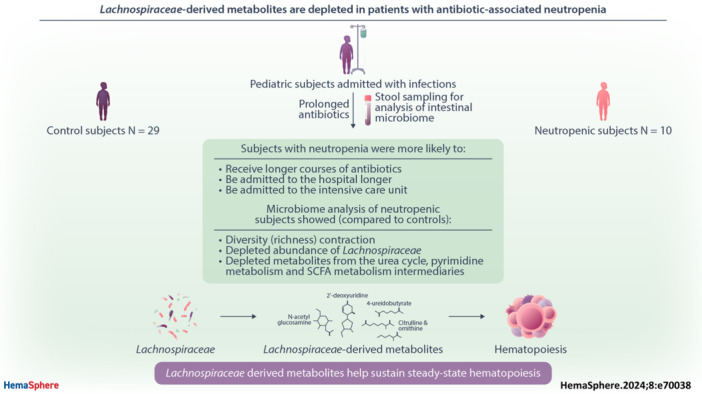
Abstract
Prolonged antibiotic exposure causes dangerous hematologic side effects, including neutropenia, in up to 34% of patients. Murine studies established a link between the intestinal microbiota and hematopoiesis. To identify factors that predispose to neutropenia in pediatric patients, we evaluated changes in microbiota‐derived metabolites and intestinal microbiota composition after prolonged courses of antibiotics. In this multi‐center study, patients with infections requiring anticipated antibiotic treatment of two or more weeks were enrolled. Stool samples were obtained at the start and completion of antibiotics or at neutropenia onset (prospective arm). Some patients were enrolled in a retrospective arm in which a stool sample was collected at the time of neutropenia during antibiotic therapy and 2–4 weeks after completion of antibiotics with recovery of blood counts. We identified 10 patients who developed neutropenia on antibiotics and 29 controls matched for age, sex, race, and ethnicity. Clinical data demonstrated no association between neutropenia and the type of infection or antibiotic used; however, patients with neutropenia were admitted to the intensive care unit more often and received longer courses of antibiotics. Reduced intestinal microbiome richness and, specifically, decreased abundance of Lachnospiraceae family members correlated with neutropenia. Untargeted stool metabolomic profiling revealed several metabolites that were depleted exclusively in patients with neutropenia, including members of the urea cycle pathway, pyrimidine metabolism, and fatty acid metabolism that are known to be produced by Lachnospiraceae. Our study shows a relationship between intestinal microbiota disruption and abnormal hematopoiesis and identifies taxa and metabolites likely to contribute to microbiota‐sustained hematopoiesis.
INTRODUCTION
More than 236 million antibiotic courses are prescribed every year in the United States, 1 providing life‐saving treatment for serious infections in children and adults. 2 Yet, antibiotic use can cause significant adverse effects, especially when prescribed for prolonged periods. Bone marrow suppression, which can present as anemia, leukopenia with neutropenia, and/or thrombocytopenia, occurs in up to 34% of patients receiving antibiotics for 2 weeks or more 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 and several reports list the hematologic system among the top systems affected by prolonged treatments in children. 3 , 4 , 5 , 8 , 10 While bone marrow suppression usually improves with early discontinuation of antibiotic therapy, 3 , 8 shortening antibiotic treatment in response to myelosuppression hampers treatment of the underlying infection. Monitoring for manifestations of bone marrow suppression represents a significant burden of morbidity and healthcare costs. Better understanding the causes of antibiotic‐associated bone marrow suppression is critical to developing prevention and treatment strategies that optimize antibiotic use.
Although antibiotic‐associated neutropenia is reported most commonly with beta‐lactam antibiotics, 11 it has been described as an adverse effect of virtually all classes of antibiotics. 4 , 8 , 9 , 12 The mechanisms underlying antibiotic‐associated neutropenia, which occurs in 10%–15% of pediatric patients receiving prolonged antibiotic courses, are not well understood. In recent preclinical work, we and others discovered that antibiotic disruption of the intestinal microbiota plays a critical role in antibiotic‐associated cytopenias. 13 In line with those findings, recent studies using murine models demonstrated that prolonged antibiotic use can suppress hematopoiesis by affecting the intestinal microbiota. 13 , 14 , 15 , 16 , 17 Consistent with this, germ‐free (GF) mice have abnormal blood counts and hematopoiesis. Further, the recolonization of GF mice with intestinal microbiota results in the expansion of the bone marrow myeloid cell pool. 14 , 18 Fecal microbiota transplantation (FMT) from young mice to aged mice has also been shown to increase lymphoid differentiation and rejuvenate aged hematopoietic stem cells (HSCs). 19
In prior work using animal models, we found that depletion of the intestinal microbiota suppresses type I interferon (IFN) signaling in the bone marrow, whereas supplementation with microbial metabolites was sufficient to restore signaling, normal hematopoiesis, and resolve low blood counts in antibiotic‐treated mice. 20 These findings support a paradigm in which intestinal microbiota products enter the bloodstream and travel to the bone marrow, where they support normal hematopoiesis. 13 In support of this, recent human studies indicate that the state of the intestinal microbiota correlates with hematopoietic stem cell transplant (HSCT) outcomes. 21 , 22 , 23 , 24 , 25
While murine studies have elucidated the mechanisms by which the microbiota contribute to normal hematopoiesis, there is a dearth of clinical research to validate findings from murine models in humans. Here, we sought to analyze the microbiota and microbiota‐derived metabolites in stool samples from pediatric patients receiving prolonged antibiotic therapy. Our study is the first to report microbiome changes in samples from human subjects without chronic or preexisting conditions and reveals key microbes and microbial metabolites depleted in children and adolescents with antibiotic‐associated neutropenia. These findings provide critical clinical data that may contribute not only to the safe administration of prolonged courses of antibiotics in healthy patients but may also positively impact outcomes from HSCT. 21
METHODS
Subjects and study design
We carried out a multi‐center, nontreatment, noninterventional case‐control study approved by Baylor College of Medicine and Affiliated Hospitals' Institutional Review Board (IRB) and by Vanderbilt University Medical Center's IRB between January 2020 and August 2022. Study sites included Texas Children's Hospital in Houston, Texas, and Monroe Carell Jr. Children's Hospital at Vanderbilt in Nashville, Tennessee. After informed consent was obtained, pediatric patients aged 0 to 18 years were enrolled. Inclusion criteria were admission to the hospital with infections requiring a planned duration of at least two weeks of antibiotic therapy administered either intravenously (IV) or orally (PO). Subjects with a prior history of bone marrow failure or neutropenia, gastrointestinal infections or syndromes, immunodeficiency, malignancy, malnutrition, genetic syndrome known to involve the gastrointestinal or hematopoietic system, antibiotic use during the week preceding hospital admission, or prophylactic antibiotics use were excluded. Complete blood counts (CBC) were obtained at the discretion of treating providers and monitored by study personnel to identify neutropenia, defined as an absolute neutrophil count (ANC) of ≤1500/mm3. When defining neutropenia, investigators considered a higher threshold of ≤1500/mm3 in contrast with that of severe neutropenia (ANC ≤ 500/mm3) perceived as more clinically relevant (1) to allow for earlier identification of cases and (2) anticipating a rather infrequent CBC monitoring after hospital discharge that potentially prevented identification of severe neutropenia developing in between infrequent blood count checks. Eighty‐one patients were enrolled (Figure 1).
Figure 1.
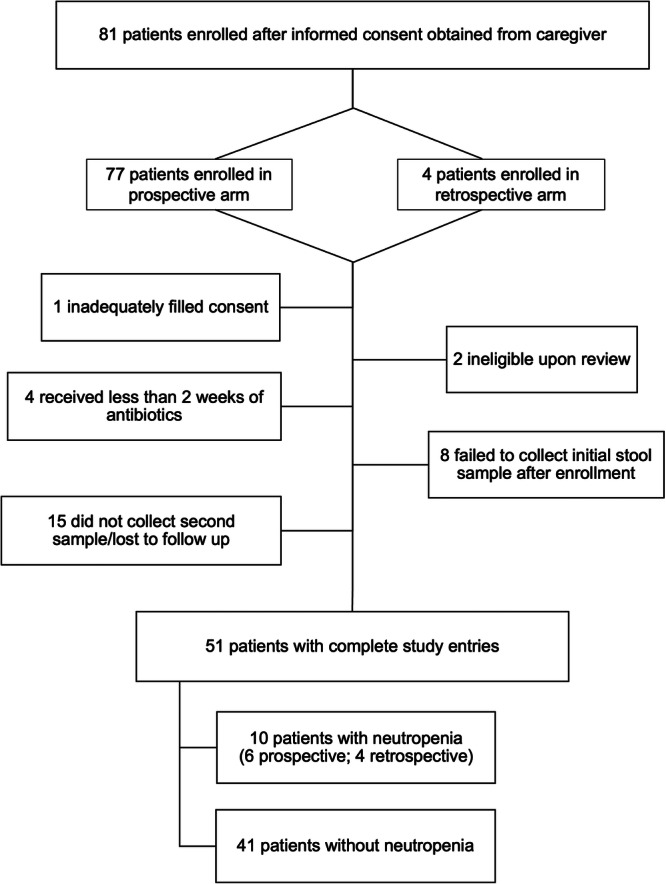
Enrollment flowchart.
The study included two arms: a prospective arm, in which patients were enrolled shortly after admission to the hospital and initiation of antibiotic therapy, and a retrospective arm, in which patients who had previously initiated antibiotic therapy were enrolled at the time of neutropenia development. Control subjects who received antibiotics but did not develop neutropenia were only selected from the prospective arm. Figure 2 illustrates the timing of stool samples' collection with respect to hospital admission, as well as antibiotic therapy initiation and completion for each subject, further detailed in Table S1. Study protocol for the prospective arm was designed so that a baseline stool sample was obtained within 7 days of antibiotic initiation and subsequent (antibiotic‐treated or ABX‐treated) stool sample was collected up to 1 week after antibiotic completion or discontinuation due to neutropenia. For the retrospective arm, stool samples were collected at the time of neutropenia development (ABX‐treated) and at least 2–4 weeks after completion of antibiotic therapy and resolution of neutropenia (accounting for “recovery” stool sample in an attempt to reflect baseline microbiome). Specific “out of window” stool sample collection timepoints were accepted per investigators discretion for several subjects. The creation of a retrospective arm allowed for increased capture of patients with neutropenia.
