Abstract
Background and objectives
Methionine adenosyltransferase I/III deficiency used to be considered a relatively benign disease. This study aims to elucidate the clinical characteristics of methionine adenosyltransferase I/III deficiency patients with neurological manifestations.
Methods
The clinical data, blood amino acids, plasma total homocysteine, gene variants, brain imaging, treatments and outcomes of 15 patients with methionine adenosyltransferase I/III deficiency were retrospectively analyzed.
Results
Of these 15 patients, 10 demonstrated neurological abnormalities, with delayed language development, learning difficulties or abnormal brain imaging findings. Eleven patients were identified by newborn screening. Patients with demyelination showed significantly higher blood methionine concentrations at baseline (1102 vs. 396 µmol/L), and their blood methionine remained markedly elevated despite a low-methionine diet. Their plasma total homocysteine was normal to moderate elevated. One patient underwent liver transplantation aged 8 years, which reduced his serum methionine concentration to normal. Compound heterozygous and homozygous MAT1A variants were identified from the patients. Among the 21 variants observed, nine have been reported previously, while 12 were novel.
Conclusions
Methionine adenosyltransferase I/III deficiency is not just a benign disease. Severe persistent hypermethioninemia can cause brain injuries, especially in the white matter. Liver transplantation may be a potential treatment option for refractory methionine adenosyltransferase I/III deficiency.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12887-024-05196-x.
Keywords: Methionine adenosyltransferase I/III deficiency, Newborn screening, Methionine, MAT1A gene, Brain damage
Introduction
Methionine adenosyltransferase (MAT) I/III deficiency is a rare inherited metabolic disorder and a primary cause of hypermethioninemia [1]. It was first identified in 1974 following newborn screening using dried blood spot analysis [2]. MAT I and III are S-adenosylmethionine (AdoMet) synthetases [3] that catalyze the transfer of an adenosyl group from ATP to methionine—the first step of the methionine cycle. They mainly function in the liver. Up to 85% of all methylation reactions and 50% of methionine catabolism occur in liver tissues [4]. MAT I/III deficiency is caused by pathogenic variants in the MAT1A gene which located in the 10q22.3 region [3], and is characterized by persistent isolated hypermethioninemia without homocystinuria [5].
The overall incidence of MAT I/III deficiency is approximately 1 in 26,000–105,000 [6, 7]. The disorder can be autosomal recessively or dominantly inherited. An increasing number of patients with homozygous or compound heterozygous MAT1A mutations have been identified during newborn screening [5]. The clinical outcomes of MAT I/III deficiency remain poorly understood. It used to be considered relatively benign [8, 9], but some clinicians have noticed the damage caused by the autosomal recessive form of this disease, and neurological sequelae of persistent hypermethioninemia have also been reported [3, 10]. Surtees et al. [10] reported that cerebrospinal fluid AdoMet deficiency was associated with demyelination among infants with inborn errors of the methyl-transfer pathway, and they showed that restoration of AdoMet was associated with remyelination. These findings generally suggest that heterozygosity for a dominant MAT1A variant does not cause a clinically significant disease phenotype, whereas homozygosity or compound heterozygosity for recessive pathogenic variants is associated with pathological hypermethioninemia and neurological symptoms [6].
The lack of specific signs and symptoms renders early diagnosis of MAT I/III deficiency challenging. Nevertheless, newborn screening represents the best approach to detect hypermethioninemia [5], while tandem mass spectrometry and the expansion of newborn screening have improved the detection of MAT I/III deficiency. The differences in clinical characteristics between benign conditions and pathological diseases remain unclear. Therefore, we aimed to elucidate the clinical characteristics and neurological outcomes of MAT I/III-deficient patients.
Materials and methods
Study participants
Fifteen patients with persistent hypermethioninemia who were diagnosed with MAT I/III deficiency at the Children’s Medical Center (Department of Pediatrics), Peking University First Hospital, between January 2014 and October 2023 were retrospectively analyzed. The data collected included clinical features, brain imaging findings, metabolic profiles, gene variants, treatments and outcomes. Evidence of brain abnormalities was also recorded, and intelligence or development quotient (IQ or DQ) assessments were performed using standardized neuropsychological tests, including the Wechsler Intelligence Scale for Children (3rd edition), the Bayley Scales of Infant Development in (2nd edition), and the Gesell Development Scale.
Biochemical assays
Blood amino acids concentrations in dried blood spots were analyzed using liquid chromatography-tandem mass spectrometry (Triple Quad 4500; AB Sciex, CA, USA). Metabolite concentrations were calculated using the Chemoview software (2.02, AB Sciex). The plasma total homocysteine was determined using a chemiluminescent immunoassay (Abbott I 2000; Abbott Park, IL, USA), and concentrations ≥ 15 µmol/L were considered elevated. Hepatic function was also evaluated.
