cancer stages (Ann Arbor stages 3 and 4) (29), where 60.88% cells were Nrf2-positive, compared with 51.26% in the early cancer stages (stages 1 and 2) of DLBCL, as determined by IHC analysis (Fig. 1B). Furthermore, when comparing the subtypes of DLBCL, it was demonstrated that the GCB subtype (84.20%) had a significantly higher frequency of Nrf2-positive cancer cells than the ABC subtype (45.32%) (P<0.0001; Fig. 1C). This trend persisted across disease stages, where the GCB subtype had a significantly greater proportion of Nrf2-positive cells compared with the ABC subtype in both the early stages (GCB, 75.98%; and ABC, 37.78%) and the late stages (GCB, 91.25%; and ABC, 49.69%) (Fig. 1D). The results revealed that Nrf2 was prominently distributed in the advanced stages of DLBCL and showed a higher frequency in the GCB subtype compared with that in the ABC subtype.
Figure 1.
Nrf2 is differentially distributed in different cancer stages and subtypes of DLBCL. (A) Elevated NFE2L2 (Nrf2) mRNA levels in DLBCL tissues compared with in normal tissues. (B) Increased % Nrf2-positive cancer cells in advanced stages of DLBCL. (C) Higher frequency of Nrf2-positive cancer cells in the GCB subtype of DLBCL compared with that in the ABC subtype. (D) Higher Nrf2-positive cancer cell numbers in the GCB subtype across all stages of DLBCL. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. All IHC sample images are ×20 magnification. ns, no significance; DLBCL, diffuse large B-cell lymphoma; Nrf2, nuclear factor erythroid 2-related factor 2; ABC, activated B-cell; GCB, germinal center B-cell; TPM, transcripts per million; IHC, immunohistochemistry.
Active Nrf2 is predominantly located in the nucleus of the DLBCL ABC subtype
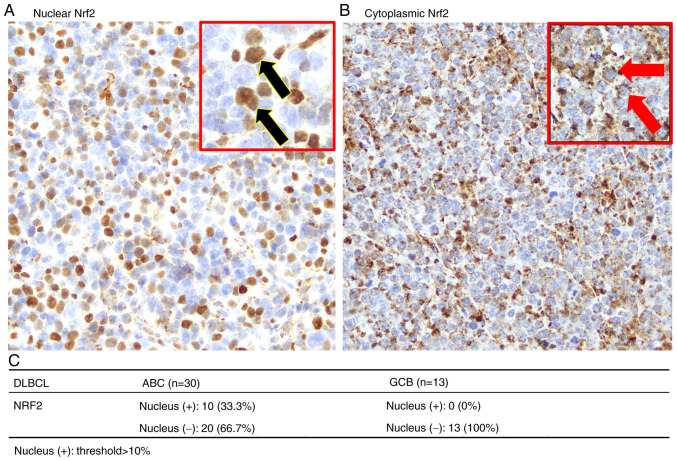
Nrf2 functions as a transcription factor activated within the cell nucleus (30). Therefore, the present study further assessed the nuclear expression of Nrf2 in DLBCL. Fig. 2A and B present images captured at ×40 magnification, of Nrf2 localization in the nucleus and cytoplasm of DLBCL cells, respectively. Notably, the analysis revealed that nuclear Nrf2 was exclusively present in the ABC subtype, accounting for 33.3% of cases (n=30), whilst no nuclear Nrf2 was observed in the GCB subtype (n=13; Fig. 2C).
Figure 2.
Immunohistochemical staining of Nrf2 localization in DLBCL. (A) Active Nrf2 is distributed in both the nucleus and cytoplasm, covering the entire cell (black arrows). (B) Nrf2 localized in the cytoplasm surrounds the nucleus (red arrows indicate cytoplasmic Nrf2; nucleus stained with hematoxylin). (C) Nuclear Nrf2 is predominantly observed in the ABC subtype of DLBCL and is absent in the GCB subtype. All IHC sample images are ×40 magnification. DLBCL, diffuse large B-cell lymphoma; Nrf2, nuclear factor erythroid 2-related factor 2; ABC, activated B-cell; GCB, germinal center B-cell.
KEAP1 mRNA and protein expression in advanced cancer stages and the DLBCL ABC subtype
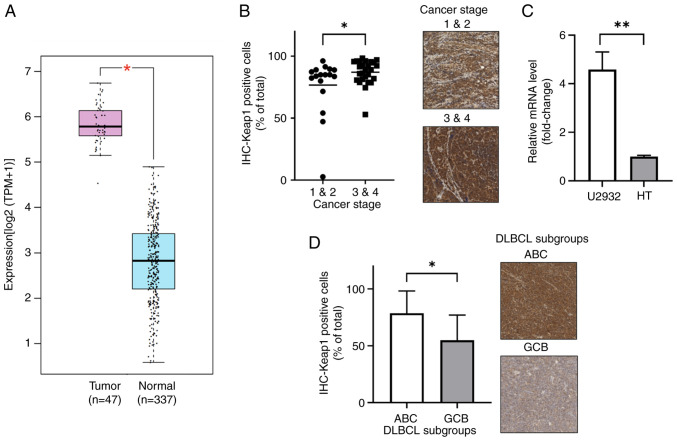
A comparative assessment of mRNA levels from the public databases of TCGA and GTEx revealed higher KEAP1 expression in tumor samples compared with that in controls. Specifically, KEAP1 exhibited an average expression of 54.07 TPM in tumor samples, significantly elevated compared with 6.1 TPM in normal controls (P<0.05; Fig. 3A). Immunohistochemical staining for Keap1 also demonstrated higher expression in advanced stages (stages 3 and 4), with 87.10% Keap1-positive cells, compared with 76.66% in the early stages (stages 1 and 2; P=0.046; Fig. 3B). Additionally, the KEAP1 expression ratios in DLBCL cell lines revealed a 4.3-fold higher expression in U2932 cells (ABC) compared with that in HT cells (GCB) when normalized to GAPDH (P<0.01; Fig. 3C). Moreover, assessment of Keap1 expression across DLBCL subtypes indicated a higher proportion of Keap1-expressing cells in tumor samples from patients with ABC (78.69%) compared with those with the GCB subtype (54.86%; P=0.014; Fig. 3D).