Figure 2.
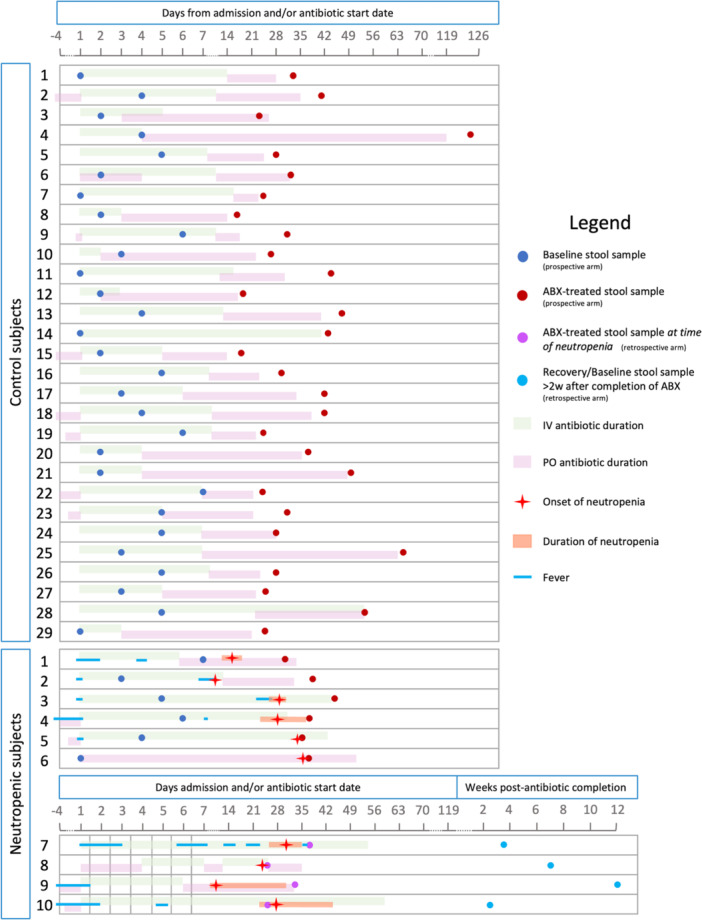
Schematic overview of sampling time and antibiotic treatment duration for each subject. Each study subject is shown on a separate line denoting: intravenous antibiotic treatment (green bar), oral antibiotic treatment (pink bar), baseline stool sample collection (dark blue dots for prospective arm), antibiotic‐treated stool sample collection (red and purple dots for prospective and retrospective arm, respectively), recovery stool sample collection (light blue dot for retrospective arm), days of fever (blue line), neutropenia onset (red star) and duration (orange bar). Timelines indicate the time since hospital admission in days.
Demographic and clinical information including but not limited to diagnosis, LOS, and laboratory results were collected via electronic medical record review.
We identified 10 subjects with neutropenia, including two whose ANC was near 1500 and trending down while antibiotics were continued without reassessment of the CBC, that is, extrapolating that ANC would have been ≤1500/mm3 if blood counts had been re‐checked. Six of these ten patients were enrolled in the prospective arm whereas four were in the retrospective arm. We used frequency matching on age and race/ethnicity to identify twenty‐nine controls. Considering the ethnicity distribution in Houston and Nashville, we combined race and ethnicity to make a single variable (Hispanic, Non‐Hispanic White, Non‐Hispanic Black, Asian, and Native American).
Statistical analysis of clinical data
For continuous variables, we first identified outliers using the ROUT method at Q = 5%; we then performed a Shapiro–Wilk normality test with a 0.05 significance. We conducted descriptive analysis by obtaining quartiles and mean/SD/SEM. We used a parametric t‐test for normal data (total length of antibiotics, length of PO antibiotics, and length of admission) and the Mann–Whitney as a nonparametric test for association between continuous and categorical variables. We used chi‐square testing for most categorical variables, except for a number of antibiotic classes used (<2 or >3) where we used Fisher's exact test.
Sample collection
Stool samples from participants were collected in sterile plastic containers. When a sample was collected while the patients remained admitted to the hospital, the sample was placed on ice and brought to the laboratory. When the sample was collected at home after discharge, it was kept in a home refrigerator and shipped on ice to the laboratory within hours. Stool samples were processed by investigators upon arrival at the laboratory by aliquoting in 1.5 mL sterile Eppendorf tubes for storage at −80°C until use. Samples were shipped in dry ice to a central laboratory that performed 16S rRNA gene sequencing and untargeted metabolomics profiling.
16S rRNA gene sequencing and untargeted metabolomic analysis
Paired stool samples from individual study subjects were compared by 16S rRNA sequencing and untargeted metabolomic analysis. The supplemental methods provide details about data collection and approaches used to analyze stool samples. The Center for Metagenomics and Microbiome Research (CMMR) core at Baylor College of Medicine performed the 16S rRNA sequencing, and Metabolon performed the untargeted metabolomic profiling of samples.
RESULTS
Patient characteristics
Fifty‐one subjects completed study enrollment and sample collection (Figure 1). Based on the definition of ANC ≤ 1500/mm3, 10 subjects developed neutropenia, 4 of whom were enrolled in the retrospective arm. Neutropenia occurred in 17% (n = 6/47) of subjects in the prospective arm, consistent with previously reported rates in pediatric patients on antibiotic therapy for greater than 2 weeks. 3 , 5 , 8 Neutropenia occurred in both male and female subjects and in non‐Hispanic White, non‐Hispanic Black, Hispanic, and Asian subjects at frequencies that roughly mirrored the population served by the enrolling sites. Twenty‐nine subjects who received antibiotics but did not develop neutropenia were selected from among prospectively enrolled subjects as controls by frequency matching for age (within ±1 year), sex, and race/ethnicity. The median age was 8.5 years for subjects with neutropenia and 8 years for control subjects, respectively. The baseline demographic characteristics of the selected cohort are shown in Table 1.
Table 1.
Baseline demographic characteristics of subjects treated with long‐term antibiotics with and without neutropenia.
| Neutropenic | Nonneutropenic | p value | |
|---|---|---|---|
| N (total) | 10 | 29 | |
| Age, median (range) | 8.5 y (6 m–17 y) | 8 y (6 m–15 y) | 0.54 |
| Gender | 0.23 | ||
| Female, N (%) | 7 (70%) | 14 (48%) | |
| Race/ethnicity | 0.77 | ||
| Hispanic (%) | 2 (20%) | 8 (28%) | |
| Non‐Hispanic Black (%) | 2 (20%) | 6 (21%) | |
| Non‐Hispanic White (%) | 3 (30%) | 12 (41%) | |
| Asian (%) | 3 (30%) | 3 (10%) | |
| Lowest ANC, median (range) | 895 (460–1570) | 3650 (1666–10,360) | <0.0001 |
Note: Statistical significance defined as p < 0.05 determined by parametric t‐test or nonparametric Mann–Whitney test for continuous variables, and Chi‐square or Fisher's exact test for categorical variables (n = 10 in neutropenic group, 29 in non‐neutropenic group).
Abbreviation: ANC, absolute neutrophil count.
Other than neonates, all ages were represented in both groups. Infants comprised 20% of subjects with neutropenia and 17% of controls, while children between 1 and 11 years old were slightly more represented in controls compared to the neutropenic group (62% vs. 40%), and adolescents were more represented in the neutropenic group (40% vs. 17%). However, no statistically significant correlation was noted between age and neutropenia development.
Timing of stool sample collection with respect to antibiotic treatment
The timing of stool sample collection with respect to hospital admission and antibiotic treatment is shown in Figure 2. A stool sample intended to represent the “baseline” intestinal microbiota was collected shortly after hospital admission for the prospective group, and 2–4 weeks after completion of antibiotic therapy and resolution of neutropenia for the retrospective group, also described as the “recovery” timepoint. However, given the complexities of study enrollment and sample collection for pediatric patients, timeframes of up to ±1 week were established for said sample collections. Considering that alteration of the intestinal microbiome occurs rapidly after antibiotic administration, even after a single dose; and recovery of the microbiome after antibiotic completion can take several months, 26 , 27 we assessed the variability between subjects of antibiotic exposure at each sample collection timepoint. There was a wide but similar variation in the number of days between the first dose of antibiotics and the baseline stool sample collection with a median of 4.5 days (range 1–10 days) for the prospective neutropenic group and a median of 3 days (range 1–10 day) for the control group; however, no statistically significant difference was found between groups for this variable (data not shown). For the retrospective arm, a “recovery” or “baseline” stool sample was collected with a median of 5.25 weeks after completion of antibiotics (range 2.5–12 weeks).
Factors associated with the development of antibiotic‐associated neutropenia
Relevant characteristics of the clinical course for subjects in each group are shown in Table 2. Neutropenia was associated with greater total length of antibiotic treatment (p = 0.0021), intravenous (IV) antibiotic treatment (p = 0.0013), length of hospital admission stay (LOS) (p = 0.0393), and admission to the ICU (p = <0.0001). Basic information regarding the clinical course of each subject is shown in Table S1, while further details of the clinical course for neutropenic subjects are provided in Table S2.
Table 2.
