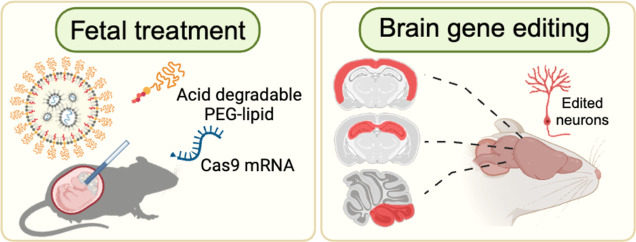
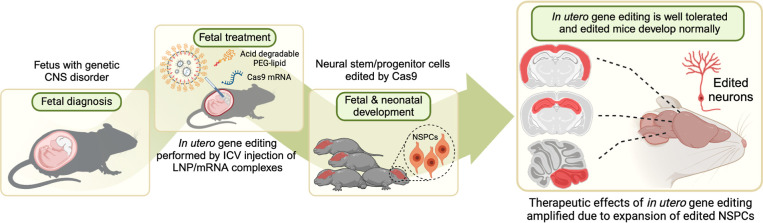
Abstract
In utero gene editing with mRNA-based therapeutics has the potential to revolutionize the treatment of neurodevelopmental disorders. However, a critical bottleneck in clinical application has been the lack of mRNA delivery vehicles that can efficiently transfect cells in the brain. In this report, we demonstrate that in utero intracerebroventricular (ICV) injection of densely PEGylated lipid nanoparticles (ADP-LNPs) containing an acid-degradable PEG–lipid can safely and effectively deliver mRNA for gene editing enzymes to the fetal mouse brain, resulting in successful transfection and editing of brain cells. ADP-LNPs containing Cre mRNA transfected 30% of the fetal brain cells in Ai9 mice and had no detectable adverse effects on fetal development and postnatal growth. In addition, ADP-LNPs efficiently transfected neural stem and progenitor cells in Ai9 mice with Cre mRNA, which subsequently proliferated and caused over 40% of the cortical neurons and 60% of the hippocampal neurons to be edited in treated mice 10 weeks after birth. Furthermore, using Angelman syndrome, a paradigmatic neurodevelopmental disorder, as a disease model, we demonstrate that ADP-LNPs carrying Cas9 mRNA and gRNA induced indels in 21% of brain cells within 7 days postpartum, underscoring the precision and potential of this approach. These findings demonstrate that LNP/mRNA complexes have the potential to be a transformative tool for in utero treatment of neurodevelopmental disorders and set the stage for a frontier in treating neurodevelopmental disorders that focuses on curing genetic diseases before birth.
Keywords: in utero, gene editing, mRNA delivery, nanoparticles, CRISPR/Cas9, CNS disorder
Introduction
Neurodevelopmental disorders are devastating diseases that impair cognitive, emotional, and motor development and affect about 15% of children and adolescents globally.1 Neurodevelopmental disorders are challenging to treat because disease onset frequently occurs early in life and damage to the brain tissue is permanent.2,3 Therefore, treating neurodevelopmental disorders during the fetal stages before disease onset occurs is desirable. However, the high sensitivity of the fetus to traditional therapeutics has limited the development of in utero therapies.4,5 Additionally, a large number of neurodevelopmental disorders are caused by relatively rare genetic mutations, which each only affects a small population of patients, and cannot be effectively treated with traditional small molecule drugs, making treatments difficult to develop from a financial perspective.6−8 For example, Angelman syndrome and Rett syndrome are devastating diseases and have an incidence of 1 in 12,000–24,000 globally.9,10 However, despite the low frequency of individual genetic neurodevelopmental disorders, collectively, these diseases affect a large number of patients and cause tremendous levels of mortality and morbidity. For instance, in 2012, the estimated total health care cost of treating pediatric genetic diseases amounted to $14–$57 billion, reflecting the urgent need for developing effective treatments for neurodevelopmental disorders.7
In utero nonviral gene editing, delivered via an intracerebroventricular (ICV) injection of LNP/mRNA complexes, holds great promise for treating neurological diseases. In utero gene editing can correct gene mutations during development in neural stem/progenitor cells (NSPCs), which can potentially generate a cure that endures for the patient’s life, due to the rapid proliferation of the corrected stem cells. In addition, ICV injection can potentially transfect large volumes of the brain tissue with minimal toxicity because the ventricles are fluid-filled organs which can receive infusions of liquids without causing brain tissue damage.11−13 In addition, since the ventricles have access to all parts of the brain, delivery via this route has the potential to transfect a much larger brain volume than a direct intracerebral injection.14,15 Direct intracerebral injections typically transfect only a small subregion of the brain, such as the left or right striatum.16,17 In addition, a direct intracerebral injection results in brain tissue damage because of the physical puncturing and high pressures needed to deliver the treatment into the tissue.18,19
However, despite its potential, very little is known about the in utero ICV delivery of LNP/mRNA complexes. Palanki et al. have demonstrated that LNPs can deliver mRNA coding for gene editing enzymes in utero after an ICV injection and can generate edits in approximately 1% of the brain tissue.20 Although these studies demonstrate that in utero brain editing is possible, the editing rates of current LNPs are not sufficient to generate therapeutic effects and it is unclear whether they can transfect NSPCs, which represent the key target cell type needed to evoke long-term curative effects.21 Finally, since LNPs are inherently inflammatory, it is imperative to uncover any safety and developmental deficits associated with in utero gene editing with LNPs.
