Abstract
The COVID-19 pandemic has created a global health crisis, with challenges arising from the ongoing evolution of the SARS-CoV-2 virus, the emergence of new strains, and the long-term effects of COVID-19. Aiming to overcome the development of viral resistance, our study here focused on developing broad-spectrum pan-coronavirus antiviral therapies by targeting host protein quality control mechanisms essential for viral replication. Screening an in-house compound library led to the discovery of three candidate compounds targeting cellular proteostasis. The three compounds are (1) the nucleotide analog cordycepin, (2) a benzothiozole analog, and (3) an acyldepsipeptide analog initially developed as part of a campaign to target the mitochondrial ClpP protease. These compounds demonstrated dose-dependent efficacy against multiple coronaviruses, including SARS-CoV-2, effectively inhibiting viral replication in vitro as well as in lung organoids. Notably, the compounds also showed efficacy against SARS-CoV-2 delta and omicron strains. As part of this work, we developed a BSL2-level cell-integrated SARS-CoV-2 replicon, which could serve as a valuable tool for high-throughput screening and studying intracellular viral replication. Our study should aid in the advancement of antiviral drug development efforts.

Subject terms: High-throughput screening, Small molecules
A Study Identifies Broad-Spectrum Pan-Coronavirus Antiviral Drugs Targeting Host Protein Quality Control Mechanisms And Reports Development Of A Bsl2-Level Cell-Integrated Sars-Cov-2 Replicon, Which Can Be Used For High-Throughput Screening.
Introduction
The emergence of the coronavirus disease 2019 (COVID-19) pandemic, caused by severe-acute-respiratory-syndrome coronavirus 2 (SARS-CoV-2), has resulted in a significant global health crisis with millions of reported cases and deaths worldwide1. The development of effective therapeutics to combat SARS-CoV-2 infection is crucial in controlling the spread of the virus and mitigating its impact on public health. Although vaccines limited the spread of SARS-CoV-2 infection2, the virus still evolves to circumvent vaccine- and antibody-induced immunity3. Small molecule therapies had moderate success with few approved antivirals, showing low efficacy, numerous side effects and subject to development of viral resistance4. The continuous evolution of the virus, the emergence of new strains with altered antigenicity and transmissibility, and the potential for the development of viral resistance pose significant challenges in managing the pandemic3,5. There are also newer aspects of concern such as long-COVID, with SARS-CoV-2 being detected for prolonged periods in the body, as well as long-term effects of the illness affecting recovery of patients, especially those with comorbid conditions6. Therefore, there is an unmet need for anti-SARS-CoV-2 drugs that are not only effective against the current virus variants, but that would continue to be effective against future coronaviruses that may emerge.
SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA virus belonging to the genus Betacoronavirus in the family Coronaviridae7. It is also only one of several pathogenic coronaviruses that have caused significant global or regional outbreaks in the past 20 years, namely, severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV)8. Other human coronaviruses, such as the alpha coronavirus HCoV-229E (229E) and the beta coronavirus HCoV-OC43 (OC43), are endemic and circulate in the population causing mild respiratory tract infections, resulting in common cold symptoms7. Infection is initiated by the binding of the coronavirus spike (S) protein to cellular entry receptors such as angiotensin-converting enzyme (ACE2) for SARS-CoV-2. Cleavage of S protein by the transmembrane protease serine 2 (TMPRSS2) triggers fusion of the viral and cell membranes. Upon entry, coronaviruses express and replicate their genomic RNA to produce full-length copies that are subsequently incorporated into newly produced viral particles. 5′ open reading frames, ORF1a and ORF1b, are immediately translated from the genomic RNA and processed into non-structural proteins forming the viral replication and transcription complexes. Viral replication organelles form near the nucleus consisting of perinuclear double-membrane vesicles (DMVs) along with other membrane networks to create a protective microenvironment for viral genomic RNA amplification and transcription of subgenomic RNAs (sgRNAs). A nested set of viral sgRNAs are then translated into structural proteins which translocate through the ER/Golgi network, where virion assembly is initiated followed by eventual release from the infected cell9.
Due to the error-prone nature of viral RNA-dependent RNA polymerase, viruses are likely to develop resistance against agents targeting virus-encoded functions. As a result, drugs targeting host mechanisms independent of the virus that are essential for viral replication represent attractive targets for broad-spectrum antiviral therapy. In particular, the proteostasis network is relied on by most viruses: the chaperones for folding and assembling viral proteins and proteases for degrading and/or processing viral proteins to aid in virus replication10. As an example related to this work, RUVBL1/2 are two AAA+ ATPases that function in a variety of cellular processes, especially when they are part of the R2TP chaperone complex, such as in chromatin remodeling, ciliogenesis, assembly, and maturation of several large macromolecular complexes including RNA polymerases, the box C/D small nucleolar ribonucleoprotein (snoRNP) and mTOR complexes11. RUVBL1/2 are involved in the assembly of the Mumps Virus (MuV) and Measles Virus (MeV) replication machineries12 and interact with the Ebola Virus (EBOV) nucleoprotein13. Also, RUVBL2 is exploited by HIV-1 to regulate its viral protein expression levels for efficient virion production14. An example of proteases influencing viral replication is the mitochondrial ClpP protease that affects mitochondrial protein quality control by degrading misfolded or damaged proteins15. Depletion of ClpP was recently found to activate the type I interferon response, generating potent antiviral phenotypes16. Hence, we undertook an effort to screen for compounds targeting the proteostasis network in mammalian cells that affect coronavirus replication.
We describe here our screening campaign for pan-coronavirus inhibitors against 229E, OC43, and SARS-CoV-2 using an in-house library of more than 500 proprietary compounds that target either the chaperone complex RUVBL1/2 or the mitochondrial protease ClpP along with general chaperone inhibitors, including ATP analogs. As part of this effort, we also describe the design, construction, and use of a SARS-CoV-2 replicon. The replicon is a mammalian cell-integrated SARS-CoV-2 replicon with several different reporters that achieved two goals: biological safety, as well as high suitability for high throughput screen (HTS). The replicon is available for use in biosafety level (BSL) 2 laboratories not only for screening but also to study the molecular mechanisms of SARS-CoV-2 replication. From our screening campaign, three hit compounds were validated using cell lines, the above-described replicon, and lung organoids.
Results
Screening of small molecules targeting host proteostasis network identified three candidate compounds effective against multiple coronaviruses
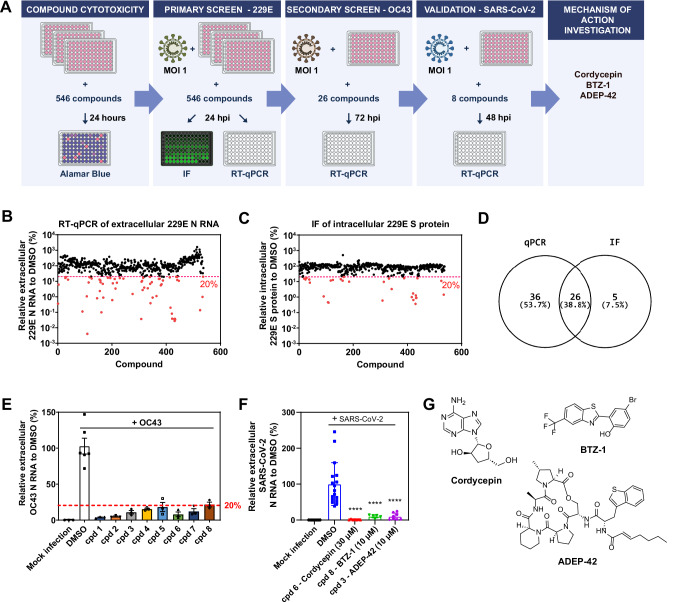
An in-house library of 546 mainly proprietary compounds, including 444 compounds targeting the ClpP protease, 93 targeting the AAA+ chaperones RUVBL1 and/or RUVBL2, and an additional 9 targeting common chaperones were selected to screen for anti-coronavirus activity in Huh-7 (human hepatocellular carcinoma) cells. ClpP targeting compounds include Activators of Cylindrical Proteases (ACPs)17,18, Acyldepsipeptides (ADEPs)19, as well as imipridones20. RUVBL1/2 targeting compounds included analogs of a known RUVBL1/2 ATPase inhibitors21–24, as well as nucleoside analogs known to interact with RUVBL1/225. Compound concentrations were adjusted for cytotoxicity effects using the alamarBlue assay26. All compounds were used at a starting concentration of 10 µM. For samples showing less than 75% of cells alive, compounds were diluted, and cytotoxicity was remeasured to ensure minimal cytotoxicity. Compounds were screened first using alpha coronavirus 229E and then beta coronavirus OC43 (Fig. 1A). Compound performance in suppressing viral replication was compared to DMSO (vehicle control) and measured by RT-qPCR of extracellular viral RNA released using primers for the N region of the viral genome unless otherwise indicated, as well as by immunofluorescence (IF) analysis of intracellular viral S protein levels where indicated. A cut-off of 20% viral RNA/protein in compound treated sample compared to DMSO was selected, and compounds passing this cut-off were moved forward in the screening pipeline and finally validated for efficacy against SARS-CoV-2 SB2 strain27. The SB2 strain of SARS-CoV-2 was used in all SARS-CoV-2 related experiments unless otherwise stated.
Fig. 1. Small molecule screen against multiple coronaviruses.
A Schematic of the workflow for the drug screening campaign. B Relative extracellular 229E N RNA levels in the media of 229E (MOI 1) infected Huh-7 cells detected by RT-qPCR after treatment with compound library compared to DMSO. Compounds reducing RNA levels below 20% are highlighted in red. C Relative intracellular 229E S protein levels in 229E (MOI 1) infected Huh-7 cells detected by IF after treatment with compound library compared to DMSO. Compounds reducing S protein levels below 20% are highlighted in red. D Overlap of compounds reducing both 229E RNA and protein levels. E Compounds that reduced extracellular OC43 N RNA levels in OC43 (MOI 1) infected Huh-7 cells below 20% compared to DMSO are shown. Mean ± SEM shown. F Compounds that reduced extracellular SARS-CoV-2 N RNA levels in SARS-CoV-2 (MOI 1) infected Huh-7-ACE2 cells below 20% compared to DMSO. cpd 6 = Cordycepin, cpd 8 = BTZ-1, cpd 3 = ADEP-42. Mean ± SEM shown generated from n = 3 independent experiments, each performed in triplicates. ****p < 0.0001. G Structures of the three candidate compounds identified in the screen.
Of 546 compounds tested, 62 compounds as measured by RT-qPCR and 31 as measured by IF were found to reduce 229E levels by more than 80% in the primary screen (Fig. 1B–D, highlighted in red). 26 compounds (around 5% of total compounds tested) inhibited accumulation of both 229E viral extracellular RNA and intracellular protein levels. These 26 compounds were then tested against OC43, out of which 8 compounds met the 20% cutoff (Fig. 1E). This set of common inhibitors of both 229E and OC43 were then subjected to further testing against SARS-CoV-2. Three of the compounds had desirable viral inhibition and cytotoxicity profiles (see below): cordycepin, BTZ-1, and ADEP-42. At 30 µM, cordycepin reduced SARS-CoV-2 RNA release by 99%, while BTZ-1 and ADEP-42 reduced viral RNA release by 90% at 10 µM (Fig. 1F) confirming their capacity to inhibit replication of a broad range of coronaviruses.
These three compounds represent varied chemical scaffolds (Fig. 1G). Cordycepin, naturally derived from the fungus Cordyceps militaris (C. militaris), is an analog of adenosine consisting of an adenine base attached to a sugar ring28. If incorporated into RNA, cordycepin can cause chain termination, inhibiting RNA synthesis29. It affects the circadian rhythm in mammals by targeting RUVBL225. A crystal structure of RUVBL1-RUVBL2 bound to cordycepin was solved showing cordycepin bound at the ATP-binding sites of the RUVBL1/2 hexamer. A systematic literature review of cordycepin suggested that it consistently repressed cell migration and cellular inflammation, mainly by inhibiting PI3K/mTOR/Akt and activating AMPK signaling30. Its potential therapeutic effects in animal models, such as reducing tumorigenesis, repressing inflammation, improving respiratory and cardiac conditions, among others, were proposed to be mainly mediated through these signaling pathways30.
BTZ-1 consists of a benzothiozole attached to a phenol group. Little is known of this compound as it was produced as an analog of an early lead compound targeting RUVBL1/2 (Houry lab). ADEP-42 contains a macrolactone core and a polyene sidechain. It is part of a class of antibiotics largely affecting Gram-positive bacteria by dysregulating the ClpP protease31. We had found certain analogs that dysregulate the human mitochondrial ClpP leading to apoptosis in several cancer cell lines32. We proposed that such ADEPs could have therapeutic potential for cancer treatment33.