Molecular testing
Genomic DNA was extracted from peripheral blood, and causative variants were determined using targeted next-generation or whole-exon sequencing. The DNA sequence was analyzed and compared to the published human genome build UCSC hg19 reference sequence. All variants were independently confirmed by Sanger sequencing, and were evaluated in accordance with the Human Gene Mutation Database (HGMD, http://www.hgmd.org) and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar). The pathogenicity of novel variants was evaluated on the basis of American College of Medical Genetics and Genomics standards and guidelines [11].
Statistical analysis
All statistical analyses were performed using SPSS version 22.0 (IBM, Armonk, NY, USA). The t-test was applied to evaluate the differences between pre- and post-treatment blood methionine levels. Statistical significance was considered as P < 0.05.
Results
Clinical data
A total of 15 patients (six male, nine female) were involved in this study (Table 1). The age at the first visit ranged between 1 month and 11 years (median, 1.5 years). Eleven patients were identified by newborn screening. Four were clinically diagnosed.
Table 1.
Clinical and genetic features of the 15 patients with hypermethioninemia due to methionine adenosyltransferase I/III deficiency
| Patient No. | Sex | NBS | Age at first visit | Current Age | Clinical features | DQ or IQ | Blood Met (µmol/L) | MAT1A variants | Brain MRI findings | CNS status | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pre-treatment | Post-treatment | Paternal | Maternal | |||||||||
| 1 | F | Y | 1 y | 8 y | No symptom | Normal | 1118 | 574–1148 | Cys120Tyr | Gly26Arg | Lesion | Abnormal |
| 2 | M | Y | 6 m | 12 y | Emotional instability, anger outbursts | UN | 1140 | 418–1200 | Tyr92His | Arg299Cys | Lesion | Abnormal |
| 3 | F | N | 3 y 8 m | 11 y | Involuntary hand-shaking, tremor, fatigue | DQ 70 | 1644 | 608–1685 | Gly253Arg | Arg292His | Lesion | Abnormal |
| 4 | M | N | 4 y 5 m | 8 y | Language delay, reduced communication, drowsiness, unstable gait, ataxia | DQ 62.1 | 1816 | 501 | Arg292Cys | Arg292Cys | Lesion | Abnormal |
| 5 | F | N | 11 m | 6 y | Language delay, learning difficulty, dizziness, fatigue |
IQ 59 DQ 54 |
1189 | 279–1088 | Arg299Cys | Tyr92His | Lesion | Abnormal |
| 6 | F | Y | 1 y 6 m | 5 y | Malodourous urine, language delay, inattention | DQ 42.6–78.1 | 982 | 567–733 | Gly336Arg | Ala157Val | Lesion | Abnormal |
| 7 | F | Y | 3 y | 5 y | Language delay | UN | - | 130 | Pro390His | Gln208Pro | Lesion | Abnormal |
| 8 | M | N | 8 y | 10 y | Fatigue, sleeping disorder | UN | - | 56–189 | Pro390His | Gln208Pro | Lesion | Abnormal |
| 9 | F | Y | 7 m | 3 y | No symptom | UN | 308 | 308–1121 | Arg299Cys | Gly63Valg | ND | Normal |
| 10 | M | Y | 1 y 6 m | 5 y | No symptom | UN | 419 | 199–1248 | Pro151Thr | Ala55Thr | ND | Normal |
| 11 | F | Y | 15 d | 3 y | No symptom | UN | 117 | 151–222 | Arg356Gln | Cys60Arg | ND | Normal |
| 12 | M | Y | 1 m | 8 y | No symptom | UN | 306 | 550–926 | Arg296Cys | Arg299Cys | Lesion | Abnormal |
| 13 | F | Y | 3 m | 8 y | No symptom | UN | 475 | 153–588 | Arg264Cys | Arg199His | ND | Normal |
| 14 | F | Y | 4 y | 7 y | No symptom | UN | 662 | - | Arg299Cys | Pro232Leu | ND | Normal |
| 15 | M | Y | 3 y 6 m | 6 y | Malodorous urine, language delay, inattention, low climbing ability | DQ 46–67 | 622 | 497–1200 | Asp191-Pro197dup | Cys60Arg | Lesion | Abnormal |
DQ /IQ = developmental quotient/intelligence quotient; CNS = central nervous system; F = female; MRI = magnetic resonance imaging; M = male; Met = methionine; N = no; NBS = newborn screening; ND = not determined; UN = unknown. Y= yes. Blood methionine: Normal range = 10–50 µmol/L, Slightly elevated = 50–300 µmol/L, Moderately elevated = 300–600 µmol/L, Severe elevated = > 600 µmol/L
Brain damage was identified in 10 patients, including neurological symptoms only (Patient 7), brain MRI abnormalities without clinical symptoms (Patient 1 and 12), or both (Patient 2–6, Patient 8, and Patient 15). Patient 7 and Patient 8 were siblings. While Patient 7 was symptomatic, brain MRI was not performed due to non-compliance. The majority of symptomatic patients demonstrated an insidious onset of language development delay and learning difficulties, with the age of onset ranging between 17 months and 5 years (average, 2.83 ± 1.15 years). IQ or DQ was slightly low in five patients (Patient 3–6 and Patient 15). Malodourous urine was observed in three patients (Patient 4, 6 and 15). The remaining five patients (Patient 9–11, 13 and 14) were clinically asymptomatic throughout the follow-up period.