In this report, we demonstrate that densely PEGylated LNPs formulated with an acid-degradable PEG–lipid (ADP-LNPs) can deliver mRNA and transfect brain cells after an in utero ICV injection, providing a potential platform for developing treatments for neurodevelopmental disorders (see Figure 1). With this approach, we were able to edit 30% of the cells throughout the brain in the mouse model due to the combination of highly efficient delivery of mRNA to NSPCs and their subsequent proliferation. Importantly, as the treated mice developed, the transfected NSPCs proliferated and distributed throughout the brain, ultimately resulting in gene editing in more than 60% of neurons in the hippocampus and 40% of neurons in the cortex at 10 weeks of age. Finally, using Angelman syndrome as an example neurodevelopmental disorder target, we demonstrated that ADP-LNPs were able to deliver Cas9 mRNA and sgRNA into fetal mouse brains and successfully edited the gene that causes Angelman syndrome at a rate that was potentially sufficient to generate therapeutic benefits. Specifically, we observed an editing efficiency of over 10% in brain cells. These experiments demonstrate that mRNA-based nonviral gene editing in NSPCs in utero with Cas9 mRNA can be achieved with high efficiency and low toxicity.
Figure 1.
In utero delivery of densely PEGylated LNPs globally transfects the brain in utero with mRNA for gene editing enzymes. In this report, we demonstrate that ADP-LNPs can safely and efficiently transfect large volumes of the fetal brain tissue with mRNA after an in utero ICV injection and have the transfection efficiency needed for treating genetic CNS disorders. We show here that an in utero ICV injection of ADP-LNPs can transfect and edit 30% of the entire fetal mouse brain cells with Cre mRNA, including the proliferating NSPCs, which populate the entire adult mouse brain as the mouse develops into adulthood. In addition, in utero ICV injection of ADP-LNPs transfected and edited 15% of the entire fetal mouse brain cells with Cas9 mRNA and efficiently edited NSPCs. These experiments demonstrate mRNA-based nonviral gene editing in NSPCs in utero using Cas9 mRNA, achieving high efficiency and minimal toxicity.
Results and Discussion
The therapeutic promise of in utero delivery of LNP/mRNA complexes is tempered by concerns over their potential toxicity and immunogenicity.22,23 In adults, intramuscular injections of LNPs have been shown to activate toll-like receptors and trigger cytokine production24 which could be detrimental to fetal development. While in utero injection of LNP/mRNA complexes has been performed, the postdelivery development of the treated fetuses remains uncertain. To address this gap, our investigation first focused on in utero toxicity of the LNP/mRNA complexes administered via ICV injection in Ai9 mice using Cre mRNA as the reporter cargo.
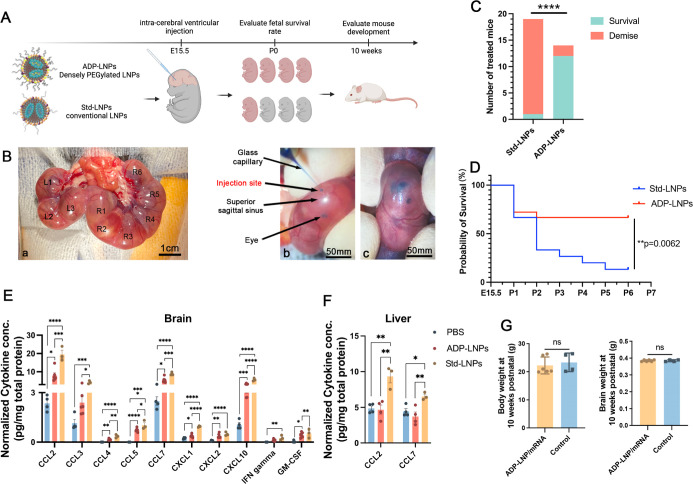
We compared two types of LNP/mRNA complexes: standard MC3-LNPs (Std-LNPs) with 1% PEG and acid-degradable PEGylated LNPs (ADP-LNPs) with 10% PEG, which are designed to retain high PEGylation and transfection efficiency as reported in our prior work25 (Figures 2A and S1). The size, zeta potential, and structure of LNPs were determined using a combination of cryogenic transmission electron microscopy (cryo-TEM) and nanoparticle tracking system (Tables S1–S3 and Figure S2). The in utero injections followed the protocol depicted in Figure 2B, with subsequent assessments of natural delivery and survival rates at birth. At embryonic day 15.5 (E15.5), we performed ICV injections on pregnant mice. Each Ai9 mouse fetus received 0.67 μg of Cre mRNA, which was delivered using Std-LNPs or ADP-LNPs. All fetuses in each pregnant mouse were given the same treatment, and each type of LNP was injected into three dams. On the day of natural delivery, we observed and counted the number of live-born pups. The Std-LNPs induced substantial in utero toxicity, with 18 out of 19 treated fetuses not surviving to birth. Conversely, ADP-LNPs demonstrated safety and tolerance with 12 out of 14 treated mouse fetuses surviving and exhibiting normal birth (Figure 2C).