Compound treatment results in a dose-dependent suppression of coronavirus replication
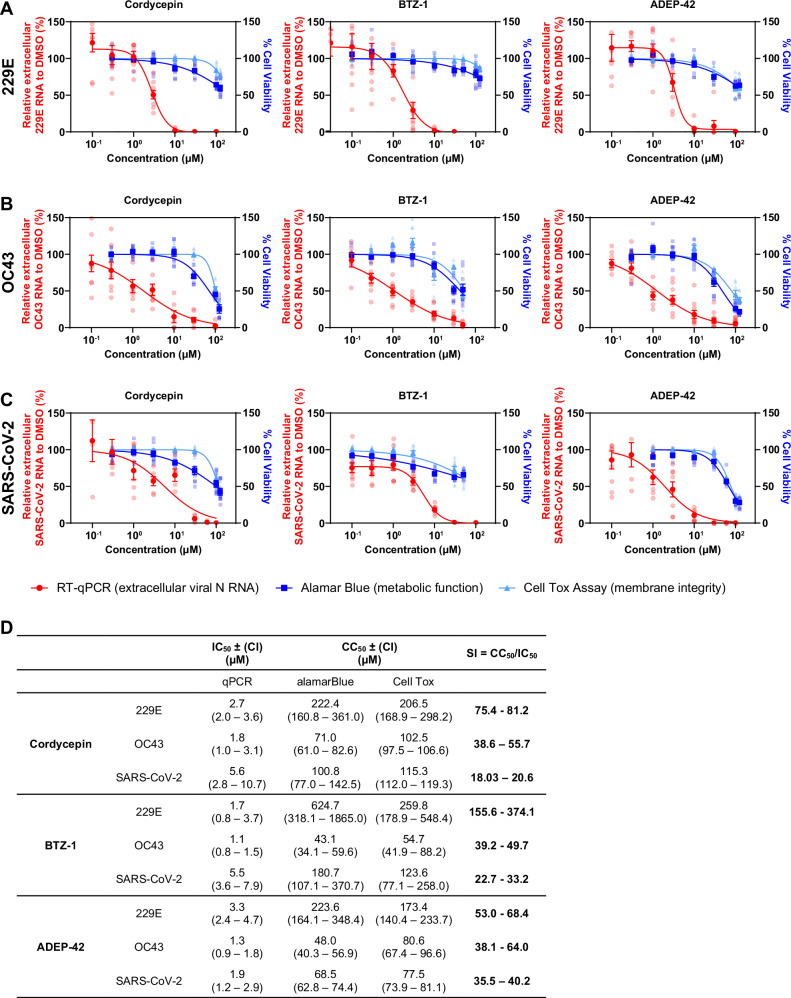
To further verify the ability of the lead compounds to inhibit viral replication, as well as to investigate their efficacy, dose responses against 229E, OC43 and SARS-CoV-2 were carried out for all three compounds with concentrations ranging from 0.03 – 100 µM (Fig. 2A–C). Candidate compounds exhibited low μM IC50 (50% inhibitory concentrations) values against the three coronaviruses, including SARS-CoV-2 (Fig. 2D), showing broad-spectrum anti-coronavirus potential. The 50% cytotoxicity concentrations (CC50) for the three compounds were also measured. Cordycepin and BTZ-1 showed a better selectivity index (SI = CC50/IC50) against 229E than the other two viruses, at more than 70 and 150, respectively. However, moderate SIs of more than 20 against SARS-CoV-2 were still achieved by both compounds. ADEP-42 showed more consistent SI against all three coronaviruses in the range of 35-70 (Fig. 2D). Our compounds are comparable to previous published host targeting compounds that inhibit SARS-CoV-2, such as the SR kinase inhibitor harmine (IC50 = 7.5 µM, CC50 ≥ 100 µM)34.
Fig. 2. Efficacy of lead compounds on 229E, OC43, and SARS-CoV-2 replication.
Concentration-response curves of indicated compounds for 229E (A) or OC43 (B) infected Huh-7 cells, or SARS-CoV-2 (C) infected Huh-7-ACE2 cells. All infections were carried out at MOI 1. Samples were collected at 24 hpi for 229E, 72 hpi for OC43, and 48 hpi for SARS-CoV-2. Viral RNA released was measured by RT-qPCR shown in red. Cell viability curves of mock-infected cells measured by alamarBlue and CellTox Green cytotoxicity assays are shown in blue and cyan, respectively. Mean ± SEM shown is generated from n = 3 independent experiments, each performed in triplicates. D Summary table of IC50, CC50, and selectivity index (SI) values for indicated compounds against different coronavirus species.
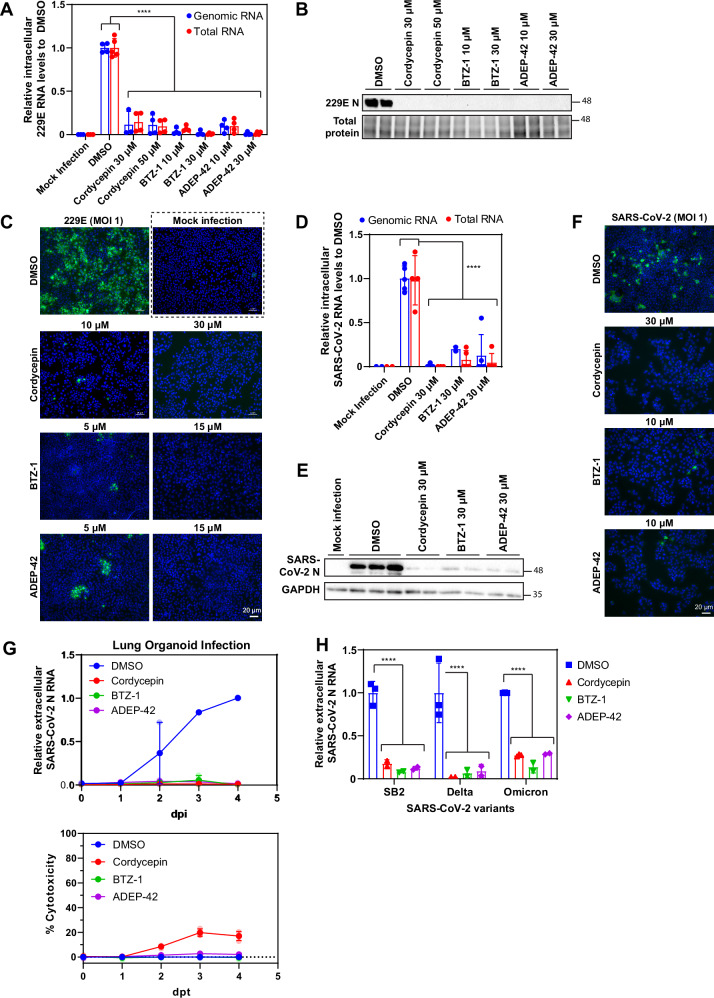
Impact of compounds on specific intracellular viral components was examined in both 229E and SARS-CoV-2 models (Fig. 3). Compound concentrations that resulted in >90% inhibition against extracellular viral RNA release were used. For both viruses, abundance of intracellular genomic RNA (RT-qPCR against Orf1a) and total RNA (includes genomic and subgenomic RNA, RT-qPCR against N) were reduced by at least 80% when treated with any of the candidate compounds (Fig. 3A, D). This finding suggests that the compounds impacted overall viral RNA replication and not specifically subgenomic RNA production. Consistent with changes in viral RNA abundance, coronavirus structural proteins levels, N and S protein, were significantly reduced by compound treatment as shown by western blotting and IF (Fig. 3B, C, E, F). It should be noted that we cannot exclude the possibility that the antiviral effect of the compounds might be post-replication.
Fig. 3. Effect of lead compounds on the expression of viral RNA and proteins.
A Relative intracellular 229E RNA levels in 229E (MOI 1) infected Huh-7 cells after treatment with cordycepin, BTZ-1 and ADEP-42 at the indicated concentrations compared to DMSO for 24 hours. Genomic RNA (blue) was measured using primers against Orf1a, and total RNA (red) was measured using primers against N, both were normalized to actin mRNA. Mean ± SD shown from 2 independent experiments, each performed in duplicates. ****p < 0.0001. B Representative western blot of 229E N protein levels in samples treated as in A. Lysates were stained with anti-coronavirus N antibody and total protein was visualized using stain-free imaging. Molecular weight (MW) markers are given on the right in kDa. C Representative images of 229E S protein levels in samples treated as in A. Cells were fixed, processed for IF, and stained with anti-229E S antibody (green) and DAPI (blue). D Relative intracellular SARS-CoV-2 RNA levels in infected Huh-7 cells after treatment with cordycepin, BTZ-1 and ADEP-42 at 30 µM compared to DMSO for 48 hours. RNA was extracted and processed as in (A). Mean ± SD shown from 2 independent experiments, each performed in duplicate. ****p < 0.0001. E Representative western blot of SARS-CoV-2 N protein levels in samples treated as in D. The lysates were stained with anti-SARS-CoV-2 N antibody and GAPDH. MW markers are given on the right in kDa. F Representative images of SARS-CoV-2 S protein in infected Huh-7 cells after treatment with cordycepin at 30 µM, BTZ-1 at 10 µM, and ADEP-42 at 10 µM. Samples were fixed 48 hpi, processed for IF, and stained with anti-SARS-CoV-2 S antibody (green) and DAPI (blue). G Top: Effects of compounds on extracellular SARS-CoV-2 N RNA levels in infected (MOI 1) 3D lung organoids measured over days post infection (dpi). Bottom: Effects of compounds on cell viability as measured by LDH release over days post-treatment (dpt). Cordycepin (30 µM), BTZ-1 (10 µM) and ADEP-42 (10 µM) were compared with DMSO treatment. H Relative extracellular SARS-CoV-2 variants N RNA levels in media of infected Huh-7 cells after treatment with cordycepin (30 µM), BTZ-1 (10 µM) and ADEP-42 (10 µM) compared to DMSO.
Compounds are effective in lung organoid model as well as against several SARS-CoV-2 strains
To examine whether our lead compounds maintain their antiviral activity in a more physiological setting, we utilized a lung organoid model35. Mature lung organoids were infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 2 and then treated with the compounds or DMSO. Extracellular media samples were harvested daily for four days after infection, with viral RNA release quantified by RT-qPCR. DMSO treated samples showed steady increases in SARS-CoV-2 viral RNA release from 1 dpi to 3 dpi, with slight plateauing by 4 dpi indicating a successful infection with viral replication maintained in the organoids (Fig. 3G, top panel). In contrast, RNA release remained low in compound treated samples. By 4 dpi, cordycepin and ADEP-42 showed 100-fold differences, while BTZ-1 showed 1000-fold differences compared to DMSO (Fig. 3G, top panel), demonstrating their ability to inhibit SARS-CoV-2 replication or infection in lung organoids. The cell viability of the organoids in the presence of compounds were assessed using LDH assay36. While cytotoxicity of BTZ-1 and ADEP-42 were comparable to DMSO control for all four days of treatment, cordycepin showed low levels of toxicity (below 20%) on days 3 and 4 (Fig. 3G, bottom panel).
Since all our screening and subsequent testing of the compounds (Figs. 1, 2, 3D–G) was done using the SB2 strain27, we also tested the compounds for efficacy against SARS-CoV-2 variants of concern. All three compounds were active against both the delta and omicron variants with an average of 80% reduction in viral RNA release (Fig. 3H). No significant differences were observed in efficacy against different SARS-CoV-2 strains. This observation provides evidence that targeting host protein homeostasis is a viable pan-anticoronavirus strategy.
Compounds can act late in 229E replication to inhibit viral infectivity
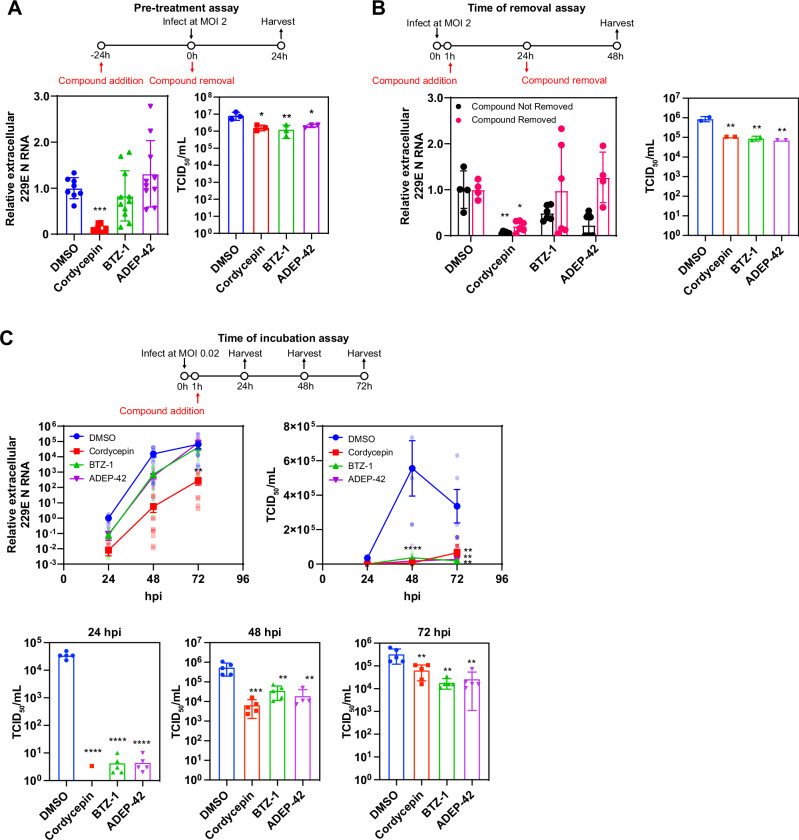
To dissect the mechanisms of inhibition by these three compounds, the fast-growing 229E was used as a model. We first characterized the kinetics of 229E replication in Huh-7 cells in the absence of any compounds. At MOI 2, 229E reached a replication plateau at 24 hpi, releasing almost 20,000-fold more viral RNA compared to 3 hpi (Fig. 4A). Viral N protein expression shows a significant increase at 12 hpi (also refer to ref. 37). A significant increase in viral RNA release was also detected 12 hpi. Utilizing this information, the addition of compounds was delayed for up to 16 hpi to examine whether compounds inhibited viral replication even after viral protein production and RNA release had initiated. All three candidate compounds reduced the release of viral RNA whether added 1, 6 or 12 hpi and samples harvested at 24 hpi (Fig. 4B) consistent with them affecting post-entry stages of virus replication and release. While some increase in viral RNA release was seen when compounds were added 16 hpi, subsequent measurement of viral infectivity, using the 50% tissue culture infectious dose (TCID50) assay, revealed that 16 hpi compound treated samples were 10-fold less infectious compared to DMSO treated samples (Fig. 4B).
