Abstract
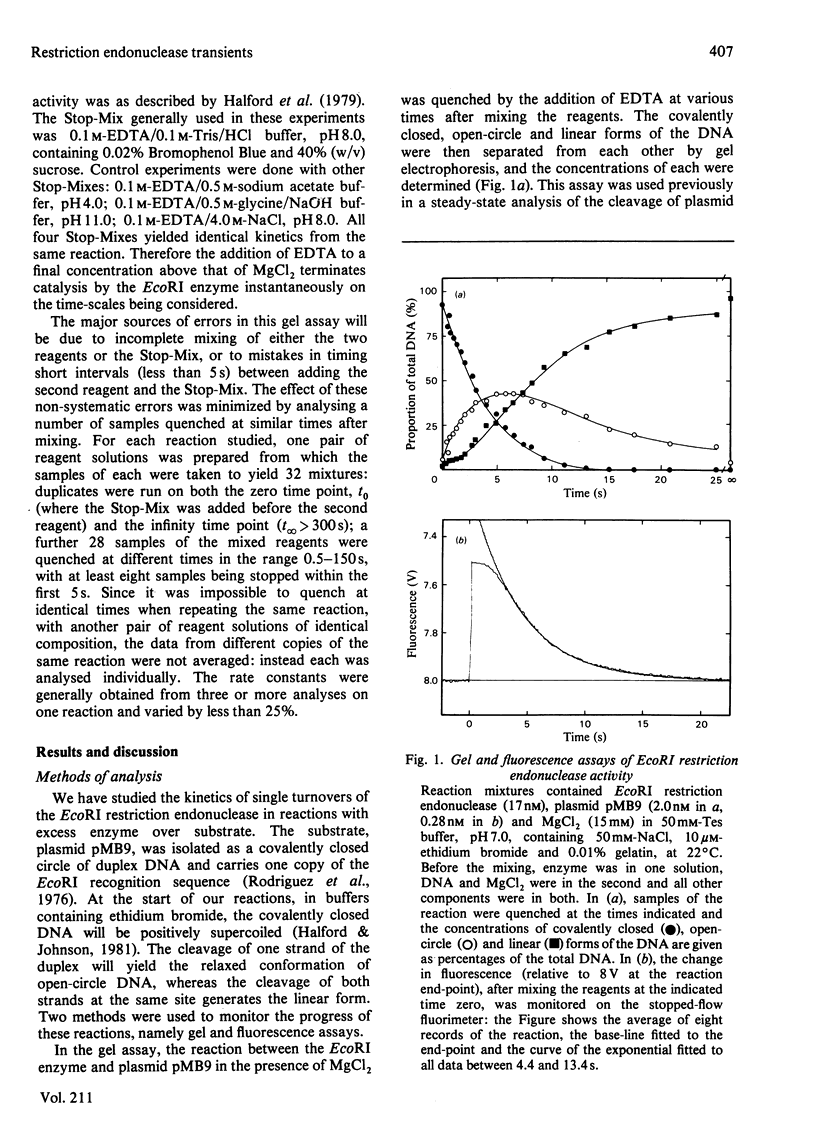
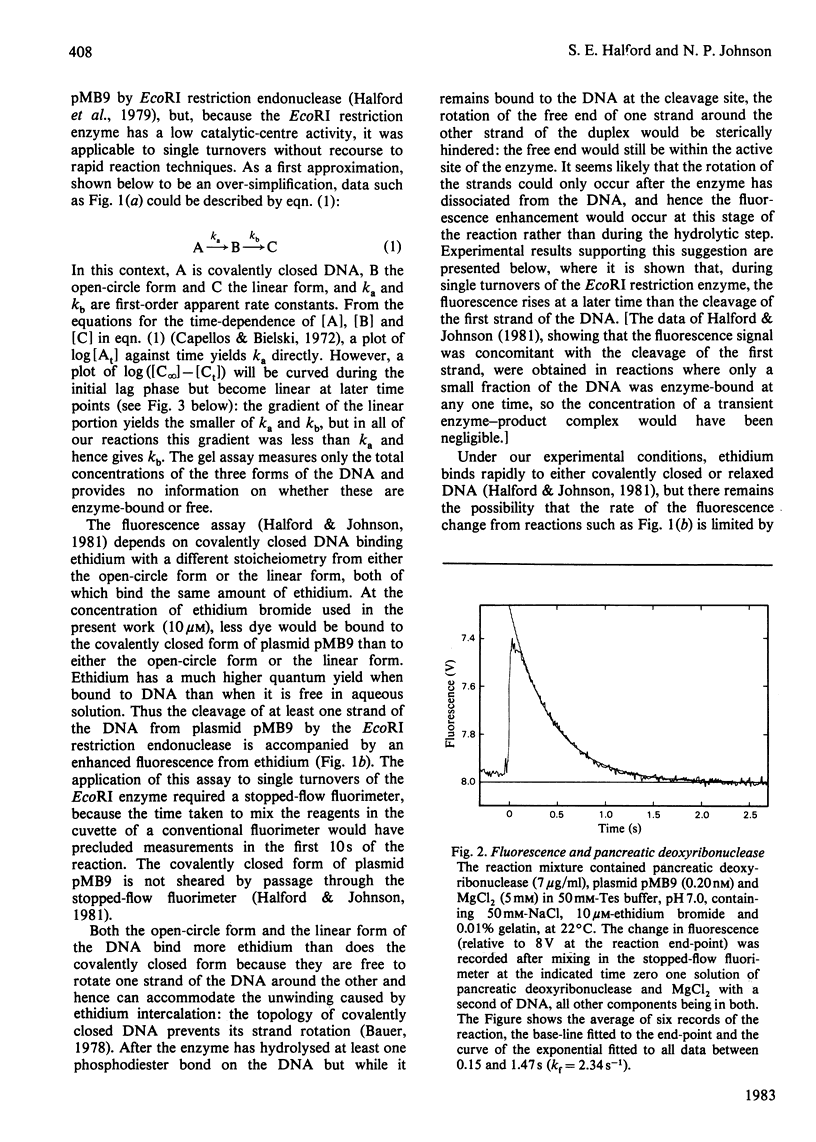
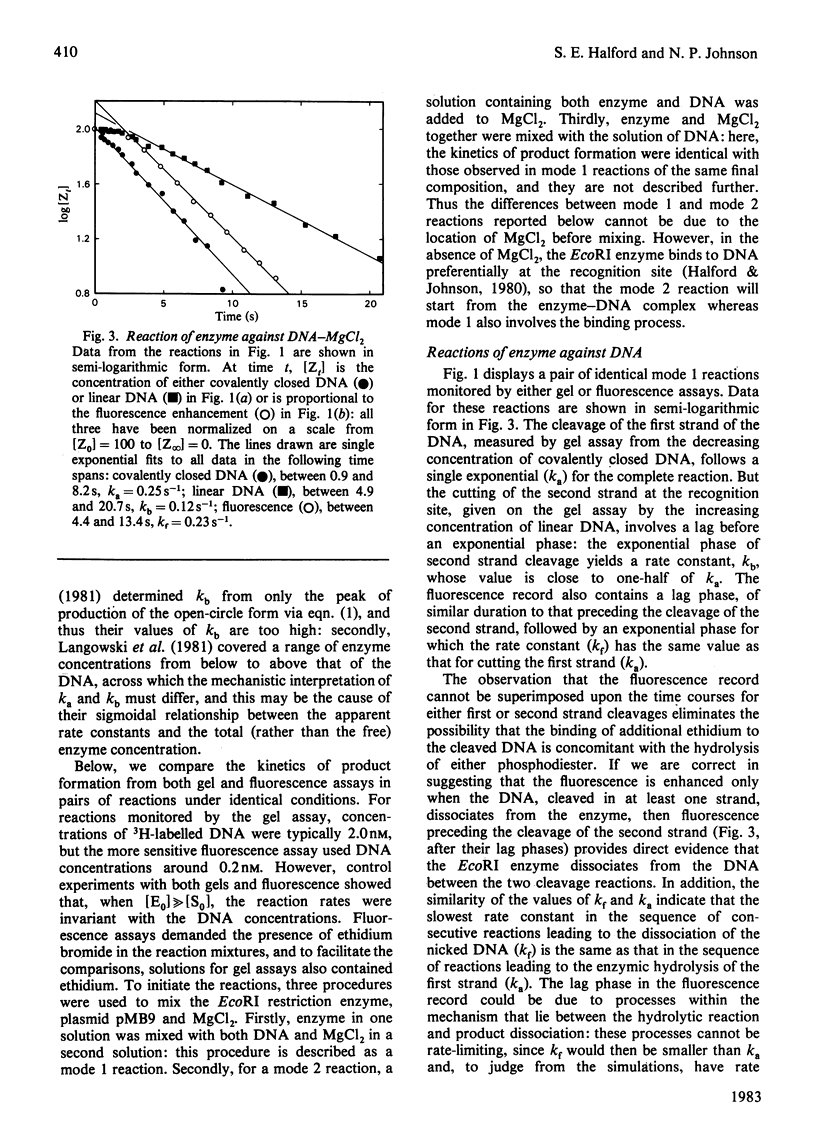
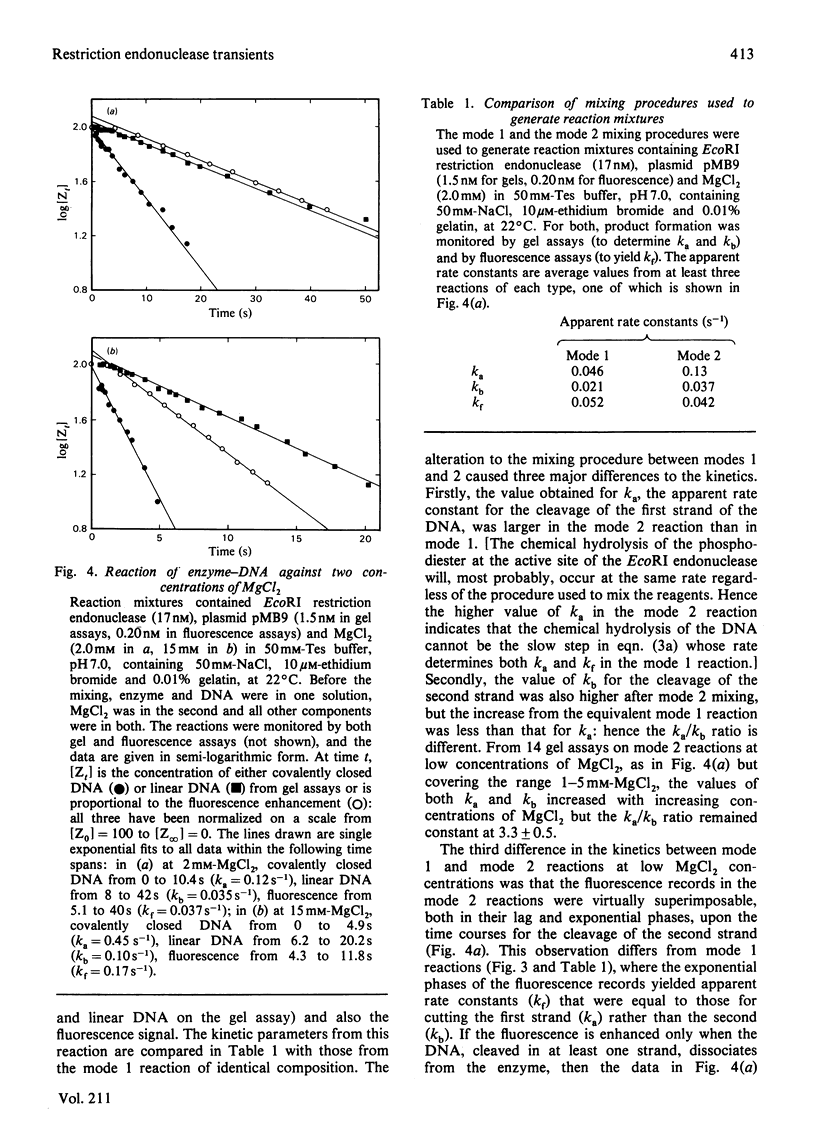
Single turnovers of the EcoRI restriction endonuclease, cleaving its recognition site on the covalently closed form of plasmid pMB9, were examined. Two methods were used to monitor the progress of the reactions: one involved quenching the reaction at various times followed by the electrophoretic separation of the products cleaved in one and in both strands of the duplex; the other employed a stopped-flow fluorimeter to measure the amount of ethidium bromide bound to the DNA as it changes when the DNA, cleaved in at least one strand, dissociates from the enzyme. Two procedures were used to initiate the reactions. For some, one solution containing the enzyme was mixed with a second containing both DNA and MgCl2: in these reactions, the fluorescence changed at the same rate as the cleavage of the first strand of the duplex. Other reactions were started by the addition of MgCl2 to a pre-equilibrium of enzyme and DNA: here, both strands of the DNA were cleaved faster than before, with the fluorescence signal now occurring at the same time as the cleavage of the second strand. The different kinetics from the two assays and the two mixing procedures are consistent with the rates of these reactions being controlled by protein conformational changes. These may affect either one subunit alone within the dimeric EcoRI enzyme, allowing the enzyme to cleave only one strand of the DNA in each turnover. Alternatively, both subunits of the dimer may change, so that the enzyme then cleaves both strands during the life-time of one enzyme-DNA complex.