Abstract
The present study aimed to evaluate the key characteristics of Toxoplasma gondii rhoptry protein 4 (TgROP4), including physicochemical parameters, structural features, immunogenic epitopes, and virtual immune simulation, using several bioinformatics-based servers and tools. Based on allergenicity and antigenicity outputs, the TgROP4 protein seemed to have an immunogenic and non-allergenic nature. The quality of the three-dimensional (3D) structure improved after refinement, according to the outcomes of the Ramachandran plot and the ProSA-web servers. ABCpred and SVMTriP web tools were used to predict linear B lymphocyte epitopes and found several promising epitopes. Acceptable antigenicity, hydrophilicity, beta-turn, Bepipred linear epitope 2.0, flexibility, and surface accessibility scores were obtained through the Immune Epitope Database (IEDB). Also, seven discontinuous B-cell epitopes ranging from scores 0.966 to 0.848 were found in the 3D model of TgROP4 via the ElliPro. The IEDB findings showed T-cell epitopes on TgROP4 protein are capable to strongly bind to the major histocompatibility complex classes. In silico immune simulation was performed using C-ImmSim server and showed three injections of TgROP4 protein at 4-week intervals is capable to elicit adequate humoral and cell-mediated immune responses.
Keywords: Rhoptry protein 4, Toxoplasma gondii, In silico, Immunoinformatics, Vaccine
Toxoplasma gondii (T. gondii) is a cosmopolitan zoonotic infection that affects a wide range of warm-blooded animals and is estimated to be over 30% of the human population to be seropositive worldwide [1,2]. Beyond the socioeconomic and health consequences of toxoplasmosis, it is also considered an important abortifacient agent in farm animals that could cause significant economic damage to the field of animal farming in various countries [1,3]. The routine medications prescribed to treat toxoplasmosis are insufficient due to their side effects and inability to remove bradyzoites within tissue cysts, thus vaccination strategies might be a good alternative to restrict T. gondii infection in domestic animals and humans, given its considerable global burden [4,5]. In the past decades, several studies have been performed to evaluate the vaccine types through different strategies against acute and chronic toxoplasmosis worldwide [5,6].
Overall, it is estimated that rhoptry proteins (ROPs) occupy about 1%–30% of the total parasite cell volume and participate in the formation and functional properties of the parasitophorous vacuole (PV) and PV membrane [7,8]. During 20 years ago, some ROPs were employed as immunogens for vaccine development to induce strong specific humoral and cellular immune responses [6,7]. Rhoptry protein 4 (ROP4), a member of the ROP2-protein family, is expressed in tachyzoite, sporozoite, and bradyzoite stage forms [9]. This protein is released from the parasite during or shortly after invasion into the PV, associates with the vacuole membrane, and becomes phosphorylated in the infected cell [8]. In addition, ROP2 and ROP4, as ligands of human lactoferrin, play a key role in the uptake of iron from the infected host, suggesting that the host lactoferrin acquisition by T. gondii is one of the vital mechanisms for parasite growth during infection [10]. Hence, the T. gondii ROP4 (TgROP4) protein can be proposed as a potential suitable vaccine candidate against both acute and chronic toxoplasmosis. The present research aimed to assess the key characteristics of TgROP4 in terms of physicochemical parameters, structural features, immunogenic epitopes, virtual immune simulation, and so forth, through several bioinformatics servers and tools.
This study received approval from the Abadan University of Medical Sciences Ethical Committee (IR.ABADANUMS.REC.1401.157). The authors have completely observed ethical issues (e.g., plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, and redundancy).
The protein sequence of TgROP4 (accession no., TGRH88_020200) was obtained from the ToxoDB and saved in FASTA format for bioinformatics analysis. Appendix 1 lists the links to all bioinformatics servers employed in this study.
The physicochemical characteristics of TgROP4 were computed through ExPASy ProtParam as follows: number of amino acids, 578; molecular weight, 64,002.63 Da; theoretical isoelectric point, 8.48; total number of negatively charged residues (Asp+Glu), 70; total number of positively charged residues (Arg+Lys), 73; chemical formula, C2832H4482N828O839S14; total number of atoms, 8,995; the estimated half-life was, 30 hours (mammalian reticulocytes, in vitro), >20 hours (yeast, in vivo), and >10 hours (Escherichia coli, in vivo); the instability index, 45.39 (This classifies the protein as unstable.); aliphatic index, 82.49; and grand average of hydropathicity, -0.337.
