Abstract
Current coronavirus disease 2019 (COVID-19) vaccines provide robust protection against severe disease but minimal protection against acquisition of infection. Intramuscularly administered COVID-19 vaccines induce robust serum neutralizing antibodies (NAbs), but their ability to boost mucosal immune responses remains to be determined. In this study, we show that the XBB.1.5 mRNA boosters result in increased serum neutralization to multiple severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants in humans, including the dominant circulating variant, JN.1. In contrast, we found that the XBB.1.5 mRNA booster did not augment mucosal NAbs or mucosal IgA responses, although acute SARS-CoV-2 XBB infection substantially increased mucosal antibody responses. These data demonstrate that current XBB.1.5 mRNA boosters substantially enhance peripheral antibody responses but do not robustly increase mucosal antibody responses. Our data highlight a separation between the peripheral and mucosal immune systems in humans and emphasize the importance of developing next-generation vaccines to augment mucosal immunity to protect against respiratory virus infections.
One sentence summary:
COVID-19 mRNA booster vaccines induce limited mucosal immunity.
Introduction
Current vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) provide robust protection against severe disease but minimal protection against acquisition of infection. One hypothesis is that intramuscular administration of messenger RNA (mRNA) vaccines may not induce robust mucosal immunity. In fall 2023, the US Food and Drug Administration authorized the use of mRNA vaccines encoding the XBB.1.5 spike protein. Recent reports indicate the ability of XBB.1.5 mRNA boosters to induce robust serum neutralizing antibody (NAb) responses against XBB.1.5 and other circulating variants (1–5), including the dominant variant JN.1 (2, 5) and the highly mutated variant BA.2.87.1 (5). However, the ability of the XBB.1.5 mRNA boosters to increase mucosal antibody responses remains unclear.
The respiratory mucosa represents the portal of entry for SARS-CoV-2, and thus mucosal antibodies and tissue-resident memory B and T cells may be required to prevent infection (6, 7). Previous reports suggest that SARS-CoV-2 infection leads to enhanced mucosal immunoglobulin (Ig)A and IgG responses (8–10). Mucosal IgA is produced as dimeric secretory IgA (sIgA) with a J-chain, the ligand for IgA transport by polymeric immunoglobulin receptor (IgR) in the mucosa (11); sIgA efficiently neutralizes virus in the upper and lower airways (12). In contrast, monomeric serum IgA lacks the J-chain and cannot transudate to the mucosa, although serum IgG can traverse mucosal barriers to a limited extent utilizing the neonatal Fc receptor (FcRn) (13–15). Although systemic B and T cell responses induced by coronavirus disease 2019 (COVID-19) vaccines have been extensively studied (16–21), mucosal immune responses induced by these vaccines have received less attention (22–25). In this study, we aimed to assess whether the current XBB.1.5 mRNA boosters delivered by the intramuscular route increased peripheral and mucosal immune responses in humans. Our findings demonstrate that the XBB.1.5 mRNA boosters robustly increased peripheral NAb and IgG antibody responses but did not substantially augment mucosal NAb or IgA responses in the nasal mucosa.
Results
Peripheral, but not mucosal, NAb responses are increased following XBB.1.5 mRNA boosting
Following the XBB.1.5 surge in 2023, subvariants emerged including EG.5.1, FL.1.5.1, HV.1, and HK.3 (Fig. 1). The highly mutated variant BA.2.86 (26) also emerged from the BA.2 lineage, and its descendant JN.1 has become the globally dominant variant (Fig. 1). To evaluate peripheral and mucosal antibody responses following XBB.1.5 mRNA boosting, we assessed NAb responses in serum and nasal swab eluates using a luciferase-based pseudovirus NAb assay. Our clinical cohort included 58 participants who did or did not receive the XBB.1.5 mRNA booster (Table 1), and we performed NAb assays at baseline prior to boosting and at week 3 following boosting. Participants had a median of four COVID-19 vaccines and approximately 80% of participants had a history of prior COVID-19 infection, although this might be an underestimate due to the high frequency of asymptomatic or minimally symptomatic infections.
Figure 1. Key spike protein mutations in the Omicron variant, XBB subvariants, and the currently circulating JN.1 SARS-CoV-2 variant.
Spike protein mutations in JN.1 and other SARS-CoV-2 variants are shown. Substitutions in the BA.1, BA.2, BA.5, XBB.1.5, EG.5.1, FL.1.5.1, HV.1, HK.3, BA.2.86, and JN.1 SARS-CoV-2 variants relative to the Wuhan/WIV04/ reference strain (https://gisaid.org/WIV04/) are indicated. Amino acid substitutions are indicated in red tiles, and deletions in blue tiles. NTD, N-terminal domain; RBD, receptor binding domain.
Table 1.
Study population. BNT, BNT162b2 Pfizer COVID-19 mRNA vaccine; 1273, mRNA-1273 Moderna COVID-19 mRNA vaccine; Ad26, Ad26.COV2.S Janssen COVID-19 viral-vector vaccine; NVX, Novavax COVID-19 vaccine (protein adjuvanted). Data are displayed as median (range or interquartile range, IQR) and n (%); BMI, body mass index; pregnant designation reflects time of last vaccine dose or time of sampling. All individuals with known prior infection had mild disease.
| XBB.1.5 Booster N = 31 | No XBB.1.5 Booster N=27 | |
|---|---|---|
| Age (years), median (range) | 48 (26–73) | 37 (22–73) |
|
| ||
| Sex at birth, Female | 24 (77) | 22 (81) |
|
| ||
| Race | ||
| White | 29 (94) | 23 (85) |
| Asian | 2 (6) | 3 (11) |
| Black | 0 | 1 (4) |
| Multiple Races | 1 (3) | 0 |
|
| ||
| Ethnicity | ||
| Non-Hispanic or Latino | 30 (97) | 23 (85) |
| Hispanic or Latino | 1 (3) | 4 (15) |
|
| ||
| Medical condition | ||