Brain MRI features
Eleven patients underwent brain MRI. Lesions were observed in nine patients (81.8%, Patient 1–6, 8, 12, and 15), as shown in Table 2; Fig. 1A–D. Cerebral white matter demyelination was observed in all of them.
Table 2.
Brain magnetic resonance imaging (MRI) abnormalities in the 9 symptomatic patients
| Patient No. | Age | Brain MRI findings |
|---|---|---|
| 1 | 3 y | Diffuse symmetrical long T1 and T2 signals, T2FLARE high signal, DWI high signal in bilateral cerebral hemisphere white matter involving the corpus callosum, bilateral globus pallidus, and the dorsal brainstem. |
| 2 | 7 y | T2 high-intensity signal in bilateral cerebral hemisphere white matter (Fig. 1A). |
| 3 | 3 y 4 m | Diffuse high-intensity T2WI and FLAIR signals in the cerebral white matter and corpus callosum, with low ADC values (Fig. 1B). |
| 4 | 4 y | Long T1 and T2 signals, and T2 FLARE high signals in bilateral cerebral hemisphere white matter area, medulla, dorsal midbrain, and dorsal pons; and increased T2WI signal in bilateral anterior limb of internal capsule, globus pallidus, and corpus callosum (Fig. 1C). |
| 5 | 11 y | Long T1 and T2 signals, T2 FLARE high signal, and DWI high signal in bilateral cerebral hemisphere white matter, bilateral internal capsule, corpus callosum; and T2 long signal in bilateral midbrain, pons, and medulla oblongata. |
| 6 | 1 y 6 m | Demyelination of subcortical white matter of the occipital lobe (Fig. 1D). |
| 8 | 3 y | Abnormal signal in white matter of the left parietal lobe. |
| 12 | 5 y | Abnormal signals in white matter, globus pallidus, and brainstem of both cerebral hemispheres. |
| 15 | 2 y | Demyelination of white matter in bilateral cerebral hemispheres. |
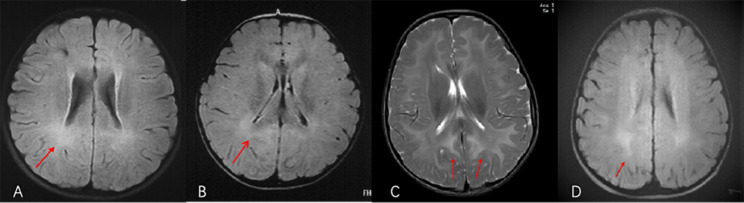
Fig. 1.
Brain MRI abnormalities observed in (A) Patient 2, (B) Patient 3, (C) Patient 4, and (D) Patient 6. (A) Brain MRI of Patient 2 (age, 7 years) showed a T2 high-intensity signal in bilateral cerebral hemisphere white matter. (B) Brain MRI of Patient 3 (age, 3 years 4 months) showed diffuse high-intensity T2-weighted imaging (T2WI) and fluid-attenuated inversion recovery (FLAIR) signals with low apparent diffusion coefficient values in the cerebral white matter and corpus callosum. (C) Brain MRI of Patient 4 showed long T1 and T2 signals and high-intensity T2 FLAIR signals in bilateral cerebral hemisphere white matter area, medulla, dorsal midbrain, and dorsal pons and increased T2WI signals in the bilateral anterior limb of the internal capsule, globus pallidus, and corpus callosum. (D) Brain MRI of Patient 6 (age, 1 year 6 months) showed demyelination of subcortical white matter of the occipital lobe
Biochemical analysis
The blood methionine concentrations of asymptomatic and symptomatic patients are shown in Tables 1 and 3. Severe hypermethioninemia was observed in eight patients (Patient 1–6 and Patient 14–15). Pre-treatment methionine concentrations ranged between 306 and 1816 µmol/L among patients with brain abnormalities (Patient 7 and 8 had missing results) and 117–475 µmol/L among those without brain abnormalities. Significant differences were observed between the mean pre-treatment methionine concentrations of these two patient groups (1102 ± 492 µmol/L vs. 396 ± 202 µmol/L; P < 0.05; Table 3). The blood methionine concentrations of all parents were normal. Plasma total homocysteine levels were normal or elevated (4.1–57.8 µmol/L) in all patients. Serum alanine aminotransferase, aspartate transaminase, total bilirubin, alkaline phosphatase, albumin, total plasma protein, and gamma glutamyl-transpeptidase levels were normal.
Table 3.
