SUMMARY
The rules of how amino acids dictate the physical properties of biomolecular condensates are still incomplete. Here, we study condensates formed by tetrapeptides of the form XXssXX. Eight peptides form four types of condensates at different concentrations and pHs: droplets (X = F, L, M, P, V, and A), amorphous dense liquids (X = L, M, P, V, and A), amorphous aggregates (X = W), and gels (X = I, V, and A). The peptides exhibit differences in phase equilibrium and material properties, including a 368-fold range in the threshold concentration for phase separation and a 3,856-fold range in viscosity. All-atom molecular dynamics simulations provide physical explanations of these results. The present work also reveals widespread critical behaviors—including critical slowing down manifested by amorphous dense liquids and critical scaling obeyed by fusion speed—with broad implications for condensate functions.
Graphical Abstract
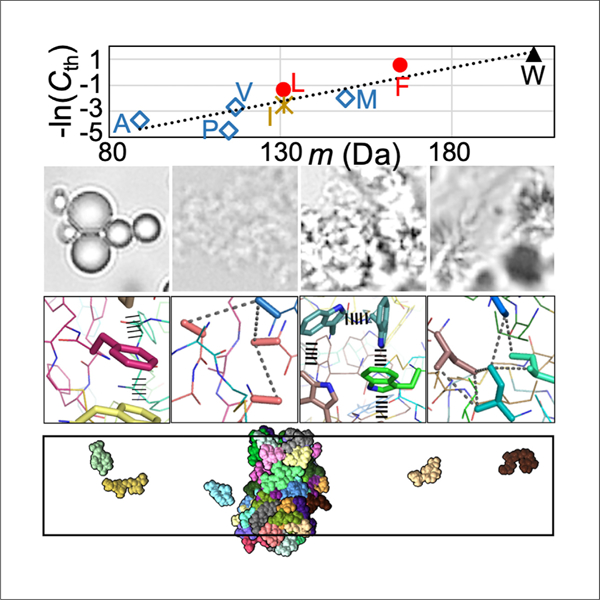
By integrating microscopy and microrheology with all-atom molecular dynamics simulations, Zhang et al. measure differing phase equilibrium and material properties of tetrapeptide condensates and provide physical explanations. The results represent a step toward quantitative understanding of the phase separation of intrinsically disordered proteins.
INTRODUCTION
Biomolecular condensates are formed via phase separation that is driven by various types of intermolecular attraction, including charge-charge, cation-, , hydrophobic, and hydrogen-bonding interactions.1,2 Many studies of the phase separation of oppositely charged intrinsically disordered proteins (IDPs) and of basic IDPs with nucleic acids and ATP have highlighted the roles of charge-charge and cation- interactions.3–12 Likewise, cation- and interactions are found to drive the phase separation of many other proteins.13–17 While 1,6-hexanediol has been widely used to disrupt hydrophobic interactions and isolate their contributions to phase separation, urea has been used analogously to probe hydrogen bonds.18,19 Coarse-grained simulations are beginning to predict well the phase-separation threshold concentrations and critical temperatures of IDPs.20,21
Biomolecular condensates can be found in different material states. A simple way to identify material states is to observe condensate morphologies under a microscope. Spherical droplets are usually assumed to be liquid-like, whereas amorphous aggregates and gels are assumed to be more solid-like. What determines condensate morphologies is still an open question. One can also characterize the material states by measuring material properties, such as the fusion speed of droplets.14,22–24 Slowed or stalled fusion is a sign that condensates are becoming solid-like. Relative to phase equilibrium properties, our understanding of the determinants of condensate material properties is lagging.2,8,25–27
Short peptides have been used to isolate the roles of different amino acids in determining phase equilibrium and condensate morphology. Yuan et al.28 observed the transition of aromatic group-blocked amino acids and dipeptides (b-X, X = A and H; b-XX, X = F) from liquid droplets to nanofibrils over a few hours. Baruch Leshem et al.29 studied W, F, and R-containing peptides (10–14 residues) and identified , cation-, and hydrogen bonding interactions by Raman and NMR spectroscopy. NMR analysis produced diffusion constants ~20 Å2/ns in the dilute phase; fluorescence recovery after photobleaching (FRAP) indicated a 10,000-fold slow-down in diffusion in the dense phase. Poudyal et al.19 studied the phase separation of homopeptides (X10, X = G and V) under crowding by PEG-8000. Twenty percent (w/v) 1,6-hexanediol significantly suppressed the phase separation of V10, but 2 M urea had little effect, indicating hydrophobic interactions as the driver for phase separation. The opposite was observed for the effects of additives on G10, suggesting that hydrogen bonding plays a major role in its phase separation. Abbas et al.18 synthesized peptides of the form XYssYX, where X, Y = W, F, and L, and ss represents the linker NH-CH2-CH2-S-S-CH2-CH2-NH. W-containing peptides formed aggregates, whereas F or L-only peptides formed droplets. FFssFF droplets exhibited an inverse fusion speed of ~1 s/μm and showed robust FRAP over 250 s. In a follow-up study, these authors showed that YFssFY formed gels containing microstructures that resemble sea urchins.30 Over a few hours, FFssFF transitioned from droplets to fibrils that are stabilized by and hydrogen-bonding interactions.31
By integrating microscopy and microrheology with all-atom molecular dynamics (MD) simulations on XYssYX condensates, we have a long-term goal of quantifying and physically interpreting the contributions of individual amino acids to the phase equilibrium and material properties of IDPs. Here, we report the physical properties of XXssXX condensates, where X is extended to all the nonpolar amino acids (Figures 1A and S1, inset). Using bright-field, confocal, and negative-staining electron microscopy (EM), we show that the condensates exhibit a variety of morphologies, including a form, referred to as amorphous dense liquid (ADL), that does not appear to have been reported previously. Using optical tweezers (OT), we measured the material properties of these condensates, which differ widely among the peptides, including a 736-fold range in fusion speed and a 3,856-fold range in viscosity. Our MD simulations provide physical explanations of these results. Importantly, the peptide condensates display widespread critical behaviors, including critical slowing down manifested by ADLs and critical scaling obeyed by fusion speed.
Figure 1. Images showing various forms of tetrapeptide condensates.
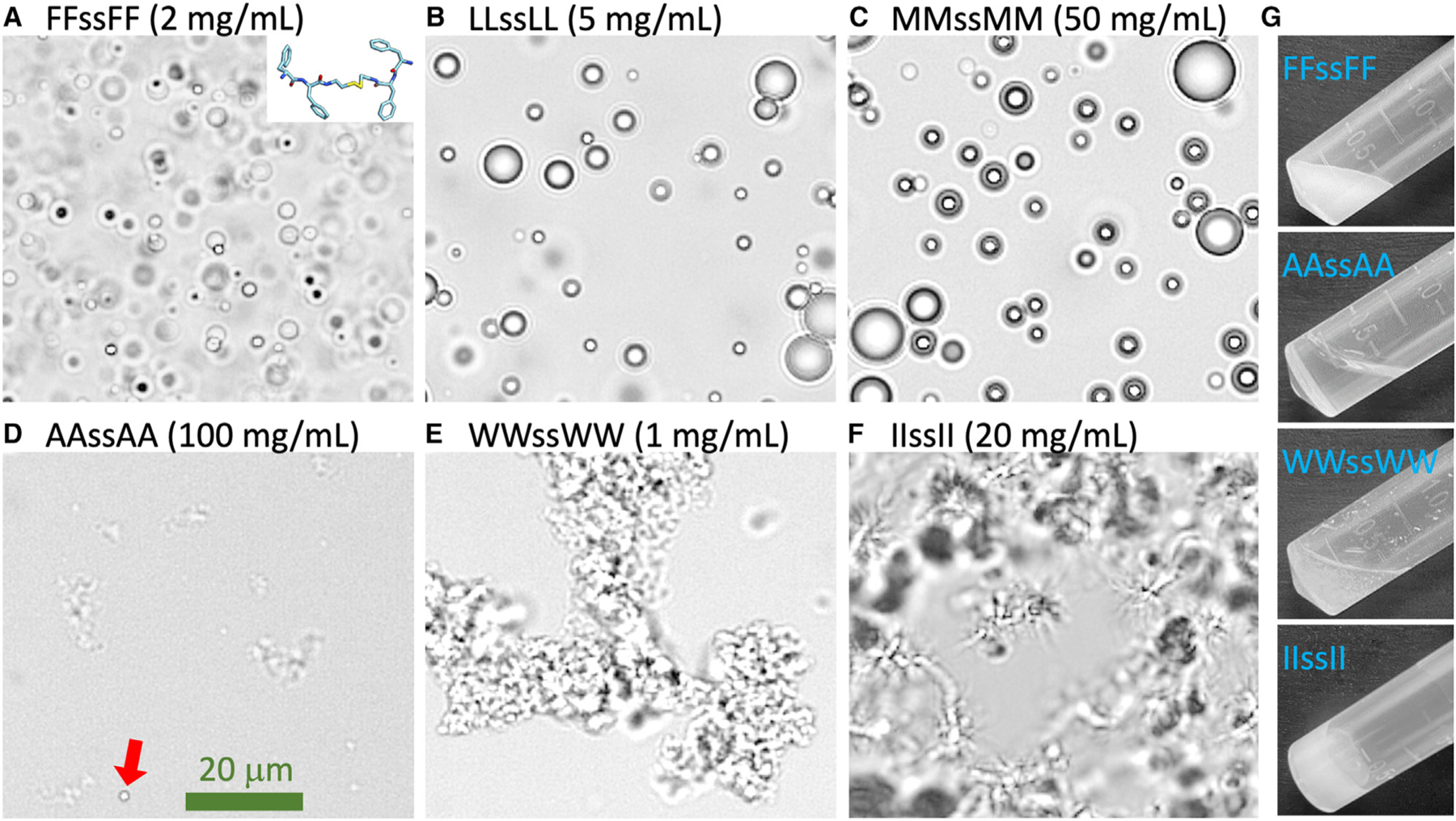
(A–F) Bright-field images of six tetrapeptide condensates formed at the indicated concentrations in Milli-Q water and at pH 13. In (D), a droplet is indicated by a red arrow.
(G) Images of 300-μL samples in a tube, all taken within 2 min of raising the pH from 2 to 13. The scale bar shown represents 20 μm.
RESULTS
We used a pH rise to trigger phase separation. As shown in Figure S1, the XXssXX peptides have two titratable terminal amines, each of which carries a positive charge at low pH (pH 2) but becomes neutral at high pH (pH 13). At low pH, the net charge repulsion between peptide molecules overrides the attraction provided by the nonpolar side chains and hence prevents phase separation. As the pH is raised, this repulsion is weakened or eliminated, and consequently, the tendency for phase separation is increased when one or both of the amines become neutral.
Tetrapeptides form a variety of condensates, including an amorphous dense liquid phase
All eight tetrapeptides (Figure S1) form condensates, but the condensates have different morphologies when observed under a bright-field microscope (Figures 1A–1F). At high pH (pH 13), condensates with X = F, L, and M are all spherical liquid droplets. The AAssAA condensate is a distinct phase that we refer to as ADL: amorphous for irregular shapes and dense liquid for the image contrast from the surrounding bulk phase. A small number of droplets may also be mixed in (one indicated by a red arrow in Figure 1D). The WWssWW condensate is an amorphous aggregate phase, while the IIssII condensate is a gel phase. Liquid droplets, amorphous aggregates, and gels have been widely reported for biomolecular condensates. ADLs have the appearance of clouds, with a morphology similar to that of amorphous aggregates but a much lower image contrast. Gels exhibit filamentous features resembling sea urchins, and in contrast to the limited size of amorphous aggregates, gel networks can grow indefinitely to span the entire field of view . ADLs were observed as precursors in the formation of crystals,32 but do not seem to have been reported previously for biomolecular condensates. Further characterizations of ADLs are presented below.