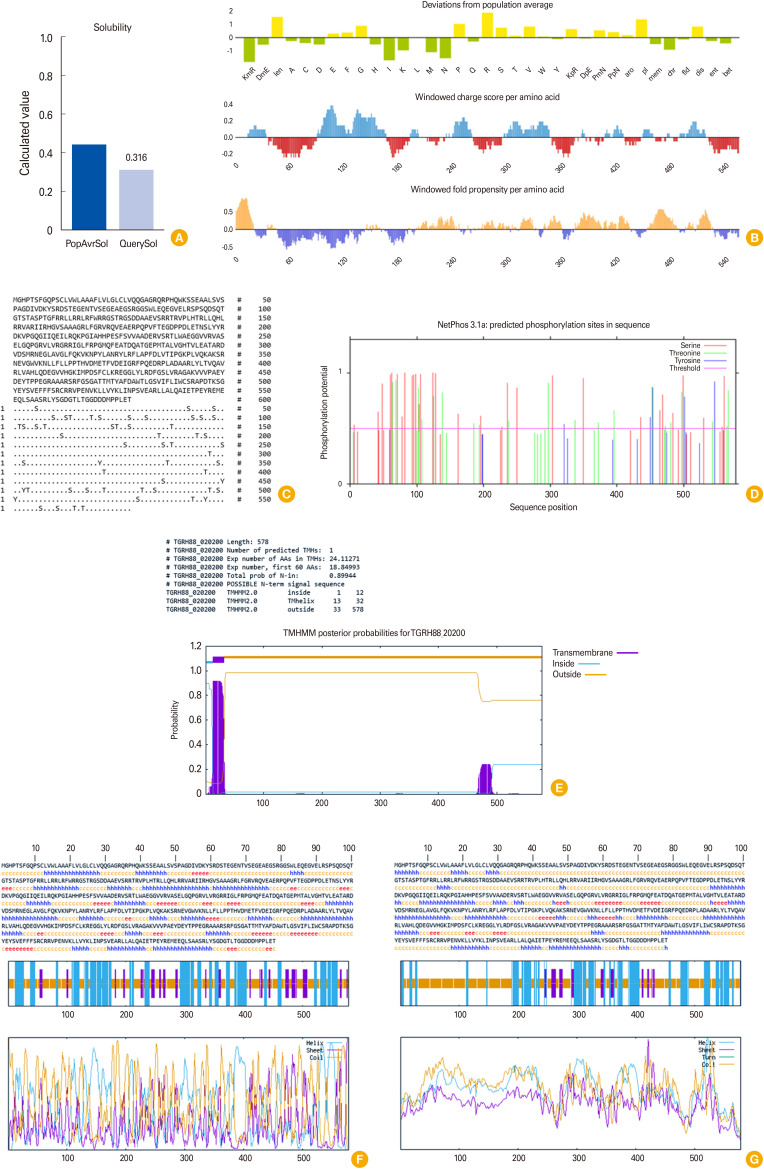
NetPhos ver. 3.1 (DTU Bioinformatics, Kongens Lyngby, Denmark) and GPS-PAIL (http://pail.biocuckoo.org/index.php) web services predicted 55 and one phosphorylation (Ser: 30, Thr: 20, and Tyr: 5) and acetylation (RQRPHQWKSSEAALS, position 41) sites, respectively (Fig. 1C, D), suggesting that these post-translational modifications might regulate the protein functions and affect their key activities. TgROP4 protein was predicted to be non-allergenic via AlgPred ver. 2.0 (https://webs.iiitd.edu.in/raghava/algpred2/) and AllerTOP ver. 2.0 (Bulgarian Academy of Sciences, Sofia, Bulgaria), while it was determined as an allergen with AllergenFP ver. 1.0 web server (Bulgarian Academy of Sciences). Also, a moderate antigenicity score was estimated using the VaxiJen ver. 2.0 (0.6327, probable antigen; http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html). Based on the outputs of the Protein-Sol (Warwicker Group, University of Manchester, Manchester, UK) (Fig. 1A, B) and SOLpro servers (https://scratch.proteomics.ics.uci.edu/), TgROP4 was determined insoluble with a probability of 0.316 and 0.633, respectively. PSORT II (Osaka University, Osaka, Japan) was employed to predict the subcellular localization of TgROP4, and the server revealed that this protein is “extracellular” (77.8%). Also, the TMHMM ver. 2.0 server (DTU Bioinformatics) showed that the ROP4 sequence consists of one transmembrane helix (position: 13 to 32) (Fig. 1E). In addition, signal peptide prediction was performed via the SignalP ver. 6.0 server (https://services.healthtech.dtu.dk/services/SignalP-6.0/), and the TgROP4 sequence contains a conventional signal peptide of the Sec/SPI form with a likelihood of 0.9998 and a cleavage site between position 33 and 34 with probability of 0.973519.
