Abstract
Background & Aims
In patients with cirrhosis, acute decompensation (AD) correlates with a hyperinflammatory state driven by mitochondrial dysfunction, which is a significant factor in the progression toward acute-on-chronic liver failure (ACLF). Elevated circulating levels of acylcarnitine, indicative of mitochondrial dysfunction, are predictors of mortality in ACLF patients. Our hypothesis posits that acylcarnitines not only act as biomarkers, but also actively exert detrimental effects on circulating immune cells.
Methods
Plasma acylcarnitine levels were measured in 20 patients with AD cirrhosis and 10 healthy individuals. The effects of selected medium- and long-chain acylcarnitines on mitochondrial function were investigated in peripheral leucocytes from healthy donors by determining mitochondrial membrane potential (Δψm) and mitochondrial respiration using the JC-1 dye and Agilent Seahorse XF technology. Changes regarding mitochondrial ultrastructure and redox systems were assessed by transmission electron microscopy and gene and protein expression analysis.
Results
Plasma levels of several acylcarnitine species were significantly elevated in patients with AD cirrhosis compared with healthy individuals, alongside increased levels of inflammatory mediators (cytokines and chemokines). Notably, the long-chain acylcarnitine palmitoylcarnitine (C16:0-carnitine, 1.51-fold higher, p = 0.0059) impaired Δψm and reduced the spare respiratory capacity of peripheral mononuclear leucocytes. Additionally, C16:0-carnitine induced mitochondrial oxidative stress, suppressed the expression of the antioxidant gene HMOX1, and increased CXCL8 expression and IL-8 release. Etomoxir, which blocks acylcarnitine entry into the mitochondria, reversed the suppression of HMOX1. Similarly, trimetazidine, a fatty acid beta-oxidation inhibitor, prevented C16:0-carnitine-induced CXCL8 expression. Importantly, oxidative stress and Δψm impairment caused by C16:0-carnitine were less severe in the presence of albumin, a standard therapy for AD cirrhosis.
Conclusions
Our findings suggest that long-chain acylcarnitines induce mitochondrial injury in immune cells, thereby contributing to the development of immune dysfunction associated with cirrhosis.
Impact and implications
Patients with acute decompensation of cirrhosis and acute-on-chronic liver failure (ACLF) display a systemic hyperinflammatory state and leukocyte mitochondrial dysfunction. We discovered that apart from being increased in the circulation of these patients, the long-chain palmitoylcarnitine is able to elicit cytokine secretion paired with mitochondrial dysfunction in leukocytes from healthy donors. In particular, we show that inhibiting the metabolism of palmitoylcarnitine could reverse these detrimental effects. Our findings underline the importance of immunometabolism as a treatment target in patients with acute decompensation of cirrhosis and ACLF.
Keywords: Immune cells, Acute decompensation of cirrhosis, Acylcarnitines, Mitochondrial dysfunction
Graphical abstract
Highlights
-
•
Palmitoylcarnitine is elevated in the plasma of patients with acute decompensation of cirrhosis.
-
•
This long-chain acylcarnitine induces mitochondrial oxidative stress in leucocytes.
-
•
Palmitoylcarnitine suppresses heme oxygenase 1 and induces the secretion of proinflammatory cytokines.
-
•
These effects were prevented by blocking the entry of palmitoylcarnitine and its oxidation in the mitochondria.
Introduction
Acylcarnitines are conjugated forms of acyl-CoA with carnitine that serve as carriers to transport medium- and long-chain fatty acids into the mitochondria via the carnitine shuttle system for subsequent β-oxidation and degradation.1 This shuttle consists of carnitine palmitoyltransferase-1 (CPT1) located at the outer mitochondrial membrane, carnitine-acylcarnitine translocase (CACT) at the inner mitochondrial membrane, and carnitine palmitoyltransferase-2 (CPT2) situated at the inner mitochondrial membrane facing the mitochondrial matrix.2 The circulating levels of acylcarnitines reflect the composition of the acylcarnitine pool within the cytoplasm, and the level of acylcarnitines in the blood mirrors the accumulation of acyl-CoA species inside the mitochondria.3 For this reason, increased levels of acylcarnitines in the circulation are regarded as biomarkers of mitochondrial dysfunction in many disease conditions.
Acylcarnitines are elevated in a murine model of acetaminophen-induced liver injury4 and in patients with hepatocellular carcinoma.5 Both medium- and long-chain acylcarnitines are also elevated in patients with sepsis and heart failure.6,7 Recently, accumulation of acylcarnitines was a finding that could be extended to patients with acutely decompensated (AD) cirrhosis and particularly AD patients developing acute-on-chronic liver failure (ACLF) in an untargeted metabolomics study of the CANONIC cohort.8 The transition from compensated to decompensated cirrhosis is accompanied by a decrease in survival (3–5 years), and once ACLF develops, the short-term mortality is very high.9 In patients with ACLF, acylcarnitines positioned among the top 50 metabolites that were elevated in comparison with healthy individuals, and in addition, the increase was much higher compared with patients with compensated cirrhosis or even AD cirrhosis.8 Specifically, acylcarnitines were part of a fingerprint of 38 metabolites which accumulated in the blood of these patients and discriminated patients with AD cirrhosis and ACLF from those without ACLF, suggesting mitochondrial dysfunction as a player in ACLF development.8 In line with this, we previously demonstrated the presence of a dysfunctional tricarboxylic acid cycle in leucocytes of patients with ACLF, but not in those with mere AD cirrhosis, indicating that mitochondrial dysfunction is a feature more prominent in ACLF.10 In the same study, we also showed acylcarnitine was predictive of mortality.10 The prognostic model CLIF-C MET, which was developed based on two large multicentre cohorts and included the medium-chain hexanoylcarnitine was superior to the model for end-stage liver disease (MELD) or MELD-sodium (MELDNa) score in predicting short-term mortality in patients with ACLF.11
There is evidence indicating that acylcarnitines are not mere biomarkers of mitochondrial dysfunction but molecules that trigger cellular damage. For instance, long-chain acylcarnitines fuel the inflammatory response and the production of reactive oxygen species (ROS) in primary human myotubes.12 In a murine macrophage cell line, medium- and long-chain acylcarnitines (i.e. C12:0-, C14:0-, and C16:0-carnitine) exert proinflammatory actions through the activation of nuclear factor kappa B (NF-κB), extracellular signal-regulated kinase and c-Jun N-terminal kinase signalling or through the induction of cyclooxygenase-2.13 However, the detrimental effects of acylcarnitines on human immune cells have not yet been studied in detail. Therefore, this study aimed at exploring the effect of acylcarnitine on mononuclear leucocytes with special emphasis on their effects on mitochondria-related functions. To investigate this, we first performed an exhaustive screening of all acylcarnitine species in patients with AD cirrhosis to test their bioactivity in representative in in vitro/ex vivo assays. In these assays, peripheral blood mononuclear cells (PBMCs) freshly isolated from healthy subjects (HS) were used because in our experience PBMCs from patients with AD cirrhosis already exhibit an array of alterations in mitochondria-related functions.10 We also explored the potential modulatory effects of the albumin molecule (a current therapy for patients with AD cirrhosis which has high avidity for fatty acids) on the effects of acylcarnitines on mitochondrial function. The ultimate goal of this study was to elucidate whether increased circulating levels of acylcarnitine might affect the homeostasis of immune cells by triggering mitochondrial dysfunction leading to an inflammatory reaction.
