Abstract
Neuroblastoma is the most common extracranial solid tumor in children, and a leading cause of childhood cancer deaths. All neuroblastomas arise from neural crest-derived sympathetic neuronal progenitors, but numerous mutations, the most common of which is MYCN amplification, give rise to these lesions. Epigenetic aberrations also play a role in oncogenesis and tumor progression. To better understand biologic diversity of neuroblastomas, we performed joint single-nucleus ATAC sequencing and single-nucleus RNA sequencing on six neuroblastoma cell lines, three of which are MYCN amplified. After standard filtering for high-quality nuclei, we obtained chromatin accessibility and transcript abundance data from 41,733 neuroblastoma tumor cells. Preliminary analysis reveals significant diversity in chromatin landscape and gene expression across neuroblastoma cell lines. This dataset is a valuable resource for studying the transcriptional and epigenetic mechanisms of this deadly childhood disease.
Subject terms: Paediatric cancer, Paediatric research
Background & Summary
Neuroblastoma is the most common extracranial solid tumor in children, with an especially high incidence among children under age 4 years1. The disease is stratified into low-risk, intermediate-risk, and high-risk categories, based on clinical and biological features. High-risk disease is fatal in over 60% of cases2. MYCN amplification is the most common mutation in neuroblastoma, and the presence of MYCN amplification is sufficient to designate a tumor as high-risk3. However, over half of high-risk lesions lack this mutation4,5, indicating significant biological diversity between neuroblastoma cases. Due to the challenges of obtaining primary tumor samples, various neuroblastoma cell lines have been established from high-risk lesions and are widely used for studying tumor biology.
There are numerous publicly-available transcriptional and epigenetic datasets derived from neuroblastoma cell lines, but information regarding heterogeneity within cell lines is not captured by the bulk methods used to generate these data6–8. The advent of single-cell sequencing tools has permitted high-parameter profiling of transcript abundance and epigenetic features in many cancers9. Several studies have reported single-cell RNA sequencing on neuroblastoma tissue, and have demonstrated considerable transcriptional heterogeneity within tumors as well as between cases10–13. Single-cell multiomic tools have been developed that measure two or more features (such as transcript abundance and chromatin accessibility) simultaneously from individual cells14. To our knowledge, however, multiome technology has not yet been applied to neuroblastoma.
We undertook the present study to better understand transcriptional and epigenetic diversity of human neuroblastoma cell lines, both within and between samples. We utilized six widely-studied neuroblastoma cell lines, features of which are displayed in Table 1, including MYCN amplification status and whether each line is known to have adrenergic or mesenchymal transcriptional circuitry15,16. One of these lines, SH-SY5Y, is a subclone derivative of another, SK-N-SH. By jointly quantifying transcript abundance and chromatin accessibility in individual nuclei, we provide a resource for studying the complex regulation of tumor cell phenotype. We anticipate these data will be extremely useful for identifying the driver genes, transcriptional regulatory networks, and transcription factor-chromatin interactions underlying neuroblastoma cell states.
Table 1.
Characteristics of cell lines studied.
| Cell Line | MYCN status | Mesenchymal vs Adrenergic | Cells retained for analysis |
|---|---|---|---|
| SH-SY5Y | Non-amplified | Adrenergic | 5,480 |
| SK-N-SH | Non-amplified | Mesenchymal | 5,977 |
| SK-N-AS | Non-amplified | Mesenchymal | 7,459 |
| SK-N-D.Z | Amplified | Adrenergic | 5,295 |
| CHP134 | Amplified | not reported in literature15,16 | 5,827 |
| Be2c | Amplified | Adrenergic | 11,695 |
Methods
Cell lines and culture
Cell lines used in this study were purchased from ATCC (Manassas, VA). All cell growth and preparation for sequencing was done at Massachusetts General Hospital. Cells were cultured under conditions recommended by ATCC. All cells were maintained in incubators a 37 °C and 5% CO2. Cells were passaged when they reached approximately 80% confluency. In all cases, cells were prepared for sequencing at a passage number less than 10.