Figure 2.
In utero ICV delivery of ADP-LNPs is safe and well tolerated. (A) Schematic diagram of the experimental protocol used to assess the safety of in utero ICV injection of ADP-LNPs and standard MC3-LNPs (Std-LNPs). (B) Intraoperative images showcasing the ICV injection procedure performed in utero. (a) All fetuses on either side of the uterine bifurcation (right or left) were noted; (b) a glass pipet was used to inject the LNPs into the fetal lateral cerebral ventricular space; and (c) successful injection into the ventricular system was determined by the spreading of the blue food dye into the ventricular zone (VZ). (C) Survival outcomes at birth for mouse fetuses subjected to LNP injections of either ADP-LNPs or Std-LNPs. ADP-LNP-treated fetuses had a significantly higher survival rate than fetuses treated with Std-LNPs (****p < 0.0001). (D) Kaplan–Meier survival curve of fetuses from E15.5 to postnatal day 7, comparing Std-LNPs and ADP-LNPs. ADP-LNPs demonstrated significantly higher survival probability (**p = 0.0062). (E,F) Normalized cytokine concentrations in brain (E) and liver (F) tissues 48 h postinjection exhibited significant differences between LNP-treated groups and the PBS control group 48 h postinjection. ADP-LNPs showed significantly lower cytokine levels compared to Std-LNPs, indicating reduced inflammatory response. (G) Body and brain weights at 10 weeks postnatal, comparing ADP-LNP/mRNA-treated mice and controls. No significant differences were observed, indicating normal postnatal development (data are represented as mean ± SEM, n = 6 for the ADP-LNP/mRNA group and n = 4 for the PBS control group).
To further investigate the toxicity of the LNPs, we performed ICV injection in C57 mouse fetuses with LNPs complexed with Luciferase mRNA. We observed and recorded their survival rates from embryonic day 15.5 (E15.5), when the ICV injections were performed, up to 7 days postnatally (P7). The Kaplan–Meier survival curves were plotted (Figure 2D) to investigate the difference between the two LNP/mRNA complex treatments. Mice treated with Std-LNPs exhibited significantly reduced survival rates, with only 2 of 15 fetus (survival rate of 13.33%) surviving to P7. The survival curve for this group shows a rapid decline, indicating acute in utero toxicity and postnatal lethality associated with the Std-LNPs. In contrast, the ADP-LNP treatment group demonstrated a substantially higher survival probability. The majority of these fetuses (12 of 18 fetus, survival rate is 66.67%) survived through the observation period, underscoring the superior safety and biocompatibility of ADP-LNPs. The statistical analysis of the Kaplan–Meier survival curves confirms a significant difference in survival outcomes between the two groups (p = 0.0062), supporting the hypothesis that ADP-LNPs have a profile for in utero applications that is safer than that of Std-LNPs. The cytokine and chemokine levels in the brain and liver of the ICV-injected fetuses were analyzed 48 h postinjection. Figures 2E and S3 show the normalized cytokine and chemokine concentrations in the brain tissue. Compared to the PBS-injected control group, both ADP-LNP and Std-LNP groups exhibited significantly elevated levels of several chemokines, including CCL2, CCL3, CCL4, CCL5, CCL7, CXCL1, CXCL2, CXCL10, IFN gamma, and GM-CSF. This indicates an inflammatory response in the brain triggered by the LNP injections. The elevation in cytokine levels was generally higher in the Std-LNP group compared to the ADP-LNP group, suggesting a greater inflammatory response and potential toxicity associated with the Std-LNPs. More importantly, Std-LNPs not only induced an immune response in the brain but also triggered a systemic immune response as evidenced by the elevated levels of CCL2 and CCL7 in the liver compared to the PBS- and ADP-LNP-injected groups (Figures 2F and S4). The ADP-LNP group showed only a modest increase in these chemokines compared with the PBS control, further highlighting the lower toxicity and better biocompatibility of ADP-LNPs. Further analysis of the naturally born ADP-LNP-treated fetuses revealed no adverse effects on growth, with their body and brain weights at 10 weeks mirroring those of untreated controls (Figure 2G).
ADP-LNPs contain two differences from the Std-LNPs, dense PEGylation and acid degradability. We performed experiments to identify which of these factors was responsible for their lower toxicity in utero. To determine this, we increased the PEG–lipid concentration to 10% in the standard LNP formulation (Std10PEG-LNPs) and tested the transfection efficiency and safety by fetal ICV injection. Figure S5A showed that the Std10PEG-LNPs complexed with Cre mRNA transfected about 10% of cells in the brain, which was significantly lower than the transfection rate of Std-LNPs and ADP-LNPs. Moreover, survival analysis showed that all mice injected with Std10PEG-LNPs died within 7 days postbirth. There was no statistically significant difference in survival rates between the Std10PEG-LNPs and the Std-LNPs containing 1% PEG (Figure S5B). The acid degradability of the PEG chains on ADP-LNPs, combined with dense PEGylation, likely contributes to their combination of reduced toxicity and high transfection efficiency. ADP-LNPs have also shown a superior capacity to deliver ribonucleoprotein (RNP) and mRNA to the liver in adult mice25,26 and may have broad utility. The findings of this study advocate for dense PEGylation combined with acid degradability as key design features for LNPs intended for in utero applications.