Fig. 1. (A) Solubility and (B) deviation from the population average, charge score, and fold propensity of TgROP4 predicted through the Protein-Sol server; (C) prediction of TgROP4 sequence phosphorylation regions in terms of serine (S), threonine (T), and tyrosine (Y) through NetPhos 3.1 server; (D) graphical diagram of phosphorylation sites; and (E) prediction of TgROP4 protein transmembrane helices. Statistics and a list of the location of the predicted transmembrane helices and the predicted location of the intervening loop regions. Length: the length of the protein sequence; number of predicted TMHs: the number of predicted transmembrane helices; Exp number of AAs in TMHs: the expected number of amino acids in the transmembrane helices. If this number is >18, it is very likely to be a transmembrane protein (or have a signal peptide). Exp number, first 60 amino acids: the expected number of amino acids in transmembrane helices in the first 60 amino acids of the protein. If this number is >a few, the predicted transmembrane helix in the N-term could be a signal peptide; total prob of N-in: the total probability that the N-term is on the cytoplasmic side of the membrane. (F, G) Graphical representations of the secondary structure prediction of TgROP4 prepared by GOR IV (F) and SOPMA online servers (G) (e=extended strand, h=helix, and c=coil).
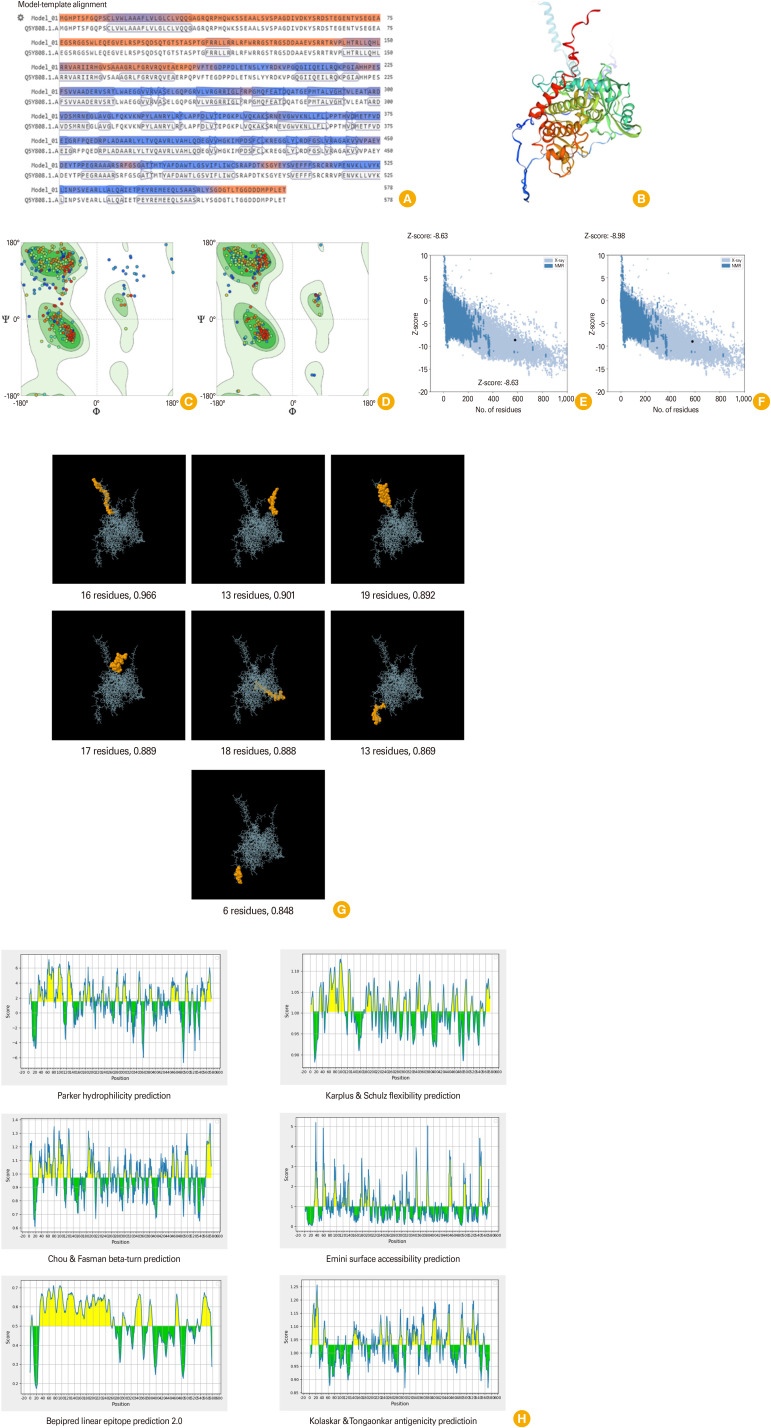
The principal biological function of each protein is focused on its spatial structure. Thus, predicting the secondary and tertiary structures of the target protein is an essential step to realizing the connections between structures and functions [11,12]. Random coils, extended strands, and alpha helices were predicted as important secondary structures in the protein sequences using two online tools, GORIV and SOPMA. According to the