Clinical characteristics of subjects treated with long‐term antibiotics with and without neutropenia.
| Neutropenic | Control | p value | |
|---|---|---|---|
| Antibiotic length, days: median (range) | |||
| Total | 38 (29−62) | 28 (14−122) | 0.0021 |
| IV | 23.5 (0−59) | 7 (2−56) | 0.0013 |
| PO | 11 (0−51) | 19 (0−119) | 0.1026 |
| Number of classes of antibiotic used, N (%) | 0.6390 | ||
| ≤2 | 2 (20%) | 4 (14%) | |
| ≥3 | 8 (80%) | 25 (86%) | |
| Class of antibiotic used, N (%) | 0.4384 | ||
| Cephalosporins | 9 (90%) | 26 (90%) | |
| Penicillin | 5 (50%) | 14 (48%) | |
| Glycopeptide | 8 (80%) | 26 (90%) | |
| Lincomycin | 5 (50%) | 10 (34%) | |
| Nitroimidazole | 5 (50%) | 15 (52%) | |
| Sulfonamides | 2 (20%) | 5 (17%) | |
| Fluoroquinolones | 1 (10%) | 1 (3%) | |
| Aminoglycosides | 2 (20%) | 0 | |
| Macrolides | 1 (10%) | 0 | |
| Carbapenems | 0 | 1 (3%) | |
| Infection type, N (%) | 0.6892 | ||
| Gram positive | 7 (70%) | 17 (59%) | |
| Gram negative | 1 (10%) | 1 (3%) | |
| Polymicrobial | 1 (10%) | 3 (10%) | |
| Culture negative | 1 (10%) | 7 (24%) | |
| Organ/system involved | 0.1279 | ||
| Soft tissue & skin | 4 (40%) | 16 (55%) | |
| Bone/Joint | 6 (60%) | 10 (34%) | |
| CNS | 2 (20%) | 1 (3%) | |
| Blood/bacteremia | 3 (30%) | 2 (7%) | |
| Heart (endocarditis) | 2 (20%) | 0 | |
| Respiratory | 3 (30%) | 13 (45%) | |
| Gastrointestinal | 0 | 1 (3%) | |
| Urogenital | 1 (10%) | 1 (3%) | |
| Length of admission, days: median (range) | 13 (3−54) | 7 (1−22) | 0.0393 |
| ICU admission, N (%) | 5 (50%) | 0 | <0.0001 |
Note: Statistical significance defined as p < 0.05 determined by parametric t‐test or nonparametric Mann–Whitney test for continuous variables, and Chi‐square or Fisher's exact test for categorical variables (n = 10 in neutropenic group, 29 in non‐neutropenic group). Some subjects received several classes of antibiotics, both IV and PO; and their infections affected more than one organ/system.
Abbreviations: CNS, Central Nervous System; ICU, Intensive Care Unit; IV, intravenous; PO, oral.
Characteristics of antibiotic regimens
The median duration of antibiotic treatment was 38 days in subjects with neutropenia and 28 days in controls. A separate analysis of antibiotic administration by mouth (PO) or IV indicated that the main difference between groups was in the duration of IV antibiotics (Table 2). Even though patients with neutropenia received longer courses of IV treatment, long courses of IV antibiotic treatment up to 50 or more days were noted in both neutropenic and control groups, indicating that prolonged IV administration does not always lead to neutropenia. Similarly, subjects that only received PO antibiotics were represented in both groups, indicating that neutropenia can occur after prolonged PO antibiotic therapy alone.
Patients were frequently started on broad‐spectrum antibiotics upon admission to provide coverage for Gram‐positive, Gram‐negative, and anaerobic pathogens such that the majority of patients in both groups received antibiotics from three or more different classes (80% and 86% in the neutropenia group and controls, respectively). Altogether, subjects received antibiotics from 10 different antibiotic classes (Table 2). Cephalosporins, glycopeptides (e.g., vancomycin), nitroimidazoles (e.g., metronidazole), penicillins, and lincomycin (e.g., clindamycin) antibiotics were used often in both groups. Sulfonamides and fluoroquinolones were used less often in both groups, while aminoglycosides, macrolides, and carbapenems were used rarely. Whereas clinicians frequently focus on penicillins and sulfonamides (e.g., trimethoprim sulfamethoxazole) as potential causes of neutropenia, we found no significant association between the antibiotic class used and neutropenia in our cohort, likely due to the small size of our cohort and selection bias toward similarly treated infections in otherwise healthy patients.
Characteristics of infectious diagnoses
Prolonged antibiotic courses were used to treat a variety of conditions, including skin and soft tissue, bone, joint, and respiratory infections. Osteomyelitis, orbital cellulitis, and sinusitis were the most common diagnoses, with the last two frequently co‐occurring. While the numbers were too small to generate any statistically significant association between the organ/system affected and neutropenia, we observed some notable trends. For example, the only two subjects enrolled in our study with infective endocarditis both developed neutropenia. In both cases, neutropenia developed after having received antibiotics for more than 30 days, consistent with long treatment durations for endocarditis. The absence of endocarditis among the control cohort is likely related to the overall rarity of infective endocarditis in children. 28 Notably, while there appeared to be an association between bone infections and neutropenia, the inverse was true for lung infections; we speculate this may be explained by treatment durations for bone infections commonly exceeding those for lung infections by weeks. The central nervous system (mostly affected by intracranial extension of orbital cellulitis), bacteremia (which in all cases co‐occurred with other infections), the urogenital system, and the gastrointestinal system were less commonly affected. The involvement of multiple organ systems in the infectious process was not associated with neutropenia in our cohort (Table 2), granting the small number of patients limits our power to detect such differences.
Individual organisms were isolated in most subjects from both groups; culture‐negative infection was slightly more common in the control group, although this was not of statistical significance. Overall, infections due to Gram‐positive microorganisms were most common, reflecting the frequency of Staphylococcal and Streptococcal infections in children. Gram‐positive infections were slightly more common in the neutropenic group than in the control group. We noted no significant association between the isolated microorganisms and neutropenia in our cohort.
Characteristics of hospital admission
Patients with neutropenia had longer admissions to the hospital and were more often admitted to the ICU (Table 2). Median admission duration was 13 days in the neutropenic group versus 7 days in the control group. Five out of 10 subjects with neutropenia required admission to the ICU; however, 3 of these 5 subjects were briefly admitted to the ICU for overnight observation after a drainage procedure was performed to control their infection and transferred to the acute care floor the next morning after an uneventful ICU course. A detailed review of the ICU admissions showed that the majority of subjects did not have prolonged mechanical ventilation, prolonged NPO (“nothing by mouth”) or enteral feeding periods, prolonged use of renal replacement therapies like hemodialysis or continuous renal replacement therapy, or sedation use. Only one subject required sedation and mechanical ventilation, this persisted for 5 days after a valvuloplasty procedure. Importantly, no subject developed neutropenia while admitted to the ICU; rather, neutropenia developed days to weeks later. These trends suggest that common factors in the ICU environment did not drive neutropenia development in our cohort. Extended details of the clinical course for each neutropenic subject are available in Table S2.
To evaluate for other potential explanations of neutropenia, we assessed inflammatory markers, fever, and other concomitant signs and symptoms, medications other than antibiotics, neutropenia onset and duration, and overall clinical status at the time of neutropenia. Most subjects became afebrile shortly after admission without fever recurrence; however, two subjects had a brief period of fever preceding neutropenia by ~3 days associated with persistent/worsening infectious abscess identified on imaging that required surgical drainage. All subjects had clinical improvement of infection and overall clinical status at the onset of neutropenia. Consistent with this, inflammatory markers had improved or normalized by neutropenia onset, though this information was not available for one subject. No other medications known to cause myelosuppression were administered concurrently with antibiotics. Neutropenia onset occurred at a median of 23 days after antibiotic initiation (range 10–37 days) with neutropenia lasting a median of 8.5 days (range 2–24 days); however, duration was only assessed in 6/10 subjects due to lack of repeat CBCs in the remainder.
Microbiota diversity contraction and decreased representation of Lachnospiraceae taxa are associated with neutropenia
Seventy‐six stool samples obtained from 28 control and 10 neutropenic subjects were analyzed by 16S ribosomal RNA gene sequencing. Beta diversity reflecting the total diversity of species represented in each group was assessed by weighted UNIFRAC distance and showed significant differences (Adonis p = 0.031) in the overall composition between groups and timepoints (all samples, Figure 3A). However, no significant difference was noted when comparing neutropenic and control subjects at the baseline/recovery or antibiotic‐treated timepoints (not shown), indicating that the major differences in beta diversity were due to antibiotic use, as expected. Changes in community composition by taxa at baseline/recovery (top) and with antibiotic treatment (bottom) in both groups are shown in Figure 3B, notably suggesting decreased representation of the Clostridia class (green) with antibiotic treatment only in neutropenic subjects. Alpha diversity assessed by richness reflects the total number of different bacterial taxa (Amplicon Sequence Variants, ASVs) detected within a sample and was significantly decreased in both groups as a result of antibiotic treatment, as expected. The decrease in variability was more significant for subjects with neutropenia (median ASV baseline/recovery = 124.5, median ASV abx‐treated = 29), suggesting increased disruption from baseline compared to the control group (median ASV baseline/recovery = 111.5, median ASV abx‐treated = 50.5; p = 0.021 vs. p = 0.043, respectively) (Figure 3C). Assessment of alpha diversity via Pielou's Evenness and Shannon Index did not show significance in either group; however, changes in the neutropenic group approached significance.
Figure 3.