ADP-LNPs Enable Extensive Transfection of the Brain Tissue with mRNA following In Utero ICV Injection
In utero ICV injection of LNPs represents a promising strategy for transfecting substantial volumes of the brain tissue with minimal toxicity. Yet, conventional LNPs often fall short in transfection efficiency and biocompatibility for in utero therapeutic applications, necessitating the development of LNP formulations optimized for fetal environments. We explored the potential of ADP-LNPs for delivering Cre mRNA to the brain tissue in utero using the Ai9 mouse model. The Ai9 system is well suited for assessing mRNA transfection rates of LNPs, because successful delivery of Cre mRNA results in the permanent expression of td-Tomato in transfected cells, facilitating both quantification of transfection efficiency and identification of transfected cell phenotypes and lineages.
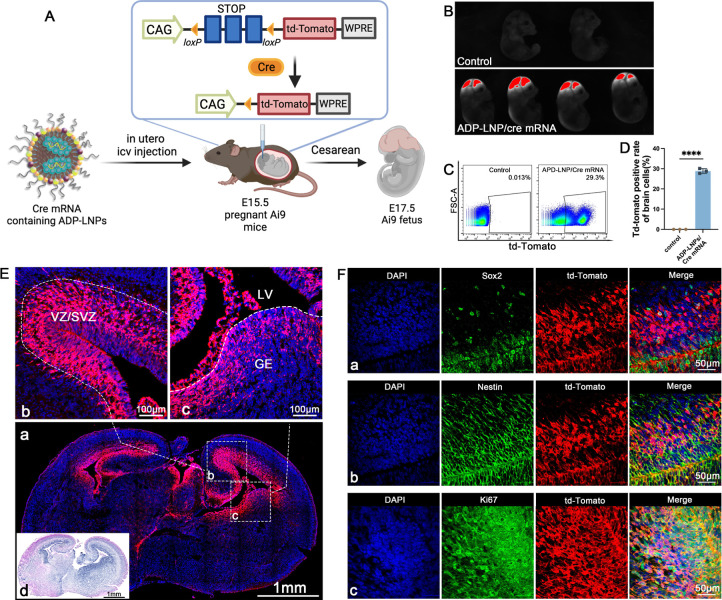
ADP-LNPs were injected in utero on embryonic age day 15.5 (E15.5) and were analyzed 48 h later for gene editing efficiency via whole fetus and organ imaging and by performing flow cytometry on the dissociated cells from the brain (Figure 3A). PBS-injected fetuses served as the control. The mouse fetuses were imaged 48 h post injection. The imaging confirmed that ADP-LNP/Cre mRNA complexes achieved efficient transfection in both fetal brain and the spinal cord. Compared to the PBS-treated control Ai9 mice, ADP-LNP/Cre mRNA-treated mice showed high levels of td-Tomato signal in the brain and spinal cord at 48 h post ICV injection (Figure 3B). Flow cytometric analysis of the entire fetal brain at 48 h after ICV injection showed that 28.87 ± 0.69% of total cells in the brain of the Ai9 mice treated with ADP-LNPs expressed td-Tomato (Figure 3C,D). In addition, we analyzed td-Tomato expression rates in the heart, lung, liver, kidney, and GI tract. Negligible td-Tomato expression was observed in all internal organs examined, suggesting that ADP-LNPs do not transfect internal organs outside the brain following in utero ICV injection (Figure S6). Brain tissue section imaging of the cells expressing td-Tomato showed that most of the td-Tomato-positive cells were adjacent to the cerebral ventricles 48 h postinjection of ADP-LNPs (Figure 3E-a). The transfected cells were primarily located in the Subventricular Zone (SVZ), which is known for its role in ongoing neurogenesis and its potential in brain repair and regeneration (Figure 3E-b) and the Ganglionic Eminence (GE), which is primarily involved in the development and regulation of interneurons (Figure 3E-c).27,28 We performed H&E staining on brain tissue sections. The tissue morphology appeared essentially normal, with no significant signs of edema, atrophy, or necrosis [Figure 3E(d)]. The Std-LNP’s transfection efficacy has also been studied. 25.00 ± 1.66% of brain cells were transfected by Std-LNPs injected at E15.5 (Figure S5A), and these transfected cells were primarily located in the SVZ and GE regions (Figure S7), showing no significant difference compared to ADP-LNPs.
Figure 3.
