Abstract
Staphylococcus aureus can cause outbreaks and becomes multi-drug resistant through gene mutations and acquiring resistance genes. However, why S. aureus easily adapts to hospital environments, promoting resistance and recurrent infections, remains unknown. Here we show that a specific S. aureus lineage evolved from a clone that expresses the accessory gene regulator (Agr) system to subclones that reversibly suppressed Agr and caused an outbreak in the hospital setting. S. aureus with flexible Agr regulation shows increased ability to acquire antibiotic-resistant plasmids, escape host immunity, and colonize mice. Bacteria with flexible Agr regulation shows altered cytosine genomic methylation, including the decreased 5mC methylation in transcriptional regulator genes (pcrA and rpsD), compared to strains with normal Agr expression patterns. In this work, we discover how altered genomic methylation promotes flexible Agr regulation which is associated with persistent pathogen colonization in the hospital environment.
Subject terms: Infection, Clinical microbiology, Bacterial infection
In this study, authors examine Staphylococcus aureus adaptability in hospitals, highlighting how a lineage evolved to reversibly suppress Agr, enhancing antibiotic resistance and colonization.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a major health problem worldwide, not only because it is highly pathogenic but also because it frequently causes infection outbreaks1. In 2019, S. aureus still caused more than 700,000 global deaths, in which around 100,000 deaths were associated with bacterial antimicrobial resistance2. MRSA is classically categorized into either community-acquired (CA)-MRSA, the most common pathogen responsible for skin and soft tissue infection among healthy individuals, or healthcare-associated (HA)-MRSA, which commonly harbors multidrug resistance and causes life-threatening infections in immunocompromised and hospitalized patients1,3. Besides being antibiotic-resistant, bacteria become antibiotic-tolerant to survive under antibiotic exposure. When bacteria face stressful environments, they form persisters; a subset of a bacterial population that becomes dormant to survive under bactericidal antibiotics4. Specific genomic changes are associated with persister phenotype during the evolution of antibiotic resistance in S. aureus infection5. Once an outbreak occurs, it is difficult to control bacterial spread because MRSA can not only cause severe infections, but also can asymptomatically colonize the anterior nares, skin, and intestine, which promotes person-to-person spread1.
One key modulator of staphylococcal behavior is the virulence accessory gene regulator (Agr)6. The Agr system is central to staphylococcal quorum sensing (QS), a system that allows bacteria to communicate and coordinate their behavior based on their population density. Staphylococci constitutively release auto-inducing peptides (AIPs) as an extracellular signal6,7. When the bacterial density is high, the amount of AIP reaches a threshold capable of stimulating its receptor on the bacterial cell surface, leading to induction of the agr operon in a positive feedback manner6,7. The Agr system contains genes to activate the operon itself (agrA, agrB, agrC, and agrD) and the regulatory RNAIII, which regulates the expression of multiple virulence genes including several toxins6–10. In most cases, CA-MRSA is associated with expression of several Agr-regulated toxins which promote infection in healthy humans, whereas a subset of HA-MRSA isolates can exhibit defective Agr which somehow confers bacterial adaptation to hospital environments6,11. However, classical CA-MRSA can also cause hospital outbreaks, but the mechanism by which CA-MRSA adapts to the hospital environment remains unclear1,12. In this work, to understand the bacterial factors that enable S. aureus to survive and become persistent in the hospital environment, we assessed the natural evolution of S. aureus by analyzing MRSA strains isolated during an infection outbreak in a neonatal intensive care unit (NICU) in a hospital in Japan.
Results
A MRSA outbreak lineage thrived in the hospital for 2 years and gained new antibiotic resistance
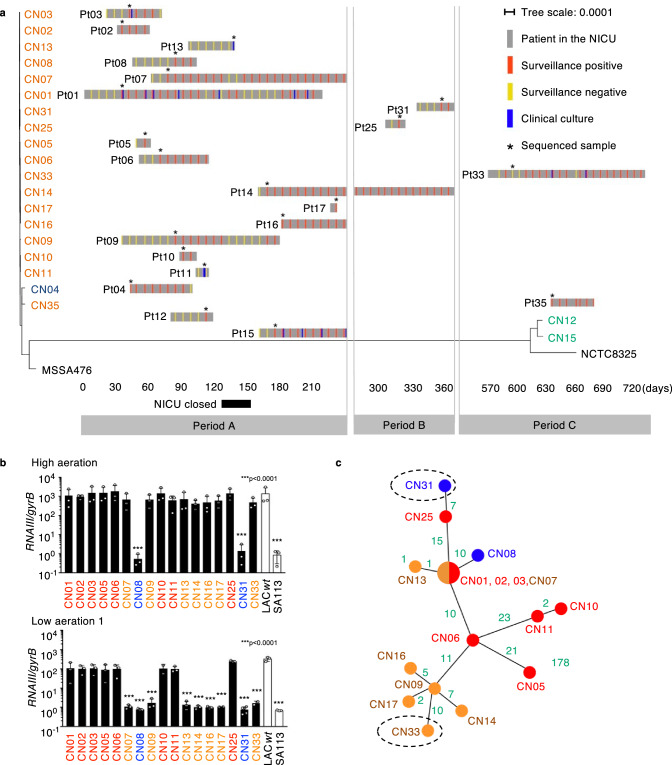
We screened all infants for six months in the NICU once a week for MRSA carriage in feces or throat swab samples and collected MRSA isolates when patients were symptomatic of MRSA infections (period A). We first performed whole-genome sequencing (WGS) to characterize 17 S. aureus isolates by multilocus sequence typing (MLST), staphylococcal cassette chromosome mec (SCCmec) typing, and agr typing. All 17 strains possessed SCCmecIV, which is characteristic of CA-MRSA (Supplementary Data 1)1. We built a maximum-likelihood phylogenetic tree based on the core genome single nucleotide variations (SNVs) and found that the 14 isolates belonging to ST1-CC1-SCCmecIV-agrIII were tightly clustered (Fig. 1a and Supplementary Data 1). Hence, we concluded that the MRSA isolates causing the outbreak at the NICU belonged to the same lineage derived from an original clone. After patient 11 died of S. aureus bacteremia, the NICU was closed for 28 days to control the S. aureus outbreak (Fig. 1a). However, MRSA was sporadically but persistently isolated from neonates for more than two years after the initial outbreak (Fig. 1a and Supplementary Data 2). For further monitoring of MRSA carriage, we also collected samples for an additional ~6 months (period B) and 1 year (period C) after the initial 6 months (period A) (Fig. 1a). Among 18 MRSA strains isolated during periods B and C, we used 3 isolates (CN25, CN31, CN33) belonging to the outbreak lineage (ST1-CC1-SCCmecIV-agrIII) for further study (Fig. 1a). CN35 differs from the other isolates by more than 170 core genome SNVs, which are much larger than what would be expected by the S. aureus mutation rate over a period of approximately two years. Therefore, it was excluded from the following analysis. The majority of the patients (70%, n = 14) were colonized asymptomatically with MRSA in their throat and intestinal tract, while 30% (n = 6) of them had symptomatic infections (Supplementary Data 2).
Fig. 1. Characterization of the MRSA lineage causing an infection outbreak at NICU.
a Timeline of the MRSA outbreak in the NICU layered with a maximum-likelihood phylogenetic tree based on the core genome SNVs. We screened all infants in the NICU for MRSA carriage in feces and throat weekly (surveillance). Some patients experienced MRSA infections (clinical culture). The sampled isolates used for analysis (*) were named CN01 to CN33, corresponding to the patient (Pt) number. Isolates are color-coded as ST1 (CC1): orange, ST2764: blue, and ST8 (CC8): green. MSSA476 (ST1) and NCTC8325 (ST8) were used as reference strains. b Normalized RNAIII expression in 4-hour cultures of MRSA outbreak lineage (CN01agr+-CN03 agr+, CN05agr+, CN06agr+, CN07EA, CN08agr–, CN09 EA, CN10agr+, CN11agr+, CN13EA, CN14EA, CN16EA, CN17EA, CN18agr–and CN33EA) under high or low aeration (n = 3, biological measurements). LAC wt and SA113 strains are shown as positive and negative controls. Data are presented as mean values +/− SEM. ***p < 0.0001, Kruskal-Wallis test with two-tailed Dunn’s post-hoc test. The dots represent the number (n) of biological measurements. c An unrooted maximum-likelihood phylogenetic tree based on the core genome SNVs of the outbreak lineage (ST1-SCCmecIV-agrIII). Green: SNV number between each isolate. Dotted circles: isolates in the late period of the study (>day 365). SNV Single nucleotide variation. b, c Red color: Agr positive subclones, Blue color: agr mutant subclones, Orange color: EA-Agr subclones.
Evaluation of antibiotic susceptibility by minimal inhibitory concentration (MIC) demonstrated that the founder CC1 outbreak clone, CN02, was generally susceptible to antibiotics other than beta-lactam antibiotics (Supplementary Data 3), which is a common trait of CA-MRSA13. However, an offspring of the CC1 outbreak lineage, subclone CN33, showed an additional resistance to gentamicin (Supplementary Data 3). Based on the WGS analyses, we found that the founder clone CN02 harbored a plasmid, which is also found in all subclones of the same CC1 outbreak lineage (Supplementary Fig. 1a and Supplementary Data 1). Subclone CN33 contained in the same plasmid an additional mobile gene element, Tn4001, which is a 4.5 kbp transposon carrying aacA-aphD, the aminoglycoside modifying enzyme gene (Supplementary Fig. 1a)14. Notably, the gentamicin resistance by Tn4001 was also found in the genome of isolate CN12 (ST8/CC8), which did not belong to the outbreak lineage (Supplementary Fig. 1a and Supplementary Data 3). These results suggest that the CC1 outbreak lineage has acquired antibiotic-resistance genes from another CC8 bacterial strain. Furthermore, a genomic insertion of a bacteriophage was detected in the chromosome of the CN09 and CN17 subclones, but not the CN11 subclone compared to the CN02 original clone in CC1 outbreak lineage (Supplementary Fig. 1b). Thus, the outbreak lineage acquired new mobile genetic elements, including resistance genes, within two years in the NICU environment.
