Abstract
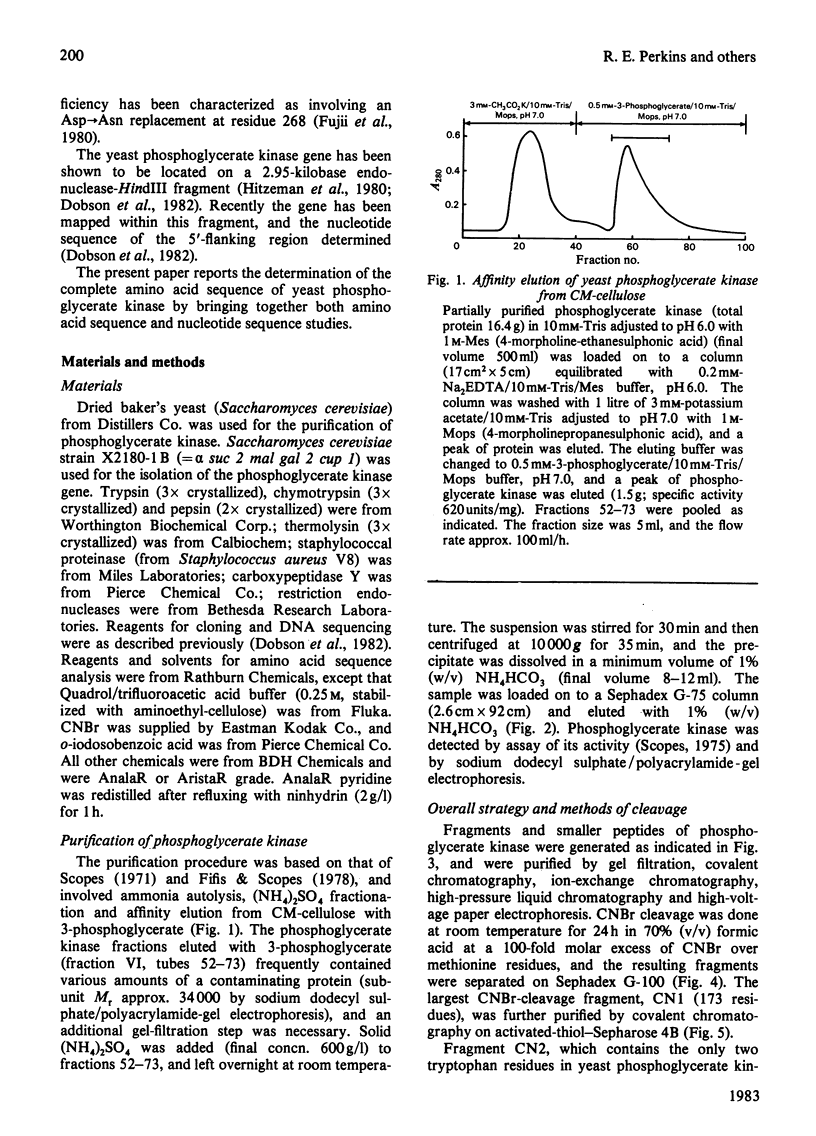
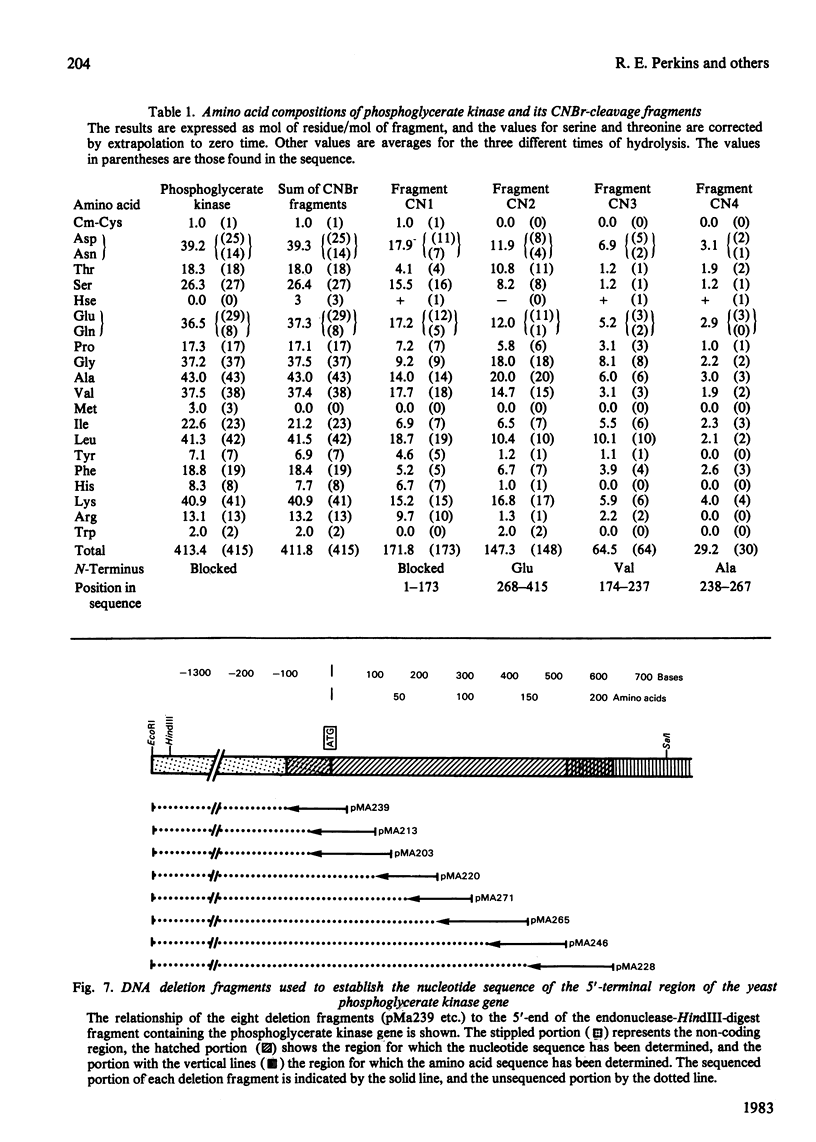
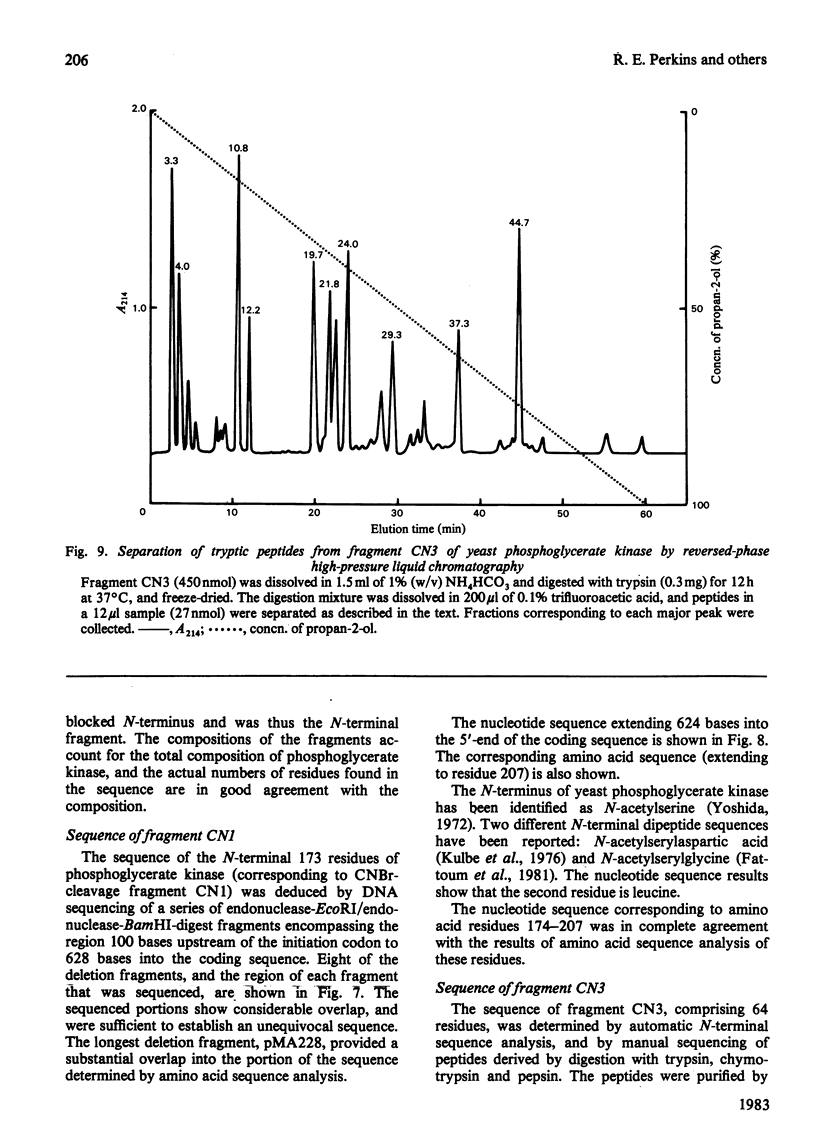
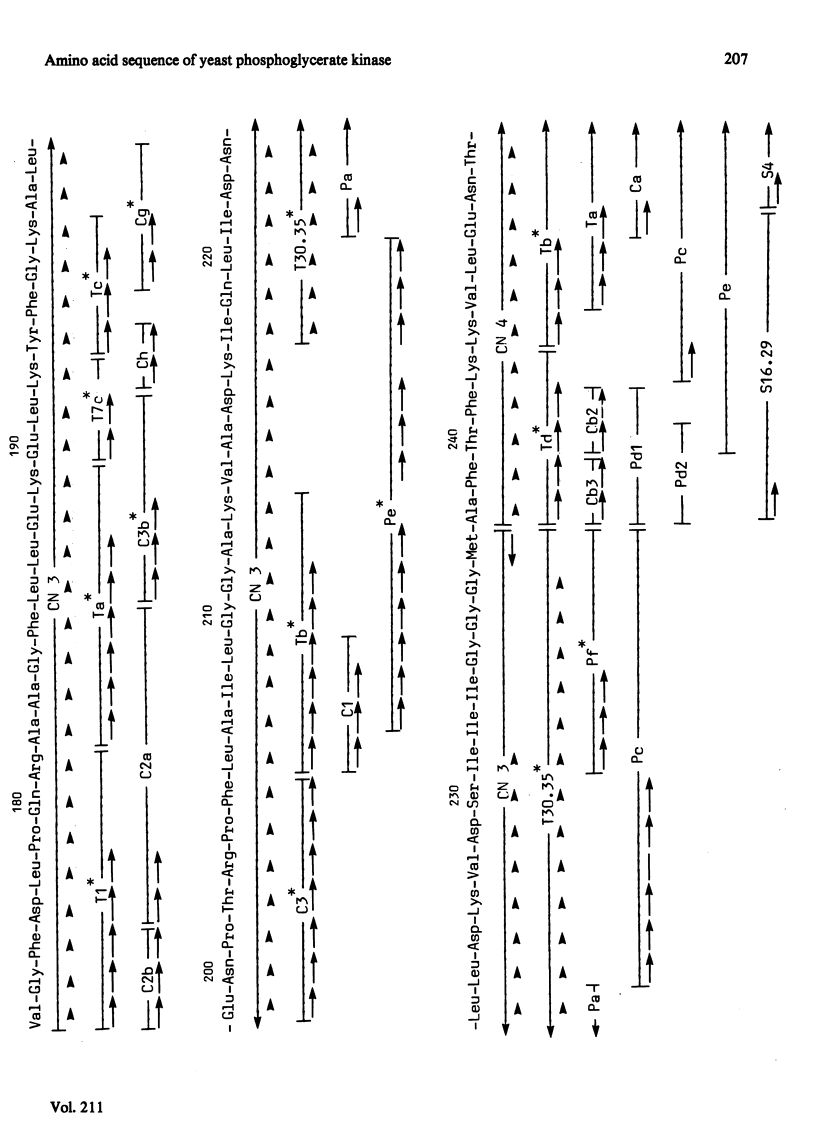
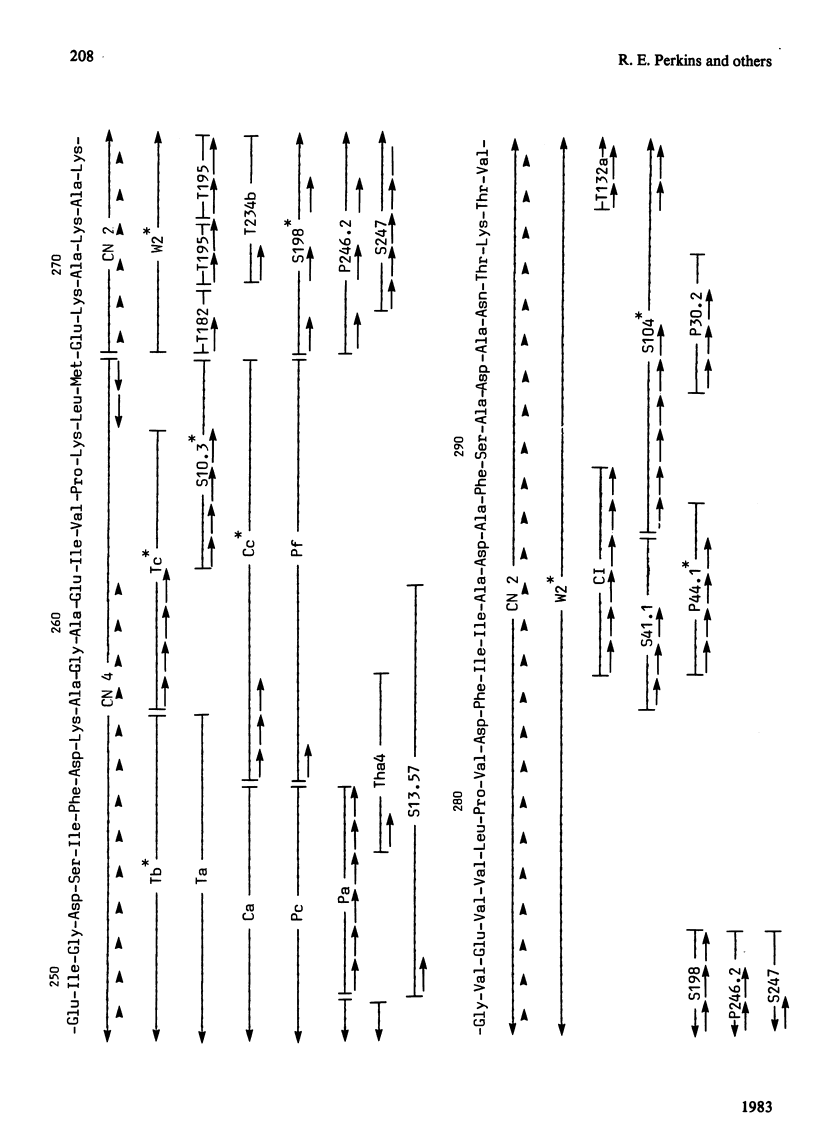
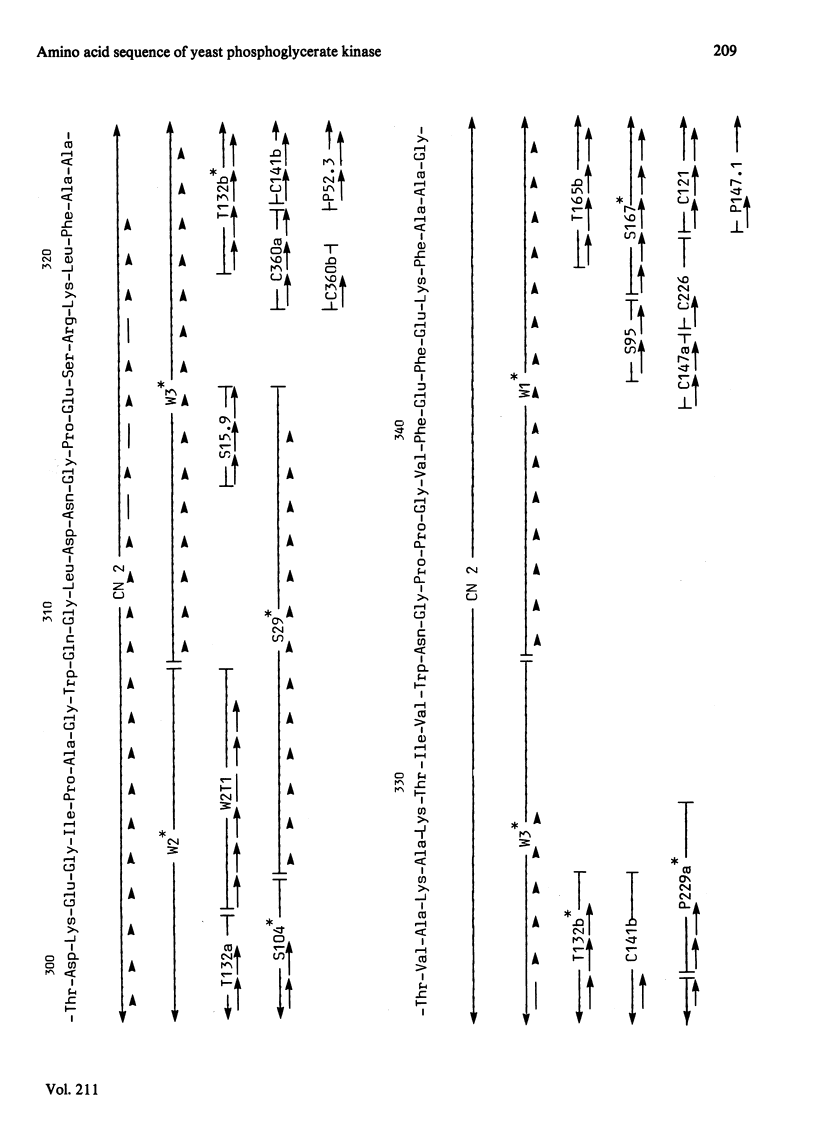
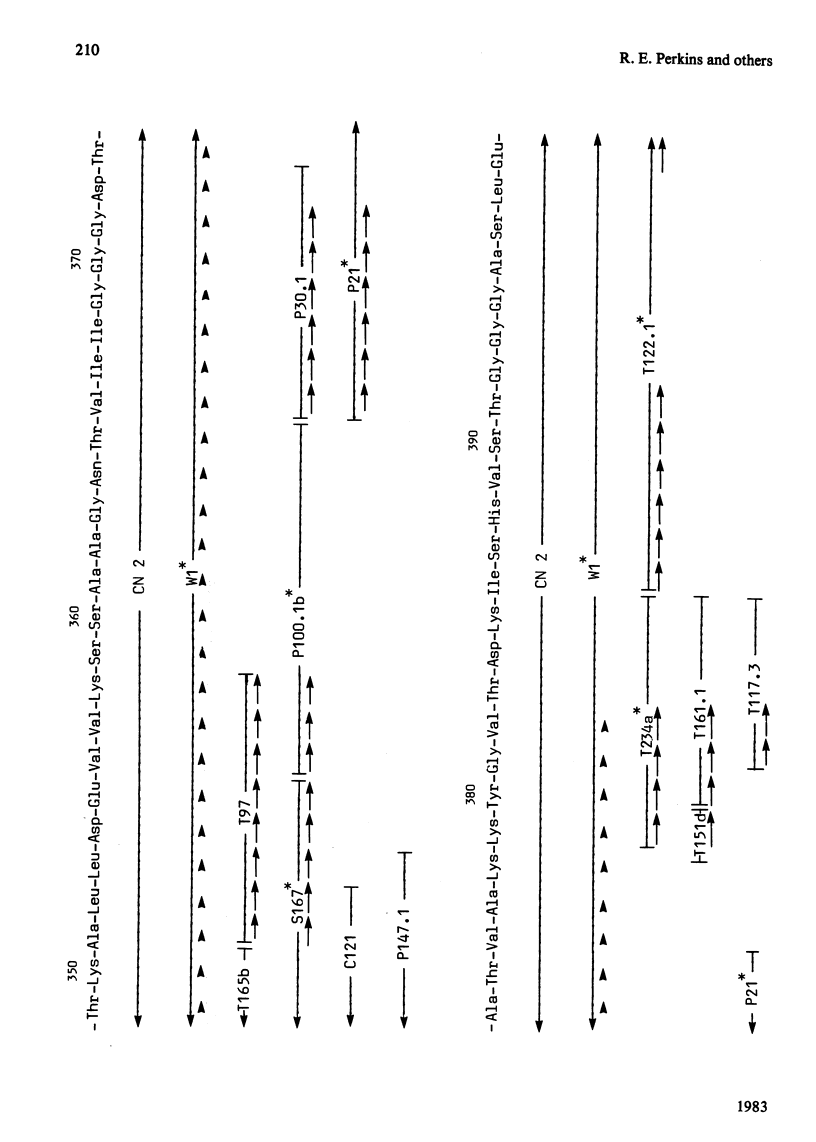
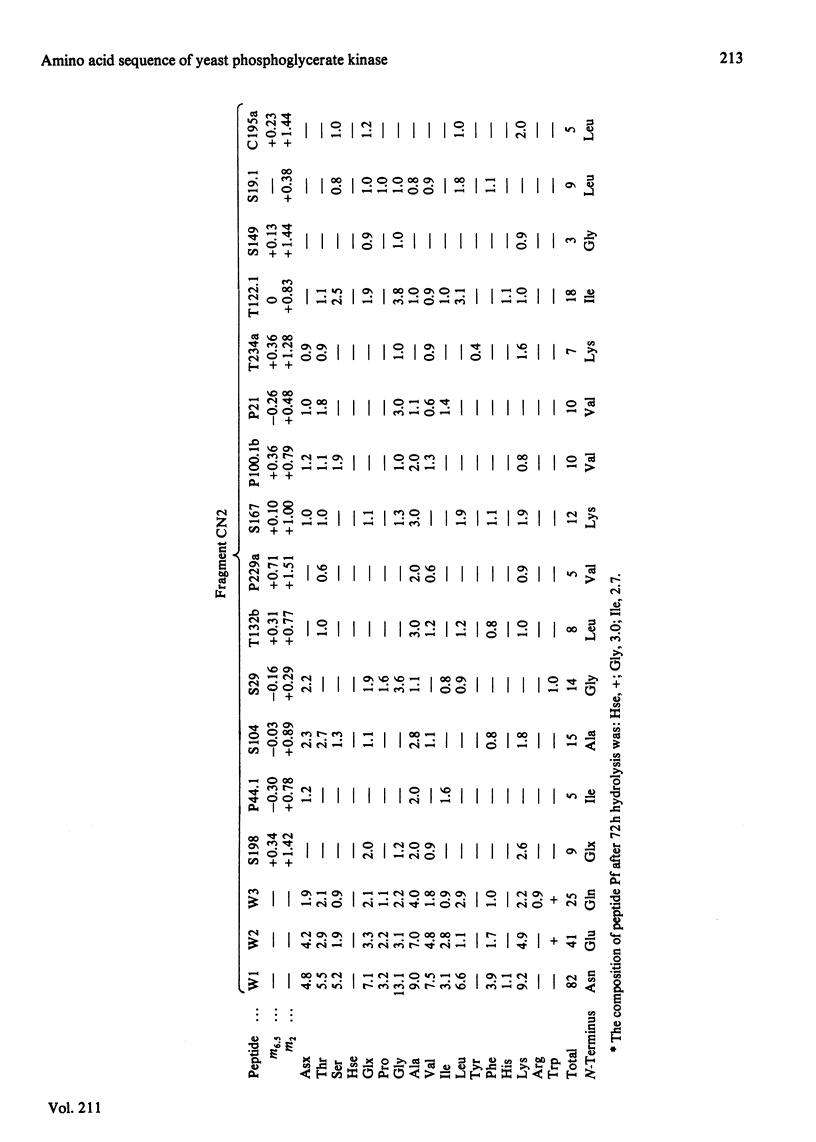