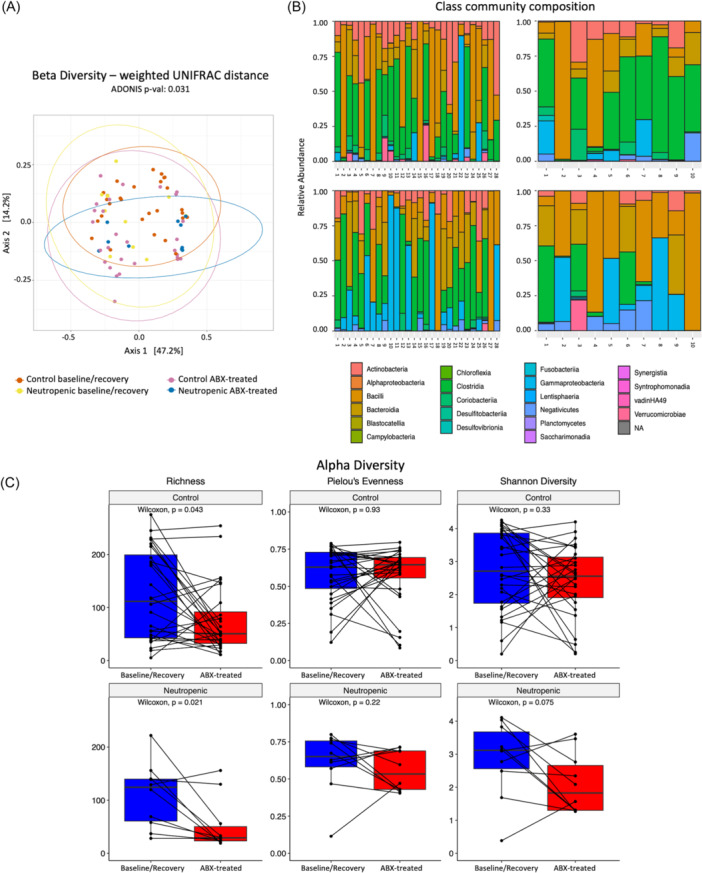
Neutropenic subjects have reduced stool microbiome diversity and decreased predominance of Lachnospiraceae by 16S sequencing. (A) Beta diversity assessed by weighted UNIFRAC distance. (B) Class community composition by groups (control on the left, neutropenic on the right) at baseline/recovery timepoint (top) and with antibiotic treatment (bottom). (C) Alpha diversity, assessed by richness, Pielou's Evenness, and Shannon diversity. Statistical significance is defined by p < 0.05 as determined by the Adonis test for beta‐diversity, and p < 0.05 determined by the Wilcoxon test for alpha‐diversity analysis (n = 10 neutropenic, 28 controls).
Differential analysis of taxa abundance across groups and timepoints was performed with DESeq2 (see Supporting Information Methods). Volcano plots showing differentially abundant taxa between groups and time points are displayed in Figure S1. Table 3 shows a list of taxa with significant +log2FC at baseline/recovery versus with antibiotic treatment uniquely in the neutropenic group (i.e., decreased representation with antibiotic treatment in neutropenic subjects but not controls), as well as taxa with significant +log2FC with antibiotic treatment in control subjects compared to neutropenic subjects (i.e., with increased representation after antibiotic treatment in control but not neutropenic subjects). Among the 17 taxa included, 10 belong to the Lachnospiraceae family, which includes Roseburia, Blautia, Anaerostipes, Eubacterium, Ruminococcus, and Clostridioides genera among others. For 2 of these 17 taxa, namely Lachnospiraceae [Eubacterium] fissicatena group (species NA) and Erysipelotrichaceae [Clostridium] innocuum group (species NA), the mean abundance significantly decreased with antibiotic treatment by Dunn's repeat comparisons test (p adj = <0.05) in patients with neutropenia, showing a near complete loss of the latter (Figure 4A,B). There was a trend toward a decreased abundance of Blautia faecis with antibiotic treatment only in neutropenic subjects (unadjusted p = 0.053; Figure 4C). Additional ground truth plots for taxa that achieved significance only by unadjusted p‐values are shown in Figures S2A–E. A comparison of the combined abundances for the ten species belonging to the Lachnospiraceae family shown in Table 3 demonstrates significantly lower mean abundance in neutropenic subjects with antibiotic treatment but not in controls (Figure 4D). Given that the relative abundance of bacteria may vary significantly between individuals, we also plotted the data as a change in average abundance between timepoints for each subject. Plots for Clostridium innocuum and Blautia faecis are shown in Figure 4E,F. Altogether, there was a striking loss of abundance of Lachnospiraceae among neutropenic but not control subjects.
Table 3.
Several species among the Lachnospiraceae family and others had significant changes in normalized abundance with antibiotic treatment in the neutropenic subjects but not in the controls based on DESeq2 analysis.
| Family | Genus | Species | log2FoldChange | p value | p adj |
|---|---|---|---|---|---|
| Differentially abundant at baseline/recovery versus on antibiotics in neutropenic but not control subjects. (positive log2FoldChange means more abundant at baseline/recovery) | |||||
| Clostridiaceae | Clostridium sensu stricto 1 | NA | 24.5728 | 9.57E−17 | 1.93E−15 |
| Lachnospiraceae | Roseburia | intestinalis | 24.2399 | 2.46E−16 | 3.95E−15 |
| Lachnospiraceae | Roseburia | inulinivorans | 24.2319 | 2.52E−16 | 3.95E−15 |
| Veillonellaceae | Dialister | invisus | 23.0949 | 5.81E−15 | 5.85E−14 |
| Ruminococcaceae | Incertae Sedis | NA | 22.1833 | 6.54E−14 | 5.42E−13 |
| Lachnospiraceae | Coprococcus | comes | 22.0889 | 8.12E−14 | 6.36E−13 |
| Lachnospiraceae | Lachnospiraceae NK4A136 group | NA | 8.3500 | 0.0034 | 0.0221 |
| Peptostreptococcaceae | Romboutsia | NA | 7.4296 | 0.0073 | 0.0445 |
| Lachnospiraceae | Anaerostipes | hadrus | 26.6619 | 3.97E−21 | 5.59E−19 |
| Ruminococcaceae | Ruminococcus | bromii | 25.9072 | 1.92E−18 | 6.76E−17 |
| Lachnospiraceae | [Eubacterium] fissicatena group | NA | 25.5143 | 6.19E−18 | 1.75E−16 |
| Lachnospiraceae | Agathobacter | NA | 23.2200 | 4.10E−15 | 4.60E−14 |
| Enterococcaceae | Enterococcus | NA | 23.2109 | 4.24E−15 | 4.60E−14 |
| Lachnospiraceae | Lachnoclostridium | NA | 22.4440 | 3.30E−14 | 2.91E−13 |
| Erysipelotrichaceae | [Clostridium] innocuum group | NA | 7.1168 | 0.0003 | 0.0024 |
| Differentially abundant with antibiotics in control subjects compared to neutropenic subjects | |||||
| Lachnospiraceae | Blautia | NA | 21.1322 | 2.49E−10 | 6.95E−09 |
| Lachnospiraceae | Blautia | faecis | 8.5864 | 0.0014 | 0.0358 |
Note: The table shows species with significant fold changes in abundance with antibiotic treatment in subjects with neutropenia that did not differ or differed in the opposite direction in controls. Two additional taxa of interest were noted to be more abundant with antibiotic treatment in controls compared to neutropenic subjects (bottom). Statistical significance is defined as p < 0.05 determined by Wald test (NA = no specie(s) was associated with the sequence by ASV dataset).
Figure 4.
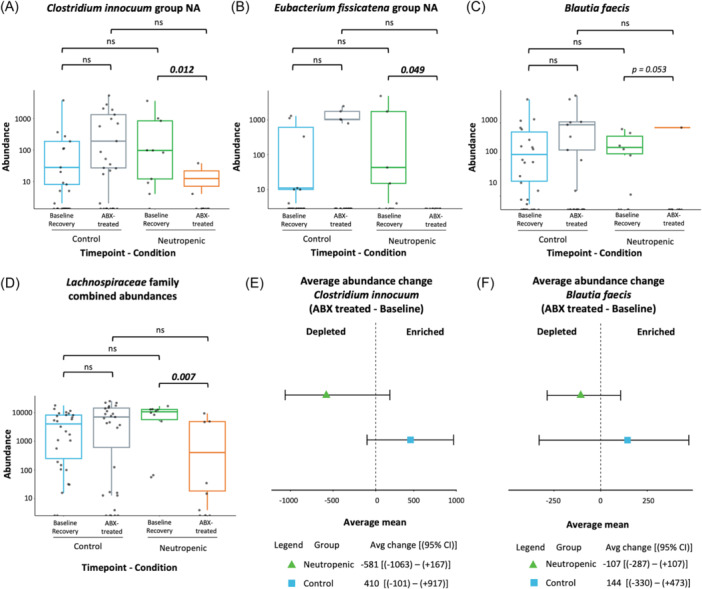
16S rRNA sequencing of stool samples reveals lower mean abundances of several bacterial species after antibiotic treatment in neutropenic subjects. (A–C) Mean abundances by groups at different timepoints. (D) Combined mean abundances for ten species of the Lachnospiraceae family by groups and timepoints. (E, F). Forest plots for Clostridium innocuum and Blautia faecis are shown. Statistical significance is defined by p adj <0.05 as determined by Dunn's multiple comparisons test for mean abundance comparisons (n = 10 neutropenic, 28 controls).
Subgroup 16S rRNA sequencing analysis of only the prospectively enrolled subjects was performed (N = 34: 28 controls and 6 neutropenic subjects). No significant differences were noted between groups for alpha and beta diversity. Notably, despite the very small sample size for this subset analysis, DESeq2 analysis for differential taxa abundance demonstrated a trend toward decreased mean abundance of Clostridium innocuum group NA after antibiotic treatment only in neutropenic subjects, approaching significance (unadjusted p = 0.051 by Dunn test, Figure S2).
Depletion of specific stool metabolites was associated with an increased likelihood of neutropenia development
Untargeted metabolomic profiling was performed on stool samples for all subjects at baseline/recovery and antibiotic‐treated timepoints. Overall, 1054 metabolites were detected, of which 849 were annotated products of endogenous or bacterial/fungal metabolism. Of these 849 metabolites, 843 were present in both groups before and after antibiotics, indicating high sample quality and adequate sensitivity of detection of all metabolites across groups. In Partial Least Squares‐Discriminant Analysis (sPLS‐DA), the metabolites contributing the most to the differences in metabolomes between groups and timepoints are represented in Components One and Two (the top 50 make up the former, and the following 50 the latter). Metabolites that contributed to Components One and Two are shown in Figure S3C and Table S3. The sPLS‐DA analysis demonstrated that metabolomes from subjects with neutropenia and controls generally overlapped both at baseline/recovery (Figure 5A) and with antibiotic treatment (Figure 5B), although there was a slight left shift along Component One in baseline/recovery samples from neutropenic subjects compared to baseline controls.