| Diabetes | 0 | 1 (4) |
| Obesity (BMI ≥30 kg/m2) | 2 (6) | 3 (11) |
| Hypertension | 1 (3) | 2 (7) |
| Asthma | 1 (3) | 3 (11) |
| Pregnant | 1 (3) | 3 (11) |
| Lactating | 1 (3) | 1 (4) |
|
| ||
| Monovalent XBB.1.5 vaccine | ||
| BNT XBB.1.5 monovalent booster | 13 (42) | N/A |
| 1273 XBB.1.5 monovalent booster | 18 (58) | N/A |
|
| ||
| Prior COVID-19 vaccine history | ||
| BNT (4 doses) | 1 (3) | 0 |
| BNT (4 doses)/1273 BA.5 (1 dose) | 1 (3) | 0 |
| BNT (3 doses) | 0 | 3 (11) |
| BNT (3 doses)/BNT BA.5 (1 dose) | 3 (10) | 2 (7) |
| BNT (3 doses)/1273 BA.5 (1 dose) | 3 (10) | 0 |
| BNT (3 doses)/1273 (1 dose)/BNT BA.5 (1 dose) | 1 (3) | 0 |
| BNT (2 dose) | 0 | 1 (4) |
| BNT (1 dose) | 0 | 1 (4) |
| BNT (2 doses)/BNT BA.5 (1 dose) | 0 | 1 (4) |
| BNT (2 doses)/1273 (1 dose)/1273 BA.5 (1 dose) | 0 | 1 (4) |
| BNT (2 doses)/1273 (1 dose)/BNT BA.5 (1 dose) | 1 (3) | 0 |
| BNT (2 doses)/1273 (2 doses)/BNT BA.5 (1 dose) | 1 (3) | 0 |
| BNT (2 doses)/1273 (2 doses)/1273 BA.5 (1 dose) | 1 (3) | 1 (4) |
| BNT (2 doses)/Ad26 (1 dose) | 0 | 1 (4) |
| BNT (2 doses)/Ad26 (1 dose)/BNT (1 dose) | 1 (3) | 0 |
| BNT (2 doses)/Ad26 (1 dose)/BNT BA.5 (1 dose) | 2 (6) | 1 (4) |
| BNT (2 doses)/Ad26 (1 dose)/1273 BA.5 (1 dose) | 0 | 1 (4) |
| BNT (2 doses)/Ad26 (1 dose)/1273 (1 dose)/BNT BA.5 (1 dose) | 2 (6) | 0 |
| BNT (2 doses)/Ad26 (1 dose)/BNT (1 dose)/BNT BA.5 (1 dose) | 1 (3) | 0 |
| 1273 (4 doses)/1273 BA.5 (1 dose) | 3 (10) | 2 (7) |
| 1273 (4 doses)/BNT BA.5 (1 dose) | 1 (3) | 0 |
| 1273 (3 doses) | 0 | 5 (19) |
| 1273 (3 doses)/1273 BA.5 (1 dose) | 4 (13) | 2 (7) |
| 1273 (3 doses)/BNT BA.5 (1 dose) | 0 | 1 (4) |
| 1273 (2 doses) | 0 | 1 (4) |
| 1273 (2 doses)/BNT (1 dose)/1273 BA.5 (1 dose) | 0 | 1 (4) |
| Ad26 (2 doses)/BNT BA.5 (1 dose) | 1 (3) | 0 |
| Ad26 (1 dose)/1273 (2 doses)/1273 BA.5 (1 dose) | 1 (3) | 0 |
| NVX (3 doses)/BNT BA.5 (1 dose) | 1 (3) | 0 |
| NVX (3 doses)/1273 BA.5 (1 dose) | 1 (3) | 0 |
| NVX (2 doses)/BNT (1 dose)/BNT BA.5 (1 dose) | 0 | 1 (4) |
| NVX (2 doses)/1273 (3 doses)/1273 BA.5 (1 dose) | 0 | 1 (4) |
| NVX (2 doses)/Ad26 (1 dose)/BNT (1 dose)/1273 BA.5 (1 dose) | 1 (3) | 0 |
|
| ||
| Days from prior vaccine to baseline sample | 345 (308–355) | 468 (342–668) |
|
| ||
| Days from XBB.1.5 vaccine to peak sample | 21 (15–33) | N/A |
|
| ||
| Days from baseline to peak sample | 50 (30–57) | 50 (43–57) |
|
| ||
| Known COVID-19 positive test * | 25(80) | 22 (81) |
| 1 prior infection | 20 (65) | 16 (59) |
| 2 prior infections | 5 (16) | 6 (22) |
|
| ||
| Days from most recent positive test to peak sample ** | 477 (395–551) | 507 (435–579) |
Reported for only those with known prior infection.
Only reported for participants who contributed a peak sample.
At baseline, median serum NAb titers to WA1/2020, BA.1, BA.5, XBB.1.5, EG.5.1, FL.1.5.1, HV.1, HK.3, and JN.1 were 3081, 3688, 2386, 114, 131, 50, 40, 66, and 184, respectively, and increased to 5289, 10,971, 9590, 1979, 2808, 1391, 1686, 2108, and 1514, respectively, by week 3 following mRNA boosting (Fig. 2A). This increase represented a 1.2-, 2.6-, 3.2-, 16.2-, 20.2-, 24.7-, 25.4-, 27.2-, and 7.5-fold increment in median NAb titers, respectively (Fig. 2B). Participants who did not receive the XBB.1.5 mRNA boosters showed median serum NAb titers to WA1/2020, BA.1, BA.5, XBB.1.5, EG.5.1, FL.1.5.1, HV.1, HK.3, and JN.1 of 2073, 4028, 2395, 158, 134, 161, 87, 103, and 149 at baseline, respectively, and at the week 3 follow up visit, the titers NAb titers were largely unchanged at 1063, 1885, 2507, 104, 75, 75, 43, 44, 74, respectively (Fig. 2C), representing a 1.2-, 0.6-, 1.3-, 0.8-, 0.7-, 0.6-, 0.7-, 0.8-, and 0.6-fold change in median NAb titers (Fig. 2D). These data demonstrate that XBB.1.5 mRNA boosting substantially increased serum NAb titers to currently circulating variants, including JN.1.
Figure 2. Peripheral and mucosal neutralizing antibody responses following XBB.1.5 mRNA boosting.
(A) Serum neutralizing antibody (NAb) titers after XBB.1.5 mRNA boosting against SARS-CoV-2 WA1/2020, BA.1, BA.5, XBB.1.5, EG.5.1, FL.1.5.1, HV.1, HK.3, and JN.1 variants by luciferase-based pseudovirus neutralization assays at baseline prior to boosting and at week 3 after boosting (n=31). NT50, half-maximal neutralizing titers. Median values are shown at the top. (B) Longitudinal paired analysis of NAb titers are shown. Median fold-change values 3 weeks after XBB.1.5 mRNA vaccination are indicated numerically at the top. (C) Serum NAb titers in participants who did not receive the XBB.1.5 mRNA boosters (n=27) presented as in (A). (D) Paired analysis of NAb titers and their fold-change values during the 3-week follow-up in participants without XBB.1.5 mRNA boosting as in (B). (E and F) Mucosal NAb titers in nasal swabs against SARS-CoV-2 WA1/2020, BA.5, XBB.1.5, HV.1, and JN.1 variants in participants with XBB.1.5 mRNA boosting (E) and without boosting (F) by luciferase-based pseudovirus neutralization assays. Median values are shown at the top. For all panels, the dotted lines reflect the limits of quantitation, and red bars reflect median responses. P values were calculated using a two-tailed Mann-Whitney test; *P≤0.05, **P≤0.01, ****P≤0.0001, and ns = not significant.
We next measured mucosal NAb titers in nasal swab eluates of participants who received the XBB.1.5 mRNA booster. Nasal swab eluates had comparable total Ig concentrations across samples (mean 226 ± 36 μg/ml). Mucosal NAb titers did not detectably increase against WA1/2020, BA.5, XBB.1.5, HV.1, and JN.1 variants following mRNA boosting (Fig. 2E, fig. S1A). We also did not detect an increment in mucosal NAb titers in participants who did not receive an mRNA booster (Fig. 2F, fig. S1B). However, in participants with acute SARS-CoV-2 infection (Table 2), we observed robust nasal NAb responses using these assays (fig. S2).
Table 2.