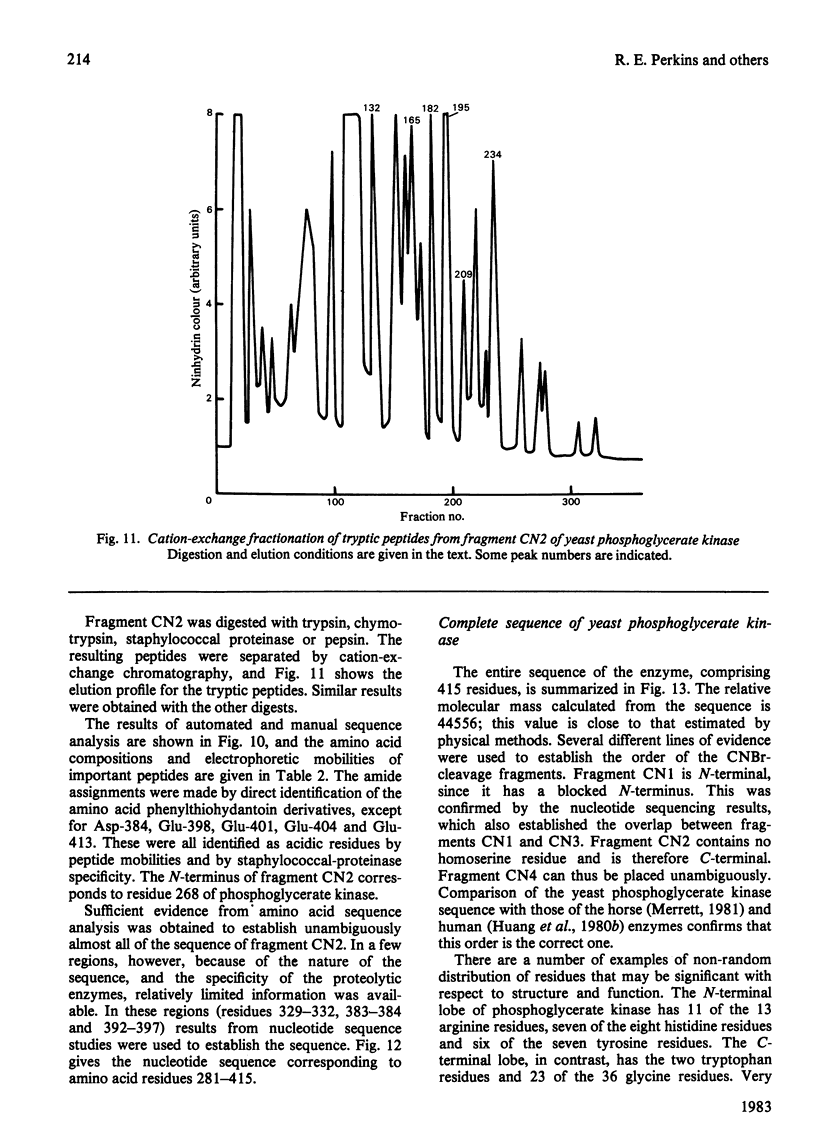
The complete amino acid sequence of yeast phosphoglycerate kinase, comprising 415 residues, was determined. The sequence of residues 1-173 was deduced mainly from nucleotide sequence analysis of a series of overlapping fragments derived from the relevant portion of a 2.95-kilobase