Fig. 4. Effect of time of addition on compound efficacy.
A Kinetics of 229E viral RNA release monitored by RT-qPCR (top panel) and by N protein expression (bottom panel) in Huh-7 cells infected at MOI 2. For viral RNA release, mean ± SD is shown from n = 4 independent experiments, each performed in triplicates. Representative western blot of N protein expression shown from n = 2 independent experiments each performed in duplicates. MW markers are given on the right in kDa. B Shown are the time of addition assays for 229E infection of Huh-7 cells at MOI 2. The assay timeline is shown as a schematic on top. Compounds were added at the indicated hpi, and released viruses were harvested 24 hpi and then analyzed by RT-qPCR (left) and TCID50 (right). Cordycepin was used at 30 μM, BTZ-1 at 10 μM, and ADEP-42 at 10 μM. Mean ± SD shown from n = 2 independent experiments, each performed in triplicates. ****p < 0.0001. C, D Huh-7 cells were infected with 229E virus at MOI 2 and treated with DMSO or compounds at 16 hpi. Cells were fixed 4.5 h post compound addition and probed for C total and D genomic viral RNA localization by FISH. Representative images of n = 3 independent experiments are shown. E Enrichment score of perinuclear foci observed in genomic DNA staining of samples in D. Foci were analyzed by CellProfiler, classified, and quantified by CellProfiler Analyst (Supplemental Data File 2). Mean ± SD shown from n = 3 independent experiments. *p < 0.05. F Relative intracellular 229E RNA levels in 229E (MOI 1) infected Huh-7 cells treated as in C and D, where compounds were added 16 hpi and samples were harvested 4.5 h post compound addition. Genomic RNA (blue) was measured using primers against Orf1a, and total RNA (red) was measured using primers against N, both were normalized to actin mRNA. Mean ± SD shown from 3 independent experiments, each performed in duplicates. G Relative extracellular 229E N RNA levels in 229E (MOI 1) infected Huh-7 cells treated as in F, where compounds were added 16 hpi and samples were harvested 4.5 h post compound addition. Mean ± SD shown from 3 independent experiments, each performed in duplicates. ****p < 0.0001, ***p < 0.001, **p 0.01.
To further investigate the basis for the antiviral effects seen, we also examined whether compound treatment affected the localization of total and genomic viral RNA inside Huh-7 cells using fluorescent in-situ hybridization (FISH). In these experiments, compounds were added 16 hpi and FISH was done at 20.5 hpi. No significant differences were observed in the distribution of total viral RNA staining (Fig. 4C). In contrast, the distribution of genomic viral RNA was clearly altered by compound treatment (Fig. 4D). Upon treatment with DMSO alone, genomic viral RNA was localized in foci bundles near the nucleus. However, upon compound treatment, genomic viral RNA became more evenly distributed within the cytoplasm. Coronavirus RNA replication is known to take place in membrane invaginations near the nucleus that form a protective microenvironment. A lack of these perinuclear foci suggests that replication sites were impacted by compound treatment. Furthermore, cytoplasmic distribution of genomic viral RNA suggests mislocalization of replication sites. Subsequent analysis using automated image analysis and pattern classification through machine learning in CellProlifer and CellProlifer Analyst38,39 support this conclusion. Nuclei were identified while cell boundaries and perinuclear space were propagated based on distance from the nuclei. Intensity and intensity distribution features were extracted from the genomic viral RNA staining. This data were then coupled with support vector machine (SVM)-based machine learning to generate a classifier capable of distinguishing cells with genomic viral RNA primarily localized to perinuclear foci from those with even distribution or cells absent of viral RNA. Enrichment scores were calculated for each sample based on comparing the probability of a specific phenotype against all samples. Compound treated samples scored much lower compared to DMSO control when enriching for perinuclear foci (Fig. 4E). We verified by RT-qPCR that, under these conditions, the compounds did not significantly change the overall levels of total or genomic viral RNA (Fig. 4F), while slight reduction of the extracellular viral RNA was observed (Fig. 4G). Taken together, this suggests that compound treatment with each of the three compounds reduced the frequency of perinuclear 229E viral replication sites under the tested conditions.
BTZ-1 and ADEP-42 suppress the formation of infectious viruses while cordycepin inhibits viral RNA release
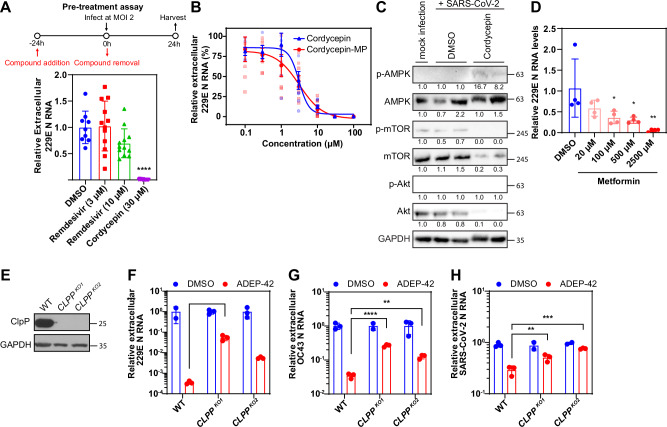
A pre-treatment assay was performed in which DMSO, 30 µM cordycepin, 10 µM BTZ-1, or 10 µM ADEP-42 were added to naïve (uninfected) Huh-7 cells for 24 hours before the compounds are then removed and washed. Cells were subsequently infected with 229E, media samples harvested 24 hpi and examined by RT-qPCR. Under these conditions, only cordycepin reduced viral RNA release, but all three compounds reduced infectious viral particles by 5-fold as quantified by TCID50 (Fig. 5A). These results suggest that all three compounds had lasting effects on host mechanisms that affect virus replication despite being removed from cells prior to virus addition.
Fig. 5. Effect of pretreatment, time of removal, and time of incubation on compound efficacy.
For all assays, cordycepin was used at 30 μM, BTZ-1 at 10 μM, and ADEP-42 at 10 μM. 229E was used to infect Huh-7 cells at MOI 2 unless otherwise indicated. Media samples harvested were analyzed for RNA release by RT-qPCR and for viral infectivity by TCID50. A Shown are pretreatment assays with the assay timeline given in the schematic. Compounds were added 24 h before infection and removed at the time of infection. Media were harvested 24 hpi; RNA release (left) and viral infectivity (right) were then analyzed. For RNA release, mean ± SD are shown from n = 3 independent experiments, each performed in triplicates, ***p < 0.001. For viral infectivity, mean ± SD are shown from n = 1 experiment, performed in triplicate, **p < 0.01, *p < 0.05. B Shown are time of removal assays with assay timeline given in schematic. Compounds were added at 1 hpi and removed or not removed at 24 hpi as indicated. Media were harvested at 48 hpi; RNA release (left) and viral infectivity (right) were then analyzed. For RNA release, mean ± SD are shown from n = 2 independent experiments, each performed in triplicates, **p < 0.01, *p < 0.05. For viral infectivity, mean ± SD are shown from n = 1 independent experiment, performed in duplicate, **p < 0.01. C Shown are the time of incubation assays with the assay timeline given in the schematic. Cells were infected at MOI 0.02, compounds were added 1 hpi, and harvested at 24, 48, as well as 72 hpi. RNA release (left) and viral infectivity (right and panels below) were analyzed for each timepoint. For both RNA release and viral infectivity, mean ± SEM are shown from n = 3 independent experiments, each performed in triplicates, ****p < 0.0001, **p < 0.01, *p < 0.05.
Time of removal assays were also carried out, where compounds were removed from the media 12 or 24 hpi, and media samples were assessed at 24 or 48 hpi of 229E viruses (Fig. 5B and Figure S1A-C). Consistent with the pre-treatment assay, all three compounds reduced the number of infectious particles produced, but only cordycepin reduced the level of viral RNA released even after being removed (Fig. 5B). When compounds are not removed, all compounds show reduction in viral replication.
To test if the compounds have a prolonged inhibitory effect on 229E replication, time of incubation assays were performed, where compounds were added 1 hpi and media samples were assessed at 24, 48, and 72 hpi. Relative to the DMSO control, cordycepin treatment dramatically reduced viral RNA release in media even up to 72 hpi, whereas suppression of viral RNA release by BTZ-1 and ADEP-42 treatment was not observed past 48 hpi (Fig. 5C). Although substantial viral RNA release was observed at 72 hpi in BTZ-1 and ADEP-42 treated samples, subsequent quantification of infectious particles by TCID50 revealed around 20-fold and more than 10-fold reduction in the presence of BTZ-1 and ADEP-42, respectively (Fig. 5C), suggesting that viral particles produced in the presence of BTZ-1 and ADEP-42 are defective. On the other hand, although cordycepin strongly suppressed viral RNA release, infectivity as measured by TCID50 was around 5-fold higher than that observed for BTZ-1 and ADEP-42 at 72 hpi (Fig. 5C). Our data suggest that cordycepin affects viral replication differently compared to BTZ-1 and ADEP-42. However, all three compounds show inhibition of virus replication whether added prior to or after infection.
Cordycepin may inhibit SARS-CoV-2 replication through AMPK and mTOR/Akt signaling
Given the distinctive characteristics of cordycepin in the experiments of Fig. 5 and Figure S1, this compound was investigated further. Cordycepin is an adenosine analog that is converted by cell metabolism into mono-, di-, and triphosphates40. Its triphosphate form (cordycepin-TP) has been suggested to directly target the RNA dependent RNA polymerase (RdRp) of SARS-CoV-2 in cell free assays41. However, cordycepin inhibited viral replication despite being removed in the pre-treatment assay (Fig. 5A) and, hence, was not directly exposed to RdRp. Thus, we initially compared cordycepin activity with remdesivir, a nucleoside inhibitor, in our pre-treatment assay to verify this removal was sufficient to eliminate exposure to the RdRp. Remdesivir has been shown to inhibit 229E to almost 100% at concentrations higher than 0.1 µM42. Using much higher concentrations of remdesivir at 3 µM and 10 µM, our results demonstrated little to no inhibition by remdesivir in the pre-treatment assay, while cordycepin maintained its strong inhibition against 229E (Fig. 6A; also refer to Fig. 5A). This suggests that cordycepin inhibits coronaviruses through mechanisms not dependent on the direct inhibition of RdRp activity. Furthermore, treating cells with cordycepin-TP only weakly inhibited viral RNA release when used at the same concentration as cordycepin (Fig. S2). Cordycepin-TP also did not impact S protein expression (Figure S2), suggesting that it is not the main active form of cordycepin affecting virus replication.
Fig. 6. Molecular basis of cordycepin and ADEP-42 activity.
A Pretreatment assay comparing cordycepin to remdesivir with assay timeline given in schematic. Compounds were added 24 h before infection and removed at time of infection. Media were harvested 24 hpi and extracellular RNA release was then analyzed. Mean ± SD are shown from n = 3 independent experiments, each performed in quadruplicates, ****p < 0.0001. B Dose-response of cordycepin and cordycepin-monophosphate (cordycepin-MP) in 229E infected Huh-7 cells (MOI 1) after 24 hpi. Extracellular viral RNA released was measured by RT-qPCR and normalized to DMSO control. Mean ± SEM shown are generated from n = 3 independent experiments, each performed in triplicates. C Western blot analysis of AMPK, mTOR, and Akt in cordycepin treated SARS-CoV-2 infected samples compared to DMSO control. Huh-7 cells were infected with SARS-CoV-2 at MOI 2, treated with DMSO or 30 µM cordycepin after 1 hour, and samples were harvested 48 hpi. Mock-infected samples were treated with DMSO. Lysates were stained with AMPKα, phospho-AMPKα (Thr172), mTOR, phospho-mTOR (Ser2448), Akt, phospho-Akt (Ser473), and GAPDH antibodies. MW markers are given on the right in kDa. D RT-qPCR quantification of extracellular 229E N RNA levels after infection (MOI 1) and treatment with various concentrations of metformin for 24 hours in Huh-7 cells. Mean ± SD shown from 4 replicates. E Representative western blot of wild type (WT) and CLPP knockout (CLPPKO) cells generated by CRISPR/Cas9. Lysates were stained with ClpP and GAPDH antibodies. MW markers are given on the right in kDa. WT and CLPPKO Huh-7 cells were infected at MOI 2 with F 229E, G OC43 or H SARS-CoV-2. After 1 hour adsorption, cells were treated with DMSO (blue), or 10 µM ADEP-42 (red). Media samples were harvested at 24, 72 or 48 hpi for (F), (G) and (H), respectively, and analyzed by RT-qPCR.