Characteristics of study participants without XBB.1.5 monovalent booster and recent infection within 90 days. Data displayed as median (range or IQR) and n (%); pregnant designation reflects time of last vaccine dose or time of sampling. All individuals with known prior infection had mild disease.
| No XBB.1.5 monovalent booster with recent infection N=5 |
|
|---|---|
| Age (years), median (range) | 29 (24–38) |
|
| |
| Sex at birth, Female | 3 (60) |
|
| |
| Race | |
| White | 3 (60) |
| Asian | 2 (40) |
| Black | 0 |
| Multiple Races | 0 |
|
| |
| Ethnicity | |
| Non-Hispanic or Latino | 5 (100) |
| Hispanic or Latino | 0 |
|
| |
| Medical condition | |
| Obesity (BMI ≥30 kg/m2) | 1 (20) |
| Hypertension | 0 |
| Asthma | 1 (20) |
| Pregnant | 0 |
| Lactating | 1 (20) |
|
| |
| Most recent vaccine dose | |
| BNT BA.5 bivalent booster | 5 (100) |
|
| |
| Prior COVID-19 vaccine history | |
| BNT (3 doses) | 3 (60) |
| BNT (2 doses)/Ad26 (1 dose)/1273 (1 dose) | 1 (20) |
| 1273 (2 doses)/Ad26 (1 dose) | 1 (20) |
|
| |
| Days from bivalent BA.5 vaccine to baseline sample | 349 (348–366) |
|
| |
| Days from bivalent BA.5 vaccine to peak sample | 405 (398–412) |
|
| |
| Days from baseline sample to peak sample | 63 (50–63) |
|
| |
| Known COVID-19 positive test * | 5 (100) |
| 1 prior infection | 3 (60) |
| 2 prior infections | 2 (40) |
|
| |
| Days from positive test to sampling * | 52 (40–64) |
Reported for only those with known prior infection.
Peripheral, but not mucosal, spike protein-binding antibody responses were enhanced following XBB.1.5 mRNA boosting
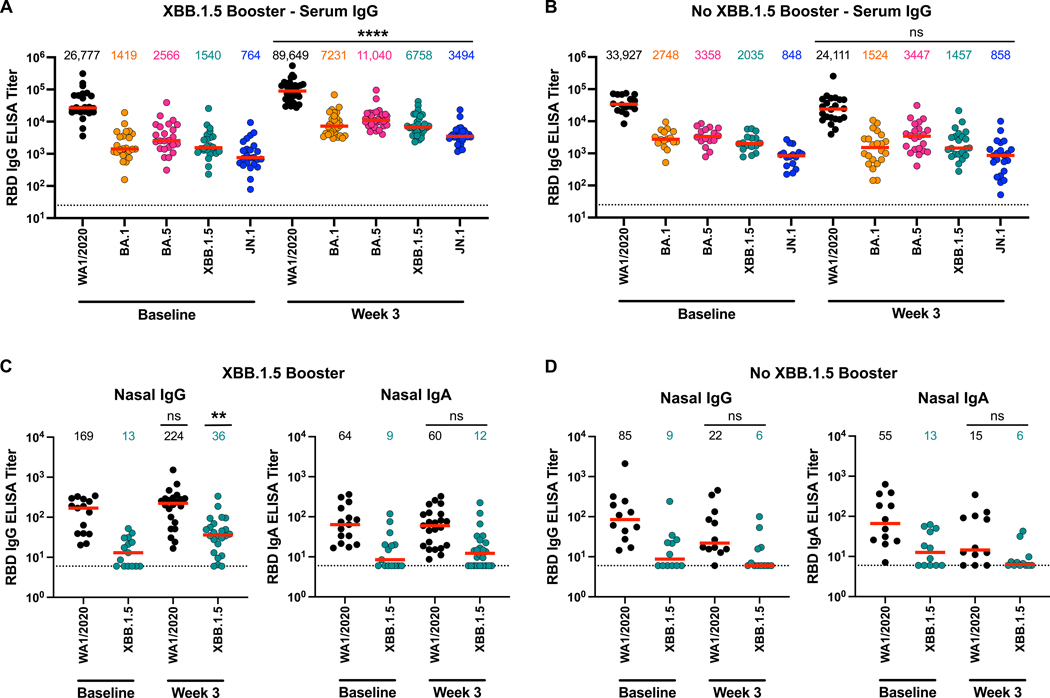
SARS-CoV-2 binding antibody responses were assessed using enzyme-linked immunosorbent assays (ELISAs) and electrochemiluminescence assays (ECLAs). In serum, receptor binding domain (RBD)-specific IgG responses by ELISA had median titers of 26,777, 1419, 2566, 1540, and 764 against WA1/2020, BA.1, BA.5, XBB.1.5, and JN.1, respectively, at baseline before boosting, and increased to 89,649, 7231, 11,040, 6758, and 3494, respectively, following mRNA boosting (Fig. 3A). No such increase was noted in participants without XBB.1.5 mRNA boosting (Fig. 3B). Spike protein-specific serum IgG responses determined by ECLA showed similar trends (fig. S3 and S4). In individuals with acute SARS-CoV-2 infection (Table 2), we observed a modest 2.1- to 2.2-fold increase in serum IgG responses to XBB.1.5 and JN.1 (fig. S5).
Figure 3. Peripheral and mucosal binding antibody responses following XBB.1.5 mRNA boosting.
(A and B) Serum IgG binding antibody titers against the RBD of SARS-CoV-2 WA1/2020, BA.1, BA.5, XBB.1.5, and JN.1 were measured at 3 weeks after XBB.1.5 mRNA boosting (A) or without XBB.1.5 mRNA boosting (B) by ELISA. (C and D) Mucosal IgG and IgA binding antibody titers in nasal swabs against the RBD of SARS-CoV-2 WA1/2020 and XBB.1.5 were measured at 3 weeks after XBB.1.5 mRNA boosting (C) or without XBB.1.5 mRNA boosting (D) by ELISA. The dotted lines reflect the limits of quantitation. Red bars reflect median responses and are shown numerically at the top. P values were calculated using a two-tailed Mann-Whitney test; **P≤0.01, ****P≤0.0001, and ns = not significant.
In nasal swab eluates, RBD-specific mucosal IgG responses showed a modest 1.3- to 2.7-fold increase against WA1/2020 and XBB.1.5, respectively, following mRNA boosting (Fig. 3C). In contrast, RBD- and spike protein-specific mucosal IgA responses did not detectably increase following mRNA boosting (Fig. 3C, fig. S6). In participants who did not receive the XBB.1.5 mRNA boosters, we also did not observe an increase in RBD- or spike protein-specific mucosal IgG and IgA responses (Fig. 3D, fig. S7). In contrast, participants with acute SARS-CoV-2 infection, median mucosal IgG titers increased 9.8- and 8.1-fold against WA1/2020 and XBB.1.5, respectively, and mucosal IgA titers increased 3.0- and 2.4-fold (fig. S8). These data demonstrate that intramuscular XBB.1.5 mRNA boosting led to minimal boosting of mucosal IgG responses and no detectable boosting of mucosal IgA responses, but natural mucosal SARS-CoV-2 infection boosted both mucosal IgG and IgA responses.
Cellular immune responses were not increased following XBB.1.5 mRNA boosting
We next assessed spike protein-specific interferon (IFN)-γ+ CD4+ and CD8+ T cell responses in peripheral blood mononuclear cells (PBMCs) using intracellular cytokine staining assays (ICS). Median spike protein-specific IFN-γ+ CD4+ T cell responses to WA1/2020, BA.5, XBB.1.5, and JN.1 were 0.094%, 0.095%, 0.101%, and 0.086% at baseline before XBB.1.5 mRNA boosting, and 0.078%, 0.070%, 0.064%, and 0.072% at week 3, respectively (Fig. 4A). Median spike protein-specific IFN-γ+ CD8+ T cell responses to WA1/2020, BA.5, XBB.1.5, and JN.1 were 0.047%, 0.029%, 0.031%, and 0.045% at baseline before boost, and 0.036%, 0.016%, 0.025%, and 0.019% at week 3, respectively, indicating no boosting of CD4+ or CD8+ T cell responses (Fig. 4A). Total IFN-γ T cell responses measured by ELISPOT also did not increase after XBB.1.5 mRNA boosting (Fig. 4B). Participants who did not receive the XBB.1.5 mRNA booster similarly did not show an increase in spike protein-specific T cell responses (Fig. 4C and D). These findings indicate that T cell responses showed substantial cross-reactivity across variants, but that XBB.1.5 mRNA boosting did not enhance these T cell responses.