ADP-LNPs efficiently deliver Cre mRNA in utero to Ai9 mice after an ICV injection. (A) Schematic describing the experimental protocol used to evaluate the transfection efficiency of Cre mRNA delivered by ADP-LNPs into the mouse brain in utero. Fetal Ai9 mice received ICV injections of LNPs on day E15.5 and were analyzed 48 h later on day E17.5. (B) Whole fetus imaging reveals significant td-Tomato expression in the brains and spinal cords of fetuses treated with ADP-LNP/Cre mRNA complexes, compared to the negligible background signal in PBS-treated control Ai9 mice, indicating efficient transfection. (C) Flow cytometry analysis of the fetal brain 48 h post ICV injection reveals an average transfection rate of 28.87 ± 0.69% (D) among total brain cells, demonstrating global transfection of the fetal brain (data are represented as mean ± SEM, n = 3, ****p < 0.0001). (E) Histological examination of brain sections indicates that the majority of td-Tomato-positive cells are localized within the GE, VZ, and SVZ (a). Higher magnification image of the VZ, SVZ (b), and GE (c) showing extensive transfection in these proliferative regions. (d) H&E staining of an adjacent coronal section showing the overall brain structure for comparison. (F) Immunohistochemical staining further characterizes the td-Tomato-positive cells and demonstrates that most transfected cells are proliferating neural stem and progenitor cells (Sox2+/Nestin+/Ki67+), confirming that ADP-LNPs can efficiently transfect stem and progenitor cell populations within the fetal brain.
A critical advantage of in utero delivery of mRNA is the abundance of NSPCs during fetal development. The potential of LNPs to specifically transfect NSPCs has not been previously explored. Our findings demonstrate that ADP-LNPs after in utero delivery transfect a significant portion of the brain tissue with a strong likelihood of targeting NSPCs. Immunohistochemistry staining of ADP-LNP/Cre mRNA-treated brain sections showed that a majority of the transfected cells were indeed NSPCs, marked by Sox2, Nestin, and Ki67 positivity (Figure 3F).
The transfection of ADP-LNPs in utero after an ICV injection is orders of magnitude higher than transfection rates achieved in adult mice after either an ICV or direct striatal injection.16 While Std-LNPs and ADP-LNPs were able to transfect neurons in adult mouse brains after an intracranial injection, their transfection efficiencies were limited and were not able to transfect large volumes of the brain tissue.16 This spatial constraint represents a significant obstacle for the treatment of CNS disorders, which often require extensive tissue transfection. Our study shows that in utero administration of ADP-LNPs overcomes this challenge and can edit cells across substantial brain tissue volumes, enabling an approach for treating CNS disorders.
Impact of ADP-LNP In Utero Transfection Is Dramatically Amplified during Development because of Proliferation of Edited NSPCs
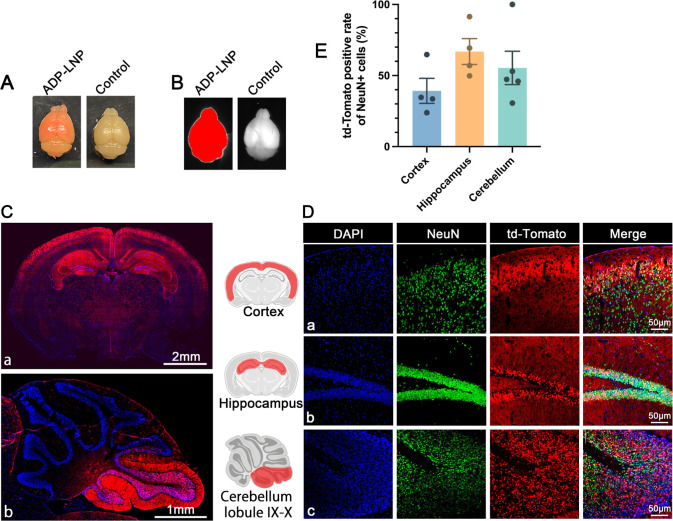
ADP-LNPs demonstrated effective transfection of NSPCs in utero. These transfected NSPCs have the potential to proliferate extensively and populate the developing mouse brain, thereby amplifying the impact of gene editing initiated during fetal development.21 To assess the long-term efficacy of this approach, we treated Ai9 fetuses with ADP-LNPs encapsulating Cre mRNA and evaluated td-Tomato expression at 10 weeks postnatal. Whole-mount view of the brains of the 10 week-old mice treated in utero with ADP-LNPs containing Cre mRNA revealed a visible red color, presumably due to efficient transfection of mRNA and the presence of a large number of td-Tomato-expressing cells (Figure 4A). In addition, the dissected brains were imaged, and the Cre mRNA/LNP-treated brains displayed a strong td-Tomato signal in the entire brain (Figure 4B).
Figure 4.
In utero ICV injection of Ai9 mice with ADP-LNPs containing Cre mRNA results in widespread transfection of the brain at 10 weeks of age. (A) A whole-mount view showcases the brain of a mouse treated with ADP-LNP/Cre mRNA at 10 weeks postnatal alongside a control. The treated brain exhibits extensive red fluorescence, indicating a high density of td-Tomato-expressing cells throughout the brain. (B) A representative image for the whole brain highlights the fluorescence in the ADP-LNP/Cre mRNA-treated brain, confirming the global expression of td-Tomato at 10 weeks postnatal. (C,D) Brain tissue section images show wide distribution of td-Tomato-expressing cells in the entire brain, especially in the cortex, hippocampus, and cerebellar lobules IX–X, displaying td-Tomato expression. The corresponding immunohistochemistry staining reveals that the majority of the td-Tomato-expressing cells are NeuN+, confirming that neuronal populations are the primary recipients of gene editing. (E) Quantitative analysis of the percentage of td-Tomato-positive NeuN+ cells within various brain regions, including the cortex, hippocampus, and cerebellar lobule IX–X (data are represented as mean ± SEM, n = 4). The cortex, hippocampus, and cerebellar lobule had greater than 40% of their total cells transfected with Cre, illustrating the efficacy of ADP-LNPs in transfecting brain cells, especially neurons, in utero.