The monophosphate form, cordycepin 5′-monophosphate (cordycepin-MP), activates 5′-adenosine monophosphate (AMP)-activated protein kinase (AMPK) by mimicking the effects of its natural activator, AMP43,44. Cordycepin can also inhibit mTOR/Akt signal transduction, which is known to control proliferation, cell adhesion, and protein synthesis45–48. This effect of cordycepin on mTOR/Akt possibly also occurs through AMPK signaling (or other upstream targets), since AMPK is an important negative regulator of mTOR signaling49. Most reported biological effects of cordycepin, such as inhibition of cell proliferation, cell migration/invasion, and inflammation, are correlated with its effects on mTOR and AMPK signaling50. Its therapeutic effects in animal models are also consistent with other AMPK activators and mTOR inhibitors30. AMPK activators such as metformin inhibit SARS-CoV-2 in vitro, as well as decrease COVID-19 severity and mortality clinically51. Thus, we hypothesized that cordycepin may inhibit coronavirus replication by activating AMPK.
As cordycepin-MP is the primary functional analog in activating AMPK, its ability to inhibit viral replication was assessed in parallel with cordycepin. As shown in Fig. 6B, cordycepin-MP suppressed 229E viral replication in a dose-dependent manner. The dose-response curve of cordycepin-MP closely resembled that of cordycepin with IC50 values of 2.5 µM and 2.1 µM, respectively – the difference falling within the 95% confidence interval. This observation suggests that cordycepin-MP may be the main active analog in inhibiting coronavirus replication.
Next, the phosphorylation of AMPK was examined after SARS-CoV-2 infection of Huh-7 cells and compound treatment to determine whether cordycepin activated AMPK and affected downstream mTOR/Akt pathways. Cordycepin addition increased the phosphorylation level of AMPK while the protein level of AMPK remained constant (Fig. 6C). In contrast, we observed a large decrease in the total levels of mTOR and Akt compared to DMSO (Fig. 6C). As a result, we cannot properly assess the phosphorylation levels of these two proteins. This has been reported in some cordycepin treated cells in the literature, suggesting that cordycepin mediated effects may be cell-type specific52. Nevertheless, these results indicate that cordycepin can affect both AMPK as well as mTOR and Akt signaling either through phosphorylation or protein expression level changes in the context of SARS-CoV-2 infection of Huh-7 cells.
Other AMPK activators such as metformin have been shown to be active against SARS-CoV-2 through a similar pathway51. To assess whether metformin activity is conserved against other coronaviruses like that for cordycepin, we tested various concentrations of metformin against 229E. Metformin inhibited 229E replication in a dose-dependent manner (Fig. 6D), suggesting that AMPK activity is indeed an important regulator of coronavirus replication.
ADEP-42 acts partly through ClpP protease to inhibit coronavirus replication
To verify that ADEP-42 inhibits virus replication by targeting mammalian mitochondrial ClpP protease, we constructed Huh-7 CLPP KO cells using CRISPR/Cas9. If ADEP-42 acts strictly by dysregulating ClpP, then viral replication upon addition of the compound to CLPP KO cells should be unaffected. KO cells were confirmed using a western blot for ClpP (Fig. 6E). Two separate CLPP KO cell lines were used to ensure that the phenotypes observed were not due to off-target activity of CRISPR/Cas9. We then compared the ability of ADEP-42 to inhibit 229E, OC43, and SARS-CoV-2 replication in WT and CLPP KOs (Fig. 6F–H). As expected, ADEP-42 significantly reduced coronavirus RNA release in WT cells. The compound partly inhibited replications of all three viruses in the CLPP KO cells. This observation suggests that ADEP-42 may have other targets in addition to ClpP, including the viral replication machinery, to inhibit coronavirus replication or infection.
Design and generation of a SARS-CoV-2 replicon integrated into human cells for use in a BSL2 lab
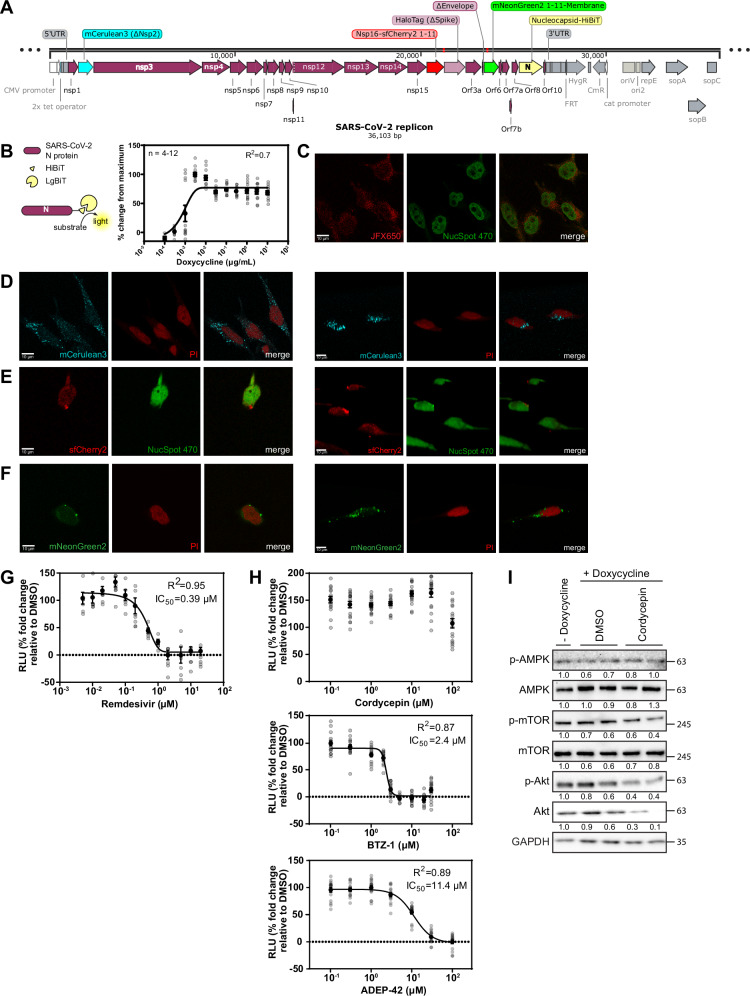
As part of our screening campaign, we also developed a human cell line containing a stably integrated SARS-CoV-2 replicon usable in a BSL2 lab. Several whole SARS-CoV-2 genome systems have recently been developed, however, they either require a BSL3 facility and/or have not been stably integrated into a mammalian cell line53. In addition, these constructs usually contain only one reporter at a single location in the viral genome. Thus, they are not amenable to HTS pipelines. Therefore, we constructed a novel human cell-integrated SARS-CoV-2 replicon with several different reporters that can be multiplexed (Fig. 7A) and that achieved two goals: biological safety and high suitability for HTS facilities.
Fig. 7. Generation and verification of the SARS-CoV-2 replicon.
A Schematic of the SARS-CoV-2 replicon. Viral genome expressed under tetracycline-inducible/CMV chimeric promoter with parts of E and S proteins deleted to attenuate virus infectivity. The replicon is stably integrated into HeLa cells through the Flp-In recombinase system. The main modifications introduced into the SARS-CoV-2 genome are highlighted. B HeLa-replicon or WT HeLa cells were exposed to doxycycline at 3 µg/mL for 24 hours, and luminescence was measured after lysis and supplementation with LgBiT and substrate furimazine. Mean ± SD are shown from 8 replicates, *p < 0.05. C Representative confocal images of the HaloTag stained with JFX650 dye and nucleus stained with NucSpot 470 in doxycycline-induced HeLa-replicon. Representative confocal images of doxycycline-induced HeLa-replicon showing expression of various markers. Left: confocal single plane images, right: 3-D rendering of z-stack. D mCerulean3 (∆Nsp2), E Nsp16-sfCherry211, F mNeonGreen211-M were co-stained with indicated nuclear dye, which is either NucSpot 470 or propidium iodide (PI). G Remdesivir inhibit doxycycline-induced HeLa-Replicon activation in a concentration-dependent manner. Replicon-containing cells were incubated with 3 µg/mL doxycycline as well as remdesivir at indicated concentrations for 24 hours before addition of LgBiT and furimazine (substrate) to quantify the generated luminescence (RLU refers to relative luminescence units). H Effects of compounds on doxycycline-induced HeLa-Replicon activation in a concentration-dependent manner. Replicon-containing cells were incubated with 3 µg/mL doxycycline as well as cordycepin, BTZ-1, and ADEP-42 at indicated concentrations for 24 hours before addition of LgBiT and furimazine (substrate) to quantify the generated luminescence (RLU refers to relative luminescence units). I Western blot analysis of AMPK, mTOR and Akt in HeLa-replicon cells treated with DMSO or cordycepin. Replicon cells were induced with 3 µg/mL doxycycline for 24 hours and treated with either DMSO or 30 µM cordycepin. Lysates were stained with AMPKα, phospho-AMPKα (Thr172), mTOR, phospho-mTOR (Ser2448), Akt, phospho-Akt (Ser473), and GAPDH antibodies. The numbers below the blots are quantified protein levels normalized against GAPDH and expressed relative to no doxycycline samples. MW markers are given on the right in kDa.
For scalability and ease of use, the replicon was encoded on a bacterial artificial chromosome (BAC) and integrated into Flp-In T-Rex HeLa cells under the control of a tetracycline-inducible promoter using the Flp recombination target (FRT) site. To ensure biological safety, two genes essential for formation of infectious SARS-CoV-2 viral particles were deleted: (1) gene encoding the S protein, essential for virus entry, and (2) the gene encoding the envelop (E) protein, required for the production of viral particles54. Lack of production of infectious virus was confirmed by a variety of assays. We showed that media collected from SARS-CoV-2 infected samples induced cytopathic effects (CPE) and apoptosis when used to infect naïve cells, while supernatant from replicon-expressing cells did not (Fig. S3A). Transmission electron microscopy (TEM) analysis of Hela-replicon supernatants revealed no viral particle formation compared to SARS-CoV-2 infected samples, where the classical rounded shape particles surrounded by S proteins can be observed (Fig. S3B). Lastly, RT-qPCR of intracellular and extracellular samples for SARS-CoV-2 N RNA was performed. As expected, N RNA was detected from intracellular samples, while no N RNA was found in the extracellular media samples (Fig. S3C). All assays established that our replicon does not produce infectious SAR-CoV-2 particles, which led to the approval of our replicon to be used in the BSL2 environment by the Public Health Agency of Canada (PHAC).
Various modifications were made in the replicon to allow the detection of various stages of replication/transcription/translation (Fig. 7A). First, the gene for Nsp2 was replaced with one coding for the blue fluorescent protein mCerulean3 to detect early transcription and translation of the opening reading frame Orf1a. Second, Nsp16 (a 2’-O-methyltransferase) was tagged on the C-terminus with the small fragment of the split superfold mCherry protein (sfCherry2 1-11), allowing detection of the subcellular localization of the transcription complex using fluorescence imaging upon transfection with the large fragment of sfCherry2. Third, the gene for the S protein was deleted and replaced with HaloTag, permitting use of various fluorescent substrates for detection. Fourth, the gene for the E protein was deleted to ensure biosafety of our replicon. Fifth, M protein was tagged on the N-terminus with the small fragment of the split mNeonGreen protein (mNeonGreen2 1-11). When co-expressed with the large fragment of mNeonGreen, it allows fluorescent detection of viral protein production and potentially viral particle assembly. Sixth, the nucleocapsid protein N was tagged on the 3′ end with the sequence encoding for HiBiT, a small high-affinity fragment of nanoLuciferase. When provided with the large fragment LgBiT and the luciferase substrate furimazine, lysed cells generate luminescence. Of note, all deletions retained 12-20 aa of the beginning and the end of the viral protein to prevent any alterations to the transcription of subgenomic RNA. The full sequence of the replicon is provided as Supplemental Data File 1 in SnapGene file format.
Subsequently, the expression of the different reporter proteins was verified by monitoring the doxycycline induction of replicon expression using the various introduced reporters (Fig. 7B–F and Figure S3D–H). Sub-micromolar concentrations of doxycycline induced concentration-dependent N protein expression using media containing tetracycline-free FBS as monitored by luminescence (Fig. 7B). However, since doxycycline-free FBS is expensive, regular FBS was also used to ensure that our replicon can be used economically. Addition of 3 µg/mL doxycycline in regular FBS resulted in a significant increase in luminescence that can be quantified using a plate reader compared to both HeLa-replicon cells without doxycycline, as well as HeLa-FlpIn TRex cells without replicon integration (Fig. S3D).