findings of the secondary structure analysis performed using the GORIV and SOPMA servers, the ROP4 protein sequence consisted of 37.37% (216 residues) and 31.66% (183 residues) alpha helices, 13.32% (77 residues) and 7.09% (41 residues) extended strands, and 49.31% (285 residues) and 61.25% (354 residues) random coils, respectively (Fig. 1F, G). The SWISS-MODEL web server (Swiss Institute of Bioinformatics, Geneva, Switzerland) was employed for the homology modeling of three-dimensional (3D) structures of the TgROP4 protein. Following the analysis, four 3D models were generated, and template 1 (Q5Y808.1.A) of the protein showed 100% sequence identity from amino acids 1 to 578 (coverage was equal to 1.00) (Fig. 2A, B). In the next step, the best-created 3D model through the SWISS-MODEL tool, was further analyzed and refined via the GalaxyRefine web server (Galux Inc. & Computational Biology Lab, Department of Chemistry, Seoul National University, Seoul, Korea). ProSA-web (https://prosa.services.came.sbg.ac.at/prosa.php) was also employed to evaluate the model’s overall quality. The outputs of GalaxyRefine revealed that the crude model included global distance test-high accuracy (1.0000), root mean square deviation (0.0000), MolProbity (1.710), the clash score (0.6), poor rotamers (3.8), and Rama favored (88.7). The scores were changed after refinement for these parameters: 0.9278, 0.545, 1.385, 7.0, 0.6, and 98.1, respectively. The quality of the 3D structure improved after refinement, according to the outcomes of the Ramachandran plot generated by the structure assessment tool (88.72% and 98.09% of residues were incorporated in the favored regions in the original and refined models, respectively.) and the ProSA-web (Z-score: –8.63 and –8.98 for the crude and refined models of TgROP4) online services (Fig. 2C–F).