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer W. R. Structure and reactions of closed duplex DNA. Annu Rev Biophys Bioeng. 1978;7:287–313. doi: 10.1146/annurev.bb.07.060178.001443. [DOI] [PubMed] [Google Scholar]
- Berg O. G., Winter R. B., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981 Nov 24;20(24):6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- Campbell V. W., Jackson D. A. The effect of divalent cations on the mode of action of DNase I. The initial reaction products produced from covalently closed circular DNA. J Biol Chem. 1980 Apr 25;255(8):3726–3735. [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M., Davies R. W. Theoretical aspects of specific and non-specific equilibrium binding of proteins to DNA as studied by the nitrocellulose filter binding assay. Co-operative and non-co-operative binding to a one-dimensional lattice. J Mol Biol. 1982 Mar 15;155(4):447–466. doi: 10.1016/0022-2836(82)90481-8. [DOI] [PubMed] [Google Scholar]
- FUORTES M. G., HODGKIN A. L. CHANGES IN TIME SCALE AND SENSITIVITY IN THE OMMATIDIA OF LIMULUS. J Physiol. 1964 Aug;172:239–263. doi: 10.1113/jphysiol.1964.sp007415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goppelt M., Pingoud A., Maass G., Mayer H., Köster H., Frank R. The interaction of the EcoRI restriction endonuclease with its substrate. A physico-chemical study employing natural and synthetic oligonucleotides and polynucleotides. Eur J Biochem. 1980 Feb;104(1):101–107. doi: 10.1111/j.1432-1033.1980.tb04405.x. [DOI] [PubMed] [Google Scholar]