The HaloTag specific fluorophore JFX650 was used to detect induction of HaloTag expression in HeLa-Replicon cells upon doxycycline induction (Figs. 7C and S3E). The HaloTag showed diffuse cytoplasmic localization, similar to that of SARS-CoV-2 infected cells stained with anti-S protein antibodies (Fig. 3F). In contrast, induction of mCerulean3 expression resulted in variably sized foci scattered throughout the cytoplasm (Figs. 7D and S3F). Though some studies have reported diffuse cytoplasmic localization for Nsp2, they used transfected plasmids to overexpress the protein in the cell55,56. The dispersed foci can be observed in cells infected with intact SARS-CoV-2 and stained with anti-Nsp2 antibodies57. It is interesting that the fluorescent markers retain the localization of SARS-CoV-2 proteins during infection when only a small sequence of the N- and C-termini remain. Lastly, HeLa-Replicon cells were transiently transfected with the large fragment of split sfCherry2 or mNeonGreen2 to complement their respective small fragments attached to either Nsp16 or M protein; these showed significant red and green fluorescence, respectively (Figs. 7E, F and S3G,H). Perinuclear foci were observed for both fluorescent markers, slightly larger in size compared to those formed by mCerulean3. These patterns are similar to literature-reported localizations for Nsp16 and M protein57. While the luminescent marker can be used as a large-scale screening tool, our set of fluorescent markers can be utilized to study the molecular mechanism of SARS-CoV-2 infection.
The effect of identified compound hits on the SARS-CoV-2 replicon
One of the main purposes of the replicon is to provide a tool for use in HTS facilities to screen large chemical libraries for anti-SARS-CoV-2 activity. It should be noted that the replicon as designed would only report on compounds modulating viral replication/transcription/translation and not viral entry or exit. We validated the efficacy of a known antiviral, remdesivir, at inhibiting replicon activity. Remdesivir shows in vitro efficacy against SARS-CoV-2 through inhibition of RNA-dependent RNA polymerase58. Originally it was widely used for seriously ill COVID-19 patients, albeit with modest clinical success in hospital stay times but not in reduction of mortality59. Remdesivir showed concentration-dependent inhibition of replicon activity with IC50 of 0.4 µM (Fig. 7G), remarkably close to the published IC50 for remdesivir efficacy against genuine SARS-CoV-2 in Vero cells60. Subsequently, we validated the efficacy of our three candidate compounds in inhibiting replicon activity. BTZ-1 and ADEP-42 both showed concentration-dependent inhibition of replicon activity as monitored by luminescence (Fig. 7H). The IC50 for both agents were 2.4 and 11.4 µM, respectively, similar to the IC50 obtained using in vitro assays described in Fig. 2. On the other hand, cordycepin did not show inhibition of replicon activity (Fig. 7H).
One possible explanation for the discrepancy for cordycepin between experimental systems is that cordycepin mediated effects are cell-type dependent. We observed that total mTOR and Akt protein levels were reduced in Huh-7 cells infected with SARS-CoV-2 upon cordycepin addition (Fig. 6C), and a similar effect was observed by others in NIH3T3 cell45. In contrast, only phosphorylation levels of mTOR and Akt were affected in other cell types30. Thus, protein and phosphorylation levels of AMPK, mTOR, and Akt were checked in the HeLa-replicon cells (Fig. 7I). Replicon expression was induced by doxycycline and cells were treated with either DMSO or cordycepin. No significant changes were observed in AMPK, mTOR or Akt phosphorylation. Interestingly, total Akt levels did decrease, but there were no changes in the levels of phosphorylated Akt, suggesting that phosphorylation may have even increased in cordycepin treated samples. On the other hand, total mTOR protein levels were not affected by cordycepin treatment. Taken together, cordycepin treatment was not effective at inducing significant changes in mTOR/Akt signal transduction, nor did it affect AMPK activity in our HeLa-replicon cells, presumably contributing to the lack of inhibition of replicon expression by cordycepin.
Discussion
In this study, we focused on targeting host processes essential for viral replication as a strategy for broad-spectrum antiviral therapy. Specifically, we targeted the chaperones RUVBL1/2 and the mitochondrial protease ClpP, which have been shown to play crucial roles in the replication of various viruses16. Screening an in-house library of compounds identified three candidate compounds, cordycepin, BTZ-1, and ADEP-42 (Fig. 1G), which demonstrated dose-dependent efficacy against multiple coronaviruses, including SARS-CoV-2, with low µM IC50 values (Fig. 2). All three compounds inhibited SARS-CoV-2 replication or infection in lung organoids (Fig. 3G), and the compounds had activity against SARS-CoV-2 delta and omicron strains (Fig. 3H).
It is important to note that the compounds identified in this study, cordycepin, BTZ-1, and ADEP-42, have varying chemical scaffolds, and their mechanisms of action likely differ. We found that cordycepin inhibits the mTOR/Akt signaling pathway and activates AMPK signaling in the context of SARS-CoV-2 infection in a cell-type-specific manner (Fig. 6C). These signaling pathways play crucial roles in cell growth, metabolism, inflammation, and immune response, and are exploited by SARS-CoV-2 to improve its survival and replication61. Overactivation of mTOR/Akt was correlated with severe cases of COVID-19 and in vitro SARS-CoV-2 infection can cause hyperactivation of mTOR61. This response eventually leads to increased protein synthesis (including viral proteins), impaired autophagy flux (preventing degradation of virions), and increased production of inflammatory cytokines. AMPK negatively regulates mTOR upstream61,62. Thus, it is not surprising that other mTOR inhibitors and AMPK activators such as rapamycin and metformin, respectively, have therapeutic potential against SARS-CoV-263. Our results suggest that cordycepin can function in the same pathway. Additionally, cordycepin was shown to inhibit inflammation in many diseases and animal models by affecting AMPK and/or mTOR64. Furthermore, C. militaris, the fungus that cordycepin is derived from, downregulates cytokines in human trials65. Given that cytokine storm and inflammation are major contributors to COVID-19 severity, cordycepin’s anti-inflammatory effects may have therapeutic implications.
Published cell-free assays provided evidence that cordycepin-TP can inhibit the activity of SARS-CoV-2 polymerase NSP1241, suggesting that cordycepin’s activity against coronaviruses may also involve direct inhibition of viral replication. However, our in vitro assays suggest that cordycepin-MP and not cordycepin-TP act as the major contributor to the inhibition of coronavirus replication (Figs. 6B and S2). In addition, cordycepin inhibited viral replication despite the lack of exposure of viral polymerase to the compound. For instance, cordycepin pre-treated cells (where compounds were removed and washed prior to infection) still showed a significant reduction of viral replication while remdesivir did not maintain its activity in this assay (Figs. 5A, 6A). In our time of removal assays, cordycepin was removed at 24 hpi, viral replication was still significantly suppressed when measured at 48 hpi, again without exposure of viral polymerase to cordycepin for 24 hours (Fig. 5B). Overall, the results support cordycepin as a promising compound for the development of antiviral therapies against coronaviruses, including SARS-CoV-2, without directly targeting the viral polymerase. Further studies are needed to elucidate the precise mechanisms of action of cordycepin against coronaviruses and to evaluate its potential as a therapeutic agent for COVID-19.
A potential clinical caveat is the bioavailability of cordycepin in vivo. Though cordycepin has been shown to have biological activity in animal models when administrated intraperitoneally, intravenously, or orally, it is rapidly deaminated by adenosine deaminase, forming 3′-deoxyinosine in blood or tissue culture media66. Some cell types can convert 3′-deoxyinosine back into cordycepin monophosphate through amination67. Thus, it is possible that cordycepin targets particular tissues because of a cell-type-dependent conversion of 3′-deoxyinosine to cordycepin triphosphate and not due to a cell-type specific molecular target. This finding may also explain the lack of viral inhibition by cordycepin in HeLa cells compared to Huh-7 cells, as well as the lack of changes to the AMPK and mTOR/Akt pathways (Figs. 6C, 7I). Therefore, it is important that biodistribution studies of cordycepin are performed for further drug development.
BTZ-1, a benzo-thiozole compound, is an analog of a proprietary compound targeting RUVBL1/2 and has limited available information. Its effect on SARS-CoV-2 warrants further research.
ADEP-42, a cyclic compound with a macrolactone core, has weak agonist activity against human ClpP protease18. Here, we show that ADEP-42 inhibition of coronavirus replication is partially mediated through ClpP (Fig. 6F–H). ClpP was proposed to function in mtDNA homeostasis as its depletion led to mtDNA instability, altered nucleoid organization, and eventual escape into the cytosol triggering anti-viral interferon responses through the cGAS-STING pathway16. In addition, ClpP deficiency caused increased mitochondrial DNA (mtDNA) abundance in both mouse models and patient fibroblasts68. Knocking out CLPP in Huh-7 cells did not unilaterally reduce SARS-CoV-2 infection or replication, suggesting that viral immune-suppressing mechanisms may overcome antiviral interferon-mediated innate immune response. Interestingly, SARS-CoV-2 infection also induces mtDNA release into the cytosol, mediated by the coordinated action of NSP4 and ORF9b69,70. However, depending on the cell type, SARS-CoV-2-dependent mtDNA release may or may not activate cGAS-STING71. In airway epithelial cells, mtDNA was shown to be secreted via mitochondrial inner membrane vesicles, escaping cytosolic DNA sensors and thus failing to activate the interferon response; this aids in SARS-CoV-2 replication and survival69. Dysregulation of ClpP by ADEP-42 may trigger mtDNA release independent of SARS-CoV-2 and allow activation of anti-viral interferon responses. Recovery of SARS-CoV-2 replication in ADEP-42 treated CLPP knockout cells is only partial, suggesting that the compound has other molecular targets yet to be elucidate.
For our compound screening, we relied on coronavirus family members that can be handled in the BSL2 environment, 229E and OC43. Our goal was to develop anti-viral compounds that targeted multiple coronaviruses. As part of this effort, we developed a first-generation SARS-CoV-2 replicon as a tool for high-throughput screening of chemical libraries and for studying all aspects of intracellular viral replication in BSL2 labs. The replicon was encoded on a bacterial artificial chromosome (BAC) and integrated into Flp-In T-Rex HeLa cells under the control of a tetracycline-inducible promoter (Fig. 7A). The replicon lacked genes for S and E proteins to prevent the production of infectious viral particles (Figure S3A–C). Various modifications enabled visual and biochemical detection of different stages of replicon replication/transcription (Fig. 7B–F). The replicon retained full functionality of SARS-CoV-2 replication and polypeptide maturation despite modifications. Utilizing the replicon, we found that remdesivir, BTZ-1, and ADEP-42 inhibited replicon activity in a concentration-dependent manner with IC50s comparable to those for intact SARS-CoV2, while cordycepin did not (Fig. 7G, H). Differences seen may be due to cell-type specific properties. Hence, future SARS-CoV-2 replicons will be generated in different cell types.
In summary, the development of effective antiviral compounds and tools is crucial for controlling the COVID-19 pandemic and preparing for future coronavirus outbreaks. The compounds identified in this study offer promising potential as broad-spectrum anti-coronavirus agents. Their effectiveness against multiple coronaviruses, including SARS-CoV-2 and its variants, and their efficacy in a lung organoid model, suggests their potential clinical relevance and validates the potential of host directed therapies. Furthermore, our replicon should provide easy access to study SARS-CoV-2 replication in a BSL2 lab, facilitating current and future antiviral drug development efforts.
Materials and Methods
Mammalian cell cultures
Huh-7 (male), HeLa Flp-In T-REx (female) and Vero E6 (female Chlorocebus sabaeus) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% Fetal Bovine Serum and 1% Penicillin/Streptomycin. Hepatocyte derived cellular carcinoma cell line overexpressing ACE2 entry receptor (Huh-7-ACE2) were established and cultured in the media described above with the presence of puromycin (1 μg/mL). All cell lines were cultured in a 5% CO2 humidified atmosphere at 37 °C and were routinely tested for mycoplasma contamination. Huh-7 cells were kindly provided by Dr. Alan Cochrane (University of Toronto), HeLa Flp-In T-REx cells were kindly provided by Dr. Liliana Attisano (University of Toronto). Vero E6 cells were obtained from ATCC.
Virus propagation
Coronaviruses HCoV-229E (https://cedarlanelabs.com/Products/Detail/VR-740?lob=AllProducts), HCoV-OC43 (https://www.cedarlanelabs.com/Products/Detail/VR-1558?lob=AllProducts), SARS-CoV-2 Delta (https://www.beiresources.org/Catalog/animalviruses/NR-55672.aspx), and SARS-CoV-2 Omicron (https://www.beiresources.org/Catalog/animalviruses/NR-56461.aspx) were obtained from the Toronto High Containment Facility. The SARS-CoV-2 SB2 was obtained from Dr. Samira Mubareka (Sunnybrook Research Institute, Toronto, Canada). To generate stocks, viruses were propagated in either Huh-7 cells for 229E and OC43 or Vero E6 cells for SARS-CoV-2. Viral titers were measured by TCID50 assays using the Spearman & Kärber method72,73. All SARS-CoV-2 work was conducted in the BSL3 facility at the University of Toronto.