Figure 3.
Keap1 expression in DLBCL, especially in the advanced staged and ABC subtype. (A) KEAP1 mRNA is significantly upregulated in DLBCL compared with that in the controls in The Cancer Genome Atlas and Genotype-Tissue Expression databases. (B) Significantly higher Keap1 expression in the advanced stages of DLBCL than in the early stages. (C) Significantly higher Keap1 expression in patients with DLBCL of the ABC subtype than the GCB subtype. (D) KEAP1 mRNA expression in DLBCL cell lines (HT and U2932). *P<0.05; **P<0.01. All IHC sample images are ×20 magnification. DLBCL, diffuse large B-cell lymphoma; Keap1, kelch-like ECH-associated protein 1; ABC, activated B-cell; GCB, germinal center B-cell; TPM, transcripts per million; IHC, immunohistochemistry.
LC3B expression and association with Nrf2 in DLBCL
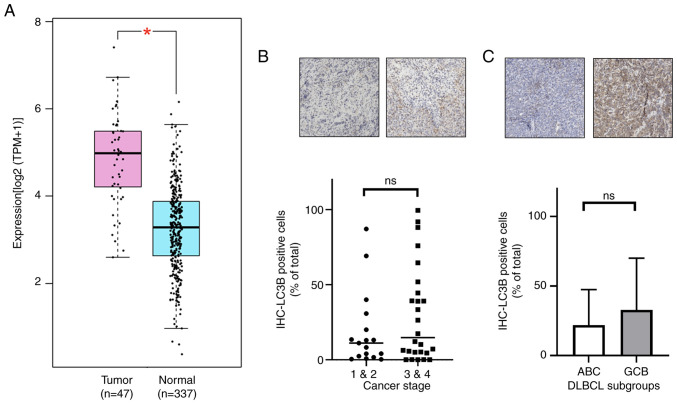
Nrf2 has been implicated in autophagy (31,32), therefore the present study evaluated the expression of the autophagosome marker LC3B in DLBCL and its relationship with Nrf2. Analysis of TCGA and GTEx data revealed that MAP1LC3B (LC3B) mRNA expression was significantly elevated in DLBCL compared with normal tissues (30.58 TPM vs. 8.77 TPM; P<0.05; Fig. 4A). In IHC-stained samples, although statistical significance was not achieved, LC3B-positive cells were notably more prevalent in advanced stages (stages 3 and 4) and in the GCB subtype of DLBCL compared with that in early stages (stages 1 and 2) and the ABC subtype, respectively (Fig. 4B and C). Furthermore, a significant positive correlation was observed between Nrf2 and LC3B expression in DLBCL samples (Pearson correlation=0.345; P=0.023; data not shown). These results indicate that LC3B expression in advanced cancer stages and the GCB subtype parallels that of Nrf2.
Figure 4.
Increased LC3B (encoded by MAP1LC3B) expression in DLBCL. (A) Elevated MAP1LC3B mRNA expression in DLBCL compared with controls in The Cancer Genome Atlas and Genotype-Tissue Expression databases. (B) LC3B expression across the early and advanced stages of DLBCL. (C) LC3B expression in the DLBCL GCB and ABC subtypes. All IHC sample images are ×20 magnification. ns, no significance; DLBCL, diffuse large B-cell lymphoma; TPM, transcripts per million; IHC, immunohistochemistry; MAP1LC3B, microtubule-associated protein 1 light chain 3β.
Nitrotyrosine expression slightly increases in advanced stages and ABC subtype of DLBCL
To evaluate whether Nrf2 suppresses ROS, nitrotyrosine was used as a marker to assess ROS levels in DLBCL. IHC analysis revealed a trend of markedly increased nitrotyrosine levels in advanced stages of DLBCL compared with that in early stages (Fig. 5A). Additionally, the frequency of nitrotyrosine-positive cells was notably higher in the ABC subtype compared with that in the GCB subtype, although this difference was not statistically significant (Fig. 5B).
Figure 5.
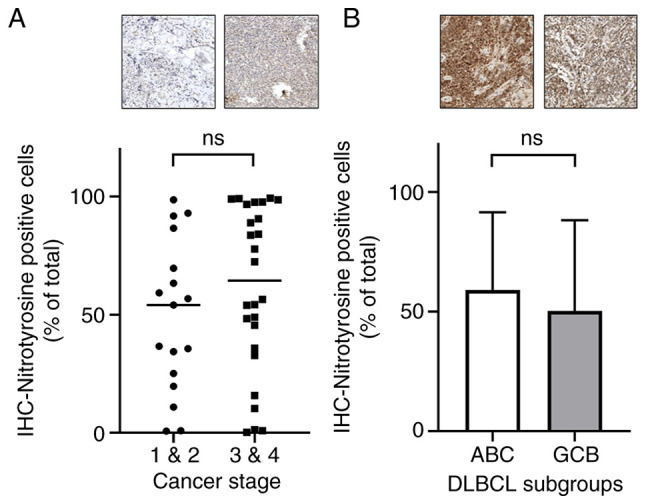
Nitrotyrosine expression in different DLBCL stages and subgroups. (A) Nitrotyrosine-positive cells were more prevalent in the advanced stages of DLBCL. (B) Higher frequency of nitrotyrosine-positive cells was observed in the ABC subtype of DLBCL. All IHC sample images are ×20 magnification. ns, no significance; DLBCL, diffuse large B-cell lymphoma; IHC, immunohistochemistry.