Figure 4. Cellular immune responses following XBB.1.5 mRNA vaccination.
(A) Spike protein-specific CD4+ and CD8+ T cell responses to SARS-CoV-2 WA1/2020, BA.5, XBB.1.5, and JN.1 following XBB.1.5 mRNA vaccination. Samples were stimulated with pooled peptides and IFN-γ production was measured by intracellular cytokine staining (ICS). (B) Spike protein-specific T cell responses to SARS-CoV-2 WA1/2020, BA.5, XBB.1.5, and JN.1 following XBB.1.5 mRNA vaccination were measured by IFN-γ ELISPOT assays after pooled peptide stimulation. Data are presented as as spot-forming cells (SFCs) per million peripheral blood mononuclear cells (PBMCs). (C and D) The ICS assay (C) and ELISPOT assay (D) were also conducted in participants without XBB.1.5 mRNA boosting and are shown as in (A) and (B), respectively. The dotted lines reflect the limits of quantitation. The red bars reflect median responses and are shown numerically at the top of each panel.
Discussion
COVID-19 vaccines have demonstrated robust protection against severe disease, but only limited prevention of acquisition of infection (27); moreover, protection against severe disease wanes over time (27, 28). In this study, we show that the XBB.1.5 mRNA boosters induced robust serum binding and NAb responses against current circulating variants including JN.1, consistent with other recent immunogenicity (1–5) and effectiveness (29) studies. However, the XBB.1.5 mRNA boosters did not induce robust mucosal NAb or IgA responses and only induced modest mucosal IgG responses. These data provide an immunologic rationale for the limited protection of current COVID-19 mRNA vaccines against infection and suggest that next generation vaccines should induce improved mucosal immunity.
Prior studies have shown minimal NAb responses in saliva and bronchoalveolar lavage (BAL) in vaccinated and convalescent individuals (9, 30). Moreover, previous reports have shown varying and limited nasal IgG (31, 32) and IgA (33, 34) responses and salivary IgG and IgA (35) responses following mRNA vaccination. We speculate that it may be easier to detect low titers of mucosal IgA in saliva than in nasal swabs given the larger sample volume. Our data extend these prior studies by showing a discordance between peripheral and mucosal antibody responses following XBB.1.5 mRNA boosting in a current population with extensive prior immunity from both vaccination and infection. We observed a modest increase in nasal IgG responses following XBB.1.5 mRNA boosting, which may reflect limited transudation of serum IgG to mucosal surfaces through FcRn (13–15), a mechanism that does not apply to IgA. Since intramuscular boosting with mRNA vaccines did not enhance mucosal NAb and IgA responses, alternative immunization strategies may be required.
Five participants in our cohort had acute SARS-CoV-2 infection at baseline and developed increased serum antibody responses (36) as well as augmented mucosal IgG and IgA responses. These data suggest that infection robustly boosts mucosal immunity. These findings are consistent with previous studies of SARS-CoV-2 infection in humans (37–40) as well as our recent study in nonhuman primates showing that mucosal boosting led to improved mucosal immunity and enhanced protection against SARS-CoV-2 challenge (41).
T cell responses are highly cross-reactive against multiple variants and likely contribute substantially to population-level immunity and protection against severe disease (42–45). However, XBB.1.5 mRNA boosting did not boost CD4+ or CD8+ T cell responses, similar to previous findings with the bivalent mRNA boosters (28). A prior study also showed that SARS-CoV-2 infection can lead to diminished CD8+ T cell responses after mRNA boosting (46). We do not exclude the possibility that the XBB.1.5 mRNA boosting may lead to modest T cell responses that are lower than the detection limit of our assays in this highly immune population.
Our study has several limitations. First, our cohort is relatively small and most participants had hybrid immunity at baseline, which may yield different results than a naïve population. In addition, our cohort involves more females than males, and thus additional studies are warranted to assess the generalizability of our findings. Finally, we were unable to measure mucosal T cell responses, since nasal swabs did not yield sufficient lymphocytes for cellular immune assays.
In summary, our data demonstrate that intramuscular XBB.1.5 mRNA boosting robustly increased serum NAb and IgG responses, but did not increase mucosal NAb and IgA responses by our assays. These findings provide an immunologic rationale for why mRNA vaccines provide robust protection against severe disease but minimal protection against infection. Moreover, the discordance between systemic and mucosal antibody responses suggests a near complete separation between the peripheral and mucosal immune systems in humans. Next generation vaccines for COVID-19 and other respiratory pathogens should focus on improving induction of mucosal immunity (47).
MATERIALS AND METHODS
Study design
We conducted an exploratory cohort study evaluating peripheral and mucosal immune responses of individuals who did or did not receive the XBB.1.5 mRNA boosters in fall 2023. During each visit, participants provided their vaccine, symptom, and testing history, as well as their race and ethnicity based on specified categories; they could select multiple race categories. The specimen biorepository at Beth Israel Deaconess Medical Center (BIDMC) obtained peripheral blood and nasal swabs from the study participants. The BIDMC institutional review board approved this study (2020P000361). All participants provided informed consent. This study included 63 individuals who did or did not receive the XBB.1.5 mRNA boosters (Pfizer-BioNTech, 30 μg; Moderna, 50 μg). Participants were excluded from the immunologic assays if they had a recent history of SARS-CoV-2 infection within 2 weeks of enrollment or if they received immunosuppressive medications. Assay operators were all blinded to participant prior infection or vaccination history.
Pseudovirus Neutralizing Antibody Assay
Neutralizing antibody (NAb) titers against SARS-CoV-2 variants were determined using pseudotyped viruses expressing a luciferase reporter gene. In brief, a luciferase reporter plasmid pLenti-CMV Puro-Luc (Addgene), packaging construct psPAX2 (AIDS Resource and Reagent Program), and spike protein-expressing pcDNA3.1-SARS-CoV-2 SΔCT were co-transfected into human embryonic kidney (HEK)293T cells (ATCC CRL_3216) with lipofectamine 2000 (Thermo Fisher Scientific). Pseudotyped viruses of SARS-CoV-2 variants were generated using the spike protein from WA1/2020 (Wuhan/WIV04/2019, GISAID accession ID: EPI_ISL_402124), Omicron BA.1 (GISAID ID: EPI_ISL_7358094.2), BA.5 (GISAID ID: EPI_ISL_12268495.2), XBB.1.5 (GISAID ID: EPI_ISL_16418320), EG.5.1 (GISAID ID: EPI_ISL_18125149), FL.1.5.1 (GISAID ID: EPI_ISL_18126515), HV.1 (GISAID ID: EPI_ISL_18592608), HK.3 (GISAID ID: EPI_ISL_18631954), and JN.1 (GISAID ID: EPI_ISL_18680594). 48 hours post-transfection, the supernatants containing the pseudotyped viruses were collected and purified by filtration with 0.45-μm filter. To determine NAb titers in serum and nasal swab eluate, HEK293T-hACE2 cells were seeded in 96-well tissue culture plates at a density of 2 × 104 cells per well overnight. Three-fold serial dilutions of heat-inactivated serum or nasal swab eluate samples were prepared and mixed with 60 μl of pseudovirus, and incubated at 37 °C for 1 hour before adding to HEK293T-hACE2 cells. 48 hours later, cells were lysed in Steady-Glo Luciferase Assay (Promega) according to the manufacturer’s instructions. SARS-CoV-2 neutralization titers were defined as the sample dilution at which a 50% reduction (NT50) in relative light units was observed relative to the average of the virus control wells. The lower limit of detection (LLOQ) was 20 for serum NAb titers and was 4 for mucosal NAb titers.