To characterize the distribution of transfected cells and quantify the transfection efficiency, we analyzed the coronal and sagittal sections of the cerebrum and cerebellum, respectively, and the cortex via flow cytometry. This analysis indicated that >40% of the cells in these structures were transfected. In addition, we also analyzed several other brain regions via histology. At 10 weeks of age, ADP-LNP-transfected cells were present throughout the entire brain and were present in the choroid plexus, cerebral ventricles, cortex, hippocampus, thalamus, and cerebellum (Figure 4C). Transfected cells were also prominently present in various layers of the cerebral cortex and the granular layer of the hippocampus (Figure 4C-a). In the cerebellum, we also found a significant number of transfected cells in certain cerebellar lobes, primarily in the granule cell layer of the nodulus and uvula (Figure 4C-b), which are generally responsible for coordination of motor and eye movements as well as overall balance and muscle tone regulation.17 Additionally, a large number of choroid plexus cells were transfected (Figure S11). These cells are primarily responsible for producing cerebrospinal fluid and constituting the blood–brain barrier.29
Immunohistochemical analysis was employed to identify the types of cells transfected by ADP-LNPs. We stained brain sections with NeuN for neurons, GFAP for astrocytes, NG2 for oligodendrocytes, and Iba-1 for microglial cells. We found that most of the td-Tomato+ cells were located in the cortex, hippocampus, and cerebellum and expressed NeuN (Figure 4D), with only a negligible amount of td-Tomato+ cells expressing GFAP, NG2, or Iba1 (Figures S8–S10). The transfected cells were counted and quantified, and quantitative analysis showed that ADP-LNPs achieved transfection rates of 39.2 ± 8.85% in cortical neurons, 66.9 ± 9.14% in hippocampal neurons, and 55.37 ± 11.75% in cerebellar neurons (Figure 4E). This position-specific neuronal transfection may be closely related to the developmental stage of the stem and progenitor cells that were transfected in utero. The E15–16 period is the peak period of neurogenesis in the hippocampal CA1 and CA2 regions, as well as in the cortical II–V layers.30 These results conclusively demonstrate that in utero ICV injection of ADP-LNPs can deliver mRNA to NSPCs with exceptional efficiency, leading to gene editing levels that far surpass those achieved in adult mice using similar methods.31 Following ICV injection at E15.5, numerous transfected NSPCs were found surrounding the lateral and third ventricles. These cells actively proliferate and migrate during brain development, ultimately forming the cerebral cortex and hippocampus.32,33 Thus, fetal ICV injection of LNPs to deliver mRNA offers therapeutic opportunities for disorders linked to abnormal neurodevelopment, such as Angelman syndrome, and widespread neurometabolic dysfunction, such as Hurler syndrome. Additionally, treating malformations of cortical development during the fetal period is crucial as it can prevent severe neurological deficits and epilepsy that arise from disrupted cortical formation.34 Furthermore, the hippocampus, crucial for learning and memory, is implicated in many neurodegenerative diseases and conditions such as Alzheimer’s, Parkinson’s, multiple sclerosis, vascular dementia, frontotemporal dementia, depression, stress, epilepsy, schizophrenia, hypertension, and Cushing’s disease.35−37
ADP-LNPs Transfect the Mouse Brain Efficiently with Cas9 mRNA after In Utero ICV Delivery
Cas9-based gene editing has tremendous potential for treating developmental neurological disorders, but delivery has been one of the key limitations of its clinical applications. Although gene editing in the adult mouse brain has been accomplished via injection of Cas9 RNP or Cas9 mRNA/gRNA in LNPs, both these approaches transfect limited volumes of the brain tissue31,38 and cannot transfect the brain volume needed to treat neurological diseases.38,39 ADP-LNPs transfected NSPCs efficiently in utero and consequently may be able to generate global levels of brain editing with Cas9 after in utero delivery. We therefore investigated if in utero gene editing with ADP-LNPs could generate global levels of gene editing in the mouse brain.