Environmentally-adapted S. aureus suppresses Agr under low aerated conditions
To assess the behavior of the CC1 outbreak lineage, we focused on the function of Agr as a critical regulator of virulence, colonization, and persistence13,15,16. While MRSA can colonize the relatively hypoxic environment of the gut17, it occasionally caused systemic infections involving several organs in our patients (Supplementary Data 2). The oxygen concentration varies significantly across different tissues and may influence bacterial behavior, although whether Agr is regulated under hypoxia remains controversial18,19. Thus, we assessed Agr expression under different aerobic conditions by measuring RNAIII levels in 4-hour cultures when bacterial concentration should reach high enough to express Agr in rich culture medium. To clarify the characteristics of the subclones in the outbreak lineage, we have included their Agr status in superscript (Agr positive: agr+, Agr mutant: agr–, EA-Agr: EA) in the names of the outbreak subclones. MRSA subclones isolated during the early period of the outbreak (CN01agr+-CN03agr+, CN05agr+, CN06agr+, CN10agr+, and CN11agr+) showed high Agr expression under all culture conditions (Fig. 1b), which is consistent with the CA-MRSA trait. However, two CC1 outbreak lineage subclones, CN08agr− and CN31agr−, showed no Agr expression in all culture conditions tested (Fig. 1b). Surprisingly, although expression of Agr typically correlates with bacterial density, some subclones (CN07EA, CN09 EA, CN13EA -CN17EA and CN33EA), mainly isolated in the late phase of the outbreak, showed positive Agr expression under highly aerated conditions but silenced Agr expression under low aerated conditions (Fig. 1b). We named these subclones environmentally-adapted-Agr (EA-Agr) subclones because they showed different behavior depending on the environmental conditions. These subclones demonstrated flexibility in Agr expression, adapting their Agr activation according to the aeration levels, which differed from the typical Agr behavior. We additionally assessed the time course of RNAIII expression in S. aureus isolates under different conditions along with their growth curve. When cultured in a low aerated environment, EA-Agr subclones suppressed Agr expression for as long as 12 h and expressed RNAIII for only a short period during the late stationary phase, irrespective of cell density (Supplementary Fig. 2a). Of note, all CC1 outbreak subclones exhibited similar growth curves under the culture conditions used in this study (Supplementary Fig. 2a, b). Consistent with these results, the expression of δ-toxin protein in S aureus EA-Agr subclone, CN17EA, was suppressed under low aeration (Supplementary Fig. 2c). We next evaluated the evolution timeline of the MRSA lineage by creating a phylogenic tree based on the core genome SNVs within the CC1 outbreak lineage. Although two agr mutants emerged in the outbreak lineage, these MRSA subclones harboring defective agr mutations did not generate descendants (Fig. 1c). Most EA-Agr subclones appeared on terminal nodes in the phylogenic tree (Fig. 1c), suggesting that the outbreak lineage evolved from a typical CA-MRSA strain into EA-Agr subclones during adaptation in the hospital setting. CN01agr+, CN02agr+ and CN03agr+ were identical in core genome SNV by Illumina sequencing data and thus were regarded as the same clone. Importantly, when comparing the HiFi sequencing (PacBio Sequel IIe) data of CN02 agr+ and CN07 EA corrected with Illumina sequencing data, the full length of the coding and regulatory sequence was completely identical between these two subclones (Fig. 1c). Therefore, we concluded that the EA-Agr phenotype was not acquired through genomic mutations. Since CN02 agr+ was genetically the closest to CN07 EA among CN01-03 agr+ and was identified the earliest, we referred to CN02 agr+ as the founder clone in this study.
To understand the regulation of Agr in S. aureus isolates of the outbreak lineage, we analyzed the genes in the agr locus and found that CN02agr+, the founder clone with positive Agr expression, had synonymous SNVs in the agr locus (Supplementary Data 4). Among the Agr-defective subclones, CN08agr− had a frameshift mutation in agrC, and CN31 agr− had a point mutation in agrC, presumably resulting in conformational changes in the cytoplasmic domain of AgrC20 (Supplementary Data 4). In contrast, we did not detect any conserved mutations in the agr locus nor in the whole genome of the EA-Agr subclones compared with the founder clone CN02agr+ (Supplementary Data 4).
EA-Agr S. aureus show persister formation, increased acquisition of antibiotic-resistance plasmids and survival inside macrophages
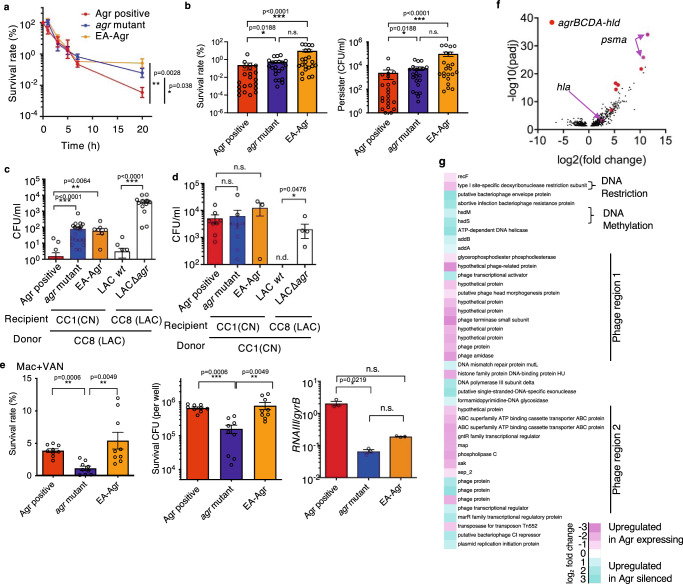
We next performed studies to understand how the flexible Agr promotes EA-Agr subclones survival in the hospital environment. Although it has been reported that Agr-dependent expressions of phenol-soluble modulins (PSMs) suppresses the formation of persisters, little is known about the molecular basis of S. aureus persister formation and its regulation by Agr system in the setting of hospital outbreaks15. To evaluate the antibiotic persistence, we cultured representative subclones under low aerated conditions, and when the bacteria was in the late growth phase (4 h culture), they were treated with a dose of vancomycin (VAN) 100-times higher than their in vitro MIC. The killing curve of agr mutant and EA-Agr subclones showed characteristic bi-phasic killing kinetics indicative of persister cell formation (Figs. 2a, b)4. The persister rate was significantly higher (p < 0.05) in strains with silenced Agr (agr mutant and EA-Agr) subclones than in Agr positive subclones (Figs. 2a, b).
Fig. 2. EA-Agr bacteria display increased persister formation, acquisition of plasmids, and survival inside macrophages.
a Time-dependent survival rate after exposure to a dose of VAN greater than 100 × MIC. b Survival rate and persister CFU at 20 h after VAN incubation. c, d Competency level of representative isolates measured as the CFU of the transformants after electroporation of pMK4 prepared from CC8 strain (C) or CC1 strain (D). e Bacterial survival rate (%), number (CFU), and relative RNAIII expression inside macrophages (Mac) 24 hrs after phagocytosis using vancomycin (VAN) to eliminate extracellular bacteria. a Agr positive: CN02agr+, agr mutant: CN08 agr–, EA-Agr: CN17EA (b) Agr positive: CN02agr+ and CN06agr+, agr mutant: CN08 and CN31 EA-Agr: CN09EA and CN17EA. Each dot represents an experimental replicate. c, d Agr positive: CN02agr+ and CN25agr+, agr mutant: CN08agr– and CN31agr–, EA-Agr: CN17EA. a n = 3 at the 0, 1, 3, 5, and 7-hour time points. n = 20 at 20-hour time point for each group. b n = 11 (c) n = 7 (d) n = 4 for each subclones. e Agr positive: CN02agr+ for, agr mutant: CN08agr–, EA-Agr: CN17EA. n = 9 for survival rate and CFU, n = 3 for RNAIII expression. a–e Data are the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, n.s. not significant, n.d. not detected. Kruskal-Wallis test with two-tailed Dunn’s post-hoc test. The dots represent the number (n) of biological measurements. f Volcano plot of RNA expression profiles of Agr-expressing and Agr-suppressed groups. Fold change values of RNA expression between Agr-expressing and Agr- suppressed groups. The adjusted p value (padj) values below 0.05 are considered significant and shown in the volcano plot. Red: agr operon genes (agrB, agrC, agrD, agrA, and hld), Pink arrows: psmα and hla. g Heatmap of the MGE-associated genes that are significantly increased in Agr-expressing (pink) or Agr-silenced (cyan) bacterial cultures (padj < 0.05). The padj calculated by two-tailed Benjamini-Hochberg method for multiple comparison testing.
We then assessed how EA-Agr subclones acquired antibiotic resistance genes in the hospital setting. The restriction-modification (RM) system is a bacterial defense mechanism to limit horizontal gene transfer (HGT). The restriction enzymes cleave foreign DNA upon recognition of specific sequence while methylases protect the host DNA from restriction clearance by methylating the genome at specific sites21. In S. aureus, the type I RM system is unique in each CC, and therefore HGT between S. aureus belonging to different CCs is rare22. In addition, the experimental plasmid transformation by electroporation was significantly affected by the RM barrier posed by Type I RM systems, which impacted the ability of plasmid acquisition23. To further characterize EA-Agr subclones, representative MRSA isolates were cultured under low aerated conditions to ensure suppression of Agr and were transformed with plasmid pMK4, a shuttle vector between Escherichia coli and S. aureus, harboring a chloramphenicol resistance gene24. When outbreak MRSA subclones (CC1) were transformed with pMK4 prepared from LAC (CC8), the subclones not expressing Agr (agr mutant and EA-Agr) showed higher transformation efficiency than subclones expressing Agr (p < 0.01, Fig. 2c and Supplementary Fig. 3a). Mutation of agr also increased the competence level in the control S. aureus LAC strain (p < 0.001, Fig. 2c and Supplementary Fig. 3a). In contrast, all outbreak subclones were transformed at high efficiency regardless of Agr expression when the plasmid was transferred from the same CC1 (Fig. 2d). Thus, Agr suppression may contribute to efficient HGT between MRSA belonging to different CCs by overcoming RM systems, and once the mobile genetic element (MGE) is integrated into the subclone, it may be conserved in the MRSA lineage independently of Agr function.
