Abstract
We describe an approach for designing high-affinity small molecule–binding proteins poised for downstream sensing. We use deep learning–generated pseudocycles with repeating structural units surrounding central binding pockets with widely varying shapes that depend on the geometry and number of the repeat units. We dock small molecules of interest into the most shape complementary of these pseudocycles, design the interaction surfaces for high binding affinity, and experimentally screen to identify designs with the highest affinity. We obtain binders to four diverse molecules, including the polar and flexible methotrexate and thyroxine. Taking advantage of the modular repeat structure and central binding pockets, we construct chemically induced dimerization systems and low-noise nanopore sensors by splitting designs into domains that reassemble upon ligand addition.
The design of small molecule–binding proteins is more challenging than the design of protein binders because there are fewer atoms to interact with; thus high–shape complementary pockets that make contact with the majority of the available atoms are required for high affinity (1). Flexible polar compounds are particularly challenging: Polar groups make hydrogen bonds to water in the unbound state, which must be replaced by hydrogen bonds to the protein for binding to be favorable, and flexible compounds lose considerable entropy upon binding, which must be compensated by extensive favorable interactions. Previous successes in small-molecule binder design that use physically based and deep-learning diffusion–based approaches have focused on rigid hydrophobic targets and have often required considerable experimental optimization to improve binding affinities from micromolar to high nanomolar levels (2–10). Beyond binding, a general unsolved challenge is to transduce binding events into downstream signals.
We set out to develop a general method for designing small molecule–binding proteins with high shape complementarity for their targets that are poised for downstream sensing applications. We hypothesized that a design approach beginning with identification of protein scaffolds with high shape complementarity (1, 9) to the targeted small molecule would be able to achieve higher-affinity binding than approaches based on fixed scaffolds (3–8) and enable binding to flexible and polar targets. We reasoned further that if the scaffolds could be split into two or more independently folded domains such that small-molecule binding drives association, then binders could be readily converted into sensors. This strategy requires that the domains be folded before association to avoid aggregation and proteolysis of the split domains in the unbound state. Thus, scaffold sets that can harbor binding pockets of widely ranging shapes and sizes and can be readily split into multiple independently folded domains could provide a general solution to the sensor-design problem (9).
Pseudocyclic scaffolds consisting of a repeating structural unit surrounding a central pore or pocket could satisfy the above requirements (9) (Fig. 1A). First, depending on the geometry of the structural unit and the number of units in the closed pseudocycle, the central pocket can have a wide range of sizes and shapes. Second, because the interactions within the protein are largely between residues close in sequence within single repeat units or between adjacent repeat units, pseudocycles can be split into multiple domains that retain almost all of the stabilizing interactions present in the unsplit protein and thus are likely to fold and to be stable on their own (9). set out to (Fig. 1A and fig. S1, A and B) develop a general approach integrating deep learning and Rosetta (10) energy-based design methods (9, 11–14) to generate high–shape complementary pseudocycle-based binding proteins for any desired small molecule (Fig. 1B).
Fig. 1. Design strategy.
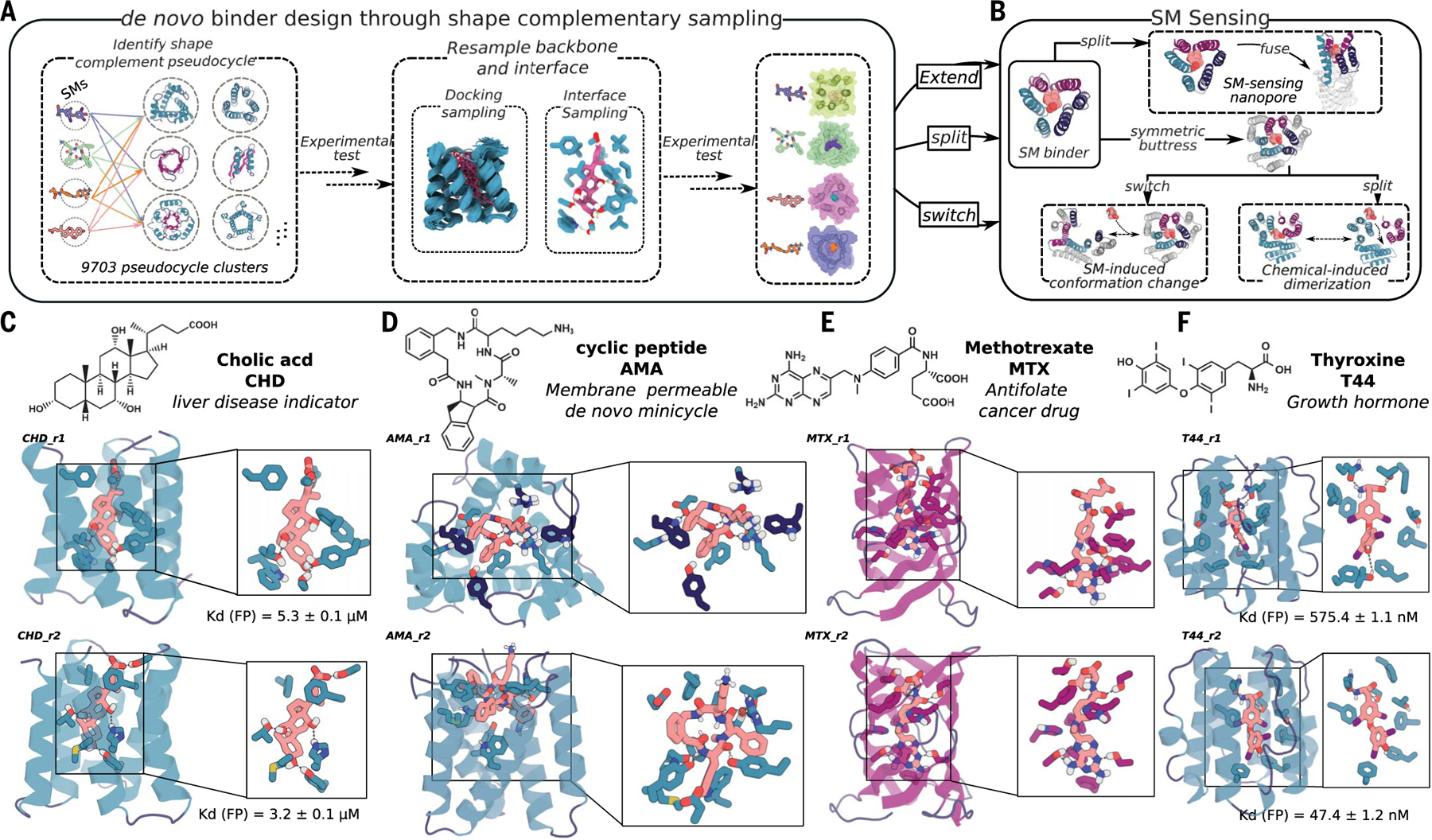
(A) Diverse conformers of the small molecule of interest are docked into deep learning–generated pseudocycles containing a wide array of central pockets, and the interface sequence is optimized for high-affinity binding with Rosetta or LigandMPNN. Top-ranked designs are tested experimentally, and the backbones of the best hits and the docked poses are extensively resampled. After sequence design, top-ranked second-round designs are experimentally tested (fig. S1, A and B). (B) Because pseudocycles are constructed from modular repeating units that surround the central binding pocket, the binders can be readily transformed into sensors through multiple strategies. (C to F) Examples of first-round design models for each target ligand.
