Abstract
Antibiotics are essential components of current medical practice, but their effectiveness is being eroded by the increasing emergence of antimicrobial-resistant infections. At the same time, the rate of antibiotic discovery has slowed, and our future ability to treat infections is threatened. Among Christopher T. Walsh’s many contributions to science was his early recognition of this threat and the potential of biosynthesis—genes and mechanisms—to contribute solutions. Here, we revisit a 2006 review by Walsh and co-workers that highlighted a major challenge in antibiotic natural product discovery: the daunting odds for identifying new naturally occurring antibiotics. The review described strategies to mitigate the odds challenge. These strategies have been used extensively by the natural product discovery community in the years since and have resulted in some promising discoveries. Despite these advances, the rarity of novel antibiotic natural products remains a barrier to discovery. We compare the challenge of discovering natural product antibiotics to the process of identifying new prime numbers, which are also challenging to find and an essential, if underappreciated, element of modern life. We propose that inclusion of filters for functional compounds early in the discovery pipeline is key to natural product antibiotic discovery, review some recent advances that enable this, and discuss some remaining challenges that need to be addressed to make antibiotic discovery sustainable in the future.
Keywords: Antibiotic discovery, Antimicrobial resistance, Natural products, Phenotypic screens, Functional compounds, Discovery pipeline
Emergence of Antibiotic Resistance and Need for New Antibiotics
Many readers of this journal are familiar with Christopher T. Walsh and the enormous body of creative research that forms his scientific legacy. If you read an article he wrote or a lecture he gave this century, you encountered the issue of antimicrobial-resistant (AMR) infections, which are a growing threat to human health. Antimicrobial resistance is inevitable, as introduction of an antibiotic creates an existential challenge for any antibiotic-sensitive bacteria, and clinical introduction creates multiple opportunities to develop resistance.1 The time it takes for this resistance to emerge varies—for example, it took less than a year between the introduction of methicillin and the identification of methicillin-resistant Staphylococcus aureus (MRSA) but a longer time, 30–44 years, between the introduction of vancomycin and the identification of vancomycin resistance in Enterococcus faecalis (VREF).1−3 Today, pathogens with resistance to nearly all antibiotics in our arsenal are already circulating among the population. Of particular concern are multi-drug-resistant pathogens, which are resistant to more than one class of antibiotic.1
One way to combat antimicrobial resistance is to discover new antibiotics. The larger the arsenal of antibiotics at our disposal, the lower the chance that a patient will acquire an infection with resistance to all known antibiotics. In addition, there is often a fitness cost for bacteria to maintain resistance in the absence of antibiotics,4−7 and there might be a theoretical limit to how many antibiotics a single strain can be resistant to, especially in cases where resistance genes or mutations have a negative epistatic relationship.8 A larger arsenal also enables other strategies that have been proposed to mitigate the development of AMR, such as reserving antibiotics as antibiotics of last resort9 and using different, or fewer, antibiotics in agricultural and human health settings.10,11 Ideally, these new antibiotics would have mechanisms of action that are not used by the current antibiotics. If the new antibiotics do not have novel mechanisms, they are more likely to be ineffective against existing pathogens already resistant to that mechanism of action. Antibiotics have historically been discovered by two approaches—either through the screening of natural or synthetic compounds in phenotypic or other screens12,13 or through structure-based drug design against a specific target thought to be essential for bacterial survival.14,15 Hits from phenotypic screens could have novel or known mechanisms of action, while screens against specific targets require a predetermined mechanism of action. Therefore, future antibiotic discovery efforts will largely rely on phenotypic screens, methods to identify previously untargeted bacterial proteins such as transposon library mutagenesis,16 or development of antibiotics that simultaneously target multiple targets.17,18
The Numbers Game of Natural Product Antibiotic Discovery
During the golden age of antibiotic discovery in the 1940s and 1950s, many natural product antibiotics were discovered through phenotypic screens for antimicrobial activity.19−21 Natural products have been modified by evolution over billions of years to help their producers adapt to and compete in their natural environments. One function of natural products is to serve as defensive agents22,23 for their producer or, in the case of symbionts, defensive agents for the producer’s host.24−27 Therefore, natural products are uniquely privileged as a source of antibiotic compounds. Screens of natural products often have higher hit rates than screens of synthetic molecule libraries,28 and natural products are more likely to succeed in later clinical trial phases29 and account for a large fraction of FDA-approved antibiotics.30 Natural products also act by various mechanisms of action; therefore, hits in phenotypic antibiotic screens of natural products can be used to discover new antibacterial targets.
Following
the end of the golden age of antibiotic discovery in
the 1960s, the rate of natural product antibiotic discovery and discovery
of novel natural products occupying novel regions of chemical space
slowed significantly.19,31 We have largely depended on modifications
of known antibiotics to provide new candidates for drug development.
A typical example is the development of azithromycin from erythromycin,
and while azithromycin extended the useful lifetime of macrolide antibiotics,
azithromycin resistance is a growing concern.32,33 While active compounds are still discovered, only a handful have
entered clinical development, and they generally fall into established
classes that were first discovered during the golden age. Only four
new classes (lipopeptides, lipiarmycins, oxazolidinones, and diarylquinolines)
that were discovered after 1970 have been approved by the FDA for
use in humans.19 This is in contrast to
the many classes of antibiotics discovered during the golden age,
which include the β-lactams, tetracyclines, macrolides, aminoglycosides,
glycopeptides, amphenicols, ansamycins, and streptogramins.19,21,34 There are multiple reasons for
this, but one is how rarely compounds with the sort of significant
activity that could lead to a clinically useful agent are encountered.
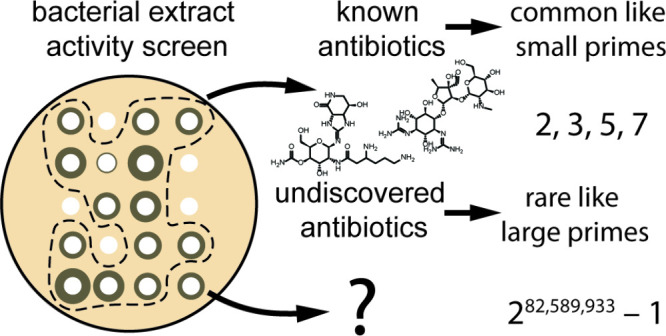
The problem of discovering new antibiotics from natural sources is
not unlike the problem of discovering new prime numbers. Small prime
numbers are relatively common, for example, 2, 3, 5, and 7, and increasingly
rare as they get higher. Specifically, prime numbers are distributed
asymptotically according to the prime number theorem; that is, the
number of prime numbers less than or equal to N is
approximately 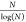 for
sufficiently large N.35 If one counts upward and notes all
prime numbers they encounter, they will have considerable success
early on compared to their later efforts, which mimics the way in
which more strains need to be screened to discover new natural product
antibiotics after the most common ones have already been found. One
possibly comforting aspect of this analogy is that there is an infinite
number of primes, but it takes considerable effort to find them. The
largest known prime has 24,862,048 digits when written in base ten.36
for
sufficiently large N.35 If one counts upward and notes all
prime numbers they encounter, they will have considerable success
early on compared to their later efforts, which mimics the way in
which more strains need to be screened to discover new natural product
antibiotics after the most common ones have already been found. One
possibly comforting aspect of this analogy is that there is an infinite
number of primes, but it takes considerable effort to find them. The
largest known prime has 24,862,048 digits when written in base ten.36
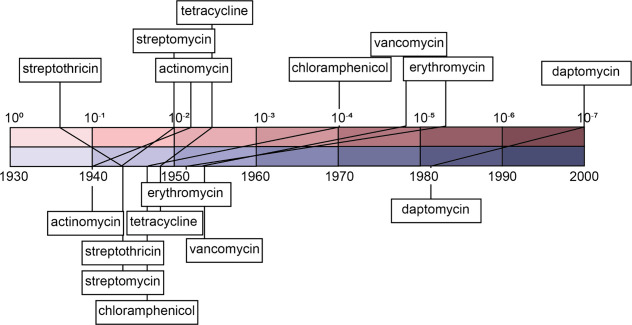