Electrochemiluminescence assay (ECLA)
ECLA plates [MesoScale Discovery (MSD); SARS-CoV-2 IgG catalog no. K15721U (panel 37)] were designed and produced with up to nine antigen spots in each well, and assays were performed. The antigens included spike proteins from WA1/2020, BA.5, XBB.1.5, XBB.1.16, XBB.1.16.6, XBB.2.3, EG.5.1, FL.1.5.1, and JN.1. The plates were blocked with 50 μl of blocker A (1% bovine serum albumin in distilled water) solution for at least 30 min at room temperature with shaking at 700 rpm with a digital microplate shaker. During blocking, serum samples were diluted to 1:5000 or nasal swab eluate samples were diluted to 1:200 in Diluent 100. The calibrator curve was prepared by diluting the calibrator mixture from MSD 1:9 in Diluent 100 and then preparing a seven-step fourfold dilution series and a blank containing only Diluent 100. The plates were then washed three times with 150 μl of wash buffer [0.5% Tween 20 in 1× phosphate-buffered saline (PBS)] and blotted dry. 50 μl of the diluted samples and calibration curve was added in duplicate to the plates and set to shake at 700 rpm at room temperature for at least 2 hours. The plates were again washed three times, and 50 μl of SULFO-TAG anti-human IgG or IgA detection antibody diluted to 1× in Diluent 100 was added to each well. Samples were incubated with shaking at 700 rpm at room temperature for at least 1 hour. Plates were then washed three times, and 150 μl of the MSD GOLD Read Buffer B was added to each well. The plates were read immediately after using a MESO QuickPlex SQ 120 machine. MSD titers for each sample were reported as relative light units, which were calculated using the calibrator. The lower limit of quantitation (LLOQ) was 100 relative light units (RLU) for serum IgG and was 40 RLU for mucosal IgG and IgA.
Enzyme-linked immunosorbent assay (ELISA)
SARS-CoV-2 RBD-specific binding antibodies in serum and nasal swab eluate were assessed by ELISA. 96-well plates were coated with 0.5 μg/mL of SARS-CoV-2 WA1/2020, Omicron BA.1, BA.5, XBB.1.5, and JN.1 RBD protein in 1× Dulbecco phosphate-buffered saline (DPBS) and incubated at 4 °C overnight. After incubation, plates were washed once with wash buffer (0.05% Tween 20 in 1× DPBS) and blocked with 350 μL of casein block solution per well for 2 to 3 hours at room temperature. Following incubation, block solution was discarded and plates were blotted dry. Serial dilutions of heat-inactivated serum or nasal swab eluate diluted in Casein block were added to wells, and plates were incubated for 1 hour at room temperature, prior to 3 more washes and a 1-hour incubation with a 1:4000 dilution of antihuman IgG horseradish peroxidase (HRP) (Invitrogen, Thermo Fisher Scientific) or 1:1000 dilution of anti-human IgA HRP (Bethyl Laboratories) at room temperature in the dark. Total Ig was comparable across nasal swab eluates. Plates were washed 3 times, and 100 μL of SeraCare KPL TMB SureBlue Start solution was added to each well; plate development was halted by adding 100 μL of SeraCare KPL TMB Stop solution per well. The absorbance at 450 nm, with a reference at 650 nm, was recorded with a VersaMax microplate reader (Molecular Devices). For each sample, the ELISA end point titer was calculated using a 4-parameter logistic curve fit to calculate the reciprocal serum dilution that yields a corrected absorbance value (450 nm-650 nm) of 0.2. The lower limit of detection (LLOQ) was 25 for serum ELISA titers and was 6 for mucosal ELISA titers.
Intracellular cytokine staining (ICS) assay
CD4+ and CD8+ T cell responses were quantitated by pooled peptide-stimulated ICS assays. Peptide pools contained 15 amino acid peptides overlapping by 11 amino acids spanning the SARS-CoV-2 WA1/2020, BA.5, XBB.1.5, and JN.1 spike proteins (21st Century Biochemicals and GenScript). 106 peripheral blood mononuclear cells were re-suspended in 100 μL of R10 media (500 ml of RPMI-1640 with 55 ml of fetal bovine serum [FBS] and 5.5 ml of 100× penicillin-streptomycin) supplemented with CD49d monoclonal antibody (1 μg/mL) and CD28 monoclonal antibody (1 μg/mL). Each sample was assessed with mock (100 μL of R10 plus 0.5% DMSO; background control), peptides (2 μg/mL), or 10 pg/mL phorbol myristate acetate (PMA) and 1 μg/mL ionomycin (Sigma-Aldrich) (100μL; positive control) and incubated at 37°C for 1 hour. After incubation, 0.25 μL of GolgiStop and 0.25 μL of GolgiPlug in 50 μL of R10 media was added to each well and incubated at 37°C for 8 hours. Samples were then held at 4°C overnight. The next day, the cells were washed twice with DPBS, stained with aqua live/dead dye (Life Technologies, 1:100 dilution) for 10 min and then stained with predetermined titers of monoclonal antibodies against CD279 (clone EH12.1, BB700; 2 μL/test), CD4 (clone L200, BV711; 0.75 μL/test), CD27 (clone M-T271, BUV563; 0.5 μL/test), CD8 (clone SK1, BUV805; 1 μL/test), CD45RA (clone 5H9, APC H7; 2.5 μL/test) for 30 min. Cells were then washed twice with DPBS containing 2% FBS and incubated for 15 min with 200 μL of BD CytoFix/CytoPerm Fixation/Permeabilization solution. Cells were washed twice with 1X Perm Wash buffer (BD Perm/Wash Buffer 10X in the CytoFix/CytoPerm Fixation/ Permeabilization kit diluted with MilliQ water and passed through 0.22μm filter) and stained intracellularly with monoclonal antibodies against Ki67 (clone B56, BB515; 0.25 μL/test), IL-21 (clone 3A3-N2.1, PE; 5 μL/test), CD69 (clone TP1.55.3, ECD; 0.75 μL/test), IL-10 (clone JES3–9D7, PE CY7; 2 μL/test), IL-13 (clone JES10–5A2, BV421; 2.5 μL/test), IL-4 (clone MP4–25D2, BV605; 2.5 μL/test), TNF-α (clone Mab11, BV650; 2.5 μL/test), IL-17 (clone N49–653, BV750; 2 μL/test), IFN-γ (clone B27; BUV395; 2.5 μL/test), IL-2 (clone MQ1–17H12, BUV737; 2.5 μL/test), IL-6 (clone MQ2–13A5, APC; 1.25 μL/test), and CD3 (clone SP34.2, Alexa 700; 0.25 μL/test) for 30 min. Cells were washed twice with 1X Perm Wash buffer and fixed with 250μL of freshly prepared 1.5% formaldehyde. Fixed cells were transferred to 96-well round bottom plate and analyzed by a BD FACSymphony system. Data were analyzed using FlowJo v9.9.