Material and methods
Patients and samples
For plasma measurements, biobanked plasma samples from 20 patients with AD cirrhosis from the PREDICT study were used. The baseline clinical and laboratory data of patients with AD cirrhosis are shown in Table S1. Plasma samples and PBMCs were obtained from HS (n = 40) recruited at the Hospital Clínic of Barcelona Blood Bank.
Study approval
All patients or their legal representatives and the ethics committee of each hospital involved in the study provided written informed consent for omics investigations in the biobanked material before participation (IRB approval #HCB/2015/0427 and #HCB/2016/0710). All research was conducted in accordance with both the Declarations of Helsinki and Istanbul.
Measurement of acylcarnitines
Acylcarnitines were measured in the plasma of patients using liquid chromatography with tandem mass spectrometry (LC-MS/MS). Methanol (450 μl) containing 100 ng of C16-carnitine-d3 (Cayman Chemical, Ann Arbor, MI, USA) as an internal standard was added to 50 μl of plasma. After mixing and centrifuging, the clear supernatant was taken and directly injected into an Agilent 6495/1290 LC/MS/MS system (Agilent Technologies, Santa Clara, CA, USA). Poroshell SB-Aq, 150 × 2.1 mm, 2.7 μm (Agilent Technologies) was used as the stationary phase and a gradient of 0.1 formic acid and methanol was used as the mobile phase. The gradient started at 0% and was increased to 95% methanol within 10 min. The flow rate was 0.4 ml/min and the injection volume was 1 μl. All solvents were Lichrosolve LC/MS grade (Merck, Darmstadt, Germany). The coupled MS was equipped with a Jet Stream electrospray ionization source (ESI) operated in positive multiple reaction monitoring (MRM) mode. Two transitions per compound were detected. Transitions with fragment ion m/z = +85 were used for quantification (Table S2).
Analysis of cytokines and chemokines
Levels of 20 cytokines/chemokines were measured in plasma and cell culture supernatants using a Milliplex MAP Human Cytokine/Chemokine/Growth factor Magnetic bead panel (MilliporeSigma, Burlington, MA, USA). Signals were read using a MAGPIX® instrument (Luminex Corp., Austin, TX, USA) and data analysis was performed using the Belysa immunoassay curve-fitting software (MilliporeSigma).
Analysis of lipid mediators by targeted lipidomics
Plasma samples were extracted using a modified Bligh and Dyer procedure and extracts were analysed using a LC-MS/MS method on an Acquity UPLC separation module coupled to an ABI Sciex 5500 QTRAP hybrid, triple quadrupole, linear ion trap mass spectrometer equipped with a Turbo V ion source, together with the Analyst 1.5.1 software package. Stable isotope labelled-internal standards were used in all sample extractions and for calibration. The quality control (QC) sample was prepared by spiking a known amount of native and labelled standards into pooled plasma from all samples, and QCs were analysed along with the samples in each batch to ensure the reliability of the method and the instrument stability.
Cell isolation
PBMCs were isolated from 20 ml EDTA blood from HS. Blood samples were centrifuged at 200 × g for 10 min to collect plasma and sedimented cells were diluted with Dulbecco’s PBS without calcium and magnesium (DBPS--) up to a volume of 20 ml. Diluted blood was layered over 13.3 ml of Ficoll-Hypaque and centrifuged at 500 × g for 25 min with the brakes off. PBMCs were obtained from the mononuclear cell layer and incubated with pre-warmed ammonium-chloride-potassium lysis buffer for 10 min at room temperature to lyse red blood cells and then centrifuged at 400 × g for 5 min. The resultant pellet was washed with DPBS--. Isolated PBMCs were enumerated and resuspended in RPMI 1640 medium containing penicillin (100 U/ml), streptomycin (100 U/ml), and l-glutamine (4 mM) without foetal bovine serum.
Cell incubation
Isolated PBMCs were cultured in RPMI 1640 as indicated above and incubated with C16:0-carnitine (2.5–25 μM), C6:0-carnitine (5–10 μM) for 4 h, with or without inhibitors of mitochondrial β-oxidation etomoxir (200 μM) and trimetazidine (100 μM) (all purchased from Sigma). Human serum albumin (HSA, 5 mg/ml) (Albutein, Grifols, Barcelona, Spain) was added in a pretreatment mode 30 min before treatment with acylcarnitine or as a co-treatment with acylcarnitine for 4 h. Carbonyl cyanide m-chlorophenyl hydrazone (CCCP) was added in a concentration of 40 μM for 5 min as a positive control for mitochondrial depolarisation as measured by JC-1. For confocal microscopy, PBMCs were incubated with 25 μM CCCP for 30 min.
Measurement of mitochondrial membrane potential (Δψm)
Δψm in PBMCs was measured using JC-1, a lipophilic dye which accumulates and forms J-aggregates in energised mitochondria. In case of low mitochondrial potential, a condition found in dysfunctional mitochondria, JC-1 is present in a monomeric form accompanied by a fluorescence emission shift from green (∼529 nm) to red (∼590 nm), which was detected on a BD FACSCanto II. Data analysis was performed with FlowJo v10 and FACSDiva v6 Analysis software.
Assessment of mitochondrial respiration in the Seahorse XF system
Extracellular flux was measured using a Seahorse XFe24 Analyzer (Agilent Technologies) using the Mito Stress Test kit. The kit uses high-throughput oximetry to simultaneously measure OCR and extracellular acidification rates. Freshly isolated PBMCs (700,000 cells/ml) with or without prior treatment were plated in poly-l-lysine-coated XF24 plates by centrifugation (200 × g with the brakes off) in XF media, composed of DMEM medium pH 7.4, 10 mM glucose, 1 mM sodium pyruvate, and 2 mM l-glutamine). Respiration was measured at baseline and in response to oligomycin A (2 μM), carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) (2 μM) and rotenone (1 μM) with antimycin A (1 μM).
Mitochondrial ROS production
PBMCs (200,000 cells/well) were seeded in technical triplicates in a black 96-well plate precoated with poly-l-lysine 0.01%. After the indicated treatments, MitoSOX Red Mitochondrial Superoxide Indicator (M36008, ThermoFisher Scientific, Waltham, MA, USA) in a final concentration of 5 μM was added to the cells and were allowed to incubate for 10 min at 37 °C in 5% CO2. Cells were washed with DPBS-- and covered with 100 μl DPBS--. The fluorescence signal was recorded using a monochrome plate reader Tecan at the excitation/emission wavelength 510/590 nm. The relative fluorescence units were normalised to protein concentration, which was measured using the MicroBCA Protein Assay Kit (ThermoFisher Scientific).
Transmission electron microscopy
For transmission electron microscopy (TEM) PBMCs were fixed in 2% paraformaldehyde and 2.5% glutaraldehyde. The samples were post-fixed with 1% osmium tetroxide and 0.8% potassium ferrocyanide, dehydrated in acetone and embedded in Spurr’s epoxy resin. Four ultrathin sections were obtained from each sample. The ultrathin sections were poststained with uranyl acetate and lead citrate and examined under a JEOL J1010 Transmission electron microscope (Akishima, Japan). From the ultrathin sections, at least 10 cells per sample were randomly selected and subjected to two-dimensional morphometric analysis at the original magnification of 50,000×. The number of mitochondria per cell, the ratio between the length and width, the cross-sectional area, and the perimeter of each mitochondrion were analysed using ImageJ (Fiji).