The rarity of naturally occurring antibacterial compounds varies widely. In their 2006 review, Walsh and co-authors assembled existing studies that sought to estimate how common various antibiotics were across actinomycetes and streptomycetes.37 In order of increasing rarity, these estimated frequencies were 2 × 10–1 (streptothricin), 1 × 10–2 (streptomycin), 4 × 10–3 (tetracyclines),1.5 × 10–5 (vancomycin), 5 × 10–6 (erythromycin), and 1 × 10–7 (daptomycin).38−40 Other antibiotics for which the order of magnitude of their frequencies have been estimated include actinomycins, which are used clinically for their antitumor activity but also have antibacterial activity (between 10–2 and 10–3), and chloramphenicol (between 10–4 and 10–5).38,39 These frequencies track roughly with the years that the antibiotics were discovered: 1940 (actinomycins), 1943 (streptothricin and streptomycin), 1947 (chloramphenicol), 1948 (tetracycline), 1952 (erythromycin), 1953 (vancomycin), and the early 1980s (daptomycin) (Figure 1).19,41 This is expected because fewer strains need to be screened to discover more common antibiotics; therefore, these more common antibiotics are discovered earlier. It should be noted that the calculated frequency values could be underestimated because of how commonly biosynthetic gene clusters (BGCs) are silent.42 But we anticipate that this causes only a minor error in the estimated frequencies, because many historical screening efforts have cultured bacteria under multiple conditions that may activate silent BGCs42−44 and different strains are known to regulate their BGCs differently,45 meaning that if a BGC is common, its product is likely to be discovered quickly, even if it is silent in some strains.46 The estimated frequencies suggest that any antibiotics with suitable activity and chemical properties to be used as therapeutics from actinomycetes that are yet to be discovered will have frequencies less than 1 × 10–7 or that they are distributed mostly in strains that have not yet been isolated or cultured. Therefore, discovery of truly new antibiotics becomes exponentially more difficult as more antibiotics are discovered before leveling off, as it is estimated that there are many antibiotics with low frequency.39,47 The 2006 Walsh review offered suggestions for tackling the problem of increasing rarity of uncharacterized natural antibiotics, including screening actinomycetes other than Streptomyces, developing new culture methods to isolate previously uncultured strains, heterologous expression of BGCs from isolated genomes or environmental metagenomes, combinatorial biosynthesis, and structure-based drug design of hybrid molecules.37
Figure 1.
Antibiotic frequency and timeline of discovery. The estimated frequency of antibiotics among Streptomyces is plotted on the top of the image on a base ten logarithmic scale; for actinomycin and chloramphenicol, only a range was reported, so the positions of those antibiotics within the range are arbitrary. The lower part of the image shows the corresponding year of discovery plotted on a linear scale.
In the 18 years since the 2006 Walsh review article, the field has generally followed all the proposed strategies while also developing some new strategies. There have been significant technological advancements in culturing technologies, heterologous expression of BGCs identified in metagenomes or eDNA, combinatorial biosynthesis, and hybrid molecules. Some notable success stories resulting from the use of these techniques include teixobactin, discovered using culturing technology,48 and hybrid molecules such as macrolones,17 although it remains to be seen if these examples can be translated into therapeutics. The dramatic reduction in costs of DNA sequencing since 2006 has enabled genomic and metagenomic mining strategies.49 An anticipated reduction in the cost of DNA synthesis50 may soon have a similar impact on heterologous expression strategies.
In addition, there has been an increasing recognition that just increasing numbers will not be a productive solution; there needs to be a filter early in the pipeline. Ideally the filter would be antibacterial activity—the filter used in the early and highly productive stages of antibiotic discovery: clear zones on a Petri dish. Paul Ehrlich, a Nobel Prize winner who worked in the field of antimicrobial chemotherapy, famously stated that “drugs do not act unless they are bound.”51 Translated into today’s terms, efficient searches for antimicrobials need to contain a filter that indicates a candidate molecule’s potential biological activity. But functional assays and their prerequisites are hard to scale to the number of candidates that need to be considered. Therefore, new strategies that scale to the number of bacteria that need to be screened to discover a new antibiotic (107) are needed.
The Importance of Function in the Discovery Pipeline
Another parallel between the search for prime numbers and antibiotic discovery is the difficulty in verifying that something is, in fact, a prime number or a therapeutically useful antibiotic. For example, the time it takes for AKS algorithm to deterministically verify that a number is prime scales proportional to (log n)6 in one implementation, where n is the prime number.52 Therefore, faster probabilistic methods that can only determine whether a number is highly likely to be prime need to be used to verify prime numbers.53 None of the necessary steps in a traditional assay scale: cultivation, extraction, assays, and purification. Fortunately, unlike prime numbers, the length of time it takes to verify that a novel molecule is an antibiotic does not depend on how many antibiotics have been discovered before. The choice of more efficient methodology in screening, akin to the use of probabilistic methods to verify likely prime numbers, incorporating the correct screens into the antibiotic discovery pipeline, is essential toward determining the success of the approach.
The classical approach to activity screens, applied extensively throughout the history of natural product discovery,54−57 is to first screen a whole extract for activity in either a phenotypic assay or an assay against a specific target. The choice of assay is essential to the likely future success of any active compounds discovered in the screen. Screening against a known target, for which resistant pathogens already exist, is probably not an ideal strategy for discovering antibiotics that will be effective against extensively drug-resistant pathogens. Screening against nonresistant strains may result in the discovery of active compounds that work through established or novel mechanisms. To make it more likely that hits in the screen will be effective against resistant pathogens, strains with multi-drug resistance can be used in phenotypic screens. Differential screening, in which activity is compared between pairs of microbes—either members of the same species, one with a target gene knocked down, or two different species—can be used for dereplication of known mechanisms or to discover narrow-spectrum antibiotics.46 In some cases, a narrow-spectrum antibiotic may be more desirable than a broad-spectrum antibiotic, and to discover these compounds, it is important to use the target pathogen in the screen.58 It is possible that some narrow-spectrum antibiotics are more common than the frequencies discussed above and have been missed due to a historical bias in which species are used during initial screens of whole extracts. Another strategy that might be effective would be to probe the mechanism of action in high throughput during the screening process to avoid discovery of compounds for which a resistance mechanism might already be widespread. High-throughput methods for mechanism-of-action screening have been applied more frequently in anticancer59−61 and antifungal62−65 drug discovery efforts but have also shown promise for antibacterial discovery.66−69
If an extract shows the desired activity, the compounds responsible for such activity can be isolated through bioactivity-guided fractionation, a process by which the complex mixture of natural products undergoes rounds of separation through chromatography or other techniques. Each fraction is tested for activity, and active fractions can be further separated until a single pure active compound remains.70 This process is time-consuming. In addition, bioactivity-guided fractionation will likely fail to identify synergistic antibiotics such as streptogramin A and B71 because unless the synergistic pair has very similar molecular properties, they will likely be in separate fractions. In this case, strong activity will be observed in the crude extract and then be lost during the fractionation process. Identifying the separated synergistic pair in this case would be very challenging. In general, bioactivity-guided fractionation is a slow process that serves as a bottleneck in the discovery process, and new strategies are needed to make the bioactive natural product discovery pipeline more efficient.
Emerging Methods for Screening for Function
There have been multiple recent innovations in the field that have the potential to improve the efficiency of screens for novel antibiotic natural products. These innovations can be broken down into bioinformatic methods that can be used to provide predictions of structural diversity of compounds being screened, bioinformatic methods that link biosynthetic genes to prospective bioactivity, and analytical techniques for dereplication of structural classes and fast linkage of BGCs to their product.
