Abstract
Purpose
Vaccinia virus is widely used as an oncolytic agent for human cancer therapy, and several versions of vaccinia virus have demonstrated robust antitumor effects in breast cancer. Most vaccinia viruses are modified by thymidine kinase (TK) deletion. The function of the cyclin-dependent kinase inhibitor p21 in breast cancer remains controversial. We explored the impact of p21 gene knockdown (KD) on breast cancer cells and whether p21 KD interferes with the antitumor effect of TK-negative vaccinia virus.
Methods
p21 KD MDA-MB-231 and p21 KD MCF-7 cells were prepared, and cell proliferation and migration rates were evaluated using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and scratch healing assays. The tumor growth of xenografts originating from p21KD MDA-MB-231 cells and control cells was compared in a mouse model. The colony formation and sphere-forming abilities of p21 KD breast cancer cells were also determined using low-melting agarose and serum-free culture. The tumor-killing effect of the vaccinia virus was determined in breast cancer cells and mouse models using an MTT assay and tumor cell xenografts.
Results
p21 KD increased the growth and migration of MDA-MB-231 and MCF-7 cells and promoted the cell growth of MDA-MB-231 cells in mice, while decreasing the colony formation and sphere formation abilities. Expression of TK was reduced in p21 KD MDA-MB-231 cells. Oncolytic effects of both wild-type and TK-deleted vaccinia viruses were attenuated in p21KD MDA-MB-231 cells. The tumor-killing effect of TK-deleted vaccinia virus was also weakened in xenografted mice bearing p21 KD MDA-MB-231 cells.
Conclusion
Targeted inhibition of p21 accelerates the proliferation and migration of breast cancer cells and impairs the tumor-killing effect of vaccinia virus, suggesting that p21 levels in cancer cells interfere with vaccinia virus oncolytic therapy.
Keywords: Breast Neoplasms, p21, Vaccinia Virus
INTRODUCTION
Breast cancer is the most common malignant tumor worldwide and is a threat to the health of women [1]. Breast cancer can be divided into several subtypes, each of which varies in its clinical treatment and prognosis. Triple-negative breast cancer (TNBC), in which the expression of estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 are lacking, accounts for approximately 10%–15% of all breast cancer cases [2] and is responsible for more than 50% of breast cancer-related deaths [3,4]. TNBC exhibits high invasiveness and metastatic potential, poor prognosis, and high relapse potential [2,5].
p21, is a 21 kDa protein that was first identified as a cyclin-dependent kinase (CDK) inhibitor [6], and also known as wild-type activating factor-1 (WAF1)/CDK inhibitory protein-1 (CIP1) [7]. p21 is important in cell cycle control and is involved in several key biological processes [8]. Interestingly, p21 has been reported to play a paradoxical dual role as a tumor repressor or oncogene depending on its subcellular localization and posttranslational modifications [8,9]. Alterations in p21 also affect targeted therapies, such as susceptibility to chemotherapy and radiation [7]. According to an online database (http://gepia2.cancer-pku.cn), the expression of p21 is lower in breast invasive carcinoma (n = 1,085) than in normal tissues (n = 291). In contrast, a previous study reported that p21 expression in breast cancer is upregulated and closely correlated with adverse pathological parameters and poor prognosis [10]. Therefore, the effects of p21 on breast cancer prognosis and therapy for breast cancer require further investigation.
Oncolytic virotherapy is an emerging treatment for multiple tumors. Among these candidates, vaccinia virus is one of the most well-developed vectors because of its advantages such as its safety, high-titer production, large gene capacity, tumor selectivity, and high immunogenicity [11]. One strategy to increase the selective replication of vaccinia virus in tumor cells is to inactivate the viral thymidine kinase (TK) through genetic modification, resulting in preferential replication of the virus in high intracellular nucleotide-level tumor cells [12]. The most well-known example is JX-594, a replication-competent vaccinia virus engineered with a deleted TK region that expresses granulocyte-macrophage colony-stimulating factor and β-galactosidase [13]. The function of p21 is related to its interactions with TK [14,15,16]. Therefore, we explored whether p21 knockdown (KD) in breast cancer cells influences the oncolytic effect of TK-deleted vaccinia virus.
Here, we demonstrated that p21 KD in the TNBC cell line MDA-MB-231 promotes cell proliferation, migration, and tumor progression, while inhibiting cell colony formation. In addition, cell proliferation and animal experiments showed that p21 KD impaired the cytotoxicity of vaccinia virus with TK deletion. Our study provides a foundation for p21-targeted oncolytic viral therapy for TNBC.
METHODS
Cell lines and virus preparation
293T cells and human breast cancer cells MDA-MB-231 and MCF-7 were obtained from the Cell Bank of the Chinese Academy of Sciences. All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, USA) containing 10% fetal bovine serum.
WR strain vaccinia virus wild-type (VV-WT) and TK-deleted vaccinia virus carrying enhanced green fluorescent protein (VV-EGFP) were preserved in our laboratory. Vaccinia virus was amplified from HEK293 cells. After three freeze-thaw cycles and centrifugation, the viral titer was determined using the 50% tissue culture infectious dose method in 293A cells.