Blood methionine concentrations of pre-treatment in symptomatic and asymptomatic patients
| CNS status | Number | Blood methionine concentrations (µmol/L) | t-value | P-value | |
|---|---|---|---|---|---|
| Mean | Standard deviation | ||||
| Abnormal | 8 | 1102 | 492 | 3.017 | 0.012 |
| Normal | 5 | 396 | 202 | ||
CNS = central nervous system
Genetic studies
Homozygous and compound heterozygous MAT1A variants were identified in all patients, and an autosomal recessive inheritance pattern was observed in all patients. No other pathogenic or likely pathogenic variant was identified related to neurodevelopmental disorders or inborn metabolic disorders. Twenty-one variants were detected, of which nine (c.895 C > T, c.874 C > T, c.274T > C, c.188G > T, c.757G > A, c.695 C > T, c.596G > A, c.1006G > A, c.790 C > T) had been reported previously [7, 12–16] and 12 variants (c.1067G > A, c.887 A > G, c.875G > A, c.359G > A, c.178T > C, c.163G > A, c.76G > A, c.572-592dupACAATGGCGCAGTCATCCTG, c.470 C > T, c.451 C > A, c.1169 C > A, c.623 A > C) were novel (Table 4). c.895 C > T was observed in five patients, while the c.274T > C and c.178T > C variants were observed in two patients from different families. c.895 C > T is a variant hotspot in our patients. The data of 12 novel variants for the MAT1A gene have been uploaded to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar, submission ID SUB12700124, SUB12913275). No autosomal dominant mutations were found.
Table 4.
21 variants identified from MAT1A *of 15 patients
| No. | Nucleotide alteration |
Protein alteration |
Exon/ Intron |
ACMG/AMP grading | Reported PMID/References | |
|---|---|---|---|---|---|---|
| Classification | Evidence code | |||||
| 1 | c.1067G > A | Arg356Gln | 8 | VUS | PM2-supporting, PP3 | This study |
| 2 | c.895 C > T | Arg299Cys | 7 | LP | PM2-supporting, PS3_supporting, PM3_Strong | 20675163[12], 26933843[7] |
| 3 | c. 887 A > G | Tyr296Cys | 7 | VUS | PM2-supporting, PP3 | This study |
| 4 | c.875G > A | Arg292His | 7 | VUS | PM2-supporting, PP3 | This study |
| 5 | c.874 C > T | Arg292Cys | 7 | VUS | PM2-supporting, PP3, PM3_supporting | 20675163[12] |
| 6 | c.359G > A | Cys120Tyr | 4 | VUS | PM2-supporting, PP3 | This study |
| 7 | c.274T > C | Tyr92His | 3 | LP | PM3-Strong, PM2-supporting, PP3 | 15569761[13], 26933843[7], Ma YY, et al.[14], 2018 |
| 8 | c.188G > T | Gly63Val | 3 | VUS | PM2-supporting, PP3 | 31851615[15] |
| 9 | c.178T > C | Cys60Arg | 3 | VUS | PM2-supporting, PP3 | This study |
| 10 | c.163G > A | Ala55Thr | 2 | VUS | PM2-supporting, PP3 | This study |
| 11 | c.76G > A | Gly26Arg | 1 | VUS | PM2-supporting, PP3 | This study |
| 12 | c.757G > A | Gly253Arg | 6 | VUS | PM2-supporting, PP3 | Ma YY, et al.[14] 2018 |
| 13 | c.695 C > T | Pro232Leu | 6 | VUS | PM2-Supporting, PM3, PP3 | 31851615 [15], Wu J, et al.[16], 2019 |
| 14 | c.596G > A | Arg199His | 6 | VUS | PM2-supporting, PM3_supporting, PP3 | 26,289,392[19] |
| 15 | c.572-592dupACAATGGCGCAGTCATCCTG | Splicing | Intron 5 | VUS | PM2-Supporting, PVS1_Supporting | This study |
| 16 | c.470 C > T | Ala157Val | 5 | VUS | PM2-Supporting, PP3 | This study |
| 17 | c.451 C > A | Pro151Thr | 5 | VUS | PM2-Supporting, PP3 | This study |
| 18 | c.1006G > A | Gly336Arg | 8 | VUS | PM2-supportting, PP3 | 10677294[3] |
| 19 | c.790 C > T | Arg264Cys | 7 | VUS | PM2-supportting, PP3 | 10677294[3] |
| 20 | c.1169 C > A | Pro390His | 9 | VUS | PM2-Moderate, PP2-Supporting, PP3-Supporting | This study |
| 21 | c.623 A > C | Gln208Pro | 6 | LP | PM1-Moderate, PM2-Moderate, PP2-Supporting, PP3 | This study |
* The transcript references were NM_000429. ACMG/AMP, American College of Medical Genetics and Genomics and the Association for Molecular Pathology; LP = likely pathogenic, VUS = uncertain significance
Treatments and follow-up
Low-methionine diet
Dietary methionine restriction was initiated after diagnosis in all patients. However, compliance was challenging among young patients, resulting in fluctuations in serum methionine concentrations. No marked reductions to normal in methionine concentrations were observed with dietary therapy.
