Abstract
Background and study aims Endoscopic ultrasound (EUS) is vital for early pancreatic cancer diagnosis. Advances in artificial intelligence (AI), especially deep learning, have improved medical image analysis. We developed and validated the Modified Faster R-CNN (M-F-RCNN), an AI algorithm using EUS images to assist in diagnosing pancreatic cancer.
Methods We collected EUS images from 155 patients across three endoscopy centers from July 2022 to July 2023. M-F-RCNN development involved enhancing feature information through data preprocessing and utilizing an improved Faster R-CNN model to identify cancerous regions. Its diagnostic capabilities were validated against an external set of 1,000 EUS images. In addition, five EUS doctors participated in a study comparing the M-F-RCNN model's performance with that of human experts, assessing diagnostic skill improvements with AI assistance.
Results Internally, the M-F-RCNN model surpassed traditional algorithms with an average precision of 97.35%, accuracy of 96.49%, and recall rate of 5.44%. In external validation, its sensitivity, specificity, and accuracy were 91.7%, 91.5%, and 91.6%, respectively, outperforming non-expert physicians. The model also significantly enhanced the diagnostic skills of doctors.
Conclusions: The M-F-RCNN model shows exceptional performance in diagnosing pancreatic cancer via EUS images, greatly improving diagnostic accuracy and efficiency, thus enhancing physician proficiency and reducing diagnostic errors.
Keywords: Endoscopic ultrasonography; Pancreas; Endoscopy Upper GI Tract; Precancerous conditions & cancerous lesions (displasia and cancer) stomach; Diagnosis and imaging (inc chromoendoscopy, NBI, iSCAN, FICE, CLE)
Introduction
Pancreatic cancer remains one of the most lethal solid malignancies, with both incidence and mortality rates on the rise 1 . Over 80% of cases are diagnosed at advanced stages involving tissue invasion or metastasis, which contributes to the disease’s high mortality. Currently, it is the fourth leading cause of cancer deaths and is projected to become the second by 2030 1 . Early diagnosis has been shown to significantly improve the 5-year survival rate 2 .
Endoscopic ultrasound (EUS) offers closer proximity to the pancreas than computed tomography (CT) or magnetic resonance imaging, reducing gas interference and providing higher-resolution images. It has become crucial in diagnosing pancreatic tumors, as well as chronic and autoimmune pancreatitis 3 4 5 . However, the quality of pancreatic scanning images and the experience level of the operating physicians can lead to diagnostic errors. Thus, there is a pressing need for more accurate diagnostic methods.
Artificial Intelligence (AI) enables machines such as computers to emulate, extend, or augment human intelligence. Machine learning is a type of AI that refers to the process by which machines can learn and improve themselves through data and algorithms 6 7 . This technology has been utilized in diagnosing and staging various cancers, including those of the lung and ovary 8 9 . However, studies focusing on pancreatic cancer, especially using EUS for image differentiation, are limited. Kuwahara and colleagues developed an AI model that detected pancreatic lesions with an accuracy of 91.0% 10 . Zhu et al. achieved a diagnostic sensitivity of 91.6%, specificity of 95.0%, and accuracy of 94.2% by analyzing EUS images to distinguish between pancreatic cancer and chronic pancreatitis 11 . Nevertheless, these studies are in the preliminary stages, often lacking external validation and human-machine comparisons, which limits their clinical application.
The Region-based Convolutional Neural Network (Faster R-CNN) is an advanced object detection framework extensively used in various domains, including medical image recognition 12 . In this research, we introduce a novel AI algorithm based on EUS images, named Modified Faster R-CNN (M-F-RCNN), aimed at detecting pancreatic cancer. This model is designed to enhance physician diagnostic capabilities and has been externally validated to assess its effectiveness.