Previous work suggested that S. aureus requires Agr activation to survive within macrophages, due to the production of various toxins by Agr activation25–28. To analyze bacterial survival within macrophages, we incubated bone marrow-derived mouse macrophages with representative outbreak MRSA subclones and eliminated extracellular bacteria by vancomycin/gentamycin protection assay. We found that the load of phagocytized bacteria inside macrophages was comparable across all subclones while the presence of extracellular bacteria was negligible after antibiotic incubation (Supplementary Fig. 3b, c). We found that Agr positive and EA-Agr subclones survived better than the agr mutant inside macrophages (Fig. 2e and Supplementary Fig. 3d, e). As expected, Agr expression in macrophages was significantly lower in the agr mutant compared to the Agr positive subclone, while no significant difference was detected between Agr positive and EA-Agr subclones (Fig. 2e). Collectively, flexible regulation of the Agr system in EA-Agr subclones is advantageous for bacterial survival in hospital environments. While Agr expression promotes survival inside macrophages, Agr silencing is beneficial for persister formation and the incorporation of antibiotic-resistance plasmids. Therefore, the ability to flexibly regulate Agr expression allows EA-Agr subclones to optimally adapt and survive under varying conditions within hospital settings.
Comparison of RNA-sequencing results from samples collected under three different culture conditions revealed that the majority of genes exhibit changes dependent on the culture conditions (Supplementary Fig. 4a, top). Therefore, to accurately identify genes that truly change downstream of Agr expression, we cultured CC1 outbreak subclones with different Agr expression profiles (Agr positive, EA-Agr, agr mutant) under various conditions. Specifically, we performed RNA-sequencing of Agr positive (CN02agr+ and CN06agr+), agr mutant (CN08agr− and CN31agr−), and EA-Agr (CN09EA and CN17EA) under three culture conditions and categorized the data into Agr-expressing and Agr-suppressed groups (Supplementary Fig. 4a, bottom). A volcano plot of differentially expressed genes (DEGs) between Agr-expressing and Agr-suppression groups revealed a clear separation of agrBCD, hld and Agr regulated (psma) genes (Fig. 2f and Supplementary Fig. 4b). As expected, the genes encoding α-hemolysin (encoded by hla) and PSMα peptides (encoded by the psma locus), two critical factors essential for S. aureus survival within macrophages and neutrophils25–27, were highly expressed under Agr-expressing conditions (Fig. 2f). In addition, most phage-related genes were upregulated in Agr-expressing conditions (Fig. 2g). Moreover, under Agr-suppression conditions, the expression of hsdM/S, genes encoding enzymes responsible for genomic methylation, and the expressions of DNA helicases, repair enzymes and exonucleases were elevated, whereas gene expression of the Type I restriction enzyme (hsdR) was downregulated (Fig. 2g). We further confirmed that the involvement of Type I restriction system by quantitative RT-PCR. The hsdR expression is significantly decreased in Agr-suppression conditions compared to Agr expressing conditions while no significant difference detected in hsdM (Supplementary Fig. 4c). These data suggest that the decreased expression of hsdR in the restriction-modification system, along with the increased expression of DNA repair enzymes and exonucleases in subclones with suppressed Agr, may contribute to the acquisition of foreign resistance genes in both agr mutant and EA-Agr subclones.
EA-Agr S. aureus is less virulent but persistently colonizes hosts
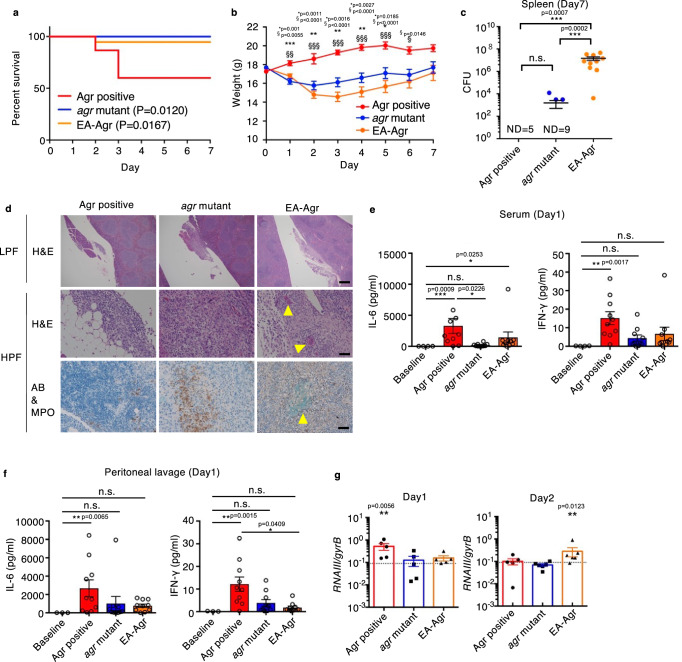
We next performed in vivo experiments to assess the ability of EA-Agr subclones to colonize hosts. We injected outbreak subclones in the intraperitoneal cavity of C57BL/6 J mice as an animal model of systemic infection29 and monitored mouse survival, body weight, bacterial loads, and inflammatory cytokines for up to 7 days. As expected, mice infected with the founder clone that exhibits normal Agr regulation (CN02agr+) showed reduced survival compared to mice infected with a subclone that harbors an agr mutation (CN08agr−) and a subclone that exhibits the EA-Agr phenotype (CN17EA) (Fig. 3a). Similar results were obtained in different subclones of Agr positive, agr-mutants, and EA-Agr subclones (CN06agr+, CN31agr−, and CN07EA) (Supplementary Fig. 5a). While not lethal, the mice infected with agr mutant and EA-Agr subclones consistently lost more body weight than mice infected with Agr positive isolates whereas mice surviving Agr positive infection recovered from weight loss (Fig. 3b and Supplementary Fig. 5b). Notably, mice that survived infection with Agr positive subclones showed variable bacterial loads in the spleen, while mice infected with EA-Agr subclones showed consistently high bacterial burden in the spleen 7 days after infection compared to agr mutant subclones (Fig. 3c and Supplementary Fig. 5c). Consistent with intraperitoneal route of inoculation, the peritoneal lavage, kidney, and liver in EA-Agr infected mice shows high bacterial burden, although this was not statistically significant compared to that observed after infection with the agr mutant subclone 7 days after infection (Supplementary Fig. 5c). In contrast, no or little amounts of bacteria were detected in the blood of all infected groups 7 days after intraperitoneal inoculation (Supplementary Fig. 5c). Histological analysis showed that EA-Agr subclones, but no other subclones, formed biofilm-like structures within the pancreaticosplenic ligament as well as the surface of the kidney and liver 7 days after infection (Fig. 3d and Supplementary Fig. 6). The levels of inflammatory cytokines, IL-6 and IFN-γ, in serum and peritoneal fluid increased markedly in mice infected with Agr positive when assessed on day 1 after infection (Fig. 3e, f, and Supplementary Fig. 5d). In contrast, infection with agr mutant and EA-Agr subclones only elicited a slight increase in these cytokines (Fig. 3e, f, and Supplementary Fig. 5d). Analysis of RNAIII in bacteria from the peritoneal fluids showed higher Agr expression in mice colonized with the Agr positive subclone on day 1, and subsequently with the EA-Agr subclone on day 2, when compared to the agr mutant subclone (Fig. 3g). Thus, EA-Agr bacteria colonizes the host persistently and the bacteria exhibit reduced induction of inflammatory responses compared to Agr positive subclones.
Fig. 3. EA-Agr bacteria exhibit reduced pathogenicity, but increased ability to colonize hosts.
a Survival rate and (b) body weight up to 7 days after C57BL/6 mice were intraperitoneally inoculated with Agr positive (CN02agr+), agr mutant (CN08agr–), or EA-Agr (CN17EA). The survival curve comparison was analyzed by two-tailed Gehan-Breslow-Wilcoxon test (CN02agr+: n = 15, CN08agr–: n = 13, CN17EA: n = 19). For each time point, a significant difference is represented as: * between Agr positive and agr mutant, and § between Agr positive and EA-Agr (CN02agr+: n = 8, CN08agr–: n = 12, CN17EA: n = 11). One-way ANOVA with Tukey’s multiple comparisons test. c Bacterial loads in the spleen at 7 days after bacterial inoculation. ND not detected. d Histological analysis of pancreaticosplenic ligament. LPF low power field, HPF high power field, H&E hematoxylin and eosin, AB alcian-blue, MPO myeloperoxidase. Scale bar; 500 µm (LPF), 100 µm (HPF). Yellow arrowheads and dotted circle indicate biofilm-like structures. e, f The inflammatory cytokine expressions of mice serum (e) and peritoneal lavage (f) at day 1 (n = 10 for infected groups, n = 3 for baseline). Representative images from three independent experiments are shown. g Normalized RNAIII expression of bacteria in peritoneal lavage fluid at day1 (CN02agr+: n = 5, CN08agr–: n = 5, CN17EA: n = 5) and 2 (CN02agr+: n = 5, CN08agr–: n = 6, CN17EA: n = 6) after infection. Each dot represents an individual mouse. Bars represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, Kruskal-Wallis test with two-tailed Dunn’s post-hoc test.