Mitochondria imaging by confocal microscopy
PBMCs were treated as indicated and then fixed, permeabilised, and stained with mouse anti-HSA (Thermofisher Scientific, HYB 192-01-02) and rabbit anti-tom20 (11802-1-AP) antibodies recognised by secondary goat anti-mouse IgG Alexa Fluor Plus 555 (Thermofisher Scientific, A32727) and goat anti-rabbit IgG Cross-Adsorbed ReadyProbes Alexa Fluor 488 (Thermofisher Scientific, R37116) antibodies, and visualised in an LSM 880 Zeiss confocal microscope.
Stress and toxicity pathway-focused gene expression array
To assess changes in mRNA expression of genes involved in response to stress and toxicity, total RNA from PBMCs was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) including the optional on-column DNase digestion. RNA quality and concentration were determined using the Agilent 4200 TapeStation (Agilent Technologies). A total of 400 ng high-quality total RNA (RNA integrity number >9.2) was reverse transcribed using the First Strand Synthesis Kit (Qiagen) and loaded on a Human Stress & Toxicity Pathway Finder RT2 profiler array of the Format E384 (4 × 96) (Qiagen). The list of genes included in the array is shown in Table S3. The following conditions were set for the ABI 7900HT instrument: hold for 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. All data were normalised to the geometric mean of five housekeeping genes (GAPDH, IL1B, RPLP0, HPRT1, and GRB2). Data analysis was conducted using Qiagen’s GeneGlobe Data Analysis Center.
Real-time PCR analysis
For the analysis of Heme oxygenase 1 (HMOX1), C-X-C motif chemokine ligand 8 (CXCL8), Nuclear factor erythroid 2-related factor 2 (NFE2L2), Activating transcription factor 4 (ATF4), and Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPARGC1A) by real-time PCR, total RNA was isolated from cells using TRI Reagent (Molecular Research Center, Cincinnati, OH, USA), retrotranscribed to complementary DNA (cDNA) using the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA) and amplified by real-time PCR in the 7900HT Fast System (Applied Biosystems) using validated and pre-designed TaqMan Gene Expression Assays: HMOX1 (ID: Hs01110250_m1), CXCL8 (Hs00174103_m1), NFE2L2 (ID: Hs00975961_g1), ATF4 (ID: Hs00909569_g1), PPARGC1A (Hs01016719_m1), and β-actin (ID: Hs99999903_m1) as endogenous control.
Protein expression by Western blotting
Total protein from PBMCs was extracted using lysis buffer containing 50 mM Hepes, 20 mM β-glycerophosphate, 2 mM EDTA, 1% Igepal, 10% (vol/vol) glycerol, 1 mM MgCl2, 1 mM CaCl2, and 150 mM NaCl supplemented with a mixture of protease (Complete Mini) and phosphatase (PhosStop) inhibitors (Roche Diagnostics, Basel, Switzerland). For protein isolation, 2 × 106 cells were resuspended in 80 μl of lysis buffer. Homogenates were incubated on ice for 20 min and centrifuged at 16,000 × g for 20 min at 4 °C. Laemmli buffer was added, and the samples were heated for 5 min at 95 °C. Electrophoresis was performed using 10–12% (vol/vol) SDS-PAGE for 90 min at 120 V, followed by transfer of proteins onto PVDF membranes using the iBlot Dry Blotting System (Invitrogen) at 20 V for 5–7 min. The membranes were blocked for 1 h at room temperature in tris-buffered saline containing 0.1% Tween 20 (T-TBS) and 5% (wt/vol) albumin. Blots were washed three times for 5 min each with T-TBS and incubated with rabbit monoclonal HO-1 (D60G11) (5853S), rabbit monoclonal β-actin (5125) (antibodies from Cell Signaling Technology, Danvers, MA, USA) and rabbit monoclonal TIM44 (Abcam, ab194829) in 0.1% T-TBS containing 5% BSA. Thereafter, the blots were washed three times for 5 min each with 0.1% T-TBS and incubated for 1 h at room temperature with a horseradish-peroxidase-linked donkey anti-rabbit antibody (1:2,000 dilution) in 0.1% T-TBS containing 5% non-fat dry milk. The bands were visualised with the EZ-ECL chemiluminescence detection kit (Biological Industries, Haemek, Israel) using ImageQuant LAS 4000 equipment (GE Healthcare Life Sciences, Little Chalfont, UK). Densitometric analysis was performed with ImageJ (version 1.54; Fiji).
Statistical analysis
Results are presented as mean ± SEM or median with IQR, as appropriate. For single comparisons, the Mann–Whitney U test was used. For multiple comparisons, the Kruskal–Wallis test was used for nonparametric distribution, followed by Dunn’s post-hoc testing. Fold changes among groups in volcano plots were calculated from the log2-transformed values. Values of p were transformed to -log10 in the representation of volcano plots. Statistical analysis and graphs/plots were obtained using GraphPad Prism v.8 (GraphPad Software, San Diego, CA, USA). Adjustment of p values was done by the determination of false discovery rate (FDR) using the Benjamini-Hochberg procedure using R v.4.1.3 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Plasma acylcarnitine levels were elevated in patients with AD cirrhosis
In comparison with HS, plasma acylcarnitine levels were significantly elevated over a wide range of concentrations (from nano- to micromolar) in patients with AD cirrhosis (Fig. 1A). Specifically, patients with AD showed significantly higher circulating levels of the short-chain (acetyl (C2:0)) and medium-chain (hexanoyl (C6:0)) acylcarnitines and especially of the long-chain myristoyl (C14:0)-, palmitoyl (C16:0)-, hexadecenoyl (C16:1)-, and octadecenoyl (C18:1)-carnitines (Fig. 1A). Analysis of sex-specific differences in the level of these acylcarnitines are reported in Fig. S1 (also see Table S4 for details of the fold changes and FDR for these acylcarnitines). We chose to focus on the long-chain C16:0-carnitine for subsequent analyses, as it was the most abundant saturated long-chain acylcarnitine in our study (Fig. 1A). Additionally, based on our experience, leucocytes from patients with AD cirrhosis actively metabolise this acylcarnitine.10 For comparison purposes, we initially performed experiments with the medium-chain C6:0-carnitine as it is part of the 38-metabolite blood fingerprint specific for the progression of patients with AD cirrhosis to ACLF described by Moreau et al.8 and a predictor of 28-day mortality.11 The concentrations used for the experiments were based on similar studies investigating the in vitro effect of acylcarnitines.[12], [13], [14] We first investigated the association of these two acylcarnitines with markers of systemic inflammation (cytokines/chemokines and lipid mediators), which, except for VEGF-A, prostaglandin E2 and thromboxane B2, were invariably elevated in patients with AD cirrhosis (Fig. 1B and Fig. S2). G-CSF, GM-CSF, and IL-7 could not be detected in the samples and therefore were not included in the analysis. The variations in cytokine levels based on sex are shown in Fig. S3. By plotting in a heatmap the Spearman’s correlation coefficients between the concentrations of C6:0-carnitine and C16:0-carnitine on the one hand and the levels of cytokines/chemokines on the other, we determined that C6:0-carnitine correlated with IL-6, IL-10, interferon-gamma (IFNγ) induced protein 10 (IP-10), and macrophage inflammatory protein (MIP)-1β, whereas C16:0-carnitine preferentially and significantly correlated with IL-8 levels, the cytokine with the second highest fold change in expression in patients after IL-6 (Fig. 1C, Fig. S4, and Table S5). Regarding lipid mediators, the pro-thrombotic factor TXB2 correlated positively with C16:0-carnitine, although correlations did not achieve statistical significance (Fig. 1D). Importantly, C6:0-carnitine displayed an inverse correlation with 5-HEPE and 18-HEPE lipid mediators with anti-inflammatory properties (see Table S6 for more details of all lipid mediators analysed).