Patients and methods
Collection of clinical data and EUS images
We collected EUS images from 103 pancreatic cancer patients and 52 non-cancer patients from three independent endoscopy centers: Main campus, East campus, and Caotang campus of Sichuan Provincial People's Hospital, from July 2022 to July 2023. This collection formed Dataset A, used for training, validation, and testing of our model. In addition, for external validation, Dataset B was created, comprising 1,000 randomly selected EUS images from patients enrolled at these centers. Inclusion criteria for pancreatic cancer patients were: (1) histopathologically confirmed pancreatic cancer and (2) underwent EUS examination before treatment. Inclusion criteria for non-pancreatic cancer patients were patients who underwent EUS examination for other diseases, with no significant abnormal image features in the pancreas section. Exclusion criteria were: (1) age <18 or >80 years and (2) nonstandard images (without anatomical markers or lesions).
Collected patient information included age, gender, symptoms, tumor size, location, EUS results, method of pathology acquisition, and histopathological results of pancreatic cancer patients. The source of information was the electronic clinical records of patients. Any data that could identify subjects were hidden and each patient was assigned a random number to ensure effective data anonymization and compliance with data protection regulations. This study adhered to the Declaration of Helsinki and received approval from the Ethics Committee of Sichuan Provincial People's Hospital.
EUS examination procedure
EUS examination was completed by an experienced EUS physician using linear EUS. EUS equipment used at the three independent endoscopy centers included SU9000 (FUJIFILM, Tokyo, Japan) and EU-ME1 (Olympus, Tokyo, Japan) models. Ultrasound probe models were EG-580UT (FUJIFILM, Tokyo, Japan) and GF-UCT260 (Olympus, Tokyo, Japan).
Development of the novel AI algorithm model
Data set and preprocessing
Dataset A comprised 414 images of cancerous areas from 103 pancreatic cancer patients and 300 images from 52 non-pancreatic cancer patients, all meeting specific inclusion criteria. To enhance the diversity and number of training samples, data augmentation techniques were employed, increasing the total to 1,500 EUS images. An experienced EUS expert pre-classified these images and marked cancerous areas. The dataset was randomly divided into training, validation, and test sets with a distribution ratio of 7:1:2. In addition, to assess model specificity, the original 300 non-cancerous images were included in a separate test set to evaluate the false detection rate.
Dataset B used for external validation contained 1,000 EUS images, including 585 pancreatic cancer images and 415 non-pancreatic cancer images.
Image processing
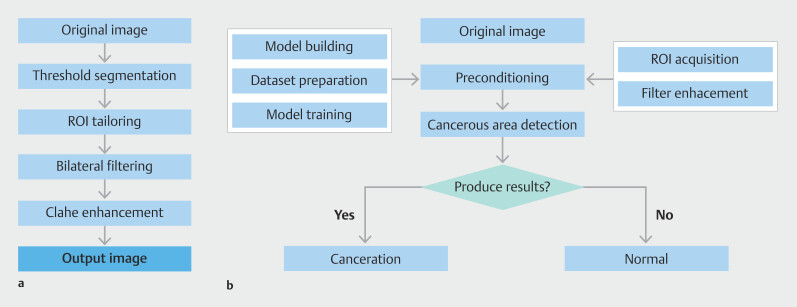
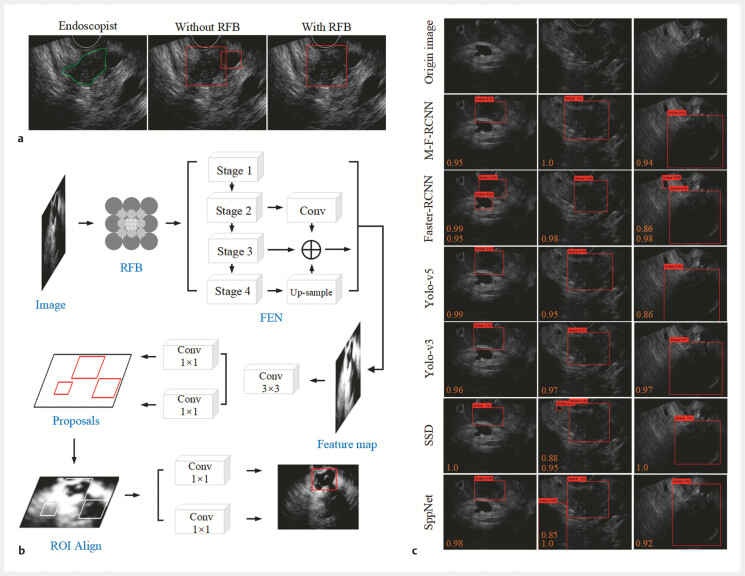
To tailor our detection algorithm specifically for pancreatic cancer, we developed a dedicated EUS image detection algorithm. This algorithm was implemented in the PyCharm (Community Edition, version 2020.1.3) environment, configured with PyTorch 7.1 and Python 3.7, and run on an NVIDIA GeForce RTX 3070 GPU. The preprocessing stage involved extracting regions of interest (ROI) and applying enhanced filtering techniques to produce high-quality images. Optimization was further achieved using the Contrast-Limited Adaptive Histogram Equalization (CLAHE) algorithm. To minimize risk of false positives, a Receptive Field Block (RFB) module was integrated at the top of the detection model, designed to emulate perceptual features of the human visual system 13 14 . Fig. 1 illustrates the complete architecture of the algorithm, while Fig. 2 a demonstrates the improvement in detection results for the same input image, pre-integration and post-integration of the RFB module.