EA-Agr phenotype is found in hospital-adapted strains but not in skin-isolated S. aureus strains
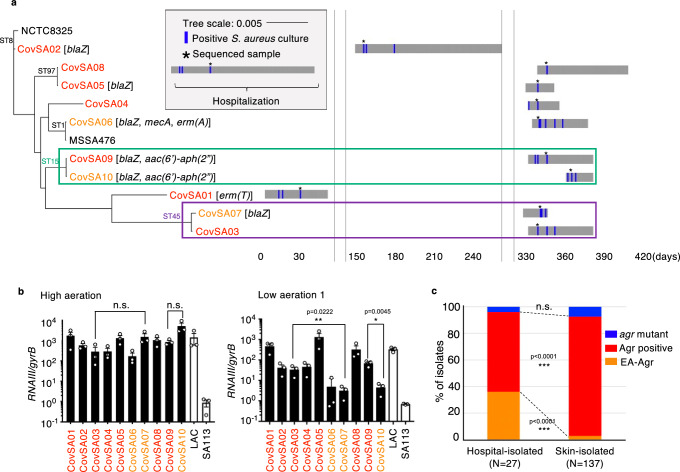
We next assessed whether the evolution of S. aureus to gain EA phenotype is specific to the environment associated with the NICU. To study this, we analyzed S. aureus strains isolated from the skin of healthy 6-month-old infants (n = 137)16 and S. aureus isolates collected from patients hospitalized for COVID-19 pneumonia in the same hospital (n = 10) (Fig. 4a and Supplementary Data 5). Only one strain (CovSA06agr+) was MRSA, whereas the other COVID-19-associated isolates were methicillin-sensitive S. aureus (Supplementary Data 6). We identified two distinct subclone pairs; CovSA03 agr+ and CovSA07 EA (ST45), CovSA09 agr+ and CovSA10 EA (ST15) (Fig. 4a). The latter subclones, CovSA07 EA and CovSA10 EA, derived from CovSA03 agr+ and CovSA09 agr+, respectively, had no mutation in agr and showed EA-Agr phenotype (Fig. 4b). Furthermore, CovSA07 EA with EA-Agr phenotype gained additional antibiotic resistance to penicillin via the acquisition of the blaZ gene during hospital colonization (Fig. 4a and Supplementary Data 6). Importantly, the EA-Agr phenotype in skin-isolated S. aureus strains was relatively rare (4 strains from 137 isolates, 2.9%) compared to the hospital isolates (9 strains from 27 isolates, 33.3%) (p < 0.001, chi square test) (Fig. 4c and Supplementary Data 7). Therefore, we concluded that S. aureus evolution to gain the EA-phenotype is not specific to the NICU but is associated with the hospital environment.
Fig. 4. S. aureus causing co-infection with COVID-19 pneumonia evolved to gain EA-Agr phenotype.
a A maximum-likelihood phylogenetic tree and isolation timeline of S. aureus causing co-infection with COVID-19 pneumonia (COVID-19-related S. aureus). The resistant genes identified in each isolate are indicated in brackets. b Normalized RNAIII expression in 4-hour cultures of COVID-19-related S. aureus under high or low aerated conditions. Red: Agr positive, Orange: EA-Agr S.aureus. n = 3 for each strain. The dots represent the number (n) of biological measurements. Data are presented as mean values +/− SEM. *p < 0.05, **p < 0.01, n.s. not significant, Kruskal-Wallis test with two-tailed Dunn’s post-hoc test. c Rate of Agr positive, agr mutant, and EA-Agr S. aureus in hospital-isolated and skin-isolated S. aureus. All clinical isolates are derived from different patients. ***p < 0.001, two-tailed chi square test.
Altered cytosine methylation controls EA-Agr phenotype in hospital-adapted S. aureus
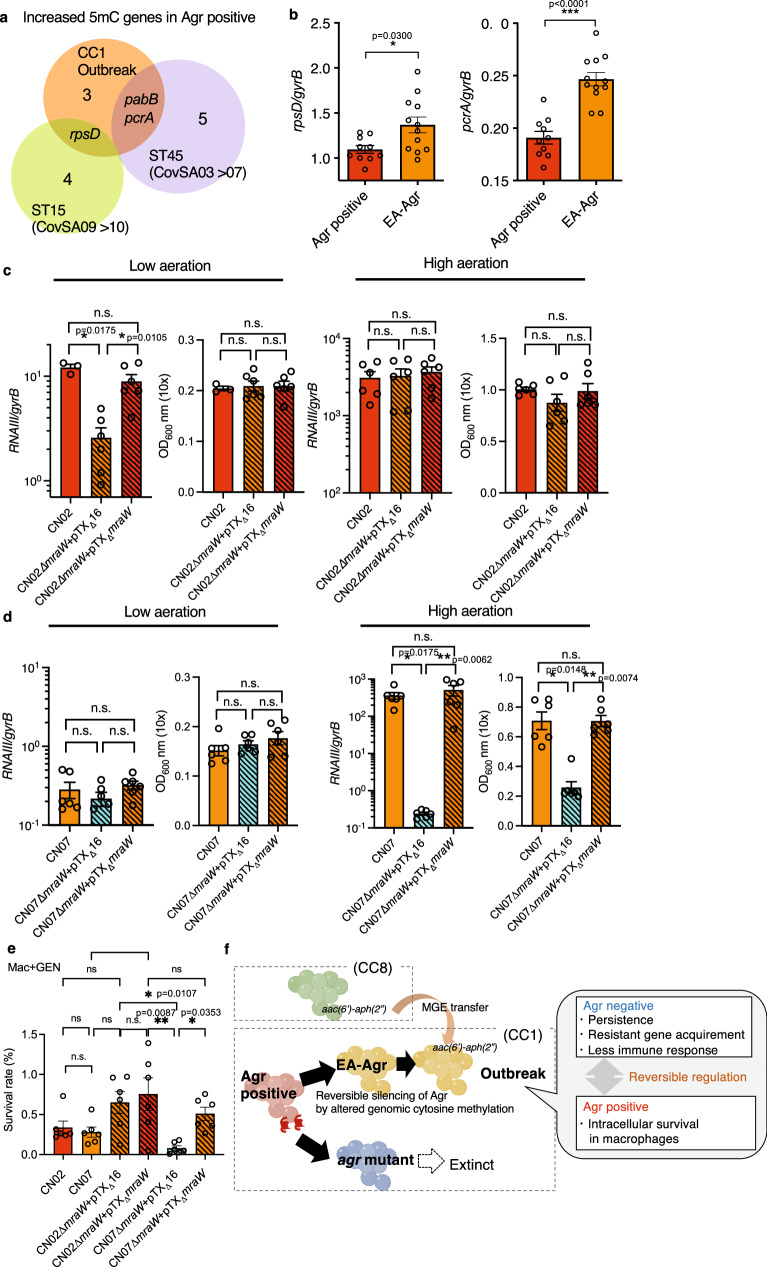
Next, we focused on the mechanism by which EA-Agr silences Agr under certain environmental conditions. Because EA-Agr subclones of the same lineage did not show any conserved genomic mutation compared to the original clone, and furthermore, the core genomes of CN02agr+ and CN07EA were completely identical, we sought to determine whether epigenetic mechanisms might explain the EA-Agr phenotype. Until now, there are only a few reports of epigenetic analysis on S. aureus. In S. aureus, most methyltransferases methylate adenine nucleotides in their target sequences to form N6-methyl-adenine (6 mA), although some methyltransferases can generate N4-methyl-cytosine (4 mC). Although several restriction enzymes are known, the methylase (s) responsible for C5-methyl-cytosine (5mC) methylation remains unknown30. To detect all three methylation patterns at the genome-wide scale, we performed methylome analysis using single-molecule real-time sequencing (SMRT) (Sequal/Sequel IIe, PacBio) for 6mA and 4 mC analysis and enzymatic methyl sequencing (EM-seq, Illumina) for 5 mC analysis. Consistent with previously reported CC1-specific methylation motifs31, the 6 mA target motifs were conserved across Agr positive subclones (CN02agr+ and CN06agr+), EA-Agr subclones (CN07EA and CN17EA), and an agr mutant subclone (CN08agr-) (Supplementary Data 8 and 9). However, CN31agr−, an agr mutant subclone that emerged in the late period of the outbreak, showed a different 6 mA methylation profile than the other strains (Supplementary Data 9). In contrast, we could not detect the 4mC motif with the default settings of the PacBio SMRT Sequel methylation analysis (Supplementary Data 9). To obtain the cleanest 4 mC motifs, we performed a PacBio Sequel IIe and optimized the PacBio SMRT methylation analysis parameters from the default settings. As a result, multiple 4 mC methylation motifs were detected in all analyzed subclones. Interestingly, while only two 4 mC motifs were conserved in CN02agr+, CN06agr+, CN07EA, and CN17EA, the other 4 mC motifs were detected as subclone-specific motifs (Supplementary Data 9). The high conservation of 6 mA motifs and the isolate-specific nature of 4 mC motifs were similarly observed between CovSA03agr+ and CovSA07EA, as well as between CovSA09agr+ and CovSA10EA (Supplementary Data 9). In 5 mC methylation analysis by EM-seq, some specific genomic regions showed differential methylation between Agr positive and EA-Agr, with most of the regions displaying increased methylation in Agr positive (Fig. 5a, Supplementary Data 10). Notably, the genes pcrA (ATP-dependent DNA helicase) and rpsD (30S ribosomal protein S4), which are involved in gene expression regulation, exhibited increased methylation in Agr positive. 5 mC methylation changes in pcrA and rpsD were preserved in the transitions from CovSA03 agr+ to CovSA07 EA (ST45) and from CovSA09agr+ to CovSA10EA (ST15), respectively (Fig. 5a). Quantitative RT-PCR analysis revealed that expressions of pcrA and rpsD were significantly increased in EA-Agr subclones compared to Agr positive subclones in the late growth phase (4 h culture) of low aeration condition (Fig. 5b).
Fig. 5. Altered cytosine DNA methylation induces EA-Agr phenotype in S. aureus.
a Venn diagram comparing identified the increased 5mC genes in Agr positive group compared to EA-Agr group. Orange: CC1 outbreak lineage (CN02agr+, CN06agr+, CN07EA, CN17EA), Green: ST15 (CovSA09agr+, CovSA10EA), Purple: ST45(CovSA03agr+, CovSA07EA). The numbers in the Venn diagram represent the number of differentially methylated genes in each subset. b RpsD and pcrA expressions of Agr positive (CN02agr+, CN06agr+, n = 5 for each) and EA-Agr (CN07 EA, CN17 EA, n = 6 for each) subclones in the late growth phase (4 h culture) of low aeration condition 1. Data are the mean ± SEM. *p < 0.05, ***p < 0.001, two-tailed Mann-Whitney test. c RNAIII expression and bacterial density (OD600) of CN02agr+ (Agr positive) subclone, CN02agr+ deficient in mraW (CN02∆mraW + pTX∆16), and CN02agr+ mutant subclone complemented with mraW plasmid (CN02∆mraW + pTX∆mraW) grown under low aeration 2 and high aeration condition for 6.5 h. OD600; the optical density (OD) at 600 nm. n = 6 for each. d RNAIII expression and bacterial density (OD600) of CN07EA (EA-Agr) subclone, CN07EA mutant subclone deficient in mraW (CN07∆mraW + pTX∆16) and CN07EA mutant subclone complemented with mraW expression plasmid (CN07∆mraW + pTX∆mraW). n = 6 for each. e Bacterial survival rate (%) inside Mac 24 h after phagocytosis using gentamycin (GEN) to eliminate extracellular bacteria. Agr positive: CN02agr+, CN02∆mraW + pTX∆mraW, EA-Agr: CN07EA, CN02∆mraW + pTX∆16 and CN07∆mraW + pTX∆mraW, Agr negative: CN07∆mraW + pTX∆16. n = 6 for each. c–e Data are the mean ± SEM. *p < 0.05, **p < 0.01, n.s. not significant. Kruskal-Wallis test with two-tailed Dunn’s post-hoc test. The dots represent the number (n) of biological measurements. f A hospital adaptation model of EA-Agr S. aureus.
