Abstract
Background
The placenta is essential for nutrient exchange and hormone production between the mother and the developing fetus and serves as an invaluable model for epigenetic research. Most epigenetic studies of the human placenta have used whole placentas from term pregnancies and have identified the presence of partially methylated domains (PMDs). However, the origin of these domains, which are typically absent in most somatic cells, remains unclear in the placental context.
Results
Using whole-genome bisulfite sequencing and analysis of histone H3 modifications, we generated epigenetic profiles of human cytotrophoblasts during the first trimester and at term, as well as human trophoblast stem cells. Our study focused specifically on PMDs. We found that genomic regions likely to form PMDs are resistant to global DNA demethylation during trophectoderm reprogramming, and that PMDs arise through a slow methylation process within condensed chromatin near the nuclear lamina. In addition, we found significant differences in histone H3 modifications between PMDs in cytotrophoblasts and trophoblast stem cells.
Conclusions
Our findings suggest that spatiotemporal genomic features shape megabase-scale DNA methylation patterns, including PMDs, in the human placenta and highlight distinct differences in PMDs between human cytotrophoblasts and trophoblast stem cells. These findings advance our understanding of placental biology and provide a basis for further research into human development and related diseases.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-10986-9.
Keywords: Human cytotrophoblasts, Human trophoblast stem cells, Partially methylated domains, Methylomes
Background
The human placenta is critical to fetal development and serves as the essential link between the mother and the fetus. It provides critical support for the transfer of nutrients, oxygen, and waste products, and produces critical pregnancy-sustaining hormones [1]. Composed primarily of cytotrophoblasts, syncytiotrophoblasts, and extravillous trophoblasts, the placenta undergoes significant cellular transformation. Cytotrophoblasts initially form a mononuclear layer that evolves into a multinucleated layer of syncytiotrophoblasts and invasive extravillous trophoblasts as the placenta matures [2]. The placenta is an important model for epigenetic studies [3]. As part of the International Human Epigenome Consortium (IHEC), our research has focused on isolating specific trophoblast subtypes for comprehensive epigenomic analysis of human cytotrophoblasts (hCTs) [4]. In addition, our pioneering work in establishing human trophoblast stem cells (hTSCs) [5] has contributed significantly to the advancement of placental research [6–16].
DNA methylation is a critical epigenetic modification that regulates cellular processes. Whole-genome bisulfite sequencing (WGBS) is the primary method for analyzing global DNA methylation patterns (i.e., methylomes) at single-base resolution [17]. Several studies have examined the methylomes of the whole human placenta during late pregnancy using WGBS [18–21]. In addition, DNA methylation in hCTs has also been studied; however, these analyses have used non-WGBS techniques that only cover regions where cytosine-guanine (CpG) dinucleotides are concentrated, such as reduced representation bisulfite sequencing (RRBS) and methylation arrays [22–27]. In contrast to these studies, our research has generated methylome profiles of hCTs using WGBS at two developmental stages: first trimester and term [9], as well as histone H3 modifications.
WGBS has revealed partially methylated domains (PMDs) that span hundreds of kilobases (kb) and exhibit moderate to low levels of methylation [28]. Despite the initial discovery of PMDs in the human placental methylome [19], the origin of these domains, which are typically absent from most somatic cells, in the placenta remains elusive [29]. PMDs have also been identified in the methylome of hTSCs [5]. In this study, we investigate the epigenetic dynamics of PMDs in hCTs and hTSCs to understand the process of PMD formation in hCTs and to elucidate the different characteristics of PMDs in hTSCs compared to those in hCTs. Through comprehensive analysis, we aim to understand the complex mechanisms of epigenetic regulation affecting placental development and trophoblast stem cell differentiation.
Results
Dynamics of PMDs during human placental development
We analyzed hCTs at two developmental stages: first trimester (7 weeks) and term pregnancy. Using WGBS, we generated high coverage (> 30x) methylome profiles for two biological replicates at each developmental stage (Additional file 1: Fig. S1A and Table S1). The global CpG methylation level increased from 52.6% at first trimester to 59.9% at term (Additional file 1: Fig. S1B). This increase in placental methylation levels during pregnancy mirrors findings from previous studies using RRBS and methylation arrays [22, 26, 27]. To track methylome progression during placental development, we incorporated published WGBS data from the human trophectoderm [30]. This early blastocyst component undergoes global waves of DNA demethylation and then de novo DNA methylation as it differentiates into the placenta [31].