Fig. 1.
Data preprocessing flow and overall algorithm architecture diagram. a Data preprocessing flow chart. b Overall algorithm architecture diagram.
Fig. 2.
Overall structure and improvement of the pancreatic lesion detection model based on EUS images. a Introduction of the Receptive Field Block (RFB) module improves detection results of the pancreatic lesion for the same input image. b Overall structure of the detection model. c Performance of each model in detecting pancreatic cancerous regions were presented with average precision (AP) values.
Cancerous region detection algorithm
The Feature Extraction Network (FEN) is a crucial component of the Faster R-CNN model, tasked with analyzing and extracting image information 15 . As depicted in Fig. 2 b , we restructured the FEN architecture and integrated ResNet-50, a Feature Fusion Module (FFM), and ROI Align into our Modified Faster R-CNN, resulting in a new model dubbed Modified Faster R-CNN (M-F-RCNN) 16 17 18 .
To assess model recognition capabilities, we compared M-F-RCNN with both single-stage (SSD, Yolo-v3, Yolo-v5) and two-stage models (Faster R-CNN, SPPNet), visualizing algorithmic differences and conducting a statistical analysis of average precision (AP), recall rate, accuracy, and loss rate.
For diagnostic validation, Dataset B was utilized to statistically analyze model sensitivity, specificity, accuracy, area under the receiver operator characteristic curve (AUC), and diagnostic time.
Human-computer competition and AI-assisted image reading effect
Five EUS physicians from the Digestive Endoscopy Centers of Sichuan Provincial People's Hospital and Chongqing University Cancer Hospital participated in this study. The group comprised two experts and three novices. Initially, they independently diagnosed images and recorded the time taken, which was then compared with the model's external validation results. After a blind initial reading, the results from M-F-RCNN were revealed, and the physicians reevaluated the images without the re-reading time being recorded, to avoid any bias in diagnostic time.
Statistical analysis
Data analysis was performed using R language (version 4.3.1). We employed two-sample t -tests or Wilcoxon-Mann-Whitney tests for analyzing patient characteristics. Model performance was evaluated using AUC for the ROC curve and AP for the Precision-Recall curve. These metrics assessed model effectiveness at varying recall levels. Sensitivity and recall rates quantified the proportion of true positives correctly identified by the model. Specificity and accuracy assessed the proportion of true negatives correctly identified and the correctness of positive predictions, respectively. Internal validation utilized AP, recall rate, accuracy, and loss rate to compare model recognition capabilities, whereas external validation quantitatively evaluated AUC, sensitivity, specificity, and accuracy. AP and AUC curves were generated using the pROC and ggplot2 packages, respectively. AUCs of different models were compared using DeLong tests, while paired chi-square tests were employed to compare sensitivity and specificity. Two-sided P <0.05 was considered statistically significant.