ELISPOT assay
ELISPOT plates were coated with mouse anti-human IFN-γ monoclonal antibody from Mabtech at 1 μg per well and incubated overnight at 4°C. Plates were washed with DPBS and blocked with R10 medium for 2 to 4 hours at 37°C. SARS-CoV-2–pooled spike peptides from WA1/2020, BA.5, XBB.1.5, and JN.1 (21st Century Biochemicals and GenScript) were prepared and plated at a concentration of 2 μg per well, and 200,000 cells per well were added to the plate. The peptides and cells were incubated for 15 to 20 hours at 37°C. All steps after this incubation were performed at room temperature. The plates were washed with ELISPOT wash buffer and incubated for 2 to 4 hours with biotinylated mouse anti-human IFN-γ monoclonal antibody from Mabtech (1 μg/ml). The plates were washed a second time and incubated for 2 to 3 hours with conjugated goat anti-biotin alkaline phosphatase from Rockland Inc. (1.33 μg/ml). The final wash was followed by the addition of Nitro blue tetrazolium chloride/5-bromo-4-chloro 3 indolyphosphate p-toludine salt (NBT/BCIP chromagen) substrate solution for 7 min. The chromagen was discarded, and the plates were washed with water and dried in a dim place for 24 hours. Plates were scanned and automatically counted on a Cellular Technologies Limited Immunospot Analyzer.
SARS-CoV-2 Variant Mutation Graph
Amino acid differences in the spike proteins of BA.1, BA.2, BA.5, XBB.1.5, EG.5.1, FL.1.5.1, HV.1, HK.3, BA.2.86, and JN.1 relative to WA1/2020 were analyzed through a Jupyter Notebook from Jesse Bloom (https://github.com/jbloom/SARS2-clade-spike-diffs). The mutation data were graphed in Adobe Illustrator.
Statistical analysis
All raw, individual-level data are presented in data file S1. Descriptive statistics were calculated using GraphPad Prism v10.2.2. All data points represent biological replicates for each group. Each immunologic assay was performed for each sample independently without technical replicates. Data are presented as median with interquartile range. Statistical comparisons for NAb and ELISA data between pre-boost and post-boost time-points were performed using a two-tailed Mann-Whitney test, and a P value of <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank the study participants for their participation and the BIDMC Clinical Research Center for assistance with this study. We thank M. Lifton and the Center for Virology and Vaccine Research Flow Cytometry core; A. Mutoni, B. Wixted, O. Heard, D. Cabrera-Barragan, A. Waller-Pulido, R. Patio, T. Anand, R. Guan, and L. Barrett from the Center for Virology and Vaccine Research for assisting with assays in this study; and the staff at MesoScale Discovery for providing the ECLA multiplexing kits used in this study.
Funding
The authors acknowledge NIH grant CA260476 and the Massachusetts Consortium for Pathogen Readiness (to D.H.B.).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Data and materials availability
All data associated with this study are present in the paper or the Supplementary Materials. Correspondence and requests for materials should be addressed to D.H.B. (dbarouch@bidmc.harvard.edu).
References
- 1.Stankov MV, Hoffmann M, Gutierrez Jauregui R, Cossmann A, Morillas Ramos G, Graalmann T, Winter EJ, Friedrichsen M, Ravens I, Ilievska T, Ristenpart J, Schimrock A, Willenzon S, Ahrenstorf G, Witte T, Förster R, Kempf A, Pöhlmann S, Hammerschmidt SI, Dopfer-Jablonka A, Behrens GMN, Humoral and cellular immune responses following BNT162b2 XBB.1.5 vaccination. The Lancet Infectious Diseases 24, e1–e3 (2024). [DOI] [PubMed] [Google Scholar]
- 2.Wang Q, Guo Y, Bowen A, Mellis IA, Valdez R, Gherasim C, Gordon A, Liu L, Ho DD, XBB.1.5 monovalent mRNA vaccine booster elicits robust neutralizing antibodies against emerging SARS-CoV-2 variants. bioRxiv, 2023.2011.2026.568730 (2023). [Google Scholar]
- 3.Chalkias S, McGhee N, Whatley JL, Essink B, Brosz A, Tomassini JE, Girard B, Edwards DK, Wu K, Nasir A, Lee D, Avena LE, Feng J, Deng W, Montefiori DC, Baden LR, Miller JM, Das R, Interim report of the reactogenicity and immunogenicity of SARS-CoV-2 XBB-containing vaccines. J Infect Dis, (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gayed J, Diya O, Lowry FS, Xu X, Bangad V, Mensa F, Zou J, Xie X, Hu Y, Lu C, Cutler M, Belanger T, Cooper D, Koury K, Anderson AS, Türeci Ö, Şahin U, Swanson KA, Modjarrad K, Gurtman A, Kitchin N, Safety and Immunogenicity of the Monovalent Omicron XBB.1.5-Adapted BNT162b2 COVID-19 Vaccine in Individuals ≥12 Years Old: A Phase 2/3 Trial. Vaccines (Basel) 12, (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lasrado N, Rössler A, Rowe M, Collier A.-r. Y., Barouch DH, Neutralization of SARS-CoV-2 Omicron subvariant BA.2.87.1. Vaccine, (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng MZM, Wakim LM, Tissue resident memory T cells in the respiratory tract. Mucosal Immunol 15, 379–388 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith N, Goncalves P, Charbit B, Grzelak L, Beretta M, Planchais C, Bruel T, Rouilly V, Bondet V, Hadjadj J, Yatim N, Pere H, Merkling SH, Ghozlane A, Kernéis S, Rieux-Laucat F, Terrier B, Schwartz O, Mouquet H, Duffy D, Di Santo JP, Distinct systemic and mucosal immune responses during acute SARS-CoV-2 infection. Nature Immunology 22, 1428–1439 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sterlin D, Mathian A, Miyara M, Mohr A, Anna F, Claër L, Quentric P, Fadlallah J, Devilliers H, Ghillani P, Gunn C, Hockett R, Mudumba S, Guihot A, Luyt C-E, Mayaux J, Beurton A, Fourati S, Bruel T, Schwartz O, Lacorte J-M, Yssel H, Parizot C, Dorgham K, Charneau P, Amoura Z, Gorochov G, IgA dominates the early neutralizing antibody response to SARS-CoV-2. Science Translational Medicine 13, eabd2223 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang J, Zeng C, Cox TM, Li C, Son YM, Cheon IS, Wu Y, Behl S, Taylor JJ, Chakaraborty R, Johnson AJ, Shiavo DN, Utz JP, Reisenauer JS, Midthun DE, Mullon JJ, Edell ES, Alameh MG, Borish L, Teague WG, Kaplan MH, Weissman D, Kern R, Hu H, Vassallo R, Liu S-L, Sun J, Respiratory mucosal immunity against SARS-CoV-2 after mRNA vaccination. Science Immunology 7, eadd4853 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler SE, Crowley AR, Natarajan H, Xu S, Weiner JA, Bobak CA, Mattox DE, Lee J, Wieland-Alter W, Connor RI, Wright PF, Ackerman ME, Distinct Features and Functions of Systemic and Mucosal Humoral Immunity Among SARS-CoV-2 Convalescent Individuals. Frontiers in Immunology 11, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandtzaeg P, Induction of secretory immunity and memory at mucosal surfaces. Vaccine 25, 5467–5484 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Viant