endonuclease-HindIII-digest fragment containing the yeast phosphoglycerate kinase gene. The sequence of residues 174-415 was deduced mainly from amino acid sequence analysis of three CNBr-cleavage fragments, and from peptides derived from these fragments after digestion by a number of proteolytic enzymes. Cleavage at the two tryptophan residues with o-iodosobenzoic acid was also used to isolate fragments suitable for amino acid sequence analysis. Determination of the complete sequence now allows a detailed interpretation of the existing high-resolution X-ray-crystallographic structure. The sequence -Ile-Ile-Gly-Gly-Gly- occurs twice in distant parts of the linear sequence (residues 232-236 and 367-371). Both these regions contribute to the nucleoside phosphate-binding site. A comparison of the sequence of yeast phosphoglycerate kinase reported here with the sequences of phosphoglycerate kinase from horse muscle and human erythrocytes shows that the yeast enzyme is 64% identical with the mammalian enzymes. The yeast has strikingly fewer methionine, cysteine and tryptophan residues.
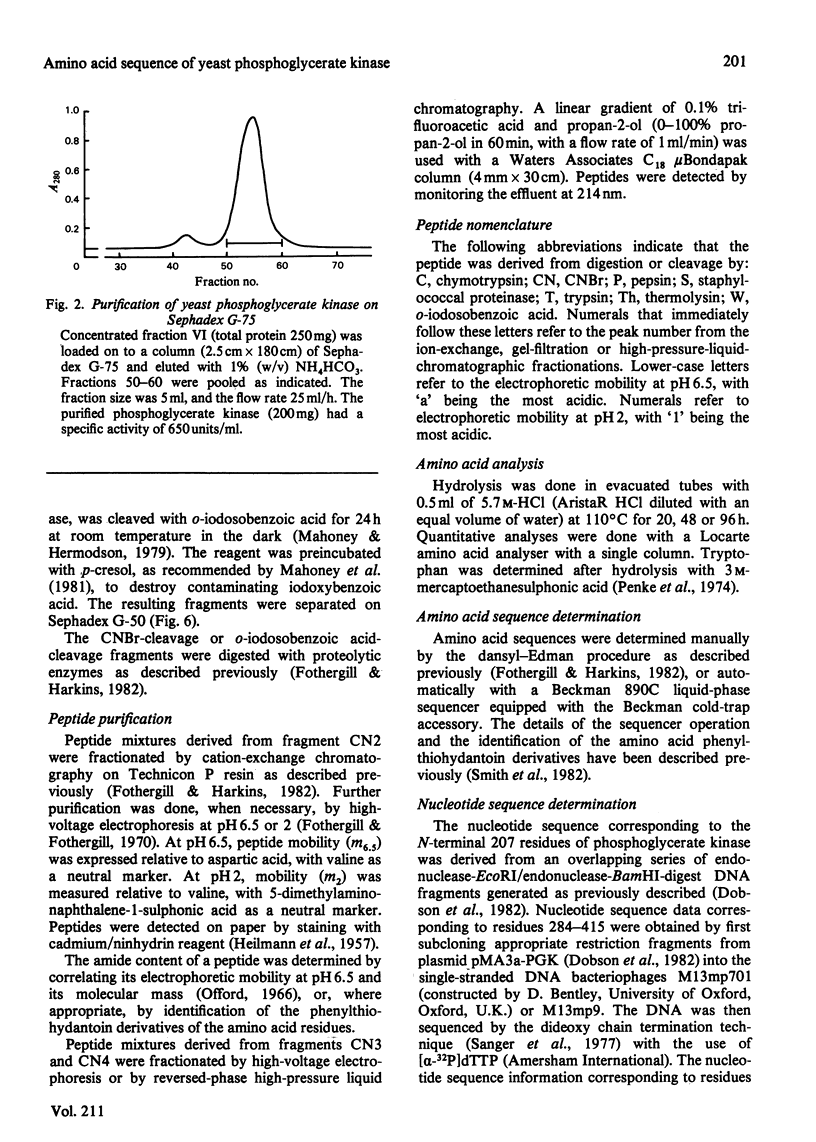
Full text
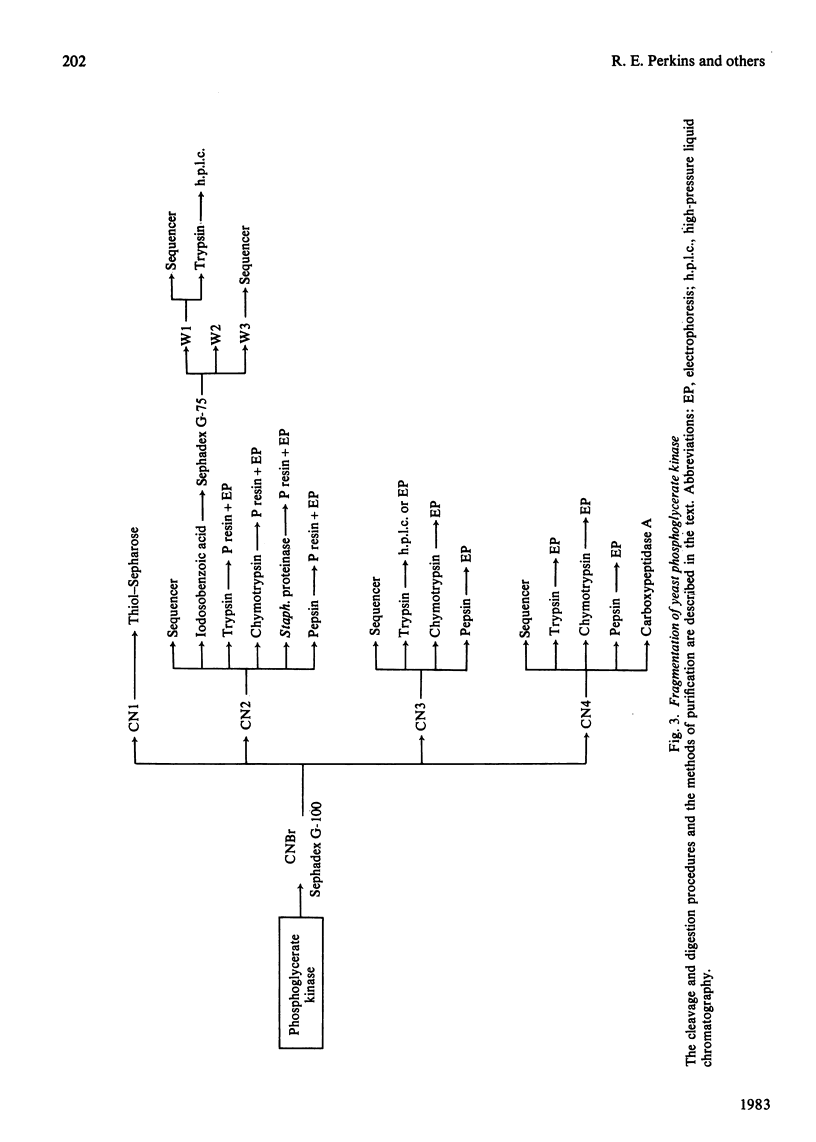
PDF







Selected References
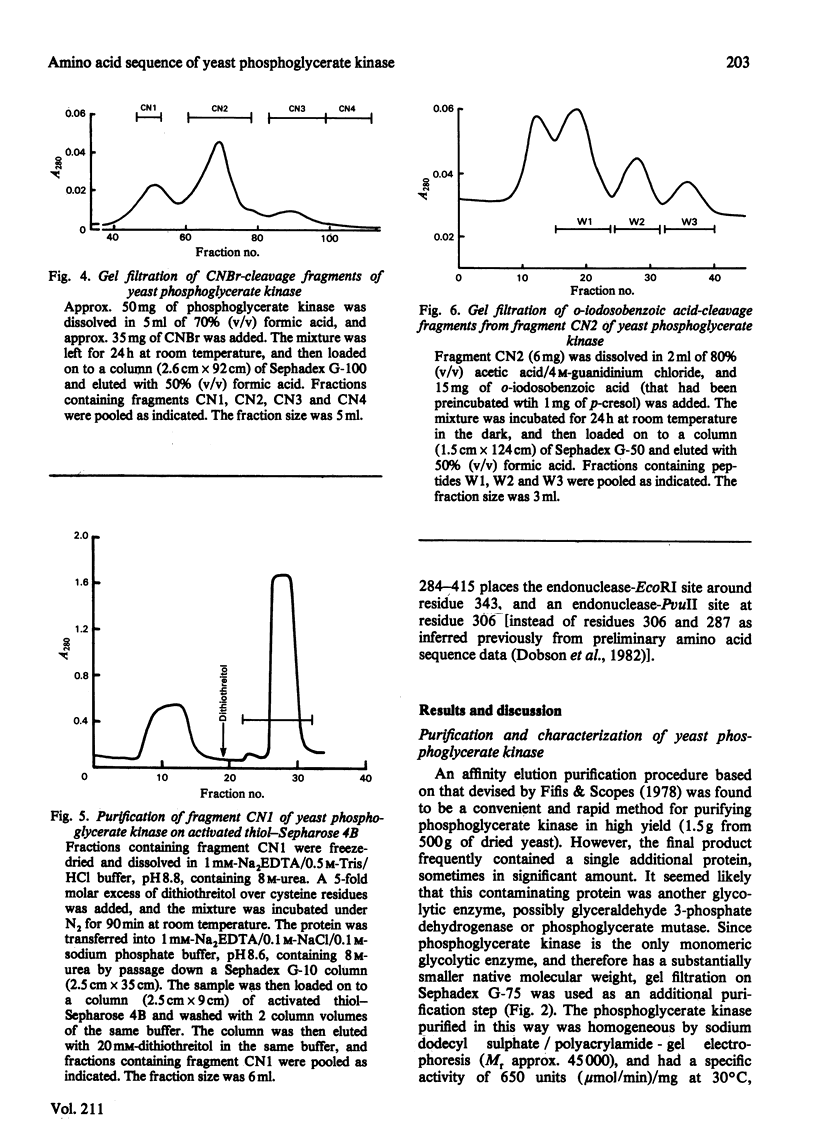
These references are in PubMed. This may not be the complete list of references from this article.