Clinical differences between ABC and GCB subtypes of DLBCL
Given the classification of DLBCL into ABC and GCB subtypes, a comparative analysis of the clinical data was performed to elucidate the differences between these subgroups. Table I presents the clinical characteristics of the ABC and GCB subtypes. The findings revealed that the ABC subtype was significantly associated with a lower white blood cell (WBC) count compared with those with the GCB subtype (P=0.0096). Moreover, patients in the ABC subtype underwent a significantly greater number of chemotherapy cycles compared with those with the GCB subtype (P=0.0173). Additionally, several clinical parameters, including Eastern Cooperative Oncology Group performance status (33), overall survival (OS), progression-free survival (PFS), platelet count, lactate dehydrogenase, albumin, glutamic oxaloacetic transaminase (GOT), glutamic-pyruvic transaminase (GPT), blood urea nitrogen, creatinine and ionized calcium levels, were markedly lower in the ABC subtype compared with that of the GCB subtype. These data indicate that the ABC subgroup presented with worse clinical values.
Table I.
Basic clinical characteristics of the diffuse large B-cell lymphoma activated B-cell and germinal center B-cell subtypes.
| Clinical characteristic | ABC (n=30) | GCB (n=13) | P-value |
|---|---|---|---|
| Sex | 0.1854 | ||
| Male | 13 | 9 | |
| Female | 17 | 4 | |
| Age, years | 58.5 | 62.5 | 0.4313 |
| Ann Arbor stage | 0.4970 | ||
| 1-2 | 10 | 4 | |
| 3-4 | 20 | 9 | |
| ECOG performance status | 0.3607 | ||
| <2 | 25 | 10 | |
| ≥2 | 3 | 3 | |
| N.D. | 2 | 0 | |
| OS, months | 33.0±30.8 | 34.1±43.0 | 0.9240 |
| PFS, months | 28.2±32.0 | 34.1±43.0 | 0.6215 |
| HBsAg | 0.6972 | ||
| Positive | 6 | 2 | |
| Negative | 19 | 10 | |
| N.D. | 5 | 1 | |
| HBcAb | 0.4340 | ||
| Positive | 15 | 8 | |
| Negative | 10 | 2 | |
| N.D. | 5 | 3 | |
| HCV | 0.6594 | ||
| Positive | 4 | 3 | |
| Negative | 21 | 9 | |
| N.D. | 5 | 1 | |
| B symptomsb | 0.2953 | ||
| Positive | 14 | 4 | |
| Negative | 11 | 8 | |
| N.D. | 5 | 1 | |
| WBC, µl | 6,189.6±1,912.2 | 9,545.0±5,543.8 | 0.0096a |
| Platelet, ×103/µl | 230.5±118.9 | 257.7±129.1 | 0.5311 |
| LDH, U/l | 378.6±437.6 | 673.5±1091.9 | 0.2459 |
| β-2 microglobulin, µg/l | 435.9±483.1 | 306.9±215.9 | 0.4055 |
| Albumin, g/dl | 3.9±0.6 | 3.9±0.4 | 0.8598 |
| GOT, U/l | 31.6±18.9 | 45.9±46.2 | 0.1860 |
| GPT, U/l | 26.9±24.7 | 30.1±19.4 | 0.6960 |
| Blood urea nitrogen, mg/dl | 16.7±9.6 | 19.4±14.4 | 0.4981 |
| Creatinine, mg/dl | 1.38±2.1 | 1.52±2.2 | 0.8558 |
| Ionized calcium, mg/dl | 4.68±0.4 | 4.94±0.4 | 0.0945 |
| Chemotherapy courses | 5.93±2.25 | 3.75±3.05 | 0.0173a |
Data are presented as n or mean ± standard deviation.
P<0.05.
B symptoms: Fevers >38°C for at least 3 consecutive days, night sweats and body weight loss >10% during the 6 months prior to diagnosis. ABC, activated B-cell; GCB, germinal center B-cell; ECOG, Eastern Cooperative Oncology Group; OS, overall survival; PFS, progression-free survival; HBsAg, hepatitis B surface antigen; HBcAb, Hepatitis B core antibody; HCV, Hepatitis C virus; WBC, white blood cell; LDH, lactate dehydrogenase; GOT, glutamate oxaloacetate transaminase; GPT, glutamate pyruvate transaminase; N.D., not detected.
Patients with nuclear expression of Nrf2 have worse clinical biomarkers
Previous findings indicated the presence of nuclear Nrf2 in the ABC subtype of DLBCL (Fig. 2C). To further assess the impact of nuclear Nrf2 expression on patient outcomes, clinical data were collected and analyzed by dividing patients into two groups based on the presence or absence of nuclear Nrf2. The comparison revealed significant associations with several clinical characteristics (Table II). Specifically, hepatitis B surface antigen (HBsAg), body weight loss (BWL) and ionized calcium were significantly higher in the nuclear Nrf2-positive group than in the Nrf2-negative group (P<0.05). Additionally, there was a marked trend towards lower WBC and platelet counts in the nuclear Nrf2-positive group compared with that of the nuclear Nrf2-negative group, whilst higher levels of GOT and GPT were observed in the nuclear Nrf2-positive group compared with that of the nuclear Nrf2-negative group. These findings demonstrate that nuclear Nrf2 expression had a notable impact on the clinical biomarkers of patients with DLBCL.
Table II.
Clinical analysis of nuclear factor erythroid 2-related factor 2 expression in diffuse large B-cell lymphoma.
| Clinical characteristic | Nuclear Nrf2-positive | Nuclear Nrf2-negative | P-value |
|---|---|---|---|
| Subtype | 0.0196a | ||
| ABC | 10 | 20 | |
| GCB | 0 | 13 | |
| HBsAg | 0.0487a | ||
| Positive | 4 | 4 | |
| Negative | 4 | 25 | |
| N.D. | 2 | 4 | |
| BWL | 0.0421a | ||
| Yes | 6 | 9 | |
| No | 2 | 20 | |
| N.D. | 2 | 4 | |
| WBC, µl | 6471.3±1290.6 | 7500.0±4220.8 | 0.5043 |
| Platelet, ×103/µl | 201.1±83.8 | 239.8±136.6 | 0.3207 |
| GOT, U/l | 37.1±29.5 | 36.0±31.5 | 0.9621 |
| GPT, U/l | 35.4±38.0 | 25.9±17.1 | 0.3044 |
| Ionized calcium, mg/dl | 5.1±0.4 | 4.7±0.4 | 0.0134a |
Data are presented as n or mean ± standard deviation.