Fig. 1.
Levels of plasma acylcarnitines and their association with markers of inflammation.
(A) Plasma levels of short-, medium- and long-chain acylcarnitines in plasma of 20 patients with acutely decompensated (AD) cirrhosis and in 10 healthy subjects (HS), displayed as median values. The Mann–Whitney U-test was used for statistical analysis. (B) Plasma levels of chemokines/cytokines in patients with AD cirrhosis in comparison with HS. (C) Correlation heatmap between acylcarnitines of different chain lengths and selected cytokines/chemokines in patients with AD cirrhosis. (D) Heatmap showing correlation between acylcarnitine and bioactive lipid mediators in patients with AD. ∗p <0.05, ∗∗p <0.01. IP-10, interferon-gamma induced protein 10; TNFα, tumour necrosis factor α.
C16:0-carnitine interfered with mitochondrial biophysics and triggers mitochondrial oxidative stress
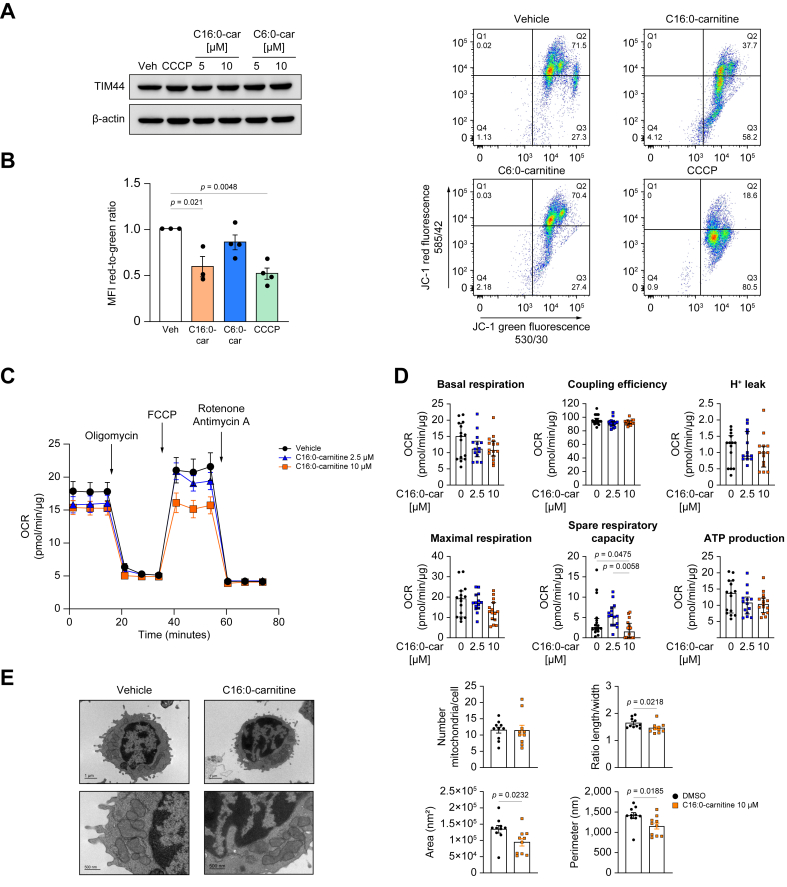
We next explored whether the presence of high plasma concentrations of acylcarnitines could interfere with mitochondrial function in immune cells circulating in the bloodstream. As mononuclear leucocytes are fundamental immune cell effectors contributing to the hyper-inflammatory and immunosuppressive landscape characteristic of cirrhosis-associated immune dysfunction, and because leucocytes from patients with AD cirrhosis already exhibit signs of mitochondrial dysfunction, we tested the effects of acylcarnitines on PBMCs freshly isolated from HS. We first assessed the effects of C6:0- and C16:0-carnitine on the protein levels of translocase of inner mitochondrial membrane (TIM) 44, which serves as a housekeeping marker of mitochondrial abundance.15 Neither C6:0- nor C16:0-carnitine altered TIM44 protein levels in PBMCs incubated in vitro with these acylcarnitines (Fig. 2A). We next assessed the effects of acylcarnitines on mitochondrial membrane potential (Δψm), a key parameter of mitochondrial biophysics.16 As shown in Fig. 2B, C16:0-carnitine but not C6:0-carnitine significantly reduced the ratio of red to green fluorescence of the JC-1 dye, indicating depolarisation of the mitochondrial membrane. This impairment was comparable to that of CCCP, a well-known mitochondrial uncoupler which served as positive control (Fig. 2B).17 As C16:0-carnitine appeared to be detrimental to the mitochondrial cell compartment, we further tested the effects of this long-chain acylcarnitine on parameters of mitochondrial function. Specifically, we measured oxidative phosphorylation (OXPHOS) using the Seahorse technology and performed a mitochondrial stress test, thereby measuring oxygen consumption rate (OCR) over time in response to inhibitors of the mitochondrial electron transport chain (ETC). In these experiments, two concentrations of C16:0-carnitine in the range of those observed in plasma of humans with pathological conditions18 were used. Representative mitochondrial respiration profiles of PBMCs incubated with vehicle (DMSO) and two increasing concentrations of C16:0-carnitine are shown in Fig. 2C. Among the mitochondrial respiratory parameters measured, C16:0-carnitine in a lower concentration increased spare respiratory capacity albeit not to a significant level, which can be explained by increased substrate availability for β-oxidation,19 whereas in a higher concentration it significantly reduced spare respiratory capacity, a marker of mitochondrial fitness,20 without altering the production of ATP (Fig. 2D). These findings were accompanied by reduced length-to-width ratio, and decreased area and perimeter of each mitochondrion as assessed by TEM, whereas the number of mitochondria per cell remained unaffected (Fig. 2E).
Fig. 2.
Long-chain palmitoylcarnitine reduces mitochondrial membrane potential (Δψm) and spare respiratory capacity.
(A) Representative Western blot of TIM44 in PBMCs incubated with C16:0- and C6:0-carnitine. (B) Analysis of Δψm in PBMCs incubated with C16:0-carnitine by measuring the ratio of the red-to-green median fluorescence intensity emitted by the voltage-sensitive dye JC-1. (C) Mitochondrial stress test measuring the OCR in PBMCs incubated with different concentrations of C16:0-carnitine using Agilent Seahorse technology. (D) The key parameters of mitochondrial respiratory function were calculated from the OCR readings. (E) Representative electron microscopy images of PBMCs incubated for 4 h with vehicle (left panel) and C16:0-carnitine 10 μM (right panel), and analysis of mitochondrial ultrastructural parameters. Statistical differences were calculated using one-way ANOVA. CCCP, carbonyl cyanide m-chlorophenyl hydrazone; OCR, oxygen consumption rate; PBMCs, peripheral blood mononuclear cells.