Cell viability assays
Huh-7 or Vero E6 were seeded in 96-well plates at 3.0 × 105 cells/mL, 100 μL per well, for 24 hours. Compounds or DMSO (0.5% final concentration) were added after mock infection for 1 hour performed as described for viral infection below without the addition of virus and incubated for different amounts of time to mimic infection conditions. Cell viability was assessed by using CellTox Green Cytotoxicity Assay (Promega) or alamarBlue (ThermoFisher Scientific) according to manufacturer’s protocol. While CellTox Green was added at the time of addition of compounds and monitored throughout the assay at the indicated times, alamarBlue was added at the endpoints. Effects of compounds on cell viability were expressed relative to cells treated with DMSO alone. Background signal was controlled by incubation of cells with 0.1% Triton for 15 min (100% death) at various endpoints. For Huh-7 cells, both CellTox and alamarBlue were measured at 24, 48, and 72 hours post treatment mimicking infection times for 229E, SARS-CoV-2, and OC43, respectively. For Vero E6 cells, alamarBlue was measured at 48 h post-treatment to mimic OC43 infection in Vero E6 cells. Measurements were taken using the EnSpire Multimode Plate Reader (PerkinElmer), 485 nm (ex) / 590 nm (em) for CellTox Green and 560 nm (ex)/590 nm (em) for alamarBlue.
For viability of organoids, proximal lung organoids were seeded as single domes in 24-well plates. Either DMSO (0.5% final concentration), 30 µM cordycepin, 10 µM BTZ-1, or 10 µM ADEP-42 were added on day 0 to three domes each. LDH release was measured for 4 days post-treatment using the CyQuant LDH Cytotoxicity Assay Kit (ThermoFisher Scientific) according to manufacturer’s instructions. Absorbances were measured at 490 and 680 nm using microplate reader VersaMax 190 (Molecular Devices), and 680 nm (background) were subtracted from 490 nm values before calculation of cytotoxicity based on the following formula:
Maximum LDH activities were obtained by completely lysing organoids at corresponding days after compound treatment.
Viral infection and inhibition assays
Cells were seeded in complete media (DMEM with 10% FBS, D10 media) to grow to 90-100% confluency the next day. 24 hours after seeding, cells were washed with DMEM without FBS (D0 media). Viruses were diluted in D0 media to specific MOIs and added at half of the seeding volume. 1 hpi, cells were washed with DMEM containing 2% FBS (D2) to remove remaining virus particles in the media. Fresh D2 media-containing compounds were then added (unless otherwise indicated) and incubated for specific times as necessary for the assay. Media containing viruses released into the extracellular space (media samples) were harvested for downstream analysis, such as TCID50 and RT-qPCR, while infected cells were harvested for intracellular RT-qPCR and western blotting as outlined in the sections below. For IF analysis, cells were fixed with ice-cold methanol and processed as outlined in the IF section.
For drug screening assays, Huh-7 cells were seeded on 96-well plates and infected at MOI 1 with either 229E, OC43, or SARS-CoV-2. Compounds were diluted in DMSO and added at final concentration of 10 µM (0.5% DMSO in media) if cell viability measured by alamarBlue was above 75%, otherwise, compound concentrations were reduced accordingly with DMSO concentration kept constant. Media samples were harvested at 24 hpi for 229E infected samples, 72 hpi for OC43 infected samples, and 48 hpi for SARS-CoV-2 infected samples as viral particle reaches maximum levels in Huh-7 at these times when examined by RT-qPCR and/or TCID50. All media samples were examined by RT-qPCR. 229E infected cells were also fixed and quantified by IF.
For dose responses in Huh-7 cells, cordycepin, cordycepin monophosphate, BTZ-1, and ADEP-42 were added at 100, 30, 10, 3, 1, 0.3, 0.1 µM final concentration to cells seeded on 96-well plates. All viruses were used at MOI 1. 229E infected media samples were harvested at 24 hpi, OC43 at 72 hpi, and SARS-CoV-2 at 48 hpi and analyzed by RT-qPCR.
To examine the impact of compounds on coronaviruses, Huh-7 cells were seeded on 6-well plates and infected with either 229E or SARS-CoV-2 at MOI 1. Cell lysates were collected for both RNA and protein analysis. RNA extraction and two-step RT-qPCR were performed to analyze intracellular viral RNA, while western blots were performed to analyze impacts on viral protein as outlined in the sections below. To investigate the effects of compounds on SARS-CoV-2 variants, Huh-7 cells were seeded on 96-well plates and infected by SB, delta or the omicron strain at an MOI 1, media were harvested 48 hpi and analyzed by RT-qPCR.
For viral kinetics time of addition, time of removal, and pre-treatment assays, Huh-7 cells were seeded on 96-well plates and infected with 229E using MOI 2, while MOI 0.02 was used for the time of incubation assay. Cordycepin was used at 30 μM, BTZ-1 at 10 μM, and ADEP-42 at 10 μM where indicated. For viral kinetics, media samples were collected at 3-hour intervals for 24 hpi and at 48 hpi and analyzed by RT-qPCR, while cells were collected at the same intervals for western blotting. For the time of addition, compounds were added at various times post virus removal, and media harvested at 24 hpi to assay viral RNA levels. For time of removal assays, compounds were removed at 12 or 24 hpi and media harvested at 24 and/or 48 hpi to assay viral levels both by RT-qPCR and TCID50. For pre-treatment assays, compounds were added 24 hours pre-infection and removed at the time of infection by washing with 1x PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4); media was then harvested 24 hpi and analyzed by RT-qPCR as well as TCID50. For the time of incubation assay, media samples were collected at 24, 48, or 72 hpi and again analyzed by both RT-qPCR and TCID50.
Determination of viral titer by TCID50
To assess viral infectivity, TCID50 was performed by seeding Huh-7 cells on 96-well plates followed by infection with 10-fold serial dilutions of the media samples, using a total of 8 dilutions with 6 replicates each. The inoculum was washed off 1 hour later and cells replenished with fresh 200 μL D2 media. After 5-10 days, wells were examined for color change macroscopically for cytopathic effect (CPE). Wells showing CPE were marked, and TCID50/mL was calculated using the Spearman & Kärber calculator72,73.
To determine infectivity of HeLa-replicon samples, Vero E6 cells were plated at 90% confluence in 96-well plates. The next day, media were inoculated with 10-fold serially diluted mock infected media, SARS-CoV-2 infected media or media harvested from HeLa-replicon cells after 24 hours incubation with 3 µg/mL doxycycline. The SARS-CoV-2 inoculum was washed off 1 hour later and cells replenished with 200 μL of D2 while media only and HeLa-replicon media samples were left on cells. On days 3 and 5, cells were examined microscopically to determined TCID50 as described before74.
RNA analysis by RT-qPCR
To analyze the level of viral RNA release in media (extracellular RNA), one-step RT-qPCR was performed directly on collected media samples. Media samples were heat-inactivated at 95 °C for 5 min, with 1 µL being added to the RT-qPCR reaction setup using the Luna Universal One-Step RT-qPCR Kit (New England Biolabs, NEB) as per the manufacturer’s instructions. Briefly, each reaction included: 5 µL Luna Universal One-Step Reaction Mix (2x), 0.5 µL Luna WarmStart RT Enzyme (20x, 0.4 µL of each forward and reverse primers at 10 µM), and 1 µL of media (template RNA) in a total reaction volume of 10 µL. The sequences used to quantify viral RNA in media are provided in Table S1. RT-qPCR by the standard curve method was performed using QuantStudio 3 Real-Time PCR System (ThermoFisher Scientific) with cycling conditions as follows: reverse transcription at 55 °C for 10 min and initial denaturation at 95 °C for 1 min, followed by 40 cycles of denaturation at 95 °C for 10 s and extension at 60 °C for 1 min. The melting curve protocol followed with 15 s at 95 °C, 1 min at 60 °C, then 15 s each at 0.1 °C increments between 60 °C and 95 °C. Melting and standard curves were generated by the QuantStudio Design & Analysis Software (version 1.5.1, ThermoFisher Scientific).
To quantify intracellular viral (229E and SARS-CoV2) genomic RNA and total RNA, cells were harvested in Total RNA Lysis Buffer (Bio-Rad), RNA was extracted using the PureLink RNA Mini Kit (Thermo-Fisher Scientific) and cDNA was synthesized from the purified RNA with GoScript Reverse Transcriptase (Promega) using a combination of Oligo(dT) and Random Primers, according to manufacturer’s protocols. cDNA reactions (20 µL) were diluted to 100 µL and quantified for 229E genomic and total RNA abundance by qPCR using QuantStudio 3 Real-Time PCR System (ThermoFisher Scientific). The comparative Ct (threshold cycle) method was used for the quantification of viral RNA levels, expressed relative to DMSO, and normalized to the housekeeping gene ß-actin. Each reaction was set up as follows: 5 µL of Luna Universal qPCR Master Mix (2x), 0.25 µL of each forward and reverse primers (10 µM), and 1 µL of diluted cDNA in a total reaction volume of 10 µL. The sequences used to quantify intracellular viral RNA are provided in Table S1. Cycling conditions are the same as outlined above with the exception of holding for 1 min at 95 °C at the first step instead of reverse transcription at 55 °C. Data was analyzed using the QuantStudio Design & Analysis Software (version 1.5.1, ThermoFisher Scientific).
Immunofluorescence analysis
For drug screening and dose-response assays with IF analysis, cells were seeded on black 96-well plates. At indicated endpoints of the assays, cells were fixed and permeabilized with ice cold methanol for 10 min, washed with 1x PBS, and blocked with 3% BSA-PBST (3% BSA, 0.1% Tween-20, 1x PBS). Cells were then stained with anti-229E S antibody (1:1000, kindly provided by Dr. James Rini, University of Toronto) overnight at 4 °C, washed in PBST, and incubated with anti-mouse-AF488 (1:1000, Jackson ImmunoResearch) for 1 h at room temperature (RT). Following subsequent washes with PBST, cells were stained with Hoechst 33342 (ThermoFisher Scientific) according to manufacturer’s instructions, and fluorescent signals were quantified with EnSpire Multimode Plate Reader (PerkinElmer), 490 nm (ex)/525 nm (em) for AF488 and 358 nm (ex)/461 nm (em) for Hoechst signals. Viral protein signals were normalized to Hoechst 33342 signals to account for cell number variation and expressed relative to DMSO.
To examine the impact of compounds on coronaviruses, Huh-7 cells were seeded on coverslips in 6-well plates. After infection and treatment with compounds, cells were processed as above. Cells were stained with anti-229E S antibody or anti-SARS-CoV-2 S antibody (1:500, Abcam ab273433) and incubated with anti-mouse-AlexaFluor488 (1:1000, Jackson ImmunoResearch 111-545-144) secondary antibody. Coverslips were then mounted with DAPI Fluoromount G (Southern Biotech) to illuminate the nuclei. Images were taken on the Nikon 80i Fluorescence Miscroscope (Nikon) equipped with the X-Cite Series 120Q excitation light source (Excelitas Technologies). Image acquisition was performed with the NIS-Elements Basic Research Software (Nikon) and analyzed using ImageJ (open source).
Western blot analysis
For western blotting, protein extracts were prepared by cell lysis in RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with Halt Phosphatase Inhibitor (ThermoFisher Scientific). For Fig. 3B, lysates were separated on 10% TGX acrylamide stain-free gels (Bio-Rad) or 12% SDS-PAGE. Stain-free gels were directly imaged on ChemiDoc MP Imager (Bio-Rad) to measure total protein levels (served as loading control) prior to transfer to PVDF. Following imaging, proteins were transferred to PVDF (0.22 µM, Bio-Rad) using Trans-blot Turbo Transfer System (Bio-Rad). For all other figures, regular 12% SDS-PAGE gels were directly transferred to PVDF membrane. Immunoblots were blocked in 5% milk-TBST (5% milk, 0.1% Tween-20, 1 X TBS) for 1 hour at room temperature (RT) prior to incubation in primary antibodies overnight at 4 °C. Blots were stained with anti-229E N (1:5000, Novus Biologicals NB10064754), Sars-CoV-2 N (1:1000, ThermoFisher Scientific MA5-29981), AMPKα [1:500, Cell Signaling Technology (CST) 2532S], Phospho-AMPKα (Thr172) (1:500, CST 2535S), mTOR (1:1000, CST 2983S), Phospho-mTOR (Ser2448) (1:500, CST 5536S), Akt (pan) (1:2000, CST 4691S), Phospho-Akt (Ser473) (1:1000, CST 4060S), and GAPDH (1:20000, Abcam ab8245) for loading control. After incubation with primary antibody, blots were washed 3x in 1x TBST and incubated in appropriate HRP-conjugated secondary antibody (1:10000) for 1 h at RT. Following subsequent washes, blots were developed using SuperSignal West Pico PLUS Chemiluminescent Substrate (ThermoFisher Scientific) and imaged on ChemiDoc MP Imager (Bio-Rad).