To continue our analogy with the search for prime numbers, we note that prime numbers are the basis for modern encryption methods. Your emails, for example, are probably encrypted by the RSA scheme, which requires two prime numbers to encrypt and decrypt. These numbers need to be large so that a computer trying all primes could not intercept the message and decrypt it. As computers get faster, numbers need to get bigger, in rough analogy to antibiotic resistance. Finding larger primes needs special methods that depend on the tendencies observed in the occurrence of prime numbers. For example, numbers with the general formula (2)n – 1 are more often prime than a randomly selected number. These numbers are called Mersenne primes, after the French friar who discovered them in the 17th century. A Mersenne prime is the current champion for the largest prime mentioned above, and it was discovered in a focused search. For the curious, it is 282,589,933 – 1.36 As useful as Mersenne primes have been to prime discovery, they highlight the limitations of finding infrequently occurring objects. While there are an infinite number of Mersenne primes, only 51 are currently known.72
As discussed above, natural products occur at different frequencies across microbes. For example, streptothricin is 6 orders of magnitude more common than daptomycin38,39 and will therefore also be at least 6 orders of magnitude more common than any antibiotics that have yet to be discovered. This will lead to frequent rediscovery of streptothricin in natural product discovery efforts if Streptomyces continues to be used as the source of extracts in screening efforts. Another issue with using only Streptomyces and other common microbes to build extract libraries for screens is that it will reduce the structural diversity of compounds contained in extract libraries. It is known from screens of synthetic compounds that more diverse libraries lead to more successful screening campaigns.73 One strategy proposed in the 2006 Walsh review is to use more diverse microbes and BGCs from uncultured microbes to improve the diversity of screened compounds. Since 2006, new bioinformatics methods have been developed that can enable a more systematic approach to ensuring libraries of diverse compounds. There are now multiple methods, including BiG-SCAPE,74 BiG-SLiCE,75 clust-o-matic,76 and lsaBGC,77 that enable clustering of BGCs by similarity. This will enable exclusion of microbes that contain BGCs similar to those known as well as selection of microbes in a way that optimizes BGC diversity, which in turn should optimize natural product structural diversity, lowering the rediscovery rate and raising the success rate of screens.
Increasing the compound diversity of extract libraries does not solve the core challenge underlying natural product antibiotic discovery, which is that novel antibiotics are rare. Therefore, strategies that enrich for antibiotic compounds in extract libraries are needed. One such strategy is to identify BGCs with likely antibiotic resistance genes. A drawback of potential antibiotics in natural compounds is the likelihood that resistance genes are also present. The strategy of looking for resistance genes turns this liability into an asset. BGCs that produce antibiotic compounds generally have one or more resistance genes that provide the producer with resistance to the compound that it produces.78 There are multiple databases of resistance genes and software for searching for these genes in BGCs, such as Resistance Gene Identifier (RGI)79 and ARTS.80 A disadvantage of this approach is that many of these resistance markers are specific to a given mechanism (e.g., rRNA methyltransferases that provide resistance to ribosome-targeting antibiotics81) or structural class (e.g., β-lactamases82). Therefore, the use of these methods may not lead to the discovery of antibiotics with novel mechanisms of action. One way to circumvent this is to focus either on resistance genes that are not related to mechanism or on structural class, such as transporters.83 Another approach is to search for duplications of essential genes that are not targeted by known antibiotics in BGCs. Duplicated essential genes in BGCs have previously been shown to be resistance genes, and this strategy was used to identify natural products that target fatty acid synthase from BGCs which contained a putative fatty acid synthase resistance gene.84
With the increasing availability of genomic data, genome mining for BGCs has emerged as a key technology in prioritizing strains and BGCs that are likely to produce novel natural products. However, most genome mining software, such as the very popular antiSMASH,85 and artificial intelligence (AI) methods for identifying BGCs86−88 do not address how likely a BGC is to produce an antibacterial compound. There are also AI methods for predicting antibacterial activity from chemical structure,89,90 but this requires accurate prediction of the structure of a BGC’s product to be useful for genome mining—which still cannot be done reliably across all BGC types. To address this gap, AI and machine learning (ML) techniques that predict the bioactivity of a product from the sequence of the BGC have been developed. At least four such methods have been reported.86,91−93 Three of these methods predict activity directly from annotations of proteins in the BGC86,91,93 and the fourth from the predicted structure of the products, which is predicted from annotations of proteins in the BGC.92 When comparing across the same method, general prediction of antibacterial activity against any bacterial target is the most accurate prediction problem, with 80% accuracy, but prediction of activity against Gram-negative bacteria, which account for most of the AMR urgent threats identified by the CDC, is significantly less accurate, at 70%.91 The lower accuracy of this prediction task is likely due to the fact that there are relatively few known compounds with activity against Gram-negative bacteria, and, in general, the main limitation of these methods is a lack of high-quality training data linking BGCs to the bioactivity of their products.91 For example, a recent study adapting activity predictions for fungal BGCs showed that accuracy was lower for fungal BGCs than bacterial BGCs, likely due to the fact that there is less data available linking fungal BGCs to their products and the products’ activity.94 The accuracy of these methods will likely continue to improve as more training data becomes available and with advancements in AI technology. Even with the limited accuracies of these methods, they only need to outperform random selection to be effective at enriching extract screening libraries for compounds with antibacterial activity. Another approach that does not rely on AI is to search for homologs of biosynthetic genes known to produce reactive groups, such as enediynes,95 which could serve as warheads that react with their molecular target. This approach has been reviewed previously.96
Even if the bioinformatic methods described above are effective at enriching extract libraries for active compounds, it is still challenging to identify and isolate those compounds. Analytical methods for linking compounds to their activity or BGC and methods for dereplication can make the isolation process more efficient. One method for linking metabolites to activity in complex mixtures is NPAnalyst, which combines assay and LC-MS data from different mixtures to identify mass features that are strongly associated with activity and are therefore likely to be responsible for the observed activity.97 Once the compound suspected of activity is known through one of these methods, it can be directly purified, avoiding the need for costly bioactivity-guided fractionation. In addition to linking mass features to likely activity, there are also methods for linking mass features to the BGCs that are likely to produce them. This approach is useful in combination with one of the bioinformatic methods for identifying promising BGCs described above, as it enables faster identification of the product of interest. These methods include IsoAnalyst, which uses different isotopically labeled precursors to identify the likely biosynthetic pathway for metabolites in crude extracts,98 and metabologenomics methods that rely on correlations between BGC presence/absence in genomes and metabolomic signals.99 There are also a number of emerging AI methods for determining the structural class of a metabolite from LC-MS/MS data which can also be used to narrow down the possible BGCs that could produce the natural product.100−104 Analytical methods for dereplicating compounds similar to known antibiotics can also improve the efficiency of screening and avoid the discovery of compounds for which resistance is already widespread. These methods include the previously mentioned structural class identification methods as well as the molecular networking method GNPS105 and methods for analyzing NMR spectra of complex mixtures, such as SMART106 and MADByTE.107,108
Remaining Challenges
Regardless of the number of antibiotics discovered, evolution will likely find a way to escape all of them. However, some antibiotics are more prone to the development of resistance than others. As described above, the amount of time it took for resistant pathogens to first emerge against various antibiotics varied significantly. Recently, there have also been several reports of antibiotics for which resistance does not emerge over the span of laboratory evolution experiments. There are a few characteristics that make an antibiotic more likely to be resistance-proof. These include targeting molecules that are not directly genetically encoded (e.g., not proteins or RNAs) or compounds that have multiple targets. One such example is the natural product teixobactin, which was isolated from a previously uncultured organism through use of a device called iChip. Teixobactin was found to inhibit cell wall synthesis through binding to the precursors, lipid II and lipid III, and binds to the pyrophosphate-sugar moiety rather than the d-Ala-d-Ala moiety targeted by vancomycin. No resistant mutants were obtained after culturing Staphylococcus aureus with sub-inhibitory concentrations over 27 days.48 But it is important to temper enthusiasm by noting that we once thought vancomycin thwarted resistance. Clovibactin is another natural product that targets cell wall precursors that did not produce resistant mutants when plating cultures with clovibactin at a concentration of 4× MIC.109
There are also several synthetic compounds designed to avoid resistance that were inspired by natural products. Macolacin was designed by predicting the most likely product structure of a BGC related to the BGC that produces colistin that showed signs of divergence that could indicate evolutionary pressure to escape resistance. As expected, macolacin was effective against colistin-resistant bacteria.110 Cresomycin is a synthetic antibiotic that targets the ribosome and escapes common resistance mechanisms for ribosome-targeting antibiotics. Cresomycin was designed such that the molecule would be preorganized into the conformation adopted by lincosamides, a class of compounds that include the natural product lincomycin, upon binding to the ribosome.111 Other examples of successful design strategies of natural-product-inspired compounds that evade resistance include rational design of compounds that avoid degradation by β-lactamases while maintaining cell permeability112 or linkage of two antibiotics that bind to different targets.17
As discussed above, the use of multi-drug-resistant organisms in screening campaigns increases the likelihood that hits will escape resistance, like the compounds discussed in this section. It remains to be seen if the lack of resistance to these compounds can hold over decades of use in clinical settings like it does in laboratory evolution experiments that span much shorter time periods. Even if pathogens cannot evolve resistance through modification of a target, it is possible that they could evolve transporters or enzymes that modify or degrade the antibiotic that provide resistance or obtain such genes through horizontal gene transfer. It should also be noted that some of the studies discussed here demonstrated only that the compound escaped known resistance mechanisms and did not perform laboratory evolution experiments to determine how easily new resistance mechanisms might arise.