We chose four small molecules as binding targets: cholic acid (CHD), methotrexate (MTX), thyroxine (T44), and a de novo–designed cell-permeable cyclic tetrapeptide, referred to as AMA (Fig. 1; fig. S4, A to D; and table S4). CHD is the primary bile acid, and detection of its free form is important for liver-disease determination (15). MTX is an antifolate cancer treatment agent, which requires regular blood monitoring to reduce adverse outcomes for patients (16). T44 is a human hormone regulating energy usage and other functions; at-home monitoring to detect free T44 levels is used for patient thyroid-condition diagnosis. The current commercially available detection methods for CHD, MTX, and T44 (15–19) cannot distinguish individual molecules from variants (CHD, T44) or bound from free forms (T44, MTX); higher-affinity and specificity binders are needed for rapid at-home sensing devices. From the design perspective, T44 and MTX are more flexible and polar than previous small-molecule targets for computational protein design (2–7), and MTX is particularly challenging because of its high polarity and flexibility (fig. S4G and table S4). To test the ability of the method to design binders to larger ligands, we also included AMA (Fig. 1 and fig. S4A), a de novo–designed cell membrane–permeable tetrapeptidic macrocycle (20); designed binders for such compounds could be turned into chemical-induced dimerization (CID) systems enabling bioorthogonal control for adoptive cell therapies and other applications. We obtained or synthesized fluorescein isothiocyanate (FITC; see supplementary materials, materials and methods) or biotin-labeled versions of all four ligands for experimental screening (fig. S4).
Identification of pseudocycle scaffolds with high shape complementarity to targets
We begin by identifying for a given ligand of interest the most shape-complementary pockets present in diverse pseudocycle scaffolds (9) generated with AlphaFold2 (AF2) and ProteinMPNN (Fig. 1A and fig. S1A; details are provided in methods and materials). We generated representative conformer ensembles for each ligand and used Rosetta to dock these into the pseudocycle pockets and rapidly identify pseudocycle scaffold-ligand pairs expected to have high shape complementarity (materials and methods). The promising docks were then sequence-designed with Rosetta and the recently developed LigandMPNN method (14). For experimental characterization, we selected 5000 to 10,000 final designs for each target predicted to fold to the designed structures by AF2 (12) and make low free-energy interactions with the ligand (as computed by Rosetta) (table S1).
The selected designs were encoded in oligonucleotide libraries and displayed on the yeast cell surface, and binding was assessed by fluorescence-activated cell sorting (FACS) and deep sequencing (materials and methods). For CHD (Fig. 1C and fig. S2, A and B), we obtained two binders based on the same scaffold with dissociation constants (Kd) of 3.2 ± 0.1 μM and 5.3 ± 0.1 μM by fluorescence polarization (FP; materials and methods); in the design models, all ligand polar atoms are contacted by side chains that are buttressed by hydrogen-bonding networks (Fig. 1C and fig. S2, A and B). For T44, we again obtained two binders based on the same scaffold with Kd of 575.4 ± 1.1 nM and 47.4 ± 2 nM by FP (Fig. 1F and fig. S2, C and D). For MTX and AMA, we identified three and four potential binding designs from two and four pseudocycle scaffolds, respectively (Fig. 1, D and E, and figs. S3 and S6); these binders were weaker than the CHD and T44 binders but still showed clear ligand-specific binding signals on FACS performed using yeast surface display (fig. S3). The contact molecular surface with ligand was generally higher for the designs with binding activity, but more data are needed to set thresholds (fig. S1C). Binders were obtained by using both the position-specific scoring matrix (PSSM)–based Rosetta design protocol and the LigandMPNN design protocol (table S2), suggesting that when there is close shape complementarity between protein and ligand, the details of the sequence design method become less critical.
We were able to obtain two crystal structures of CHD binders: CHD_r1 with partial electron density in the ligand binding pocket (2.0-Å resolution, with the ligand not included in the final model; Fig. 2, A and B, and fig. S7, A, C, and D) and a buttressed version with the same ligand-protein–interface design in which we modeled cholic acid into density within the ligand binding pocket (3.5-Å resolution, see below) (Fig. 2, C to F, and fig. S7. B, E, and F). The crystal structures of CHD_r1 and CHD_ buttress matched the computational design models with 0.80- and 0.92-Å Cα-RMSD (root mean square deviation), respectively (Fig. 2, A to D, and fig. S7). In the CHD_buttress structure, all ligand-protein hydrogen-bond interactions and hydrogen-bond networks were clearly recapitulated, including two hydrogen bonds to the side hydroxyl groups of CHD and a hydrogen-bonding network around the CHD head hydroxyl group (Fig. 2, D to F, and fig. S7F).