Figure 5.
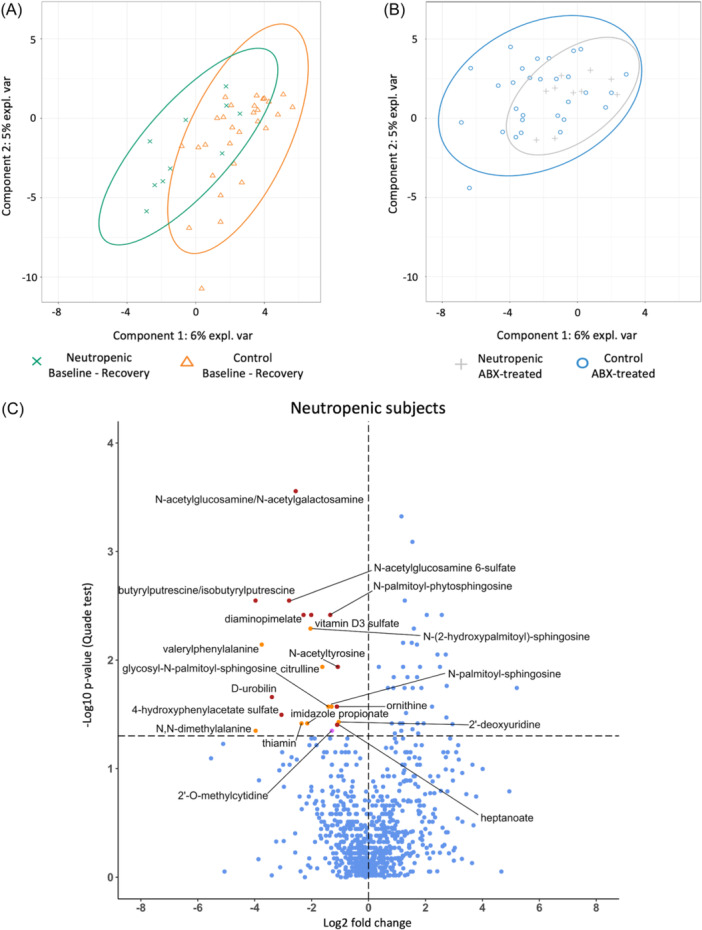
Twenty stool metabolites were depleted after antibiotic treatment in neutropenic subjects but not in controls. Stool samples were assessed by untargeted metabolomic analysis (A, B). sPLS‐DA analysis shows that metabolomes between groups generally overlap between groups at baseline (and/or recovery) (A) and during (B) antibiotic treatment. (C) Volcano plot with annotations shows the twenty‐one metabolites that were significantly depleted with antibiotic treatment in neutropenic but not control subjects (orange dots represent metabolites that contribute to sPLS‐DA Component One, purple dot for Component Two; remainder of metabolites shown with red dots). Horizontal dotted line indicates p = 0.05, statistical significance is defined as p < 0.05 determined by Quade test. (n = 10 neutropenic, 29 controls).
We further conducted a subset sPLS‐DA analysis using only samples from the prospectively enrolled subjects. In this reduced dataset, the two pretreatment groups were nearly indistinguishable, whereas the posttreatment neutropenic group was clearly separated from the rest (Figure S3). In this subset analysis, Component One largely distinguished pre‐treatment from posttreatment groups, whereas Component Two identified a separate metabolomic signature that separates neutropenic posttreatment samples from nonneutropenic post‐samples.
To further understand relationships between metabolites altered with antibiotic treatment in the study samples, we performed over‐representation analysis (ORA) including samples from all 10 neutropenic subjects and 29 controls using sPLS‐DA component loadings. This analysis indicated that Component One was enriched for metabolites involved in pantothenate and coenzyme A biosynthesis, whereas Component Two demonstrated enrichment for metabolites involved in sphingolipid metabolism, pyrimidine metabolism (upstream of urea cycle), butyrate metabolism, and thiamine metabolism (Figure S3).
Using the sPLS‐DA component loadings for the metabolites contributing to Components One and Two, logistic regression models were computed to establish odds of neutropenia relative to either metabolite abundance at the baseline/recovery timepoint or the difference in abundance with antibiotic treatment (the latter adjusted for baseline abundance) (Tables 4 and S5). According to this analysis, a baseline OR <1 indicates that low baseline/recovery abundance (>1 SD) of that metabolite was associated with neutropenia. At baseline, no metabolites with low abundance were associated with neutropenia. However, high abundances of N(6)‐methyllysine, alanylproline, phenylacetyltaurine, 3‐carboxy‐4‐methyl‐5‐pentyl‐2‐furanpropionate, diacetylspermidine, and N‐palmitoyltaurine were each associated with neutropenia (aOR >1). We also assessed whether a change in metabolite abundance was associated with neutropenia, with a change in aOR <1, indicating that a decrease in abundance of that metabolite (>1 SD) from the untreated to treated timepoint was associated with neutropenia (Table 4, far right column). Decreases in 4‐ureidobutyrate and citrulline after antibiotic treatment were associated with increased odds of neutropenia (aOR, change <1), whereas increases in N‐lactoyl phenylalanine and fructose were associated with increased odds of neutropenia (aOR, change >1). All metabolites associated with neutropenia by baseline or change in abundance in this logistic regression analysis were part of Component One except for citrulline, which is part of Component Two. Notably, when subset analysis was completed using the prospectively obtained samples, alanylproline, phenylacetyltaurine, and 3‐carboxy‐4‐methyl‐5‐pentyl‐2‐furanpropionate were no longer significantly associated with neutropenia, but the remaining results were unchanged.
Table 4.
Several metabolites mapped to PLS‐DA components one or two demonstrated significant (p < 0.05) associations with neutropenia based on the change in abundance between timepoints adjusted for baseline abundance (aOR change) or based on the baseline abundance (OR baseline).
| Metabolite | Metabolic pathway annotationa | PLS‐DA component | Component rankingb | OR (95% CI), baselinec | aOR (95% CI), changed |
|---|---|---|---|---|---|
| Citrulline | Urea cycle; Arginine and Proline Metabolism | 2 | 12 | 1.19 (0.61–2.30) | 0.08 (0.01–0.67) |
| 4‐ureidobutyrate | Pyrimidine Metabolism, Uracil containing | 1 | 2 | 1.59 (0.60–4.19) | 0.21 (0.05–0.77) |
| Fructose | Fructose, Mannose and Galactose Metabolism | 1 | 43 | 0.93 (0.79–1.09) | 2.47 (1.05–5.84) |
| N‐lactoyl phenylalanine | Lactoyl Amino Acid | 1 | 33 | 0.55 (0.19–1.56) | 3.89 (1.07–14.2) |
| Diacetylspermidine | Polyamine Metabolism | 1 | 32 | 1.46 (1.00–2.14) | 0.76 (0.47–1.24) |
| Alanylproline | Dipeptide | 1 | 8 | 2.35 (1.07–5.15) | 0.60 (0.24–1.50) |
| N6‐methyllysine | Lysine Metabolism | 1 | 3 | 3.10 (1.16–8.28) | 0.57 (0.22–1.50) |
| N‐palmitoyltaurine | Endocannabinoid | 1 | 34 | 3.40 (1.15–10.1) | 0.47 (0.12–1.91) |
| 3‐carboxy‐4‐methyl‐5‐pentyl‐2‐furanpropionate | Fatty Acid, Dicarboxylate | 1 | 26 | 3.47 (1.1–10.93) | 1.12 (0.67–1.87) |
| Phenylacetyltaurine | Acetylated Peptides | 1 | 22 | 12.8 (1.13–146.2) | 0.31 (0.03–3.29) |
| Ribitol | Pentose Metabolism | 1 | 38 | 0.37 (0.12–1.13) | 1.20 (0.69–2.08) |
| N‐acetyl‐1‐methylhistidine | Histidine Metabolism | 1 | 28 | 0.65 (0.18–2.41) | 0.43 (0.16–1.17) |
| Nicotinate | Nicotinate and Nicotinamide Metabolism | 1 | 30 | 1.36 (1.00–1.84) | 1.02 (0.75–1.38) |
| 4‐hydroxyphenylacetate | Phenylalanine Metabolism | 1 | 15 | 1.40 (0.78–2.52) | 0.17 (0.03–1.06) |
| N2‐acetyllysine | Lysine Metabolism | 2 | 16 | 1.45 (1.00–2.10) | 0.79 (0.49–1.27) |
| N‐acetyltaurine | Methionine, Cysteine, SAM and Taurine Metabolism | 1 | 37 | 1.48 (0.96–2.31) | 0.84 (0.61–1.17) |
| 3‐hydroxyvalerate | Fatty Acid, Monohydroxy | 2 | 29 | 1.57 (0.92–2.67) | 1.06 (0.52–2.14) |
| Pantothenate | Pantothenate and CoA Metabolism | 1 | 13 | 1.89 (0.98–3.63) | 0.74 (0.39–1.38) |
| Lactosyl‐N‐stearoyl‐sphingosine (d18:1/18:0) | Lactosylceramides | 1 | 47 | 2.54 (0.87–7.42) | 0.60 (0.22–1.59) |
| N6‐acetyllysine | Lysine Metabolism | 1 | 7 | 2.66 (0.86–8.21) | 0.84 (0.38–1.87) |
| N6‐carboxyethyllysine | Lysine Metabolism | 1 | 6 | 2.99 (0.96–9.33) | 0.83 (0.41–1.68) |
| Tyrosol | Tyrosine Metabolism | 1 | 9 | 3.10 (0.76–12.6) | 0.21 (0.04–1.12) |
| Beta‐sitosterol | Sterol | 1 | 45 | 3.44 (0.95–12.5) | 1.09 (0.40–2.98) |
| 2‐hydroxysebacate | Fatty Acid, Dicarboxylate | 1 | 18 | 4.51 (0.84–24.2) | 0.73 (0.29–1.81) |
Abbreviations: aOR, adjusted odds ratio; CI, confidence interval; PLS‐DA, partial least squares‐discriminant analysis; OR, odds ratio.