Results
Basic patient information
Dataset A comprised 1,800 EUS images, which included 414 original cancerous images from 103 pancreatic cancer patients. Following data augmentation, this number was increased to 1,500 pancreatic cancer EUS images, alongside 300 non-pancreatic cancer EUS images. Detailed demographics and clinical characteristics of these patients are presented in Table 1 . The cohort consisted of 63 males (61.2%) and 40 females (38.8%), with ductal adenocarcinoma being the predominant lesion type, observed in 91 cases (88.3%). The most common tumor location was the pancreatic head, accounting for 68 cases (66%). Surgical operations and EUS-FNA were the primary methods for pathology acquisition, represented in 47 cases (45.6%) and 56 cases (54.4%), respectively. The most frequently reported symptom was abdominal pain, occurring in 55 cases (53.4%). The average age of participants was 67.51 years (SD = 9.90), and the average tumor size was 3.53 cm (SD = 1.00).
Table 1 Description of various parameters in 103 pancreatic cancer patients.
| Parameter | Category | Description [ n (%),x±s] |
| EUS-FNA, endoscopic ultrasound fine-needle aspiration. | ||
| Age (years) | 67.51 ± 9.90 | |
| Gender | Male | 63 (61.2 |
| Female | 40 (38.3) | |
| Symptoms | Abdominal pain | 55 (53.4) |
| Jaundice | 48 (46.6) | |
| Tumor size(cm) | 3.53 ± 1.00 | |
| Location | Pancreatic neck | 12 (11.7) |
| Pancreatic body | 14 (13.6) | |
| Pancreatic head | 68 (66.0) | |
| Pancreatic tail | 9 (8.7) | |
| Pathological method | Surgical resection | 47 (45.6) |
| EUS-FNA | 56 (54.4) | |
| Nature of lesion | Ductal adenocarcinoma | 91 (88.3) |
| Cystic adenocarcinoma | 3 (2.9) | |
| IPMN carcinogenesis | 4 (3.9) | |
| Adenocarcinoma with squamous | 2 (1.9) | |
| Adenopapillary carcinoma | 3 (2.9) | |
Dataset B contained 1,000 EUS images, including 585 pancreatic cancer images and 415 non-pancreatic cancer images, with no detailed patient information collected.
M-F-RCNN performance
In Dataset A, performance of M-F-RCNN was compared with classic object detection algorithms, as detailed in Table 2 . This included comparisons with single-stage models (SSD, Yolo-v3, Yolo-v5) and two-stage models (Faster R-CNN, SPPNet). M-F-RCNN demonstrated superior detection performance, achieving an AP of 97.35% and improving accuracy to 96.49%. This model showed a notable reduction in misdiagnoses and an increase in recall rate by 5.44%, indicating fewer missed detections. The model required an average of 98 ms to detect tumors in a single image, fulfilling real-time diagnostic requirements for EUS. Fig. 2 c illustrates the enhanced performance of M-F-RCNN in identifying cancerous features in EUS images compared to other models.
Table 2 Performance comparison of cross-validation models.
| Number | Model | Recall/(%) | Precision/(%) | AP/(%) | Rate of missed detections/(%) |
| AP, average precision; M-F-RCNN, Modified Faster Region-based Convolutional Neural Network; Faster R-CNN, Region-based Convolutional Neural Network; SPPNet, Spatial Pyramid Pooling Network; SSD, Single Shot MultiBox Detector. | |||||
| 1 | M-F-RCNN | 98.72 | 96.49 | 97.35 | 3.51 |
| 2 | Faster-RCNN | 93.28 | 86.65 | 91.16 | 13.35 |
| 3 | SPPNet | 84.89 | 82.26 | 83.44 | 17.74 |
| 4 | SSD | 86.21 | 91.86 | 87.58 | 8.14 |
| 5 | Yolo-v3 | 88.56 | 89.47 | 89.22 | 10.53 |
| 6 | Yolo-v5 | 90.24 | 91.68 | 91.22 | 8.32 |
External validation using Dataset B demonstrated that M-F-RCNN achieved a sensitivity of 91.7%, specificity of 91.5%, and accuracy of 91.6%, with an average detection time of 113 ms ( Table 3 ).