Fig. 2. X-ray crystallography demonstrates accuracy of design approach.
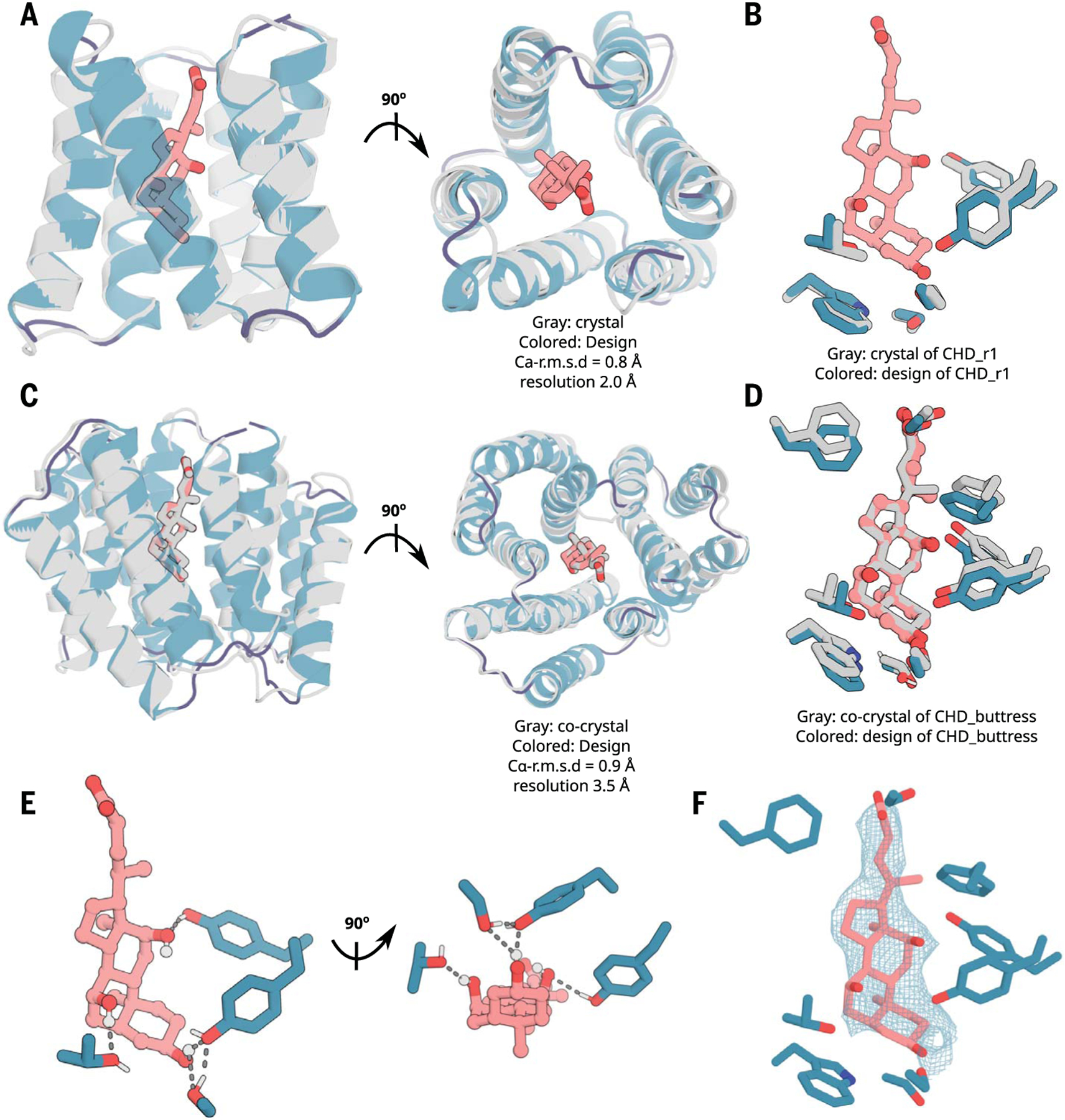
(A and B) The crystal structure of CHD_r1 (gray; ligand not modeled because of partial ligand electron density) is very similar to the computational design model (colored). (C and D) The crystal structure and the binding interface of CHD_buttress (gray) is very similar to the computational design model (colored). (E) The key polar- and hydrogen-bonding networks at the designed interface. (F) Composite omit map of interface region; the 2mFo-DFc electron density map at 1 σ level for CHD_buttress matches the design closely. Density maps are colored in teal. The protein backbone is shown in cartoons, and CHD and the key interacting side chains are shown in sticks. Pink, ligand carbon atoms; red, oxygen; blue, nitrogen; white, polar hydrogen. Also see fig. S7.
Generating higher-affinity designs with scaffold resampling
We next sought to generate higher-affinity designs by sampling new backbones around the pseudocycle scaffolds, which gave rise to the first-round hits. We generated ~5000 backbones with 0.5- to 3-Å Cα-RMSD from the first-round binding scaffolds (fig. S1B) and used Rosetta to generate 10 to 30 million new docks, which were filtered down to 50,000 to 500,000 on the basis of predicted shape complementarity (materials and methods). After sequence design based on Rosetta or LigandMPNN, 5000 to 15,000 designs (table S1) were selected for a second round of binder screening. As expected, the designs from the second round showed higher shape complementarity to the ligand than the first-round designs (fig. S1C).