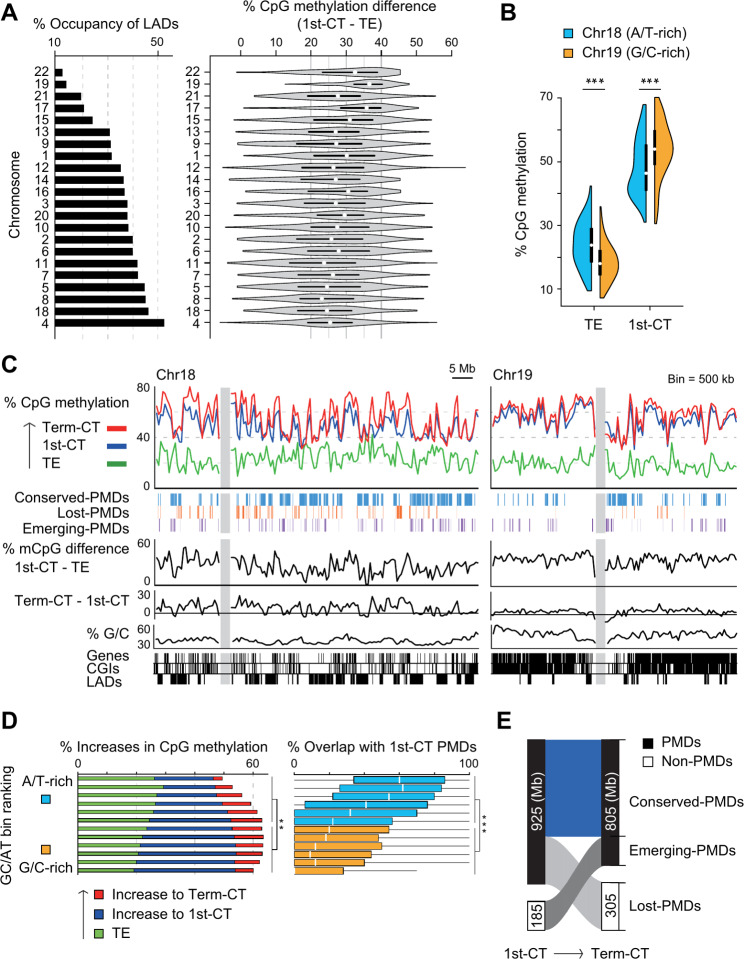
Chromosomes within the nucleus occupy specific regions called chromosome territories [32]. We examined the relationship between location in the cell nucleus and DNA methylation levels, using lamina-associated domains (LADs) [33] as markers to infer the spatial arrangement of genomic regions. Our analysis revealed that chromosomes positioned near the nuclear center, such as chromosome 22 (Chr22), Chr19, and Chr17, showed a greater increase in methylation levels during the transition from trophectoderm to first-trimester hCTs compared to those positioned more peripherally, such as Chr4, Chr18, and Chr8 (Fig. 1A, Additional file 1: Fig. S1C). In addition, we focused on Chr18 and Chr19, which are similar in size but differ significantly in guanine/cytosine (G/C) content and location in the nucleus [34–36]. At the trophectoderm stage, the G/C-rich Chr19 was more demethylated than the adenine/thymine (A/T)-rich Chr18 (Fig. 1B, C). Conversely, during the transition from trophectoderm to first trimester hCTs, Chr19 was more methylated than Chr18 (Fig. 1B, C).
Fig. 1.
Tracking methylome dynamics and the emergence of PMDs during human placental development. (A) Correlation between position in the nucleus and DNA methylation levels. Bar plots show the LAD occupancy for each chromosome, ordered from top to bottom by decreasing LAD occupancy, with LAD data taken from Guelen et al. (2008) [33]. Violin plots illustrate the differences in methylation levels between trophectoderm and first trimester hCTs for each chromosome. TE, trophectoderm; 1st-CT, first trimester hCTs (B) The transition of DNA methylation levels from trophectoderm to first trimester hCTs. The methylation level of the 500-kb bins in Chr18 and Chr19 is shown by half-violin plots. Trophectoderm data are from Zhu et al. (2018) [30]. (C) DNA methylation levels in trophectoderm and hCTs across Chr18 and Chr19. CpG methylation levels and PMDs are shown as lines and rectangles, respectively. G/C content, RefSeq genes, CpG islands (CGIs), and LADs are shown at the bottom, and gray bars indicate centromere positions. Term-CT, term hCTs (D) Relationship between G/C content and DNA methylation levels during placental development. All non-overlapping 500 kb bins were sorted by G/C content and divided into 12 groups in order of increasing G/C content. Stacked bar graphs (left) show increases in DNA methylation levels across developmental stages. Baseline CpG methylation levels in the trophectoderm are shown in green, and additional methylation levels are shown in blue and red. Increases (indicated by blue) in methylation levels from trophectoderm to first trimester hCTs were evaluated for statistical significance. Box plots (right) show the overlap rate between PMDs in first trimester hCTs and bins for each group (E) Transitional dynamics of PMDs from first trimester to term hCTs. The numbers indicate the total PMD size (in megabases) for each category Statistical significance for changes in methylation levels and PMD overlap was assessed using the Wilcoxon rank sum test. Significance is indicated by double asterisks (**) for P < 0.005 and triple asterisks (***) for P < 0.0001 across all relevant panels
This trend of G/C-rich regions showing higher methylation levels early in placental development was also observed genome-wide (Fig. 1D), similar to patterns observed in the mouse placenta [37]. In our approach, PMDs were defined relative to the average global methylation level of each sample, rather than absolute levels. This method identified 4,086 PMDs in first trimester hCTs and 2,244 in term hCTs. Thus, PMDs were identified predominantly in A/T-rich genomic regions of first trimester hCTs (Fig. 1D). In addition, 81% of these PMDs in term hCTs overlapped with those previously identified in the term whole placenta [19] (Additional file 1: Fig. S1D). PMDs were further classified into three categories–conserved, lost, and emerging–based on their persistence or change from first trimester to term hCTs (Fig. 1E).