- Bacharach A. D., Markland F. S., Pellino A., Weber B. H. Modification of yeast 3-phosphoglycerate kinase: isolation and sequence determination of a nitrated active-site peptide and isolation of a carboxyl modified active-site peptide. Biochem Biophys Res Commun. 1977 Jan 10;74(1):165–171. doi: 10.1016/0006-291x(77)91389-4. [DOI] [PubMed] [Google Scholar]
- Banks R. D., Blake C. C., Evans P. R., Haser R., Rice D. W., Hardy G. W., Merrett M., Phillips A. W. Sequence, structure and activity of phosphoglycerate kinase: a possible hinge-bending enzyme. Nature. 1979 Jun 28;279(5716):773–777. doi: 10.1038/279773a0. [DOI] [PubMed] [Google Scholar]
- Brevet A., Roustan C., Desvages G., Pradel L. A., van Thoai N. Yeast 3-phosphoglycerate kinase: evidence for a glutamyl residue in the phosphoryl transfer. Eur J Biochem. 1973 Nov 1;39(1):141–147. doi: 10.1111/j.1432-1033.1973.tb03112.x. [DOI] [PubMed] [Google Scholar]
- Bryant T. N., Watson H. C., Wendell P. L. Structure of yeast phosphoglycerate kinase. Nature. 1974 Jan 4;247(5435):14–17. doi: 10.1038/247014a0. [DOI] [PubMed] [Google Scholar]
- Burgess R. J., Pain R. H. The reversible unfolding of yeast 3-phosphoglycerate kinase [proceedings]. Biochem Soc Trans. 1977;5(3):692–694. doi: 10.1042/bst0050692. [DOI] [PubMed] [Google Scholar]
- Desvages G., Roustan C., Fattoum A., Pradel L. A. Structural studies on yeast 3-phosphoglycerate kinase. Identification by immuno-affinity chromatography of one glutamyl residue essential for yeast 3-phosphoglycerate kinase activity. Its location in the primary structure. Eur J Biochem. 1980 Apr;105(2):259–266. doi: 10.1111/j.1432-1033.1980.tb04496.x. [DOI] [PubMed] [Google Scholar]
- Dobson M. J., Tuite M. F., Roberts N. A., Kingsman A. J., Kingsman S. M., Perkins R. E., Conroy S. C., Fothergill L. A. Conservation of high efficiency promoter sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1982 Apr 24;10(8):2625–2637. doi: 10.1093/nar/10.8.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattoum A., Roustan C., Feinberg J., Desvages G., Pradel L. A. Structural studies on yeast 3-phosphoglycerate kinase. Isolation by affinity chromatography and characterization of the peptides produced by cyanogen bromide cleavage. Location of the single cysteinyl residue in the primary structure. Eur J Biochem. 1978 Jan 2;82(1):161–167. doi: 10.1111/j.1432-1033.1978.tb12007.x. [DOI] [PubMed] [Google Scholar]
- Fattoum A., Roustan C., Karoui D., Feinberg J., Pradel L. A., Gregoire J., Rochat H. Structural studies on yeast 3-phosphoglycerate kinase. Linear arrangement of the CNBr fragments, partial amino acid sequence of the inner part of the polypeptide chain, and analyses of the N-terminal domain of the protein. Int J Pept Protein Res. 1981 Mar;17(3):393–400. doi: 10.1111/j.1399-3011.1981.tb02006.x. [DOI] [PubMed] [Google Scholar]
- Fifis T., Scopes R. K. Purification of 3-phosphoglycerate kinase from diverse sources by affinity elution chromatography. Biochem J. 1978 Oct 1;175(1):311–319. doi: 10.1042/bj1750311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fothergill L. A., Fothergill J. E. Structural comparison of ovalbumins from nine different species. Eur J Biochem. 1970 Dec;17(3):529–532. doi: 10.1111/j.1432-1033.1970.tb01196.x. [DOI] [PubMed] [Google Scholar]
- Fothergill L. A., Harkins R. N. The amino acid sequence of yeast phosphoglycerate mutase. Proc R Soc Lond B Biol Sci. 1982 Apr 22;215(1198):19–44. doi: 10.1098/rspb.1982.0026. [DOI] [PubMed] [Google Scholar]
- Fujii H., Krietsch W. K., Yoshida A. A single amino acid substitution (Asp leads to Asn) in a phosphoglycerate kinase variant (PGK München) associated with enzyme deficiency. J Biol Chem. 1980 Jul 10;255(13):6421–6423. [PubMed] [Google Scholar]
- HEILMANN J., BARROLLIER J., WATZKE E. Beitrag zur Aminosäurebestimmung auf Papierchromatogrammen. Hoppe Seylers Z Physiol Chem. 1957;309(4-6):219–220. [PubMed] [Google Scholar]
- Hardy G. W., Darbre A., Merrett M. Primary structure of 3-phosphoglycerate kinase from horse muscle. I. Purification of cyanogen bromide peptides and amino acid sequence of peptide CB5 (104 residues). J Biol Chem. 1981 Oct 25;256(20):10284–10292. [PubMed] [Google Scholar]
- Hitzeman R. A., Clarke L., Carbon J. Isolation and characterization of the yeast 3-phosphoglycerokinase gene (PGK) by an immunological screening technique. J Biol Chem. 1980 Dec 25;255(24):12073–12080. [PubMed] [Google Scholar]
- Hjelmgren T., Arvidsson L., Larsson-Raźnikiewicz M. Spectrophotometric pH titrations and nitration with tetranitromethane of the tyrosyl residues in yeast phosphoglycerate kinase. Biochim Biophys Acta. 1976 Sep 14;445(2):342–349. doi: 10.1016/0005-2744(76)90088-7. [DOI] [PubMed] [Google Scholar]