Fig. 2. (A) Model-template alignment and (B) three-dimensional (3D) model structure predicted for TgROP4 protein, provided by SWISS-MODEL server. (C–F) Validation of the 3D model of the TgROP4 protein via structure assessment tool and ProSA-web. Following analysis, the structure assessment tool of the SWISS-MODEL generated Ramachandran plots. It estimated that 88.72% and 98.09% of residues were located in favored regions prior to (C) and post-refinement (D), respectively. Based on the ProSA-web server, Z-score was determined to be –8.63 and –8.98 in the crude (E) and refined (F) 3D models, respectively, indicating that the 3D structure’s quality was enhanced post-refinement procedure. NMR, nuclear magnetic resonance. (G) Seven conformational B-cell epitopes on TgROP4 protein predicted by ElliPro. The white rods and orange domains show the TgROP4 protein and discontinuous B-cell epitopes, respectively. (H) The results of the IEDB online server were visualized for six variables, including hydrophilicity (average: 1.526), beta-turn (average: 0.970), Bepipred linear epitope 2.0 (average: 0.530), flexibility (average: 1.003), surface accessibility (average: 1.000), and antigenicity (average: 1.031). The x-axes represent the residue positions in the sequence, while the y-axes represent the corresponding score for each residue; the yellow color represents that the residue might have a greater probability of being a part of the epitope, and the green color represents the unfavorable regions relevant to the properties of interest.
Predicting B and T cell epitopes could suggest useful information that helps in determine the immunogenic peptides. Thus, recognition of the prominent epitopes capable of activating the immune system is considered an important step in the field of reverse vaccinology through immunoinformatics approaches [11,13]. ABCpred (https://webs.iiitd.edu.in/raghava/abcpred/index.html) and SVMTriP web tools (http://sysbio.unl.edu/SVMTriP/index.php) were used to predict linear B lymphocyte epitopes for this aim. SVMTriP server was predicted three potential epitopes (16-mer) as follows: (1) HTRLLQHLRRVARIIR, score: 1.000, location: 143–158; (2) AWTLGSVIFLIWCSRA, score: 0.850, location: 479–494; and (3) PSCLVWLAAAFLVLGL, score: 0.814, location: 10–25. Additionally, using the ABCpred online web server, we found 36 promising epitopes (threshold: 0.75), which the five best epitopes were including, AHLQDEGVVHGKIMPD (start position: 404, score: 0.94), TVLEATARDVDSMRNE (start position: 292, score: 0.94), PAEYDEYTPPEGRAAA (start position: 447, score: 0.89), QGIIQEILRQKPGIAH (start position: 206, score: 0.89), and RGSDDAAEVSRRTRVP (start position: 126, score: 0.89). The higher score of the peptide means the higher probability of being an epitope [14]. Furthermore, we recruited the Immune Epitope Database (IEDB) tool (http://tools.iedb.org/bcell/) to predict six variables: antigenicity, hydrophilicity, beta-turn, Bepipred linear epitope 2.0, flexibility, and surface accessibility. According to the results, the above items’ average scores (threshold) were computed as 1.031, 1.526, 0.970, 0.530, 1.003, and 1.000, respectively (Fig. 2H). Prediction of discontinuous epitopes is a valuable step that is needed for antibody-antigen interaction. For this aim, the ElliPro tool in the IEDB server (http://tools.iedb.org/ellipro/) was used, and seven conformational B-cell epitopes ranging from scores 0.966 to 0.848 were found in the 3D model of TgROP4 (Fig. 2G). The binding of peptides to the major histocompatibility complex (MHC) classes (MHC-I and MHC-II) is necessary to present target antigens to T-cells. It is the main step in choosing potential epitopes [13,15,16]. The IEDB findings showed that T-cell epitopes on TgROP4 protein are capable of binding to both MHC classes strongly.