Chromatin states of PMDs in human cytotrophoblasts
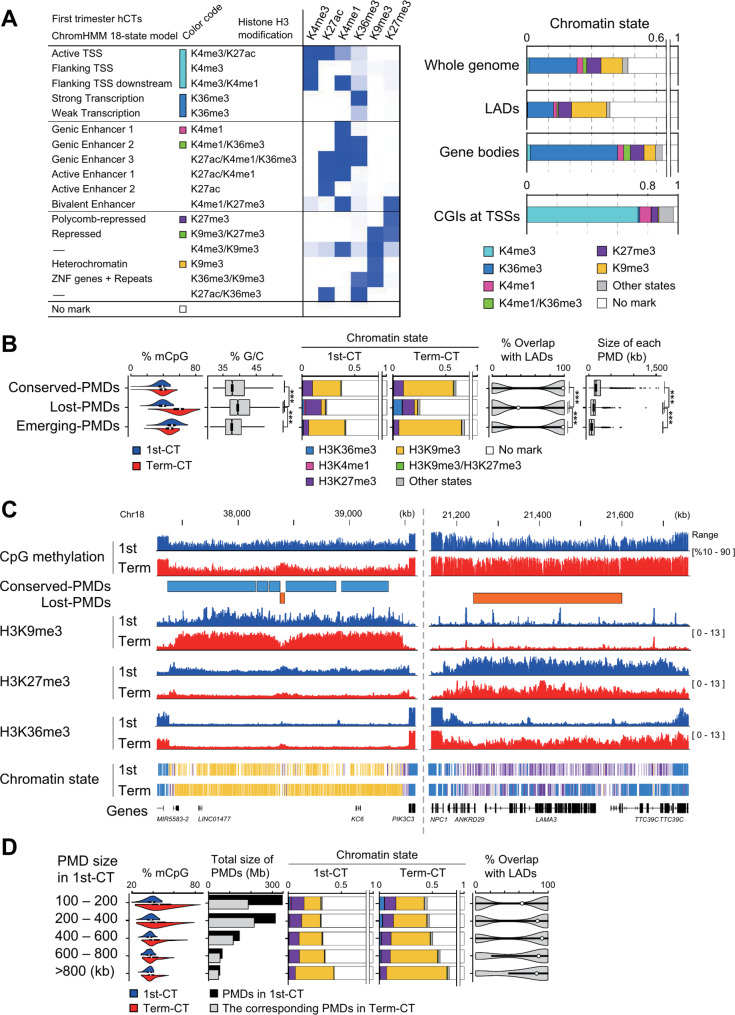
To examine the relationship between PMDs and histone H3 modifications in hCTs, we used chromatin immunoprecipitation sequencing (ChIP-seq) to determine the genome-wide distribution of six different histone H3 modifications (Additional file 1: Table S1). Using ChromHMM [38], we defined chromatin states across the hCT genome based on these modifications (Fig. 2A). Regions categorized as “Polycomb-repressed (H3K27me3)” were consistently present in all PMDs in both first trimester and term hCTs. In addition, regions defined as “Heterochromatin (H3K9me3)” were present in all PMDs of term hCTs and in all but four small PMDs of first trimester hCTs. Notable differences were observed between conserved-PMDs and lost-PMDs, with conserved-PMDs exhibiting similar chromatin characteristics to emerging-PMDs. Throughout placental development, from first trimester to term, conserved-PMDs predominantly overlapped with constitutive heterochromatic H3K9me3 marks and showed an increase in these marks (Fig. 2B, C, Additional file 1: Fig. S2A). In contrast, lost-PMDs were less frequently associated with H3K9me3 and more frequently with H3K27me3 marks. In addition, H3K27me3 marks decreased slightly from first trimester to term, whereas H3K36me3 marks increased over time. (Fig. 2B, C, Additional file 1: Fig. S2A). This suggests that changes in histone modifications precede changes in DNA methylation in placental PMDs.
Fig. 2.
Epigenetic dynamics in PMDs of human cytotrophoblasts. (A) A chromatin state model specific for hCTs generated with ChromHMM. The model is based on the enrichment levels of six different histone H3 modifications, and the blue intensity in the left panel reflects the histone H3 enrichment at different chromatin states. The right panel shows the distribution of these chromatin states across the genome, as well as in LADs, gene bodies, and CpG islands (CGIs) at transcription start sites (TSSs) in first trimester hCTs. The histone H3 modifications analyzed include K4me3 (lysine 4 trimethylation), K4me1 (lysine 4 monomethylation), K9me3 (lysine 9 trimethylation), K27me3 (lysine 27 trimethylation), K36me3 (lysine 36 trimethylation), and K27ac (lysine 27 acetylation) (B) Characteristics of three PMD categories across developmental stages in hCTs. mCpG: Violin plots illustrate the distribution of CpG methylation level, highlighting changes from first trimester to term. G/C: Box plots the G/C content. Chromatin state: This panel illustrates the distribution of chromatin states for both developmental stages, using the chromatin state model from panel A for clarity. Overlap with LADs: Violin plots show the percent overlap of PMDs with LADs. Size of each PMD: Box plots illustrate the size range of PMDs. Statistical significance was assessed using the Wilcoxon rank sum test, with triple asterisks (***) indicating P < 0.0001. 1st-CT, first trimester hCTs; Term-CT, term hCTs (C) A genome browser view of regions containing conserved-PMDs (left) and lost-PMDs (right). DNA methylation levels and histone H3 modification signals for first trimester hCTs and term hCTs are shown. Plots are provided for each histone H3 modification from one of the individual biological replicates, with PMDs marked as rectangles. Three chromatin states (H3K9me3, H3K27me3, H3K36me3) in ChromHMM were shown at the bottom. The color code for chromatin states is the same as in panel A (D) Comparative analysis of PMD characteristics by size across developmental stages in hCTs. mCpG: Violin plots illustrate the distribution of CpG methylation levels, highlighting changes from first trimester to term. Total size of PMDs: Black bars illustrate the total size of PMDs during the first trimester, while lighter shaded bars indicate the total size of the corresponding PMDs at term. Chromatin states: This section shows the distribution of chromatin states in PMDs at both stages of development. The color code for chromatin states is the same as in panel B. Overlap with LADs: Violin plots show the percent overlap of PMDs with LADs
This study showed that conserved-PMDs are more A/T rich, overlap more with LADs, and are longer compared to lost-PMDs (Fig. 2B). In addition, the characteristics of PMDs in first trimester hCTs vary with their length. Longer PMDs showed more H3K9me3 marks and fewer H3K27me3 marks (Fig. 2D). In contrast, shorter PMDs showed higher levels of DNA methylation by term hCTs and less overlap with LADs. Consequently, approximately 46% of the total area of short PMDs (< 200 kb) was absent in term hCTs, whereas most (90%) of the long PMDs (> 800 kb) remained as PMDs in term hCTs (Fig. 2D). In addition, 912 (74%) of the total 1,239 LADs overlapped with PMDs in first trimester hCTs. The remaining 327 LADs that did not overlap with PMDs were generally smaller and had higher G/C content (Additional file 1: Fig. S2B).