In order to determine whether cytosine methylation impacts Agr expression, we conducted RNA-sequencing of representative Agr positive (CN02agr+, CN05agr+, CN06agr) and EA-Agr (CN07EA, CN09EA, CN17EA) subclones cultured under low and high aeration conditions. In high aeration condition, mraW was the only cytosine methylase gene differently expressed between Agr positive and EA-Agr (p = 0.0116, padj = 0.0600 by DESeq2, Supplementary Data 11). Interestingly, compared to the low aeration condition, decreased expressions of mraW were observed in the high aeration condition in the Agr positive subclones. In contrast, elevated expressions were observed in the high aeration condition in EA-Agr subclones. Therefore, we expected that Agr expression in EA-Agr subclones would have a high dependency on cytosine methylation for Agr expression (Supplementary Data 11). The other three cytosine methylation genes were expressed at very low levels and their expression was comparable in Agr positive and EA-Agr subclones (Supplementary Data 11). Thus, we hypothesized that impaired mraW expression would reproduce the same flexible Agr regulation in Agr-positive subclones. To test this hypothesis, we generated a mraW mutant in CN02agr+, a subclone that was isolated early in the infection outbreak and exhibits normal Agr regulation. We found that the CN02agr+ mraW-deficient mutant, CN02 agr+∆mraW + pTX∆16, showed Agr suppression under low aeration conditions which was rescued by genetic complementation of the mraW deletion mutant (Fig. 5c). The wild-type and mraW mutant CN02 agr+ bacteria showed comparable Agr expression under high aeration conditions (Fig. 5c). In contrast, deletion of mraW in CN07EA, EA-Agr subclone showed very low Agr expression which was not affected by mraW deletion or mraW expression under low aeration conditions (Fig. 5d). Notably, the mraW CN07EA mutant showed markedly impaired Agr expression with severe growth retardation under high aeration conditions which was rescued by mraW complementation (Fig. 5d). Similar to the Agr positive and EA-Agr subclones, CN02agr+∆mraW + pTX∆16 and CN02agr+∆mraW + pTX∆mraW exhibited conserved 6 mA motifs but subclone-specific unique 4 mC motifs (Supplementary Data 9).
MraW, originally identified as a 16S rRNA methyltransferase was recently found to be a DNA methylase32, is conserved as a core genome element within S. aureus. To determine whether DNA and/or rRNA cytosine methylation regulate EA-Agr phenotype, we performed rRNA methylation analyses on CC1 outbreak subclones and the mraW mutants. The pattern of rRNA cytosine methylation was highly conserved in CC1 outbreak subclones, regardless of Agr positive or EA-Agr status. However, rRNA cytosine methylation was severely defective in CN02agr+∆mraW + pTX∆16 and partially rescued in CN02agr+∆mraW + pTX∆mraW (Supplementary Fig. 7).
To test the functional importance of cytosine methylation, we performed macrophage phagocytosis assay using mraW mutant and mraW Agr-positive (CN02 agr+) and EA-Agr (CN07EA) subclones. EA-Agr phenotype subclones (mraW mutant CN02agr+ and mraW reconstituted CN07EA) as well as Agr positive subclone (mraW reconstituted CN02agr+) survived better than Agr negative subclone (mraW mutant CN07EA) (Fig. 5e). These results indicate that genomic cytosine methylation may potentially control the EA-Agr phenotype in the outbreak lineage.
Discussion
In this study, we found that a specific S. aureus lineage thrived in the hospital environment to cause an infection outbreak and eventually gained resistance to antibiotics and environmental stress. Although the original clone in the outbreak had a functional Agr system, its offspring subclones gained flexible Agr-QS regulation associated with differential signatures of cytosine genomic methylation, resistance to antibiotics, and increased ability to acquire antibiotic resistance plasmids and persist in the host.
The agr locus is prone to mutations, and Agr variation has been linked to the adaption of emerging MRSA lineages to distinct ecological niches16,33. In a previous infant cohort study, we chronologically analyzed S. aureus strains on human skin and showed that retention of a functional Agr system was associated with the development of atopic dermatitis16. Other studies showed that Agr is necessary for S. aureus to compete with other commensal bacteria in an intradermal skin model34,35. Thus, a functional Agr system appears to be essential for bacteria to colonize hosts or cause infections in immunocompetent humans or on animal skin. On the other hand, HA-MRSA strains occasionally display Agr defects11, suggesting that the Agr-QS system is not necessary for bacteria to survive in hospitalized patients, and defective Agr may somehow contribute to pathogen survival in the hospital environment36–38. Indeed, EA-Agr and agr mutant S. aureus showed more persistence against antibiotic treatment and did not induce overt immune responses in infected mice compared to Agr positive subclones. Consistent with animal studies, our clinical data showed that some of the Agr positive subclones could induce symptomatic bacterial infections in neonates while EA-Agr and agr mutant subclones did not. In addition, similar to the Agr positive subclones, EA-Agr subclones showed higher survival than agr mutant S. aureus subclones inside macrophages. Collectively, these results suggest that EA-Agr subclones with flexible regulation of Agr-QS are better adapted to survive in the host compared to Agr positive and agr mutant S. aureus (Fig. 5f).
The persister phenotype provides a bacterial population an increased opportunity to acquire resistance by promoting bacterial survival under stressful environments39. Our experiments show that MRSA persistence and competence to acquire plasmid DNA are simultaneously associated with Agr suppression in EA-Agr subclones, thus enabling a subpopulation of bacteria to function as an infection reservoir that maintains and fosters MRSA outbreaks in the hospital environment. Our transcriptome analysis revealed that flexible Agr suppression is associated with suppression of Type I DNA restriction enzymes, which may result in increased acquisition of foreign antibiotic resistant genes in outbreak isolates.
Recent advances in methylation sequencing have revealed that bacterial epigenetics functions as a critical force in species evolution40. Our data indicate that altered genomic cytosine methylation contributes to the flexible regulation of Agr in EA-Agr subclones. In the 5mC methylation analysis, key 5mC methylation signatures (pcrA and rpsD), essential for gene transcription, differed between Agr-positive and EA-Agr and were consistently preserved across STs. This suggests that the altered cytosine genomic methylation may play a crucial role in regulating various gene expression, including the Agr phenotype in EA-Agr (Supplementary Fig. 8). However, no mutations in these methylases have been detected in EA-Agr subclones, leaving open the question of how cytosine methylases contribute to this phenotype. The mechanisms by which cytosine methylation is controlled and the upstream factors that regulate cytosine methylation remain elusive. Collectively, our results suggest that epigenetic changes S. aureus can be a key event for adaptation of S. aureus to the hospital environment.
Methods
Clinical study design and bacterial isolation
We screened all infants in the NICU at Chiba University Hospital, Japan, weekly for MRSA carriage in feces or throat swab samples. This screening is performed as a routine measure since before the outbreak. We also collected MRSA isolates when patients were symptomatic for MRSA infections. The patients’ symptoms and signs indicating infections are described in Table S1. We analyzed 21 isolates (all 17 isolates from April 2016 to October 2016, two from November 2016 to April 2017, and two from October 2017 to February 2018). All isolates from Period A were included in the analysis without exclusion. We excluded bacterial isolates not belonging to the outbreak lineage in the latter periods of the outbreak (Period B and Period C). The Biomedical Research Ethics Committee of the Graduate School of Medicine, Chiba University approved the study protocol (ID: 3250). We additionally collected S. aureus isolated from patients hospitalized for COVID-19 infection from August 2021 to September 2021 (Approved M10368). These studies were part of a surveillance program conducted at the hospital during an outbreak and were not initially intended for research purposes. Therefore, an opt-out consent system was utilized, whereby guardians of infants or patients were informed that study information was available on the university hospital website. Guardians or patients could decline participation if desired. Skin adapted strains isolated from healthy infant skin were previously reported (14).
Bacterial strains and culture conditions
All S. aureus strains were stored in tryptic soy broth (TSB) containing 40% glycerol at −80 °C. The bacterial strains used in this study can be provided following the internal university review for pathogen distribution and the subsequent review and approval of the Material Transfer Agreement between institutions. Bacteria were grown in TSB overnight at 37 °C, and were used for subsequent experiments. For high aeration, low aeration 1, and low aeration 2 culture conditions, the strains were cultured in L-shaped glass tubes using a Monod culture apparatus, in plastic tubes using a Stack shaker (Waken, Tokyo, Japan) with constant 180 rpm shaking, and in plastic tubes standing still in an incubator, respectively. We defined this Monod culture apparatus as high aeration because the use of the Monod culture apparatus relatively increases aeration compared to the plastic tube culture conditions. The control S. aureus strains LACwt, LACΔagr, LACΔpsmα and SA113 were described previously41. CN02agr+ and CN07 strains deficient in mraW were generated as previously described42. Briefly, upstream and downstream regions were PCR-amplified from S. aureus CN02agr+ or CN07EA genomic DNA using primer pairs MraW_1/MraW_2 and MraW_3/MraW_4, respectively (Supplementary Data 12). PCR products were resolved on a 1% agarose gel and purified from the gel using Minelute PCR Purification columns (QIAGEN, Hilden, Germany) according to manufacturer specifications. To fuse the upstream and downstream homologous regions, the purified PCR products were mixed 1:1 and used as template in the fusion PCR reaction using primers MraW_1 and MraW_4 to amplify the fused PCR product. The resulting ~2 kBP PCR product was purified as previously described and phosphorylated using phosphonucleotide kinase (New England Biolabs, Ipswich, Massachusetts, USA). The phosphorylated PCR product was purified from the reaction mix using Minelute PCR Purification columns (QIAGEN, Hilden, Germany). The purified PCR product was then ligated into pIMAY which was linearized by digestion with SmaI and dephosphorylated using calf intestinal phosphatase (New England Biolabs, Ipswich, Massachusetts, USA). The ligation mix was used to transform E. coli DC10B. The resulting pIMAYΔmraW plasmid was used to transform S. aureus strains as previously described42. Allelic replacement and counter selection were performed as previously described42. After counter selection, large clones were screened by analytical PCR using primers MraW_1 and MraW_4. Plasmid pTX∆mraW was constructed by cloning the mraW coding sequence containing the ribosomal binding site region in the BamH I/Mlu I sites of plasmid pTXΔ1616,41 (Supplementary Data 12). All these mraW mutants and complemented strains do not have agr mutations, which was confirmed by HiFi sequencing.