Future
The largest searchable database containing information about what microbes are capable of making is the collection of genomes in NCBI’s Genbank, with over 3 million prokaryotic sequences of varying quality. More curated databases with similar functionality include NCBI’s Refseq database (over 315,000 prokaryotic genomes)113 and the Joint Genome Institute’s (JGI) GOLD data set (219,320 prokaryotic genomes).114 There are also specific collections of precomputed BGCs, such as the antiSMASH database,115 the JGI’s IMG-ABC116 and Secondary Metabolism Collaboratory (SMC),117 and the metagenome-focused BGC-ATLAS.118 The Natural Product Discovery Center (NPDC) is another database focused on genomes and BGCs of Actinobacteria and is unique among the genomic resources in that it also houses and distributes strains.119 As described above, numerous researchers have used these resources to identify BGCs or other encoded traits. But, they contain no useful information about antimicrobial function. You can find BGCs that are likely to make new molecules, or BGCs that make variations of known antibiotics, or a resistance gene. But you cannot search genomes for antibacterial activity. So, the challenge for the future is to create a pathway, or even pathways, from sequence to function, for then the task begun by Chris Walsh and others to connect genes, enzymes, and biosynthesis to the antibiotic resistance crisis can begin on the needed scale.
Acknowledgments
A.S.W. acknowledges National Institutes of Health, grant number R35GM146987. J.C. acknowledges the National Institutes of Health, grant number R01AT009708. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interest.
Special Issue
Published as part of Biochemistryspecial issue “A Tribute to Christopher T. Walsh”.
References
- U.S. Department of Health and Human Services, Centers for Disease Control and Prevention . Antibiotic Resistance Threats in the United States, 2019. CDC, December 2019. 10.15620/cdc:82532 [DOI]
- Chambers H. F.; Deleo F. R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7 (9), 629. 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agata E. M.; Li H.; Gouldin C.; Tang Y. W. Clinical and molecular characterization of vancomycin-resistant Enterococcus faecium strains during establishment of endemicity. Clin. Infect. Dis. 2001, 33 (4), 511. 10.1086/322615. [DOI] [PubMed] [Google Scholar]
- Herren C. M.; Baym M. Decreased thermal niche breadth as a trade-off of antibiotic resistance. ISME J. 2022, 16 (7), 1843. 10.1038/s41396-022-01235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajer F.; Sandegren L. The Role of Antibiotic Resistance Genes in the Fitness Cost of Multiresistance Plasmids. mBio 2022, 13 (1), e0355221 10.1128/mbio.03552-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk A. H.; Wong A.; Kassen R. The fitness costs of antibiotic resistance mutations. Evol. Appl. 2015, 8 (3), 273. 10.1111/eva.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury F. R.; Findlay B. L. Fitness Costs of Antibiotic Resistance Impede the Evolution of Resistance to Other Antibiotics. ACS Infect. Dis. 2023, 9 (10), 1834. 10.1021/acsinfecdis.3c00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porse A.; Jahn L. J.; Ellabaan M. M. H.; Sommer M. O. A. Dominant resistance and negative epistasis can limit the co-selection of de novo resistance mutations and antibiotic resistance genes. Nat. Commun. 2020, 11 (1), 1199. 10.1038/s41467-020-15080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon-Bingham G. M.; Hedderwick S. A.; McKeating C. M.; McKee P. M.; McNally J. C.; Lennon L. M.; McGivern O.; Lewis K.; McKenna D.; Lattyak E. A.; Lattyak W. J.; Aldeyab M. A. Preserving last resort antibiotics: A Meropenem reduction strategy. Infect. Control Hosp. Epidemiol. 2022, 43 (10), 1516. 10.1017/ice.2021.276. [DOI] [PubMed] [Google Scholar]
- Manyi-Loh C.; Mamphweli S.; Meyer E.; Okoh A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23 (4), 795. 10.3390/molecules23040795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B.; Bhat A.; Ravi K. Antibiotics Misuse and Antimicrobial Resistance Development in Agriculture: A Global Challenge. Environ. Health 2024, 2, 618. 10.1021/envhealth.4c00094. [DOI] [Google Scholar]
- Baranova A. A.; Alferova V. A.; Korshun V. A.; Tyurin A. P. Modern Trends in Natural Antibiotic Discovery. Life (Basel) 2023, 13 (5), 1073. 10.3390/life13051073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J. G.; Vincent F.; Lee J. A.; Eder J.; Prunotto M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 2017, 16 (8), 531. 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- Landeta C.; Mejia-Santana A. Union Is Strength: Target-Based and Whole-Cell High-Throughput Screens in Antibacterial Discovery. J. Bacteriol. 2022, 204 (4), e0047721 10.1128/jb.00477-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer S. B.; Brown E. D. New screens and targets in antibacterial drug discovery. Curr. Opin. Microbiol. 2009, 12 (5), 497. 10.1016/j.mib.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Trauner A.; Sassetti C. M.; Rubin E. J. Genetic Strategies for Identifying New Drug Targets. Microbiol. Spectr. 2014, 2 (4), MGM2-0030-2013. 10.1128/microbiolspec.MGM2-0030-2013. [DOI] [PubMed] [Google Scholar]
- Aleksandrova E. V.; Ma C. X.; Klepacki D.; Alizadeh F.; Vazquez-Laslop N.; Liang J. H.; Polikanov Y. S.; Mankin A. S. Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms. Nat. Chem. Biol. 2024, 10.1038/s41589-024-01685-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhzem A. H.; Woodman T. J.; Blagbrough I. S. Design and synthesis of hybrid compounds as novel drugs and medicines. Rsc Adv. 2022, 12 (30), 19470. 10.1039/D2RA03281C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings M. I.; Truman A. W.; Wilkinson B. Antibiotics: past, present and future. Curr. Opin. Microbiol. 2019, 51, 72. 10.1016/j.mib.2019.10.008. [DOI] [PubMed] [Google Scholar]
- Lewis K. Antibiotics: Recover the lost art of drug discovery. Nature 2012, 485 (7399), 439. 10.1038/485439a. [DOI] [PubMed] [Google Scholar]
- Lyddiard D.; Jones G. L.; Greatrex B. W. Keeping it simple: lessons from the golden era of antibiotic discovery. FEMS Microbiol. Lett. 2016, 363 (8), fnw084 10.1093/femsle/fnw084. [DOI] [PubMed] [Google Scholar]
- Granato E. T.; Meiller-Legrand T. A.; Foster K. R. The Evolution and Ecology of Bacterial Warfare. Curr. Biol. 2019, 29 (11), R521. 10.1016/j.cub.2019.04.024. [DOI] [PubMed] [Google Scholar]
- Traxler M. F.; Kolter R. Natural products in soil microbe interactions and evolution. Nat. Prod. Rep. 2015, 32 (7), 956. 10.1039/C5NP00013K. [DOI] [PubMed] [Google Scholar]
- Shi Y. M.; Bode H. B. Chemical language and warfare of bacterial natural products in bacteria-nematode-insect interactions. Nat. Prod. Rep. 2018, 35 (4), 309. 10.1039/C7NP00054E. [DOI] [PubMed] [Google Scholar]