Sample preparation
Nuclei were isolated from individual cells using the 10X Genomics (Pleasanton, CA) Demonstrated Protocol CG000365: Nuclei Isolation for Single Cell Multiome ATAC + Gene Expression Sequencing. Briefly, cells were washed with cold PBS and trypsinized to a single-cell suspension. After pelleting at 300 rcf in a tabletop centrifuge in 15 mL conical tubes, cells were washed twice in 1 mL cold PBS supplemented with 0.04% BSA. Cells were then passed through a 40 μm strainer and counted using a standard hemocytometer. A total of 1,000,000 cells were transferred to 2 mL microcentrifuge tubes. Cells were resuspended in 100 mL of ice-cold Lysis Buffer (10 mM Tris-HCl at pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20, 0.1% NP-40, 0.01% digitonin, 1% BSA, 1 mM DTT, 1 U/mL RNase inhibitors, all in nuclease-free water) and incubated for 4 minutes on ice, followed by addition of 1 mL of Wash Buffer (10 mM Tris-HCl at pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1% BSA, 0.1% Tween-20, 1 mM DTT, and 1 U/mL RNase inhibitors in nuclease-free water). Cells were centrifuged at 500 rcf for 5 minutes at 4 °C to repellet. A total of 3 washes in Wash Buffer were performed. Cells were then suspended in 1 mL of 1x Nuclei Buffer (provided at 20x concentration by 10X Genomics in Chromium Next GEM Single Cell Multiome ATAC Kit A, PN-1000280) supplemented with 1 mM DTT and 1 U/mL RNase inhibitors. GEM generation and cell barcoding were immediately performed using 10X Genomics Chromium Controller and 10X Genomics Next GEM Chip J. ATAC and gene expression library construction was performed per the 10X Genomics Next GEM Single Cell Multiome ATAC + Gene Expression User Guide, with reagents purchased from 10X Genomics. Sequencing was performed on the Illumina NovaSeq platform at the Harvard University Bauer Core facility.
Data analysis
ATAC and gene expression FASTQ output files were demultiplexed and mapped to the Genome Refence Consortium hg38 reference genome with Cell Ranger ARC (10X Genomics) software on the Harvard University Bauer Core’s computing cluster. The resulting fragment files and count matrices were processed further using Signac version 1.1.0 and Seurat version 4.3.0.1, and implemented in R version 4.3.2 in the Rstudio computing environment version 2023.03.1 + 446. High-quality nuclei were selected based on the following criteria: ATAC counts > 2500 and <1000000, RNA counts > 2000 and <10000, nucleosome signal <2, transcriptional start site enrichment > 1, and mitochondrial RNA < 5% of total transcripts. Table 1 shows the number of cells from each line retained after qualiy filtering. ATAC peak calling was performed using the Signac “CallPeaks” function and MACS2 version 2.2.7.1. The standard Seurat and Signac workflows were then used to analyze data. Marker peaks for each cell line with the ATAC data were identified using the Seurat “FindAllMarkers” function with default arguments, except for the following: only.pos = “TRUE”, test.use = “LR”, and latent.vars = “nCount_ATAC”. Similarly, marker genes for each cell line were identified with the same function, with the following adjustments to default arguments: only.pos = “TRUE”, logfc.threshold = 0.5, and min.pct = 0.2.
Data Records
Raw FASTQ files, count matrices in CSV fomat, and key Cell Ranger output files (filtered gene expression feature marices and ATAC fragment files) have been uploaded to the NCBI Gene Expression Omnibus, with accession number GSE26218917. RDS files containing Seurat objects derived from analyzing each cell line as described here have been uploaded to Mendeley Data, with the following DOIs and corresponding URLs: 10.17632/wvzz6hbttg.2 (https://data.mendeley.com/datasets/wvzz6hbttg/1)18, 10.17632/29g4826npf.1 (https://data.mendeley.com/datasets/29g4826npf/1)19, 10.17632/9yc8d8bnss.1 (https://data.mendeley.com/datasets/9yc8d8bnss/1)20, 10.17632/cp4d7t74vb.1 (https://data.mendeley.com/datasets/cp4d7t74vb/1)21.
Technical Validation
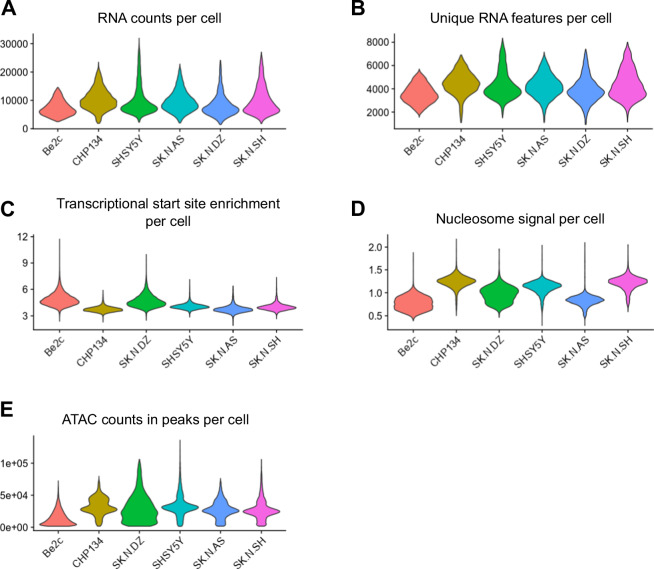
We evaluated standard quality control metrics for snRNA-seq and snATAC-seq datasets. After filtering for high-quality nuclei, as described above, our dataset consistent of 41,733 nuclei. The gene expression data showed consistency of counts per nucleus and unique features per nucleus across the six cell lines studied (Fig. 1A,B). Similarly, ATAC data was consistent across cell lines with respect to transcriptional start site enrichment, nucleosome signal, and the number of counts in peaks (Fig. 1C–E).