One way to further distinguish the four different forms of tetrapeptide condensates is to observe them in a tilted test tube with the naked eye (Figure 1G). Droplet, ADL, and aggregate samples are all liquid-like in the sense that the surface of the solution remains horizontal when the test tube is tilted. In each of these three forms, the individual condensate particles are invisible to the naked eye; the surrounding bulk phase explains the horizontal surface of the solution in a tilted test tube. Droplet samples, as illustrated by FFssFF prepared at 2 mg/mL, have a milky appearance. ADL samples, as represented by AAssAA at 100 mg/mL, are transparent and look almost like a homogeneous solution, consistent with the low contrast in the bright-field image (Figure 1D). For amorphous aggregates, as shown by WWssWW at 1 mg/mL, sedimentation ensues quickly so that the top of the solution becomes transparent while the bottom remains turbid; the tendency of sedimentation can be attributed to denser packing (relative to droplets). In contrast to these liquid-like samples, gel samples are solid-like. At lower concentrations, gels are visible as suspended, turbid particles in the test tube; at higher concentrations, the entire sample is turbid, suggesting that the gels become a single densely connected mesh that sticks to the test tube wall. The sample does not move relative to the tilted test tube, making its surface slanted (Figure 1G).
The bright-field images in Figure 1 show the contrast order ADL < droplet < aggregate. This order is confirmed by negative-stain EM (Figure 2A). At pH 13, VVssVV starts to form gels at 5 mg/mL, as indicated by the filamentous appearance in a bright-field image. EM images further demonstrated the filamentous feature. The condensate morphologies of the eight peptides are summarized in Figure 2B.
Figure 2. Four types of condensates formed by tetrapeptides.

(A) Contrasting the four types of condensates by bright-field (BF) and negative-stain electron microscopy (EM). Samples were prepared at the indicated concentrations in Milli-Q water at pH 13. The green scale bar applies to all the BF images and represents 20 μm; all the brown scale bars represent 200 nm.
(B) Summary of tetrapeptides that occur in each condensate morphology. Single arrows mean roughly one-half of the phase-separation pH range; double arrows mean a small fraction of the phase-separation pH or concentration range. Arrows in the upward direction mean high pH or concentration; arrows in the downward direction mean low pH.
As one more way to distinguish the four condensate morphologies, we stained the samples with a viscosity-sensitive dye (Figure S2); the fluorescence intensity provides a measure of the local peptide density. As expected, the fluorescence intensity is very uniform inside FFssFF droplets. It is also uniform inside WWssWW aggregates, such that regions showing intense fluorescence match those that are in focus in the corresponding bright-field image; these regions have rough boundaries instead of the smooth, circular ones of droplets. By contrast, the dye appears to stain only dense cores in AAssAA ADLs and IIssII gels. These core regions are tiny for ADLs but more spread out for gels.
We used FRAP to probe the molecular mobility inside condensates (Figure S3). A 200 × 200 nm square region inside condensates was photobleached; the subsequent recovery was fit to an exponential function to obtain the mobile fraction of dye molecules. This fraction is 61% inside FFssFF droplets (comparable to what was reported by Abbas et al.18) and reduces to 29% inside WWssWW aggregates and only 18% inside IIssII gels. The last result suggests that the gel networks are largely solid-like. Related to these FRAP results, IIssII gels take minutes to fully form when started inside the phase-separation region but close to the phase boundary, whereas WWssWW aggregates, similar to droplets and ADLs, form instantaneously (Figure S4).
The morphology results summarized in Figure 2B were based on the phase diagrams of the peptides, determined by scanning pH and peptide concentration and observing the sample at the given pH and under a bright-field microscope. We dissolved the peptides in 50 mM imidazole buffer to better tune the pH, at least for the pH 6.2–7.8 operating range of this buffer. The outcome is either no phase separation (‘‘×’’ in Figures 3A–3E) or, when phase separation occurs, the condensate morphology (‘‘●’’ for droplets, ‘‘◇’’ for ADLs, and ‘‘✲’’ for gels). Only a single condensate morphology is observed for three peptides: WWssWW as amorphous aggregates, IIssII as gels, and FFssFF as droplets (Figure 3A). For LLssLL (Figure 3B) and MMssMM (Figure 3C), droplets are observed in most of the phase-separated region of the phase diagram, whereas ADLs are observed at low pH; for PPssPP (not shown), ADLs are observed in most of the phase-separated region, while droplets are observed at high pH. For VVssVV (Figure 3D), the phase-separated region is divided into a gel subregion (higher pH) and an ADL subregion (lower pH). AAssAA is similar (Figure 3E), except that its gel subregion is limited to very high pH and very high peptide concentration.
Figure 3. Phase diagrams and threshold concentrations at pH 7.
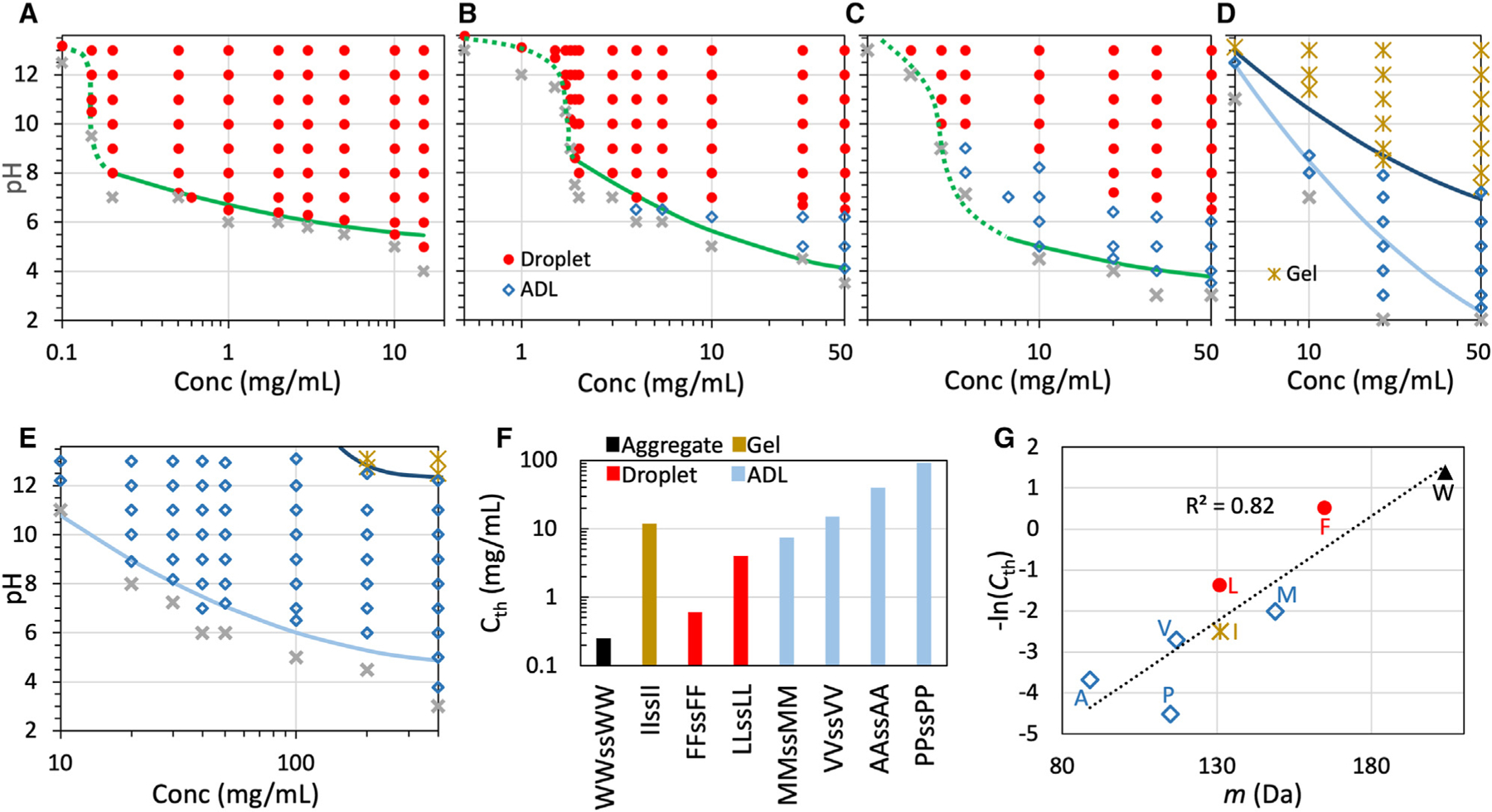
(A–E) Phase diagrams of tetrapeptides with X = F, L, M, V, and A in 50 mM imidazole buffer. Solid curves are fits to a parabolic function (Equation 1); dotted curves are drawn to guide the eye. For LLssLL and MMssMM, the transition from ADL to droplets may span a finite interval of pH, which expands with increasing peptide concentration to ~1 pH unit; in the transition region, the morphology is assigned to the dominant species.
(F) Threshold concentrations for phase separation at pH 7.
(G) Correlation between threshold concentration and amino acid molecular mass.
Phase-separation threshold concentration correlates with side-chain interaction strength
To compare the drive for phase separation among the peptides, we measured the minimum, or threshold, concentration for phase separation at pH 7 (Figure 3F). The values of range from 0.25 mg/mL for WWssWW to 92 mg/mL for PPssPP. This 368-fold difference demonstrates the enormous disparity between amino acids in their drive for phase separation. Because the amino acids in all the eight homopeptides are nonpolar and the strengths of side chain-side chain interactions are expected to scale with the amino acid sizes, we tested for correlation between and the amino acid molecular mass (Figure 3G). There is a strong correlation between these two properties, with the coefficient of determination at 0.82. A slightly higher correlation, , is obtained with a compound factor, , where is the Lennard-Jones diameter and is the ‘‘stickiness’’ parameter of a coarse-grained model for IDPs (one bead per amino acid).20 This compound factor was devised to capture the strengths of nonpolar interactions between amino acids.2
We wondered whether backbone hydrogen bonding also contributed to the drive for phase separation. Urea was used as a reporter for backbone hydrogen bonding in previous studies.18,19 Consistent with the observation of Abbas et al.,18 urea had a modest effect on droplet formation of 0.25 mg/mL FFssFF at pH 13 (Figure S5a). In contrast, droplet formation of 1.5 mg/mL LLssLL is inhibited by 2 M urea (Figure S5b). Likewise, gel formation of 1.5 mg/mL IIssII is greatly suppressed by 2 M urea and blocked by 3 M urea (Figure S5c). Interestingly, 2 M urea turns gels formed by 6 mg/mL VVssVV into ADLs; the ADL morphology is maintained at 3 M urea, though the amount of ADL is reduced (Figure S5d). These observations suggest that backbone hydrogen bonding plays an appreciable role in the phase separation of LLssLL, IIssII, and VVssVV.