Short hairpin RNA (shRNA) and expression plasmid transfection
shRNAs targeting p21 (5′-GACACCACTGGAGGGTGACTTCTC-3′) and control shRNAs were synthesized and ligated into the pLKO.1-puro vector. The shuttle plasmid and package plasmids pMD2.G and psPAX2 were co-transfected into 293T cells (ratio: 4:3:1). Forty-eight hours later, the supernatant (containing the lentivirus) was filtered through a 0.45 μm filter and transferred into MDA-MB-231 and MCF-7 cells. At 24 hours post-infection, the culture medium was replaced with fresh medium containing puromycin (1 µg/mL) to screen for stable shRNA-expressing cell lines.
Sphere formation and cloning formation assay
Breast cancer cells were seeded into an ultra-low adsorption 6-well plate (2,000 cells/well). To induce cell sphere formation, the cells were cultured in serum-free stem cell culture medium (DMEM-F12 supplemented with insulin-like growth factor, epidermal growth factor, fibroblast growth factor, B-27, and penicillin/streptomycin). Images were captured using a microscope (IX71; Olympus, Tokyo, Japan). The number of spheres was counted, and the relative sphere formation efficiency was quantified and statistically analyzed.
For the cloning assay, 500 cells were seeded into each well of a 6-well plate. The cells were cultured and observed daily for 20 days; the culture medium was replenished during culture as appropriate. Cells were fixed with 70% ethanol and stained with crystal violet to photograph the colonies. The colonies were counted.
RNA isolation and reverse transcription-quantitative polymerase chain reaction analysis
Cellular RNA was extracted using TRIzol (Ambion, Austin, USA) and reverse-transcribed using a reverse transcription kit (Generay, Shanghai, China). The cDNA was subjected to qPCR amplification using a SYBR Green kit (TOYOBO, Osaka, Japan). The 2−△△Ct method was used to calculate the expression of target genes [17]. The primers (5′–3′) were as follows: CD133-F, GTCAGTTCTCCAGCTCTCCC; CD133-R, TCATGGCGACCTTTAACCCA; CD90-F, AACAGTTTGCCCCCAGGAAA; CD90-R, GAAGGACTCGTTGCTGGTGA; Oct4-F, GTGTTCAGCCAAAAGACCATCT; Oct4-R, GGCCTGCATGAGGGTTTCT; NANOG-F, AAGGCCTCAGCACCTACCTA; NANOG-R, TGCACCAGGTCTGAGTGTTC; glyceraldehyde 3-phosphate dehydrogenase (GAPDH)-F, CTGGGCTACACTGAGCACC; GAPDH-R, AAGTGGTCGTTGAGGGCAATG.
Western blot analysis
The cells were rinsed with phosphate-buffered saline (PBS), lysed in lysis buffer, and denaturized at 95°C for 5 minutes. Proteins were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electro-transferred onto polyvinylidene fluoride membranes. After blocking and incubation with antibodies, protein expression was visualized using a Tanon 5,500 gel imaging system (Tanon Science & Technology Co., Ltd., Shanghai, China). Antibodies against the following proteins were used: p21 (WAF1/CIP1) (2947S; Cell Signaling Technology, Danvers, USA), vimentin (5741S; Cell Signaling Technology), E-cadherin (3195S; Cell Signaling Technology), proliferating cell nuclear antigen (PCNA) (WL01804; Wanleibio, Shenyang, China), CDK2 (WL01543; Wanleibio), cyclin E (WL01072; Wanleibio), CDKN1B (E1A3325; EnoGENE, Nanjing, China), and GAPDH (R1210-1; HuaAnBio, Jinan, China).
Cell number analysis
Cell viability was analyzed using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell activity assay. The cells were plated in 96-well plates (5,000 cells/well). At termination (24 hours, 48 hours, 72 hours, 96 hours, and 120 hours), MTT was added to the wells (20 µL/well), and the plates were incubated in a cell incubator (37°C) for four hours. After incubation, the medium was aspirated carefully, and 150 µL dimethyl sulfoxide was added to each well. The plates were placed on a shaker for 10 minutes at 18°C–25°C. Absorbance was measured at 490 nm.
Scratch healing assay
The cells were plated in 6-well plates (5 × 105 cells/well). When the cells reached 90%–100% confluence, the medium was replaced with fresh DMEM + 3% fetal bovine serum. Scratches were made with a 200 µL pipette tip, and the scratches were photographed every six hours. The scratch areas were determined using ImageJ 1.53e software (NIH, Bethesda, USA). The average migration rate was calculated by dividing the change in scratch area over time.
Animal models
All animal studies were performed in accordance with the “Guide for the Care and Use of Laboratory Animals” and approved by the Laboratory Animal Ethics Committee of Zhejiang Sci-Tech University (approval number 2022-0105-05). Four-week-old female nude mice were used as orthotopic xenograft recipients (n = 6–8 per group). MDA-MB-231 shRNA targeting p21 (shp21) cells and MDA-MB-231 shRNA control (shCtrl) cells (1 × 106 per mouse) were implanted into the fourth mammary fat pad of anesthetized mice. The long diameter (LD) and short diameter (SD) of the tumors were measured, and (LD × SD × SD)/2 was used to calculate the tumor volume. The animals were euthanized under deep anesthesia when the tumor size reached 20 mm.