Fig. 1.
Quality control analysis of single-nucleus datasets. (A) Violin plot showing RNA counts in each cell in the snRNA-seq dataset, with the cell line indicated on the X-axis. (B) Violin plot showing unique RNA features identified in each cell in the snRNA-seq dataset, with the cell line indicated on the X-axis. (C) Violin plot showing transcriptional start site enrichment in each cell in the snATAC-seq dataset, with the cell lines indicated on the X-axis. (D) Violin plot showing nucleosome signal in each cell in the snATAC-seq dataset, with the cell lines indicated on the X-axis. (E) Violin plot showing ATAC counts in MACS2-identified peaks in each cell in the snATAC-seq dataset, with the cell lines indicated on the X-axis.
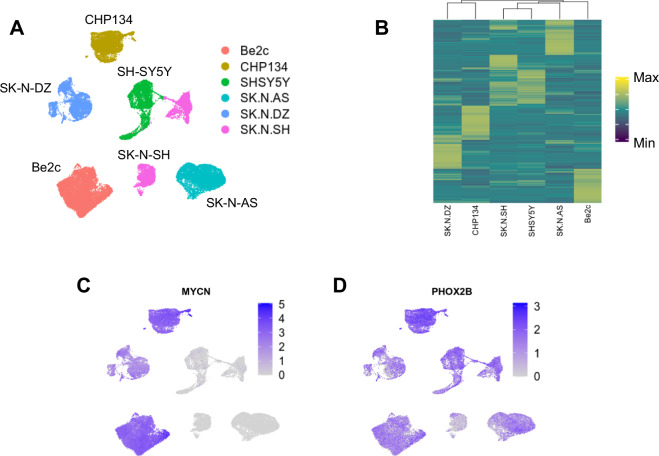
We performed basic dimensional reduction, clustering, and analysis using transcriptional data. As expected, there was more variation between cell lines that within cell lines, as demonstrated on a UMAP projection (Fig. 2A). A subset of SK-N-SH cells does appear closely related to SH-SY5Y cells (Fig. 2A), which may be expected since SH-SY5Y cells are a subclose of the SK-N-SH line. We identified the top 250 marker genes for each cell line, relative to all other cell lines. Hierarchical clustering based on scaled expression of these 250 genes shows that the non-MYCN-amplified cell lines (SHSY5Y, SK-N-SH, and SK-N-AS) were most similar to one another, with SK-N-SH and SH-SY5Y being most similar, while the MYCN-amplified cell lines (CHP134, Be2c, and SK-N-DZ) diverged from the non-amplified lines (Fig. 2B). Next, we assessed gene expression of two important neuroblastoma driver genes: MYCN and PHOX2B. As anticipated, our data show markedly higher expression of MYCN transcripts in the MYCN-amplified cell lines (Fig. 2C), while PHOX2B transcripts are more uniformly distributed across all six cell lines (Fig. 2D).
Fig. 2.
Basic analysis of snRNA-seq data. (A) UMAP projection showing cells from each cell line largely cluster together based on transcript abundance. (B) Heatmap showing mean expression in each cell line of the top 250 marker genes identified for each cell line. (C) Transcript abundance of MYCN within each cell, overlayed on the UMAP projection shown in (A). As anticipated, MYCN-amplified cell lines have markedly higher MYCN transcript levels. (D) Transcript abundance of PHOX2B within each cell, overlayed on the UMAP projection shown in (A). In contrast with MYCN, PHOX2B transcripts are abundant in all cell lines.