Table 3 Comparison of diagnostic performance between M-F-RCNN and physicians.
| AUC (95%CI) | P value | Sensitivity (95%CI) | P value | Specificity (95%CI) | P value | Accuracy | P value | Average time per image | |
| M-F-RCNN, Modified Faster Region-based Convolutional Neural Network; AUC, area under the receiver operating characteristic; AI, artificial intelligence. | |||||||||
| AI | 0.916 (0.898, 0.933) | Ref | 0.917 (0.894, 0.939) | Ref | 0.915 (0.888, 0.942) | Ref | 0.916 | Ref | 113 ms |
| Expert 1 | 0.891 (0.871, 0.910) | 0.096 | 0.905 (0.881, 0.928) | 0.463 | 0.877 (0.845, 0.908) | 0.066 | 0.893 | 0.080 | 10 s |
| Expert 2 | 0.913 (0.895, 0.931) | 0.894 | 0.918 (0.896, 0.940) | 0.917 | 0.908 (0.880, 0.936) | 0.710 | 0.914 | 0.873 | 8 s |
| Novice 1 | 0.704 (0.675, 0.733) | <0.001 | 0.698 (0.661, 0.736) | <0.001 | 0.709 (0.666, 0.753) | <0.001 | 0.703 | <0.001 | 20 s |
| Novice 2 | 0.730 (0.707, 0.752) | <0.001 | 0.508 (0.467, 0.548) | <0.001 | 0.952 (0.931, 0.972) | 0.039 | 0.691 | <0.001 | 19 s |
| Novice 3 | 0.594 (0.563, 0.625) | <0.001 | 0.617 (0.577, 0.656) | <0.001 | 0.571 (0.524, 0.619) | <0.001 | 0.598 | <0.001 | 23 s |
Human-computer competition comparison
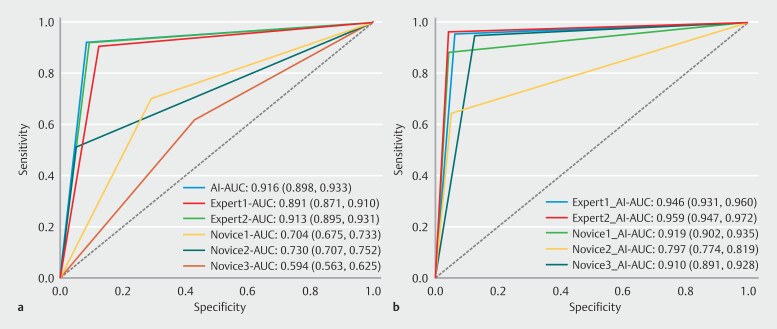
Diagnostic performance of five physicians was compared with the external validation results of M-F-RCNN, as shown in Fig. 3 a and Table 3 . M-F-RCNN matched or slightly exceeded the performance of two expert physicians and demonstrated significant advantages over the three novices in sensitivity, specificity, accuracy, and efficiency, with the differences being statistically significant ( P <0.01). Modes detection time was markedly less, averaging 98 ms per image, compared with the significantly longer time taken by the experts and novices (10 s, 8 s, 20 s, 19 s, and 23 s, respectively).
Fig. 3.
Receiver operating characteristic curves for the M-F-RCNN model, experts, novices, and model-assisted. a The model outperforms both novices and experts in terms of sensitivity, specificity, accuracy, and efficiency. b With the assistance of the M-F-RCNN model, the area under the curve (AUC), sensitivity, specificity, and accuracy of experts and novices were significantly improved, while minimizing the gaps among the physicians.