We observed improvement in binder quality and quantity in the second design round (tables S1 and S2). For CHD, the highest affinity of the 37 verified binders (as measured by FP) improved by ~700-fold from 3.2 and 5.3 μM (Fig. 1C and fig. S2, A and B) in the first round to 4.7 nM in the second round (Fig. 3, A to D, and see full binder list in figs. S8 to S10). Site-saturation mutagenesis (SSM) of the five highest-affinity second-round binders (CHD-d1 to d5) confirmed that the key interactions in the design model were essential for binding (fig. S11). For T44, the affinity by FP improved from 47.4 to 18.2 nM (Fig. 3, G to H, and figs. S14 and S15). For AMA and MTX, the binding affinities in the first round were too weak to be measured off the yeast surface confidently (where there is very high avidity), likely in the high-micromolar-to-millimolar range. In the second round, we obtained 870-nM binders [measured by surface plasmon resonance (SPR)] for AMA (Fig. 3E and fig. S12). For MTX, the more flexible and polar target (Fig. 1E and fig. S6A), we obtained a 6.9-μM binder (MTX-d1) from one of the β barrel–like scaffolds (Fig. 3F), despite the challenges associated with binding this polar and flexible ligand (fig. S4E and table S4). The designed binding mode of MTX-d1 is supported by SSM analysis and competition assays (fig. S16). As in the first round, binders were obtained by using both the Rosetta-based and LigandMPNN-based interface design protocols (table S2), but the LigandMPNN-based method generated generally tighter binders. Control experiments for CHD and MTX in which the backbone and docking pose were fixed did not yield improvements in affinity (14), highlighting the importance of resampling for increasing shape complementarity and binding affinity. Cross-binding studies (fig. S17) showed that the designs are selective for their targets (materials and methods), and circular dichroism (CD; fig. S18) spectra were consistent with the designed secondary-structure composition and indicated that most of the designs have high thermostability (materials and methods).
Fig. 3. Experimental characterization of selected round-two designs.
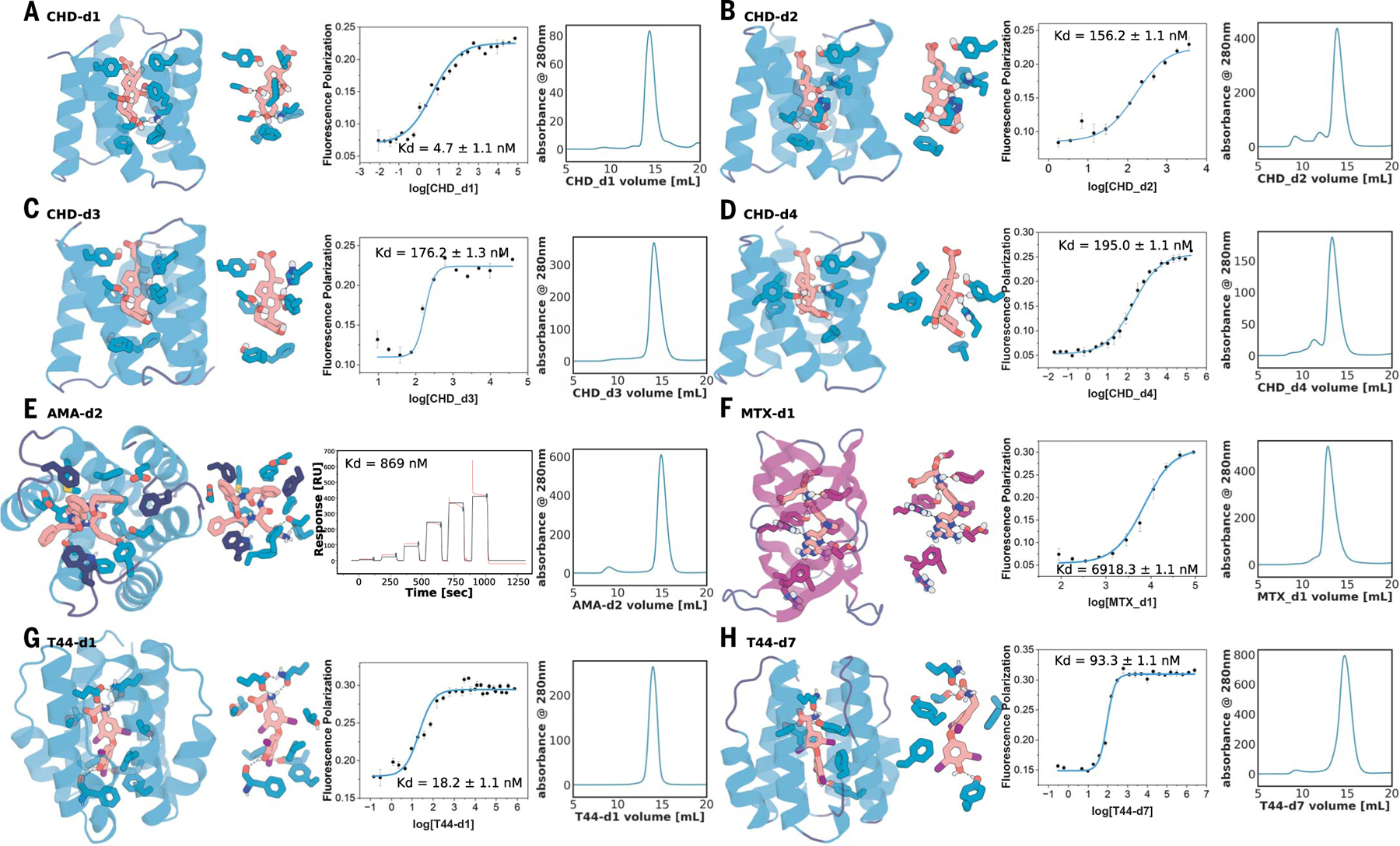
(A to D) Nanomolar affinity CHD binders CHD_d1 to d4 (full list in fig. S9). (E) Nanomolar binder for AMA (see fig. S12). (F) Micromolar methotrexate binder. (G and H) Two nanomolar T44 binders (full list in fig. S14). For each panel, from left to right: the design model, zoom-in on the side-chain–ligand interactions, FP (or SPR in the case of AMA) binding measurements, and SEC traces. Kd values and error bars are from two independent experiments. Interacting side chains and ligands are shown in sticks, with oxygen, nitrogen, iodine, and polar hydrogen colored in red, blue, purple, and white, respectively. Key interactions are indicated by gray dashed lines. Cartoons and sticks for helixes, sheets, and loops are colored in teal, magenta, and dark blue, respectively.