C16:0-carnitine triggered mitochondrial oxidative stress and cytokine release
We next investigated whether the long-chain C16:0-carnitine, in addition to compromising Δψm and spare mitochondrial respiratory capacity, induced mitochondrial oxidative stress in immune cells. In fact, using the MitoSOX dye assay, we identified that C16:0-carnitine triggered mitochondrial oxidative stress in a concentration-dependent manner (Fig. 3A). As mitochondrial oxidative stress is often linked to an inflammatory response,21 we then measured the levels of 20 cytokines in the supernatant of PBMCs treated with different concentrations of C16:0-carnitine. As shown in Fig. 3B, a stepwise increase in IL-8 levels accompanied by IFNγ, IL-4, MIP-1α and MIP-1β was observed after exposing the PBMCs to C16:0-carnitine. TNFα did not reach the level of significance; other cytokines/chemokines that were not affected by C16:0-carnitine treatment are shown in Fig. S5.
Fig. 3.
Palmitoylcarnitine induces ROS production and release of pro-inflammatory cytokines.
(A) Mitochondrial superoxide production in PBMCs incubated with different concentrations of C16:0-carnitines for 4 h using the indicator MitoSOX. Results are expressed as mean ± SEM. (B) Cytokines and chemokines that are significantly increased in the supernatant of PBMCs incubated with C16:0-carnitine. DMSO in vehicles 0.25% v/v. Results are expressed as median with interquartile range. Statistical differences were determined using the Kruskal–Wallis test. IFNγ, interferon-gamma; PBMCs, peripheral blood mononuclear cells; ROS, reactive oxygen species.
C16:0-carnitine suppressed the antioxidant defence enzyme HMOX1
To obtain mechanistic insights into the induction of oxidative stress and cytokine release by C16:0-carnitine, we used an 84-gene oxidative stress pathway-focused expression array. The volcano plot comparing the gene expression values of PBMCs incubated with C16:0-carnitine with respect to PBMCs incubated with vehicle is shown in Fig. 4A. The incubation of cells with C16:0-carnitine was associated with a significant downregulation of HMOX1 expression, the gene coding for heme oxygenase-1 (HO-1), a protein implicated in the cellular anti-oxidative system with dual localisation in the cytosol and mitochondria.22 The transcriptomic analysis also revealed that incubation of PBMCs with C16:0-carnitine was associated with a significant upregulation of CXCL8, the gene coding for the proinflammatory chemokine IL-8 (Fig. 4A). These results were confirmed by real-time PCR, which demonstrated the suppression of HMOX1 at the transcriptional level by C16:0-carnitine to a similar extent as TNFα, selected as positive control known to repress HMOX1 in PBMCs (Fig. 4B).23 Interestingly, the suppression of the antioxidant defence HO-1 system occurred in a concentration-dependent manner (Fig. 4C). Densitometric analysis is shown in Fig. S6A. Another important mitochondrial antioxidant system, namely MnSOD, which encodes the antioxidant enzyme superoxide dismutase 2, was not modified by C16:0-carnitine (Fig. 4D). Conversely, the upregulation of the IL-8 coding gene CXCL8 by C16:0-carnitine was also confirmed at both mRNA (real-time PCR) (Fig. 4E) and protein (Luminex assay) levels (Fig. 3B, first panel).
Fig. 4.
Palmitoylcarnitine suppresses the expression of the gene coding for heme oxygenase-1 (HMOX1).
(A) Volcano plot representing the pair-wise comparison of the expression level of the 84 genes related to stress and toxicity pathways in PBMCs incubated with C16:0-carnitine 10 μM for 4 h with respect to that in PBMCs incubated with control vehicle. (B) Real-time PCR confirms C16:0-carnitine as a repressor of HMOX1. (C) C16:0-carnitine suppresses heme oxygenase-1 (HO-1) protein levels in a concentration-dependent manner. (D) MnSOD mRNA expression in PBMCs incubated with increasing concentrations of C16:0-carnitine. (E) C16:0-carnitine increases the mRNA level of CXCL8 in a concentration-dependent manner. DMSO in vehicle conditions did not exceed 0.25% v/v. Results are displayed as mean ± SEM, and statistical differences were determined using the Kruskal–Wallis test. CXCL8, C-X-C Motif Chemokine Ligand 8; PBMCs, peripheral blood mononuclear cells.
Etomoxir reversed the inhibitory effect of C16:0-carnitine on HMOX-1 expression and its proinflammatory actions
To further investigate the mechanisms by which C16:0-carnitine is linked to induction of oxidative stress and cytokine release in PBMCs, we interrogated several cytoplasmic transcription factors that could be dysregulated by this acylcarnitine, including the transcription factor nuclear factor erythroid 2-related factor 2 (Nrf2, encoded by NFE2L2), the activating transcription factor 4 (ATF4), a key regulator of mitochondrial function in the context of the integrated stress response24 and PGC1a (encoded by PPARGC1A), the master regulator of mitochondrial biogenesis.25 As shown in Fig. S6B, none of these transcription factors that regulate the expression of HMOX-1 were modified during the incubation of PBMCs with C16:0-carnitine. In view of these findings, we then hypothesised that the pro-oxidant effects of C16:0-carnitine could be secondary to entry into the mitochondria for its β-oxidation. To test this hypothesis, we performed experiments with etomoxir, an irreversible inhibitor of CPT1, which prevents the entry of long-chain acylcarnitines into the mitochondria (Fig. 5A). As shown in Fig. 5B, etomoxir was able to fully restore the levels of HMOX1 suppressed by C16:0-carnitine treatment. In addition, etomoxir partially ameliorated CXCL8 expression induced by C16:0-carnitine (Fig. 5C). As the detrimental actions of C16:0-carnitine were prevented by etomoxir, which does not allow C16:0-carnitine to be transported into the mitochondrial matrix, the deleterious actions of this long-chain acylcarnitine are likely related to its accumulation in the mitochondrial compartment. To investigate whether other mitochondrial processes were implicated in the detrimental effects of C16:0-carnitine, we performed experiments with trimetazidine, which blocks long-chain fatty acid β-oxidation by inhibiting 3-ketoacyl-CoA thiolase.26 Trimetazidine did not restore HMOX1 expression suppressed by C16:0-carnitine (Fig. 5D) but abolished C16:0-acylcarnitine-induced CXCL8 expression (Fig. 5E). In the absence of C16:0-carnitine, etomoxir and trimetazidine alone had no effect on HMOX-1 or CXCL8 expression (Fig. S7). Together, these findings suggest that the suppressive effect of C16:0-carnitine on HMOX1 (pro-oxidant action) was dependent on its accumulation inside the mitochondria, whereas its stimulatory effect on CXCL8 expression (pro-inflammatory action) more likely depended on its β-oxidation in the mitochondrial matrix.
Fig. 5.
Etomoxir reverses the suppressive effect of palmitoylcarnitine on heme oxygenase-1.