To evaluate the tumor-killing effect of vaccinia virus on p21 KD cells, different numbers of MDA-MB-231 shCtrl (8 × 106 per mouse) and MDA-MB-231 shp21 (5 × 106 per mouse) cells were subcutaneously injected into the underarms of mice to minimize the influence of the different growth rates of MDA-MB-231 shCtrl and MDA-MB-231 shp21 cells. When the tumor size reached approximately 500 mm3, the mice injected with MDA-MB-231shCtrl and MDA-MB-231 shp21cells were randomly divided into two treatment groups (n = 6 per group). Accordingly, 1 × 106 VV-EGFP virus/100 μL in PBS or 100 μL PBS was injected into the tumor of the mice. The tumors were measured every five days. When the tumors reached 20 mm in any dimension, the animals were euthanized.
Immunohistochemical staining analysis
Tumors were removed, fixed in formalin, embedded in paraffin wax, and cut into sections. After standard basic protocols, including deparaffinization, rehydration, antigen retrieval, and blocking, the tissue sections were stained with anti-p21 primary antibody and horseradish peroxidase-conjugated secondary antibody and observed and photographed under a microscope.
Statistical analysis
GraphPad Prism (version 8.3.0) software (GraphPad, Inc., La Jolla, USA) was used to analyze the data. Student's t-test or two-way analysis of variance was used to determine statistical significance. The data are shown as the mean ± standard deviation. The p-values considered statically significant are indicated in the figures as follows: p < 0.05, p < 0.01, p < 0.001, and p < 0.0001.
RESULTS
p21 inhibits the proliferation and migration of breast cancer cells
Previous studies demonstrated that p21 is a tumor suppressor protein; however, evidence indicates that p21 may also act as an antiapoptotic molecule [9]. To identify the role of p21 in breast cancer cells, we constructed MDA-MB-231 p21 KD and MCF-7 p21 KD cells using a lentivirus carrying shRNA targeting shp21 and confirmed the expression of p21 in the two cell lines (Figure 1A). We compared the proliferation of p21 KD breast cancer and control cells. The results of the MTT assay showed that the viability of MDA-MB-231 shp21 and MCF-7 shp21 cells was significantly increased compared with that of control cells (Figure 1B). Next, we examined the effects of p21 KD on cell migration. As shown in Figure 1C and D, the migration rates of MDA-MB-231 shp21 and MCF-7 shp21 cells were enhanced.
Figure 1. p21 knockdown promotes the proliferation and migration of breast cancer cells. (A) Western blot result of p21 expression in shp21 and shCtrl breast cancer cells. p21 expression was reduced in shp21 cells. (B) MTT results showed proliferation of MDA-MB-231 shp21 cells, MDA-MB-231 shCtrl cells, MCF-7 shp21cells, and MCF-7 shCtrl cells. The cell viability of shp21 breast cancer cells was increased compared with that of control cells. (C) MDA-MB-231 shp21cell and MDA-MB-231 shCtrl cell migration photos in the scratch healing assay and quantification of average migration ratio. (D) MCF-7 shp21 cell and MCF-7 shCtrl cell migration photos in the scratch healing assay and quantification of average migration ratio. The migration rate of p21 knockdown breast cancer cells was significantly increased. Student’s t-test was used to compare the two groups. Data are shown as the mean ± standard deviation.
shCtrl = short hairpin RNA control; shp21 = short hairpin RNA targeting p21; MTT = 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
*p < 0.05, †p < 0.001.
To identify the contribution of p21 KD to the expression of proteins involved in cell proliferation and tumor invasion, several proteins were detected using western blotting in p21 KD cells. PCNA plays a role in regulating DNA replication, the cell cycle, and DNA repair [18,19], and interacts with p21 in DNA replication [18]. PCNA is regarded as a standard marker of proliferation for assessing the growth fraction of a cell population in the diagnosis of breast cancer [20]. PCNA expression was increased in MDA-MB-231 shp21 and MCF-7 shp21 cells (Figure 2A). Vimentin is an intermediate filament protein recognized as a marker for epithelial-mesenchymal transition [21]. Vimentin overexpression is closely related to accelerated tumor growth, invasion, and poor prognosis [22]. E-cadherin, a protein normally expressed in breast epithelial tissue as a tumor suppressor; its loss promotes carcinogenesis, invasion, and epithelial-mesenchymal transition-associated metastasis [23,24]. Immunoblot analysis also demonstrated that p21 KD upregulated the expression of vimentin and downregulated that of E-cadherin (Figure 2A-C). These results support the role of p21 in inhibiting the proliferation and invasion of breast cancer cells.