Characteristics of genomic regions prone to placental PMDs
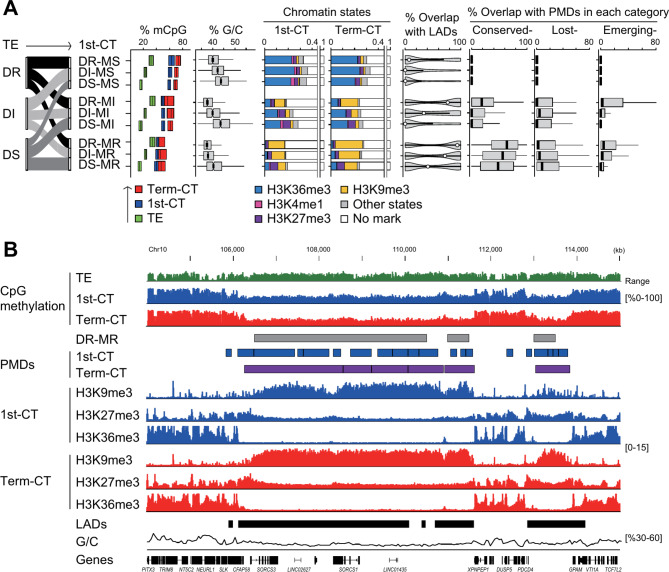
We analyzed the dynamic epigenetic changes in 500-kb genomic bins during the phases of global DNA demethylation and methylation in the placental lineage. To effectively capture the variation in epigenetic response, we classified these regions based on their susceptibility to epigenetic modifications. Thus, these bins were categorized into demethylation-resistant, demethylation-intermediate, and demethylation-susceptible groups based on their behavior during trophectoderm DNA demethylation and similarly for de novo DNA methylation sensitivity in first trimester hCTs (see the Methods section for more details). This tripartite classification facilitates detailed analysis of genomic regions ranging from those highly resistant to those highly susceptible to methylation changes during developmental transitions. This classification yielded nine distinct groups by combining each of the three groups from both stages (Fig. 3A), highlighting regions (DR-MR) that were both demethylation-resistant and methylation-resistant, often A/T-rich, closer to the nuclear lamina, enriched in H3K9me3 marks, and associated with conserved-PMDs (Fig. 3B).
Fig. 3.
Characteristics of genomic regions prone to placental PMDs. (A) Dynamic epigenetic changes in 500-kb genomic bins during the phases of global DNA demethylation and methylation in the placental lineage. Bin groups are categorized based on their resistance or susceptibility to global DNA demethylation during trophectoderm reprogramming (demethylation-resistant [DR], demethylation-intermediate [DI], demethylation-susceptible [DS]) and their sensitivity to methylation during the global de novo DNA methylation phase in first trimester hCTs (methylation-susceptible [MS], methylation-intermediate [MI], methylation-resistant [MR]). Details of the bin classification methods are described in the Methods section. The first flowchart illustrates the transition of genomic bins from trophectoderm to first trimester hCTs. mCpG: Box plots show the CpG methylation levels across the three developmental stages. G/C: Box plots show the distribution of G/C content. Chromatin states: This section shows the distribution of chromatin states in bins at both stages of development. Color codes in chromatin states correspond to those defined in Fig. 2A. Overlap with LADs: Violin plots show the percent overlap of bins with LADs. Overlap with PMDs: Box plots show the percent overlap of bins with PMDs in each category. TE, trophectoderm; 1st-CT, first trimester hCTs; Term-CT, term hCTs (B) A genome browser view of regions resistant to both demethylation and methylation (DR-MR). DNA methylation levels are shown for trophectoderm, first trimester hCTs, and term hCTs. Histone H3 modification signals for first trimester hCTs and term hCTs from one of the individual biological replicates are also shown. The positions of DR-MR, PMDs, and LADs are indicated by rectangles. G/C content and RefSeq genes are shown at the bottom
Features of PMDs in human trophoblast stem cells
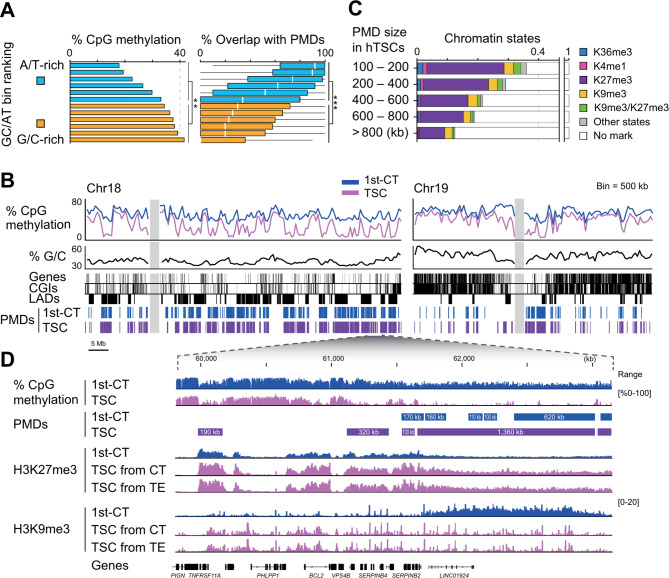
We established hTSCs from hCTs and trophectoderm [5], and generated comprehensive methylome profiles and six different histone H3 modifications in hTSCs (Additional file 1: Table S1). We observed that G/C-rich genomic regions were more methylated in hTSCs (Fig. 4A, B), consistent with observations in first trimester hCTs (Fig. 1D). We identified PMDs predominantly in A/T-rich genomic regions of hTSCs (Fig. 4A), located at genomic positions analogous to PMDs in first trimester hCTs, although with lower methylation levels (Fig. 4B, Additional file 1: Fig. S2C). Using ChromHMM [38], we categorized the chromatin states in the hTSC genome based on these histone H3 marks (Fig. 4C). Thus, significant differences in the organization of the histone H3 modification were observed in PMDs between hTSCs and hCTs. In contrast to PMDs in first trimester hCTs, which showed an enrichment for H3K9me3 marks (Fig. 2D), PMDs in hTSCs were predominantly enriched for H3K27me3 marks (Fig. 4C). Furthermore, longer PMDs in hTSCs were associated with fewer histone H3 modifications (Fig. 4C, D) compared to longer PMDs in first trimester hCTs, which showed a higher presence of these modifications (Fig. 2D).
Fig. 4.