Converting pseudocycle binders into sensors
In pseodycycles, the fold-stabilizing interactions are primarily within repeat units and between adjacent units, thus the binders by construction can be split into two or more chains that are likely to fold at least in part in isolation (Fig. 4, A and G, and fig. S18). This contrasts with most native small molecule–binding proteins, which, like most globular proteins, often have extensive contacts between residues distant along the protein chain (21), such that split fragments cannot fold in isolation and thus are prone to aggregation and proteolysis.
Fig. 4. Conversion of pseudocycle binders into ligand-gated channels and CID systems.
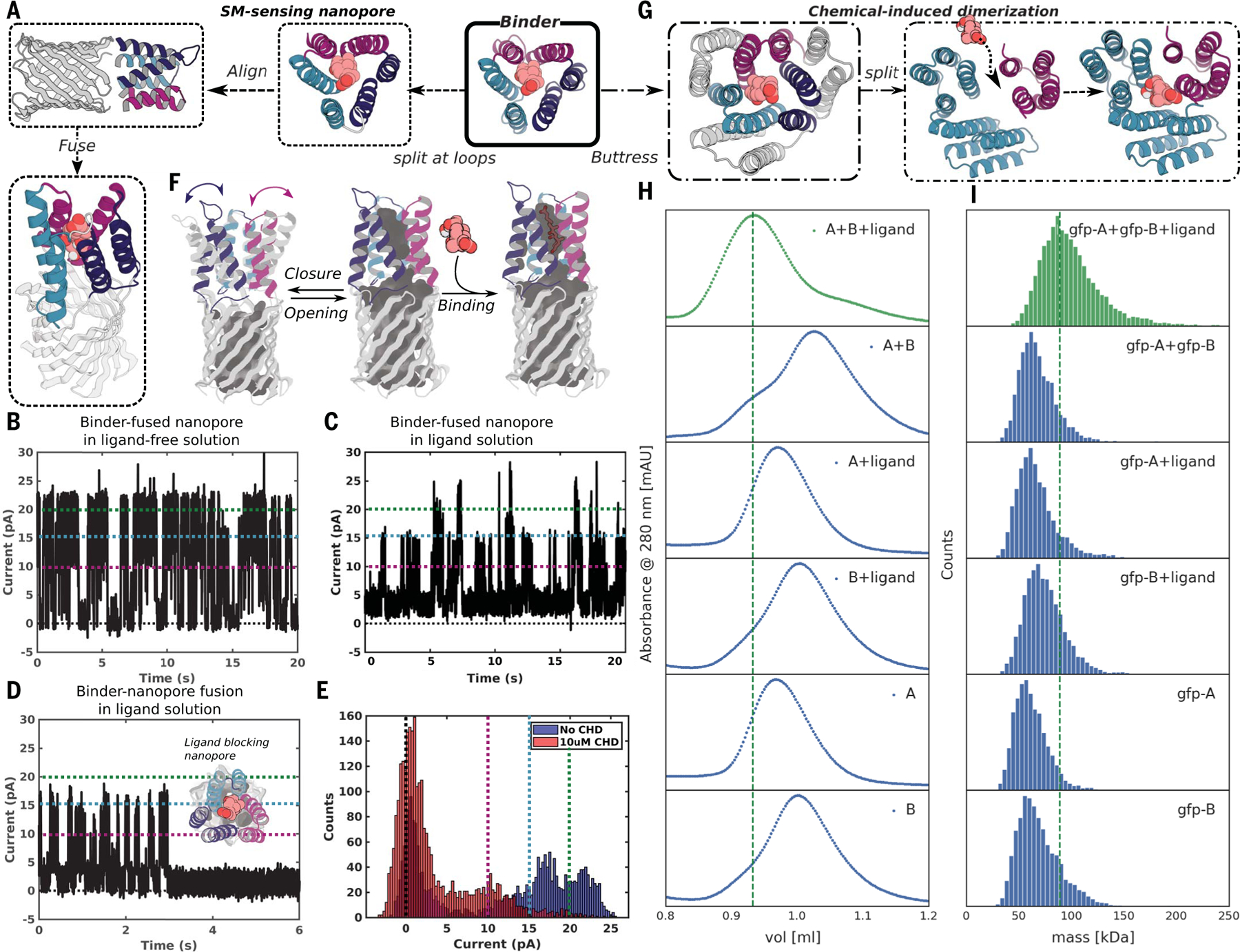
(A to F) Ligand-sensing de novo nanopore construction. (A) shows how the three structural repeat units of CHD_r1 were inserted into three different loops in a 12-stranded de novo nanopore by using inpainting to join the chains, such that the central axes of binder and nanopore are aligned. The conductance of the original nanopore is ~220 pS (fig. S17A) and is not influenced by CHD. The conductance of binder-fused nanopore in the absence (B) and presence of CHD [(C) to (E)] are shown for comparison. In the absence of CHD (B), the pore fluctuates between a state with high conductance very similar to the unmodified pore and a low-conductance state (C); in the presence of CHD, the duration of the low-conductance states is greatly increased [(C) to (E)]; the longer record (C) and a single closure event (D) are shown for clarity, and the histogram of the current with and without ligand is shown in (E). Different currents (0, 10, 15, and 20 pA) are marked out for clarity using dashed lines in black, magenta, teal, and green, respectively in (B) to (E). The gated nanopores are robust through multiple cycles of opening and closure (C). Upon reversal of the voltage, the original high-conductance state is restored (fig. S19B). As indicated schematically in (F), the conductance fluctuations of the binder-fused pore in absence of ligand likely reflect transient association of the three subunits; ligand binding stabilizes the associated state leading to prolonged blocking of the pore. (G to I) CID system construction. The CHD binder, CHD_r1, was buttressed by diffusion of an outer ring of helices to increase the stability of split protein fragments (Fig. 2, C to F, and figs. S7B and S20). To create a CID system, we split the buttressed binders into halves and redesign the protein-protein interface to increase solubility of the fragments and disfavor association in the absence of ligand. Characterization of CHD induced association of the split fragments by SEC (H) and mass photometry (I). Dimerization of the two split domains (A and B) in presence (first trace from top), but not the absence, (second trace from top) of ligand. The individual monomers do not dimerize in the presence (third and fourth trace from top) or absence (fifth and sixth traces from the top) of ligands. N-terminal GFP tags were fused to the monomers to facilitate detection by mass photometry.