Impact of AI assistance on diagnostic performance
With M-F-RCNN assistance, both experts and novices exhibited significant improvements in AUC, sensitivity, specificity, and accuracy, effectively reducing discrepancies in diagnostic performance among different experience levels, as illustrated in Fig. 3 and Table 4 . All improvements were statistically significant ( P <0.01), except for the AUC and specificity in Novice 2's assessments. Application of M-F-RCNN not only enhanced the diagnostic capabilities of the physicians, but in some cases, also led to performance indicators exceeding those of the model itself, likely due to mode guidance prompting a more thorough review of the diagnoses.
Table 4 Comparison of diagnostic performance of physicians with and without M-F-RCNN assistance.
| AUC (95%CI) | P value | Sensitivity (95%CI) | P value | Specificity (95%CI) | P value | Accuracy | P value | |
| AUC, area under the receiver operating characteristic; M-F-RCNN, Modified Faster Region-based Convolutional Neural Network; AI, artificial Intelligence. | ||||||||
| Expert 1+AI | 0.946 (0.931,0.960) | <0.001 | 0.954 (0.937, 0.971) | <0.001 | 0.937 (0.914,0.960) | <0.001 | 0.947 | <0.001 |
| Expert 2+AI | 0.959 (0.947,0.972) | <0.001 | 0.963 (0.947, 0.978) | <0.001 | 0.956 (0.937, 0.976) | <0.001 | 0.960 | <0.001 |
| Novice 1+AI | 0.919 (0.902,0.935) | <0.001 | 0.881 (0.855, 0.907) | <0.001 | 0.956 (0.937, 0.976) | <0.001 | 0.912 | <0.001 |
| Novice 2+AI | 0.797 (0.774,0.819) | 0.008 | 0.644 (0.605, 0.683) | <0.001 | 0.949 (0.928, 0.970) | 0.317 | 0.770 | <0.001 |
| Novice 3+AI | 0.910 (0.891,0.928) | <0.001 | 0.945 (0.927, 0.964) | <0.001 | 0.874 (0.842, 0.906) | <0.001 | 0.916 | <0.001 |
Discussion
AI image recognition technology has significantly advanced in fields like CT and ultrasonography, aiding clinicians in delineating target areas and identifying lesions 19 20 21 . This study leveraged the Faster R-CNN object detection framework to develop the Modified Faster R-CNN (M-F-RCNN) model, which demonstrated robust performance in EUS diagnosis of pancreatic cancer through internal testing, external validation, and a human-computer competition.
The inherent challenges of high noise, low contrast, and non-uniformity in EUS images necessitate effective denoising solutions. While various models have been proposed for ultrasound image denoising across different diseases, none fully met our criteria for EUS image denoising 22 23 24 . This study assessed two histogram equalization methods: Enhanced Contrast Histogram Equalization (ECHE) and CLAHE. Our findings confirmed that CLAHE markedly improved image enhancement, making EUS image features more discernible and improving hierarchical differentiation, thereby becoming the preferred method for subsequent image processing.
For AI-based EUS diagnosis of pancreatic cancer, we refined the Faster R-CNN model to include anchor boxes, classification, and regression branches, which improved the algorithm's ability to discern high-level features and distinguish between foreground and background, ultimately enhancing object detection. Adoption of the ResNet-50 backbone for feature extraction addressed the issue of blurry and unclear image edges, which are prevalent in EUS images 14 . Its residual structure helps mitigate degradation problems associated with increased network depth. Integrating a FFM and a RFB further enhanced feature representation and aligned the model more closely with human perceptual processes. The use of ROI Align over ROI Pooling, which employs bilinear interpolation to handle floating-point coordinates, helped avoid misalignment issues, culminating in a sophisticated M-F-RCNN model.
AI has seen promising research in EUS diagnosis of pancreatic cancer. For instance, Kim et al. demonstrated the efficacy of four deep learning models in segmenting pancreatic cystic lesions, with several models showing excellent results 25 . Kuwahara et al. distinguished between pancreatic and non-pancreatic cancer with high sensitivity and specificity using a CNN 10 . A meta-analysis reported combined sensitivity, specificity, and AUC scores of 93%, 90%, and 0.95, respectively 26 . In this study, the M-F-RCNN model identified pancreatic cancer with sensitivity, specificity, and accuracy of 91.7%, 91.5%, and 91.6%, respectively, and compared with the diagnostic capabilities of EUS physicians, the indicators were comparable or even superior, proving the model's diagnostic capability has the potential for clinical use.