Figure 2. p21 knockdown (interferes with the expressions of proteins involved in cell proliferation and tumor invasion and induces rapid growth of breast cancer cells. (A) Western blot showing the expression of proteins related to cell growth and invasion. (B, C) Western blot images were quantified and statistically analyzed using Student’s t-test. (D) Tumor formation after implantation of MDA-MB-231 shp21 cells and MDA-MB-231 shCtrl cells into the fourth mammary fat pads of nude mice. (E) Tumor growth curve. Tumor growth is shown as the mean ± standard deviation. (F) MDA-MB-231 shCtrl or MDA-MB-231 shp21 tumors were surgically removed. After fixation and sectioning, the tumor tissue sections were analyzed by immunohistochemistry using p21 monoclonal antibody.
shCtrl = short hairpin RNA control; shp21 = short hairpin RNA targeting p21; PCNA = proliferating cell nuclear antigen; GAPDH = glyceraldehyde 3-phosphate dehydrogenase.
*p < 0.01, †p < 0.001, ‡p < 0.0001.
To further determine the role of p21 KD in vivo, equal numbers of MDA-MB-231 shCtrl and MDA-MB-231 shp21 cells were implanted into the mammary fat pads of nude mice, and tumor xenograft growth was detected. The results showed that the tumor growth rate and volume in the MDA-MB-231 shp21 group were significantly faster and larger, respectively, than those in the MDA-MB-231 shCtrl group (Figure 2D and E). Immunohistochemical staining analysis confirmed that p21 levels were much lower in the MDA-MB-231 shp21 group than in the MDA-MB-231 shCtrl group (Figure 2F), suggesting that p21 downregulation triggered the growth of breast cancer cells in vivo.
Downregulation of p21 leads to reduced colony formation and sphere formation of breast cancer cells
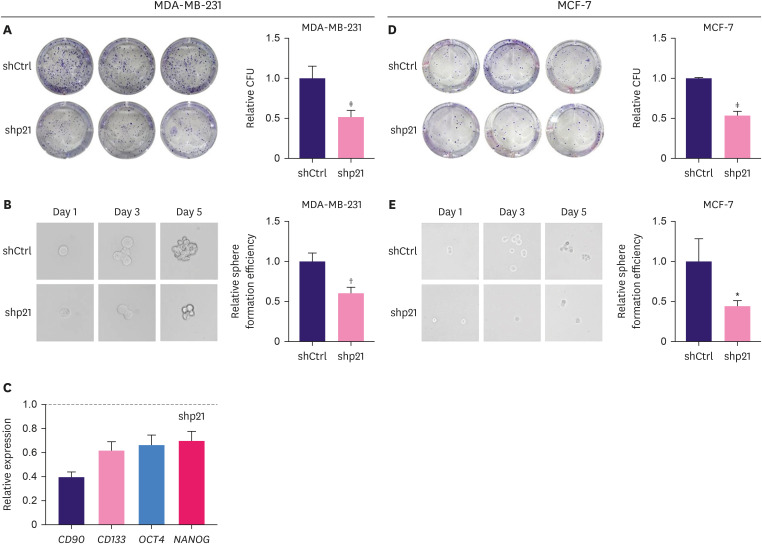
The colony formation assay is a widely used method to identify and quantify the self-renewal capacity of mammalian cells cultured in vitro [25]. Because the proliferation of MDA-MB-231 shp21 cells was increased and a previous study reported that p21 can improve the colony formation of murine myeloid progenitor cells [26], we speculated that the colony formation of p21KD breast cancer cells would be increased. Unexpectedly, as demonstrated in the colony formation assay (Figure 3A and D), the colony-forming units of MDA-MB-231 shp21 and MCF-7 shp21 cells were significantly lower than those of control cells. The sphere formation assay (Figure 3B and E) consistently showed that spheres formed by p21KD breast cancer cells were smaller than those formed by control cells. We then determined the relative RNA levels of stem cell-related genes in MDA-MB-231 shp21 cells. We found that the relative mRNA levels of CD90, CD133, OCT4, and NANOG were decreased after p21KD (Figure 3C), suggesting that p21 KD plays a role in maintaining breast cancer cell stemness.
Figure 3. Inhibition of p21 decreased breast cancer cells’ clonogenic and sphere formation abilities. (A) Colony formation photos and quantification of CFU of MDA-MB-231 shCtrl cells and MDA-MB-231 shp21 cells. (B) Sphere formation photos and quantification of the sphere formation efficiency of MDA-MB-231 shCtrl and MDA-MB-231shp21 cells. (C) Relative mRNA levels of stem-related genes CD90, CD133, OCT4, and NANOG were decreased in MDA-MB-231 shp21 cells. (D) Colony formation photos and quantification of CFU of MCF-7 shCtrl cells and MCF-7 shp21 cells. (E) Sphere formation photos and quantification of the sphere formation efficiency of MCF-7 shCtrl cells and MCF-7 shp21 cells. Student’s t-test was used to compare the two groups. Data are shown as the mean ± standard deviation.
shCtrl = short hairpin RNA control; shp21 = short hairpin RNA targeting p21; CFU = colony-forming units.
*p < 0.05, †p < 0.01, ‡p < 0.001.