We took advantage of this property by integrating CHD_r1 into a de novo–designed 12-stranded β-barrel nanopore (22), such that reconstitution of the complete pseudocycle from the split domains would block ion conductance through the pore (Fig. 4A). We aligned the central axis of CHD_r1 with the central axis of the designed nanopore (TMB12_3) (22) and inserted the three helical hairpins of the binder between strands 3 and 4, 7 and 8, and 11 and 12 of the β barrel by using RoseTTAfold joint inpainting (23) to build short connectors (5 to 8 Å) between the binder hairpins and the nanopore (Fig. 4A and materials and methods).
We expressed 12 integrated CHD binder–nanopore fusions in Escherichia coli and purified them from inclusion bodies (materials and methods). Five designs with monomeric size-exclusion chromatography (SEC) peaks were integrated into planar lipid bilayers, and ionic conductances were measured under an applied voltage of 100 mV. The open-pore conductance of the original nanopore is constant at ~220 pS and was not affected by CHD (fig. S19A). Two of the binder-nanopore fusions had similar open-pore conductances (Fig. 4B) to the original nanopore (Fig. S19A) but showed frequent transitions to two lower-conductance states, which likely reflect transient association of two or three helical hairpins in the absence of the ligand (Fig. 4F, left). In the presence of CHD, the dwell time of the nanopore in the closed state increased for the most sensitive design (Fig. 4, C to E), likely reflecting stabilization of the trimeric pseudocycle by the ligand (Fig. 4F, right). The current amplitudes approached near zero for ~0.4 to 3 s on average before returning to higher conductance (Fig. 4, C to E); each fluctuation to a low-conductance state likely corresponds to an independent CHD binding event, indicating that the pores are robust to multiple cycles of CHD binding and release (Fig. 4C). The limited number of observed states enabled quantification of the pore blocking and release dwell-time distributions, which were consistent with the binding affinity of the unfused CHD_r1 (fig. S19, C and D). In some cases, the blocked state was very long-lived (Fig. 4D), switching back to fluctuating between open and closed states only upon reversal of the applied-voltage polarity without compromising the stability or the conductance of the nanopore (fig. S19D). The lower noise and considerable increase in duration of the closed-state conductances at longer time-scales (100 ms to s) in the presence of the ligand (the fraction of transitions to closed states with dwell times of 1 s or more increased from 1 to 6.5% upon addition of CHD) leads to lower sampling frequency requirements compared with previously engineered sensors (24–26). The simplicity of constructing nanopore sensors by incorporating designed pseudocycles that bind targets of interest into de novo–designed quiet nanopores should enable the generation of a wide variety of sensors with superior properties.
We next sought to convert the pseudocycle binders into chemically induced dimerization (CID) systems (Fig. 4G). There has been considerable interest in CID systems in protein engineering and synthetic biology for small molecule–inducible switches for regulating protein association (27), signaling, or enzymatic activity (28). Despite the potential, almost all work has utilized a small number of CID systems, such as rapamycin-FKBP-FRB (29), and involved natural proteins as one (30) or both partners (31). Synthetic biology approaches would benefit considerably from systematic approaches for designing CID systems for new ligands, for example, for feedback control based on product levels in metabolic engineering.
We first stabilized the pseudocycles by building a second ring of stabilizing structural elements around the inner ring, which forms the binding interface (Fig. 4G and fig. S20A). The interactions between the inner and outer rings should contribute sufficient stabilization to make up for the loss in interactions between the N- and C-terminal repeats upon splitting. We chose the CHD binder, CHD_r1, as a proof of concept and incorporated a second ring of helices by using RFDiffusion (32); the inner and outer helices packed closely around a hydrophobic core (fig. S20A). As noted above, we obtained a co-crystal structure of the buttressed binder with CHD bound (Fig. 2, C to F and fig. S7, B and E to F) with the outer buttressing ring of helices, with the ligand conformation and key interacting side chains very similar to the design model (Fig. 2, D to F, and fig. S7F). The buttressed binder was more stable and bound to CHD slightly more tightly, likely owing to rigidification of the target-binding conformation (fig. S20B). This approach of testing binding first at the single-layer stage and then adding an outer layer by buttressing to stabilize the highest-affinity binders has the considerable advantage of reducing the difficulty and cost of design testing: The single-ring structures are less than 120 amino acids long and can be encoded on two 250-to-300–nucleotide commercially available oligonucleotides from an oligo array, whereas the two-ring structures are longer than 200 amino acids, for which gene synthesis is far more expensive and lower throughput.