Drug therapy
Eight patients received vitamin B6 before genetic analysis, but no significant effects were observed. Blood methionine levels decreased in six children who received oral ademethionine 1,4-butanedisulfonate (500–1000 mg/day) in addition to a low-methionine diet. Their blood methionine concentrations decreased from 926 to 1685 µmol/L to 502–757 µmol/L after 1 month.
Liver transplantation
One patient (Patient 2) benefited from newborn screening and was initially managed with dietary control and ademethionine 1,4-butanedisulfonate. However, development and progression of white matter lesions were observed when he was 7 years old. After specialist consultation and obtaining consent from his family, segmental living-donor liver transplantation from his father was performed at the age of 8 years. His blood methionine concentration decreased from 1200 µmol/L to 108 µmol/L one week after liver transplantation, and remained stable thereafter (Fig. 2). At present, he is 11 years old and studying in primary school. His physical and psychomotor development have remained good, and the white matter of his brain has shown significant improvement.
Fig. 2.
Blood methionine concentrations of Patient 2 before and after liver transplantation. Before liver transplantation, the blood methionine concentrations of Patient 2 were markedly elevated (418–1200 µmol/L). After liver transplantation, the patient’s blood methionine concentration decreased rapidly to the normal level (10–50 µmol/L)
Discussion
While MAT I/III deficiency is a rare disease, it is the most common cause of persistent isolated hypermethioninemia. Expanded newborn screening with tandem mass spectrometry represents the gold-standard approach for early detection of hypermethioninemia [17], and increasing numbers of patients with MAT I/III deficiency have been diagnosed by newborn screening [1, 5, 18]. While the exact incidence of MAT I/III deficiency remains unknown, the incidence of persistent hypermethioninemia has been reported to be 1 in 23,470 and 1 in 30,893 in Galicia [6] and Suzhou [17], China, respectively. In our study, 11 patients were identified by expanded newborn screening, and four were clinically diagnosed by selective screening for neurological symptoms. Blood amino acids analysis was considered the first-line diagnostic approach for hypermethioninemia in both patient groups.
MAT I/III deficiencies, particularly those caused by heterozygous MAT1A variants, were previously considered to be a clinically benign condition [19]. However, the clinical manifestations of MAT I/III deficiency are heterogenous and poorly understood [3]. Neurological manifestations include intellectual disability, dystonia, and reduced IQ [20], while extra-CNS manifestations include growth retardation, anorexia, gastrointestinal disturbances, and malodourous urine and sweat smelling like boiled cabbage [21]. In our data, brain damage was observed in 10 patients (66.7%), including intellectual disability, language development delays, abnormal psychomotor development or intelligence, emotional instability, ataxia, and low IQ/DQ scores. Delayed language development and learning difficulties were the main neurological manifestations of this disease. Malodourous urine was only reported in three patients, while epilepsy was not observed. Thus, our findings indicate that blood amino acids and acylcarnitine analysis should be performed for all patients presenting with delayed development and low IQ/DQ scores.
Demyelination as a consequence of severe hypermethioninemia has been reported in MAT I/III deficient patients. In the study by Mudd et al. [22], brain MRI abnormalities indicative of cerebral edema were observed in the cerebral cortex and posterior brainstem of two patients with severe hypermethioninemia. In our study, white matter demyelination was the main MRI feature. Abnormal MRI signals were also observed in the white matter area of both cerebral hemispheres, corpus callosum, globus pallidus, brain stem, medulla, dorsal midbrain, dorsal pons, bilateral anterior limb of the internal capsule, and bilateral internal capsule. However, the clinical symptoms were inconsistent with brain MRI findings, with two patients showing evidence of abnormal myelination demonstrating normal intelligence and neurological function. Chien et al. [19] similarly reported the presence of brain MRI lesions despite the lack of CNS abnormalities. Regardless, brain MRI remains an important examination for evaluating the degree of abnormal myelination, even in asymptomatic patients. Close follow-up and brain MRI are therefore required for all patients with persistent hypermethioninemia. Furthermore, brain MRI and metabolic studies should be considered in all patients showing development delay or retrogression.