P<0.05. Nrf2, nuclear factor erythroid 2-related factor 2; ABC, activated B-cell; GCB, germinal center B-cell; HBsAg, hepatitis B surface antigen; BWL, body weight loss; WBC, white blood cell; GOT, glutamate oxaloacetate transaminase; GPT, glutamate pyruvate transaminase; N.D., not detected.
Discussion
DLBCL, the most common subtype of non-Hodgkin's lymphoma in Asia, is regarded as a severe form of non-Hodgkin's lymphoma and is increasing in incidence (34). DLBCL can be further categorized into ABC and GCB subtypes based on cell of origin and gene expression analysis (35). The frequency of the ABC subtype is 60–70% in Asian countries, which is markedly higher than the 37–40% observed in Western countries, and it is associated with a worse prognosis (36,37). The distinction between ABC and GCB subtypes notably impacts the prognosis of patients with DLBCL (38), as the expression of antioxidant genes in cancer cells can attenuate the efficacy of drug therapy, leading to drug resistance and relapse (39). Despite extensive research on DLBCL, there are limited studies focusing on the association between DLBCL subtypes, antioxidant genes like Nrf2 and Keap1, and clinical data. Therefore, the present study assessed the relationship between the different DLBCL subtypes (ABC and GCB), clinical data and the expression of Nrf2, Keap1, nitrotyrosine and LC3B.
Nrf2, a transcription factor with antioxidant capabilities, activates in the nucleus and is associated with worse treatment outcomes and prognosis in cancer therapy (30). In the dataset in the present study, ~23.26% (10/43) of DLBCL tissue specimens exhibited nuclear Nrf2 expression in tumor cells, particularly in the ABC subtype with worse prognosis, where it accounted for 33.33% (10/33), potentially contributing to chemotherapy resistance and worsening treatment outcomes. Previous studies have reported an association between Nrf2 expression and drug resistance in cancers, as well as its association with clinical characteristics (40). Generally, higher Nrf2 expression is associated with a worse patient prognosis, suggesting its potential as a cancer biomarker (30,39,41). The clinical data analysis in the present study revealed a higher prevalence of HBsAg in patients with nuclear Nrf2 expression, indicating a potential link between Nrf2 activation and HBsAg presence in DLBCL. Previous studies have also reported that Hepatitis B virus (HBV) regulatory proteins hepatitis B virus X protein (HBx) and large hepatitis B virus surface protein activate Nrf2 via the c-Raf and MEK pathways, thereby protecting HBV-positive cells from oxidative damage (42–44). Moreover, previous studies reported that a marked reduction in HBsAg release was associated with decreased Nrf2 activity (45,46). Conversely, other research suggested that during HBV infection, ROS production was induced, with HBsAg contributing to ROS formation. HBx further activated Nrf2 by increasing its protein levels and enhancing nuclear localization. However, excessive Nrf2 expression markedly suppressed HBV core promoter activity, leading to reduced viral replication (47,48). These findings suggest that nuclear Nrf2 activation, associated with HBsAg presence in DLBCL, may protect cancer cells from oxidative stress, indicating its potential as a biomarker for identifying patients with HBsAg-positive DLBCL. Furthermore, in the present study, patients with DLBCL with nuclear Nrf2 expression exhibited significant weight loss, suggesting that Nrf2 activation may contribute to this outcome. Previous studies using a diabetic mouse (db/db) model reported that Nrf2 activation by the inducer CDDO-Im markedly suppressed high-fat diet-induced obesity and alleviated diabetes, leading to weight loss. This is a process reversed by Nrf2 gene disruption (49,50). This mechanism involves insulin secretion by β-cells, where ROS generated from glucose metabolism activate Nrf2. In turn, Nrf2 stimulates the expression of downstream genes such as glucose-6-phosphate dehydrogenase and phosphogluconate dehydrogenase in the pentose phosphate pathway, producing NADPH, which enhances insulin secretion, glucose metabolism and energy expenditure, ultimately affecting body weight (51,52). The association between nuclear Nrf2 expression and weight loss in patients with DLBCL suggests that Nrf2 activation may influence metabolism, reinforcing its potential as a biomarker for assessing clinical outcomes.