Features of PMDs of human trophoblast stem cells. (A) PMD distribution in genomic regions with different G/C content in hTSCs. Bars (left) show total CpG methylation levels in 12 bin groups defined in Fig. 1D. Statistical significance was assessed using the exact Wilcoxon rank sum test; double asterisks (**) indicates P < 0.005. Box plots (right) show the overlap between PMDs in hTSCs and each bin group. Statistical significance was assessed using the exact Wilcoxon rank sum test; triple asterisks (***) indicate P < 0.0001 (B) Bulk CpG methylation levels in hTSCs plotted over Chr18 and Chr19. The format of the plots is consistent with Fig. 1C. PMDs are shown as blue and purple rectangles. 1st-CT, first trimester hCTs; TSC, hTSCs (C) Distribution of chromatin states within PMD groups categorized by size in hTSCs. ChromHMM was used to generate a chromatin state model for hTSCs based on the enrichment levels of six histone H3 modifications, similar to the analysis shown in Fig. 2A (D) Zoomed view of the locus containing both longer and shorter PMDs. CpG methylation levels and histone H3 modification signals are plotted for this region. PMDs are indicated by rectangles with sizes given in kilobases. CT, hCTs; TE, trophectoderm
Discussion
The human placenta has a unique methylome profile characterized by widespread hypomethylation and the presence of PMDs. This study investigates the epigenomic changes of PMDs in hCTs and hTSCs, emphasizing the spatiotemporal aspects of genomic regions to understand the mechanisms behind PMD formation and variability.
Our results show that G/C-rich genomic regions are significantly more susceptible to both global DNA demethylation and de novo DNA methylation during human placental lineage reprogramming compared to A/T-rich genomic regions. This observation is consistent with our previous research [37] suggesting that G/C-rich genomic regions, generally located closer to the nuclear center, are more susceptible to global methylation changes during both embryonic and germline reprogramming. This means that during human placental lineage reprogramming, the machinery responsible for DNA methylation and demethylation is more accessible to euchromatic G/C-rich regions close to the nuclear center.
Since the first publication of the human placental methylome profile [19], the unique presence of PMDs in the placenta, which are uncommon in other somatic cells, has been a topic of considerable intrigue [29]. Our findings, combined with our previous analysis of over 500 human and mouse WGBS datasets [37], suggest that PMD formation in the human placenta mirrors the patterns observed in early epiblasts, visceral endoderm, and prospermatogonia. This suggests that global DNA methylation events are infrequent in the heterochromatic A/T-rich regions near the nuclear lamina in the human placenta, resulting in the formation of PMDs due to a slow methylation process. As a result, genes within PMDs are expressed at low levels [19], likely due to the heterochromatic nature of these regions.
PMDs in hTSCs occur at genomic sites comparable to those in the placenta, although with lower methylation levels. This similarity may be due to the conservation of specific genomic structures that are less accessible to methylation processes in both the placenta and hTSCs. The reduced methylation levels of hTSCs compared to those in the placenta may reflect the distinct epigenetic and transcriptional states of hTSCs. Their early developmental stage and increased pluripotency require a more open and dynamic chromatin state, which may limit DNA methylation and lead to reduced methylation levels in PMDs.
In the placenta, PMDs predominantly have H3K9me3, a mark associated with heterochromatin formation and gene silencing, in contrast to the prevalence of H3K27me3 in PMDs in hTSCs, indicative of Polycomb-mediated repression common in the regulation of developmental genes [39]. This difference underscores the distinct epigenetic landscapes between these cell types. Larger PMDs in the placenta enriched for H3K9me3 suggest an advanced state of differentiation with stabilized gene expression necessary for placental function. This relationship between longer PMDs and increased H3K9me3 accumulation is consistent with observations in multiple cell types [40–43]. Conversely, the enrichment of H3K27me3 in smaller PMDs within hTSCs indicates their role in stem cell pluripotency and early differentiation, demonstrating a fine-tuned control of gene expression critical for developmental responsiveness.