The boundary of the phase-separated region moves to higher peptide concentrations as the is lowered. As noted above, determines the charge states of the terminal amines of each peptide and, hence, the extent of net charge repulsion between peptide molecules. Peptide concentration controls the density of intermolecular interactions. Greater net charge repulsion must be balanced by a higher density of interactions between amino acids for phase separation to occur. The phase boundary can be fit to a parabolic function:
| Equation 1 |
where is the minimum or critical at which phase separation is still observed, is the corresponding critical concentration, and is a constant. The critical values are 5.3, 3.8, 3.5, 0.3, and 4.8 for peptides with X = F, L, M, V, and A, respectively. The corresponding critical concentrations are 66, 148, 154, 245, and 665 mg/mL, respectively. These critical values can be taken only as qualitative indications, as we were limited by the highest concentrations at which samples could be prepared for phase-boundary determination. The phase boundaries of peptides with X = F, L, and M appear to show a transition above 8, with the caveat that the regulation capacity of the imidazole buffer is weak in this range.
At 7 and the respective threshold concentrations, the condensates formed are amorphous aggregates (X = W), gels (X = I), liquid droplets (X = F and L), or ADLs (X = M, P, V, and A) (Figures 3F and S4). While the threshold concentrations correlate well with the strengths of side-chain nonpolar interactions, the rules governing condensate morphology are more complex. Side-chain interaction strength appears to explain three of the four morphologies (Figure 3F), as amorphous aggregates, droplets, and ADLs are favored by stronger (X = W), intermediate (X = F and L), and weaker (X = M, P, V, and A) interactions, respectively. This is also the contrast order exhibited by bright-field and EM images of the condensates. However, it is difficult to predict when gels will form. For example, L and I are very similar chemically, but LLssLL favors droplets, whereas IIssII forms only gels.
We studied the phase behavior of some mixtures of different tetrapeptides. The 1:1 (molar ratio) mixture of IIssII and AAssAA forms only gels (Figures S6a and S6b). At 7, the threshold concentration is 13 mg/mL (Figure S6c), intermediate between those of pure IIssII (12 mg/mL) and pure AAssAA (40 mg/mL) but very close to the former. The mixture of these two peptides forms gels even at a 1:3 molar ratio (Figure S6d). That the mixtures take the condensate morphology of IIssII instead of AAssAA perhaps reflects the dominance of IIssII self-interactions over AAssAA self-interactions.
ADLs are characterized by weak intermolecular interactions and low peptide densities and may implicate critical slowing down
As noted above, ADLs are favored by peptides with weaker intermolecular interactions. Further accentuating this point is the fact that, for a given peptide at a fixed concentration, ADLs are observed at lower pHs, but droplets or gels are observed at higher pHs (Figures 3B–3E; also Figure S4 for PPssPP at pH 7 and 13). Lower pHs correspond to greater net charge repulsion and hence a lower net strength of interactions; conversely, higher pHs correspond to a higher net strength of interactions. Moreover, the ADL subregion as a fraction of the entire phase-separated region follows the order L < M < V < P < A, which is precisely the order of decreasing interaction strength (as measured by ). This trend neatly illustrates the point that weak interactions are inducive of ADL formation. As one more piece of supporting evidence, we point out that weakening of intermolecular interactions by adding urea converts VVssVV gels into ADLs (Figure S5d).
Sometimes ADLs are mixed with a small number of droplets (Figures 1D and S4). For LLssLL, MMssMM, and PPssPP, ADL-droplet mixtures occur at the boundary between the pure ADL and the pure droplet regions. At a fixed concentration, the transition from the ADL region to the droplet region may not be sharp but span a finite interval of pHs (Figure S7a). This interval expands with increasing peptide concentration. To gain insight into the transition zone, we introduced pH jumps that either bypass the transition zone or start or end inside it. Conversions between ADLs and droplets are completely reversible when the pH jump bypasses the transition zone, as illustrated for 10 mg/mL MMssMM by a pH jump between 9.0 and 8.0 (Figure S7b), 10 mg/mL LLssLL by a pH jump between 7.5 and 6.5 (Figure S7c), and 50 mg/mL LLssLL bya pH jump between 6.8 and 5.0 (Figure S7d). In contrast, while a pH increase starting from the transition zone (50 mg/mL LLssLL from pH 6.0 to 6.8) results in complete conversion from an ADL to droplets, a pH decrease ending in the transition zone (from pH 6.8 to 6.0) encounters hysteresis, i.e., many more droplets are now observed than in a sample prepared by raising the pH to 6.0 from an initial baseline value of 2.0 (Figure S7e). The hysteresis indicates that the ADL phase is separated from the droplet phase by a high free energy barrier.
To explore the conversion from ADLs to droplets in the transition zone, we asked whether raising the temperature could speed up the conversion. Abbas et al.18 already showed that the phase separation of XXssXX peptides had an upper critical solution temperature (UCST), meaning that raising the temperature decreases the drive for phase separation. We confirmed the UCST character by a decreasing number of FFssFF droplets as the temperature was raised from 4°C to 25°C and 100°C (Figure S8a). Still, we reasoned that a high temperature, by enhancing thermal fluctuations, could speed up the kinetics of phase transition. We checked the temperature dependence of the condensate morphologies of peptides with X = L (50 mg/mL and pH 6.2), V (50 mg/mL and pH 7.2), and A (100 mg/mL and pH 13) (Figures S8b–S8d). Whereas droplets are rare to find among ADLs at 4°C and 25°C, they are readily seen along with ADLs at 100°C. Once formed as droplets at 100°C, they remain as droplets even when the samples are cooled down to 25°C, supporting a high free energy barrier separating ADLs from droplets.
Low contrast of bright-field and EM images and low fluorescence intensity (Figures 1, 2A, and S2) all point to the low peptide densities inside ADLs relative to inside droplets. Therefore, for ADLs to convert into droplets, they have to further condense. However, because of the weak intermolecular interactions, further condensation is unfavorable, leading to a high free energy barrier. Indeed, our MD simulations show that ADLs are highly dynamic and resist further condensation (see below). Based on the fact that ADLs are always observed around the critical point (, ), we speculate that the slow conversion between ADLs and droplets might be a form of critical slowing down. The general idea is that, near the critical point, the correlation length increases indefinitely, and correspondingly, the characteristic relaxation time for phase transition grows to infinity.33
Both ADLs and aggregates are amorphous, but as already stated, they can be easily distinguished by a significant difference in image contrast and fluorescence intensity (Figures 1, 2A, and S2). Moreover, relative to droplet formation, aggregate formation requires a higher interaction strength, whereas ADL formation requires a lower interaction strength. Conforming to these requirements, for a peptide at a fixed concentration, ADLs are observed at lower pHs, whereas droplets are observed at higher pHs, as emphasized above. If ADLs were actually aggregates, they would have to occur at higher pHs than droplets, in contrast to actual observations.
Droplet fusion speed exhibits critical scaling with pH
Next, we report the material properties of tetrapeptide droplets, including fusion speed, interfacial tension , and zero-shear viscosity , all measured on a LUMICKS dual-trap OT instrument. OT-directed fusion was set up by trapping two equal-sized droplets and bringing them into contact (Figure 4A).24,34 The subsequent spontaneous fusion process was monitored by the small force signal due to the retraction of the droplets from the fixed traps. The force trace was fit to a stretched exponential to obtain the fusion time (Figure 4B). For LLssLL droplets of similar sizes (initial radius ), the fusion time has a clear dependence, increasing with decreasing (Figure 4B). By fitting the dependence of on to a proportional relation, we obtained inverse fusion speed as the slope, (Figures 4C and S9). We verified that the initial peptide concentration did not affect the fusion speed (Figure S9b).
Figure 4. OT-directed droplet fusion.
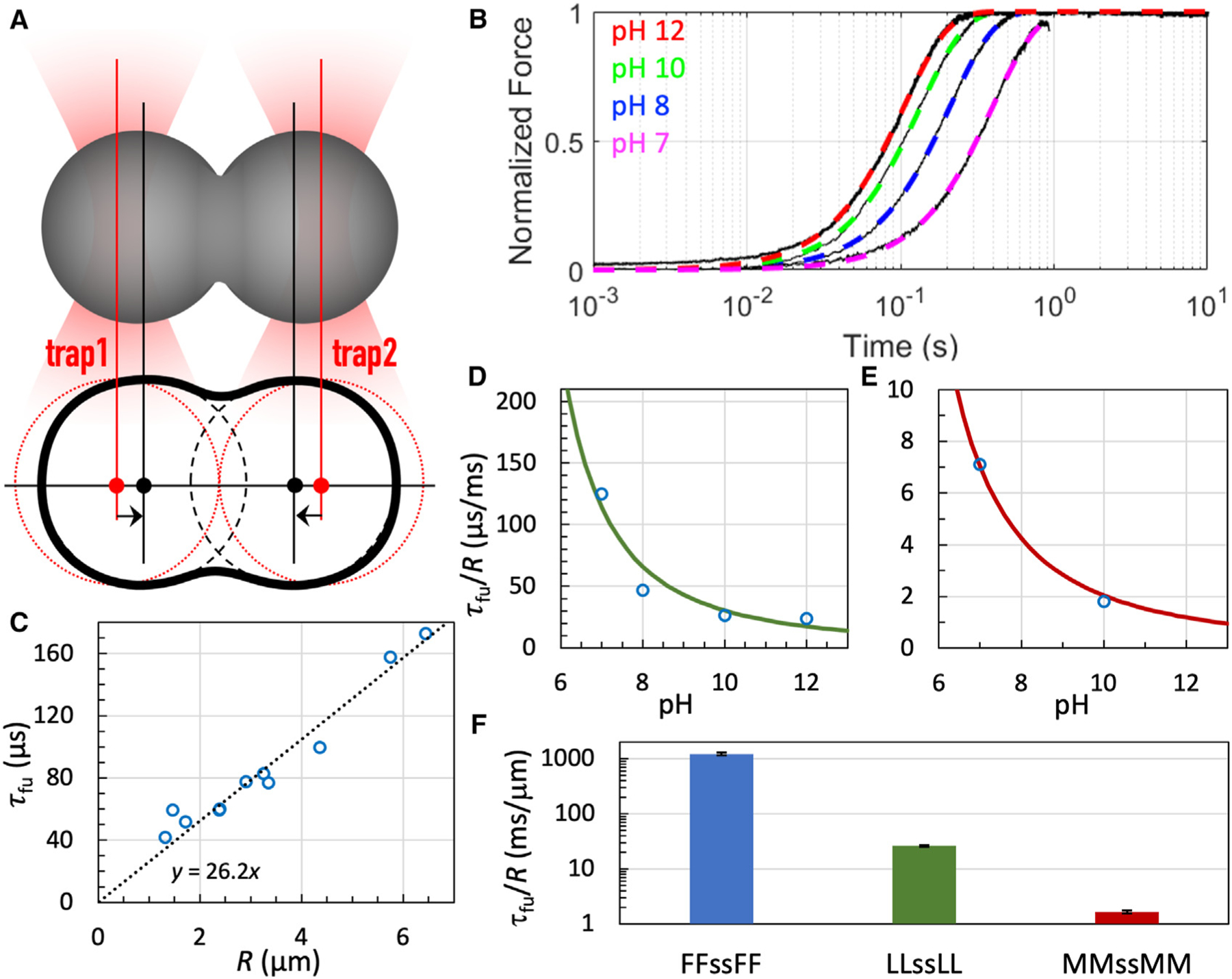
(A) Illustration of directed fusion.