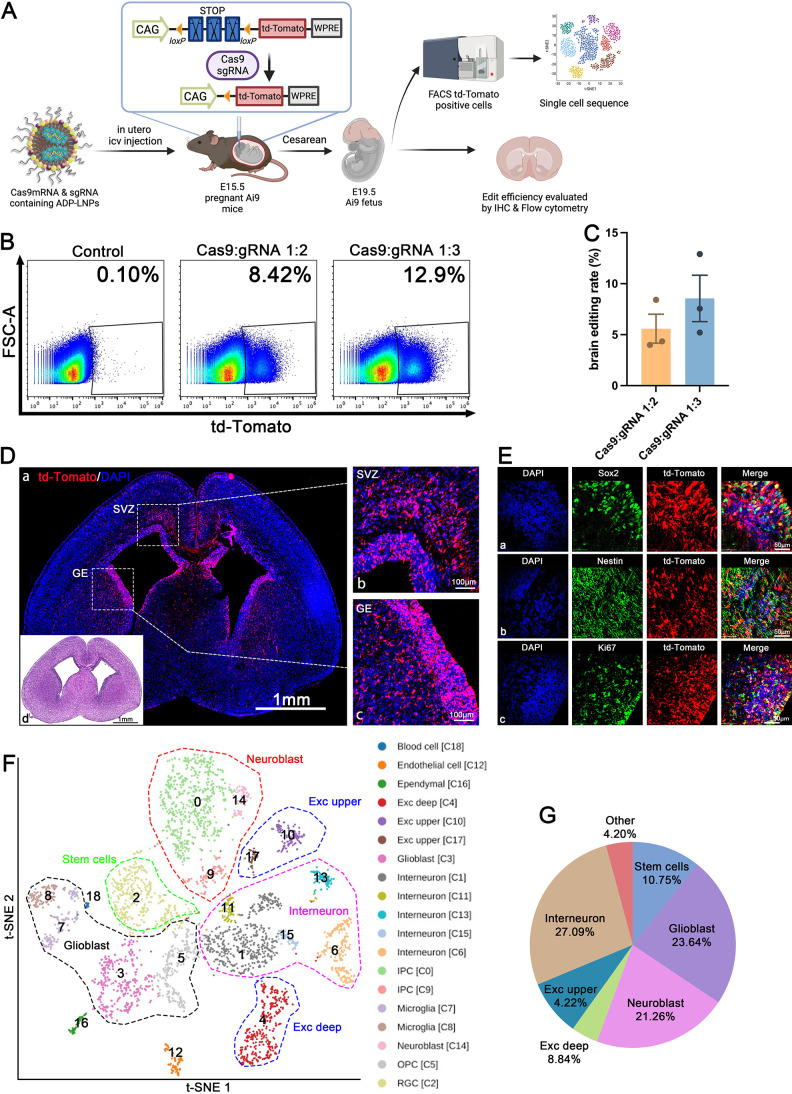
ADP-LNPs were formulated with Cas9 mRNA and gRNA targeting the Ai9 locus and were injected into Ai9 mice at E15.5 and analyzed 96 h later for brain editing (Figure 5A). Each mouse fetus received 0.5 μg of Cas9 mRNA. The harvested brain tissue was dissociated into single cells and the gene editing rate was quantified by flow cytometry. We formulated LNPs with various Cas9 mRNA to sgRNA ratios, and we found that a ratio of 1:3 demonstrated superior editing efficiency compared to 1:2. Specifically, at a 1:3 ratio of Cas9 mRNA to sgRNA, ADP-LNPs edited 8.56 ± 2.23% of the total brain cells (Figure 5B,C). This level of Cas9 editing was 1–2 orders of magnitude higher than the editing levels observed following ICV or intracranial injection in adult mice.31 Immunofluorescent imaging of td-Tomato demonstrated widespread gene editing within germinal regions of the brain, including the GE, VZ, and SVZ at 96 h postinjection (Figure 5D). Immunohistochemistry staining indicated a high proportion of td-Tomato-positive cells that coexpressed NSPC markers Sox2, Nestin, and Ki67, suggesting that the primary targets of ADP-LNPs/Cas9 mRNA and gRNA gene editing were proliferating NSPCs (Figure 5E). ADP-LNPs combined with in utero delivery can achieve the gene editing efficiency needed to treat a wide variety of developmental fetal disorders in the brain.
Figure 5.
In utero delivery of Cas9 mRNA/gRNA with ADP-LNPs results in widespread editing of multiple neural cell populations. (A) Schematic diagram describing the experimental protocol used to determine the cell types edited by ADP-LNPs with Cas9 mRNA and gRNA. 96 h after in utero delivery, cell sorting of td-Tomato-positive cells was performed and the edited cells were analyzed via single-cell sequencing. (B) Flow cytometry analysis of the entire fetal brain at 96 h after in utero ICV injection. (C) Quantification analysis of the efficiency of in utero gene editing. At the Cas9/gRNA ratio of 1:3, 8.56 ± 2.23% of total brain cells were edited by a single injection (data are represented as mean ± SEM, n = 3). (D) Representative brain section images display the distribution of td-Tomato expression, particularly within the SVZ (b) and GE (c). (d) H&E staining of an adjacent coronal section showing the overall brain structure for comparison. (E) Detailed immunohistochemistry in the SVZ and GE regions reveals that td-Tomato expression colocalizes with the NSPC markers Sox2, Nestin, and Ki67. ADP-LNPs were able to edit NSPCs with Cas9 mRNA and gRNA. (F) The edited cells were sorted and the cell types edited were identified via single-cell sequencing. t-SNE analysis of single-cell sequencing data illustrates the diverse neural cell populations edited by ADP-LNPs containing Cas9 mRNA and gRNA. (G) Pie chart summarizing the proportions of different cell types expressing td-Tomato, indicating a wide range of edited cells, with notable fractions of neuron-related cells like interneurons, neuroblasts, and glioblasts.