Although only two replicates of hCTs and hTSCs were analyzed in this study, the strong correlation observed in DNA methylation levels across these replicates suggests robustness in our data, allowing us to discern meaningful biological insights regarding PMD differences. Future studies with a larger cohort may further refine these findings and validate our observations. The trophectoderm data, likely derived from fertility clinic samples, may not be representative of the general population experiencing typical pregnancies. In addition, our study did not experimentally verify the persistence of chromosomal territories within the human placenta. Despite these limitations, our detailed study of hCTs and hTSCs will advance our understanding of epigenetic regulation in early human development.
Conclusions
This study highlights the role of spatiotemporal genomic features in shaping megabase-scale DNA methylation patterns in the human placenta, particularly in the formation of PMDs. These PMDs arise through a slow methylation process within heterochromatic regions near the nuclear lamina during placental lineage reprogramming. In addition, our analysis distinguishes the PMD patterns observed in cytotrophoblasts from those in trophoblast stem cells. These findings enhance our understanding of placental biology and provide the basis for further studies of human development and related diseases.
Methods
Sample Collection
Human placentas were obtained from healthy women who gave informed consent after approval by the ethics committees of Tohoku University School of Medicine and Kyushu University School of Medicine. hCTs were isolated from fresh placental tissue at two developmental stages: first trimester (7 weeks) and term pregnancy, as previously described [4]. Genomic DNA was extracted from individual donors at each time point, with two biological replicates (one male and one female) for each sample. hTSCs were prepared according to established protocols [5]. First trimester hCTs (female, 6 weeks) and trophectoderm (male) used to establish hTSCs was obtained from single donors.
Sequencing
Epigenomic profiling of hCTs and hTSCs was performed using WGBS and ChIP-seq protocols approved by the IHEC. For each sample, data from the sequenced biological replicates were merged for analysis to increase statistical power and coverage. For WGBS, libraries were prepared using the post-bisulfite adapter tagging (PBAT) method [44]. Sequencing was performed on an Illumina HiSeq 1500/2500 platform using HCS v2.0.5 and RTA v1.17.20 base calling software, which is suitable for PBAT-based WGBS [45]. For ChIP-seq, six H3 histone modifications were analyzed in hCTs and hTSCs using ChIP reagents (Nippon Gene) and monoclonal antibodies against K4me3 (CMA304), K4me1 (CMA302 or D1A9), K27ac (CMA309), K27me3 (CMA323), K9me3 (CMA318), and K36me3 (CMA333). Libraries were constructed using the Ovation Ultralow System V2 (NuGEN) and sequenced on an Illumina HiSeq 2500 platform. Reads were aligned to the human reference genome (hg19) using Bowtie2 v4.1.2 [46]. The 18-state chromatin model was generated from these modifications using ChromHMM v1.12 [38].
DNA methylation analysis
Methylome maps were constructed using the human reference genome (hg19) and associated metadata, including CpG islands and LADs, from the UCSC Genome Browser. Raw fastq files were processed by trimming low quality base sequences (< Q30) from the 3’ ends, retaining reads longer than 50 bases. These were aligned to the reference genome (hg19) using Bismark v0.10.0 [47] with specific parameters including a seed length of 28, a maximum of one mismatch in the seed, and the “--pbat” option for PBAT sequences.
Methylation analysis focused only on uniquely aligned reads, taking into account autosomal CpGs. CpG methylation levels were calculated by combining the counts from both DNA strands. Global CpG methylation levels for each sample were determined by summing the number of methylated and unmethylated CpGs on autosomes, considering only CpGs with a sequencing depth of five or more. Methylation at non-CpG sites was not included in this analysis. WGBS data for the human trophectoderm and term whole placenta from the Gene Expression Omnibus (accession GSE81233 and GSE39775, respectively) were analyzed for methylation levels using identical methods. All box and violin plots in the figures were generated using R v3.6.0.
A bin size of 500 kb was chosen to effectively observe megabase-scale methylome patterns. This approach ensures that methylation levels are confidently determined even in regions of low sequence depth, while capturing significant megabase-scale changes in DNA methylation. Chromosomes were divided into 5,413 non-overlapping 500-kb bins, which were then sorted and ranked by their CpG methylation levels from highest to lowest. During the global hypomethylation phase of the trophectoderm, shown in Fig. 3A, these bins were categorized into groups: demethylation-resistant (top 1,800 bins), intermediate (middle 1,800 bins), and susceptible (bottom 1,800 bins). A similar categorization occurred during the first trimester hCTs stage into methylation-susceptible (top 1,800 bins), intermediate (middle 1,800 bins), and resistant (bottom 1,800 bins) groups. This categorization across both stages formed nine distinct groups, allowing for a comprehensive analysis of methylation dynamics.
In this study, PMDs were identified using the following protocol. Chromosomes were segmented into non-overlapping 10-kb bins, and each bin was scored for its CpG methylation level. Bins with methylation levels below the global average of the sample were classified as small PMDs. These small PMDs were merged into larger PMDs if they were adjacent. The analysis focused only on PMDs larger than 100 kb.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Hiroshi Kimura (Institute of Science Tokyo) for providing the necessary antibodies and Wan Kin Au Yeung, Tomomi Akinaga, Miho Miyake, and Junko Oishi (Kyushu University) for technical assistance.