(B) Fusion progress curves of LLssLL droplets. Black traces show raw data; colored curves are fits to Equation 9. Droplets were prepared at the indicated pH and grown to 3 to 4 μm in radius before fusion.
(C) Fusion time of LLssLL droplets as a function of initial droplet radius. Each circle represents a fusion event. A line displays the proportional relation between fusion time and droplet radius. Droplets were prepared with 5.5 mg/mL LLssLL at pH 10.
(D) Inverse fusion speed of LLssLL vs. pH. The curve displays a fit to Equation 2 with fixed at 2, fixed at 3.8, and adjusted to 1,165.
(E) Inverse fusion speed of MMssMM vs. pH. The curve displays a fit to Equation 2 with fixed at 2, fixed at 3.5, and adjusted to 86.
(F) Inverse fusion speeds of droplets with X = F, L, and M at pH 10. Error bars represent the standard error in as a fitting parameter.
The effect of on can be modeled by a scaling law,
| (Equation 2) |
where is a critical exponent and is a constant. For purely viscous droplets, one expects24
| (Equation 3) |
obeys the scaling law:
| (Equation 4a) |
| (Equation 4b) |
where 35 and Equation 1 has been used ongoing from the first line to the second line. does not exhibit critical singularity (i.e., it remains finite at the critical point). Equations 3 to 4b predict . For both LLssLL and MMssMM droplets, the dependence of follows the scaling law with the critical exponent (Figures 4D and 4E). However, although Equation 3 serves as a motivation for the scaling law for , it breaks down for the peptide condensates studied here, as shown below, because of their viscoelastic nature.12,25
Fusion speeds of tetrapeptide droplets span three orders of magnitude
We compare the inverse fusion speeds of droplets with X = F, L, and M in Figure 4F. For this comparison as well as comparisons of other material properties below, we selected pH 10, at which the peptides readily form a large number of droplets. MMssMM droplets fuse at . The corresponding fusion speed is 16-fold faster than that of LLssLL droplets and 736-fold faster than that of FFssFF droplets. (The inverse fusion speed measured here for FFssFF is comparable to that reported by Abbas et al.18) The fusion speeds follow the same order as the threshold concentrations (Figures 3A–3C), suggesting side-chain interaction strength as a determinant for fusion speed. However, whereas the threshold concentrations differ by only 20-fold, the fusion speeds differ by 736-fold.
At 10, PPssPP forms a significant portion of ADLs along with droplets, making it difficult to study droplet fusion. We thus measured the fusion speed at pH 13 (Figure S9f), where only droplets are formed. The inverse fusion speed is . When the scaling law is used to predict an inverse fusion speed for MMssMM, a value of 0.95 ms/μm is obtained. This value indicates that PPssPP has a higher fusion speed than MMssMM, which is consistent with the order in threshold concentration between these two peptides (Figure 3F) and further supports side-chain interaction strength as a determinant for fusion speed.
Interfacial tensions fall into a relatively narrow range
As Equation 3 indicates, droplet fusion is driven by interfacial tension and retarded by viscosity. By using two optically trapped beads to stretch a droplet (Figure 5A) and monitoring the forces at successive extensions (Figure 5B), we measured the interfacial tension.25,36 For droplets formed by peptides with X = F, L, and M, the interfacial tensions are , , and (Figure 5C). The values differ by only 3-fold. Interfacial tensions of various biomolecular condensates all fall into a relatively narrow range, mostly from 20 to 200 pN/μm.2
Figure 5. Interfacial tensions of tetrapeptide droplets.
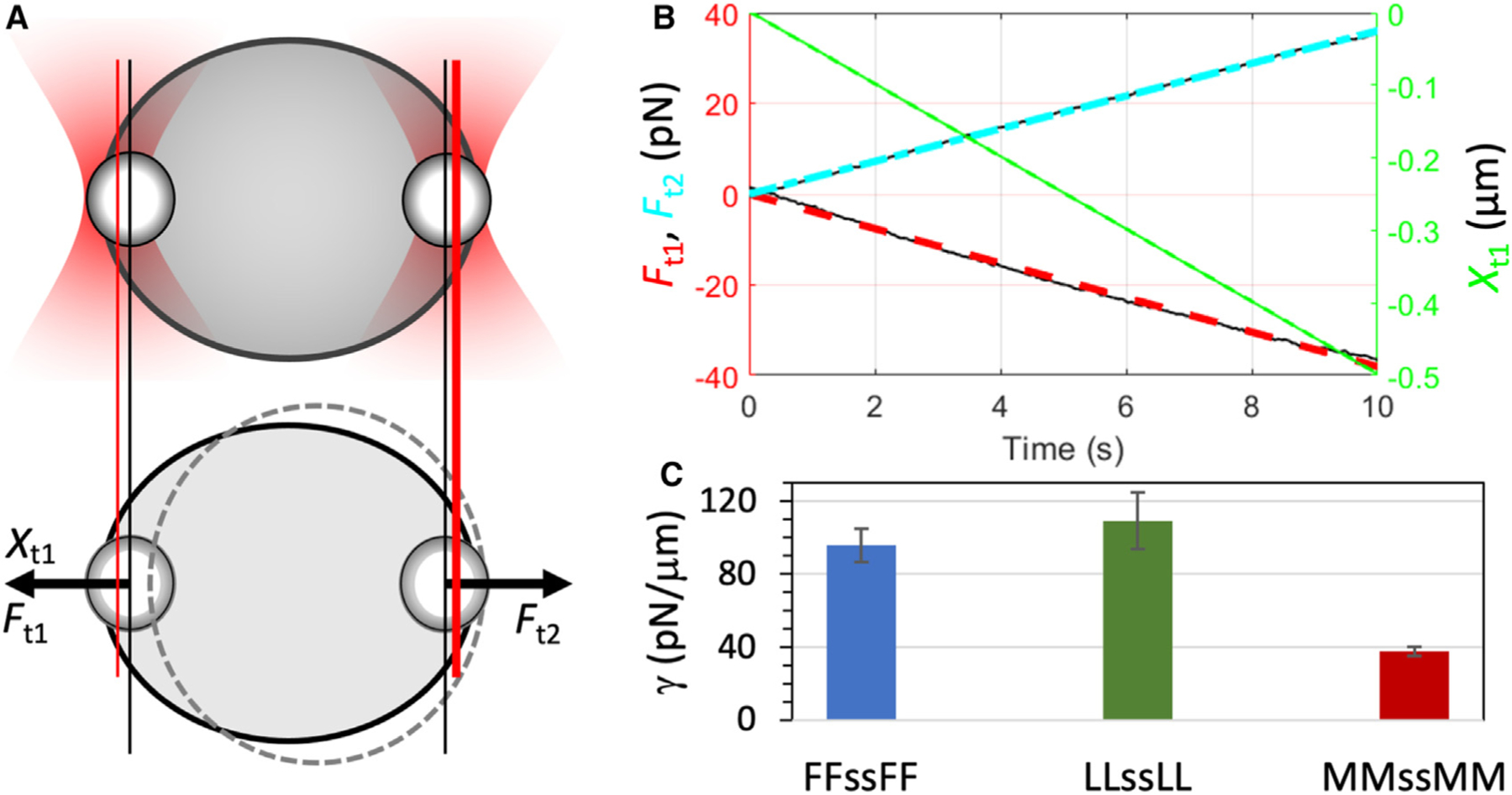
(A) Stretching of a droplet by two optically trapped beads (1.13 μm in radius).
(B) Stretching forces and extensions measured on an LLssLL droplet formed at 5.5 mg/mL and pH 10.
(C) Interfacial tensions of tetrapeptide droplets with X = F, L, and M at pH 10. Error bars represent standard deviations among four droplets with ~6 μm radii.
Effective viscosity during droplet fusion is much lower than zero-shear viscosity
With the fusion speed and interfacial tension at hand, we can deduce the effective viscosity, , in the fusion process. is defined by inverting Equation 3:
| (Equation 5) |
The values for droplets formed by peptides with X = F, L, and M are 59, 1.5, and 0.032 Pa s. IDP condensates have a tendency to exhibit effective viscosities much lower than their zero-shear viscosities, a phenomenon called shear thinning.2,25,37
To test whether tetrapeptide droplets also exhibit shear thinning, we measured their zero-shear viscosities using a bright-field camera to track a free bead inside a droplet that was settled on a coverslip (Figures 6A and 6B).37 By fitting the mean-square displacement (MSD) of the tracked bead to a linear function of time (Figures 6C and S10), one can find the zero-shear viscosity from the slope. The results are , , and Pa s, respectively, for peptides with X = F, L, and M. These values are 7- to 85-fold higher than the corresponding effective viscosities, thereby implicating substantial shear thinning. The extent of shear thinning exhibited by the tetrapeptides here is much greater than that by IDPs25 but some-what less than that by ATP-IDP mixtures.12 The enormous shear thinning in the latter systems was partly attributed to the small size of ATP, allowing it to quickly re-form bridges between IDP chains during droplet fusion. Given the relatively small size of the tetrapeptides, perhaps a similar mechanism is at play here.
Figure 6. Zero-shear viscosities of tetrapeptide droplets.
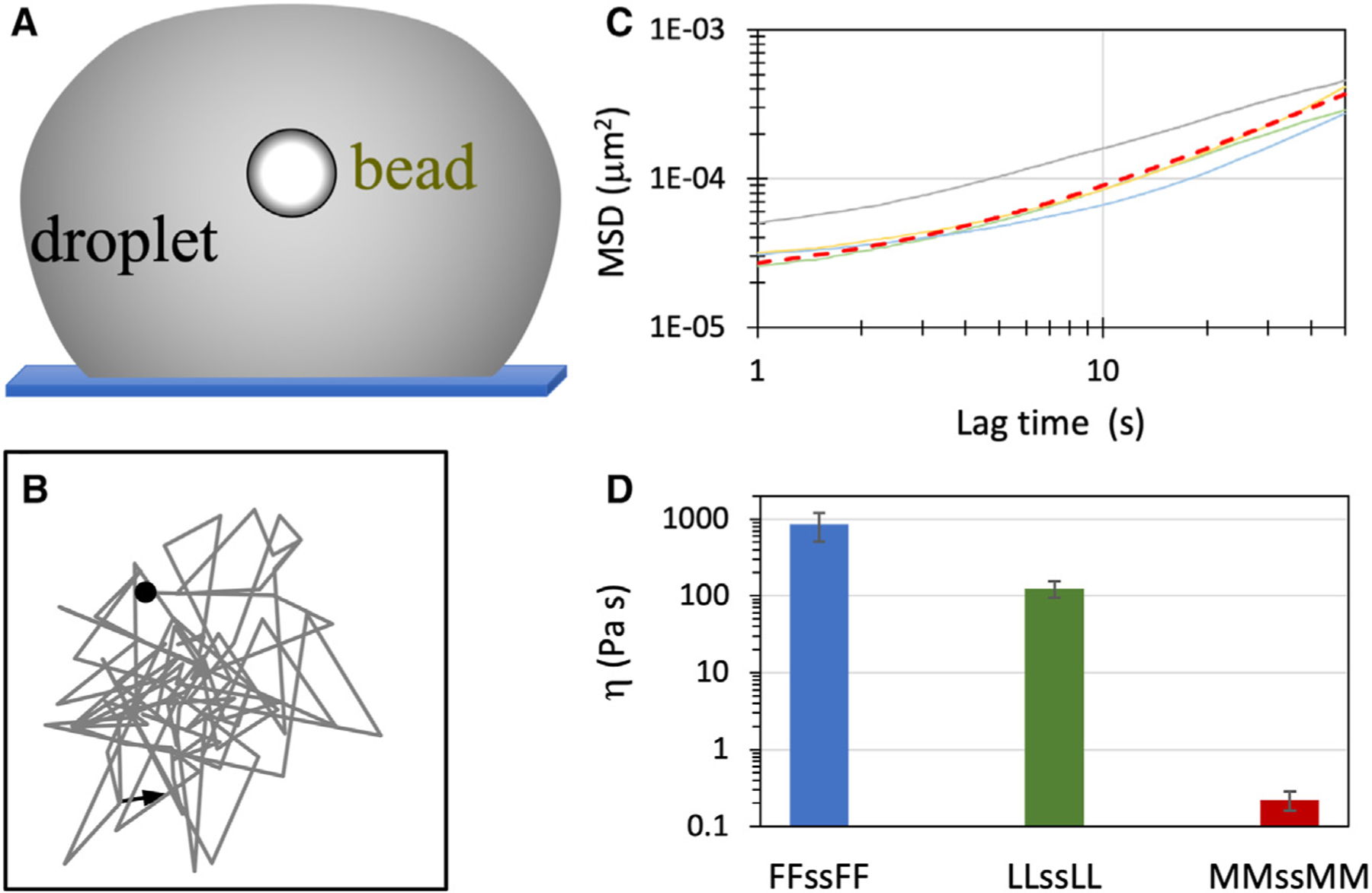
(A) Tracking of a free bead inside a settled droplet.