Blood methionine levels may serve as the best indicator for the severity of MAT I/III deficiency. Cases of mild-to-moderate hypermethioninemia due to dominant variant c.791G > A (p.Arg264His) on MAT1A have been reported. In the study by Couce et al. [6] involving five MAT I/III deficiency patients with heterozygous c.791G > A variants, blood methionine concentrations ranged between 164 and 573 µmol/L, with no neurological symptoms observed. CNS abnormalities often occur in patients with substantially elevated methionine levels. Chien et al. [19] reported that plasma methionine concentrations > 800 µmol/L were likely to be associated with CNS abnormalities. Consistent with this finding, Hirabayashi et al. [23] reported neurological complications in nine MAT I/III deficiency patients with plasma methionine concentrations of 600–1870 µmol/L, while Mudd et al. [24] reported the association of extreme hypermethioninemia (> 1000 µmol/L, and certainly > 2000 µmol/L) with severe neurological disorders such as cerebral edema. The lack of AdoMet-dependent methylated products may be a cause of CNS demyelination [25]; thus, the lack of AdoMet analyses was a limitation of the present study. However, we observed that pre-treatment methionine concentrations were higher in patients with CNS abnormalities than those who were asymptomatic, suggesting that higher methionine concentrations may increase the risk for CNS complications. Additional studies are warranted to determine the cutoff value for such risk.
MAT1A variants mostly show autosomal recessive inheritance. Several autosomal dominant mutations have also been observed, including c.776 C > T (p.Ala259Val), c.791G > A (p.Arg264His) [26], c.746G > A (p.Arg249Gln), and c.838G > A (p.Gly280Arg) [7]. Insights into the relationship between genotype and phenotype are currently lacking. The c.791G > A mutation represents the most prevalent variant in Asian populations, such as those in Japan and Taiwan [1, 27], and is associated with the development of mild hypermethioninemia [28]. Among 13 patients with hypermethioninemia in Suzhou, China, 10 carried the c.791G > A mutation and were free of CNS abnormalities [17]. Similarly, none of the 13 patients with the dominant form of MAT I/III deficiency in Michigan demonstrated any neurological signs [29]. These findings support the benign nature of persistent isolated hypermethioninemia due to heterozygous MAT1A variants. However, no dominant variants, including c.791G > A, were observed in our patients. c.895 C > T is the most frequent variant in our population. All of our patients demonstrated autosomal recessive mutations with two variants. Besides one patient who carried a duplication (Patient 15), the remaining patients demonstrated missense variants. No exonic deletions or truncating mutations were detected, as reported previously [24]. Brain damage was observed in the majority of our patients (66.7%). As such, we speculated that CNS manifestations may be more prevalent among patients with autosomal recessive MAT I/III deficiency. Large-scale studies are thus warranted to investigate the relationship between the genotype and phenotype of this disease.
There is no consensus regarding the treatment for MAT I/III deficiency. Although some studies have suggested strict dietary methionine restriction in early infancy to maintain plasma methionine levels of < 750 µmol/L and prevent neurological complications [23], evidence regarding the efficacy of such interventions is limited. Furthermore, strict dietary control may be unfeasible during the neonatal period, and only regular clinical monitoring can be undertaken. Other studies have recommended ademethionine 1,4-butanedisulfonate supplementation or protein restriction [30]. Furujo et al. [31] reported neurological improvement after ademethionine 1,4-butanedisulfonate supplementation alone, and they omitted dietary methionine restrictions based on the premise that reduced methionine intake may be associated with reduced AdoMet synthesis. Dietary control was recommended to our patients, but their blood methionine levels remained elevated during treatment and cerebral white matter lesions could not be avoided. Ademethionine 1,4-butanedisulfonate was used in five cases during our study, and these patients’ blood methionine levels decreased slightly. The long-term effectiveness of ademethionine 1,4-butanedisulfonate supplementation as a treatment for MAT I/III deficiency remains to be studied.
The outcomes of liver transplantation in MAT I/III deficiency have not yet been elucidated. In the present study, liver transplantation was performed in one patient (Patient 2), which resulted in drastically reduced methionine concentrations. To our knowledge, this is the first study reporting the effects of liver transplantation on hypermethioninemia in a MAT I/III deficiency patient. In the report by Strauss et al. [32] describing the findings for a child with severe S-adenosylhomocysteine hydrolase deficiency, segmental liver transplantation from a healthy, unrelated living donor following failure of dietary therapy resulted in normal and stable blood methionine and AdoMet levels on an unrestricted diet throughout the 6-month follow-up period. The patient also showed promising improvements in gross motor, language, and social skills [32]. Since MAT1A, which encodes MAT I and III, is only expressed in non-fetal liver tissues [13, 23], any healthy liver tissue should be able to promote the synthesis of sufficient amounts of MAT I and III to increase overall MAT activity. Indeed, liver transplantation has been widely used in the treatment of inherited metabolic liver disorders in China, and its efficacy is now well-established [33, 34].
Conclusions
Hypermethioninemia due to autosomal recessive MAT I/III deficiency is a disorder which can have significant clinical consequences. The clinical phenotypes vary greatly. The possibility of CNS abnormalities should be considered in patients with MAT I/III deficiency and severe hypermethioninemia. These patients should receive close follow-up and brain MRI examinations. Liver transplantation may be a treatment alternative following failure of dietary and drug therapy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank all patients and their families for their participation. We are grateful to the Euler Genomics (Beijing, China), the Berry Genomics Corporation (Beijing, China), and the Running Gene Inc. (Beijing, China) for providing professional genetic sequencing and analyzing.