(A) Schematic diagram of the intracellular actions of etomoxir and trimetazidine. Created with BioRender.com. (B) Effect of co-treatment of C16:0-carnitine and etomoxir on HMOX1 expression. (C) Effect of co-treatment of C16:0-carnitine and etomoxir on CXCL8 expression. (D) Gene expression analysis of HMOX1 under co-treatment of C16:0-carnitine and trimetazidine. (E) Gene expression analysis of CXCL8 under co-treatment of C16:0-carnitine and trimetazidine. Results are expressed as mean ± SEM, and the Mann–Whitney U-test was applied to detect statistical differences. CPT1A, carnitine palmitoyltransferase 1A; CXCL8, C-X-C Motif Chemokine Ligand 8; HMOX1, heme oxygenase 1; TMZ, trimetazidine.
C16:0-carnitine-induced mitochondrial oxidative stress is lessened by the albumin molecule
We then explored whether the albumin could interfere with C16:0-carnitine and modulate its damaging effects on circulating PBMCs. The rationale for exploring albumin in this setting was based on three premises: (1) albumin has high affinity for lipids, especially for fatty acids, which are transported in the circulation attached to its molecule;27 (2) albumin can also bind acylcarnitines to which fatty acids are attached to carnitine;28 and (3) albumin is used in the clinical management of patients with AD cirrhosis.29 We incubated PBMCs with albumin at similar concentrations to those encountered by patients with AD cirrhosis receiving albumin infusions, and then exposed these cells to C16:0-carnitine. Notably, albumin was able to prevent two detrimental actions of C16:0-carnitine, namely the impairment of Δψm (Fig. 6A) and the induction of mitochondrial oxidative stress (Fig. 6B). Although albumin was not able to restore the C16:0-induced suppression of HMOX1 (Fig. 6C), albumin restricted to some extent C16:0-carnitine-induced CXCL8 expression (Fig. 6D). Finally, given that albumin is internalised by immune and parenchymal cells,29,30 we eventually sought to evaluate the possibility that the protective actions of albumin against C16:0-carnitine impairment of mitochondrial function were related to its internalisation into the mitochondrial compartment. However, confocal microscopy experiments revealed that albumin did not colocalise with the mitochondrial marker Tom20, neither under resting conditions nor after mitochondrial membrane depolarisation with CCCP (Fig. 6E).
Fig. 6.
Albumin restores mitochondrial membrane potential (Δψm) and abolishes ROS production elicited by palmitoylcarnitine.
(A) Analysis of Δψm in PBMCs incubated with C16:0-carnitine in the presence or absence of albumin (HSA, 15 mg/ml) by measuring the ratio of the red-to-green median fluorescence intensity emitted by JC-1. (B) Mitochondrial superoxide production in PBMCs co-incubated with C16:0-carnitine and HSA. (C) HMOX1 expression in PBMCs pre-incubated with HSA and then exposed to C16:0-carnitine. (D) CXCL8 expression in PBMCs pre-incubated with HSA and then exposed to C16:0-carnitine. (E) Representative confocal images of the mitochondrial marker tom20 and HSA, with or without CCCP challenge, and quantitative analysis of co-localisation represented by Pearson’s correlation coefficient. Results are displayed as mean ± SEM, and statistical differences were detected by Kruskal–Wallis test. Scale bar: 10 μm. CCCP, carbonyl cyanide m-chlorophenyl hydrazone; CXCL8, C-X-C Motif Chemokine Ligand 8; HMOX1, heme oxygenase 1; HSA, human serum albumin; PBMCs, peripheral blood mononuclear cells; ROS, reactive oxygen species.
Discussion
This is the first study to demonstrate the proinflammatory and pro-oxidant effects of long-chain C16:0-carnitine in peripheral blood leucocytes. These findings suggest that its elevated plasma levels in patients with AD cirrhosis could contribute to the systemic hyperinflammatory and pro-oxidant stress responses observed in these patients, which are part of the so-called cirrhosis-associated immune dysfunction.31 These bioactions in circulating immune cells complement the function of acylcarnitines as mere biomarkers of mitochondrial dysfunction in this condition.
As acylcarnitines have to be transported into the mitochondrial matrix to be metabolised via β-oxidation, we focused on the effects of the long-chain C16:0-carnitine on several parameters of mitochondrial function. We found that C16:0-carnitine reduced the spare respiratory capacity, which is considered a particularly robust functional parameter to estimate mitochondrial reserve, that is, the mitochondrial capacity to meet extra energy required during a response to acute stress.20,32 The response to C16:0-carnitine that led to reduced spare respiratory capacity was not attributed to differences in the cellular mitochondrial content, as TIM44 protein was not affected by C16:0-carnitine treatment.
The major finding of our study was that C16:0-carnitine increased mitochondrial ROS production. Consistent with this finding, we observed a significant downregulation of HMOX1, the gene coding for the antioxidant enzyme HO-1, by C16:0-carnitine, which seems to be counterintuitive in light of the enhanced oxidative stress level in cells exposed to C16:0-carnitine. HO-1, initially classified as a heat shock protein, is an enzyme upregulated during inflammation via a region in the promoter which contains NF-κB and AP-2 binding sequences.33 It degrades pro-oxidant free heme into equimolar quantities of carbon monoxide (CO), biliverdin and iron, which exert cytoprotective and immunomodulatory properties. HO-1 is also involved in regulating mitochondrial quality control by promoting biogenesis in response to mitochondrial injury.34 Mitochondrial HO-1 has an important role in regulating mitochondrial heme turnover and mitochondrial-targeted NO synthase expression, thereby exerting a protective role against hypoxia, neurodegenerative diseases, and sepsis.35 Therefore, inadequate HO-1 levels may result in a wide spectrum of severe side effects which might affect many organs.36 In contrast to the large number of inducers of HO-1 transcription, only few inhibitors of HO-1 expression are known including IFNγ. We propose that C16:0-carnitine is a novel suppressor of HO-1 in immune cells with proinflammatory actions. Thus, immune cells depleted of HO-1 display reduced mitochondrial oxidative capacity, paralleled by an increase in mitochondrial ROS production, very similar to the processes seen in lung epithelial cells lacking HO-1.37 Others have ascribed these alterations in mitochondrial function to the accumulation of heme in mitochondria.35,37 However, the exact mechanisms by which C16:0-carnitine suppresses HO-1 and whether this holds true for other cell types will be the focus of future research.
The unfavourable actions of C16:0-carnitine on HMOX1 transcription could be effectively reversed by etomoxir, which prevents medium- and long-chain fatty acids from being catabolised by mitochondrial β-oxidation, meaning that C16:0-carnitine needs to enter mitochondria to interfere with HMOX1 expression. We can think of two possible explanations for the fact that trimetazidine, in contrast to etomoxir, could not rescue the expression of HMOX1. First, trimetazidine is known to only partially inhibit fatty acid oxidation and to interfere with other cellular functions,38 and second, the effect of C16:0-carnitine on HMOX1 might be only dependent on C16:0-carnitine accumulation in mitochondria and not its downstream oxidation.