Although AI alone may not replace diagnostic processes, it can significantly enhance accuracy of clinical diagnoses. For example, a DLR model distinguishing between pancreatic cancer and chronic pancreatitis provided AI scores and highlighted key areas, improving physician diagnostic sensitivity without compromising specificity 27 . In our study, model-assisted improvements in diagnostic AUC, sensitivity, specificity, and accuracy were substantial, particularly in bridging the gap between novice and expert physicians. Given the growing demand for EUS examinations and the lengthy training required for EUS physicians, this model offers substantial support, reducing the likelihood of diagnostic errors and fostering novice physician development.
However, this study has limitations. The external validation performance of M-F-RCNN was slightly below internal test results, potentially due to limited diversity in the training dataset. Clinically, distinguishing pancreatic cancer from other conditions like solid pancreatic inflammations or neuroendocrine tumors remains challenging. Plans include expanding the training dataset to cover a broader range of conditions and conducting subgroup analyses. Although the model achieved impressive single-image recognition speeds, video validation was not included in this study; thus, future clinical applications should ideally involve prospective video validation to fully harness the capabilities of the new model.
Footnotes
Conflict of Interest The authors declare that they have no conflict of interest.
References
- 1.Sung H, Ferlay J, Siegel RL et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Khalaf N, El-Serag HB, Abrams HR et al. Burden of pancreatic cancer: from epidemiology to practice. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc. 2021;19:876–884. doi: 10.1016/j.cgh.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King D, Kamran U, Dosanjh A et al. Rate of pancreatic cancer following a negative endoscopic ultrasound and associated factors. Endoscopy. 2022;54:1053–1061. doi: 10.1055/a-1784-1661. [DOI] [PubMed] [Google Scholar]
- 4.Guo T, Xu T, Zhang S et al. The role of EUS in diagnosing focal autoimmune pancreatitis and differentiating it from pancreatic cancer. Endosc Ultrasound. 2021;10:280. doi: 10.4103/EUS-D-20-00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rana SS. Evaluating the role of endoscopic ultrasound in pancreatitis. Expert Rev Gastroenterol Hepatol. 2022;16:953–965. doi: 10.1080/17474124.2022.2138856. [DOI] [PubMed] [Google Scholar]
- 6.Bhinder B, Gilvary C, Madhukar NS et al. Artificial intelligence in cancer research and precision medicine. Cancer Discov. 2021;11:900–915. doi: 10.1158/2159-8290.CD-21-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greener JG, Kandathil SM, Moffat L et al. A guide to machine learning for biologists. Nat Rev Mol Cell Biol. 2022;23:40–55. doi: 10.1038/s41580-021-00407-0. [DOI] [PubMed] [Google Scholar]
- 8.Gao Q, Yang L, Lu M et al. The artificial intelligence and machine learning in lung cancer immunotherapy. J Hematol OncolJ Hematol Oncol. 2023;16:55. doi: 10.1186/s13045-023-01456-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim M, Chen C, Wang P et al. Detection of ovarian cancer via the spectral fingerprinting of quantum-defect-modified carbon nanotubes in serum by machine learning. Nat Biomed Eng. 2022;6:267–275. doi: 10.1038/s41551-022-00860-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuwahara T, Hara K, Mizuno N et al. Artificial intelligence using deep learning analysis of endoscopic ultrasonography images for the differential diagnosis of pancreatic masses. Endoscopy. 2023;55:140–149. doi: 10.1055/a-1873-7920. [DOI] [PubMed] [Google Scholar]