Per Metabolon.
From 1 (highest) to 50 (lowest), based on the absolute value of the loading score.
OR and 95% CI of neutropenia at the abx‐treated timepoint per one standard deviation increase in metabolite abundance at the baseline‐recovery timepoint (baseline).
OR and 95% CI of neutropenia at the abx‐treated timepoint per one standard deviation increase in metabolite abundance from the baseline‐recovery timpoint (baseline) to the abx‐treatment (follow‐up) timepoint. Adjusted for baseline metabolite abundance.
Several metabolites were uniquely depleted after antibiotic treatment in subjects with neutropenia
We further conducted a Quade test to identify changes in specific stool metabolites with antibiotic treatment in neutropenic versus control subjects. This analysis capitalizes on the paired longitudinal sampling in our study by accounting for subject‐level effects. Quade analysis revealed that 21 metabolites were significantly depleted with antibiotic treatment in subjects with neutropenia but not in controls (Figure 5C). These metabolites include the bacterial cell wall component N‐acetylglucosamine, the urea cycle intermediates citrulline and ornithine; as well as the pyrimidine metabolism intermediary 2′‐deoxyuridine. Results of the statistical analysis by Quade test for these metabolites are shown in Table S4. When a similar analysis was applied to only the prospectively enrolled subset of subjects, N‐acetylglucosamine remained significantly depleted in only the neutropenic subjects. Whereas urea cycle intermediate ornithine was no longer significant, N‐delta‐acetyl‐ornithine emerged. Further, whereas 2′‐deoxyuridine was no longer significant, another pyrimidine metabolism intermediate 2′‐O‐methylcytidine emerged. Altogether these results indicate a strong significant depletion in N‐acetylglucosamine in neutropenic subjects as well as changes in urea cycle and pyrimidine synthesis metabolites.
Notably, urea cycle intermediary citrulline was significantly depleted with antibiotic treatment only in neutropenic subjects by both sPLS‐DA analysis and Quade test (Figure 5C, Tables 4 and S5). Metabolites related to pyrimidine and butyrate metabolism surfaced in both the ORA analysis and the logistic regression of PLS‐DA analysis (Figure S3). Altogether, these analyses identified several microbially‐derived metabolites specifically changed in neutropenic subjects but not controls.
DISCUSSION
In this study, we enrolled subjects receiving prolonged courses of antibiotics and compared clinical and biological characteristics between subjects who developed neutropenia with controls who maintained normal neutrophil counts despite antibiotic treatment. In the prospective arm of our study, 17% of patients developed neutropenia, similar to the previously reported literature. 3 , 5 , 8 Despite a small sample size and high clinical variability between study subjects, neutropenia was associated with contraction of gastrointestinal microbiome diversity, and taxonomic analysis revealed that decreased abundance of several species within the Lachnospiraceae family was associated with neutropenia after prolonged antibiotic treatment. Stool metabolites, including bacterial cell wall component N‐acetylglucosamine/N‐acetylgalactosamine and urea cycle metabolites citrulline and ornithine, were depleted only in subjects with neutropenia. Additionally, sPLS‐DA and ORA analysis suggested that decreases in 4‐ureidobutyrate and citrulline after antibiotics were associated with increased odds of neutropenia. Interestingly, citrulline and butyrate levels have been reported to correlate with the prevalence of Lachnospiraceae in prior studies. 29 Altogether, this study provides critical clinical data supporting published preclinical studies that (1) commensal bacteria support steady‐state hematopoiesis by supporting the production of specific metabolites and (2) their disruption by prolonged antibiotic exposure may result in antibiotic‐associated cytopenias. Our data suggest that loss of Lachnospiraceae among patients with neutropenia leads to the depletion of metabolites critical for steady‐state hematopoiesis, resulting in antibiotic‐associated neutropenia (Figure 6).
Figure 6.
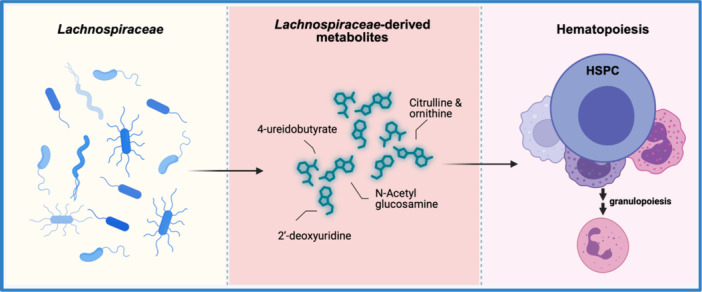
Proposed model of Lachnospiraceae ‐sustained hematopoiesis. Disruption of the intestinal microbiome with depletion of Lachnospiraceae contributes to antibiotic‐associated neutropenia. We hypothesize that Lachnospiraceae‐derived metabolites including citrulline, 4‐ureidobutyrate, 2′‐deoxyuridine, and N‐acetylglucosamine are absorbed from the intestine and travel to the bone marrow where they promote steady‐state hematopoiesis. Figure was created using BioRender.
The literature has long described bone marrow suppression as a result of prolonged antibiotic therapy. 30 , 31 , 32 This clinical problem continues to interfere with the treatment of infections and increase the risks of morbidity and mortality, as well as healthcare costs. 15 Pre‐clinical findings from murine studies support the idea of microbiota‐sustained steady‐state hematopoiesis and our recent work establishes that type I IFN‐STAT1 signaling is critical in this interaction. 20 Our current study focuses on adding to these preclinical data in the clinical setting by analyzing both the intestinal microbiome in stool samples from otherwise healthy human patients and disruption patterns in the microbiome after antibiotic therapy. In addition, we investigated the correlation between disruption in the microbiome to the development of antibiotic‐associated neutropenia.
Our cohort was very heterogeneous in that a wide array of antibiotic classes, infective microorganisms, and organ systems infected were represented. While recognizing this limitation, we did not find an association between these characteristics and neutropenia. We identified associations between neutropenia and the total length of antibiotic therapy, length of IV antibiotic therapy, length of admission to the hospital, and admission to the ICU (Table 2). Previous reports support the increased risk of neutropenia with longer treatment courses, as we saw in our cohort. 33 , 34 While standard intravenous and oral dosing for many common antibiotics (e.g., clindamycin) results in similar blood concentrations, prolonged oral antibiotic administration did not correlate with neutropenia in our cohort. One possible explanation is that more severe infections that tend to require a longer IV treatment duration also have a greater impact on hematopoiesis. In particular, we wonder whether the impact of osteomyelitis on bone marrow physiology, when combined with microbiota‐mediated effects, may increase the risk of cytopenia. Even though some studies suggest that PO antibiotics impact the gut microbiome in a direct or local manner and that IV antibiotics have an impact only through biliary excretion 35 ; recent studies typically link both IV and PO antibiotics with intestinal microbiota changes, including the well‐studied influence of intrapartum IV antibiotic prophylaxis in neonatal microbiota composition. 26 , 27 , 36 , 37 Our detailed review did not reveal ICU‐specific events that could further disrupt the intestinal microbiota and explain the correlation noted in our cohort. Further studies in larger cohorts could help elucidate whether common aspects of admission to the ICU, such as stress, anesthesia, or nutritional status, increase the likelihood of antibiotic‐associated neutropenia. While alternative mechanisms of neutropenia such as drug‐induced, immune‐mediated, or sepsis and cytokine storm‐induced myelosuppression are plausible 11 , 12 ; our comprehensive review of fever curve, concomitant symptoms, inflammatory markers, clinical signs and symptoms, other medications administered, as well as overall improvement of infection (versus lack thereof) suggest that those mechanisms are not at play in our cohort. Interestingly, both in the literature and in our cohort, neutropenia resolves shortly after discontinuation of antibiotics, and in some cases, even while continued exposure to antibiotics exists, 3 , 8 when the microbiome composition remains altered. The resolution of antibiotic‐associated neutropenia while the microbiome has not yet recuperated is interesting and represents an area for future investigation. The ability of HSCs to go from “resting” steady‐state blood production to “emergency” hematopoiesis upon different stressors, including infection and inflammation is an area of active research. 38 , 39 , 40 We speculate that the prompt recovery of antibiotic‐associated neutropenia derives from the activation of these mechanisms of emergency hematopoiesis, bypassing the effect of the depletion of intestinal bacteria and its metabolites.
We did not identify a correlation between neutropenia and antibiotic class, number of antibiotic classes used, organ/system involved, or isolated microorganisms. Although beta‐lactams are classically recognized to cause neutropenia, several other classes of antibiotics were represented in our neutropenic cohort, which coincides with what has been previously described in the literature. 4 , 8 , 9 , 12 Even though no specific class of microorganism was associated with neutropenia, the predominance of gram‐positive infections reflects the overall high prevalence of these infections in the pediatric population. 41 , 42 However, our study was not powered to detect such differences and larger prospective studies will be informative to address these questions.
As expected, microbiome richness was decreased in both groups with antibiotic treatment. However, this depletion was more significant in subjects with neutropenia, suggesting that more profound microbiome disruption alters steady‐state hematopoiesis and increases the likelihood of developing cytopenias. Interestingly, differential taxa abundance analysis showed a decreased predominance of several species within the Lachnospiraceae family and other Clostridia (Table 3). This family is a phylogenetically and morphologically heterogeneous taxon belonging to the clostridial cluster XIVa of the phylum Firmicutes and is among the top short‐chain fatty acid (SCFA) producers. Within the Lachnospiraceae family, Blautia, Coprococcus, Dorea, Lachnospira, Oribacterium, Roseburia, and L‐Ruminococcus are the main genera that metagenomic analyses have detected in the human intestinal microbiota. 29 Changes in Lachnospiraceae abundance have been linked to both health and disease, including metabolic syndrome, obesity, diabetes, liver diseases, inflammatory bowel disease, and chronic kidney disease. 29 Lower abundance of Lachnospiraceae was associated with febrile neutropenia in hematopoietic stem cell transplant patients in a recent study. 43 Interestingly, Jenq et al noted that depleting the genus Blautia was associated with worse outcomes after HSC transplantation and higher rates of GVHD. 23 Notably, in our cohort, Blautia faecis was significantly more represented by DESEQ analysis in samples from control subjects with antibiotics when compared to samples from neutropenic subjects during antibiotic treatment (Table 3).