More importantly, the alterations in mitochondrial bioenergetics elicited by C16:0-carnitine were paralleled by an increase in CXCL8 gene expression and increased secretion of IL-8 by mononuclear cells, thus reinforcing the paradigm that mitochondrial dysfunction is intricately linked with inflammation. Importantly, IL-8 was the second most increased cytokine in patients with AD cirrhosis, supporting the hypothesis that C16:0-carnitine in part contributes to the proinflammatory environment in the systemic circulation of these patients. The increase could be attributed to enhanced superoxide production through oxidation of C16:0-carnitine in the mitochondrial β-oxidation, which is converted to other oxygen radicals such as H2O2 with known NF-κB-mediated CXCL8 transcription-promoting effects.39
In summary, our findings unravel for the first time the damaging actions of C16:0-carnitine on mitochondrial antioxidant responses in circulating immune cells. Because C16:0-carnitine as well as other medium- and long-chain acylcarnitines are dramatically increased in the systemic circulation of patients with acute decompensation of liver cirrhosis, our findings suggest that acylcarnitines represent a synergistic mechanism contributing to the exaggerated hyperinflammatory and pro-oxidant state characteristic of this condition.
Abbreviations
ACLF, acute-on-chronic liver failure; AD, acute decompensation; ATF4, Activating transcription factor 4; CACT, carnitine-acylcarnitine translocase; CCCP, carbonyl cyanide m-chlorophenyl hydrazone; CPT, carnitine palmitoyltransferase; CXCL8, C-X-C Motif Chemokine Ligand 8; ETC, electron transport chain; FCCP, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone; FDR, false discovery rate; HMOX1, heme oxygenase 1; HS, healthy subjects; HSA, human serum albumin; IP-10, induced protein 10; LC-MS/MS, liquid chromatography with tandem mass spectrometry; MELD, model for end-stage liver disease; MELDNa, model for end-stage liver disease-sodium; NF-κB, nuclear factor kappa B; NFE2L2, Nuclear factor erythroid 2-related factor 2; OCR, oxygen consumption rate; OXPHOS, oxidative phosphorylation; PBMCs, peripheral blood mononuclear cells; PPARGC1A, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha; QC, quality control; ROS, reactive oxygen species; TEM, transmission electron microscopy; T-TBS, 0.1% Tween 20.
Financial support
Supported by the Plan Nacional de I+D funded by the Agencia Estatal de Investigación (grants PID2019-105240RB-I00 and PID2022-138970OB-I00 [to JC] and PID2019-111669RB-100 and PID2020-115055RB-I00 [to JCF-C]). JC laboratory is a Consolidated Research Group recognised by the Generalitat de Catalunya (2021 SGR 01323) and receives funds from EF CLIF and European Union’s Horizon 2020 research and innovation programme (MICROB-PREDICT [project ID 825694], DECISION [project ID 847949]). JCF-C receives funds from the center grant P50AA011999 Southern California Research Center for ALPD and Cirrhosis funded by NIAAA/NIH, the Project 201916/31 by Fundació Marató TV3 and the European Horizon´s research and innovation program HORIZON-HLTH-2022-STAYHLTH-02 under agreement No 101095679. JT is supported by the German Research Foundation project ID 403224013 – SFB 1382 (A09), by the German Federal Ministry of Education and Research for the DEEP-HCC project and by the Hessian Ministry of Higher Education, Research, the Arts for the ENABLE and ACLF-I cluster projects and the European Union’s Horizon 2020 research and innovation program for MICROB-PREDICT (project ID 825694), DECISION (project ID 847949), GALAXY (project ID 668031), LIVERHOPE (project ID 731875), and IHMCSA (project ID 964590) projects. IWZ was supported by the Sheila Sherlock Post Graduate Programme of the European Association for the Study of the Liver.
Conflicts of interest
JT received speaking and/or consulting fees from Versantis, Gore, Boehringer-Ingelheim, Falk, Grifols, Genfit and CSL Behring. JC received speaking and/or consulting fees from Grifols and Genfit.
Please refer to the accompanying ICMJE disclosure forms for further details.
Authors’ contributions
Study concept and design: IZ, JC. Acquisition of samples and data: IZ, MS-R, FA, JT. Contribution to experimental procedures: MS-R, CL-V, MC, MD-G, RF-C, MR, PS, CG-R, JF-C. Statistical analysis: IZ. Interpretation of data: IZ, JC. Drafting and writing of the final manuscript: IZ, JC. Critical revision of the manuscript for important intellectual content: VA. Study supervision: JC.
Data availability statement
The data that support the findings of this study will be openly available in a repository on acceptance for publication.
Acknowledgements
We are indebted to Eva Prats from the Electron Microscopy Department of the University of Barcelona and the Cytometry and cell sorting facility of the IDIBAPS for technical help.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2024.101187.
Contributor Information
Ingrid Wei Zhang, Email: ingrid-wei.zhang@charite.de.
Joan Clària, Email: jclaria@clinic.cat.
Supplementary data
The following are the Supplementary data to this article:
References
- 1.Longo N., Frigeni M., Pasquali M. Carnitine transport and fatty acid oxidation. Biochim Biophys Acta. 2016;1863:2422–2435. doi: 10.1016/j.bbamcr.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGarry J.D., Brown N.F. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur J Biochem. 1997;244:1–14. doi: 10.1111/j.1432-1033.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- 3.Reuter S.E., Evans A.M. Carnitine and acylcarnitines: pharmacokinetic, pharmacological and clinical aspects. Clin Pharmacokinet. 2012;51:553–572. doi: 10.1007/BF03261931. [DOI] [PubMed] [Google Scholar]
- 4.McGill M.R., Li F., Sharpe M.R., et al. Circulating acylcarnitines as biomarkers of mitochondrial dysfunction after acetaminophen overdose in mice and humans. Arch Toxicol. 2014;88:391–401. doi: 10.1007/s00204-013-1118-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enooku K., Nakagawa H., Fujiwara N., et al. Altered serum acylcarnitine profile is associated with the status of nonalcoholic fatty liver disease (NAFLD) and NAFLD-related hepatocellular carcinoma. Sci Rep. 2019;9 doi: 10.1038/s41598-019-47216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter W.G., Kelly J.P., McGarrah R.W., 3rd, et al. Metabolomic profiling identifies novel circulating biomarkers of mitochondrial dysfunction differentially elevated in heart failure with preserved versus reduced ejection fraction: evidence for shared metabolic impairments in clinical heart failure. J Am Heart Assoc. 2016;5 doi: 10.1161/JAHA.115.003190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langley R.J., Tsalik E.L., van Velkinburgh J.C., et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreau R., Clària J., Aguilar F., et al. Blood metabolomics uncovers inflammation-associated mitochondrial dysfunction as a potential mechanism underlying ACLF. J Hepatol. 2020;72:688–701. doi: 10.1016/j.jhep.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Arroyo V., Moreau R., Kamath P.S., et al. Acute-on-chronic liver failure in cirrhosis. Nat Rev Dis Primers. 2016;2 doi: 10.1038/nrdp.2016.41. [DOI] [PubMed] [Google Scholar]