(B) A short two-dimensional trajectory of a bead (1.13 μm in radius) in an LLssLL droplet prepared at 5.5 mg/mL and pH 10, tracked by a bright-field camera.
(C) MSDs (solid curves) of beads inside four LLssLL droplets. The fit to a linear function for one MSD curve (orange) is displayed as a red dashed curve.
(D) Zero-shear viscosities of tetrapeptide droplets with X = F, L, and M at pH 10. Error bars represent standard deviations among four beads.
It is worth noting that the viscosity measured here for the FFssFF condensate appears to be higher than any previously reported value for biomolecular condensates.2 It is higher than the LLssLL viscosity by 7-fold and the MMssMM viscosity by nearly 4,000-fold.
Molecular dynamics simulations of tetrapeptides recapitulate condensate morphologies
Previously we observed spontaneous condensate formation in MD simulations of 64 copies of FFssFF in explicit water.38 Here, we changed to a force-field combination that was found to model well IDPs in explicit water39 and expanded the simulations to all eight tetrapeptides. For each tetrapeptide, 64 loosely packed copies of the form with both terminal amines neutral (modeling high pH) were solvated in a cubic simulation box. Four different fractions of water molecules were removed to concentrate the peptide to a range of initial concentrations; duplicate simulations of each of these systems were carried out at constant temperature (294 K) and pressure (1 bar). Within 1 μs of the simulations, condensates start to take shape. For peptides with X = F, M, L, I, and V, the condensates in at least one of the eight simulations are a stable slab (Figures 7A and S11), which corresponds to a spherical droplet in a bulk solution. The peptide molecules in these condensates exhibit a moderate level of dynamics (Video S1 for LL), with 20%–40% of molecules surrounding a tagged molecule moving away from that molecule after 1 μs (Figure S12a). In comparison, WWssWW condenses only into an amorphous aggregate (Figures 7A and S11), which remains static except for a few molecules on the surface (Video S2 for WW; Figures S12a and S12b). By contrast, AAssAA and PPssPP also form slabs, but the boundaries of the slabs are typically rough (Figures 7A and S11) and change rapidly. In 1 μs, ~80% of molecules surrounding a tagged molecule move away from that molecule (Video S3 for AA; Figures S12a and S12c).
Figure 7. Condensate morphologies and intermolecular interactions in MD simulations.
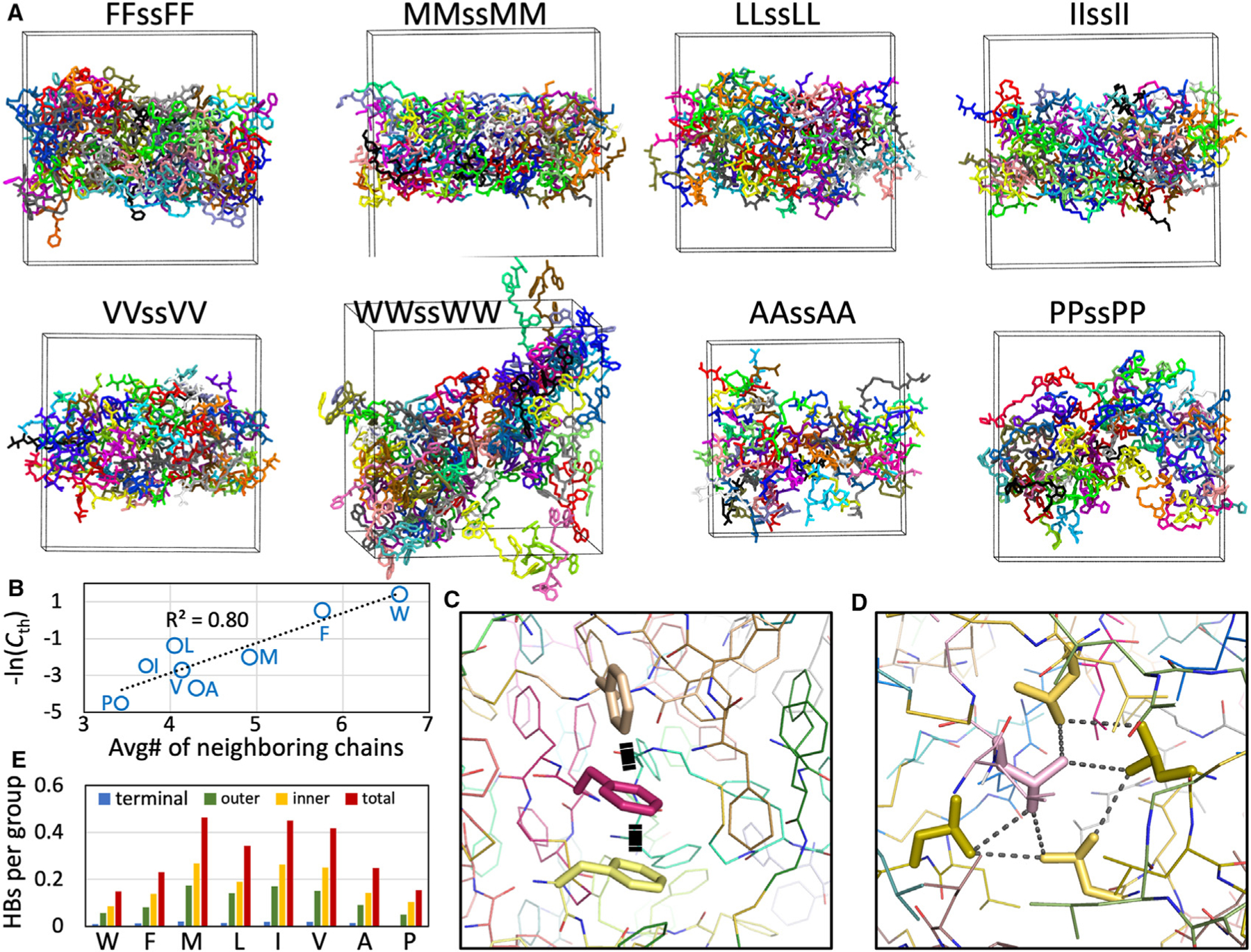
(A) Morphologies of the eight tetrapeptides.
(B) Correlation of the average number of chain neighbors with phase-separation threshold concentration.
(C) interactions in the FFssFF condensate.
(D) A hydrophobic cluster in the LLssLL condensate.
(E) Average numbers of hydrogen bonds formed by the terminal amine, the outer peptide group, or the inner peptide group.
Whereas peptides with X = F, M, L, A, and P form slabs in four or five of the eight simulations, IIssII and VVssVV do so in only one and two, respectively, of the eight simulations. In the remaining simulations, the condensates have many holes, which perhaps resemble gels (Figure S13). Overall, the simulations show that peptides with X = F, M, and L favor droplets, peptides with X = A and P form condensates that partly resemble droplets and partly resemble ADLs, peptides with X = I and V prefer gel-like condensates, and WWssWW forms only aggregates. These results match well with the high-pH condensate morphologies observed under a microscope and highlight peptide interaction strength as a major determinant of condensate morphology.
The threshold concentration quantifies the propensity to phase separate: a lower corresponds to a higher propensity. A determinant of this propensity is the ability of the component molecules to network with one another. As a measure of this property, we calculated the number of neighboring chains of each peptide molecule in the condensate (Figure S14). The average number of neighboring chains ranges from 6.7 for X = W to 3.4 for X = P. A strong correlation is found between the average number of neighboring chains and , with at 0.80 (Figure 7B).
Intermolecular networks in the condensates are stabilized by interactions for X = W and F and hydrophobic clusters for the other peptides (Figures 7C, 7D, and S15). The backbones of the peptides also exhibit varying levels of hydrogen bonding. LLssLL, IIssII, and VVssVV condensates have much higher levels of backbone hydrogen bonding than FFssFF condensates (Figure 7E), consistent with the experimental results reported by adding urea (Figure S5). The lower backbone hydrogen-bonding level of FFssFF is due to steric interference of the bulky F side chain. Interestingly, the hydrogen-bonding levels of both IIssII and VVssVV are particularly high, on average forming close to one hydrogen bond per chain. Backbone hydrogen bonding perhaps contributes to gel formation.
To study the equilibration between the dense and the dilute phases of FFssFF and LLssLL peptides, we placed the dense slab formed in the simulations with a cubic box to an elongated box and also varied the charge states of the terminal amines to model different values (Figure 8A). The numbers of chains with zero, single, and double charges were determined by assuming values of 7 and 6 for the two amines in each chain. For example, at 7.3, half of the 64 copies are neutral at both amines and the other half are charged at one amine. With decreasing , the dilute-phase concentrations increase significantly (Figures 8A–8C), due to net charge repulsion in the dense phase. The dilute-phase arm of the FFssFF and LLssLL binodals is qualitatively similar to the experimental phase boundaries of Figures 3A and 3B. The dilute-phase concentrations from the simulations of FFssFF and LLssLL at pH 7.3 are 2 and 7 mg/mL, respectively, comparable to the experimental values of 0.5 and 3 mg/mL (Figure 3F). In comparison, the dense-phase concentrations decrease, but only slightly, with decreasing pH.
Figure 8. Properties of the dilute and dense phases of FFssFF and LLssLL.