Downregulation of p21 results in suppression of the tumor-killing effect of VV-EGFP
Vaccinia virus has been widely studied as an oncolytic therapy. To enhance its selectivity for replication in tumor cells, deletion of the TK region of the vaccinia virus is a commonly used genetic modification strategy. As several studies have shown that the level or function of p21 may be related to TK1 [14,15,16], we hypothesized that p21 KD may interfere with the effect of vaccinia virus with inactivated TK. We tested the tumor-killing effect of VV-EGFP. The cell viability results showed that at a multiplicity of infection of 1–8, the cytotoxicity of VV-EGFP in MDA-MB-231 shp21 cells was alleviated compared to that in MDA-MB-231 shCtrl cells (Figure 4A). Because VV-EGFP is a TK-deleted vaccinia virus, we predicted that the difference in TK expression between cells would influence cell survival after VV-EGFP infection. Therefore, TK expression levels were measured. As shown in Figure 4B, the relative expression of TK was reduced in MDA-MB-231 shp21 cells compared to that in shCtrl cells.
Figure 4. p21 knockdown impairs the oncolytic effect of the vaccinia virus. (A) Tumor-killing effect of TK-deleted vaccinia virus on MDA-MB-231 shp21 cells was reduced compared to that on MDA-MB-231 shCtrl cells. (B) Relative mRNA level of TK in MDA-MB-231 shp21 cells was lower than that in MDA-MB-231 shCtrl cells. (C) Tumor-killing effect of wild-type vaccinia virus on MDA-MB-231 shp21 was greater than that on MDA-MB-231 shCtrl cells. (D) Replication ability of TK-deleted vaccinia virus in MDA-MB-231 shCtrl cells and MDA-MB-231 shp21 cells. (E) Photographs showing that TK-deleted VV-EGFP successfully infected MDA-MB-23 shCtrl and MDA-MB-231 shp21cells. (F) p21 KD in MDA-MB-231 cells inhibited the oncolytic effect of vaccinia virus in mouse tumor burden experiments. MDA-MB-231 shp21-derived cells (5 × 106 per mouse) and MDA-MB-23 shCtrl-derived cells (8 × 106 per mouse) were injected into the mice subcutaneously. Once the tumor reached approximately 500 mm3, 1 × 106 VV-EGFP virus/100 μL in PBS or 100 μL PBS was injected into the tumor of the mice according to the group. Student’s t-test was used to compare two groups, and two-way analysis of variance was used to compare multiple groups. Data are shown as the mean ± standard deviation.
shCtrl = short hairpin RNA control; shp21 = short hairpin RNA targeting p21; TK = thymidine kinase; VV-WT = vaccinia virus wild-type; NS = not significant; EGFP = enhanced green fluorescent protein; VV-EGFP= vaccinia virus carrying enhanced green fluorescent protein; PBS = phosphate-buffered saline.
*p < 0.05, †p < 0.001.
Furthermore, VV-WT was used to determine whether TK inactivation in vaccinia virus induced a difference in the tumor-killing effect between p21 KD and control cells. The cell viability assay showed that VV-WT containing the TK region still induced a weaker tumor-killing effect in MDA-MB-231 shp21 cells compared with in MDA-MB-231 shCtrl cells (Figure 4C), suggesting that the oncolytic effect of the vaccinia virus was weakened in p21-downregulated cancer cells. We further investigated the replication efficiency of VV-EGFP in MDA-MB-231 shp21 and MDA-MB-231 shCtrl cells. The results showed that the viral replication efficiency in p21KD TNBC cells was lower than that in control cells. However, this difference was not significant (Figure 4D). Accordingly, based on the successful delivery of VV-EGFP into MDA-MB-231 shCtrl and shp21 cells (Figure 4E), the in vivo animal experiments showed that downregulation of p21 in MDA-MB-231 cells inhibited the oncolytic effect of vaccinia virus (Figure 4F). Taken together, p21 KD in MDA-MB-231 cells impaired the tumor-killing effect of vaccinia virus, indicating that the p21 level and p21-related pathway should be considered when using vaccinia virus as a therapeutic vector. The underlying mechanism of p21 downregulation, which interferes with the oncolytic effect of vaccinia virus, should be further explored.
DISCUSSION
The high heterogeneity of breast cancer affects patient prognosis and treatment options [27]. Understanding the genetic and non-genetic variability of breast cancer will contribute to the development of cancer-targeting therapeutics. p21 has been reported to play paradoxical roles in the regulation of breast cancer [7,9] and has become a potential target for the treatment of this disease. Although an online database (http://gepia2.cancer-pku.cn) revealed lower expression of p21 in invasive breast carcinoma, some studies reported that increased p21 expression in breast cancer is significantly correlated with larger tumor volume and poor prognosis [10]. Antisense oligodeoxynucleotides to p21 inhibit the growth of the highly metastatic breast cancer cell line Met-1 and tumor angiogenesis [28] and cause apoptosis in MCF7 and T47D breast cancer cells [29]. A recent study showed that p53-p21 plays a novel role in encoding DNA damage and fine-tuning proliferation and might contribute to maintaining proliferation heterogeneity [30]. In the present study, we investigated the function of p21 in the TNBC cell line MDA-MB-231 using a lentivirus carrying shRNA targeting p21. In vivo and in vitro experiments showed that p21 KD significantly promoted the development and migration of breast cancer cells, indicating the tumor-suppressive activity of p21 in TNBC cells (Figure 5).