- Greene P. J., Gupta M., Boyer H. W., Brown W. E., Rosenberg J. M. Sequence analysis of the DNA encoding the Eco RI endonuclease and methylase. J Biol Chem. 1981 Mar 10;256(5):2143–2153. [PubMed] [Google Scholar]
- Halford S. E., Johnson N. P., Grinsted J. The EcoRI restriction endonuclease with bacteriophage lambda DNA. Kinetic studies. Biochem J. 1980 Nov 1;191(2):581–592. doi: 10.1042/bj1910581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S. E., Johnson N. P., Grinsted J. The reactions of the EcoRi and other restriction endonucleases. Biochem J. 1979 May 1;179(2):353–365. doi: 10.1042/bj1790353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S. E., Johnson N. P. The EcoRI restriction endonuclease with bacteriophage lambda DNA. Equilibrium binding studies. Biochem J. 1980 Nov 1;191(2):593–604. doi: 10.1042/bj1910593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S. E., Johnson N. P. The EcoRI restriction endonuclease, covalently closed DNA and ethidium bromide. Biochem J. 1981 Dec 1;199(3):767–777. doi: 10.1042/bj1990767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgpeth J., Goodman H. M., Boyer H. W. DNA nucleotide sequence restricted by the RI endonuclease. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3448–3452. doi: 10.1073/pnas.69.11.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imber R., Bickle T. A. Purification and properties of the restriction endonuclease BglII from Bacillus globigii. Eur J Biochem. 1981 Jul;117(2):395–399. doi: 10.1111/j.1432-1033.1981.tb06351.x. [DOI] [PubMed] [Google Scholar]