C, Gaebler C, Cipolla M, Hoffmann HH, Oliveira TY, Oren DA, Ramos V, Nogueira L, Michailidis E, Robbiani DF, Gazumyan A, Rice CM, Hatziioannou T, Bieniasz PD, Caskey M, Nussenzweig MC, Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci Transl Med 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshida M, Masuda A, Kuo TT, Kobayashi K, Claypool SM, Takagawa T, Kutsumi H, Azuma T, Lencer WI, Blumberg RS, IgG transport across mucosal barriers by neonatal Fc receptor for IgG and mucosal immunity. Springer Semin Immunopathol 28, 397–403 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Spiekermann GM, Finn PW, Ward ES, Dumont J, Dickinson BL, Blumberg RS, Lencer WI, Receptor-mediated immunoglobulin G transport across mucosal barriers in adult life: functional expression of FcRn in the mammalian lung. J Exp Med 196, 303–310 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida M, Claypool SM, Wagner JS, Mizoguchi E, Mizoguchi A, Roopenian DC, Lencer WI, Blumberg RS, Human Neonatal Fc Receptor Mediates Transport of IgG into Luminal Secretions for Delivery of Antigens to Mucosal Dendritic Cells. Immunity 20, 769–783 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Chandrashekar A, Sellers D, Barrett J, Jacob-Dolan C, Lifton M, McMahan K, Sciacca M, VanWyk H, Wu C, Yu J, Collier A.-r. Y., Barouch DH, Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 Omicron. Nature 603, 493–496 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vidal SJ, Collier A.-r. Y., Yu J, McMahan K, Tostanoski LH, Ventura JD, Aid M, Peter L, Jacob-Dolan C, Anioke T, Chang A, Wan H, Aguayo R, Ngo D, Gerszten RE, Seaman MS, Barouch DH, Correlates of Neutralization against SARS-CoV-2 Variants of Concern by Early Pandemic Sera. Journal of Virology 95, e00404–00421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collier A.-r. Y., Yu J, McMahan K, Liu J, Chandrashekar A, Maron JS, Atyeo C, Martinez DR, Ansel JL, Aguayo R, Rowe M, Jacob-Dolan C, Sellers D, Barrett J, Ahmad K, Anioke T, VanWyk H, Gardner S, Powers O, Bondzie EA, Wan H, Baric RS, Alter G, Hacker MR, Barouch DH, Differential Kinetics of Immune Responses Elicited by Covid-19 Vaccines. New England Journal of Medicine 385, 2010–2012 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Painter MM, Mathew D, Goel RR, Apostolidis SA, Pattekar A, Kuthuru O, Baxter AE, Herati RS, Oldridge DA, Gouma S, Hicks P, Dysinger S, Lundgreen KA, Kuri-Cervantes L, Adamski S, Hicks A, Korte S, Giles JR, Weirick ME, McAllister CM, Dougherty J, Long S, D’Andrea K, Hamilton JT, Betts MR, Bates P, Hensley SE, Grifoni A, Weiskopf D, Sette A, Greenplate AR, Wherry EJ, Rapid induction of antigen-specific CD4(+) T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity 54, 2133–2142.e2133 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goel RR, Apostolidis SA, Painter MM, Mathew D, Pattekar A, Kuthuru O, Gouma S, Hicks P, Meng W, Rosenfeld AM, Dysinger S, Lundgreen KA, Kuri-Cervantes L, Adamski S, Hicks A, Korte S, Oldridge DA, Baxter AE, Giles JR, Weirick ME, McAllister CM, Dougherty J, Long S, D’Andrea K, Hamilton JT, Betts MR, Luning Prak ET, Bates P, Hensley SE, Greenplate AR, Wherry EJ, Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci Immunol 6, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A, Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 181, 1489–1501.e1415 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darwich A, Pozzi C, Fornasa G, Lizier M, Azzolini E, Spadoni I, Carli F, Voza A, Desai A, Ferrero C, Germagnoli L, Mantovani A, Rescigno M, Alessio A, Clement A, Salvatore B, Cristina B, Alice B, Sara B, Paola B, Francesca C, Michela C, Assunta C, Arianna C, Claudia C, Sara C, Silvia C, Valentina C, Maurizio C, Michele C, Nicolò C, Abbass D, Lleo De Nalda A, Federica DP, Rachele DD, Elisabeth D, Barbara D, Maria FF, Valentina F, Giulia F, Sara F, Antonio GG, Silvia G, Gomes AR, Michela L, Antonino LC, Alessia M, Alessandro M, Ilaria M, Bianca O, Fabio P, Anna P, Erica P, Chiara P, Chiara P, Valeria R, Monica R, Alice S, Carlo S, Alessandra S, Marina S, Ilaria S, Salvatore S, Gianmarco S, Domenico S, Paolo T, Aldo U, Sonia V, Antonio V, Elisa Z, Veronica Z, BNT162b2 vaccine induces antibody release in saliva: a possible role for mucosal viral protection? EMBO Molecular Medicine 14, e15326 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cagigi A, Yu M, Österberg B, Svensson J, Falck-Jones S, Vangeti S, Åhlberg E, Azizmohammadi L, Warnqvist A, Falck-Jones R, Gubisch PC, Ödemis M, Ghafoor F, Eisele M, Lenart K, Bell M, Johansson N, Albert J, Sälde J, Pettie DD, Murphy MP, Carter L, King NP, Ols S, Normark J, Ahlm C, Forsell MN, Färnert A, Loré K, Smed-Sörensen A, Airway antibodies emerge according to COVID-19 severity and wane rapidly but reappear after SARS-CoV-2 vaccination. JCI Insight 6, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aksyuk AA, Bansal H, Wilkins D, Stanley AM, Sproule S, Maaske J, Sanikommui S, Hartman WR, Sobieszczyk ME, Falsey AR, Kelly EJ, AZD1222-induced nasal antibody responses are shaped by prior SARS-CoV-2 infection and correlate with virologic outcomes in breakthrough infection. Cell Reports Medicine 4, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stolovich-Rain M, Kumari S, Friedman A, Kirillov S, Socol Y, Billan M, Pal RR, Das K, Golding P, Oiknine-Djian E, Sirhan S, Sagie MB, Cohen-Kfir E, Gold N, Fahoum J, Kumar M, Elgrably-Weiss M, Zhou B, Ravins M, Gatt YE, Bhattacharya S, Zelig O, Wiener R, Wolf DG, Elinav H, Strahilevitz J, Padawer D, Baraz L, Rouvinski A, Intramuscular mRNA BNT162b2 vaccine against SARS-CoV-2 induces neutralizing salivary IgA. Frontiers in Immunology 13, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lasrado N, Collier AY, Hachmann NP, Miller J, Rowe M, Schonberg ED, Rodrigues SL, LaPiana A, Patio RC, Anand T, Fisher J, Mazurek CR, Guan R, Wagh K, Theiler J, Korber BT, Barouch DH, Neutralization escape by SARS-CoV-2 Omicron subvariant BA.2.86. Vaccine 41, 6904–6909 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin D-Y, Xu Y, Gu Y, Zeng D, Sunny SK, Moore Z, Durability of Bivalent Boosters against Omicron Subvariants. New England Journal of Medicine 388, 1818–1820 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lasrado N, Collier A.