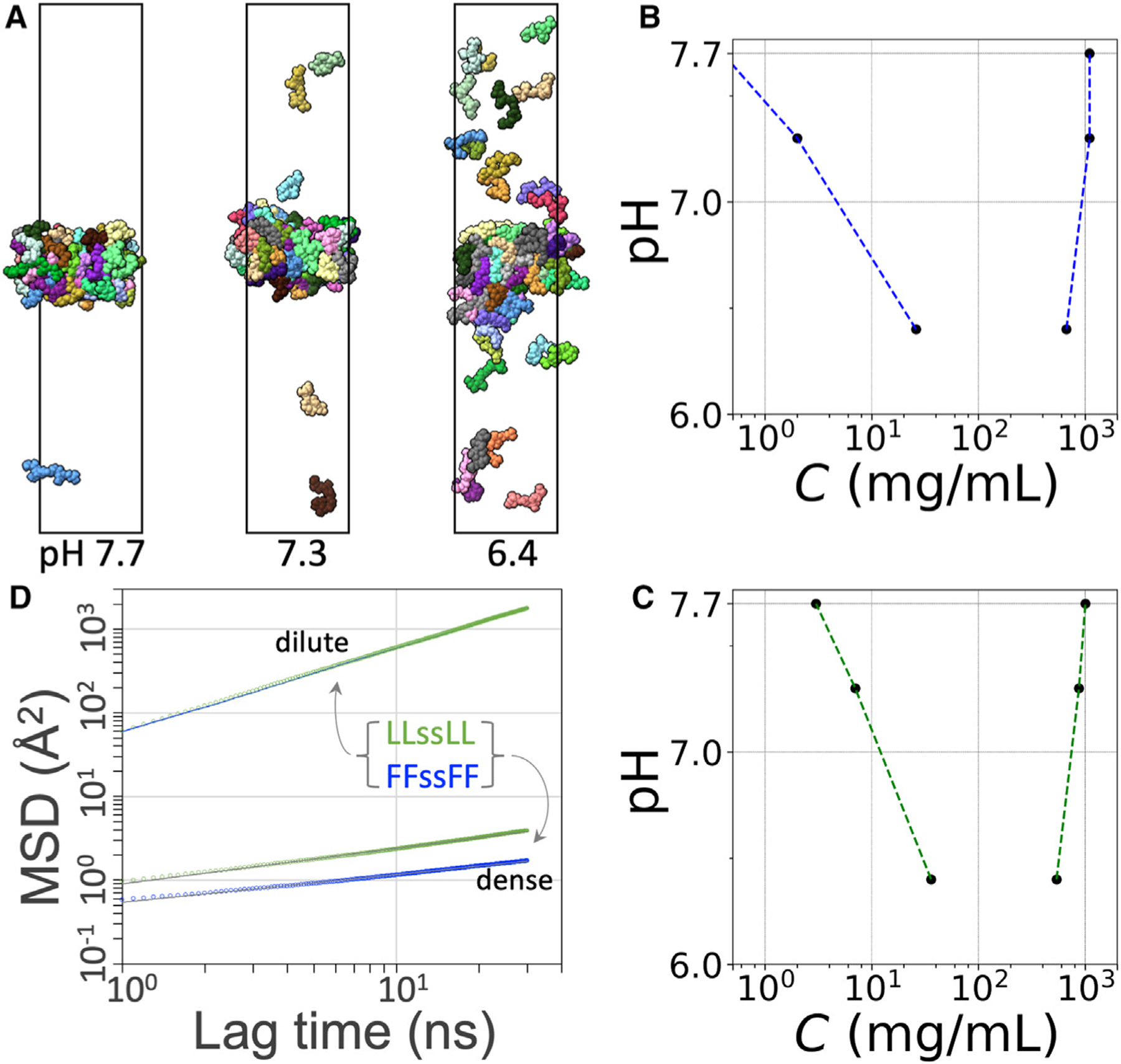
(A) Phase coexistence of LLssLL at different values.
(B) Binodal of FFssFF.
(C) Binodal of LLssLL.
(D) MSDs of tetrapeptide molecules in the dilute and dense phases at pH 7.3. For the dilute phase, the results for FFssFF and LLssLL are displayed as a blue line and green circles, respectively; for the dense phase, the raw data are displayed as blue or green circles, and a fit to Equation 6 is shown as a black line.
To probe the dynamics within the condensates, we calculated the diffusion constants of peptide molecules that stayed either in the dilute phase or in the dense phase in a 1.04-μs period (Figure 8D). In the dilute phase, the MSDs of FFssFF and LLssLL were essentially identical and conformed to a Brownian behavior, with time dependence given by and a diffusion constant . This has the same order of magnitude as that for dilute-phase short peptides measured by NMR.29 In the dense phase, the MSDs exhibit subdiffusion, which is characteristic of molecular motions hindered by mobile obstacles.40 The time dependence of the MSDs fits well to
| (Equation 6) |
with for FFssFF and 0.42 for LLssLL. The subdiffusion can be viewed as a manifestation of condensate viscoelasticity at the submicrosecond timescale.41 Subdiffusion of a protein inside postsynaptic condensates has recently been observed using superresolution imaging.42 We define an effective diffusion coefficient as
| (Equation 7) |
The difference in between FFssFF and LLssLL grows with and is 2.3-fold at . The theoretical expectation is that would become a constant at much longer times. If Equation 6 is extrapolated to 0.5 ms, it would predict for FFssFF and for LLssLL. The corresponding values of would yield dense-phase viscosities, 700 and 126 Pa s, that match well with the measured values (Figure 6D), with the caveat that such extrapolation carries significant uncertainty.
Two interrelated factors explain the lower diffusion coefficient and hence higher viscosity of FFssFF relative to LLssLL. First, as noted above, the amino acid F has a greater side-chain interaction strength than L. This difference is reflected by the higher average number of neighboring chains for FFssFF than for LLssLL. Second, as shown in Figures 8B and 8C, the FFssFF dense phase has higher densities than the LLssLL counterpart.
The difference in dense-phase molecular diffusion coefficient is consistent with the dispersivity defined as the fraction of surrounding molecules that moved away after 1 ms of simulation (Figure S12a). This parameter may also serve as an indicator of the fusion speed, as fusion, similar to dispersion, involves breaking and re-forming intermolecular contacts. The dispersivities have the orderF<L<M< P, which is precisely the order of the measured fusion speeds (Figure 4F).
DISCUSSION
By combining bright-field, confocal, and negative-staining electron microscopy, OT-based microrheology, and all-atom MD simulations, we have characterized the condensates formed by homotetrapeptides of eight nonpolar amino acids. The condensates have a variety of morphologies, including droplets, amorphous aggregates, gels, and ADLs; the last appear to have not been reported previously. The phase-separation threshold concentrations of these peptides differ by 368-fold, from 0.25 mg/mL for WWssWW to 92 mg/mL for PPssPP. The droplets formed by three of the tetrapeptides span a wide range of material properties, including a 736-fold difference in fusion speed and a 3,856-fold difference in viscosity.
For these homotetrapeptides of nonpolar amino acids, we have found that the size of the amino acid, by serving as a measure of side-chain interaction strength, is a strong determinant of the phase-separation threshold concentration, fusion speed, and viscosity. An important advantage of the short peptides is that their small sizes enable all-atom MD simulations of spontaneous phase separation and two-phase equilibrium and the calculation of equilibrium and dynamic properties in the separated phases. The calculated phase-separation threshold concentrations are comparable to the experimental counterparts, and the differences in calculated dispersivities and diffusion coefficients qualitatively explain the observed disparities in fusion speed and viscosity. Expanding the present approach to all 20 natural amino acids may ultimately allow us to precisely determine the contributions of individual amino acids to phase equilibrium and material properties of IDP condensates.
The side-chain interaction strength, as indicated by the size of the amino acid, is also a contributing factor to condensate morphology. Specifically, the tendency to form three of the condensate morphologies, amorphous aggregate, droplet, and ADL, appears to be correlated with interaction strength. Strong interactions (as in WWssWW) favor amorphous aggregates, medium interactions (as in FFssFF, LLssLL, and MMssMM) favor droplets, and weak interactions (as in AAssAA and PPssPP) favor ADLs. However, the fourth morphology observed here, gel, cannot be solely explained by interaction strength. Backbone hydrogen bonding may possibly be an additional determinant for condensate morphology, in particular for gel formation. Our study raises a number of questions about gels. Why are amorphous aggregates self-limiting in size but gels grow indefinitely? What are the molecular structures that underlie the filamentous features of gels?
Our study also highlights the importance of criticality in biomolecular condensates, which has received only scant attention.21,35,43 We have shown that five of the tetrapeptides can form ADLs, suggesting that this morphology may occur widely for biomolecular condensates. ADLs are characterized by weak intermolecular interactions and low peptide densities, are highly dynamic, and may implicate critical slowing down. We suspect that some in vivo biomolecular condensates may include ADLs in their mix. For example, live-cell superresolution imaging showed that the Mediator coactivator and RNA polymerase II form both large stable clusters and small transient clusters.44 The stable clusters were characterized as transcriptional condensates. The material state of the transient clusters remains unknown, but perhaps they bear some resemblance to ADLs, e.g., by their smaller sizes in fluorescence images (to be compared with the first two images in the middle row of our Figure S2) and their dynamic nature (to be compared with our Video S3). More generally, ADLs represent a condensed phase distinct from liquid droplets for cellular functions. Last, the fact that fusion speeds exhibit critical scaling behavior means that environmental factors such as pH may sensitively tune material properties.
In short, the present study of tetrapeptide condensates breaks new ground in three directions. First, we quantitatively measure the phase equilibrium and material properties for a series of peptides and provide physical explanations for these results. In particular, we show that the threshold concentration for condensate formation is explained by the strength of side-chain interactions. Second, we integrate all-atom MD simulations with the experimental observations to reach a deep and quantitative understanding. Third, we characterize the critical behaviors of peptide condensates. These include ADL and critical scaling.
EXPERIMENTAL PROCEDURES
Synthesis, purification, and characterization
The procedure for the tetrapeptides with X = F, M, L, I, V, and W followed Abbas et al.18; modifications were introduced for X = A and P. Details are described in the supplemental experimental procedures, including Figures S16–S31 for 1H and 13C NMR and mass spectra. The procedure for the dye molecule, 2-[4-(diethylamino) styryl]-3,3-dimethyl-1-octadecyl-3H-indol-1-ium iodide, was as described by Wang et al.45 Details are found in the supplemental experimental procedures, including Figure S32 for 1H and 13C NMR spectra.
Preparation of tetrapeptide condensate samples
The stock solution of a known concentration was prepared by weighing the white powder of a peptide on an analytical balance and dissolving it in Milli-Q water or 50 mM imidazole buffer. A few drops of 37% (w/w) HCl were added to lower the initial pH to 2. The stock solution was filtered with a Millex polyethersulfone syringe filter (0.22-μm pore size; Millipore Sigma cat. no. SLGPR33RS) and stored at room temperature. An aliquot was diluted to the desired concentration (typically 20 μL final volume) with either Milli-Q water or the buffer; condensate formation was induced by raising pH with the addition of a small amount of 5 M NaOH.
Bright-field microscopy
A 2-μL drop was placed on a glass slide and observed under an Olympus BX53 bright-field microscope using a 40× objective. Images were exported in .tiff format and processed using ImageJ. For studying the effects of urea, condensate samples were prepared with urea added before raising the pH.
Negative-staining electron microscopy
A 20-μL drop of tetrapeptide sample, a 20-μL drop of 1% phosphotungstic acid (Electron Microscopy Sciences SKU 19502–1), and three 20-μL drops of deionized water were sequentially placed on a piece of parafilm. Using forceps, a Formvar-coated grid (Ted Pella, product no. 01753-F) was positioned onto the sample drop for 2 min. Excess liquid was removed by gently touching the edge of the forceps with the ragged edge of a piece of filter paper. The grid was transferred to the phosphotungstic acid drop for 2 min and then rinsed in the deionized water drops for 10 s each. Surplus liquid was again removed by filter paper. The grid was left to dry for 10 min in a covered Petri dish. EM images were acquired on a JEM-1400 Flash transmission electron microscope (JEOL) equipped with a BIOSPRINT 12M-B camera (AMT) operating at 80 kV.