We identified many metabolites that were depleted after antibiotic treatment only in subjects with neutropenia (Figure 5C, Table S4) and that correlated with increased likelihood of neutropenia (Table 4) based on regression models performed with sPLS‐DA component loadings. Notably, several of these metabolites were also detected in serum and/or stool from leukopenic mice in our prior murine work, with significant changes in pyrimidine metabolites such as orotidine and 3‐Amino‐2‐piperidone in the serum of leukopenic mice. 20 The sPLS‐DA analysis of metabolomes showed a slight shift in the metabolomic profile of subjects who went on to develop neutropenia (Figure 5A,B), suggesting lower abundance of certain Component One metabolites at baseline may predispose to antibiotic‐associated neutropenia.
Excitingly, a number of metabolites identified in our metabolomic study are related to bacterial species identified orthogonally through our microbiome analysis. Among these, low plasma levels of citrulline have been associated with decreased abundance of Lachnospiraceae in severely malnourished patients with anorexia nervosa receiving enteral nutrition. 44 We also observed decreased 4‐ureidobutyrate, an intermediary of pyrimidine metabolism, in neutropenic subjects. Pyrimidine supplementation has been reported to correlate with the restoration of SCFA‐producing bacteria, including Lachnospiraceae, in a number of studies. 29 , 45 Finally, N‐acetylglucosamine is a bacterial cell wall component which, while not specific to Lachnospiraceae, is known to support interferon responses by catalyzing phosphorylation of interferon regulatory factor 3 (IRF3). 46 This provides a potential direct link between microbiome changes and tonic interferon signaling identified as a mediator of microbiome‐sustained hematopoiesis in our prior preclinical studies. Based on these findings, we propose a model for Lachnospiraceae‐sustained hematopoiesis that builds on our previous findings derived from murine work. 20 Specifically, we propose that bacterial metabolites such as N‐acetylglucosamine and urea cycle metabolites from Lachnospiraceae are absorbed into the circulation and travel to the bone marrow where they potentiate tonic interferon signaling to support normal blood production (Figure 6). We speculate that these microbial metabolites may bind to intracellular receptors such as Rig‐I‐like receptors to trigger Type I interferon production and, in preliminary work, we have found that rescue of Type I interferon signaling restores normal blood counts in antibiotic‐treated mice. Depletion of Lachnospiraceae by antibiotic therapy would thus contribute to cytopenias by reducing the availability of these key metabolites. Further confirmation of this model in animal experiments may help elucidate the exact mechanisms through which microbiome‐derived metabolites exert their impact on normal hematopoiesis.
There were many limitations to our study. First, it was limited by a small sample size of neutropenic subjects. Further, the subject cohort was very heterogeneous in race, sex, age, and clinical characteristics. This introduces significant limitations as it is well known that changes in the diet through development from infancy to adulthood impact microbiome composition. Additionally, some of the antibiotic classes were rarely used to treat subjects in our cohort—specifically aminoglycosides, macrolides, and carbapenems. These limitations increase the risk of misleading or overstated conclusions. In the future, larger sample sizes may increase our ability to detect differences between groups and minimize the effect of age and diet by being able to perform subgroup analyses. While we analyzed the total length of antibiotic therapy and the number of classes used, a more in‐depth analysis accounting for the treatment duration of each antibiotic and timing of administration with respect to neutropenia development could unveil specific associations.
The low frequency of antibiotic‐associated neutropenia motivated the creation of a retrospective arm to facilitate study enrollment. Coanalyzing data from subjects who were enrolled prospectively and retrospectively in a single group could introduce bias to our analyses. Additionally, the study design allowed for stool samples intended to represent the baseline intestinal microbiota to be collected after several days of antibiotic exposure (in the prospective arm) or a few weeks after completion of antibiotic treatment (in the retrospective arm), which affects our ability to interpret the findings at the baseline/recovery timepoint. However, the lack of significant differences in beta diversity analysis between groups, as well as the overlapped detection of 843 out of 849 metabolites in all groups suggests that baseline/recovery samples were affected similarly by recent antibiotic exposure. Furthermore, the fact that numerous findings, including the decreased prevalence of Lachnospiraceae and reduced prevalence of metabolites involved in the urea cycle and pyrimidine synthesis, remained true in a subset analysis of only prospectively enrolled subjects lends strength to these findings. Finally, the identification of consistent differences across a highly diverse group of subjects supports the relevance of these findings in real‐world clinical conditions.
To our knowledge, this is the first study to analyze microbiome composition changes and intestinal microbiota‐derived metabolites after prolonged antibiotic therapy and their relationship to antibiotic‐associated neutropenia in a pediatric population. Our study provides the first evidence for the importance of Lachnospiraceae and its metabolites in steady‐state hematopoiesis and reveals bacterial species and metabolites of interest that may be predictive of neutropenia risk or therapeutically useful in preventing and treating neutropenia. Improved definition and validation of these findings will help us develop prediction models that will identify patients at risk of developing antibiotic‐associated neutropenia. This will be of particular importance in immunologically fragile populations such as HSCT recipients for whom neutropenia is especially dangerous and in whom strategies to prevent the microbiome‐driven impact in hematopoiesis could help shorten periods of neutropenia and lessen the risk of infection.
AUTHOR CONTRIBUTIONS
Hannah Yan, Katherine Y. King, and Megan T. Baldridge designed the study protocol. Robert R. Jenq and Pavan Reddy provided critical insights. Josaura Fernandez‐Sanchez and Katherine Y. King proposed and reviewed subsequent protocol amendments. Hannah Yan, Josaura Fernandez‐Sanchez, and Nusrat Shaikh enrolled subjects and, together with Arushana A. Maknojia, coordinated sample collection and processing; Hope Hendricks and Ritu Banerjee enrolled subjects at participating site. Josaura Fernandez‐Sanchez collected, analyzed, and interpreted clinical data. Jeremy M. Schraw analyzed and interpreted metabolomics results; Marlyd E. Mejia, Rachel Rodgers, and Robert R. Jenq analyzed and interpreted 16S rRNA sequencing results; Josaura Fernandez‐Sanchez and Katherine Y. King wrote the manuscript; all authors have read and agreed to the published version of the manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
FUNDING
This project was supported by the National Institutes of Health (NIH) R01 AI141716 (M.T.B. and K.Y.K.), R35HL155672 (K.Y.K.), F31HL168921 (A.M.), T32GM136554 (A.M.), F31HL147514 A1 (H.Y.) and F31AI167538 (M.E.M). J.F.S. was supported by the Baylor College of Medicine Comprehensive Cancer Training Program via the Cancer Prevention & Research Institute of Texas Training Award RP210027.
Supporting information
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
The authors thank the Division of Pediatrics Center for Research Advancement at Texas Children's Hospital—Baylor College of Medicine for their support with subject screening and enrollment and the Center for Metagenomics and Microbiome Research at Baylor College of Medicine. We acknowledge Enrico Moiso PhD for his contribution with the initial analysis of 16S rRNA sequencing data. We thank Catherine Gillespie for editing the manuscript.
DATA AVAILABILITY STATEMENT
Data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/bioproject/PRJEB72348/, reference number PRJEB72348. Original 16S rRNA gene sequencing data and untargeted metabolomics data provided by the BCM CMMR core and Metabolon are available through NCBI ENA accession PRJEB72348 and Mendeley/Digital Common data doi:10.17632/38z7h96km4.1, respectively.
REFERENCES
- 1. CDC . Antibiotic Use in the United States, 2022 Update: Progress and Opportunities. Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Division of Healthcare Quality Promotion (DHQP); 2022.