To verify the conservation of mraW within the species, the complete genomes of 860 S. aureus strains were downloaded from GenBank. The quality of these genomes was assessed using CheckM43, which evaluates genome completeness and contamination. Gene prediction was performed with Prokka, followed by pangenome analysis using Panaroo44 with the default settings. The genomic sequence of mraW is conserved between these 860 S. aureus strains.
Antibiotic susceptibility testing
The minimum inhibitory concentration (MIC) test was performed according to the Clinical and Laboratory Standards Institute (CLSI M100 ED30-2020).
Bacterial DNA isolation, WGS, and analysis
Bacterial genomic DNA was extracted using NucleoSpin Tissue kit (Macherey-Nagel, Germany) according to the manufacturer’s instructions. The KAPA HyperPlus Kit (Sigma Aldrich, St. Louis, MO, USA) was used to prepare multiplexed shotgun libraries of DNA samples according to the manufacturer’s instructions. The quality of all libraries was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and Qubit 3 Quantitation Starter Kit (Life Technologies, Carlsbad, CA, USA). Whole-genome sequencing (WGS) was performed using a MiSeq (300 bp paired-end) (Illumina, San Diego, CA, USA) platform according to the manufacturer’s instructions. All raw data were deposited in the Sequence Read Archive (DNA Data Bank of Japan BioProject ID: PRJDB5246). All Illumina data sets were cleaned using Trimmomatic v.0.3345. Trimmed reads were used to generate de novo assemblies of the draft genomes using SPAdes v.3.11.1 with default parameters other than ‘--cov-cutoff auto’ and ‘--careful’46. Annotations of all predicted open reading frames of the draft genomes were performed using Prokka v.1.1447. SNVs of other ST1 strain isolates based on the genome of CN02agr+ were identified using SAMtools v. 0.1.19-44428 cd48, an in-house Python script49, and visually checked using IGV (Integrative Genomics Viewer) v.2.10.050. The functional effects of SNVs and small INDELs were annotated using SNVEff v.4.1 l51.
For genome-wide phylogenetic analysis shown in Fig. 1a, a total of 21 S. aureus strains isolated in the current study and 2 reference strains, MSSA476 (accession ID: NC_002953) and NCTC8325 (accession ID: NC_007795), downloaded from the GenBank database (National institute of health, Bethesda, MD, USA) were used to assemble the core genome. A total of 2264 core genes, 666 shell genes and 447 cloud genes were defined using Roary with default parameters, ‘-i 95’52. A maximum-likelihood tree of 23 S. aureus isolates was constructed on the basis of 177 SNVs in the core genes using RAxML v.8.2.10 with 1000 bootstraps and visualized using iTOL53 or PHYLOViZ online54. For Fig. 1c, SNVs were calculated within the core genes of CC1 outbreak lineage. A total of 2523 core genes, 143 shell genes and 248 cloud genes were identified using Roary within CC1 outbreak lineage. A background analysis of 164 S. aureus isolates from the same region (the same hospital and prefecture in Japan). We identified 1769 core genes, 140 soft core genes, 1188 shell genes, and 3784 cloud genes within the background of isolates by using Roary. The nucleotide sequences of seven genes (arc, aro, glp, gmk, pta, tpi, and yqi) were compared to the S. aureus MLST database for MLST typing55. STs were derived from the assemblies, and CCs were assigned.
We performed MinION (Oxford Nanopore Technologies, UK) to combined data with Miseq data and assemble complete genomes for CN02agr+, CN09EA, CN11agr+, CN12, CN17EA, CN33EA, and CN35EA. High molecular weight DNA was isolated using a modified DNA extraction protocol using cetyltrimethylammonium bromide (CTAB) and polyvinylpyrrolidone (PVPP) as previously described56. DNA libraries were prepared using a ligation sequencing kit and a native barcoding expansion (Oxford Nanopore Technologies), and the prepared library was subsequently loaded into a MinION flow cell (R9.4; Oxford Nanopore Technologies). The MinION sequencing run was performed for 12 h. The base call for MinION data was performed using Guppy v.4.2.357. The data was trimmed using Porechop v.0.2.4 (https://github.com/rrwick/Porechop), Nanofilt v.2.7.158, and SeqKit v.0.8.159. The assembly of MinION reads was performed using Canu v.2.1.160 with default settings and annotated using Prokka v.1.14.6 (35). The data was polished and finalized using Pilon v.1.2261, bwa v.0.7.1762, Circlator v.1.5.563, and Gepard v.1.4064. A plasmid map was generated using BLAST Ring Image Generator v.0.95-dev.000465. The ResFinder web server (https://cge.cbs.dtu.dk/services/ResFinder/) was used to identify antimicrobial resistance genes.
The HiFi sequencing (PacBio Sequel II) was performed to analyze methylome of representative isolates (CN02agr+, CN06agr+, CN07EA, CN17EA, CN02∆mraW + pTX∆mraW and CN02∆mraW + pTX∆16). Samples were cultured under low aerated conditions 2 for 5 h (CN02agr+, CN06agr+, CN07EA, CN17EA) or for 6.5 h (CN02agr+∆mraW + pTX∆mraW and CN02agr+∆mraW + pTX∆16) before sampling. The genomic DNA were extracted by CTAB-PVPP method as described above and then libraries were prepared according to the manufacturer’s instructions (Pacific bioscience, California, USA). The 6 mA and 4 mC types of methylation were detected by Microbial Genome Analysis and Base Modification Analysis applications of SMRT Link v.11.1.0.166339 with the option of the function motifMaker ‘--minScore 35’ or ‘--minScore 100’. All the raw SMRT sequencing data presented in this paper are available in the DDBJ/EMBL/GenBank databases under the accession numbers DRR378798–DRR378822, and SMRT analysis data are available under accession numbers DRZ078082–DRZ078093. For sequencing of agr region, the genomes of outbreak lineage were aligned to the reference genome RN8465 as the S. aureus agr-III prototype.
The HiFi sequencing reads of CN02 was assembled by using SMRT LINK PacBio. The assembled sequences were polished by using the Illumina reads with bwa and HyPo v. 1.0.366. The gene predictions were performed using Prokka. By using Roary, a set of coding genes in CN02 was identical to that in CN07.
Bacterial RNA extraction and quantitative PCR
Bacterial RNA was extracted using the E.Z.N.A. Bacterial RNA Kit (Omega Bio-tek, Norcross, GA, USA) and subjected to reverse transcription using a PrimeScript RT Master Mix (Takara Bio, Shiga, Japan), according to the manufacturer’s instructions. RNAIII and gyrB expression were measured by quantitative PCR (qPCR) as described previously13,15,16. All qPCR analysis were repeated for a total of three independent experiments with each experiment containing at least two technical replicates for each condition. All primers are described in Supplementary Data 12.
Western blot analysis
δ-toxin and SpA levels in 12-hour Staphylococcus culture supernatants were analyzed by previously described polyclonal anti-δ-toxin antibody67 and anti-protein A antibody (# ab60206, Abcam, Cambridge, United Kingdom). The antibodies were used at a 1:1000 (anti-protein A) and 1:250 (anti-δ-toxin) dilution. Western blot analysis was repeated for a total of three independent experiments.
Persister cell assay
Overnight cultures of S. aureus strains were washed twice with sterile PBS, diluted to an optical density of 600 nm (OD600) of 0.1 in 4 ml of TSB and incubated for 4 h. The cultures were diluted (1:100) in TSB to approximately 1 × 107 CFU per ml immediately before incubation with vancomycin (VAN) at 100 µg per ml and cultured at low oxygenic 2 conditions. At 1, 3, 5, 7, and 20 h after VAN incubation, 1 ml of culture was sampled, washed with 1 ml of sterile PBS, appropriately diluted in PBS, and plated on TSB agar to determine colony forming units (CFU). The killing curve was calculated by dividing the value of the CFU at the indicated time by the value of CFU at the time of VAN incubation. Persister assays were repeated for a total of three independent experiments with each experiment containing at least six technical replicates for each condition.
Electroporation
Electroporation was conducted essentially as described42. In detail, the overnight cultures of S. aureus were grown in 4 ml BHI medium (in 14 ml tubes) at low aeration condition 1 and then washed thrice with sterile PBS. The cultures were diluted to an OD600 of 0.5 in 30 ml fresh BHI medium. The cultures were incubated for 90 min with 210 rpm horizontal shaking using StackShake (WAKO). After harvested by centrifugation at 4000 × g for 10 min, the cells were resuspended in an equal volume of autoclaved ice-cold water. The cells were then repeatedly centrifuged and resuspended first in 1/3rd, then in 1/12th, 1/230th, and finally in 1/300th volume of autoclaved ice-cold 10% (w/v) glycerol. Aliquots of 100 µl were frozen at −80 °C. For electroporation, the cells were thawed on ice for 5 min and then left at room temperature for 5 min before undergoing centrifugation (4000 × g for 3 min), followed by resuspension in 50 µl of 10% glycerol and 500 mM sucrose (filter sterilized). After incubated with the plasmid DNA (5 µg) on ice for 10 min, samples were transferred to a 2-mm electroporation cuvette (Bio-Rad) at room temperature, and pulsed at 9 kV per cm. The cells were incubated in 1 ml of BHI supplemented with 500 mM sucrose (filter sterilized) at 37 °C for 2 h before plating on BHI agar containing 10 µg of chloramphenicol, and incubated at 37 °C overnight for CFU count. Transformation assays were repeated for a total of three independent experiments with each experiment containing at least two technical replicates for each strain.