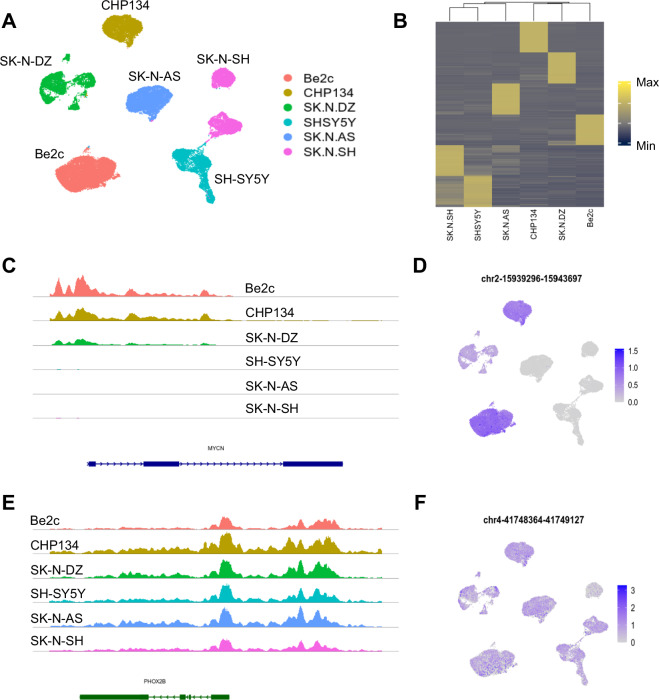
Similarly, we assessed the chromatin accessibility dataset in each cell line. Just as for gene expression data, we found that the greatest variability was between cell lines, as visualized on a UMAP projection (Fig. 3A). The top 1000 marker peaks for each cell line were identified and used to generate a heatmap (Fig. 3B). Hierarchical clustering of this data shows that the three non-MYCN-amplified lines cluster together and maintain open chromatin at similar genomic regions. Again, both the UMAP projection and hierarchical clustering of the heatmap suggest a close relationship between the SH-SY5Y and SK-N-SH lines (Fig. 3A,B). In contrast, MYCN-amplified lines show more divergence in their chromatin accessibility patterns. Due to MYCN amplification, we expect the CHP134, Be2c, and SK-N-DZ cell lines to have far more counts mapped to the MYCN locus than the non-amplified lines. A tracks plot of the MYCN gene shows these results (Fig. 3C). Similarly, counts within the ATAC peak found at chromosome 2, bases 15,939,296-15,943,697, which lies within the MYCN coding sequence, are seen exclusively in the MYCN-amplified lines (Fig. 3D). In contrast, there is a similar chromatin signal in both the coding sequence and upstream promoter region of PHOX2B, including at the peak at chromosome 4, bases 41,748,364-41,749,127 (Fig. 3E,F).
Fig. 3.
Basic analysis of snATAC-seq data. (A) UMAP projection showing cells from each cell line largely cluster together based on chromatin accessibility. (B) Heatmap showing mean expression in each cell line of the top 1000 marker peaks identified for each cell line. (C,E) Track plots showing ATAC counts in and around the MYCN and PHOX2B coding sequences in the indicated cell lines. (D,F) ATAC count abundance within each cell for the indicated MACS2 peaks, which lie within the MYCN (D) and PHOX2B (F) coding regions, overlayed on the UMAP projection shown in (A).
Acknowledgements
This work was supported by NIDDK F32DK121440 to RAG, NIDDK F32DK131792 to JLM, NIDDK R01DK119210 to AMG, and by the Massachusetts General Hospital Department of Surgery Patricia K. Donahoe Resident Research Catalyst Award to RAG. The authors thank the staff at the Harvard University Bauer Core for their expertise and assistance.
Author contributions
R.A.G. and A.M.G. conceived and designed this project. R.A.G., J.L.M. and N.B. performed cell culture, sample preparation, and library construction. R.A.G. did all data analysis and processing. R.A.G. wrote all code used for data analysis. Funding for the project was obtained by R.A.G. and A.M.G. R.A.G. drafted the manuscript and prepared figures. All authors participated in manuscript editing, and all authors have approved the version of the manuscript submitted for publication.