Confocal microscopy
All other experiments were conducted at room temperature on a LUMICKS C-Trap instrument, which includes a bright-field camera, a confocal fluorescence imaging module, and dual-trap OT. Confocal scanning over a field of view was carried out with excitation wavelength at 532 nm (20% laser power; 100 nm pixel size; 0.0128 or 0.1 ms pixel dwell time). A bright-field image of the scanned area was also taken for comparison.
For experiments, a square region inside condensates was bleached for 5 or 10 s with 100% laser power. Fluorescence recovery was then monitored with 20% laser power and 0.05 ms pixel dwell time. The scanned area was . After shifting and normalization by the pre- and post-bleaching fluorescence intensities, the recovery curve was fit to
| (Equation 8) |
where denotes the mobile fraction and denotes the recovery time. When spontaneous photobleaching occurred, the decrease in fluorescence in a region away from the laser-bleached region was fit to a linear function of time and FRAP was further normalized by this function to correct for spontaneous photobleaching.
OT-directed droplet fusion
Fusion speed was measured according to Ghosh and Zhou.24 Two droplets of equal size were trapped using 2%–10% overall power and approximately 50:50 split. The trap 1 droplet was moved toward the trap 2 droplet in 10-nm steps until the droplets came into contact. The trapping forces were recorded at a sampling rate of 78,125 Hz. The entire process was recorded by the bright-field camera; the video was analyzed using ImageJ to determine the radii of the droplets before and after fusion. The trap 2 force trace was fit to
| (Equation 9) |
where denotes the maximum force and is the fusion time.
Interfacial tension measurement by droplet stretching
This measurement followed Zhou36 and Ghosh et al.25 Droplet samples containing uncoated polystyrene beads (1.13-μm radius) were prepared. Two droplets, each containing a single bead, were trapped and fused to generate a configuration where a single droplet was suspended by two trapped beads at the opposite poles. The overall trapping power was 10%–40% and split 50:50 between the two traps. While trap 2 was fixed, trap 1 was pulled at a low speed of 0.05 μm/s for a distance of 0.5 μm. The pulling process was recorded by the bright-field camera; the video was analyzed using ImageJ to obtain the radius of the unstretched droplet and to verify that droplet stretching proceeded without interference. The spring constant of the droplet-trap system was obtained from the ratios of the trapping forces over the extension of the droplet. Using the stiffnesses of the two traps to remove their contributions, the spring constant, , of the droplet was calculated. Last, the interfacial tension of the droplet was found as
| (Equation 10) |
where is the radius of the unstretched droplet and is the radius of the beads.
Condensate viscosity measurement by bead tracking
This measurement followed Kota and Zhou.37 Large droplets (>5 μm in radius), each containing a single bead (1.13-μm radius) and settled on a coverslip coated with polyethylene glycol,46 were used for bead tracking. The bead was trapped and moved to the center of the droplet and then released. After 5–10 min, a bright-field image of the bead was taken to serve as the template, and the bead movement was then tracked by template matching at a 15-Hz frame rate. The time trace of the bead’s and positions was exported. The was calculated and fit to
| (Equation 11) |
where denotes the two-dimensional diffusion constant of the bead and is a constant that arises from the uncertainty in locating the bead position.47 The zero-shear viscosity was then obtained according to the Stokes-Einstein relation:
| (Equation 12) |
where is the Boltzmann constant, is the absolute temperature, and again is the bead radius.
Molecular dynamics simulations
The tetrapeptides, denoted as XXssXX, consist of two dipeptides linked by NH-CH2-CH2-S-S-CH2-CH2-NH (Figure S1). We generated force-field parameters for the linker using Amber tools48 and Gaussian 16.49 More specifically, we parameterized one-half of the linker and connected the two halves by a disulfide bond. The half-linker patched with terminal hydrogens, NH2-CH2-CH2-SH, was generated using Pymol (https://pymol.org); its initial coordinate file for parametrization was prepared using Antechamber. Geometry optimization and partial charge calculation were performed at the HF/6–31G* level in Gaussian 16. Other parameters were from the Parm10 parameter file. prepgen was used to prepare the half-linker as an amino acid-like residue and connect it to a dipeptide to generate the reduced form, XXs-H, of the tetrapeptide. Any missing parameters were completed by parmchk. Last, a disulfide bond between two copies of the reduced form was imposed in tleap to form a tetrapeptide. The parameters for the link residues were appended to Amberff14SB50 to form the force field of the tetrapeptides; parameters for neutral terminal residues were modified from the charged versions. The water model was TIP4PD.51
Preparation of a dense solution of the tetrapeptides and subsequent simulations were done in AMBER1852 as described previously.38 Energy minimization (2,000 steps of steepest descent and 3,000 steps of conjugate gradient) was carried out using sander. All MD simulations were carried out on GPUs using pmemd.-cuda.53 Long-range electrostatic interactions were treated using the particle mesh Ewald method54 with a nonbonded cutoff of 10 Å. The Langevin thermostat with a damping constant of 3 ps−1 was used to maintain temperature, while the Berendsen barostat55 was used to regulate the pressure. All bonds connected with the hydrogen atoms were constrained using the SHAKE algorithm.56
To start, eight copies of the tetrapeptide with both termini neutral (to model high pH) were randomly inserted into a cubic box with a side length of 30 Å and solvated with 500–600 waters. After energy minimization, a short simulation of 100 ps was performed at constant NVT (particle numbers, volume, and temperature) and a time step of 1 fs, with temperature ramping from 0 to 294 K over the first 40 ps and maintained at 294 K for the remaining 60 ps. The eight copies of the peptide in the last snapshot were duplicated in each of three orthogonal directions to build 64 copies in a cubic box with a side length of 60 Å, solvated again with 3,800–5,100 water molecules. To condense the system, different levels of water molecules (1/4, 1/3, 1/2, and 2/3) were randomly removed, which generated four independent systems. These systems were energy minimized again and first simulated for 500 ps at constant NVT (temperature ramped from 0 to 294 K for 40 ps and then maintained at 294 K for 460 ps; 1-fs time step). The simulations then continued in duplicates at constant NPT (pressure at 1 bar and temperature at 294 K) for 3.6–6.8 μs at a time step of 2 fs. The cubic simulation boxes settled to ~54 Å in side length for the systems with 1/4 water removal to ~47 Å with 2/3 water removal.
Slab configurations from the preceding simulations of FFssFF and LLssLL were modified to lower values and simulated in an elongated box . The values of the terminal amines were assumed to 7 and 6, respectively; the lower second models an increased tendency to be neutral once the first terminus is already charged. For pH 7.7, 18 copies of the peptide had one terminal amine changed from neutral to charged; this number increased to 32 copies at 7.3. For pH 6.4, 48 copies had one charged amine and 10 copies had two charged amines. The 64 copies of the peptides were solvated with 18,400–22,000 water molecules and neutralized with Cl− ions. Each system was energy minimized and simulated first at constant NVT (500 ps at 1-fs time step) and then at constant NPT (100 ns at 2-fs time step). Last, the simulations continued at constant NVT for 4.5–4.9 μs each at a 2-fs time step. Snapshots were saved at 100-ps intervals for analysis.
Simulation data analyses
Backbone hydrogen bonds and the number of neighboring or surrounding chains (3.5 or 4 Å cutoff between heavy atoms) were calculated using CPPTRAJ57 on a 1,000-ns portion of the cubic-box simulations. This portion was chosen based on well-formed slabs for peptides that did form slabs. The calculations were done by centering the periodic system on each copy of the peptide and then averaging over all the 64 copies.
For the elongated-box simulations, binodals were calculated using tcl scripts and MSDs were calculated using CPPTRAJ and python scripts. For binodals, the simulation box was divided into 2-Å-thick slices along the axis, and the number of peptide heavy atoms in each slice was counted. The results were averaged over the snapshots in the 1,000- to 4,500-ns portion of the simulations. The heavy-atom count as a function of the coordinate was fit to a hyperbolic tangent function to obtain the densities in the dense and dilute phases. MSDs were calculated over the 3,490- to 4,530-ns portion of the simulations at pH 7.3. For the dilute phase, calculations were done on the unwrapped trajectories of the peptide molecules that stayed in the dilute phase over the considered time period. For the dense phase, after unwrapping, the center of the chains that remained in the slab was removed before calculating MSDs. VMD58 and ChimeraX59 were used for rendering images and making movies.
RESOURCE AVAILABILITY
Lead contact
The lead contact on this article is Huan-Xiang Zhou (e-mail: hzhou43@uic.edu).
Materials availability
Synthesis procedures of peptide and dye molecules reported here are fully described below, including starting materials. Requests for materials will be addressed by the lead contact.
Supplementary Material
Highlights.
Tetrapeptides form a variety of condensates, including amorphous dense liquids
Phase and material properties differ by orders of magnitude among amino acids
Molecular dynamics simulations provide physical explanations of measured properties
Condensates exhibit rich critical behaviors
ACKNOWLEDGMENTS
We thank Divya Kota for assistance with confocal microscopy, Fidha Muhammedkutty for assistance with simulation data analysis, Andy Nguyen for the use of an HPLC instrument, and Jan-Hendrik Spille for discussion. This work was funded in part by National Institutes of Health grant R35 GM118091 (H.-X.Z.).
Footnotes
DECLARATION OF INTERESTS
The authors declare that they have no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.xcrp.2024.102218.
Data and code availability
All data are presented in the main text and supplemental figures. Data analyses used widely available codes. Any questions or clarifications needed to reanalyze the data will be answered by the lead contact on reasonable request.