Fluorescent in situ hybridization and quantification
Huh-7 cells seeded on coverslips in 12-well dish were infected with 229E (MOI 2) for an hour after which the virus inoculum was washed off and cells treated with DMSO, 30 µM cordycepin, 10 µM BTZ-1, or 10 µM ADEP-42 at 16 hpi. Cells were fixed with 4% paraformaldehyde in 1x PBS 4.5 h post-compound treatment and processed for fluorescence in situ hybridization (FISH). Fixed cells were dehydrated in 70% ethanol, then rehydrated in hybridization buffer (10% formamide, 2X SSPE: 300 mM NaCl, 20 mM NaH2PO4·H2O, 2 mM EDTA, pH 7.4). Hybridization was performed using 229E genomic or total RNA oligonucleotides spanning the regions of the virus as detailed for RT-qPCR. Following washing with 10% formamide, 2x SSPE was used to remove unbound probe. Nuclei were DAPI stained, and slides were mounted on glass slides using ProLong Gold Antifade Mountant (ThermoFisher Scientific). Images were acquired using Zeiss microscope with AxioCamICc 5 camera at 40x oil immersion using Zen software.
Quantification of genomic viral RNA localization was performed using CellProfiler image analysis and CellProfiler Analyst following published guidelines38,39 – pipeline is available as Supplemental Data File 2. Briefly, DAPI-stained nuclei were identified as primary objects and cytoplasm propagated using distance from nuclei with edge nuclei being excluded. Perinuclear space was arbitrarily defined as a 30 pixels radius away from the nucleus due to the accumulation of GFP signals in DMSO samples in this area (Fig. S4A). Intensity and radial intensity distribution of genomic RNA staining along with various associated features were measured in the perinuclear space as well as cytoplasm. Ratio of intensity was calculated by division of cytoplasmic intensity versus intensity in perinuclear space. Classification of cells by phenotype was performed using CellProfiler Analyst, consisting of at least 50 cells per phenotype. Three bins were created: perinuclear foci (positive bin), diffuse distribution (negative bin) and cells containing no virus. Following training, Random Forest Classifier was used to make label predictions for all identified cells in three independent experiments based on features measured with an average of 87.5% accuracy on training set. Classification of perinuclei foci can be scored with 94% accuracy (Fig. S4B). Enrichment scores were calculated based on prior and posterior distributions. Prior probability was computed from Dirichlet-Multinomial distribution from the full experiment and posterior was computed for each group independently.
Generation of human lung organoids
For SARS-CoV-2 infection assays, human lung organoids were generated at the Applied Organoid Core Facility (Donnelly Centre, University of Toronto) with a modified protocol35 using H9 human embryonic stem cells (hPSCRegID: WAe009-A) provided by Dr. James A. Thomson, University of Wisconsin-Madison to the WiCell Research Institute National Stem Cell Bank, which obtained informed donor consent. The use of these cells was approved by the Research Ethics Board at the University of Toronto, Canada. All ethical regulations relevant to human research participants were followed. In brief, H9s were plated in V-bottom plates and differentiated into endoderm in DMEM/F12 (Gibco), supplemented with 20% KnockOut Serum Replacement (KSR, Life Technologies), 2% MEM-NEAA, 55 μM β-mercaptoethanol (Life Technologies), 50 μM Y-27632 (Selleck Chem) using 3 mM CHIR, and 100 ng/mL Activin A. To generate anterior foregut endoderm from H9s, cells were transferred to Serum Free media [SFM: DMEM/F12 (HAM) (Gibco), supplemented with N2 (Gibco), B27 with Vitamin A (Gibco), ascorbic acid (Thermo Scientific Chemicals), Glutamax (Gibco), N-acetyl-L-cysteine (Thermo Scientific Chemicals)] supplemented with 300 nM LDN (Tocris) and 10 μM SB431542 (Tocris). After 24 hours, media were replaced with SFM supplemented with 1 μM IWP-2 (Tocris) and 10 μM SB431542 (Tocris) for another 24 hours. For the next 2 days, cells were incubated with SFM supplemented with SAG, CHIR, BMP4 and all-trans retinoic acid to induce growth of lung progenitors. The early lung organoids were then embedded in Matrigel in Maturation media (MM) comprised of SFM supplemented with CHIR, FGF7, FGF10, BMP4 and all-trans retinoic acid for 7-10 days. Media was replaced every 3 days to ensure replenishment of nutrients and signals. Lung organoids were then excised and maintained in Air-liquid Interface (ALI) cultures until Day 30.
SARS-CoV-2 infection and inhibition assays in organoids were similar to those used for cell lines. Matured organoids were infected at MOI 2, estimated for ~2 g of mass, for 1 hour. Organoids were then washed twice with 1x PBS and incubated in media with various treatment conditions: 0.5% DMSO, 50 μM cordycepin, 30 μM BTZ-1, or 30 μM ADEP-42. Media samples were harvested daily for 4 dpi and assessed using RT-qPCR.
For organoid viability assays, human airway organoids were generated from embryonic stem cells (ESC) using an established protocol75. In brief, fetal lung progenitor cells were generated following a 3-stage directed differentiation protocol in which cells are induced into definitive endoderm using the STEMDiff definitive endoderm kit (STEMCELL Technologies; cat. no. 05110). This is followed by the anterior ventral foregut endoderm stage, which expresses NKX2-1 indicating lung lineages, then fetal lung progenitors. They are then expanded in matrigel in Airway Epithelial Spheroid (AES) expansion media. For functional assays, organoids were plated into a matrigel dome in one well of a 24-well plate. Each dome (50 μL) contained approximately 2000 organoids. Viabilities were assessed by LDH release (see cell viability assays section) for 4 days of compound treatment.
Replicon generation
BAC encoding replicon was generated from RT-qPCR of SARS-CoV-2 genome and modified using molecular cloning approaches in collaboration with Codex DNA (currently Telesis Bio). Full sequencing of BAC clone was performed to verify modifications (Supplemental Data File 1). List of features and modifications is available in Table S2.
For integration of the replicon into HeLa-FlpIn T-Rex cells, the cells were plated at 80% confluence in 1 well of 6-well plate. After 24 hours, cells were transfected using Lipofectamine 3000 (Thermo Scientific) with equimolar amounts of BAC encoding replicon and pOG44 following manufacturer’s instructions for the transfection reagent. After 24 hours, cells were re-plated in T-175 flasks. After additional 24 hours, HeLa cells were exposed to hygromycin B at 200 µg/mL for two weeks, followed by 300 µg/mL for another two weeks.
Electron microscopy imaging
Coronaviral concentration for electron microscopy was done as described before76,77. Briefly, HeLa-replicon cells were incubated with 3 µg/mL doxycycline and, after 24 hours, cells were centrifuged at 10,000 g for 20 mins. Media were then slowly and carefully overlayed on a cushion of 20% sucrose in 1x PBS containing 0.01 M HEPES, pH 7 followed by ultracentrifugation in Sorvall centrifuge with SW32 rotor at 27,000 g for 90 mins at 4 °C. Supernatant was discarded and pellet was resuspended in 100 µL 1x PBS and flash frozen in liquid nitrogen followed by electron microscopy imaging using T20 Thermo transmission electron microscope housed at SickKids.
Replicon activity assays
Cells were plated at 10% confluence in 96-well black plates and, after 24 hours, exposed to doxycycline at 3 µg/mL in regular FBS. After 16-24 hours, replicon activity was measured using Nano-Glo HiBiT Lytic Detection kit (Promega) following manufacturer’s instructions. Briefly, a buffer containing a mild detergent, the large nanoLuciferase fragment termed LgBiT, which complements HiBiT-tagged nucleoprotein, and furimazine (the nanoLuciferase substrate) were added in a volume equal to media in 96-well plate containing HeLa-Replicon cells plated as described above. Plates were orbitally shaken at 300 RPM for 10 mins and then luminescence was measured using EnSpire Multimode Plate Reader. In cases where drug assays were conducted, HeLa-FlpIn T-Rex cells without integrated replicon were used as background control, and compound activity at various concentrations were compared to DMSO.
For confocal images of the SARS-CoV-2 replicon markers, HeLa-replicon cells were seeded on 24-well culture plates. The next day, for visualization of sfCherry2 and mNeonGreen2, cells were transfected with pSFFV_sfCherry2(1-10) and pSFFV_mNG2(11)1-1078, the respective large fragments complement the integrated small fragments expressed from the replicon. 24 hours after transfection, cells were reseeded on acid-washed coverslips in 24-well culture plates at 10% confluency and induced with 3 µg/mL doxycycline or equal volume 1x PBS for another 24 hours. Cells were then washed and fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 in 1x PBS, and stained with appropriate nuclear dye: propidium iodide (PI) for mNeonGreen2, and NucSpot 470 for sfCherry2. Coverslips were mounted using ProLong Gold Antifade Mountant (ThermoFisher Scientific) on glass slides. For visualization of mCerulean3, cells were directly seeded on acid-washed coverslips without transfection, induced and processed as above. For HaloTag visualization, cells were directly seeded on acid-washed coverslips at 10% confluency and induced with 3 µg/mL doxycycline for 24 hours. Before fixation, cells were incubated with 200 nM JFX650 HaloTag ligand for 15 min in warm media in the incubator at 37 °C. Fixed cells were processed as above. Images were taken on a TCS SP8 Confocal microscope (Leica) using a 63x (1.4NA) Plan-Apochromatic oil immersion objective and 1 AU (95.8 µm) pinhole. Fluorophores were laser-excited sequentially using a combination of wavelengths at 405 nm, 488 nm, and 552 nm, and emissions were collected using PMT detectors with band-pass filters at 410–495 nm, 410–495 nm, and 578–726 nm, respectively. Images were acquired at 2048 × 2048 resolution and 400 Hz scanning speed. Z-stacks were collected at 512 × 512 resolution and 400 Hz scanning speed. Image analysis and z-stack processing was performed using the LAS X software (Leica) where 3D renderings were produced.
Statistics and Reproducibility
Graphpad Prism version 8 was used for all statistical analyses. Statistical tests used and sample sizes are described in figure legends. Generally, all quantitative data were collected from three independent experiments, each done in triplicates unless otherwise indicated. Dose responses and trend plots are presented as means ± SEM while bar graphs are presented as means ± SD unless otherwise indicated. ANOVA with Dunnett’s multiple comparison test assuming equal variances was used for multi-sample comparison and Student’s two-tailed paired or non-paired t-test assuming equal variances was used for two-sample comparisons. Asterisks correspond to p values as follows ****< 0.0001, ***< 0.001, **< 0.01, *< 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors thank Professor Samira Mubareka (Sunnybrook Research Institute, Canada) and Professor Arinjay Banerjee (Vaccine and Infectious Disease Organization, University of Saskatchewan, Canada) for providing the SB2 virus and for helpful discussions. YQM was supported by a fellowship from the Centre for Pharmaceutical Oncology (University of Toronto), a University of Toronto Fellowship, Jaro Sodek Award Ontario Student Opportunity Trust Fund, and by a Mitacs Research Training Award (Mitacs, Canada). SJ was supported by a Precision Medicine Initiative (PRiME) postdoctoral fellowship (University of Toronto). DAJVO was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Undergraduate Student Research Award. YW was supported by NSERC Undergraduate Student Research Award and by a Biochemistry Summer Student Undergraduate Research Opportunity Program at the University of Toronto. TMM was funded by a Mitacs Accelerate award. SHWC was supported by Graduate and Life Sciences Education Undergraduate Summer Research Project Studentship Award from the University of Toronto and by NSERC Undergraduate Student Research Award. VP was supported by a Biochemistry Summer Student Undergraduate Research Opportunity Program at the University of Toronto. FL was supported by Ontario Graduate Scholarship and PRiME graduate fellowship (University of Toronto). Research was supported by funding from the Canadian Institutes of Health Research (PJT-173345) to WAH, the CIHR COVID-19 Rapid Research response funding opportunity to WAH and MB (VR2-172717), the Toronto COVID-19 Action Initiative to AC and WAH, and CIHR Foundation grant and a Medicine by Design, CFREF grant to LA.
Author contributions
W.A.H. and A.C. conceived and supervised this project. Y.Q.M. and R.M. initiated the project with the compound screening campaign. Y.Q.M., R.M., and T.B. performed initial assay development and optimization under supervision of A.C. and W.A.H. Y.Q.M., with the help of Y.W. and R.M., performed the initial compound screening assays. N.C.H. under supervision of S.D.G.O. maintained and distributed the BSL3 viral stocks. S.J., R.M., and C.A. carried out all BSL3 experiments involving SARS-CoV-2 SB2, and S.J. further conducted the experiments involving the SARS-CoV-2 delta and omicron variants. S.D. performed the FISH experiments. Y.Q.M. performed the IF experiments. Y.Q.M. and S.J. with help of D.A.J.V.O., S.H.W.C., V.P., and T.M.M. performed all other BSL2 experiments. R.M. and W.A.H. conceived and designed the replicon. F.L. synthesized ADEP-42 under the supervision of R.A.B. M.M. and J.N. at the Applied Organoid Core Facility and generated lung organoids under the supervision of L.A. Y.S. under the supervision of A.P.W. and performed the organoid experiments of Fig. 3G, bottom panel. S.J. performed all other organoid experiments. Y.Q.M., S.J., D.A.J.V.O., and Y.W. performed the data analyses. R.M. under the supervision of M.B. contributed to the data analyses. Y.Q.M. and W.A.H. wrote the first draft of the manuscript. The draft was edited with the help of everyone.