Code availability
R scripts use for processing and analysis of this data are publicly available at a Mendeley Data repository with 10.17632/s2fcfb8phh.1 (https://data.mendeley.com/datasets/s2fcfb8phh/1)22.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Richard A. Guyer, Email: rguyer2@jh.edu
Allan M. Goldstein, Email: agoldstein@mgb.org
References
- 1.Li, J., Thompson, T. D., Miller, J. W., Pollack, L. A. & Stewart, S. L. Cancer Incidence Among Children and Adolescents in the United States, 2001–2003. Pediatrics121, e1470–e1477 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Newman, E. A. et al. Update on neuroblastoma. Journal of Pediatric Surgery54, 383–389 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Cohn, S. L. et al. The International Neuroblastoma Risk Group (INRG) Classification System: An INRG Task Force Report. JCO27, 289–297 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee, J. W. et al. Clinical significance of MYCN amplification in patients with high-risk neuroblastoma. Pediatr Blood Cancer65, e27257 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Yanishevski, D. et al. Impact of MYCN status on response of high-risk neuroblastoma to neoadjuvant chemotherapy. Journal of Pediatric Surgery55, 130–134 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Guyer, R. A. et al. Differentiated neuroblastoma cells remain epigenetically poised for de-differentiation to an immature state. Disease Models & Mechanisms16, dmm049754 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Upton, K. et al. Epigenomic profiling of neuroblastoma cell lines. Sci Data7, 116 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harenza, J. L. et al. Transcriptomic profiling of 39 commonly-used neuroblastoma cell lines. Sci Data4, 170033 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kashima, Y. et al. Single-cell sequencing techniques from individual to multiomics analyses. Exp Mol Med52, 1419–1427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, R. et al. Single-Cell Characterization of Malignant Phenotypes and Developmental Trajectories of Adrenal Neuroblastoma. Cancer Cell38, 716–733.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Mercatelli, D. et al. Single-Cell Gene Network Analysis and Transcriptional Landscape of MYCN-Amplified Neuroblastoma Cell Lines. Biomolecules11, 177 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansky, S. et al. Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma. Nature Genetics53, 683–693, 10.1038/s41588-021-00806-1 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Kildisiute, G. et al. Tumor to normal single-cell mRNA comparisons reveal a pan-neuroblastoma cancer cell. Science Advances7, eabd3311 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma, S. et al. Chromatin Potential Identified by Shared Single-Cell Profiling of RNA and Chromatin. Cell183, 1103–1116.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boeva, V. et al. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nature Genetics49, 1408–1413, 10.1038/ng.3921 (2017). [DOI] [PubMed] [Google Scholar]
- 16.van Groningen, T. et al. Neuroblastoma is composed of two super-enhancerassociated differentiation states. Nature Genetics49, 1261–1266, 10.1038/ng.3899 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Guyer, R. A. & Goldstein, A. M. GEOhttps://identifiers.org/geo/GSE262189 (2024).
- 18.Guyer, R. A. & Goldstein, A. M. Seurat objects for multiome analysis of neuroblastoma cell lines - 1/4. Mendeley Data V1 10.17632/wvzz6hbttg.1 (2024).
- 19.Guyer, R. A. & Goldstein, A. M. Seurat objects for multiome analysis of neuroblastoma cell lines - 2/4. Mendeley Data V1 10.17632/29g4826npf.1 (2024).
- 20.Guyer, R. A. & Goldstein, A. M. Seurat objects for multiome analysis of neuroblastoma cell lines - 3/4. Mendeley Data V1 10.17632/9yc8d8bnss.1 (2024).
- 21.Guyer, R. A. & Goldstein, A. M. Seurat objects for multiome analysis of neuroblastoma cell lines - 4/4. Mendeley Data V1 10.17632/cp4d7t74vb.1 (2024).
- 22.Guyer, R. A. & Goldstein, A. M. Analysis of multiome sequencing data from neuroblastoma cell lines. Mendeley Data V1 10.17632/s2fcfb8phh.1 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Guyer, R. A. & Goldstein, A. M. Seurat objects for multiome analysis of neuroblastoma cell lines - 1/4. Mendeley Data V1 10.17632/wvzz6hbttg.1 (2024).
- Guyer, R. A. & Goldstein, A. M. Seurat objects for multiome analysis of neuroblastoma cell lines - 2/4. Mendeley Data V1 10.17632/29g4826npf.1 (2024).
- Guyer, R. A. & Goldstein, A. M. Seurat objects for multiome analysis of neuroblastoma cell lines - 3/4. Mendeley Data V1 10.17632/9yc8d8bnss.1 (2024).
- Guyer, R. A. & Goldstein, A. M. Seurat objects for multiome analysis of neuroblastoma cell lines - 4/4. Mendeley Data V1 10.17632/cp4d7t74vb.1 (2024).
- Guyer, R. A. & Goldstein, A. M. Analysis of multiome sequencing data from neuroblastoma cell lines. Mendeley Data V1 10.17632/s2fcfb8phh.1 (2024).
Data Availability Statement
R scripts use for processing and analysis of this data are publicly available at a Mendeley Data repository with 10.17632/s2fcfb8phh.1 (https://data.mendeley.com/datasets/s2fcfb8phh/1)22.