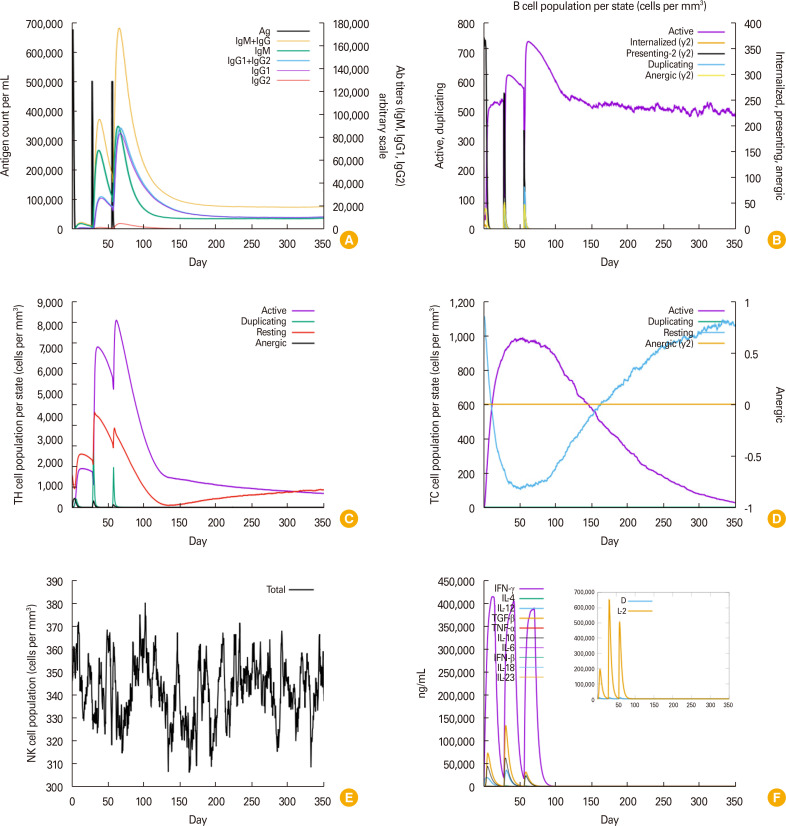
In silico immune simulation is a critical step that analyzes humoral and cell-mediated immune responses of the mammalian immune system to a certain antigen [17]. The C-ImmSim server (https://kraken.iac.rm.cnr.it/C-IMMSIM/index.php) was used to simulate immune responses. The following settings were applied: simulation volume, 10; random seed, 12345; simulation steps, 1,050; three injections at 4-week intervals (lipopolysaccharide free) with time series of 1, 84, and 168; number of antigens to inject, 1,000; and host human leukocyte antigen selection, default. Following injection, sufficient titers of immunoglobulin M (IgM), immunoglobulin G (IgG)1, and a combination of both IgG+IgM were secreted (Fig. 3A). Approximately 65 days post-immunization, active B lymphocytes reached a peak state (approximately 720 cells/mm3), which subsequently remained for 350 days (approximately 450 cells/mm3) (Fig. 3B). The active T-CD4+ and T-CD8+ cells started duplicating just a few days after TgROP4 immunization and remained for up to 350 days (Fig. 3C, D). Furthermore, interferon-γ (IFN-γ) and interleukin-2 cytokine levels were increased after injection (Fig. 3F), indicating strong induction of the T helper 1 immune responses required for the clearance of invasive free or intracellular tachyzoites.
Fig. 3. Prediction of immune profile through in silico immune simulation approach using the C-ImmSim server. (A) Immunoglobulin secretion in response to TgROP4; (B) B-cell population per state (cells/mm3); (C) TH cell (CD4+) population per state (cells/mm3); (D) TC cell (CD8+) population per state (cells/mm3); (E) Natural killer (NK) cell population (cells/mm3); and (F) level of cytokines production (ng/mL) by TgROP4. Ag, antigen; Ab, antibody; IgM, immunoglobulin M; IgG, immunoglobulin G; IFN-γ, interferon-gamma; IL, interleukin; TGF-β, transforming growth factor-beta; TNF-β, tumor necrosis factor-beta.
In conclusion, in the present study, immunoinformatics-based servers and tools recognized that the TgROP4 protein had many potential T and B-cell epitopes. Also, based on allergenicity and antigenicity outputs, the TgROP4 protein seemed to have an immunogenic and non-allergenic nature, indicating its potential use for inclusion in vaccine formulations. In silico immune simulation predicted that humoral responses, including specific IgM, IgG1, and a combination of both IgG+IgM titers along with cell-mediated cytokines, particularly IFN-γ, raised to significant levels following three injections of TgROP4 by 4-week intervals. In the next step, it is highly recommended that a novel multi-epitope vaccine be constructed and tested through in silico and in vivo approaches in suitable animal models.
Appendix 1
The links and description of all bioinformatics online servers used in the research
Footnotes
No potential conflict of interest relevant to this article was reported.
This work was supported by the Abadan University of Medical Sciences, Abadan, Iran (grant/award no., 1603).