-r. Y., Miller J, Hachmann NP, Liu J, Anand T, Bondzie EA, Fisher JL, Mazurek CR, Patio RC, Rodrigues SL, Rowe M, Surve N, Ty DM, Wu C, Chicz TM, Tong X, Korber B, McNamara RP, Barouch DH, Waning immunity and IgG4 responses following bivalent mRNA boosting. Science Advances 10, eadj9945 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Link-Gelles R CA, Mak J, et al. , Early Estimates of Updated 2023–2024 (Monovalent XBB.1.5) COVID-19 Vaccine Effectiveness Against Symptomatic SARS-CoV-2 Infection Attributable to Co-Circulating Omicron Variants Among Immunocompetent Adults — Increasing Community Access to Testing Program, United States, September 2023–January 2024. MMWR Morb Mortal Wkly Rep 73, 77–83 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winklmeier S, Rübsamen H, Özdemir C, Wratil PR, Lupoli G, Stern M, Schneider C, Eisenhut K, Ho S, Wong HK, Taskin D, Petry M, Weigand M, Eichhorn P, Foesel BU, Mader S, Keppler OT, Kümpfel T, Meinl E, Intramuscular vaccination against SARS-CoV-2 transiently induces neutralizing IgG rather than IgA in the saliva. Frontiers in Immunology 15, (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puhach O, Bellon M, Adea K, Bekliz M, Hosszu-Fellous K, Sattonnet P, Hulo N, Kaiser L, Eckerle I, Meyer B, SARS-CoV-2 convalescence and hybrid immunity elicits mucosal immune responses. eBioMedicine 98, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishizaka A, Koga M, Mizutani T, Uraki R, Yamayoshi S, Iwatsuki-Horimoto K, Yamamoto S, Imai M, Tsutsumi T, Suzuki Y, Kawaoka Y, Yotsuyanagi H, Antibody induction and immune response in nasal cavity by third dose of SARS-CoV-2 mRNA vaccination. Virology Journal 20, 146 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheikh-Mohamed S, Isho B, Chao GYC, Zuo M, Cohen C, Lustig Y, Nahass GR, Salomon-Shulman RE, Blacker G, Fazel-Zarandi M, Rathod B, Colwill K, Jamal A, Li Z, de Launay KQ, Takaoka A, Garnham-Takaoka J, Patel A, Fahim C, Paterson A, Li AX, Haq N, Barati S, Gilbert L, Green K, Mozafarihashjin M, Samaan P, Budylowski P, Siqueira WL, Mubareka S, Ostrowski M, Rini JM, Rojas OL, Weissman IL, Tal MC, McGeer A, Regev-Yochay G, Straus S, Gingras A-C, Gommerman JL, Systemic and mucosal IgA responses are variably induced in response to SARS-CoV-2 mRNA vaccination and are associated with protection against subsequent infection. Mucosal Immunology 15, 799–808 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao KT, Cobos-Uribe C, Knight N, Jonnalagadda R, Robinette C, Jaspers I, Rebuli ME, SARS-CoV-2 mRNA vaccination induces an intranasal mucosal response characterized by neutralizing antibodies. Journal of Allergy and Clinical Immunology: Global 2, 100129 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorochov G, Ropers J, Launay O, Dorgham K, da Mata-Jardin O, Lebbah S, Durier C, Bauer R, Radenne A, Desaint C, Vieillard L-V, Rekacewicz C, Lachatre M, Parfait B, Batteux F, Hupé P, Ninove L, Lefebvre M, Conrad A, Dussol B, Maakaroun-Vermesse Z, Melica G, Nicolas J-F, Verdon R, Kiladjian J-J, Loubet P, Schmidt-Mutter C, Dualé C, Ansart S, Botelho-Nevers E, Lelièvre J-D, de Lamballerie X, Kieny M-P, Tartour E, Paul S, Serum and Salivary IgG and IgA Response After COVID-19 Messenger RNA Vaccination. JAMA Network Open 7, e248051-e248051 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lasrado N, Barouch DH, SARS-CoV-2 Hybrid Immunity: The Best of Both Worlds. J Infect Dis 228, 1311–1313 (2023). [DOI] [PubMed] [Google Scholar]
- 37.Mitsi E, Diniz MO, Reiné J, Collins AM, Robinson RE, Hyder-Wright A, Farrar M, Liatsikos K, Hamilton J, Onyema O, Urban BC, Solórzano C, Belij-Rammerstorfer S, Sheehan E, Lambe T, Draper SJ, Weiskopf D, Sette A, Maini MK, Ferreira DM, Respiratory mucosal immune memory to SARS-CoV-2 after infection and vaccination. Nature Communications 14, 6815 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collier AY, Brown CM, McMahan KA, Yu J, Liu J, Jacob-Dolan C, Chandrashekar A, Tierney D, Ansel JL, Rowe M, Sellers D, Ahmad K, Aguayo R, Anioke T, Gardner S, Siamatu M, Bermudez-Rivera L, Hacker MR, Madoff LC, Barouch DH, Characterization of immune responses in fully vaccinated individuals after breakthrough infection with the SARS-CoV-2 delta variant. Sci Transl Med 14, eabn6150 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mucosal IgA against SARS-CoV-2 Omicron Infection. New England Journal of Medicine 387, e55 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Nantel S, Sheikh-Mohamed S, Chao GYC, Kurtesi A, Hu Q, Wood H, Colwill K, Li Z, Liu Y, Seifried L, Bourdin B, McGeer A, Hardy WR, Rojas OL, Al-Aubodah T-A, Liu Z, Ostrowski MA, Brockman MA, Piccirillo CA, Quach C, Rini JM, Gingras A-C, Decaluwe H, Gommerman JL, Comparison of Omicron breakthrough infection versus monovalent SARS-CoV-2 intramuscular booster reveals differences in mucosal and systemic humoral immunity. Mucosal Immunology, (2024). [DOI] [PubMed] [Google Scholar]
- 41.McMahan K, Wegmann F, Aid M, Sciacca M, Liu J, Hachmann NP, Miller J, Jacob-Dolan C, Powers O, Hope D, Wu C, Pereira J, Murdza T, Mazurek CR, Hoyt A, Boon ACM, Davis-Gardner M, Suthar MS, Martinot AJ, Boursiquot M, Cook A, Pessaint L, Lewis MG, Andersen H, Tolboom J, Serroyen J, Solforosi L, Costes LMM, Zahn RC, Barouch DH, Mucosal boosting enhances vaccine protection against SARS-CoV-2 in macaques. Nature 626, 385–391 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wherry EJ, Barouch DH, T cell immunity to COVID-19 vaccines. Science 377, 821–822 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Barouch DH, Covid-19 Vaccines - Immunity, Variants, Boosters. N Engl J Med 387, 1011–1020 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moss P, The T cell immune response against SARS-CoV-2. Nature Immunology 23, 186–193 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Yu J, McMahan K, Jacob-Dolan C, He X, Giffin V, Wu C, Sciacca M, Powers O, Nampanya F, Miller J, Lifton M, Hope D, Hall K, Hachmann NP, Chung B, Anioke T, Li W, Muench J, Gamblin A, Boursiquot M, Cook A, Lewis MG, Andersen H, Barouch DH, CD8 T cells contribute to vaccine protection against SARS-CoV-2 in macaques. Science Immunology 7, eabq7647 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao F, Mallajosyula V, Arunachalam PS, van der Ploeg K, Manohar M, Röltgen K, Yang F, Wirz O, Hoh R, Haraguchi E, Lee JY, Willis R, Ramachandiran V, Li J, Kathuria KR, Li C, Lee AS, Shah MM, Sindher SB, Gonzalez J, Altman JD, Wang TT, Boyd SD, Pulendran B, Jagannathan P, Nadeau KC, Davis MM, Spheromers reveal robust T cell responses to the Pfizer/BioNTech vaccine and attenuated peripheral CD8(+) T cell responses post SARS-CoV-2 infection. Immunity 56, 864–878.e864 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Becerra X, Jha A, Project NextGen - Defeating SARS-CoV-2 and Preparing for the Next Pandemic. N Engl J Med 389, 773–775 (2023). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in the paper or the Supplementary Materials. Correspondence and requests for materials should be addressed to D.H.B. (dbarouch@bidmc.harvard.edu).