Peer review
Peer review information
Communications Biology thanks Lucia Carrau and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editors: Emily Lee and David Favero. A peer review file is available.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. Numerical source data for graphs and charts are available in the supplemental excel file entitled “Supplemental Data File 3”. Uncropped and unedited blots and gels are in the supplemental pdf file. If required, all data are also available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yu-Qian Mao, Shahrzad Jahanshahi, Ramy Malty.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07143-z.
References
- 1.World Health Organization. WHO Coronavirus (COVID-19) Dashboard. World Health Organization (2023).
- 2.Li, M. et al. COVID-19 vaccine development: milestones, lessons and prospects. Signal Transduct. Target Ther.7, 146 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabbatucci, M. et al. Omicron variant evolution on vaccines and monoclonal antibodies. Inflammopharmacology31, 1779–1788 (2023). [DOI] [PMC free article] [PubMed]
- 4.Li, G., Hilgenfeld, R., Whitley, R. & De Clercq, E. Therapeutic strategies for COVID-19: progress and lessons learned. Nat. Rev. Drug Discov.22, 449–475 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li, J. et al. SARS-CoV-2 omicron BA.1.1 is highly resistant to antibody neutralization of convalescent serum from the origin strain. Virus Res.332, 199131 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canas, L. S. et al. Profiling post-COVID-19 condition across different variants of SARS-CoV-2: a prospective longitudinal study in unvaccinated wild-type, unvaccinated alpha-variant, and vaccinated delta-variant populations. Lancet Digit Health, (2023). [DOI] [PMC free article] [PubMed]
- 7.Coronaviridae Study Group of the International Committee on Taxonomy of V. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol5, 536–544 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabaan, A. A. et al. SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Infez. Med28, 174–184 (2020). [PubMed] [Google Scholar]
- 9.V’Kovski, P., Kratzel, A., Steiner, S., Stalder, H. & Thiel, V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol19, 155–170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thaingtamtanha, T. & Baeurle, S. A. Study of protease-mediated processes initiating viral infection and cell-cell viral spreading of SARS-CoV-2. J. Mol. Model28, 224 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao, Y. Q. & Houry, W. A. The Role of Pontin and Reptin in cellular physiology and cancer etiology. Front Mol. Biosci.4, 58 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katoh, H. et al. The R2TP complex regulates paramyxovirus RNA synthesis. PLoS Pathog.15, e1007749 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morwitzer, M. J. et al. Identification of RUVBL1 and RUVBL2 as novel cellular interactors of the Ebola virus nucleoprotein. Viruses11, 372 (2019). [DOI] [PMC free article] [PubMed]
- 14.Mu, X. et al. HIV-1 Exploits the Host Factor RuvB-like 2 to balance viral protein expression. Cell Host Microbe18, 233–242 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Mabanglo, M. F., Bhandari, V. & Houry, W. A. Substrates and interactors of the ClpP protease in the mitochondria. Curr. Opin. Chem. Biol.66, 102078 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Torres-Odio, S. et al. Loss of Mitochondrial Protease CLPP Activates Type I IFN responses through the mitochondrial DNA-cGAS-STING signaling axis. J. Immunol.206, 1890–1900 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung, E. et al. Activators of cylindrical proteases as antimicrobials: identification and development of small molecule activators of ClpP protease. Chem. Biol.18, 1167–1178 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Binepal, G. et al. Development of antibiotics that dysregulate the Neisserial ClpP Protease. ACS Infect. Dis.6, 3224–3236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brotz-Oesterhelt, H. & Vorbach, A. Reprogramming of the Caseinolytic Protease by ADEP Antibiotics: Molecular Mechanism, Cellular Consequences, Therapeutic Potential. Front Mol. Biosci.8, 690902 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen, J. E. et al. Discovery and clinical introduction of first-in-class imipridone ONC201. Oncotarget7, 74380–74392 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Assimon, V. A. et al. CB-6644 Is a Selective Inhibitor of the RUVBL1/2 Complex with Anticancer Activity. ACS Chem. Biol.14, 236–244 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Yenerall, P. et al. RUVBL1/RUVBL2 ATPase activity drives PAQosome maturation, DNA replication and radioresistance in lung cancer. Cell Chem. Biol.27, 105–121.e114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nano, N. et al. Sorafenib as an Inhibitor of RUVBL2. Biomolecules10, 605 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hattori, A. et al. Genetic and chemical targeting of the ATPase complex TIP48 and 49 impairs acute myeloid leukemia. Leukemia37, 1812–1829 (2023). [DOI] [PubMed] [Google Scholar]
- 25.Ju, D. et al. Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals. Sci. Transl. Med.12, eaba0769 (2020). [DOI] [PubMed]
- 26.Rampersad, S. N. Multiple applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors12, 12347–12360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banerjee, A. et al. Isolation, sequence, infectivity, and replication kinetics of severe acute respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis.26, 2054–2063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuli, H. S., Sharma, A. K., Sandhu, S. S. & Kashyap, D. Cordycepin: a bioactive metabolite with therapeutic potential. Life Sci.93, 863–869 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Muller, W. E. et al. Effect of cordycepin on nucleic acid metabolism in L5178Y cells and on nucleic acid-synthesizing enzyme systems. Cancer Res37, 3824–3833 (1977). [PubMed] [Google Scholar]
- 30.Radhi, M. et al. A systematic review of the biological effects of Cordycepin. Molecules26, 5886 (2021). [DOI] [PMC free article] [PubMed]
- 31.Brotz-Oesterhelt, H. et al. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med11, 1082–1087 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Wong, K. S. et al. Acyldepsipeptide analogs dysregulate human mitochondrial ClpP protease activity and cause apoptotic cell death. Cell Chem. Biol.25, 1017–1030 e1019 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Wong, K. S. & Houry, W. A. Chemical modulation of human mitochondrial ClpP: Potential application in cancer therapeutics. ACS Chem. Biol.14, 2349–2360 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Dahal, S. et al. On a path toward a broad-spectrum anti-viral: inhibition of HIV-1 and coronavirus replication by SR kinase inhibitor harmine. J. Virol.97, e0039623 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han, Y. et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature589, 270–275 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, S. M., Wunder, M. B., Norris, D. A. & Shellman, Y. G. A simple protocol for using a LDH-based cytotoxicity assay to assess the effects of death and growth inhibition at the same time. PLoS One6, e26908 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jahanshahi, S. et al. Broad spectrum post-entry inhibitors of coronavirus replication: Cardiotonic steroids and monensin. Virology589, 109915 (2024). [DOI] [PubMed] [Google Scholar]
- 38.Stirling, D. R. et al. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinforma.22, 433 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stirling, D. R., Carpenter, A. E. & Cimini, B. A. CellProfiler Analyst 3.0: accessible data exploration and machine learning for image analysis. Bioinformatics37, 3992–3994 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klenow, H. Formation of the Mono-, Di- and Triphosphate of Cordycepin in Ehrlich Ascites-tumor cells in vitro. Biochim Biophys. Acta76, 347–353 (1963). [PubMed] [Google Scholar]
- 41.Daikopoulou, V. et al. Targeting SARS-CoV-2 polymerase with new nucleoside analogues. Molecules26, 3461 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown, A. J. et al. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir. Res.169, 104541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, C. et al. Cordycepin activates AMP-activated protein kinase (AMPK) via interaction with the gamma1 subunit. J. Cell Mol. Med18, 293–304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawley, S. A. et al. Mechanism of Activation of AMPK by Cordycepin. Cell Chem. Biol.27, 214–222 e214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong, Y. Y. et al. Cordycepin inhibits protein synthesis and cell adhesion through effects on signal transduction. J. Biol. Chem.285, 2610–2621 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuo, S. Q. et al. Cordycepin inhibits cell senescence by ameliorating lysosomal dysfunction and inducing autophagy through the AMPK and mTOR-p70S6K pathway. FEBS Open Bio.11, 2705–2714 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao, Y. et al. Cordycepin enhances the chemosensitivity of esophageal cancer cells to cisplatin by inducing the activation of AMPK and suppressing the AKT signaling pathway. Cell Death Dis.11, 866 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, W. D. et al. Cordycepin down-regulates multiple drug resistant (MDR)/HIF-1alpha through regulating AMPK/mTORC1 signaling in GBC-SD gallbladder cancer cells. Int J. Mol. Sci.15, 12778–12790 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin, S. C. & Hardie, D. G. AMPK: Sensing Glucose as well as Cellular Energy Status. Cell Metab.27, 299–313 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Wei, C. et al. Cordycepin inhibits drug-resistance non-small cell lung cancer progression by activating AMPK signaling pathway. Pharm. Res144, 79–89 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Parthasarathy, H., Tandel, D., Siddiqui, A. H. & Harshan, K. H. Metformin suppresses SARS-CoV-2 in cell culture. Virus Res323, 199010 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao, H. L., Liu, Z. J. & Chang, Z. Cordycepin induces apoptosis in human bladder cancer cells via activation of A3 adenosine receptors. Tumour Biol.39, 1010428317706915 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Kurhade, C., Xie, X. & Shi, P. Y. Reverse genetic systems of SARS-CoV-2 for antiviral research. Antivir. Res210, 105486 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen, H. T., Falzarano, D., Gerdts, V. & Liu, Q. Construction of a noninfectious SARS-CoV-2 Replicon for antiviral-drug testing and gene function studies. J. Virol.95, e0068721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, J. et al. A systemic and molecular study of subcellular localization of SARS-CoV-2 proteins. Signal Transduct. Target Ther.5, 269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee, J. G. et al. Characterization of SARS-CoV-2 proteins reveals Orf6 pathogenicity, subcellular localization, host interactions and attenuation by Selinexor. Cell Biosci.11, 58 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi, F. S. et al. Expression profile and localization of SARS-CoV-2 nonstructural replicase proteins in infected cells. Microbiol Spectr.10, e0074422 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gordon, D. E. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature583, 459–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Consortium WHOST. et al. Repurposed antiviral drugs for Covid-19 - Interim WHO solidarity trial results. N. Engl. J. Med384, 497–511 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sheahan, T. P. et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med.12, eabb5883 (2020). [DOI] [PMC free article] [PubMed]
- 61.Zambalde, E. P. et al. Increased mTOR Signaling and Impaired Autophagic Flux Are Hallmarks of SARS-CoV-2 Infection. Curr. Issues Mol. Biol.45, 327–336 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mullen, P. J. et al. SARS-CoV-2 infection rewires host cell metabolism and is potentially susceptible to mTORC1 inhibition. Nat. Commun.12, 1876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khalid, T. et al. Therapeutic role of mTOR inhibitors in control of SARS-CoV-2 viral replication. Mol. Biol. Rep.50, 2701–2711 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen, J. et al. Cordycepin alleviated metabolic inflammation in Western diet-fed mice by targeting intestinal barrier integrity and intestinal flora. Pharm. Res178, 106191 (2022). [DOI] [PubMed] [Google Scholar]
- 65.Sun, Y. et al. Regulation of human cytokines by Cordyceps militaris. J. Food Drug Anal.22, 463–467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niramitranon, J. & Pongprayoon, P. Exploring the binding modes of cordycepin to human adenosine deaminase 1 (ADA1) compared to adenosine and 2’-deoxyadenosine. J. Mol. Model26, 29 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Lee, J. B. et al. A novel nucleoside rescue metabolic pathway may be responsible for therapeutic effect of orally administered cordycepin. Sci. Rep.9, 15760 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Key, J. et al. Inactivity of Peptidase ClpP causes primary accumulation of mitochondrial disaggregase ClpX with its interacting nucleoid proteins, and of mtDNA. Cells10, 3354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Faizan, M. I. et al. NSP4 and ORF9b of SARS-CoV-2 induce pro-inflammatory mitochondrial DNA release in inner membrane-derived vesicles. Cells11, 2969 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Archer, S. L. et al. SARS-CoV-2 mitochondriopathy in COVID-19 pneumonia exacerbates hypoxemia. Redox Biol.58, 102508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Domizio, J. D. et al. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature603, 145–151 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kärber, G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn Schmiedebergs Arch. Exp. Pathol. Pharmakol.162, 480–483 (1931). [Google Scholar]
- 73.Karakus, U., Crameri, M., Lanz, C. & Yanguez, E. Propagation and titration of influenza viruses. Methods Mol. Biol.1836, 59–88 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Hierholzer J. C., Killington R. A. 2 - Virus isolation and quantitation. In: Virology Methods Manual (ed^(eds Mahy B. W. J., Kangro H. O.). Academic Press (1996).
- 75.Ngan, S. Y. et al. Stage-specific generation of human pluripotent stem cell derived lung models to measure CFTR function. Curr. Protoc.2, e341 (2022). [DOI] [PubMed] [Google Scholar]
- 76.Ke, Z. et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature588, 498–502 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Neuman, B. W., Adair, B. D., Yeager, M. & Buchmeier, M. J. Purification and electron cryomicroscopy of coronavirus particles. Methods Mol. Biol.454, 129–136 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feng, S. et al. Improved split fluorescent proteins for endogenous protein labeling. Nat. Commun.8, 370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data supporting the findings of this study are available within the paper and its Supplementary Information. Numerical source data for graphs and charts are available in the supplemental excel file entitled “Supplemental Data File 3”. Uncropped and unedited blots and gels are in the supplemental pdf file. If required, all data are also available from the corresponding author on reasonable request.