References
- 1.Robert-Gangneux F, Darde ML. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin Microbiol Rev. 2012;25:264–296. doi: 10.1128/CMR.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rostami A, Riahi SM, Gamble HR, et al. Global prevalence of latent toxoplasmosis in pregnant women: a systematic review and meta-analysis. Clin Microbiol Infect. 2020;26:673–683. doi: 10.1016/j.cmi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Dubey JP. The history of Toxoplasma gondii: the first 100 years. J Eukaryot Microbiol. 2008;55:467–475. doi: 10.1111/j.1550-7408.2008.00345.x. [DOI] [PubMed] [Google Scholar]
- 4.Antczak M, Dzitko K, Dlugonska H. Human toxoplasmosis: searching for novel chemotherapeutics. Biomed Pharmacother. 2016;82:677–684. doi: 10.1016/j.biopha.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 5.Wang JL, Zhang NZ, Li TT, He JJ, Elsheikha HM, Zhu XQ. Advances in the development of anti-Toxoplasma gondii vaccines: challenges, opportunities, and perspectives. Trends Parasitol. 2019;35:239–253. doi: 10.1016/j.pt.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Foroutan M, Ghaffarifar F, Sharifi Z, Dalimi A, Jorjani O. Rhoptry antigens as Toxoplasma gondii vaccine target. Clin Exp Vaccine Res. 2019;8:4–26. doi: 10.7774/cevr.2019.8.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dlugonska H. Toxoplasma rhoptries: unique secretory organelles and source of promising vaccine proteins for immunoprevention of toxoplasmosis. J Biomed Biotechnol. 2008;2008:632424. doi: 10.1155/2008/632424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carey KL, Jongco AM, Kim K, Ward GE. The Toxoplasma gondii rhoptry protein ROP4 is secreted into the parasitophorous vacuole and becomes phosphorylated in infected cells. Eukaryot Cell. 2004;3:1320–1330. doi: 10.1128/EC.3.5.1320-1330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Hajj H, Demey E, Poncet J, et al. The ROP2 family of Toxoplasma gondii rhoptry proteins: proteomic and genomic characterization and molecular modeling. Proteomics. 2006;6:5773–5784. doi: 10.1002/pmic.200600187. [DOI] [PubMed] [Google Scholar]
- 10.Dziadek B, Dziadek J, Dlugonska H. Identification of Toxoplasma gondii proteins binding human lactoferrin: a new aspect of rhoptry proteins function. Exp Parasitol. 2007;115:277–282. doi: 10.1016/j.exppara.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Wang G, Cai J, Yin H. Review on the identification and role of Toxoplasma gondii antigenic epitopes. Parasitol Res. 2016;115:459–468. doi: 10.1007/s00436-015-4824-1. [DOI] [PubMed] [Google Scholar]
- 12.Foroutan M, Ghaffarifar F, Sharifi Z, Dalimi A, Pirestani M. Bioinformatics analysis of ROP8 protein to improve vaccine design against Toxoplasma gondii. Infect Genet Evol. 2018;62:193–204. doi: 10.1016/j.meegid.2018.04.033. [DOI] [PubMed] [Google Scholar]
- 13.Kazi A, Chuah C, Majeed AB, Leow CH, Lim BH, Leow CY. Current progress of immunoinformatics approach harnessed for cellular- and antibody-dependent vaccine design. Pathog Glob Health. 2018;112:123–131. doi: 10.1080/20477724.2018.1446773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foroutan M, Ghaffarifar F, Sharifi Z, Dalimi A. Vaccination with a novel multi-epitope ROP8 DNA vaccine against acute Toxoplasma gondii infection induces strong B and T cell responses in mice. Comp Immunol Microbiol Infect Dis. 2020;69:101413. doi: 10.1016/j.cimid.2020.101413. [DOI] [PubMed] [Google Scholar]
- 15.El-Kady IM. T-cell immunity in human chronic toxoplasmosis. J Egypt Soc Parasitol. 2011;41:17–28. [PubMed] [Google Scholar]
- 16.Foroutan M, Ghaffari AD, Soltani S, Majidiani H, Taghipour A, Sabaghan M. Bioinformatics analysis of calcium-dependent protein kinase 4 (CDPK4) as Toxoplasma gondii vaccine target. BMC Res Notes. 2021;14:50. doi: 10.1186/s13104-021-05467-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rapin N, Lund O, Bernaschi M, Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS One. 2010;5:e9862. doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]