Phagocytosis assay
Phagocytosis assay was performed as described before68. Briefly, bone-marrow-derived macrophages (BMDM) taken from C57BL/6 J mice were seeded in a 24-well plate (5.0 × 105 cells per well, 500 µl) and incubated at 37 °C for 24 h. The cell culture medium was then washed twice with sterile PBS. The overnight culture of each strain was incubated for 5 h at low aeration condition 1 (CN02agr+, CN07EA, and CN08agr−) or for 6.5 h at low aeration condition 2 (CN02agr+∆mraW + pTX∆mraW and CN02agr+∆mraW + pTX∆16) or for 6.5 h at high aeration condition (CN07EA∆mraW + pTX∆mraW and CN07EA∆mraW + pTX∆16) before harvested by centrifugation (2000 × g, 15 min, 4 °C). The pellets were then washed twice with sterile PBS, resuspended in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% Fetal bovine serum (FBS) to a cell density of 1 × 108 CFU per ml. BMDMs were infected with 250 µl of the bacterial cells at a multiplicity of infection (MOI) of 50. At 30 min post infection, the culture medium was washed twice with sterile PBS, replaced with 10% FBS-DMEM supplemented with 300 µg per ml vancomycin (VAN) or 30 µg per ml gentamicin (GEN), and incubated at 37 °C for 24 h to kill non-phagocytosed bacteria. Macrophages were lysed with 200 µl of cold PBS supplemented with 0.02% Triton X-100, and the samples were subjected to quantitative PCR analysis as described above or plated on TSB agar plates to count CFU after overnight incubation. CFUs were counted in the supernatant at 30 min post-infection and subtracted from applied CFU to assess phagocyted bacteria. To assess extracellular persisters, 2.5 × 107 CFU per well of bacterial culture was incubated at 37 °C in 24-well plate for 30 min and 300 µg per ml VAN or 30 µg per ml GEN was added. The percentage of extracellular persisters was calculated 24 h later by dividing the remaining CFU in 24-well plate by the original applied CFU and multiplying by 100. Phagocytosis assays were repeated for a total of three independent experiments with each experiment containing at least two technical replicates for each condition.
RNA sequencing and analysis
2 ml of the 4-hour bacterial cultures were collected, suspended in 2 ml of Buffer A (1% SDS, 0.1 M NaCl, and 8 mM EDTA) and incubated at 100 °C for 2 min. The lysates were suspended in 4 ml of Acid-Phenol: Chloroform, pH 4.5 (with 3-Methl-1-butanol, 125:24:1) at 100 °C for 5 min. The suspension was centrifuged at 12,000 × g for 10 min. The aqueous phase was used for RNA extraction using the Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, CA, USA), according to the manufacturer’s instructions. rRNA was removed using the MICROBExpress kit (Ambion, Austin, TX, USA) in accordance with the manufacturer’s instructions. The KAPA Stranded RNA-seq Library Preparation Kit (Kapa Biosystems) or NEBNext UltraTM II RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) was used to prepare libraries of RNA samples. The quality of all libraries was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies). RNA sequencing was performed using an HiSeq platform (60 bp paired-end) (Illumina) according to the manufacturer’s instructions. All raw data were deposited in the Sequence Read Archive (DDBJ Bioproject ID: PRDJDB5246). All Illumina data sets were cleaned using Trimmomatic45. TPMs (transcript per million) were calculated using Salmon v.1.10.2. To find differentially expressed genes and pathways and the adjusted p-value/False Discovery Rate (FDR) calculated by Benjamini-Hochberg method for multiple comparison testing which is performed by DESeq2 package69.
Mouse systemic infection model
After overnight culture at 37 °C with shaking, S. aureus was cultured in fresh TSB medium for 4 h at 37 °C with shaking at 180 rpm, washed and resuspended in PBS. 6 to 8-week-old female C57BL/6 J wild-type mice were infected with S. aureus by intra-peritoneal injection (3.0 × 108 CFU equivalents per animal). The sample size was calculated by the resource equation method with consideration of the expected death of animals. Before starting the experiment, mice with body weights outside two standard deviations from the group mean were excluded. Animal experiments were repeated for a total of three independent experiments. Survival and body weight were monitored daily, and Kaplan Meier survival graphs were generated (Prism, GraphPad). At specific time points (Day 1, 2, and 7), mice were sacrificed to assess bacterial loads, cytokine production and tissue histology. Peritoneal fluids were isolated by lavage of the peritoneal cavity with 5 ml sterile PBS, centrifuged, and supernatants were stored at −80 °C for subsequent cytokine analysis. The spleens, kidneys, and livers were homogenized in 500 μl sterile PBS. Total tissue (or right lobe of liver) bacterial burden was determined by plating serial dilutions of peritoneal lavage fluid or organ homogenates on mannitol salt phenol red agar plates for 24 h at 37 °C. Results were expressed as CFU per milliliter or CFU per organ. Mouse tissue samples were fixed in formalin, embedded in paraffin, and sectioned for hematoxylin and eosin staining. The samples underwent immunohistochemical staining for myeloperoxidase and were double-stained with alcian blue as described before70. All experimental protocols were approved by the Animal Experiment Committee at Osaka University (ID: 01-027-016).
Cytometric bead assay
Levels of IL-6, IFN-γ, IL-17A and IL-10 proteins in serum and peritoneal fluid were measured using BD CBA Flex Sets according to the manufacturer’s instructions (BD Biosciences, San Jose, CA). The results were analyzed using FCAP Array version 3.0 software (BD Bioscience).
Mass spectrometry of rRNA modifications
rRNAs were prepared from the ribosomes by phenol/chloroform extraction and ethanol precipitation. The 57-mer fragment covering positions 1386–1442 was carved out from the 16S rRNA of S. aureus using a complementary oligodeoxynucleotide (CTTCGGGTGTTACAAACTCyyTCGTGGTGTGACGGGCGGTGTGTACAAGACCCGGGAAC) as previously described71. The purified DNA/RNA heteroduplex [1 pmol] was digested with 20 units of RNase T1 in 20 mM NH4OAc (pH 5.3) at 37 °C for 60 min. Digested RNA was mixed with an 1/10 volume of 10 mM triethylamine acetate and subjected to capillary LC/nano ESI-MS on a Q Exactive hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with a splitless nanoflow high-performance LC (HPLC) system (DiNa, Techno Alpha) as previously described71.
Enzymatic methyl-seq
100 ng of S. aureus genomic DNA was made up to 50 µL with 10 mM Tris 0.1 mM EDTA (pH 8.0). The DNA was transferred to a Covaris microTUBE (Covaris) and sheared to around 150 bp using the Covaris S220 instrument. Prepared DNA libraries using the NEBNext enzymatic methyl-seq kit (New England Biolabs) following the manufactural protocol. Sequencing was performed on the NovaSeq X Plus Sequencing Systems (Illumina). The raw reads were trimmed using Trim Galore v.0.6.4_dev with the options ‘--clip_R1 10 --clip_R2 10 --three_prime_clip_R1 10 --three_prime_clip_R2 10’. The trimmed reads were aligned with the reference genome of either CN02, CovSA03, or CovSA09 using Bismark v.0.23.1dev72, followed by deduplication of the reads and extraction of methylation information. The differentially methylated loci were detected based on Wald test using R packages DSS and bsseq73,74.
Statistical analysis
All analyses were performed using GraphPad Prism and R statistical language75. Differences were considered significant when p < 0.05. The specific statistical tests are mentioned in Figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank M. Tanaka, N. Saito, A. Mizuno, Y. Nomura and M. Zen for technical assistance. Some of the computational analysis was performed using the NIG supercomputer at the ROIS National Institute of Genetics. This work was supported by grants from JSPS KAKENHI 20H03701 (Y.N.), JST FOREST JPMJFR200Y (Y.N.), JSPS KAKENHI 18K07102 (A.T.), JSPS KAKENHI 22K16278 (Y.Y.), MEXT KAKENHI JP22H04925 (H.T.), AMED-PRIME JP 18gm6010016h0002 (Y.N.), AMED-CREST 23gm1610004h0003 (Y.N., H.T., A.T.), The Uehara Memorial Foundation (A.T.), Joint Usage/Research Program of Medical Mycology Research Center, Chiba University (Y.N. and H.T.), Environmental Restoration and Conservation Agency of Japan (N.S.), Institute for Global Prominent Research, Chiba University (Y.N., H.T., A.T., S.N. and N.S.), JSPS KAKENHI Grant Number 16H06279 (PAGS) (Y.N.), Intramural Research Program of the National Institute of Allergy and Infectious Diseases, Project number ZIA-AI000904 (M.O.).
Author contributions
Conceptualization: Y.N., A.T. Methodology: Y.N., A.T., H.T., G.N. Resources: S.M., K.M., A.M., Y.O., N.S., H. Hishiki, H. Hamada, T.N., N.I., T.T., H.I. Investigation: Y.Y., M. Tamai, T.S., S.N., N.I., T.I., S.T., M. Taniguchi, Y.K., R.A., K.I., H.T., A.T., Y.N., A.V., B.S., S.L., K.I., A.S., C.K.-T. Visualization: Y.Y., H.T., A.T., Y.N. Funding acquisition: Y.Y., Y.N., A.T., H.T., N.S., M.O. Project administration: Y.N., A.T., H.T. Supervision: Y.N., A.T., H.T., H.M., M.F., G.N., M.O., T.S. Writing—original draft: Y.Y., H.T., A.T., Y.N. Writing—review & editing: Y.Y., H.T., A.T., G.N., M.O., Y.N.
Peer review
Peer review information
Nature Communications thanks Anne Jamet, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available
Data availability
All sequence data generated in this study were deposited in the Sequence Read Archive (DNA Data Bank of Japan BioProject ID: PRJDB5246). MSSA476 (accession ID: NC_002953 [https://www.ncbi.nlm.nih.gov/nuccore/NC_002953]) for ST1 and NCTC8325 (accession ID: NC_007795 [https://www.ncbi.nlm.nih.gov/nuccore/NC_007795]) were used as reference sequence. Source Data are provided with this paper. All graph data generated in this study are provided in the Source Data file. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yuriko Yamazaki, Tomoka Ito, Seitaro Nakagawa.