REFERENCES
- 1.Das S, Lin YH, Vernon RM, Forman-Kay JD, and Chan HS (2020). Comparative Roles of Charge, Pi, and Hydrophobic Interactions in Sequence-Dependent Phase Separation of Intrinsically Disordered Proteins. Proc. Natl. Acad. Sci. USA 117, 28795–28805. 10.1073/pnas.2008122117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou HX, Kota D, Qin S, and Prasad R (2024). Fundamental Aspects of Phase-Separated Biomolecular Condensates. Chem. Rev 124, 8550–8595. 10.1021/acs.chemrev.4c00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340. 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal A, Ali R, Yunus AA, Liu DR, Pappu RV, and Rosen MK (2016). Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Mol. Cell 63, 72–85. 10.1016/J.MOLCEL.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakashima KK, Baaij JF, and Spruijt E (2018). Reversible generation of coacervate droplets in an enzymatic network. Soft Matter 14, 361–367. 10.1039/c7sm01897e. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh A, Mazarakos K, and Zhou HX (2019). Three archetypical classes of macromolecular regulators of protein liquid-liquid phase separation. Proc. Natl. Acad. Sci. USA 116, 19474–19483. 10.1073/pnas.1907849116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher RS, and Elbaum-Garfinkle S (2020). Tunable multiphase dynamics of arginine and lysine liquid condensates. Nat. Commun 11, 4628. 10.1038/s41467-020-18224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alshareedah I, Moosa MM, Pham M, Potoyan DA, and Banerjee PR (2021). Programmable viscoelasticity in protein-RNA condensates with disordered sticker-spacer polypeptides. Nat. Commun 12, 6620. 10.1038/s41467-021-26733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toyama Y, Rangadurai AK, Forman-Kay JD, and Kay LE (2022). Mapping the per-residue surface electrostatic potential of CAPRIN1 along its phase-separation trajectory. Proc. Natl. Acad. Sci. USA 119, e2210492119. 10.1073/pnas.2210492119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farag M, Borcherds WM, Bremer A, Mittag T, and Pappu RV (2023). Phase separation of protein mixtures is driven by the interplay of homotypic and heterotypic interactions. Nat. Commun 14, 5527. 10.1038/s41467-023-41274-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galvanetto N, Ivanović MT, Chowdhury A, Sottini A, Nüesch MF, Nettels D, Best RB, and Schuler B (2023). Extreme dynamics in a biomolecular condensate. Nature 619, 876–883. 10.1038/s41586-023-06329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kota D, Prasad R, and Zhou HX (2024). Adenosine Triphosphate Mediates Phase Separation of Disordered Basic Proteins by Bridging Intermolecular Interaction Networks. J. Am. Chem. Soc 146, 1326–1336. 10.1021/jacs.3c09134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, Farber P, Lin H, and Forman-Kay JD (2018). Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife 7, e31486. 10.7554/ELIFE.31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. (2018). A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 174, 688–699.e16. 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, Grace CR, Soranno A, Pappu RV, and Mittag T (2020). Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699. 10.1126/science.aaw8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bremer A, Farag M, Borcherds WM, Peran I, Martin EW, Pappu RV, and Mittag T (2022). Deciphering how naturally occurring sequence features impact the phase behaviours of disordered prion-like domains. Nat. Chem 14, 196–207. 10.1038/s41557-021-00840-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamps J, Bader V, Winklhofer KF, and Tatzelt J (2024). Liquid-liquid phase separation of the prion protein is regulated by the octarepeat domain independently of histidines and copper. J. Biol. Chem 300, 107310. 10.1016/j.jbc.2024.107310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abbas M, Lipiński WP, Nakashima KK, Huck WTS, and Spruijt E (2021). A Short Peptide Synthon for Liquid–Liquid Phase Separation. Nat. Chem 13, 1046–1054. 10.1038/s41557-021-00788-x. [DOI] [PubMed] [Google Scholar]
- 19.Poudyal M, Patel K, Gadhe L, Sawner AS, Kadu P, Datta D, Mukherjee S, Ray S, Navalkar A, Maiti S, et al. (2023). Intermolecular interactions underlie protein/peptide phase separation irrespective of sequence and structure at crowded milieu. Nat. Commun 14, 6199. 10.1038/s41467-023-41864-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tesei G, and Lindorff-Larsen K (2022). Improved predictions of phase behaviour of intrinsically disordered proteins by tuning the interaction range. Open Res. Eur 2, 94. 10.12688/openreseurope.14967.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maristany MJ, Gonzalez AA, Collepardo-Guevara R, and Joseph JA (2023). Universal predictive scaling laws of phase separation of prion-like low complexity domains. bioRxiv. 10.1101/2023.06.14.543914. [DOI] [Google Scholar]
- 22.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. (2015). A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077. 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 23.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, and Brangwynne CP (2016). Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cellam 165, 1686–1697. 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh A, and Zhou HX (2020). Determinants for Fusion Speed of Biomolecular Droplets. Angew. Chem. Int. Ed 59, 20837–20840. 10.1002/anie.202006711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh A, Kota D, and Zhou HX (2021). Shear relaxation governs fusion dynamics of biomolecular condensates. Nat. Commun 12, 5995. 10.1038/s41467-021-26274-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benayad Z, von Bülow S, Stelzl LS, and Hummer G (2021). Simulation of FUS Protein Condensates with an Adapted Coarse-Grained Model. J. Chem. Theory Comput 17, 525–537. 10.1021/acs.jctc.0c01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sundaravadivelu Devarajan D, Wang J, Szała-Mendyk B, Rekhi S, Nikoubashman A, Kim YC, and Mittal J (2024). Sequence-dependent material properties of biomolecular condensates and their relation to dilute phase conformations. Nat. Commun 15, 1912. 10.1038/s41467-024-46223-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan C, Levin A, Chen W, Xing R, Zou Q, Herling TW, Challa PK, Knowles TPJ, and Yan X (2019). Nucleation and Growth of Amino Acid and Peptide Supramolecular Polymers through Liquid–Liquid Phase Separation. Angew. Chem. Int. Ed 58, 18116–18123. 10.1002/ANIE.201911782. [DOI] [PubMed] [Google Scholar]
- 29.Baruch Leshem A, Sloan-Dennison S, Massarano T, Ben-David S, Graham D, Faulds K, Gottlieb HE, Chill JH, and Lampel A (2023). Biomolecular condensates formed by designer minimalistic peptides. Nat. Commun 14, 421. 10.1038/s41467-023-36060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abbas M, Law JO, Grellscheid SN, Huck WTS, and Spruijt E (2022). Peptide-Based Coacervate-Core Vesicles with Semipermeable Membranes. Adv. Mater 34, 2202913. 10.1002/adma.202202913. [DOI] [PubMed] [Google Scholar]
- 31.Lipiński WP, Zehnder J, Abbas M, Güntert P, Spruijt E, and Wiegand T (2023). Fibrils Emerging from Droplets: Molecular Guiding Principles behind Phase Transitions of a Short Peptide-Based Condensate Studied by Solid-State NMR. Chem. Eur J. 29, e202301159. 10.1002/chem.202301159. [DOI] [PubMed] [Google Scholar]
- 32.Smeets PJM, Finney AR, Habraken WJEM, Nudelman F, Friedrich H, Laven J, De Yoreo JJ, Rodger PM, and Sommerdijk NAJM (2017). A classical view on nonclassical nucleation. Proc. Natl. Acad. Sci. USA 114, E7882–E7890. 10.1073/pnas.1700342114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hohenberg PC, and Halperin BI (1977). Theory of dynamic critical phenomena. Rev. Mod. Phys 49, 435–479. 10.1103/RevModPhys.49.435. [DOI] [Google Scholar]
- 34.Ghosh A, Kota D, and Zhou HX (2023). Determining Thermodynamic and Material Properties of Biomolecular Condensates by Confocal Microscopy and Optical Tweezers. Methods Mol. Biol 2563, 237–260. 10.1007/978-1-0716-2663-4_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazarakos K, and Zhou HX (2021). Macromolecular regulators have matching effects on the phase equilibrium and interfacial tension of biomolecular condensates. Protein Sci 30, 1360–1370. 10.1002/pro.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou HX (2020). Determination of Condensate Material Properties from Droplet Deformation. J. Phys. Chem. B 124, 8372–8379. 10.1021/acs.jpcb.0c06230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kota D, and Zhou HX (2022). Macromolecular Regulation of the Material Properties of Biomolecular Condensates. J. Phys. Chem. Lett 5285–5290. 10.1021/acs.jpclett.2c00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazarakos K, Prasad R, and Zhou HX (2022). SpiDec: Computing binodals and interfacial tension of biomolecular condensates from simulations of spinodal decomposition. Front. Mol. Biosci 9, 1021939. 10.3389/fmolb.2022.1021939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dey S, MacAinsh M, and Zhou HX (2022). Sequence-Dependent Backbone Dynamics of Intrinsically Disordered Proteins. J. Chem. Theory Comput 18, 6310–6323. 10.1021/acs.jctc.2c00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berry H, and Chaté H (2014). Anomalous diffusion due to hindering by mobile obstacles undergoing Brownian motion or Orstein-Ulhenbeck processes. Phys. Rev. E 89, 022708. 10.1103/PhysRevE.89.022708. [DOI] [PubMed] [Google Scholar]
- 41.Grimm M, Jeney S, and Franosch T (2011). Brownian motion in a Maxwell fluid. Soft Matter 7, 2076–2084. 10.1039/C0SM00636J. [DOI] [Google Scholar]
- 42.Shen Z, Jia B, Xu Y, Wessén J, Pal T, Chan HS, Du S, and Zhang M (2023). Biological Condensates Form Percolated Networks with Molecular Motion Properties Distinctly Different from Dilute Solutions. Elife 12, e81907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pyo AGT, Zhang Y, and Wingreen NS (2023). Proximity to criticality predicts surface properties of biomolecular condensates. Proc. Natl. Acad. Sci. USA 120, e2220014120. 10.1073/pnas.2220014120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, and Cisse II (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415. 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y-N, Xu B, Qiu L-H, Sun R, Xu Y-J, and Ge J-F (2021). Viscosity sensitive fluorescent dyes with excellent photostability based on hemicyanine dyes for targeting cell membrane. Sens Actuators B Chem 337, 129787. 10.1016/j.snb.2021.129787. [DOI] [Google Scholar]
- 46.Zhang Z, Park S, Pertsinidis A, and Revyakin A (2016). Cloud-point PEG Glass Surfaces for Imaging of Immobilized Single Molecules by Total-internal-reflection Microscopy. Bio-Protocol 6, e1784. 10.21769/BioProtoc.1784. [DOI] [Google Scholar]
- 47.Michalet X (2010). Mean square displacement analysis of single-particle trajectories with localization error: Brownian motion in an isotropic medium. Phys. Rev. E 82, 041914. 10.1103/PhysRevE.82.041914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Case DA, Aktulga HM, Belfon K, Cerutti DS, Cisneros GA, Cruzeiro VWD, Forouzesh N, Giese TJ, Götz AW, Gohlke H, et al. (2023). AmberTools. J. Chem. Inf. Model. 63, 6183–6191. 10.1021/acs.jcim.3c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Petersson GA, Nakatsuji H, et al. (2016). Gaussian 16 Rev. C.01 (Gaussian, Inc; ). https://gaussian.com/gaussian16/. [Google Scholar]
- 50.Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, and Simmerling C (2015). ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput 11, 3696–3713. 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piana S, Donchev AG, Robustelli P, and Shaw DE (2015). Water Dispersion Interactions Strongly Influence Simulated Structural Properties of Disordered Protein States. J. Phys. Chem. B 119, 5113–5123. 10.1021/jp508971m. [DOI] [PubMed] [Google Scholar]
- 52.Case DA, Ben-Shalom IY, Brozell SR, Cerutti DS, Cheatham TE I.I.I., Cruzeiro VWD, Darden TA, Duke RE, Ghoreishi D, Gilson MK, et al. (2018). AMBER 2018. [Google Scholar]
- 53.Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, and Walker RC (2013). Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput 9, 3878–3888. 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 54.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, and Pedersen LG (1995). A smooth particle mesh Ewald method. J. Chem. Phys 103, 8577–8593. 10.1063/1.470117. [DOI] [Google Scholar]
- 55.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, and Haak JR (1984). Molecular-Dynamics with Coupling to an External Bath. J. Chem. Phys 81, 3684–3690. 10.1063/1.448118. [DOI] [Google Scholar]
- 56.Ryckaert J-P, Ciccotti G, and Berendsen HJ (1977). Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys 23, 327–341. 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 57.Roe DR, and Cheatham TE (2013). PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput 9, 3084–3095. 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 58.Humphrey W, Dalke A, and Schulten K (1996). VMD: visual molecular dynamics. J. Mol. Graph 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 59.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are presented in the main text and supplemental figures. Data analyses used widely available codes. Any questions or clarifications needed to reanalyze the data will be answered by the lead contact on reasonable request.