Under normal conditions, the transcription factor Nrf2 is bound by the Keap1-dependent E3 ubiquitin ligase complex, which includes Keap1, Cullin 3 and RING box protein 1, along with an E2 ubiquitin-conjugating enzyme. This interaction facilitates the ubiquitination and subsequent proteasomal degradation of Nrf2. However, under stress conditions, sensor cysteine residues within Keap1 are modified, allowing Nrf2 to escape degradation, translocate to the nucleus and initiate its antioxidant transcriptional program (53). The findings of the present study, consistent with those reported by Yi et al (41), demonstrated significant Keap1 expression in DLBCL, particularly in advanced stages of the disease. In the ABC subtype, which is characterized by elevated oxidative stress (54), Nrf2 predominantly localized to the nucleus and was positively associated with HBsAg expression and BWL, suggesting increased oxidative stress in this subtype. Furthermore, despite elevated Keap1 expression in ABC subtype, its regulatory function appeared to be impaired, resulting in enhanced nuclear translocation of Nrf2. This dysregulation likely contributed to the activation of aberrant metabolic and stress response pathways in ABC, negatively influencing clinical outcomes. In contrast, Nrf2 was primarily confined to the cytoplasm in the GCB subtype, indicating that the inhibitory function of Keap1 remained intact. This suggests that the Keap1-Nrf2 axis was more effectively regulated in GCB, allowing for a more controlled cellular response to oxidative stress. These findings highlight substantial differences in the regulation of the Nrf2-Keap1 pathway between the ABC and GCB subtypes of DLBCL. In ABC, the compromised inhibitory function of Keap1 may serve a pivotal role in disease progression by promoting oxidative stress-induced cellular damage, whereas in GCB, Keap1 retained its suppressive role, potentially contributing to the less aggressive disease phenotype observed in this subtype. Previous studies have reported abnormal expression of Keap1 and Nrf2 in solid tumors, possibly due to mutations in the NFE2L2 or KEAP1 genes, preventing Keap1 from binding to Nrf2, thereby allowing Nrf2 to enter the nucleus and be activated (55,56). Additionally, abnormalities in NFE2L2 and KEAP1 in DLBCL may not solely result from genetic mutations but may involve other factors. Both GCB and ABC DLBCL cell lines exhibit autophagy-dependent characteristics when treated with autophagy inhibitors, with no difference in LC3B expression between subtypes (57). The results of the present study also indicated a positive correlation between Nrf2 and LC3B. As a major autophagy-related gene, LC3B expression is regulated by p62, which promotes autophagy generation and regulates LC3B expression, whilst also activating Nrf2 by bypassing Keap1 through non-canonical pathways (31,32,58). In summary, abnormalities in the NFE2L2 or KEAP1 genes may lead to Nrf2 activation without Keap1 inhibition and may also be regulated by non-canonical pathways involving sequestosome 1 (SQSTM1/p62). These possibilities suggest the activation of nuclear Nrf2 and cytoplasmic Keap1 expression in ABC subtype DLBCL. Nrf2 induction is associated with ROS generation. In the present study, immunohistochemical analysis revealed no differences in ROS levels across different cancer stages or DLBCL subtypes. This may be due to ROS being detected by nitrotyrosine, which specifically represents tyrosine nitration and may not comprehensively reflect total ROS, potentially introducing measurement bias (59). Additionally, the similar ROS expression indicates that the amount of ROS generated under oxidative stress is consistent across different DLBCL subtypes. Alternatively, whilst the ROS levels appear comparable, the activated Nrf2 in the ABC subtype may have already suppressed a portion of the ROS. Consequently, the original ROS levels in the ABC subtype would be higher; however, the retrospective data of the present study cannot demonstrate this.
Patients with DLBCL of the ABC subtype have a worse prognosis, as confirmed by the analysis of patient medical records in the present study. Despite being older on average, patients with the DLBCL GCB subtype demonstrated better OS and PFS compared with those with the ABC subtype (Table I). Furthermore, patients with GCB exhibited higher WBC counts and required fewer chemotherapy sessions, indicating better treatment outcomes in patients with DLBCL GCB than in patients with DLBCL ABC. The worse prognosis of patients with the DLBCL ABC subtype compared with those with the GCB subtype may be attributed to differences in nuclear Nrf2 expression. Whilst the GCB subtype exhibited cytoplasmic accumulation of Nrf2, this inactive form did not influence the disease progression in GCB patients. This hypothesis is supported by the clinical data in Table II, which demonstrates that patients with nuclear Nrf2 expression have worse clinical outcomes.
The present study has certain limitations. The small sample size of patients with the GCB subtype limits the generalizability of the conclusions regarding Nrf2 expression. The difficulty in obtaining pathological specimens and the lack of complete clinical records had also contributed to the inability of the present study to collect a larger sample size. Additionally, in Asian countries, the ratio of the DLBCL ABC subtype to GCB subtype is ~2:1 (37), which further explains the limited number of GCB subtype samples in the present study. Despite the smaller sample size, the present study demonstrated certain trends in Nrf2 expression in patients with ABC subtype DLBCL were associated with clinical data, warranting further investigation. However, there was no associated demonstrated between distinct genes, such as Nrf2 and Keap1, and there were no statistically significant differences in OS between the DLBCL ABC and GCB subtypes in the cohort due to the limited sample size and salvage therapy. Typically, patients with the ABC subtype receive salvage therapy after disease progression, and the diverse treatment options, including novel pharmacological agents, hematopoietic stem cell transplantation and cellular therapy, may also contribute to the absence of statistically significant differences in OS between the subtypes (9,10,13). Furthermore, whilst IHC staining was performed using automated staining equipment with control groups for comparison, the staining intensity remains subject to interpretation. Moreover, gene expression levels were analyzed by calculating the % stained cells, and human subjective decisions were still required to define the regions of interest for calculating changes in cell staining even with automated quantification using the HistoQuest software. Efforts were made to reduce bias by randomly selecting ≥3 regions of interest for averaging. Future directions should focus on efficient computer-based identification to reduce subjectivity and potential bias. In future studies, multi-center and prospective research should be performed to collect a larger sample size and more diverse patient populations. This will enable a more detailed analysis of the differences in Nrf2 expression in DLBCL subtypes and clinical characteristics, and will help mitigate the impact of sample size on the interpretation of results, thereby validating and expanding upon the findings of the present study.
In conclusion, the proportion of Nrf2-positive cells was predominantly distributed in advanced stages of DLBCL and the GCB subtype, correlating with LC3B expression, whereas activated nuclear Nrf2 was exclusively detected in the nuclei of cells within the ABC subtype. Patients with nuclear Nrf2-positive DLBCL were more frequently associated with clinical symptoms such as HBsAg positivity and significant BWL. Additionally, Keap1 expression increased with disease progression and was significantly elevated in the GCB subtype. These findings suggest that the oxidative stress marker Nrf2 may serve as a potential biomarker for the ABC subtype in DLBCL, aiding in the identification of therapeutic targets.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Ming-Yen Lin from the Division of Medical Statistics and Bioinformatics, Department of Medical Research, Kaohsiung Medical University Hospital, Kaohsiung Medical University for assisting with correctly applying statistical methods. The authors would also like to thank Ms. Li Li Lin from the Biobank, Kaohsiung Medical University Hospital, Department of Medical Research, Kaohsiung Medical University Hospital, Kaohsiung Medical University, for her assistance with the IHC stain.