Abbreviations
- AdoMet
S-adenosylmethionine
- DQ
Development quotient
- IQ
Intelligence quotient
- MAT
Methionine adenosyltransferase
Author contributions
Y.Y. conceived the study. X.M. and M.L. drafted the manuscript. Z.C., H.Z., J.S., H.D., Y.J., M.L., R.H., L.K., Y.L., Y.C., Z.Z. and L.S. participated in the clinical management and patient data collection. Y.Z. checked the genetic data. All authors read and approved the final version of the manuscript.
Funding
This work was supported by grants from the National Key Research and Development Program of China (grant numbers 2021YFC2700903, 2022YFC2703401) and the Xiamen Science and Technology Guiding Project (grant number 3502Z20199078 to M.L.).
Data availability
The data of novel variants for the MAT1A gene analyzed during the current study have been uploaded to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar, submission ID SUB12700124, SUB12913275).
Declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Peking University First Hospital Institutional and was performed in accordance with the Declaration of Helsinki. Written informed consent was obtained from the parents of the patients for collection of samples and publication of medical data.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Conflict of interest
All authors declare no potential conflicts of interest with any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xue Ma and Mei Lu contributed equally to this work.
References
- 1.Chien YH, Chiang SC, Huang A, Hwu WL. Spectrum of hypermethioninemia in neonatal screening. Early Hum Dev. 2005;81(6):529–33. [DOI] [PubMed] [Google Scholar]
- 2.Gaull GE, Tallan HH. Methionine adenosyltransferase deficiency: new enzymatic defect associated with hypermethioninemia. Science. 1974;186(4158):59–60. [DOI] [PubMed] [Google Scholar]
- 3.Chamberlin ME, Ubagai T, Mudd SH, et al. Methionine adenosyltransferase I/III deficiency: novel mutations and clinical variations. Am J Hum Genet. 2000;66(2):347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mudd SH, Poole JR. Labile methyl balances for normal humans on various dietary regimens. Metabolism. 1975;24(6):721–35. [DOI] [PubMed] [Google Scholar]
- 5.Chadwick S, Fitzgerald K, Weiss B, Ficicioglu C. Thirteen patients with MAT1A mutations detected through newborn screening: 13 years’ experience. JIMD Rep. 2014;14:71–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couce ML, Bóveda MD, Castiñeiras DE, et al. Hypermethioninaemia due to methionine adenosyltransferase I/III (MAT I/III) deficiency: diagnosis in an expanded neonatal screening programme. J Inherit Metab Dis. 2008;31(Suppl 2):S233–9. [DOI] [PubMed] [Google Scholar]
- 7.Kim YM, Kim JH, Choi J, et al. Determination of autosomal dominant or recessive methionine adenosyltransferase I/III deficiencies based on clinical and molecular studies. Mol Med. 2016;22:147–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez Mato IP, Sanchez del Pino MM, Chamberlin ME, et al. Biochemical basis for the dominant inheritance of hypermethioninemia associated with the R264H mutation of the MAT1A gene. A monomeric methionine adenosyltransferase with tripolyphosphatase activity. J Biol Chem. 2001;276(17):13803–9. [DOI] [PubMed] [Google Scholar]
- 9.Chamberlin ME, Ubagai T, Mudd SH, et al. Dominant inheritance of isolated hypermethioninemia is associated with a mutation in the human methionine adenosyltransferase 1A gene. Am J Hum Genet. 1997;60(3):540–6. [PMC free article] [PubMed] [Google Scholar]
- 10.Surtees R, Leonard J, Austin S. Association of demyelination with deficiency of cerebrospinal-fluid S-adenosylmethionine in inborn errors of methyl-transfer pathway. Lancet. 1991;338(8782–8783):1550–4. [DOI] [PubMed] [Google Scholar]
- 11.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of. Sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernández-Irigoyen J, Santamaría E, Chien YH, et al. Enzymatic activity of methionine adenosyltransferase variants identified in patients with persistent hypermethioninemia. Mol Genet Metab. 2010;101(2–3):172–7. [DOI] [PubMed] [Google Scholar]
- 13.Tada H, Takanashi J-i, Barkovich AJ, et al. Reversible white matter lesion in methionine adenosyltransferase I/III deficiency. AJNR Am J Neuroradiol. 2004;25(10):1843–5. [PMC free article] [PubMed] [Google Scholar]
- 14.Ma YY, Li DX, Li XY, et al. Hypermethioninemia caused by deficient activity of methionine adenosyltransferase. J Clin Pediatr. 2018;36(01):57–60. [Google Scholar]
- 15.Zhang Z, Wang Y, Ma D, et al. Analysis of five cases of hypermethioninemia diagnosed by neonatal screening. J Pediatr Endocrinol Metab. 2020;33(1):47–52. [DOI] [PubMed] [Google Scholar]