- Jack W. E., Terry B. J., Modrich P. Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4010–4014. doi: 10.1073/pnas.79.13.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Smith H. O. A restriction enzyme from Hemophilus influenzae. II. J Mol Biol. 1970 Jul 28;51(2):393–409. doi: 10.1016/0022-2836(70)90150-6. [DOI] [PubMed] [Google Scholar]
- Langowski J., Pingoud A., Goppelt M., Maass G. Inhibition of Eco RI action by polynucleotides. A characterization of the non-specific binding of the enzyme to DNA. Nucleic Acids Res. 1980 Oct 24;8(20):4727–4736. doi: 10.1093/nar/8.20.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langowski J., Urbanke C., Pingoud A., Maass G. Transient cleavage kinetics of the Eco RI restriction endonuclease measured in a pulsed quench-flow apparatus: enzyme concentration-dependent activity change. Nucleic Acids Res. 1981 Jul 24;9(14):3483–3490. doi: 10.1093/nar/9.14.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Jack W. E., Modrich P. DNA determinants important in sequence recognition by Eco RI endonuclease. J Biol Chem. 1981 Dec 25;256(24):13200–13206. [PubMed] [Google Scholar]
- Maxwell A., Halford S. E. The SalGI restriction endonuclease. Mechanism of DNA cleavage. Biochem J. 1982 Apr 1;203(1):85–92. doi: 10.1042/bj2030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P., Zabel D. EcoRI endonuclease. Physical and catalytic properties of the homogenous enzyme. J Biol Chem. 1976 Oct 10;251(19):5866–5874. [PubMed] [Google Scholar]
- Newman A. K., Rubin R. A., Kim S. H., Modrich P. DNA sequences of structural genes for Eco RI DNA restriction and modification enzymes. J Biol Chem. 1981 Mar 10;256(5):2131–2139. [PubMed] [Google Scholar]