- Wakimoto T.; Egami Y.; Nakashima Y.; Wakimoto Y.; Mori T.; Awakawa T.; Ito T.; Kenmoku H.; Asakawa Y.; Piel J.; Abe I. Calyculin biogenesis from a pyrophosphate protoxin produced by a sponge symbiont. Nat. Chem. Biol. 2014, 10 (8), 648. 10.1038/nchembio.1573. [DOI] [PubMed] [Google Scholar]
- Florez L. V.; Biedermann P. H.; Engl T.; Kaltenpoth M. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat. Prod. Rep. 2015, 32 (7), 904. 10.1039/C5NP00010F. [DOI] [PubMed] [Google Scholar]
- Van Arnam E. B.; Currie C. R.; Clardy J. Defense contracts: molecular protection in insect-microbe symbioses. Chem. Soc. Rev. 2018, 47 (5), 1638. 10.1039/C7CS00340D. [DOI] [PubMed] [Google Scholar]
- Ayon N. J. High-Throughput Screening of Natural Product and Synthetic Molecule Libraries for Antibacterial Drug Discovery. Metabolites 2023, 13 (5), 625. 10.3390/metabo13050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Fernández D.; Gadiya Y.; Preto A.; Krettler C. A.; Mubeen S.; Allen A.; Healey D.; Colluru V. Natural Products Have Increased Rates of Clinical Trial Success throughout the Drug Development Process. J. Nat. Prod. 2024, 87, 1844. 10.1021/acs.jnatprod.4c00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. J.; Cragg G. M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod 2020, 83 (3), 770. 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- Pye C. R.; Bertin M. J.; Lokey R. S.; Gerwick W. H.; Linington R. G. Retrospective analysis of natural products provides insights for future discovery trends. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (22), 5601. 10.1073/pnas.1614680114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou C. S.; Hong Y. P.; Wang Y. W.; Chen B. H.; Teng R. H.; Song H. Y.; Liao Y. S. Antimicrobial Resistance and Mechanisms of Azithromycin Resistance in Nontyphoidal Salmonella Isolates in Taiwan, 2017 to 2018. Microbiol Spectr 2023, 11 (1), e0336422 10.1128/spectrum.03364-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z.; Tadi D. A.; Fu J.; Azizian K.; Kouhsari E. Global status of Azithromycin and Erythromycin Resistance Rates in Neisseria gonorrhoeae: A Systematic Review and Meta-analysis. Yale J. Biol. Med. 2022, 95 (4), 465. [PMC free article] [PubMed] [Google Scholar]
- Mahajan G. B. Antibacterial agents from actinomycetes - a review. Front. Biosci. 2012, E4 (1), 240. 10.2741/e373. [DOI] [PubMed] [Google Scholar]
- Goldstein L. J. A History of the Prime Number Theorem. American Mathematical Monthly 1973, 80 (6), 599. 10.1080/00029890.1973.11993338. [DOI] [Google Scholar]
- Great Internet Mersenne Prime Search, Dec 21, 2018. https://www.mersenne.org/ [PubMed]
- Clardy J.; Fischbach M. A.; Walsh C. T. New antibiotics from bacterial natural products. Nat. Biotechnol. 2006, 24 (12), 1541. 10.1038/nbt1266. [DOI] [PubMed] [Google Scholar]
- Baltz R. H. Antibiotic discovery from actinomycetes: will a renaissance follow the decline and fall?. SIM News 2005, 55, 186. [Google Scholar]
- Baltz R. H. Marcel Faber Roundtable: is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration?. J. Ind. Microbiol Biotechnol 2006, 33 (7), 507. 10.1007/s10295-005-0077-9. [DOI] [PubMed] [Google Scholar]
- Arai T.Actinomycetes: The Boundary Microorganisms; Toppan Company, 1976. [Google Scholar]
- Waksman S. A.; Woodruff H. B. Bacteriostatic and Bactericidal Substances Produced by a Soil Actinomyces. Proceedings of the Society for Experimental Biology and Medicine 1940, 45 (2), 609. 10.3181/00379727-45-11768. [DOI] [Google Scholar]
- Covington B. C.; Xu F.; Seyedsayamdost M. R. A Natural Product Chemist’s Guide to Unlocking Silent Biosynthetic Gene Clusters. Annu. Rev. Biochem. 2021, 90, 763. 10.1146/annurev-biochem-081420-102432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermolen K. M.; Raja H. A.; El-Elimat T.; Oberlies N. H. Evaluation of culture media for the production of secondary metabolites in a natural products screening program. AMB Express 2013, 3 (1), 71. 10.1186/2191-0855-3-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J. C. Media and growth conditions for induction of secondary metabolite production. Methods Mol. Biol. 2012, 944, 47. 10.1007/978-1-62703-122-6_3. [DOI] [PubMed] [Google Scholar]
- Shima J.; Hesketh A.; Okamoto S.; Kawamoto S.; Ochi K. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol. 1996, 178 (24), 7276. 10.1128/jb.178.24.7276-7284.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. The Science of Antibiotic Discovery. Cell 2020, 181 (1), 29. 10.1016/j.cell.2020.02.056. [DOI] [PubMed] [Google Scholar]
- Watve M. G.; Tickoo R.; Jog M. M.; Bhole B. D. How many antibiotics are produced by the genus Streptomyces?. Arch. Microbiol. 2001, 176 (5), 386. 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- Ling L. L.; Schneider T.; Peoples A. J.; Spoering A. L.; Engels I.; Conlon B. P.; Mueller A.; Schaberle T. F.; Hughes D. E.; Epstein S.; Jones M.; Lazarides L.; Steadman V. A.; Cohen D. R.; Felix C. R.; Fetterman K. A.; Millett W. P.; Nitti A. G.; Zullo A. M.; Chen C.; Lewis K. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517 (7535), 455. 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P. R. Natural Products and the Gene Cluster Revolution. Trends Microbiol 2016, 24 (12), 968. 10.1016/j.tim.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D.; Church G.; Hessel A.; Kelley N. J.; Arkin A.; Cai Y.; Carlson R.; Chakravarti A.; Cornish V. W.; Holt L.; Isaacs F. J.; Kuiken T.; Lajoie M.; Lessor T.; Lunshof J.; Maurano M. T.; Mitchell L. A.; Rine J.; Rosser S.; Sanjana N. E.; Silver P. A.; Valle D.; Wang H.; Way J. C.; Yang L. GENOME ENGINEERING. The Genome Project-Write. Science 2016, 353 (6295), 126. 10.1126/science.aaf6850. [DOI] [PubMed] [Google Scholar]
- Bosch F.; Rosich L. The contributions of Paul Ehrlich to pharmacology: a tribute on the occasion of the centenary of his Nobel Prize. Pharmacology 2008, 82 (3), 171. 10.1159/000149583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AbuDaqa A.; Abu-Hassan A.; Imam M. Taxonomy and Practical Evaluation of Primality Testing Algorithms. arXiv Preprint 2020, 08444. 10.48550/arXiv.2006.08444. [DOI] [Google Scholar]
- Rabin M. O. Probabilistic algorithm for testing primality. Journal of Number Theory 1980, 12 (1), 128. 10.1016/0022-314X(80)90084-0. [DOI] [Google Scholar]
- Katz L.; Baltz R. H. Natural product discovery: past, present, and future. J. Ind. Microbiol Biotechnol 2016, 43 (2–3), 155. 10.1007/s10295-015-1723-5. [DOI] [PubMed] [Google Scholar]