- Huang I. Y., Rubinfien E., Yoshida A. Complete amino acid sequence of human phosphoglycerate kinase. Isolation and amino acid sequence of tryptic peptides. J Biol Chem. 1980 Jul 10;255(13):6408–6411. [PubMed] [Google Scholar]
- Huang I. Y., Welch C. D., Yoshida A. Complete amino acid sequence of human phosphoglycerate kinase. Cyanogen bromide peptides and complete amino acid sequence. J Biol Chem. 1980 Jul 10;255(13):6412–6420. [PubMed] [Google Scholar]
- Larsson-Raźnikiewicz M., Jansson J. R. Activation of yeast phosphoglycerate kinase by salts of monovalent cations. FEBS Lett. 1973 Feb 1;29(3):345–347. doi: 10.1016/0014-5793(73)80055-9. [DOI] [PubMed] [Google Scholar]
- Mahoney W. C., Hermodson M. A. High-yield cleavage of tryptophanyl peptide bonds by o-iodosobenzoic acid. Biochemistry. 1979 Aug 21;18(17):3810–3814. doi: 10.1021/bi00584a026. [DOI] [PubMed] [Google Scholar]
- Markland F. S., Bacharach A. D., Weber B. H., O'Grady T. C., Saunders G. C., Umemura N. Chemical modification of yeast 3-phosphoglycerate kinase. J Biol Chem. 1975 Feb 25;250(4):1301–1310. [PubMed] [Google Scholar]
- Offord R. E. Electrophoretic mobilities of peptides on paper and their use in the determination of amide groups. Nature. 1966 Aug 6;211(5049):591–593. doi: 10.1038/211591a0. [DOI] [PubMed] [Google Scholar]
- Penke B., Ferenczi R., Kovács K. A new acid hydrolysis method for determining tryptophan in peptides and proteins. Anal Biochem. 1974 Jul;60(1):45–50. doi: 10.1016/0003-2697(74)90129-8. [DOI] [PubMed] [Google Scholar]
- Philips M., Roustan C., Fattoum A., Pradel L. A. Yeast 3-phosphoglycerate kinase. Essential arginyl residues at the 3-phosphoglycerate binding site. Biochim Biophys Acta. 1978 Apr 12;523(2):368–376. doi: 10.1016/0005-2744(78)90039-6. [DOI] [PubMed] [Google Scholar]
- Pickover C. A., McKay D. B., Engelman D. M., Steitz T. A. Substrate binding closes the cleft between the domains of yeast phosphoglycerate kinase. J Biol Chem. 1979 Nov 25;254(22):11323–11329. [PubMed] [Google Scholar]
- Rogers K., Weber B. H. Evidence for an essential role for arginyl residues for yeast phosphoglycerate kinase. Arch Biochem Biophys. 1977 Apr 15;180(1):19–25. doi: 10.1016/0003-9861(77)90003-0. [DOI] [PubMed] [Google Scholar]
- Roustan C., Fattoum A., Jeanneau R., Pradel L. A. Yeast 3-phosphoglycerate kinase: sulfate and substrate binding, their effect on the conformational state of the enzyme. Biochemistry. 1980 Nov 11;19(23):5168–5175. doi: 10.1021/bi00564a003. [DOI] [PubMed] [Google Scholar]
- Roustan C., Fattoum A., Pradel L. A. Physicochemical and kinetic properties of iodinated yeast 3-phosphoglycerate kinase. Biochemistry. 1976 May 18;15(10):2172–2177. doi: 10.1021/bi00655a022. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes R. K. 3-phosphoglycerate kinase of skeletal muscle. Methods Enzymol. 1975;42:127–134. doi: 10.1016/0076-6879(75)42105-x. [DOI] [PubMed] [Google Scholar]
- Scopes R. K. An improved procedure for the isolation of 3-phosphoglycerate kinase from yeast. Biochem J. 1971 Mar;122(1):89–92. doi: 10.1042/bj1220089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes R. K. The steady-state kinetics of yeast phosphoglycerate kinase. Anomalous kinetic plots and the effects of salts on activity. Eur J Biochem. 1978 Apr 17;85(2):503–516. doi: 10.1111/j.1432-1033.1978.tb12266.x. [DOI] [PubMed] [Google Scholar]
- Smith M. A., Gerrie L. M., Dunbar B., Fothergill J. E. Primary structure of bovine complement activation fragment C4a, the third anaphylatoxin. Purification and complete amino acid sequence. Biochem J. 1982 Nov 1;207(2):253–260. doi: 10.1042/bj2070253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanswell P., Westhead E. W., Williams R. J. Nuclear-magnetic-resonance study of the active-site structure of yeast phosphoglycerate kinase. Eur J Biochem. 1976 Mar 16;63(1):249–262. doi: 10.1111/j.1432-1033.1976.tb10227.x. [DOI] [PubMed] [Google Scholar]
- Watson H. C., Walker N. P., Shaw P. J., Bryant T. N., Wendell P. L., Fothergill L. A., Perkins R. E., Conroy S. C., Dobson M. J., Tuite M. F. Sequence and structure of yeast phosphoglycerate kinase. EMBO J. 1982;1(12):1635–1640. doi: 10.1002/j.1460-2075.1982.tb01366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A. Micro method for determination of blocked NH 2 - terminal amino acids of protein: application to identification of acetylserine of phosphoglycerate kinase and pyroglutamic acid of glucose 6-phosphate dehydrogenase. Anal Biochem. 1972 Oct;49(2):320–325. doi: 10.1016/0003-2697(72)90434-4. [DOI] [PubMed] [Google Scholar]