Funding Statement
The present study was supported by the Kaohsiung Medical University Hospital [grant nos. KMUH109-M907, KMUH111-1R16, KMUH111-1M17, KMUH112-2M12, KMUH112-2M13 and KMUH-DK(B)111002-3] and the Ministry of Health and Welfare (grant no. MOHW112-TDU-B-212-144006).
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
CMH, KCC and HHH designed the present study and contributed to the conceptualization. HHH contributed to the project administration. CMH and CHY cultured the DLBCL cell lines. CHY, YT and LCH performed qPCR. CHY, YT, SYH and LCH scanned the IHC-stained samples using TissueFAXS PLUS. SYH, LCH, SYK, CEH, YT and KCC quantified the IHC staining through HistoQuest analysis. SFC, HCW, TJY, JSD, MHW, TYH and YCL collected the clinical data. SYK, HCW, KCC and HHH organized the patient data. SFC, HCW, TJY, JSD, MHW, TYH and YCL analyzed the patient data and performed statistical analyses. SYK, CEH, SYH, LCH, KCC and HHH revised and edited the manuscript. CMH, YT and HHH validated the data. CMH wrote the original draft. HHH and CMH confirm the authenticity of all the raw data. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was performed in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Kaohsiung Medical University Chung-Ho Memorial Hospital [approval nos. KMUHIRB-E(I)-20210119 and KMUHIRB-E(I)-20220298]. Due to the retrospective nature of the study, the requirement for informed patient consent was waived.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Park C, Lee HS, Kang KW, Lee WS, Do YR, Kwak JY, Shin HJ, Kim SY, Yi JH, Lim SN, et al. Combination of acalabrutinib with lenalidomide and rituximab in relapsed/refractory aggressive B-cell non-Hodgkin lymphoma: A single-arm phase II trial. Nat Commun. 2024;15:2776. doi: 10.1038/s41467-024-47198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y, Barta SK. Diffuse large B-cell lymphoma: 2019 update on diagnosis, risk stratification, and treatment. Am J Hematol. 2019;94:604–616. doi: 10.1002/ajh.25460. [DOI] [PubMed] [Google Scholar]
- 3.Silkenstedt E, Salles G, Campo E, Dreyling M. Lancet; 2024. B-cell non-Hodgkin lymphomas. [DOI] [PubMed] [Google Scholar]
- 4.Sehn LH, Salles G. Diffuse Large B-Cell Lymphoma. N Engl J Med. 2021;384:842–858. doi: 10.1056/NEJMra2027612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, Araujo IBO, Berti E, Bhagat G, Borges AM, Boyer D, Calaminici M, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia. 2022;36:1720–1748. doi: 10.1038/s41375-022-01620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eriksen PRG, de Groot F, Clasen-Linde E, de Nully Brown P, de Groen R, Melchior LC, Maier AD, Minderman M, Vermaat JSP, von Buchwald C, et al. Sinonasal DLBCL: Molecular profiling identifies subtypes with distinctive prognosis and targetable genetic features. Blood Adv. 2024;8:1946–1957. doi: 10.1182/bloodadvances.2023011517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia M, David L, Teater M, Gutierrez J, Wang X, Meydan C, Lytle A, Slack GW, Scott DW, Morin RD, et al. BCL10 mutations define distinct dependencies guiding precision therapy for DLBCL. Cancer Discov. 2022;12:1922–1941. doi: 10.1158/2159-8290.CD-21-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowakowski GS, LaPlant B, Macon WR, Reeder CB, Foran JM, Nelson GD, Thompson CA, Rivera CE, Inwards DJ, Micallef IN, et al. Lenalidomide combined with R-CHOP overcomes negative prognostic impact of Non-germinal center B-cell phenotype in newly diagnosed diffuse Large B-Cell lymphoma: A phase II study. J Clin Oncol. 2015;33:251–257. doi: 10.1200/JCO.2014.55.5714. [DOI] [PubMed] [Google Scholar]
- 9.Lenz G, Hawkes E, Verhoef G, Haioun C, Thye Lim S, Seog Heo D, Ardeshna K, Chong G, Haaber J, Shi W, et al. Single-agent activity of phosphatidylinositol 3-kinase inhibition with copanlisib in patients with molecularly defined relapsed or refractory diffuse large B-cell lymphoma. Leukemia. 2020;34:2184–2197. doi: 10.1038/s41375-020-0743-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson WH, Young RM, Schmitz R, Yang Y, Pittaluga S, Wright G, Lih CJ, Williams PM, Shaffer AL, Gerecitano J, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21:922–926. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson WH, Gerecitano JF, Goy A, de Vos S, Kenkre VP, Barr PM, Blum KA, Shustov AR, Advani RH, Lih J. The Bruton's tyrosine kinase (BTK) inhibitor, ibrutinib (PCI-32765), has preferential activity in the ABC subtype of relapsed/refractory de novo diffuse large B-cell lymphoma (DLBCL): Interim results of a multicenter, open-label, phase 2 study. Blood. 2012;120:686. doi: 10.1182/blood.V120.21.686.686. [DOI] [Google Scholar]
- 12.Goy A, Ramchandren R, Ghosh N, Munoz J, Morgan DS, Dang NH, Knapp M, Delioukina M, Kingsley E, Ping J, et al. Ibrutinib plus lenalidomide and rituximab has promising activity in relapsed/refractory non-germinal center B-cell-like DLBCL. Blood. 2019;134:1024–1036. doi: 10.1182/blood.2018891598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czuczman MS, Trneny M, Davies A, Rule S, Linton KM, Wagner-Johnston N, Gascoyne RD, Slack GW, Brousset P, Eberhard DA, et al. A Phase 2/3 multicenter, randomized, open-label study to compare the efficacy and safety of lenalidomide versus Investigator's choice in patients with relapsed or refractory diffuse large B-Cell lymphoma. Clin Cancer Res. 2017;23:4127–4137. doi: 10.1158/1078-0432.CCR-16-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitamura H, Motohashi H. NRF2 addiction in cancer cells. Cancer Sci. 2018;109:900–911. doi: 10.1111/cas.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong Z, Xue L, Li H, Fan S, van Hasselt CA, Li D, Zeng X, Tong MCF, Chen GG. Targeting Nrf2 to treat thyroid cancer. Biomed Pharmacother. 2024;173:116324. doi: 10.1016/j.biopha.2024.116324. [DOI] [PubMed] [Google Scholar]