- 16.Wu J, Lu AD, Zhang LP, et al. Study of clinical outcome and prognosis in pediatric core binding factor-acute myeloid leukemia. Zhonghua Xue Ye Xue Za Zhi. 2019;40(1):52–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T, Ma J, Zhang Q, et al. Expanded newborn screening for inborn errors of metabolism by tandem mass spectrometry in Suzhou, China: disease spectrum, prevalence, genetic characteristics in a Chinese population. Front Genet. 2019;10:1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Couce ML, Bóveda MD, García-Jimémez C, et al. Clinical and metabolic findings in patients with methionine adenosyltransferase I/III deficiency detected by newborn screening. Mol Genet Metab. 2013;110(3):218–21. [DOI] [PubMed] [Google Scholar]
- 19.Chien YH, Abdenur JE, Baronio F, et al. Mudd’s disease (MAT I/III deficiency): a survey of data for MAT1A homozygotes and compound heterozygotes. Orphanet J Rare Dis. 2015;10:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mudd SH, Levy HL, Tangerman A, et al. Isolated persistent hypermethioninemia. Am J Hum Genet. 1995;57(4):882–92. [PMC free article] [PubMed] [Google Scholar]
- 21.Gout JP, Serre JC, Dieterlen M, et al. Une nouvelle cause d’hyperméthioninémie de l’enfant: le deficit en S-adenosyl-méthionine-synthétase [Still another cause of hypermethioninemia in children: S-adenosylmethionine synthetase deficiency]. Arch Fr Pediatr. 1977;34(5):416–23. [PubMed] [Google Scholar]
- 22.Harvey Mudd S, Braverman N, Pomper M, et al. Infantile hypermethioninemia and hyperhomocysteinemia due to high methionine intake: a diagnostic trap. Mol Genet Metab. 2003;79(1):6–16. [DOI] [PubMed] [Google Scholar]
- 23.Hirabayashi K, Shiohara M, Yamada K, et al. Neurologically normal development of a patient with severe methionine adenosyltransferase I/III deficiency after continuing dietary methionine restriction. Gene. 2013;530(1):104–8. [DOI] [PubMed] [Google Scholar]
- 24.Mudd SH. Hypermethioninemias of genetic and non-genetic origin: a review. Am J Med Genet C Semin Med Genet. 2011;157 C(1):3–32. [DOI] [PubMed] [Google Scholar]
- 25.Chamberlin ME, Ubagai T, Mudd SH, et al. Demyelination of the brain is associated with methionine adenosyltransferase I/III deficiency. J Clin Invest. 1996;98(4):1021–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muriello MJ, Viall S, Bottiglieri T, et al. Confirmation that MAT1A p.Ala259Val mutation causes autosomal dominant hypermethioninemia. Mol Genet Metab Rep. 2017;13:9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagao M, Tanaka T, Furujo M. Spectrum of mutations associated with methionine adenosyltransferase I/III deficiency among individuals identified during newborn screening in Japan. Mol Genet Metab. 2013;110(4):460–4. [DOI] [PubMed] [Google Scholar]
- 28.Chou JY. Molecular genetics of hepatic methionine adenosyltransferase deficiency. Pharmacol Ther. 2000;85(1):1–9. [DOI] [PubMed] [Google Scholar]
- 29.Sen K, Felice MD, Bannick A, et al. Mild persistent isolated hypermethioninemia identified through newborn screening in Michigan. J Pediatr Genet. 2019;8(2):54–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furujo M, Kinoshita M, Nagao M, Kubo T. S-adenosylmethionine treatment in methionine adenosyltransferase deficiency, a case report. Mol Genet Metab. 2012;105(3):516–8. [DOI] [PubMed] [Google Scholar]
- 31.Furujo M, Kinoshita M, Nagao M, Kubo T. Methionine adenosyltransferase I/III deficiency: neurological manifestations and relevance of S-adenosylmethionine. Mol Genet Metab. 2012;107(3):253–6. [DOI] [PubMed] [Google Scholar]
- 32.Strauss KA, Ferreira C, Bottiglieri T, et al. Liver transplantation for treatment of severe S-adenosylhomocysteine hydrolase deficiency. Mol Genet Metab. 2015;116(1–2):44–52. [DOI] [PubMed] [Google Scholar]
- 33.Jiang YZ, Zhou GP, Wu SS, et al. Safety and efficacy of liver transplantation for methylmalonic acidemia: a systematic review and meta-analysis. Transpl Rev (Orlando). 2021;35(1):100592. [DOI] [PubMed] [Google Scholar]
- 34.Chu TH, Chien YH, Lin HY, et al. Methylmalonic acidemia/propionic acidemia - the biochemical presentation and comparing the outcome between liver transplantation versus non-liver transplantation groups. Orphanet J Rare Dis. 2019;14(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data of novel variants for the MAT1A gene analyzed during the current study have been uploaded to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar, submission ID SUB12700124, SUB12913275).