- 10.Zhang I.W., Curto A., López-Vicario C., et al. Mitochondrial dysfunction governs immunometabolism in leukocytes of patients with acute-on-chronic liver failure. J Hepatol. 2022;76:93–106. doi: 10.1016/j.jhep.2021.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Weiss E., de la Peña-Ramirez C., Aguilar F., et al. Sympathetic nervous activation, mitochondrial dysfunction and outcome in acutely decompensated cirrhosis: the metabolomic prognostic models (CLIF-C MET) Gut. 2023;72:1581–1591. doi: 10.1136/gutjnl-2022-328708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguer C., McCoin C.S., Knotts T.A., et al. Acylcarnitines: potential implications for skeletal muscle insulin resistance. FASEB J. 2015;29:336–345. doi: 10.1096/fj.14-255901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutkowsky J.M., Knotts T.A., Ono-Moore K.D., et al. Acylcarnitines activate proinflammatory signaling pathways. Am J Physiol Endocrinol Metab. 2014;306:E1378–E1387. doi: 10.1152/ajpendo.00656.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbull P.C., Hughes M.C., Perry C.G. The fatty acid derivative palmitoylcarnitine abrogates colorectal cancer cell survival by depleting glutathione. Am J Physiol Cel Physiol. 2019;317:C1278–C1288. doi: 10.1152/ajpcell.00319.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neupert W., Brunner M. The protein import motor of mitochondria. Nat Rev Mol Cel Biol. 2002;3:555–565. doi: 10.1038/nrm878. [DOI] [PubMed] [Google Scholar]
- 16.Bazhin A.A., Sinisi R., De Marchi U., et al. A bioluminescent probe for longitudinal monitoring of mitochondrial membrane potential. Nat Chem Biol. 2020;16:1385–1393. doi: 10.1038/s41589-020-0602-1. [DOI] [PubMed] [Google Scholar]
- 17.Heytler P.G. Uncoupling of oxidative phosphorylation by carbonyl cyanide phenylhydrazones. I. Some characteristics of m-Cl-CCP action on mitochondria and chloroplasts. Biochemistry. 1963;2:357–361. doi: 10.1021/bi00902a031. [DOI] [PubMed] [Google Scholar]
- 18.Costa C.G., Struys E.A., Bootsma A., et al. Quantitative analysis of plasma acylcarnitines using gas chromatography chemical ionization mass fragmentography. J Lipid Res. 1997;38:173–182. [PubMed] [Google Scholar]
- 19.Sansbury B.E., Jones S.P., Riggs D.W., et al. Bioenergetic function in cardiovascular cells: the importance of the reserve capacity and its biological regulation. Chem Biol Interact. 2011;191:288–295. doi: 10.1016/j.cbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchetti P., Fovez Q., Germain N., et al. Mitochondrial spare respiratory capacity: mechanisms, regulation, and significance in non-transformed and cancer cells. FASEB J. 2020;34:13106–13124. doi: 10.1096/fj.202000767R. [DOI] [PubMed] [Google Scholar]
- 21.López-Armada M.J., Riveiro-Naveira R.R., Vaamonde-García C., et al. Mitochondrial dysfunction and the inflammatory response. Mitochondrion. 2013;13:106–118. doi: 10.1016/j.mito.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Bansal S., Biswas G., Avadhani N.G. Mitochondria-targeted heme oxygenase-1 induces oxidative stress and mitochondrial dysfunction in macrophages, kidney fibroblasts and in chronic alcohol hepatotoxicity. Redox Biol. 2014;2:273–283. doi: 10.1016/j.redox.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirino Y., Takeno M., Murakami S., et al. Tumor necrosis factor alpha acceleration of inflammatory responses by down-regulating heme oxygenase 1 in human peripheral monocytes. Arthritis Rheum. 2007;56:464–475. doi: 10.1002/art.22370. [DOI] [PubMed] [Google Scholar]
- 24.Quirós P.M., Prado M.A., Zamboni N., et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cel Biol. 2017;216:2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Marcos P.J., Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr. 2011;93:884S. doi: 10.3945/ajcn.110.001917. 90S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kantor P.F., Lucien A., Kozak R., et al. The antianginal drug trimetazidine shifts cardiac energy metabolism from fatty acid oxidation to glucose oxidation by inhibiting mitochondrial long-chain 3-ketoacyl coenzyme A thiolase. Circ Res. 2000;86:580–588. doi: 10.1161/01.res.86.5.580. [DOI] [PubMed] [Google Scholar]
- 27.Spector A.A. Fatty acid binding to plasma albumin. J Lipid Res. 1975;16:165–179. [PubMed] [Google Scholar]
- 28.Casulleras M., Flores-Costa R., Duran-Güell M., et al. Albumin lipidomics reveals meaningful compositional changes in advanced cirrhosis and its potential to promote inflammation resolution. Hepatol Commun. 2022;6:1443–1456. doi: 10.1002/hep4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casulleras M., Flores-Costa R., Duran-Güell M., et al. Albumin internalizes and inhibits endosomal TLR signaling in leukocytes from patients with decompensated cirrhosis. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.aax5135. [DOI] [PubMed] [Google Scholar]
- 30.Duran-Güell M., Flores-Costa R., Casulleras M., et al. Albumin protects the liver from tumor necrosis factor alpha-induced immunopathology. FASEB J. 2021;35 doi: 10.1096/fj.202001615RRR. [DOI] [PubMed] [Google Scholar]
- 31.Albillos A., Martin-Mateos R., Van der Merwe S., et al. Cirrhosis-associated immune dysfunction. Nat Rev Gastroenterol Hepatol. 2022;19:112–134. doi: 10.1038/s41575-021-00520-7. [DOI] [PubMed] [Google Scholar]
- 32.Nicholls D.G. Spare respiratory capacity, oxidative stress and excitotoxicity. Biochem Soc Trans. 2009;37:1385–1388. doi: 10.1042/BST0371385. [DOI] [PubMed] [Google Scholar]
- 33.Lavrovsky Y., Schwartzman M.L., Levere R.D., et al. Identification of binding sites for transcription factors NF-kappa B and AP-2 in the promoter region of the human heme oxygenase 1 gene. Proc Natl Acad Sci U S A. 1994;91:5987–5991. doi: 10.1073/pnas.91.13.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hull T.D., Boddu R., Guo L., et al. Heme oxygenase-1 regulates mitochondrial quality control in the heart. JCI Insight. 2016;1 doi: 10.1172/jci.insight.85817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Converso D.P., Taillé C., Carreras M.C., et al. HO-1 is located in liver mitochondria and modulates mitochondrial heme content and metabolism. FASEB J. 2006;20:1236–1238. doi: 10.1096/fj.05-4204fje. [DOI] [PubMed] [Google Scholar]
- 36.Loboda A., Jazwa A., Grochot-Przeczek A., et al. Heme oxygenase-1 and the vascular bed: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. 2008;10:1767–1812. doi: 10.1089/ars.2008.2043. [DOI] [PubMed] [Google Scholar]
- 37.Carr J.F., Garcia D., Scaffa A., et al. Heme oxygenase-1 supports mitochondrial energy production and electron transport chain activity in cultured lung epithelial cells. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21186941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renaud J.F. Internal pH, Na+, and Ca2+ regulation by trimetazidine during cardiac cell acidosis. Cardiovasc Drugs Ther. 1988;1:677–686. doi: 10.1007/BF02125756. [DOI] [PubMed] [Google Scholar]
- 39.DeForge L.E., Preston A.M., Takeuchi E., et al. Regulation of interleukin 8 gene expression by oxidant stress. J Biol Chem. 1993;268:25568–25576. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study will be openly available in a repository on acceptance for publication.