To generate a CID system, we split the buttressed binder into two parts (Fig. 4G). Using tied-position ProteinMPNN design (materials and methods), we optimized the newly created interfaces to increase the solubility of the split domains while retaining the ability to form the holocomplex in the presence of the ligand. We expressed, purified, and tested 34 designed CID protein pairs and determined their oligomerization state by SEC (Fig. 4H) and mass photometry (Fig. 4I) in the presence and absence of CHD. One of the designed pairs showed clear CID behavior: The individual domains were monomeric in isolation but associated to form the heterodimer in the presence of 10 μM CHD. Both split domains were well-expressed and soluble alone, with highly helical CD spectra as in the design models (fig. S18). Because the individual subdomains (A is 9.8 kDa; B is 18.9 kDa) are smaller than the detection limit of mass photometry (around 30 kDa), we generated N-terminal fusions to green fluorescent protein. Using both SEC and mass photometry, dimerization was observed with (Fig. 4I, first lane from top) but not without ligand (Fig. 4I, second lane from top). The individual domains remained monomeric with (Fig. 4I, third and fourth lane from top) or without (Fig. 4I, fifth and sixth lane from top) CHD. The mass of the dimer peak derived from mass photometry, 89.0 ± 22.0 kDa, was close to the expected 83 kDa for the gfp-A-ligand-gfp-B complex (gfp, green fluorescent protein). Because protein association can be readily coupled to site-specific transcriptional activation (for example by linking a DNA binding module with an activator module) and other cellular readouts, the ability to generate CID systems for potentially any small molecule of interest could have very broad impact for metabolic pathway engineering by enabling feedback control based on levels of the desired product and/or unwanted or intermediate species.
Discussion
Our pseudocycle-based design approach goes beyond previous design studies (2–8, 33) in generating binders to large, polar, and/or flexible small molecules, such as MTX, T44, and AMA, with affinities in the range for use in diagnostics without experimental affinity maturation. The crystal structures of the CHD binders display a network of side chain–ligand interactions nearly identical to the computational design model, highlighting the accuracy of the design approach. Although the importance of shape complementarity for binding has been long understood (1), our direct optimization of shape complementarity by design would have been difficult before the recent advances in deep learning–based protein structure prediction and design (11–13), which enabled rapid de novo sampling and evaluation of pseudocycle topologies (9), resampling of the best solutions for a given ligand, and design of interactions with the ligand with LigandMPNN. The ability to iteratively resample the backbones and docking poses was essential for binding-affinity improvement. The computational and experimental effort required to generate the designs [30 central processing unit (CPU) min per design plus 20 graphic processing unit (GPU) s for validation, and 3 weeks for one round of experimental screening and characterization; see materials and methods] will likely decrease considerably as protein design methods continue to rapidly improve.
Our pseudocycle-based approach has advantages for ligand binder and sensor design. First, centrally located and high–shape complementary binding pockets can be generated with high affinity for very diverse small molecules. The variety of possible individual repeat unit structures is almost unlimited, and the number of repeat units can be readily varied, leading to vast numbers of possible pseudocycle structures, all harboring central pockets. Because the structures are largely locally encoded with few long-range contacts, interactions with each portion of the ligand can subsequently be optimized independently. Second, the ability to test small–genetic footprint single rings forming the binding pocket in a first step, and to then buttress the best designs in a second step, enables rapid, low-cost gene synthesis for exploring diverse design solutions without compromising the robustness and stability of the final binding modules. Third, because of the local encoding of the structure, designed binders can be readily integrated into ligand-gated channels and CID systems by splitting into subdomains that retain most of the interactions present in the full protein. The power of de novo design is highlighted by our ability to integrate the three repeat units of the CHD binder into three loops of a robust designed nanopore to create a CHD-gated nanopore with almost complete gating of current by CHD; because of the high signal-to-noise ratio, minimal post-processing of the acquired signal is required compared with previously engineered pores based on native proteins, which typically exhibit high levels of noise in the absence of ligand owing to the presence of a multitude of partially occluded states (24, 34). In this work, we used RFdiffusion (32) to build the outer ring to buttress the inner pseudocycle; it should be straightforward to adapt RFdiffusion All-Atom (33) to generate single-ring pseudocycles directly around target ligands. We anticipate that our shape-complementary pseudocycle-based approach should enable generation of robust, ligand-responsive channels and sensors for a wide variety of molecules of biological interest.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Pellock and I. Kalvet for providing general discussions for laboratory settings, A. Swartz and P. Erickson for chromatography assistance, and B. Huang and R. Ragotte for SPR assistance. Crystallographic diffraction data was collected at the Northeastern Collaborative Access Team beamlines at the Advanced Photon Source and at CBMS/NSLS2. NECAT is funded by the National Institute of General Medical Sciences from the NIH (P30 GM124165). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract DE-AC02–06CH11357. The Center for Bio-Molecular Structure (CBMS) is primarily supported by the NIH-NIGMS through a Center Core P30 grant (P30GM133893) and by the DOE Office of Biological and Environmental Research (KP1607011). NSLS2 is a US DOE Office of Science User Facility operated under contract DE-SC0012704. This publication resulted from the data collected using the beamtime obtained through NECAT BAG proposal no. 311950. M.S., L.T., and S.M. are co-second authors; they contributed equally, and everyone agrees that their authorship orders can be exchanged to benefit their own career development.
Funding:
This research was supported by the Audacious Project at the Institute for Protein Design (L.A., M.S., S.M., I.G., G.R.L., D.J., J.D., I.A., B.C., A.K.B., A.K., P.M.L., V.A., P.S., D.Z., D.H., X.L., M.G.S., D.K.V., and D.B.); the Innovation Fellows Program (L.A. and G.R.L) and the Translational Research Fund (L.A.); the Bill and Melinda Gates Foundation grant OPP1156262 (L.A., A.K., and X.L.); the Higgins Family and the Defense Threat Reduction Agency (DTRA) grant HDTRA1–19-1–0003 (M.S.); DTRA grant HDTRA1–21-1–0007 (I.A.); DTRA grant HR0011–21-2–0012 under the HEALR program (A.B. and A.K.); DTRA grant HDTRA1–19-1–0003 (P.L.); DTRA grant HR0011–21-2–0012 under the HEALR program (X.L.); the Air Force Office of Scientific Research under award FA9550–22-1–0506 (S.M.); the Washington Research Foundation (G.R.L. and A.P.); a gift from Microsoft (D.J., I.A., D.F, and J.D.); Schmidt Futures funding from Eric and Wendy Schmidt by recommendation of the Schmidt Futures program (D.J.); the Open Philanthropy Project Improving Protein Design Fund (J.D. and B.C.); Spark Therapeutics/Computational Design of a Half Size Functional ABCA4 project (I.A.); the SSGCID, which is supported by NIAID federal contract HHSN272201700059C (I.A.); NIH grant 75N93022C00036 under NIAID contract HHSN272201700059C (I.A.); NIH grant R01AG063845 (B.C. and D.H.); NIH grant R0AI160052 (A.K.B.); NIH grant U19AG065156 (D.H.); the Howard Hughes Medical Institute (HHMI) (B.C., A.B., D.H., and D.B); NSF grant CHE-1629214 (A.K.B.); the Juvenile Diabetes Research Foundation International (JDRF) grant 2-SRA-2018–605-Q-R (P.S. and X.L.); Novo Nordisk Foundation grant NNF18OC0030446 (C.N.); and the Helmsley Charitable Trust Type 1 Diabetes (T1D) Program grant 2019PG-T1D026 (X.L.).