Figure 5. Experimental flowchart. p21 knockdown in breast cancer cells increased cell proliferation and migration, decreased cell sphere formation and cloning formation, triggered the growth of triple-negative breast cancer cells, and weakened the oncolytic effect of the TK-deleted vaccinia virus.
shRNA = short hairpin RNA; shp21 = short hairpin RNA targeting p21; TNBC = triple-negative breast cancer; TK = thymidine kinase.
The colony formation assay, also known as the clonogenic assay, was introduced several decades ago and is used to assess the viability and reproducibility of single cells. This method is widely used in cancer biology to evaluate cellular growth and stemness because stem cells have the potential for continuous proliferation. p21 increases colony formation in murine myeloid progenitor cells [26]. Combining our cell proliferation test and animal experimental results, we predicted that p21 KD could increase the colony size or count. Unexpectedly, sphere formation and colony formation assays revealed a reduction in both the size and number of colonies and spheres in p21 KD breast cancer cells, consistent with a previous study showing suppression of tumor sphere formation in PyMT mammary tumor cells with p21 knockout [31]. Similarly, another study using the human colon cancer cell line HCT116 revealed that the colony number and size of p21 null cells were smaller than those of p21wt cells. Their nude mouse xenograft results also showed that p21-deficient xenografts grew faster at two weeks post-inoculation and that the tumors were larger and heavier than p21wt tumors [32]. As the underlying mechanism, p21 defect triggers autophagy, which inhibits initial tumorigenesis but fuels tumor growth in established tumors [32]. In addition, considering the mRNA levels of stem cell-related genes in our results, the stemness of p21 KD cells may be impaired. Previous research showed that for mesenchymal stem cells, colony-forming efficiency is independent of in vivo survival and cannot be used as a metric of in vivo survival [33]. Measurement of the colony-forming ability of human breast carcinomas in vitro lacks prognostic significance [34]. It has also been reported that sphere culture is optional for three-dimensional cell culture rather than enriching stem cells in breast cancer cell lines [35]. In the sphere formation assay, the primarily formed spheres do not truly identify stem cells, as progenitor cells can also form short-lived spheres [36]. Therefore, further investigation of the propagation of these spheres may contribute to the understanding of their self-renewal ability. Collectively, the colony and sphere formation assays may not be warranted to evaluate the tumorigenic ability of TNBC cells, and further investigation of the relationship between p21, autophagy, and tumor progression in TNBC is needed.
Oncolytic virotherapy is an effective candidate for tumor therapy. Vaccinia virus, a member of the oncolytic poxvirus family, is a double-stranded DNA virus with an important role in eradicating smallpox and has been developed as an oncolytic agent based on its inherent biological properties [11]. The oncolytic effects of several vaccinia viruses on breast cancer have been previously studied. Deng et al. [37] reported that the vaccinia virus VG9-IL-24, in which the TK gene region is disrupted and armed with the IL-24 gene, exhibits enhanced antitumor effects in human breast cancer. The vaccinia virus GLV-1h153 has shown promising therapeutic effects in the prevention and treatment of metastatic TNBC [38]. GLV-1h68 shows preferential replication and cytotoxicity in breast cancer stem-like cells [39]. All vaccinia viruses mentioned above are TK-deleted, which results in preferential replication of vaccinia virus in cancerous cells with high intracellular nucleotide pools [12]. Some studies reported a relationship between TK and p21 expression. The molecular function of p21 is disrupted by its interaction with TK1 [14]. TK1 silencing upregulates p21 and suppresses the proliferation of pancreatic ductal adenocarcinoma cells [15]. In p53+/+ cells, DNA damage-induced p21 expression affects the cell cycle and TK1 levels [16]. Considering the interaction between TK and p21, we examined the effect of p21 KD on the tumor-killing effect of vaccinia virus with TK deletion. Cell and animal experiments demonstrated that p21 KD weakens the oncolytic effect of TK-deleted vaccinia virus. p21 KD in TNBC cells causes decreased expression of TK, which may disturb the replication of TK-deleted vaccinia virus. Further investigation of other mechanisms that contribute to the reduced tumor-killing effect of TK-deleted vaccinia virus in p21 KD breast cancer cells is warranted.
It is undeniable that there are some limitations for our study. We only investigated the effect of p21 in p21 KD cells. The underlying mechanism of p21 KD impairing the oncolytic effect of TK-deleted vaccinia virus in TNBC cells was not fully elucidated. Overexpressing p21 in TNBC cells and in-depth studying the mechanism of p21 KD influencing the tumor-killing effect of vaccinia virus will help to fully understanding the function of p21.
Taken together, we found that p21 KD weakened the oncolytic effect of TK-deleted vaccinia virus in TNBC cells (Figure 5). Our study provides a reference for application of TK-deleted vaccinia virus in the treatment of tumors with differential expression of p21.