- 16.Barrera-Rodriguez R. Importance of the Keap1-Nrf2 pathway in NSCLC: Is It a possible biomarker? Biomed Rep. 2018;9:375–382. doi: 10.3892/br.2018.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao H, Zhou Q, Zhang Z, Wang Q, Sun Y, Yi X, Feng Y. NRF2 is overexpressed in ovarian epithelial carcinoma and is regulated by gonadotrophin and sex-steroid hormones. Oncol Rep. 2012;27:1918–1924. doi: 10.3892/or.2012.1700. [DOI] [PubMed] [Google Scholar]
- 18.Glorieux C, Enriquez C, Gonzalez C, Aguirre-Martinez G, Buc Calderon P. The Multifaceted Roles of NRF2 in Cancer: Friend or Foe? Antioxidants (Basel) 2024;13:70. doi: 10.3390/antiox13010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker A, Singh A, Tully E, Woo J, Le A, Nguyen T, Biswal S, Sharma D, Gabrielson E. Nrf2 signaling and autophagy are complementary in protecting breast cancer cells during glucose deprivation. Free Radic Biol Med. 2018;120:407–413. doi: 10.1016/j.freeradbiomed.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Liu Z, Hu T, Han L, Yu S, Yao Y, Ruan Z, Tian T, Huang T, Wang M, et al. Nrf2 promotes progression of non-small cell lung cancer through activating autophagy. Cell Cycle. 2017;16:1053–1062. doi: 10.1080/15384101.2017.1312224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang T, Harder B, Rojo de la Vega M, Wong PK, Chapman E, Zhang DD. p62 links autophagy and Nrf2 signaling. Free Radic Biol Med. 2015;88:199–204. doi: 10.1016/j.freeradbiomed.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartolini D, Dallaglio K, Torquato P, Piroddi M, Galli F. Nrf2-p62 autophagy pathway and its response to oxidative stress in hepatocellular carcinoma. Transl Res. 2018;193:54–71. doi: 10.1016/j.trsl.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: An enhanced web server for Large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne U, et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. doi: 10.1016/j.neo.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz AD, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu CM, Chang KC, Chuang TM, Chu ML, Lin PW, Liu HS, Kao SY, Liu YC, Huang CT, Wang MH, et al. High G9a expression in DLBCL and its inhibition by niclosamide to induce autophagy as a therapeutic approach. Cancers (Basel) 2023;15:4150. doi: 10.3390/cancers15164150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu CM, Tsai Y, Wan L, Tsai FJ. Bufalin induces G2/M phase arrest and triggers autophagy via the TNF, JNK, BECN-1 and ATG8 pathway in human hepatoma cells. Int J Oncol. 2013;43:338–348. doi: 10.3892/ijo.2013.1942. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Armitage JO. Staging non-Hodgkin lymphoma. CA Cancer J Clin. 2005;55:368–376. doi: 10.3322/canjclin.55.6.368. [DOI] [PubMed] [Google Scholar]
- 30.Rojo de la Vega M, Chapman E, Zhang DD. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 32.Zhang C, Ma S, Zhao X, Wen B, Sun P, Fu Z. Upregulation of antioxidant and autophagy pathways via NRF2 activation protects spinal cord neurons from ozone damage. Mol Med Rep. 2021;23:428. doi: 10.3892/mmr.2021.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azam F, Latif MF, Farooq A, Tirmazy SH, AlShahrani S, Bashir S, Bukhari N. Performance status assessment by using ECOG (Eastern Cooperative Oncology Group) score for cancer patients by oncology healthcare professionals. Case Rep Oncol. 2019;12:728–736. doi: 10.1159/000503095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang SS. Epidemiology and etiology of diffuse large B-cell lymphoma. Semin Hematol. 2023;60:255–266. doi: 10.1053/j.seminhematol.2023.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 36.Nowakowski GS, Chiappella A, Witzig TE, Scott DW, Spina M, Gascoyne RD, Zhang L, Russo J, Kang J, Zhang J, et al. Variable global distribution of cell-of-origin from the ROBUST phase III study in diffuse large B-cell lymphoma. Haematologica. 2020;105:e72–e75. doi: 10.3324/haematol.2019.220475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiozawa E, Yamochi-Onizuka T, Takimoto M, Ota H. The GCB subtype of diffuse large B-cell lymphoma is less frequent in Asian countries. Leuk Res. 2007;31:1579–1583. doi: 10.1016/j.leukres.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 38.Pileri SA, Tripodo C, Melle F, Motta G, Tabanelli V, Fiori S, Vegliante MC, Mazzara S, Ciavarella S, Derenzini E. Predictive and prognostic molecular factors in diffuse large B-Cell Lymphomas. Cells. 2021;10 doi: 10.3390/cells10030675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yen CH, Hsiao HH. NRF2 is one of the players involved in bone marrow mediated drug resistance in multiple myeloma. Int J Mol Sci. 2018;19:3503. doi: 10.3390/ijms19113503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi X, Zhao Y, Xue L, Zhang J, Qiao Y, Jin Q, Li H. Expression of Keap1 and Nrf2 in diffuse large B-cell lymphoma and its clinical significance. Exp Ther Med. 2018;16:573–578. doi: 10.3892/etm.2018.