- Harvey A. L.; Edrada-Ebel R.; Quinn R. J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov 2015, 14 (2), 111. 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- Atanasov A. G.; Zotchev S. B.; Dirsch V. M.; Supuran C. T. International Natural Product Sciences, T.; Supuran, C. T. Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discov 2021, 20 (3), 200. 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey A. L. Natural products in drug discovery. Drug Discov Today 2008, 13 (19–20), 894. 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Lewis K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov 2013, 12 (5), 371. 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- Balsamo J. A.; Penton K. E.; Zhao Z.; Hayes M. J.; Lima S. M.; Irish J. M.; Bachmann B. O. An immunogenic cell injury module for the single-cell multiplexed activity metabolomics platform to identify promising anti-cancer natural products. J. Biol. Chem. 2022, 298 (9), 102300 10.1016/j.jbc.2022.102300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacace E.; Kritikos G.; Typas A. Chemical genetics in drug discovery. Curr. Opin Syst. Biol. 2017, 4, 35. 10.1016/j.coisb.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muellner M. K.; Uras I. Z.; Gapp B. V.; Kerzendorfer C.; Smida M.; Lechtermann H.; Craig-Mueller N.; Colinge J.; Duernberger G.; Nijman S. M. A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat. Chem. Biol. 2011, 7 (11), 787. 10.1038/nchembio.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.; Zhao M.; Braun D. R.; Ericksen S. S.; Piotrowski J. S.; Nelson J.; Peng J.; Ananiev G. E.; Chanana S.; Barns K.; Fossen J.; Sanchez H.; Chevrette M. G.; Guzei I. A.; Zhao C.; Guo L.; Tang W.; Currie C. R.; Rajski S. R.; Audhya A.; Andes D. R.; Bugni T. A marine microbiome antifungal targets urgent-threat drug-resistant fungi. Science 2020, 370 (6519), 974. 10.1126/science.abd6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyche T. P.; Piotrowski J. S.; Hou Y.; Braun D.; Deshpande R.; McIlwain S.; Ong I. M.; Myers C. L.; Guzei I. A.; Westler W. M.; Andes D. R.; Bugni T. S. Forazoline A: marine-derived polyketide with antifungal in vivo efficacy. Angew. Chem., Int. Ed. Engl. 2014, 53 (43), 11583. 10.1002/anie.201405990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue A.; Robbins N.; Cowen L. E. Advances in fungal chemical genomics for the discovery of new antifungal agents. Ann. N.Y. Acad. Sci. 2021, 1496 (1), 5. 10.1111/nyas.14484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins N.; Cowen L. E. Genomic Approaches to Antifungal Drug Target Identification and Validation. Annu. Rev. Microbiol. 2022, 76, 369. 10.1146/annurev-micro-041020-094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintses B.; Jangir P. K.; Fekete G.; Szamel M.; Mehi O.; Spohn R.; Daruka L.; Martins A.; Hosseinnia A.; Gagarinova A.; Kim S.; Phanse S.; Csorgo B.; Gyorkei A.; Ari E.; Lazar V.; Nagy I.; Babu M.; Pal C.; Papp B. Chemical-genetic profiling reveals limited cross-resistance between antimicrobial peptides with different modes of action. Nat. Commun. 2019, 10 (1), 5731. 10.1038/s41467-019-13618-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Poulton N. C.; Chang J. S.; Azadian Z. A.; DeJesus M. A.; Ruecker N.; Zimmerman M. D.; Eckartt K. A.; Bosch B.; Engelhart C. A.; Sullivan D. F.; Gengenbacher M.; Dartois V. A.; Schnappinger D.; Rock J. M. CRISPRi chemical genetics and comparative genomics identify genes mediating drug potency in Mycobacterium tuberculosis. Nat. Microbiol 2022, 7 (6), 766. 10.1038/s41564-022-01130-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrier T.; Romano K. P.; Clatworthy A. E.; Hung D. T. Integrated genomics and chemical biology herald an era of sophisticated antibacterial discovery, from defining essential genes to target elucidation. Cell Chem. Biol. 2022, 29 (5), 716. 10.1016/j.chembiol.2022.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonejuie P.; Burkart M.; Pogliano K.; Pogliano J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (40), 16169. 10.1073/pnas.1311066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller M. G. A unifying review of bioassay-guided fractionation, effect-directed analysis and related techniques. Sensors (Basel) 2012, 12 (7), 9181. 10.3390/s120709181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston N. J.; Mukhtar T. A.; Wright G. D. Streptogramin antibiotics: mode of action and resistance. Curr. Drug Targets 2002, 3 (4), 335. 10.2174/1389450023347678. [DOI] [PubMed] [Google Scholar]
- Mersenne Prime. MathWorld, 2024. https://mathworld.wolfram.com/MersennePrime.html.
- Brenk R.; Schipani A.; James D.; Krasowski A.; Gilbert I. H.; Frearson J.; Wyatt P. G. Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. Chemmedchem 2008, 3 (3), 435. 10.1002/cmdc.200700139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Munoz J. C.; Selem-Mojica N.; Mullowney M. W.; Kautsar S. A.; Tryon J. H.; Parkinson E. I.; De Los Santos E. L. C.; Yeong M.; Cruz-Morales P.; Abubucker S.; Roeters A.; Lokhorst W.; Fernandez-Guerra A.; Cappelini L. T. D.; Goering A. W.; Thomson R. J.; Metcalf W. M.; Kelleher N. L.; Barona-Gomez F.; Medema M. H. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 2020, 16 (1), 60. 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautsar S. A.; van der Hooft J. J. J.; de Ridder D.; Medema M. H. BiG-SLiCE: A highly scalable tool maps the diversity of 1.2 million biosynthetic gene clusters. Gigascience 2021, 10 (1), giaa154 10.1093/gigascience/giaa154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavriilidou A.; Kautsar S. A.; Zaburannyi N.; Krug D.; Muller R.; Medema M. H.; Ziemert N. Compendium of specialized metabolite biosynthetic diversity encoded in bacterial genomes. Nat. Microbiol 2022, 7 (5), 726. 10.1038/s41564-022-01110-2. [DOI] [PubMed] [Google Scholar]
- Salamzade R.; Cheong J. Z. A.; Sandstrom S.; Swaney M. H.; Stubbendieck R. M.; Starr N. L.; Currie C. R.; Singh A. M.; Kalan L. R. Evolutionary investigations of the biosynthetic diversity in the skin microbiome using lsaBGC. Microb Genomics 2023, 9 (4), 000988 10.1099/mgen.0.000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.; Liu N.; Tang Y. Recent developments in self-resistance gene directed natural product discovery. Nat. Prod. Rep. 2020, 37 (7), 879. 10.1039/C9NP00050J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock B. P.; Raphenya A. R.; Lau T. T. Y.; Tsang K. K.; Bouchard M.; Edalatmand A.; Huynh W.; Nguyen A. V.; Cheng A. A.; Liu S.; Min S. Y.; Miroshnichenko A.; Tran H.; Werfalli R. E.; Nasir J. A.; Oloni M.; Speicher D. J.; Florescu A.; Singh B.; Faltyn M.; Hernandez-Koutocheva A.; Sharma A. N.; Bordeleua E.; Pawlowski A. C.; Zubyk H. L.; Dooley D.; Griffiths E.; Maguire F.; Winsor G. L.; Beiko R. G.; Brinkman F. S. L.; Hsiao W. W. L.; Domselaar G. V.; McArther A. G. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48 (D1), D517. 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungan M. D.; Blin K.; Ziemert N. ARTS-DB: a database for antibiotic resistant targets. Nucleic Acids Res. 2022, 50 (D1), D736. 10.1093/nar/gkab940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long K. S.; Poehlsgaard J.; Kehrenberg C.; Schwarz S.; Vester B. The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob. Agents Chemother. 