Footnotes
Funding: This work was supported by the Natural Science Foundation of Zhejiang Province (Grant number LGF22C010005); Science Foundation of Zhejiang Sci‐Tech University (grant number 18042291-Y); Public Welfare Technology Project of Zhejiang Province (grant number LGF21H160033); Zhejiang Medical Technology Plan Project (grant number 2021KY047); Hangzhou Science and Technology Bureau (grant number 20201203B44); and Hangzhou Medical Health Science and Technology Project (grant number B20220173).
Conflict of Interest: The authors declare that they have no competing interests.
Data Availability: The data generated in the current study are available from the corresponding author upon reasonable request.
- Conceptualization: Huang F, Wang Y.
- Data curation: Jia X, Zhao Y, Xu Y.
- Formal analysis: Jia X, Lu X.
- Funding acquisition: Wang X, Wang H, Huang F, Jia X.
- Investigation: Zhao Y, Li Q, Lu X, Wang X, Wang H, Shi Z.
- Methodology: Huang B.
- Project administration: Wang Y.
- Writing - original draft: Jia X.
- Writing - review & editing: Jia X, Huang F, Wang Y.
References
- 1.Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Primers. 2019;5:66. doi: 10.1038/s41572-019-0111-2. [DOI] [PubMed] [Google Scholar]
- 2.Chavez KJ, Garimella SV, Lipkowitz S. Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2010;32:35–48. doi: 10.3233/BD-2010-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simu S, Marcovici I, Dobrescu A, Malita D, Dehelean CA, Coricovac D, et al. Insights into the behavior of triple-negative MDA-MB-231 breast carcinoma cells following the treatment with 17β-ethinylestradiol and levonorgestrel. Molecules. 2021;26:2776. doi: 10.3390/molecules26092776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Treeck O, Schüler-Toprak S, Ortmann O. Estrogen actions in triple-negative breast cancer. Cells. 2020;9:2358. doi: 10.3390/cells9112358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin L, Duan JJ, Bian XW, Yu SC. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020;22:61. doi: 10.1186/s13058-020-01296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 7.Shamloo B, Usluer S. p21 in cancer research. Cancers (Basel) 2019;11:1178. doi: 10.3390/cancers11081178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreis NN, Louwen F, Yuan J. The multifaceted p21 (Cip1/Waf1/CDKN1A) in cell differentiation, migration and cancer therapy. Cancers (Basel) 2019;11:1220. doi: 10.3390/cancers11091220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zohny SF, Al-Malki AL, Zamzami MA, Choudhry H. p21Waf1/Cip1: its paradoxical effect in the regulation of breast cancer. Breast Cancer. 2019;26:131–137. doi: 10.1007/s12282-018-0913-1. [DOI] [PubMed] [Google Scholar]
- 10.Wei CY, Tan QX, Zhu X, Qin QH, Zhu FB, Mo QG, et al. Expression of CDKN1A/p21 and TGFBR2 in breast cancer and their prognostic significance. Int J Clin Exp Pathol. 2015;8:14619–14629. [PMC free article] [PubMed] [Google Scholar]
- 11.Kirn DH, Thorne SH. Targeted and armed oncolytic poxviruses: a novel multi-mechanistic therapeutic class for cancer. Nat Rev Cancer. 2009;9:64–71. doi: 10.1038/nrc2545. [DOI] [PubMed] [Google Scholar]
- 12.Chan WM, McFadden G. Oncolytic poxviruses. Annu Rev Virol. 2014;1:119–141. doi: 10.1146/annurev-virology-031413-085442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breitbach CJ, Burke J, Jonker D, Stephenson J, Haas AR, Chow LQ, et al. Intravenous delivery of a multi-mechanistic cancer-targeted oncolytic poxvirus in humans. Nature. 2011;477:99–102. doi: 10.1038/nature10358. [DOI] [PubMed] [Google Scholar]
- 14.Huang DY, Chang ZF. Interaction of human thymidine kinase 1 with p21(Waf1) Biochem J. 2001;356:829–834. doi: 10.1042/0264-6021:3560829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu X, Shi C, Peng Y, Yin L, Tu M, Chen Q, et al. Thymidine kinase 1 silencing retards proliferative activity of pancreatic cancer cell via E2F1-TK1-P21 axis. Cell Prolif. 2018;51:e12428. doi: 10.1111/cpr.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen YL, Eriksson S, Chang ZF. Regulation and functional contribution of thymidine kinase 1 in repair of DNA damage. J Biol Chem. 2010;285:27327–27335. doi: 10.1074/jbc.M110.137042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Mansilla SF, de la Vega MB, Calzetta NL, Siri SO, Gottifredi V. CDK-independent and PCNA-dependent functions of p21 in DNA replication. Genes (Basel) 2020;11:593. doi: 10.3390/genes11060593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González-Magaña A, Blanco FJ. Human PCNA structure, function and interactions. Biomolecules. 2020;10:570. doi: 10.3390/biom10040570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juríková M, Danihel Ľ, Polák Š, Varga I. Ki67, PCNA, and MCM proteins: markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118:544–552. doi: 10.1016/j.acthis.