- 2. Tice AD, Rehm SJ, Dalovisio JR, et al. Practice guidelines for outpatient parenteral antimicrobial therapy. Clin Infect Dis. 2004;38(12):1651‐1671. 10.1086/420939 [DOI] [PubMed] [Google Scholar]
- 3. Gomez M, Maraqa N, Alvarez A, Rathore M. Complications of outpatient parenteral antibiotic therapy in childhood. Pediatr Infect Dis J. 2001;20(5):541‐543. 10.1097/00006454-200105000-00015 [DOI] [PubMed] [Google Scholar]
- 4. Same RG, Hsu AJ, Cosgrove SE, et al. Antibiotic‐associated adverse events in hospitalized children. J Pediatric Infect Dis Soc. 2021;10(5):622‐628. 10.1093/jpids/piaa173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Olson SC, Smith S, Weissman SJ, Kronman MP. Adverse events in pediatric patients receiving long‐term outpatient antimicrobials. J Pediatric Infect Dis Soc. 2015;4(2):119‐125. 10.1093/jpids/piu037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Furtek KJ, Kubiak DW, Barra M, Varughese CA, Ashbaugh CD, Koo S. High incidence of neutropenia in patients with prolonged ceftaroline exposure. J Antimicrob Chemother. 2016;71(7):2010‐2013. 10.1093/jac/dkw062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. LaVie KW, Anderson SW, O'Neal HR, Rice TW, Saavedra TC, O'Neal CS. Neutropenia associated with long‐term ceftaroline use. Antimicrob Agents Chemother. 2016;60(1):264‐269. 10.1128/AAC.01471-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Madigan T, Banerjee R. Characteristics and outcomes of outpatient parenteral antimicrobial therapy at an academic children's hospital. Pediatr Infect Dis J. 2013;32(4):346‐349. 10.1097/INF.0b013e31827ee1c2 [DOI] [PubMed] [Google Scholar]
- 9. Tamma PD, Avdic E, Li DX, Dzintars K, Cosgrove SE. Association of adverse events with antibiotic use in hospitalized patients. JAMA Intern Med. 2017;177(9):1308‐1315. 10.1001/jamainternmed.2017.1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fernandes P, Milliren C, Mahoney‐West HM, Schwartz L, Lachenauer CS, Nakamura MM. Safety of outpatient parenteral antimicrobial therapy in children. Pediatr Infect Dis J. 2018;37(2):157‐163. 10.1097/INF.0000000000001716 [DOI] [PubMed] [Google Scholar]
- 11. Cimino C, Allos BM, Phillips EJ. A review of β‐lactam‐associated neutropenia and implications for cross‐reactivity. Ann Pharmacother. 2021;55(8):1037‐1049. 10.1177/1060028020975646 [DOI] [PubMed] [Google Scholar]
- 12. Lam PW, Leis JA, Daneman N. Antibiotic‐induced neutropenia in patients receiving outpatient parenteral antibiotic therapy: a retrospective cohort study. Antimicrob Agents Chemother. 2023;67(3):e0159622. 10.1128/aac.01596-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yan H, Baldridge MT, King KY. Hematopoiesis and the bacterial microbiome. Blood. 2018;132(6):559‐564. 10.1182/blood-2018-02-832519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iwamura C, Bouladoux N, Belkaid Y, Sher A, Jankovic D. Sensing of the microbiota by NOD1 in mesenchymal stromal cells regulates murine hematopoiesis. Blood. 2017;129(2):171‐176. 10.1182/blood-2016-06-723742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Josefsdottir KS, Baldridge MT, Kadmon CS, King KY. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood. 2017;129(6):729‐739. 10.1182/blood-2016-03-708594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han H, Yan H, King KY. Broad‐spectrum antibiotics deplete bone marrow regulatory T cells. Cells. 2021;10(2):277. 10.3390/cells10020277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kennedy EA, King KY, Baldridge MT. Mouse microbiota models: comparing germ‐free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol. 2018;9:1534. 10.3389/fphys.2018.01534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Balmer ML, Schürch CM, Saito Y, et al. Microbiota‐derived compounds drive steady‐state granulopoiesis via MyD88/TICAM signaling. J Immunol. 2014;193(10):5273‐5283. 10.4049/jimmunol.1400762 [DOI] [PubMed] [Google Scholar]
- 19. Zeng X, Li X, Li X, et al. Fecal microbiota transplantation from young mice rejuvenates aged hematopoietic stem cells by suppressing inflammation. Blood. 2023;141(14):1691‐1707. 10.1182/blood.2022017514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yan H, Walker FC, Ali A, et al. The bacterial microbiota regulates normal hematopoiesis via metabolite‐induced type 1 interferon signaling. Blood Adv. 2022;6(6):1754‐1765. 10.1182/bloodadvances.2021006816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fernandez Sanchez J, Maknojia AA, King KY. Blood and guts: how the intestinal microbiome shapes hematopoiesis and treatment of hematologic disease. Blood. 2024;143(17):1689‐1701. 10.1182/blood.2023021174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weber D, Jenq RR, Peled JU, et al. Microbiota disruption induced by early use of broad‐spectrum antibiotics is an independent risk factor of outcome after allogeneic stem cell transplantation. Biol Blood Marrow Transplant. 2017;23(5):845‐852. 10.1016/j.bbmt.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jenq RR, Taur Y, Devlin SM, et al. Intestinal blautia is associated with reduced death from graft‐versus‐host disease. Biol Blood Marrow Transplant. 2015;21(8):1373‐1383. 10.1016/j.bbmt.2015.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peled JU, Devlin SM, Staffas A, et al. Intestinal microbiota and relapse after hematopoietic‐cell transplantation. J Clin Oncol. 2017;35(15):1650‐1659. 10.1200/JCO.2016.70.3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peled JU, Gomes ALC, Devlin SM, et al. Microbiota as predictor of mortality in allogeneic hematopoietic‐cell transplantation. N Engl J Med. 2020;382(9):822‐834. 10.1056/NEJMoa1900623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Doan T, Liu Z, Sié A, et al. Gut microbiome diversity and antimicrobial resistance after a single dose of oral azithromycin in children: a randomized placebo‐controlled trial. Am J Trop Med Hyg. 2024;110(2):291‐294. 10.4269/ajtmh.23-0651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gough EK. The impact of mass drug administration of antibiotics on the gut microbiota of target populations. Infect Dis Poverty. 2022;11(1):76. 10.1186/s40249-022-00999-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vicent L, Luna R, Martínez‐Sellés M. Pediatric infective endocarditis: a literature review. J Clin Med. 2022;11(11):3217. 10.3390/jcm11113217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vacca M, Celano G, Calabrese FM, Portincasa P, Gobbetti M, De Angelis M. The controversial role of human gut lachnospiraceae. Microorganisms. 2020;8(4):573. 10.3390/microorganisms8040573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Olaison L, Belin L, Hogevik H, Alestig K. Incidence of β‐lactam–induced delayed hypersensitivity and neutropenia during treatment of infective endocarditis. Arch Intern Med. 1999;159(6):607‐615. 10.1001/archinte.159.6.607 [DOI] [PubMed] [Google Scholar]
- 31. Shah I, Kumar KS, Lerner AM. Agranulocytosis associated with chronic oral administration of cloxacillin for suppression of staphylococcal osteomyelitis. Am J Hematol. 1982;12(2):203‐206. 10.1002/ajh.2830120213 [DOI] [PubMed] [Google Scholar]
- 32. Neftel KA, Hauser SP, Muller MR. Inhibition of granulopoiesis in vivo and in vitro by β‐lactam antibiotics. J Infect Dis. 1985;152(1):90‐98. 10.1093/infdis/152.1.90 [DOI] [PubMed] [Google Scholar]
- 33. Battini V, Mari A, Gringeri M, et al. Antibiotic‐induced neutropenia in pediatric patients: new insights from pharmacoepidemiological analyses and a systematic review. Front Pharmacol. 2022;13:877932. 10.3389/fphar.2022.877932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Solis K, Dehority W. Antibiotic‐induced neutropenia during treatment of hematogenous osteoarticular infections in otherwise healthy children. J Pediatr Pharmacol Ther. 2019;24(5):431‐437. 10.5863/1551-6776-24.5.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang L, Huang Y, Zhou Y, Buckley T, Wang HH. Antibiotic administration routes significantly influence the levels of antibiotic resistance in gut microbiota. Antimicrob Agents Chemother. 2013;57(8):3659‐3666. 10.1128/AAC.00670-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ainonen S, Tejesvi MV, Mahmud MR, et al. Antibiotics at birth and later antibiotic courses: effects on gut microbiota. Pediatr Res. 2022;91(1):154‐162. 10.1038/s41390-021-01494-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xue L, Ding Y, Qin Q, et al. Assessment of the impact of intravenous antibiotics treatment on gut microbiota in patients: Clinical data from pre‐ and post‐cardiac surgery. Front Cell Infect Microbiol. 2023;12:1043971. 10.3389/fcimb.2022.1043971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang J, Erlacher M, Fernandez‐Orth J. The role of inflammation in hematopoiesis and bone marrow failure: what can we learn from mouse models? Front Immunol. 2022;13:951937. 10.3389/fimmu.2022.951937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hormaechea‐Agulla D, Le DT, King KY. Common sources of inflammation and their impact on hematopoietic stem cell biology. Curr Stem Cell Rep. 2020;6(3):96‐107. 10.1007/s40778-020-00177-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Olson OC, Kang YA, Passegué E. Normal hematopoiesis is a balancing act of self‐renewal and regeneration. Cold Spring Harbor Perspect Med. 2020;10(12):a035519. 10.1101/cshperspect.a035519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Babay HA, Twum‐Danso K, Kambal AM, Al‐Otaibi FE. Bloodstream infections in pediatric patients. Saudi Med J. 2005;26(10):1555‐1561. [PubMed] [Google Scholar]
- 42. Kaplan SL. Implications of methicillin‐resistant Staphylococcus aureus as a community‐acquired pathogen in pediatric patients. Infect Dis Clin North Am. 2005;19(3):747‐757. 10.1016/j.idc.2005.05.011 [DOI] [PubMed] [Google Scholar]
- 43. Sardzikova S, Andrijkova K, Svec P, et al. Gut diversity and the resistome as biomarkers of febrile neutropenia outcome in paediatric oncology patients undergoing hematopoietic stem cell transplantation. Sci Rep. 2024;14(1):5504. 10.1038/s41598-024-56242-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hanachi M, Manichanh C, Schoenenberger A, et al. Altered host‐gut microbes symbiosis in severely malnourished anorexia nervosa (AN) patients undergoing enteral nutrition: an explicative factor of functional intestinal disorders? Clin Nutr. 2019;38(5):2304‐2310. 10.1016/j.clnu.2018.10.004 [DOI] [PubMed] [Google Scholar]
- 45. Niu K, Bai P, Zhang J, Feng X, Qiu F. Cytidine alleviates dyslipidemia and modulates the gut microbiota composition in ob/ob mice. Nutrients. 2023;15(5):1147. 10.3390/nu15051147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang S, Jin S, Xian H, et al. Metabolic enzyme UAP1 mediates IRF3 pyrophosphorylation to facilitate innate immune response. Mol Cell. 2023;83(2):298‐313. 10.1016/j.molcel.2022.12.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Data Availability Statement
Data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/bioproject/PRJEB72348/, reference number PRJEB72348. Original 16S rRNA gene sequencing data and untargeted metabolomics data provided by the BCM CMMR core and Metabolon are available through NCBI ENA accession PRJEB72348 and Mendeley/Digital Common data doi:10.17632/38z7h96km4.1, respectively.