- 11.Zhu M, Xu C, Yu J et al. Differentiation of pancreatic cancer and chronic pancreatitis using computer-aided diagnosis of endoscopic ultrasound (EUS) Images: A diagnostic test. PLoS ONE. 2013;8:e63820. doi: 10.1371/journal.pone.0063820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karako K, Mihara Y, Arita J et al. Automated liver tumor detection in abdominal ultrasonography with a modified faster region-based convolutional neural networks (Faster R-CNN) architecture. Hepatobiliary Surg Nutr. 2022;11:675–683. doi: 10.21037/hbsn-21-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dollar P, Appel R, Belongie S et al. Fast Feature pyramids for object detection. IEEE Trans Pattern Anal Mach Intell. 2014;36:1532–1545. doi: 10.1109/TPAMI.2014.2300479. [DOI] [PubMed] [Google Scholar]
- 14.Pan J, Sun H, Song Z et al. Dual-resolution dual-path convolutional neural networks for fast object detection. Sensors. 2019;19:3111. doi: 10.3390/s19143111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren S, He K, Girshick R et al. Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell. 2017;39:1137–1149. doi: 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Jiang L, Yang D et al. Urine sediment recognition method based on multi-view deep residual learning in microscopic image. J Med Syst. 2019;43:325. doi: 10.1007/s10916-019-1457-4. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Lv C, Wang W et al. Generalized focal loss: Towards efficient representation learning for dense object detection. IEEE Trans Pattern Anal Mach Intell. 2023;45:3139–3153. doi: 10.1109/TPAMI.2022.3180392. [DOI] [PubMed] [Google Scholar]
- 18.Lin T-Y, Goyal P, Girshick R et al. Focal loss for dense object detection. IEEE Trans Pattern Anal Mach Intell. 2020;42:318–327. doi: 10.1109/TPAMI.2018.2858826. [DOI] [PubMed] [Google Scholar]
- 19.Cao K, Xia Y, Yao J et al. Large-scale pancreatic cancer detection via non-contrast CT and deep learning. Nat Med. 2023;29:3033–3043. doi: 10.1038/s41591-023-02640-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drukker L. Real-time identification of fetal anomalies on ultrasound using artificial intelligence: what’s next? Ultrasound Obstet Gynecol. 2022;59:285–287. doi: 10.1002/uog.24869. [DOI] [PubMed] [Google Scholar]
- 21.Bera K, Braman N, Gupta A et al. Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nat Rev Clin Oncol. 2022;19:132–146. doi: 10.1038/s41571-021-00560-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marya NB, Powers PD, Chari ST et al. Utilisation of artificial intelligence for the development of an EUS-convolutional neural network model trained to enhance the diagnosis of autoimmune pancreatitis. Gut. 2021;70:1335–1344. doi: 10.1136/gutjnl-2020-322821. [DOI] [PubMed] [Google Scholar]
- 23.Yoon H, Zhu YI, Yarmoska SK et al. Design and demonstration of a configurable imaging platform for combined laser, ultrasound, and elasticity imaging. IEEE Trans Med Imaging. 2019;38:1622–1632. doi: 10.1109/TMI.2018.2889736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu X, Liu X, Xiao Z et al. Self-supervised dual-head attentional bootstrap learning network for prostate cancer screening in transrectal ultrasound images. Comput Biol Med. 2023;165:107337. doi: 10.1016/j.compbiomed.2023.107337. [DOI] [PubMed] [Google Scholar]
- 25.Oh S, Kim Y-J, Park Y-T et al. Automatic pancreatic cyst lesion segmentation on EUS images using a deep-learning approach. Sensors. 2021;22:245. doi: 10.3390/s22010245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin H, Yang X, Sun L et al. The value of artificial intelligence techniques in predicting pancreatic ductal adenocarcinoma with EUS images: A meta-analysis and systematic review. Endosc Ultrasound. 2023;12:50–58. doi: 10.4103/EUS-D-21-00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong T, Gu J, Xu D et al. Deep learning radiomics based on contrast-enhanced ultrasound images for assisted diagnosis of pancreatic ductal adenocarcinoma and chronic pancreatitis. BMC Med. 2022;20:74. doi: 10.1186/s12916-022-02258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]