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Arrindell J, Desnues B. Vimentin: from a cytoskeletal protein to a critical modulator of immune response and a target for infection. Front Immunol. 2023;14:1224352. doi: 10.3389/fimmu.2023.1224352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68:3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Na TY, Schecterson L, Mendonsa AM, Gumbiner BM. The functional activity of E-cadherin controls tumor cell metastasis at multiple steps. Proc Natl Acad Sci U S A. 2020;117:5931–5937. doi: 10.1073/pnas.1918167117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horne HN, Oh H, Sherman ME, Palakal M, Hewitt SM, Schmidt MK, et al. E-cadherin breast tumor expression, risk factors and survival: Pooled analysis of 5,933 cases from 12 studies in the Breast Cancer Association Consortium. Sci Rep. 2018;8:6574. doi: 10.1038/s41598-018-23733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brix N, Samaga D, Belka C, Zitzelsberger H, Lauber K. Analysis of clonogenic growth in vitro. Nat Protoc. 2021;16:4963–4991. doi: 10.1038/s41596-021-00615-0. [DOI] [PubMed] [Google Scholar]
- 26.Braun SE, Mantel C, Rosenthal M, Cooper S, Liu L, Robertson KA, et al. A positive effect of p21cip1/waf1 in the colony formation from murine myeloid progenitor cells as assessed by retroviral-mediated gene transfer. Blood Cells Mol Dis. 1998;24:138–148. doi: 10.1006/bcmd.1998.0181. [DOI] [PubMed] [Google Scholar]
- 27.Lüönd F, Tiede S, Christofori G. Breast cancer as an example of tumour heterogeneity and tumour cell plasticity during malignant progression. Br J Cancer. 2021;125:164–175. doi: 10.1038/s41416-021-01328-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss RH, Marshall D, Howard L, Corbacho AM, Cheung AT, Sawai ET. Suppression of breast cancer growth and angiogenesis by an antisense oligodeoxynucleotide to p21(Waf1/Cip1) Cancer Lett. 2003;189:39–48. doi: 10.1016/s0304-3835(02)00495-0. [DOI] [PubMed] [Google Scholar]
- 29.Fan Y, Borowsky AD, Weiss RH. An antisense oligodeoxynucleotide to p21(Waf1/Cip1) causes apoptosis in human breast cancer cells. Mol Cancer Ther. 2003;2:773–782. [PubMed] [Google Scholar]
- 30.Gutu N, Binish N, Keilholz U, Herzel H, Granada AE. p53 and p21 dynamics encode single-cell DNA damage levels, fine-tuning proliferation and shaping population heterogeneity. Commun Biol. 2023;6:1196. doi: 10.1038/s42003-023-05585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benard O, Qian X, Liang H, Ren Z, Suyama K, Norton L, et al. p21CIP1 promotes mammary cancer-initiating cells via activation of Wnt/TCF1/CyclinD1 signaling. Mol Cancer Res. 2019;17:1571–1581. doi: 10.1158/1541-7786.MCR-18-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maheshwari M, Yadav N, Hasanain M, Pandey P, Sahai R, Choyal K, et al. Inhibition of p21 activates Akt kinase to trigger ROS-induced autophagy and impacts on tumor growth rate. Cell Death Dis. 2022;13:1045. doi: 10.1038/s41419-022-05486-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Connor K. A cautionary tale about the use of colony-forming efficiency as a proxy for the survival of mesenchymal stem cells. Stem Cell Res Ther. 2020;11:292. doi: 10.1186/s13287-020-01805-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ottestad L, Tveit KM, Hannisdal E, Skrede M, Nesland JM, Gundersen S, et al. Colony forming ability of human breast carcinomas: lack of prognostic significance. Br J Cancer. 1989;60:216–219. doi: 10.1038/bjc.1989.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smart CE, Morrison BJ, Saunus JM, Vargas AC, Keith P, Reid L, et al. In vitro analysis of breast cancer cell line tumourspheres and primary human breast epithelia mammospheres demonstrates inter- and intrasphere heterogeneity. PLoS One. 2013;8:e64388. doi: 10.1371/journal.pone.0064388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bahmad HF, Cheaito K, Chalhoub RM, Hadadeh O, Monzer A, Ballout F, et al. Sphere-formation assay: three-dimensional in vitro culturing of prostate cancer stem/progenitor sphere-forming cells. Front Oncol. 2018;8:347. doi: 10.3389/fonc.2018.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng L, Fan J, Ding Y, Yang X, Huang B, Hu Z. Target therapy with vaccinia virus harboring IL-24 for human breast cancer. J Cancer. 2020;11:1017–1026. doi: 10.7150/jca.37590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gholami S, Chen CH, Lou E, De Brot M, Fujisawa S, Chen NG, et al. Vaccinia virus GLV-1h153 is effective in treating and preventing metastatic triple-negative breast cancer. Ann Surg. 2012;256:437–445. doi: 10.1097/SLA.0b013e3182654572. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Chen NG, Minev BR, Szalay AA. Oncolytic vaccinia virus GLV-1h68 strain shows enhanced replication in human breast cancer stem-like cells in comparison to breast cancer cells. J Transl Med. 2012;10:167. doi: 10.1186/1479-5876-10-167. [DOI] [PMC free article] [PubMed] [Google Scholar]