Footnotes
Competing interests: L.A., M.S., L.T., S.M., and D.B. are the authors of a provisional patent application (63/610,726) submitted by the University of Washington for the design, composition, and function of the proteins created in this study. S.M. and V.A. are the authors of a provisional patent application submitted by the VIB-VUB Center for Structural Biology, for the design, composition, and function of the nanopore (TMB12_3) used in this study. C.N. serves on a scientific advisory board of Monod Bio.
Data and materials availability:
All scripts for stepwise sampling are available at https://github.com/LAnAlchemist/Pseudocycle_small_molecule_binder. All scripts for CID design are available at https://github.com/iamlongtran/pseudocycle_paper. All sequencing data were analyzed using an in-house script available at https://github.com/feldman4/ngs_app. These repositories are archived at Zenodo (35). The coordinates and structure factors have been deposited at the Protein Data Bank (https://www.rcsb.org/) under accession numbers 8VEI (CHD_r1), and 8VEJ (CHD_buttress).
REFERENCES AND NOTES
- 1.Gao M, Skolnick J, Proc. Natl. Acad. Sci. U.S.A . 109, 3784–3789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dou J et al. , Protein Sci. 26, 2426–2437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dou J et al. , Nature 561, 485–491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bick MJ et al. , eLife 6, e28909 (2017).28925919 [Google Scholar]
- 5.Polizzi NF, DeGrado WF, Science 369, 1227–1233 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tinberg CE et al. , Nature 501, 212–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee GR et al. , Small-molecule binding and sensing with a designed protein family. bioRxiv 2023.11.01.565201 [Preprint] (2023); 10.1101/2023.11.01.565201. [DOI] [Google Scholar]
- 8.Lu L et al. , Science 384, 106–112 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.An L et al. , Nat. Struct. Mol. Biol. 30, 1755–1760 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leaver-Fay A et al. , in Computer Methods, Part C, Johnson ML, Brand L, Eds., vol. 487 of Methods in Enzymology (Elsevier, 2011), pp. 545–574. [Google Scholar]
- 11.Dauparas J et al. , Science 378, 49–56 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jumper J et al. , Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baek M et al. , Science 373, 871–876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dauparas J et al. , Atomic context-conditioned protein sequence design using LigandMPNN. bioArxiv 2023.12.22.573103 [Preprint] (2023). 10.1101/2023.12.22.573103. [DOI] [Google Scholar]
- 15.Zhao X et al. , Front. Physiol. 13, 826740 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shadeed A, Kattach L, Sam S, Flora K, Farah Z, Rheumatol. Adv. Pract 6, rkac100 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spencer CA, “Assay of Thyroid Hormones and Related Substances” in Endotext, Feingold KR et al. , Eds. (MDText.com, 2017). https://www.ncbi.nlm.nih.gov/books/NBK279113/. [Google Scholar]
- 18.Welsh KJ, Soldin SJ, Eur. J. Endocrinol. 175, R255–R263 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tripathy NK et al. , J. Lab. Physicians 15, 344–353 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salveson PJ et al. , Science 384, 420–428 (2024). [DOI] [PubMed] [Google Scholar]
- 21.Plaxco KW, Simons KT, Baker D, J. Mol. Biol. 277, 985–994 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Berhanu S et al. , Science 385, 276–282 (2024).39024436 [Google Scholar]
- 23.Wang J et al. , Science 377, 387–394 (2022).35862514 [Google Scholar]
- 24.Ahmad M et al. , Nat. Commun. 14, 1374 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fahie MA, Chen M, J. Phys. Chem. B 119, 10198–10206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foster JC et al. , Angew. Chem. Int. Ed. 62, e202214566 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greene BL et al. , Annu. Rev. Biochem. 89, 45–75 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rappas M, Niwa H, Zhang X, Curr. Protein Pept. Sci. 5, 89–105 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Banaszynski LA, Liu CW, Wandless TJ, J. Am. Chem. Soc. 127, 4715–4721 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Foight GW et al. , Nat. Biotechnol. 37, 1209–1216 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanton BZ, Chory EJ, Crabtree GR, Science 359, eaao5902 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watson JL et al. , Nature 620, 1089–1100 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishna R et al. , Science 384, eadl2528 (2024). [DOI] [PubMed] [Google Scholar]
- 34.Shen J et al. , Nat. Commun. 13, 5985 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An L, Tran L, Feldman D, “Binding and sensing diverse small molecules using shape complementary pseudocycles,” v1, Zenodo (2024); 10.5281/zenodo.11406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scripts for stepwise sampling are available at https://github.com/LAnAlchemist/Pseudocycle_small_molecule_binder. All scripts for CID design are available at https://github.com/iamlongtran/pseudocycle_paper. All sequencing data were analyzed using an in-house script available at https://github.com/feldman4/ngs_app. These repositories are archived at Zenodo (35). The coordinates and structure factors have been deposited at the Protein Data Bank (https://www.rcsb.org/) under accession numbers 8VEI (CHD_r1), and 8VEJ (CHD_buttress).