2006, 50 (7), 2500. 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooke C. L.; Hinchliffe P.; Bragginton E. C.; Colenso C. K.; Hirvonen V. H. A.; Takebayashi Y.; Spencer J. beta-Lactamases and beta-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431 (18), 3472. 10.1016/j.jmb.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crits-Christoph A.; Bhattacharya N.; Olm M. R.; Song Y. S.; Banfield J. F. Transporter genes in biosynthetic gene clusters predict metabolite characteristics and siderophore activity. Genome Res. 2021, 31 (2), 239. 10.1101/gr.268169.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.; Li J.; Millan-Aguinaga N.; Zhang J. J.; O’Neill E. C.; Ugalde J. A.; Jensen P. R.; Mantovani S. M.; Moore B. S. Identification of Thiotetronic Acid Antibiotic Biosynthetic Pathways by Target-directed Genome Mining. ACS Chem. Biol. 2015, 10 (12), 2841. 10.1021/acschembio.5b00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K.; Shaw S.; Augustijn H. E.; Reitz Z. L.; Biermann F.; Alanjary M.; Fetter A.; Terlouw B. R.; Metcalf W. W.; Helfrich E. J. N.; van Wezel G. P.; Medema M. H.; Weber T. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51 (W1), W46. 10.1093/nar/gkad344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan G. D.; Prihoda D.; Palicka A.; Soukup J.; Klempir O.; Rampula L.; Durcak J.; Wurst M.; Kotowski J.; Chang D.; Wang R.; Piizzi G.; Temesi G.; Hazuda D. J.; Woelk C. H.; Bitton D. A. A deep learning genome-mining strategy for biosynthetic gene cluster prediction. Nucleic Acids Res. 2019, 47 (18), e110 10.1093/nar/gkz654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll L. M.; Larralde M.; Fleck J. S.; Ponnudurai R.; Milanese A.; Cappio E.; Zeller G. Accurate de novo identification of biosynthetic gene clusters with GECCO. bioRxiv Preprint 2021, 442509. 10.1101/2021.05.03.442509. [DOI] [Google Scholar]
- Sanchez S.; Rogers J. D.; Rogers A. B.; Nassar M.; McEntyre J.; Welch M.; Hollfelder F.; Finn R. D. Expansion of novel biosynthetic gene clusters from diverse environments using SanntiS. bioRxiv Preprint 2023, 540769. 10.1101/2023.05.23.540769. [DOI] [Google Scholar]
- Stokes J. M.; Yang K.; Swanson K.; Jin W.; Cubillos-Ruiz A.; Donghia N. M.; MacNair C. R.; French S.; Carfrae L. A.; Bloom-Ackermann Z.; Tran V. M.; Chiappino-Pepe A.; Badran A. H.; Andrews I. W.; Chory E. J.; Curch G. M.; Brown E. D.; Jaakkola T. S.; Barzilay R.; Collins J. J. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 181 (2), 475. 10.1016/j.cell.2020.01.021. [DOI] [PubMed] [Google Scholar]
- Wong F.; Zheng E. J.; Valeri J. A.; Donghia N. M.; Anahtar M. N.; Omori S.; Li A.; Cubillos-Ruiz A.; Krishnan A.; Jin W.; Manson A. L.; Friedrichs J.; Helbig R.; Hajian B.; Fiejtek D. K.; Wagner F. F.; Soutter H. H.; Earl A. M.; Stokes J. M.; Renner L. D.; Collins J. J. Discovery of a structural class of antibiotics with explainable deep learning. Nature 2024, 626 (7997), 177. 10.1038/s41586-023-06887-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A. S.; Clardy J. A Machine Learning Bioinformatics Method to Predict Biological Activity from Biosynthetic Gene Clusters. J. Chem. Inf Model 2021, 61 (6), 2560. 10.1021/acs.jcim.0c01304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinnider M. A.; Johnston C. W.; Gunabalasingam M.; Merwin N. J.; Kieliszek A. M.; MacLellan R. J.; Li H.; Ranieri M. R. M.; Webster A. L. H.; Cao M. P. T.; Pfeifle A.; Spencer N.; To Q. H.; Wallace D. P.; Dejong C. A.; Magarvey N. A. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 2020, 11 (1), 6058. 10.1038/s41467-020-19986-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei S. A Multi-Label Learning Framework for Predicting Chemical Classes and Biological Activities of Natural Products from Biosynthetic Gene Clusters. J. Chem. Ecol 2023, 49 (11–12), 681. 10.1007/s10886-023-01452-z. [DOI] [PubMed] [Google Scholar]
- Riedling O.; Walker A. S.; Rokas A. Predicting fungal secondary metabolite activity from biosynthetic gene cluster data using machine learning. Microbiol. Spectr. 2024, 12 (2), e0340023 10.1101/2023.09.12.557468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf J. D.; Yan X.; Shen B. Genome neighborhood network reveals insights into enediyne biosynthesis and facilitates prediction and prioritization for discovery. J. Ind. Microbiol Biotechnol 2016, 43 (2–3), 261. 10.1007/s10295-015-1671-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman K. D.; Butler K. S.; Moore B. S.; Chekan J. R. Genome mining methods to discover bioactive natural products. Nat. Prod. Rep. 2021, 38 (11), 2100. 10.1039/D1NP00032B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.; van Santen J. A.; Farzaneh N.; Liu D. Y.; Pye C. R.; Baumeister T. U. H.; Wong W. R.; Linington R. G. NP Analyst: An Open Online Platform for Compound Activity Mapping. Acs Central Sci. 2022, 8 (2), 223. 10.1021/acscentsci.1c01108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughey C. S.; van Santen J. A.; van der Hooft J. J. J.; Medema M. H.; Linington R. G. An isotopic labeling approach linking natural products with biosynthetic gene clusters. Nat. Chem. Biol. 2022, 18 (3), 295. 10.1038/s41589-021-00949-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering A. W.; McClure R. A.; Doroghazi J. R.; Albright J. C.; Haverland N. A.; Zhang Y.; Ju K. S.; Thomson R. J.; Metcalf W. W.; Kelleher N. L. Metabologenomics: Correlation of Microbial Gene Clusters with Metabolites Drives Discovery of a Nonribosomal Peptide with an Unusual Amino Acid Monomer. ACS Cent Sci. 2016, 2 (2), 99. 10.1021/acscentsci.5b00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwen J. J. R.; van der Hooft J. J. J. Comprehensive Large-Scale Integrative Analysis of Omics Data To Accelerate Specialized Metabolite Discovery. mSystems 2021, 6 (4), e0072621 10.1128/mSystems.00726-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge N. F.; Louwen J. J. R.; Chekmeneva E.; Camuzeaux S.; Vermeir F. J.; Jansen R. S.; Huber F.; van der Hooft J. J. J. MS2Query: reliable and scalable MS(2) mass spectra-based analogue search. Nat. Commun. 2023, 14 (1), 1752. 10.1038/s41467-023-37446-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjorleifsson Eldjarn G.; Ramsay A.; van der Hooft J. J. J.; Duncan K. R.; Soldatou S.; Rousu J.; Daly R.; Wandy J.; Rogers S. Ranking microbial metabolomic and genomic links in the NPLinker framework using complementary scoring functions. PLoS Comput. Biol. 2021, 17 (5), e1008920 10.1371/journal.pcbi.1008920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhrkop K.; Fleischauer M.; Ludwig M.; Aksenov A. A.; Melnik A. V.; Meusel M.; Dorrestein P. C.; Rousu J.; Bocker S. SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16 (4), 299. 10.1038/s41592-019-0344-8. [DOI] [PubMed] [Google Scholar]