Abbreviations
- A/T
Adenine/thymine
- ChIP-seq
Chromatin immunoprecipitation sequencing
- Chr
Chromosome
- CpG
Cytosine-guanine
- G/C
Guanine/cytosine
- hCTs
Human cytotrophoblasts
- hTSCs
Human trophoblast stem cells
- IHEC
International Human Epigenome Consortium
- LADs
Lamina-associated domains
- PBAT
Post-bisulfite adapter tagging
- PMDs
Partially methylated domains
- RRBS
Reduced representation bisulfite sequencing
- WGBS
Whole-genome bisulfite sequencing
Author contributions
H.T. designed the study, curated the data, performed formal analyses, conducted the study, developed the methodology, validated the results, created the visualizations, and contributed to drafting the original manuscript and revising and editing the manuscript. H.O. curated the data, performed formal analysis, conducted the study, developed the methodology, and provided resources. K.S. contributed to the conduct of the study and development of the methodology. T.S. contributed to the formal analysis and software development. H.H. provided resources. C.K. and D.S. were responsible for data curation. T.A. acquired funding, provided resources, and managed the project. H.S. acquired funding and supervised the team. M.S. acquired funding, managed the project, supervised the team, and revised and edited the manuscript.
Funding
This study was supported in part by grants from the Japan Agency for Medical Research and Development (AMED) under grant numbers JP17gm0510011 and JP19gm1310001, the Medical Research Center Initiative for High Depth Omics (Kyushu University MIB), and the MEXT Coalition of Universities for Research Excellence Program (JPMXP1323015486, Kyushu University MIB).
Data availability
All raw sequencing data from WGBS and ChIP-seq of human cytotrophoblasts and trophoblast stem cells have been deposited and made publicly available at the National Bioscience Database Center under the accession numbers hum0086 and hum0112.
Declarations
Ethics approval and consent to participate
Human placentas were obtained from healthy women who gave informed consent after approval by the ethics committees of the Tohoku University School of Medicine (research licenses 2015-1-219 and 2017-1-200) and Kyushu University School of Medicine (research licenses 526-00 and 723-01). We confirm that all experiments were performed in accordance with relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hidehiro Toh, Email: toh@nig.ac.jp.
Hiroyuki Sasaki, Email: hsasaki@bioreg.kyushu-u.ac.jp.
Mikita Suyama, Email: mikita@bioreg.kyushu-u.ac.jp.
References
- 1.Aplin JD, Myers JE, Timms K, Westwood M. Tracking placental development in health and disease. Nat Rev Endocrinol. 2020;16:479–94. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Li ZH, Wang YX, Liu TH. A comprehensive review of human trophoblast fusion models: recent developments and challenges. Cell Death Discov. 2023;9:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pastor WA, Kwon SY. Distinctive aspects of the placental epigenome and theories as to how they arise. Cell Mol Life Sci. 2022;79:569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamada H, Okae H, Toh H, Chiba H, Hiura H, Shirane K, et al. Allele-specific methylome and transcriptome analysis reveals widespread imprinting in the human placenta. Am J Hum Genet. 2016;99:1045–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okae H, Toh H, Sato T, Hiura H, Takahashi S, Shirane K, et al. Derivation of human trophoblast stem cells. Cell Stem Cell. 2018;22:50–63. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi S, Okae H, Kobayashi N, Kitamura A, Kumada K, Yaegashi N, et al. Loss of p57KIP2 expression confers resistance to contact inhibition in human androgenetic trophoblast stem cells. Proc Natl Acad Sci USA. 2019;116:26606–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharya B, Home P, Ganguly A, Ray S, Ghosh A, Islam MR, et al. Atypical protein kinase C iota (PKCλ/ι) ensures mammalian development by establishing the maternal-fetal exchange interface. Proc Natl Acad Sci USA. 2020;117:14280–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castel G, Meistermann D, Bretin B, Firmin J, Blin J, Loubersac S, et al. Induction of human trophoblast stem cells from somatic cells and pluripotent stem cells. Cell Rep. 2020;33:108419. [DOI] [PubMed] [Google Scholar]
- 9.Muto M, Chakraborty D, Varberg KM, Moreno-Irusta A, Iqbal K, Scott RL, et al. Intersection of regulatory pathways controlling hemostasis and hemochorial placentation. Proc Natl Acad Sci USA. 2021;118:e2111267118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi N, Okae H, Hiura H, Kubota N, Kobayashi EH, Shibata S, et al. The microRNA cluster C19MC confers differentiation potential into trophoblast lineages upon human pluripotent stem cells. Nat Commun. 2022;13:3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeyarajah MJ, Jaju Bhattad G, Kelly RD, Baines KJ, Jaremek A, Yang FP, et al. The multifaceted role of GCM1 during trophoblast differentiation in the human placenta. Proc Natl Acad Sci USA. 2022;119:e2203071119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frost JM, Amante SM, Okae H, Jones EM, Ashley B, Lewis RM, et al. Regulation of human trophoblast gene expression by endogenous retroviruses. Nat Struct Mol Biol. 2023;30:527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuna M, Dhakal P, Iqbal K, Dominguez EM, Kent LN, Muto M, et al. CITED2 is a conserved regulator of the uterine-placental interface. Proc Natl Acad Sci USA. 2023;120:e2213622120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimizu T, Oike A, Kobayashi EH, Sekiya A, Kobayashi N, Shibata S, et al. CRISPR screening in human trophoblast stem cells reveals both shared and distinct aspects of human and mouse placental development. Proc Natl Acad Sci USA. 2023;120:e2311372120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varberg KM, Dominguez EM, Koseva B, Varberg JM, McNally RP, Moreno-Irusta A, et al. Extravillous trophoblast cell lineage development is associated with active remodeling of the chromatin landscape. Nat Commun. 