These authors jointly supervised this work: Hiroki Takahashi, Akiko Takaya, Yuumi Nakamura.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54033-3.
References
- 1.Turner, N. A. et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat. Rev. Microbiol.17, 203–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antimicrobial Resistance, C. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet399, 629–655 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naimi, T. S. et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA290, 2976–2984 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Balaban, N. Q. et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol.17, 441–448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu, J., Gefen, O., Ronin, I., Bar-Meir, M. & Balaban, N. Q. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science367, 200–204 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Le, K. Y. & Otto, M. Quorum-sensing regulation in staphylococci-an overview. Front. Microbiol.6, 1174 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayville, P. et al. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl Acad. Sci. USA96, 1218–1223 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morfeldt, E., Taylor, D., von Gabain, A. & Arvidson, S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J.14, 4569–4577 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novick, R. P. et al. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet.248, 446–458 (1995). [DOI] [PubMed] [Google Scholar]
- 10.Novick, R. P. et al. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J.12, 3967–3975 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuji, B. T., MacLean, R. D., Dresser, L. D., McGavin, M. J. & Simor, A. E. Impact of accessory gene regulator (agr) dysfunction on vancomycin pharmacodynamics among Canadian community and health-care associated methicillin-resistant Staphylococcus aureus. Ann. Clin. Microbiol. Antimicrob.10, 20 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carey, A. J. et al. Changes in the molecular epidemiological characteristics of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Infect. Control Hosp. Epidemiol.31, 613–619 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otto, M. Community-associated MRSA: what makes them special? Int. J. Med. Microbiol.303, 324–330 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahairas, G. G., Lyon, B. R., Skurray, R. A. & Pattee, P. A. Genetic analysis of Staphylococcus aureus with Tn4001. J. Bacteriol.171, 3968–3972 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bojer, M. S., Lindemose, S., Vestergaard, M. & Ingmer, H. Quorum sensing-regulated phenol-soluble modulins limit persister cell populations in Staphylococcus aureus. Front Microbiol9, 255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura, Y. et al. Staphylococcus Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci. Transl. Med.12, eaay4068 (2020). [DOI] [PMC free article] [PubMed]
- 17.Marteyn, B., Scorza, F. B., Sansonetti, P. J. & Tang, C. Breathing life into pathogens: the influence of oxygen on bacterial virulence and host responses in the gastrointestinal tract. Cell Microbiol.13, 171–176 (2011). [DOI] [PubMed] [Google Scholar]
- 18.George, S. E. et al. Oxidative stress drives the selection of quorum sensing mutants in the Staphylococcus aureus population. Proc. Natl Acad. Sci. USA116, 19145–19154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilde, A. D. et al. Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of staphylococcus aureus invasive infection. PLoS Pathog.11, e1005341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sloan, T. J. et al. Timing is everything: Impact of naturally occurring Staphylococcus aureus AgrC cytoplasmic domain adaptive mutations on autoinduction. J. Bacteriol.201, e00409-19 (2019). [DOI] [PMC free article] [PubMed]
- 21.Corvaglia, A. R. et al. A type III-like restriction endonuclease functions as a major barrier to horizontal gene transfer in clinical Staphylococcus aureus strains. Proc. Natl Acad. Sci. USA107, 11954–11958 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper, L. P. et al. DNA target recognition domains in the Type I restriction and modification systems of Staphylococcus aureus. Nucleic Acids Res.45, 3395–3406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monk, I. R. & Foster, T. J. Genetic manipulation of Staphylococci-breaking through the barrier. Front. Cell Infect. Microbiol.2, 49 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan, M. A., Yasbin, R. E. & Young, F. E. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene29, 21–26 (1984). [DOI] [PubMed] [Google Scholar]
- 25.Kubica, M. et al. A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS ONE3, e1409 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Surewaard, B. G. et al. Staphylococcal alpha-phenol soluble modulins contribute to neutrophil lysis after phagocytosis. Cell Microbiol.15, 1427–1437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grosz, M. et al. Cytoplasmic replication of Staphylococcus aureus upon phagosomal escape triggered by phenol-soluble modulin alpha. Cell Microbiol.16, 451–465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tranchemontagne, Z. R., Camire, R. B., O’Donnell, V. J., Baugh, J. & Burkholder, K. M. Staphylococcus aureus strain USA300 perturbs acquisition of lysosomal enzymes and requires phagosomal acidification for survival inside macrophages. Infect. Immun.84, 241–253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rauch, S. et al. Abscess formation and alpha-hemolysin induced toxicity in a mouse model of Staphylococcus aureus peritoneal infection. Infect. Immun.80, 3721–3732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu, S. Y., Corvaglia, A. R., Chan, S. H., Zheng, Y. & Linder, P. A type IV modification-dependent restriction enzyme SauUSI from subsp USA 300. Nucleic Acids Res.39, 5597–5610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monk, I. R., Tree, J. J., Howden, B. P., Stinear, T. P. & Foster, T. J. Complete bypass of restriction systems for major Staphylococcus aureus lineages. mBio6, e00308–e00315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu, X. et al. Beyond a ribosomal RNA methyltransferase, the wider role of MraW in DNA methylation, motility and colonization in Escherichia coli O157:H7. Front. Microbiol.10, 2520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huber, C. et al. Silence as a way of niche adaptation: mecC-MRSA with variations in the accessory gene regulator (agr) functionality express kaleidoscopic phenotypes. Sci. Rep.10, 14787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He, L. et al. Resistance to leukocytes ties benefits of quorum sensing dysfunctionality to biofilm infection. Nat. Microbiol.4, 1114–1119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paharik, A. E. et al. Coagulase-negative staphylococcal strain prevents staphylococcus aureus colonization and skin infection by blocking quorum sensing. Cell Host Microbe22, 746–756 e745 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He, L. et al. Key role of quorum-sensing mutations in the development of Staphylococcus aureus clinical device-associated infection. Clin. Transl. Med.12, e801 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar, K., Chen, J., Drlica, K. & Shopsin, B. Tuning of the lethal response to multiple stressors with a single-site mutation during clinical infection by Staphylococcus aureus. mBio8, e01476-17 (2017). [DOI] [PMC free article] [PubMed]
- 38.Paulander, W. et al. Antibiotic-mediated selection of quorum-sensing-negative Staphylococcus aureus. mBio3, e00459–00412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levin-Reisman, I. et al. Antibiotic tolerance facilitates the evolution of resistance. Science355, 826–830 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Sanchez-Romero, M. A. & Casadesus, J. The bacterial epigenome. Nat. Rev. Microbiol.18, 7–20 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Wang, R. et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med.13, 1510–1514 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Monk, I. R., Shah, I. M., Xu, M., Tan, M. W. & Foster, T. J. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio3, e00277-11 (2012). [DOI] [PMC free article] [PubMed]
- 43.Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res.25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tonkin-Hill, G. et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol.21, 180 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol.19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hagiwara, D. et al. Whole-genome comparison of Aspergillus fumigatus strains serially isolated from patients with aspergillosis. J. Clin. Microbiol.52, 4202–4209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thorvaldsdottir, H., Robinson, J. T. & Mesirov, J. P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform.14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly.6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics31, 3691–3693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics23, 127–128 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Ribeiro-Goncalves, B., Francisco, A. P., Vaz, C., Ramirez, M. & Carrico, J. A. PHYLOViZ Online: web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res.44, W246–W251 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aanensen, D. M. & Spratt, B. G. The multilocus sequence typing network: mlst.net. Nucleic Acids Res.33, W728–W733 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chapaval, L., Moon, D. H., Gomes, J. E., Duarte, F. R. & Tsai, S. M. An alternative method for Staphylococcus aureus DNA isolation. Arq. Bras. Med. Vet. Zootec.60, 10.1590/s0102-09352008000200004 (2008).
- 57.Wick, R. R., Judd, L. M. & Holt, K. E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol.20, 129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Coster, W., D’Hert, S., Schultz, D. T., Cruts, M. & Van Broeckhoven, C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics34, 2666–2669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen, W., Le, S., Li, Y. & Hu, F. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE11, e0163962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res.27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hunt, M. et al. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol.16, 294 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krumsiek, J., Arnold, R. & Rattei, T. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics23, 1026–1028 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Alikhan, N. F., Petty, N. K., Ben Zakour, N. L. & Beatson, S. A. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genom.12, 402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kundu, R., Casey, J. & Sung, W. K. HyPo: super fast & accurate polisher for long read genome assemblies. bioRxiv10.1101/2019.12.19.882506 (2019).
- 67.Nakamura, Y. et al. Staphylococcus delta-toxin induces allergic skin disease by activating mast cells. Nature503, 397–401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gor, V. et al. Finding of agr phase variants in Staphylococcus aureus. mBio10, 10.1128/mBio.00796-19 (2019). [DOI] [PMC free article] [PubMed]
- 69.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed]
- 70.Jensen, L. K., Henriksen, N. L., Bjarnsholt, T., Kragh, K. N. & Jensen, H. E. Combined staining techniques for demonstration of Staphylococcus aureus biofilm in routine histopathology. J. Bone Jt Infect.3, 27–36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ishiguro, K., Arai, T. & Suzuki, T. Depletion of-adenosylmethionine impacts on ribosome biogenesis through hypomodification of a single rRNA methylation. Nucleic Acids Res.47, 4226–4239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feng, H., Conneely, K. N. & Wu, H. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res.42, e69 (2014). [DOI] [PMC free article] [PubMed]
- 74.Hansen, K. D., Langmead, B. & Irizarry, R. A. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol.13, R83 (2012). [DOI] [PMC free article] [PubMed]
- 75.Team, R. C. R.: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available online at https://www.R-project.org/ (2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All sequence data generated in this study were deposited in the Sequence Read Archive (DNA Data Bank of Japan BioProject ID: PRJDB5246). MSSA476 (accession ID: NC_002953 [https://www.ncbi.nlm.nih.gov/nuccore/NC_002953]) for ST1 and NCTC8325 (accession ID: NC_007795 [https://www.ncbi.nlm.nih.gov/nuccore/NC_007795]) were used as reference sequence. Source Data are provided with this paper. All graph data generated in this study are provided in the Source Data file. Source data are provided with this paper.