- Duhrkop K.; Nothias L. F.; Fleischauer M.; Reher R.; Ludwig M.; Hoffmann M. A.; Petras D.; Gerwick W. H.; Rousu J.; Dorrestein P. C.; Bocker S. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 2021, 39 (4), 462. 10.1038/s41587-020-0740-8. [DOI] [PubMed] [Google Scholar]
- Wang M.; Carver J. J.; Phelan V. V.; Sanchez L. M.; Garg N.; Peng Y.; Nguyen D. D.; Watrous J.; Kapono C. A.; Luzzatto-Knaan T.; Porto C.; Bouslimani A.; Melnik A. V.; Meehan M. J.; Liu W.; Crüsemann M.; Boudreau P. D.; Esquenazi E.; Sandoval-Calderón M.; Kersten R. D.; Pace L. A.; Quinn R. A.; Duncan K. R.; Hsu C.; Floros D. J.; Gavilan R. G.; Kleigrewe K.; Northen T.; Dutton R. J.; Parrot D.; Carlson E. E.; Aigle B.; Michelsen C. F.; Jelsbak L.; Sohlenkamp C.; Pevzner P.; Edlund A.; McLean J.; Piel J.; Murphy B. T.; Gerwick L.; Liaw C.; Yang Y.; Humpf H.; Maansson M.; Keyzers R. A.; Sims A. C.; Johnson A. R.; Sidebottom A. M.; Sedio B. E.; Klitgaard A.; Larson C. B.; Boya P C. A.; Torres-Mendoza D.; Gonzalez D. J.; Silva D. B.; Marques L. M.; Demarque D. P.; Pociute E.; O’Neill E. C.; Briand E.; Helfrich E. J. N.; Granatosky E. A.; Glukhov E.; Ryffel F.; Houson H.; Mohimani H.; Kharbush J. J.; Zeng Y.; Vorholt J. A.; Kurita K. L.; Charusanti P.; McPhail K. L.; Fog Nielsen K.; Vuong L.; Elfeki M.; Traxler M. F.; Engene N.; Koyama N.; Vining O. B.; Baric R.; Silva R. R.; Mascuch S. J.; Tomasi S.; Jenkins S.; Macherla V.; Hoffman T.; Agarwal V.; Williams P. G.; Dai J.; Neupane R.; Gurr J.; Rodríguez A. M. C.; Lamsa A.; Zhang C.; Dorrestein K.; Duggan B. M.; Almaliti J.; Allard P.; Phapale P.; Nothias L.; Alexandrov T.; Litaudon M.; Wolfender J.; Kyle J. E.; Metz T. O.; Peryea T.; Nguyen D.; VanLeer D.; Shinn P.; Jadhav A.; Muller R.; Waters K. M.; Shi W.; Liu X.; Zhang L.; Knight R.; Jensen P. R.; Palsson B. O.; Pogliano K.; Linington R. G.; Gutiérrez M.; Lopes N. P.; Gerwick W. H.; Moore B. S.; Dorrestein P. C.; Bandeira N. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34 (8), 828. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Idelbayev Y.; Roberts N.; Tao Y.; Nannapaneni Y.; Duggan B. M.; Min J.; Lin E. C.; Gerwick E. C.; Cottrell G. W.; Gerwick W. H. Small Molecule Accurate Recognition Technology (SMART) to Enhance Natural Products Research. Sci. Rep 2017, 7 (1), 14243 10.1038/s41598-017-13923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan J. M.; van Santen J. A.; Liu D. Y.; Linington R. G. Development of an NMR-Based Platform for the Direct Structural Annotation of Complex Natural Products Mixtures. J. Nat. Prod 2021, 84 (4), 1044. 10.1021/acs.jnatprod.0c01076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Bocanegra L.; Al Subeh Z. Y.; Egan J. M.; El-Elimat T.; Raja H. A.; Burdette J. E.; Pearce C. J.; Linington R. G.; Oberlies N. H. Dereplication of Fungal Metabolites by NMR-Based Compound Networking Using MADByTE. J. Nat. Prod 2022, 85 (3), 614. 10.1021/acs.jnatprod.1c00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla R.; Peoples A. J.; Ludwig K. C.; Maity S.; Derks M. G. N.; De Benedetti S.; Krueger A. M.; Vermeulen B. J. A.; Harbig T.; Lavore F.; Kumar R.; Honorato R. V.; Grein F.; Nieselt K.; Liu Y.; Bonvin A. M. J. J.; Baldus M.; Kubitscheck U.; Breukink E.; Achorn C.; Nitti A.; Schwalen C. J.; Spoering A. L.; Ling L. L.; Hughes D.; Lelli M.; Roos W. H.; Lewis K.; Schneider T.; Weingarth M. An antibiotic from an uncultured bacterium binds to an immutable target. Cell 2023, 186 (19), 4059. 10.1016/j.cell.2023.07.038. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Koirala B.; Hernandez Y.; Zimmerman M.; Park S.; Perlin D. S.; Brady S. F. A naturally inspired antibiotic to target multidrug-resistant pathogens. Nature 2022, 601 (7894), 606. 10.1038/s41586-021-04264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K. J. Y.; Tresco B. I. C.; Ramkissoon A.; Aleksandrova E. V.; Syroegin E. A.; See D. N. Y.; Liow P.; Dittemore G. A.; Yu M.; Testolin G.; Mitcheltree M. J.; Liu R. Y.; Svetlov M. S.; Polikanov Y. S.; Myers A. G. An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance. Science 2024, 383 (6684), 721. 10.1126/science.adk8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Reville T. F.; Miller A. A.; O’Donnell J. P.; Wu X.; Sylvester M. A.; Guler S.; Iyer R.; Shapiro A. B.; Carter N. M.; Velez-Vega C.; Moussa S. H.; McLeod S. M.; Chen A.; Tanudra A. M.; Zhang J.; Comita-Prevoir J.; Romero J. A.; Huynh H.; Ferguson A. D.; Horanyi P. S.; Mayclin S. J.; Heine H. S.; Drusano G. L.; Cummings J. E.; Slayden R. A.; Tommasi R. A. Rational design of a new antibiotic class for drug-resistant infections. Nature 2021, 597 (7878), 698. 10.1038/s41586-021-03899-0. [DOI] [PubMed] [Google Scholar]
- Haft D. H.; Badretdin A.; Coulouris G.; DiCuccio M.; Durkin A. S.; Jovenitti E.; Li W.; Mersha M.; O’Neill K. R.; Virothaisakun J.; Thibaud-Nissen F. RefSeq and the prokaryotic genome annotation pipeline in the age of metagenomes. Nucleic Acids Res. 2024, 52 (D1), D762. 10.1093/nar/gkad988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S.; Stamatis D.; Li C. T.; Ovchinnikova G.; Bertsch J.; Sundaramurthi J. C.; Kandimalla M.; Nicolopoulos P. A.; Favognano A.; Chen I. A.; Kyrpides N. C.; Reddy T. B. K. Twenty-five years of Genomes OnLine Database (GOLD): data updates and new features in v.9. Nucleic Acids Res. 2023, 51 (D1), D957. 10.1093/nar/gkac974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K.; Medema M. H.; Kottmann R.; Lee S. Y.; Weber T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2017, 45 (D1), D555. 10.1093/nar/gkw960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniappan K.; Chen I. A.; Chu K.; Ratner A.; Seshadri R.; Kyrpides N. C.; Ivanova N. N.; Mouncey N. J. IMG-ABC v.5.0: an update to the IMG/Atlas of Biosynthetic Gene Clusters Knowledgebase. Nucleic Acids Res. 2020, 48 (D1), D422. 10.1093/nar/gkz932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secondary Metabolism Collaboratory, Joint Genome Institute, https://jgi.doe.gov/data-and-tools/data-systems/secondary-metabolism-collaboratory/.
- Bağcı C.; Nuhamunada M.; Goyat H.; Ladanyi C.; Sehnal L.; Blin K.; Kautsar S. A.; Tagirdzhanov A.; Gurevich A.; Mantri S.; von Mering C.; Udwary D.; Medema M. H.; Weber T.; Ziemert N. BGC Atlas: A Web Resource for Exploring the Global Chemical Diversity Encoded in Bacterial Genomes. bioRxiv Preprint 2024, 609335. 10.1101/2024.08.23.609335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkreuter E.; Kautsar S. A.; Yang D.; Bader C. D.; Teijaro C. N.; Fluegel L. L.; Davis C. M.; Simpson J. R.; Lauterbach L.; Steele A. D.; Gui S.; Meng S.; Li G.; Vierhrig K.; Ye F.; Su P.; Kiefer A. F.; Nichols A.; Cepeda A. J.; Yan W.; Fan B.; Jiang Y.; Adhikari A.; Zheng C.; Schuster L.; Cowan T. M.; Smanski M. J.; Chevrette M. G.; de Carvalho L. P. S.; Shen B. The Natural Products Discovery Center: Release of the First 8490 Sequenced Strains for Exploring Actinobacteria Biosynthetic Diversity. bioRxiv Preprint 2024, 571759. 10.1101/2023.12.14.571759. [DOI] [Google Scholar]