2023;14:4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hori T, Okae H, Shibata S, Kobayashi N, Kobayashi EH, Oike A, et al. Trophoblast stem cell-based organoid models of the human placental barrier. Nat Commun. 2024;15:962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olova N, Krueger F, Andrews S, Oxley D, Berrens RV, Branco MR, et al. Comparison of whole-genome bisulfite sequencing library preparation strategies identifies sources of biases affecting DNA methylation data. Genome Biol. 2018;19:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roadmap Epigenomics Consortium. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroeder DI, Blair JD, Lott P, Yu HO, Hong D, Crary F, et al. The human placenta methylome. Proc Natl Acad Sci USA. 2013;110:6037–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Court F, Tayama C, Romanelli V, Martin-Trujillo A, Iglesias-Platas I, Okamura K, et al. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014;24:554–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Fan Y, Li G, Xu X, Duan J, Li R, et al. DNA methylation reprogramming of functional elements during mammalian embryonic development. Cell Discov. 2018;4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim YC, Li J, Ni Y, Liang Q, Zhang J, Yeo GS, et al. A complex association between DNA methylation and gene expression in human placenta at first and third trimesters. PLoS ONE. 2017;12:e0181155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nordor AV, Nehar-Belaid D, Richon S, Klatzmann D, Bellet D, Dangles-Marie V, et al. The early pregnancy placenta foreshadows DNA methylation alterations of solid tumors. Epigenetics. 2017;12:793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gamage TK, Schierding W, Tsai P, Ludgate JL, Chamley LW, Weeks RJ, et al. Human trophoblasts are primarily distinguished from somatic cells by differences in the pattern rather than the degree of global CpG methylation. Biology Open. 2018;7:bio034884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gamage TK, Schierding W, Hurley D, Tsai P, Ludgate JL, Bhoothpur C, et al. The role of DNA methylation in human trophoblast differentiation. Epigenetics. 2018;13:1154–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan V, Hui D, Yin Y, Peñaherrera MS, Beristain AG, Robinson WP. Cell-specific characterization of the placental methylome. BMC Genomics. 2021;22:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang B, Kim MY, Elliot G, Zhou Y, Zhao G, Li D, et al. Human placental cytotrophoblast epigenome dynamics over gestation and alterations in placental disease. Dev Cell. 2021;56:1238–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schroeder DI, LaSalle JM. How has the study of the human placenta aided our understanding of partially methylated genes? Epigenomics. 2013;5:645–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu P, Guo H, Ren Y, Hou Y, Dong J, Li R, et al. Single-cell DNA methylome sequencing of human preimplantation embryos. Nat Genet. 2018;50:12–9. [DOI] [PubMed] [Google Scholar]
- 31.Shibata S, Kobayashi EH, Kobayashi N, Oike A, Okae H, Arima T. Unique features and emerging in vitro models of human placental development. Reprod Med Biol. 2020;19:301–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cremer M, von Hase J, Volm T, Brero A, Kreth G, Walter J, et al. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–67. [DOI] [PubMed] [Google Scholar]
- 33.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. [DOI] [PubMed] [Google Scholar]
- 34.Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M, et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell. 2015;163:134–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan L, Xing D, Chang CH, Li H, Xie XS. Three-dimensional genome structures of single diploid human cells. Science. 2018;361:924–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girelli G, Custodio J, Kallas T, Agostini F, Wernersson E, Spanjaard B, et al. GPSeq reveals the radial organization of chromatin in the cell nucleus. Nat Biotechnol. 2020;38:1184–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toh H, Sasaki H. Spatiotemporal DNA methylation dynamics shape megabase-scale methylome landscapes. Life Sci Alliance. 2024;7:e202302403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicetto D, Zaret KS. Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr Opin Genet Dev. 2019;55:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hovestadt V, Jones DT, Picelli S, Wang W, Kool M, Northcott PA, et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510:537–41. [DOI] [PubMed] [Google Scholar]
- 41.Bartholdy B, Lajugie J, Yan Z, Zhang S, Mukhopadhyay R, Greally JM, et al. Mechanisms of establishment and functional significance of DNA demethylation during erythroid differentiation. Blood Adv. 2018;2:1833–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salhab A, Nordström K, Gasparoni G, Kattler K, Ebert P, Ramirez F, et al. A comprehensive analysis of 195 DNA methylomes reveals shared and cell-specific features of partially methylated domains. Genome Biol. 2018;19:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Y, Wang P, Zhou W, Luo M, Xu Z, Cheng R, et al. Comprehensive analysis of partial methylation domains in colorectal cancer based on single-cell methylation profiles. Brief Bioinform. 2021;22:bbab267. [DOI] [PubMed] [Google Scholar]
- 44.Miura F, Enomoto Y, Dairiki R, Ito T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 2012;40:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toh H, Shirane K, Miura F, Kubo N, Ichiyanagi K, Hayashi K, et al. Software updates in the Illumina HiSeq platform affect whole-genome bisulfite sequencing. BMC Genomics. 2017;18:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing data from WGBS and ChIP-seq of human cytotrophoblasts and trophoblast stem cells have been deposited and made publicly available at the National Bioscience Database Center under the accession numbers hum0086 and hum0112.