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaedler S, Krause J, Himmelsbach K, Carvajal-Yepes M, Lieder F, Klingel K, Nassal M, Weiss TS, Werner S, Hildt E. Hepatitis B virus induces expression of antioxidant response element-regulated genes by activation of Nrf2. J Biol Chem. 2010;285:41074–41086. doi: 10.1074/jbc.M110.145862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Severi T, Vander Borght S, Libbrecht L, VanAelst L, Nevens F, Roskams T, Cassiman D, Fevery J, Verslype C, van Pelt JF. HBx or HCV core gene expression in HepG2 human liver cells results in a survival benefit against oxidative stress with possible implications for HCC development. Chem Biol Interact. 2007;168:128–134. doi: 10.1016/j.cbi.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Basic M, Thiyagarajah K, Glitscher M, Schollmeier A, Wu Q, Gorgulu E, Lembeck P, Sonnenberg J, Dietz J, Finkelmeier F, et al. Impaired HBsAg release and antiproliferative/antioxidant cell regulation by HBeAg-negative patient isolates reflects an evolutionary process. Liver Int. 2024;44:2773–2792. doi: 10.1111/liv.16048. [DOI] [PubMed] [Google Scholar]
- 45.Peiffer KH, Akhras S, Himmelsbach K, Hassemer M, Finkernagel M, Carra G, Nuebling M, Chudy M, Niekamp H, Glebe D, et al. Intracellular accumulation of subviral HBsAg particles and diminished Nrf2 activation in HBV genotype G expressing cells lead to an increased ROI level. J Hepatol. 2015;62:791–798. doi: 10.1016/j.jhep.2014.11.028. [DOI] [PubMed] [Google Scholar]
- 46.Kalantari L, Ghotbabadi ZR, Gholipour A, Ehymayed HM, Najafiyan B, Amirlou P, Yasamineh S, Gholizadeh O, Emtiazi N. A state-of-the-art review on the NRF2 in Hepatitis virus-associated liver cancer. Cell Commun Signal. 2023;21:318. doi: 10.1186/s12964-023-01351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ariffianto A, Deng L, Abe T, Matsui C, Ito M, Ryo A, Aly HH, Watashi K, Suzuki T, Mizokami M, et al. Oxidative stress sensor Keap1 recognizes HBx protein to activate the Nrf2/ARE signaling pathway, thereby inhibiting hepatitis B virus replication. J Virol. 2023;97:e0128723. doi: 10.1128/jvi.01287-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bender D, Hildt E. Effect of hepatitis viruses on the Nrf2/Keap1-signaling pathway and its impact on viral replication and pathogenesis. Int J Mol Sci. 2019;20:4659. doi: 10.3390/ijms20184659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uruno A, Furusawa Y, Yagishita Y, Fukutomi T, Muramatsu H, Negishi T, Sugawara A, Kensler TW, Yamamoto M. The Keap1-Nrf2 system prevents onset of diabetes mellitus. Mol Cell Biol. 2013;33:2996–3010. doi: 10.1128/MCB.00225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin S, Wakabayashi J, Yates MS, Wakabayashi N, Dolan PM, Aja S, Liby KT, Sporn MB, Yamamoto M, Kensler TW. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-imidazolide. Eur J Pharmacol. 2009;620:138–144. doi: 10.1016/j.ejphar.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baumel-Alterzon S, Katz LS, Brill G, Garcia-Ocana A, Scott DK. Nrf2: The master and captain of beta cell fate. Trends Endocrinol Metab. 2021;32:7–19. doi: 10.1016/j.tem.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.David JA, Rifkin WJ, Rabbani PS, Ceradini DJ. The Nrf2/Keap1/ARE pathway and oxidative stress as a therapeutic target in type II diabetes mellitus. J Diabetes Res. 2017;2017:4826724. doi: 10.1155/2017/4826724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baird L, Yamamoto M. The molecular mechanisms regulating the KEAP1-NRF2 Pathway. Mol Cell Biol. 2020;40:e00099–20. doi: 10.1128/MCB.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mai Y, Yu JJ, Bartholdy B, Xu-Monette ZY, Knapp EE, Yuan F, Chen H, Ding BB, Yao Z, Das B, et al. An oxidative stress-based mechanism of doxorubicin cytotoxicity suggests new therapeutic strategies in ABC-DLBCL. Blood. 2016;128:2797–2807. doi: 10.1182/blood-2016-03-705814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zavitsanou AM, Pillai R, Hao Y, Wu WL, Bartnicki E, Karakousi T, Rajalingam S, Herrera A, Karatza A, Rashidfarrokhi A, et al. KEAP1 mutation in lung adenocarcinoma promotes immune evasion and immunotherapy resistance. Cell Rep. 2023;42:113295. doi: 10.1016/j.celrep.2023.113295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mandhair HK, Radpour R, Westerhuis M, Banz Y, Humbert M, Arambasic M, Dengjel J, Davies A, Tschan MP, Novak U. Analysis of autophagy in DLBCL reveals subtype-specific differences and the preferential targeting of ULK1 inhibition in GCB-DLBCL provides a rationale as a new therapeutic approach. Leukemia. 2024;38:424–429. doi: 10.1038/s41375-024-02147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V, et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856–880. doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cadenas-Garrido P, Schonvandt-Alarcos A, Herrera-Quintana L, Vazquez-Lorente H, Santamaria-Quiles A, Ruiz de Francisco J, Moya-Escudero M, Martin-Oliva D, Martin-Guerrero SM, Rodriguez-Santana C, et al. Using redox proteomics to gain new insights into neurodegenerative disease and protein modification. Antioxidants (Basel) 2024;13:127. doi: 10.3390/antiox13010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in the present study may be requested from the corresponding author.