To further understand the cell types transfected in the SVZ and GE regions and their cell fate as they develop, we conducted single-cell transcriptome sequencing (scRNA-seq) analyses of the brains of fetuses injected with ADP-LNPs complexed with Cas9 mRNA. We isolated td-Tomato positive cells using flow cytometry sorting and analyzed the transcriptomes from 3893 high-quality single cells. From the flow sorted cells, we obtained a median depth of 300,000 reads per cell, detecting a median of ∼8054 transcripts and 2960 genes per cell. Principal component analysis (PCA), clustering analysis, and t-SNE visualization were used to unbiasedly group cells based on gene expression patterns. From this analysis, we identified 19 distinct cell populations which were characterized by specific differentially expressed genes (Figure 5F).
We found that a significant number of cells edited by Cas9 were neural stem cells in various developmental stages. These clusters were reliably assigned based on known differentially expressed genes. The retinal ganglion cell population, characterized by high expression of Pax6, Id4, Top2a, and Ube2c (C2, C3), includes dynamically expressed genes associated with cell cycle and progenitor identity, such as Mki67, Aspm, Cenpe, Cenpf, Id4, and Hes5 (C2, C3)40,41 (Figure S12).
The IPC population (C0, C9) shows high Eomes expression, derived from NECs and RGCs, crucial for generating most cortical neurons. Eomes-positive cells, originating from apical progenitors produce over 80% of cortical projection neurons across all layers.42,43 Notably, Eomes silencing is linked to microcephaly in humans, while its deletion in mice impairs IPC proliferation.44 We identified cells in early differentiation stages like C0, then we determined cell clusters as neuronal or non-neuronal based on Stmn2 expression levels.42 Clusters marked as differentiated or mature neurons, including C1, C6, C9, C11, C14, and C15, are distributed across various neocortical locations. Most interneurons, primarily GABAergic (inhibitory), are widespread in the neocortex, with Gad2 expression marking interneuron clusters C1 and C6.45,46 In contrast, excitatory projection neurons, generated by progenitors in the dorsal telencephalic radial glia, are identified by Neurod1 and Neurod6 expression in proliferating neurons (C0, C14).43,45 These findings align with our previous observations in Cre-transfected fetal brains where numerous cortical and hippocampal neurons were LNP transfected. Beyond neurons and neural stem cells, ADP-LNP editing also targets non-neuronal cell types, including OPCs marked by Olig2 expression (C5),43,45 microglial clusters identified by Trem2 expression (C7, C8),43 ependymal cells specifically expressing Rsph1 (C16),47 and endothelial cells marked by Col3a1 expression (C12).43
Sc-RNA seq analysis aids in understanding the populations of cell types edited by ADP-LNPs. In all brain cells edited by Cas9 mRNA, 10.8% are stem cells with the ability for proliferation and multilineage differentiation. Among the already differentiated cells, the majority are neurons, including 27.1% intermediate neurons and 13% excitatory neurons (4.2% in the upper cortex and 8.8% in the deep cortex). Various types of glial cells, including OPCs and astrocytes, together constitute 23.6% of all edited cells. In addition, there are small numbers of endothelial cells and choroid plexus cells, totaling 4.2% (Figure 5G). These results suggest that in utero ICV injection of ADP-LNPs complexed with Cas9 mRNA and gRNA mainly edits cells destined for neuronal fate, indicating potential applications in genetic diseases affecting neurons. This technique’s utility for editing astrocytes, OPCs, microglia, and endothelial cells, which can spread across different brain regions, provides potential for treating neurodevelopmental and genetic disorders related to these cell types.
In Utero ICV Delivery of ADP-LNPs Containing Cas9 mRNA/sgRNA Targeting the Angelman Syndrome Region Edits the UBE3A-ATS Locus in the Mouse Brain
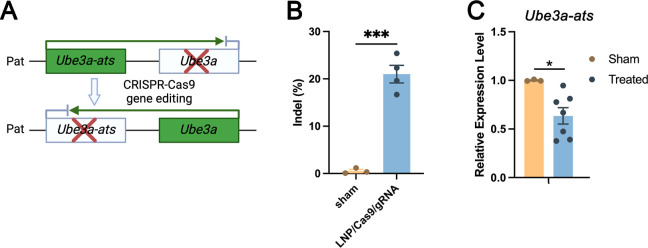
We further investigated if ADP-LNPs could edit a disease-causing gene in utero in the CNS. We selected Angelman syndrome as an example disease target because of the significant unmet medical need and the clinically relevant potential to treat this disease in utero. Angelman syndrome is a neurodevelopmental disorder caused by a deficiency in expression of the maternal Ube3a gene which encodes the UBE3A ubiquitin ligase.48 Angelman syndrome symptoms include severe cognitive impairment, seizures, and motor dysfunction.10,49 Previous studies by Silva-Santos et al. have demonstrated a distinct developmental window for treatment of Angelman syndrome, establishing the need for early therapeutic intervention.149 In most cases, the paternal allele encodes the wildtype Ube3a sequence but this gene is not translated into protein in the CNS because neuronal cell types also express a Ube3a antisense transcript (Ube3a-ats). Therefore, we aimed to correct the Angelman syndrome phenotype by knocking down the expression of Ube3a-ats via CRISPR Cas9 (Figure 6A). ADP-LNPs were complexed with Cas9 mRNA and gRNA targeting the Ube3a-ats locus and injected into fetal mice at E15.5. The brain tissue was analyzed 10 days postinjection (seventh day postnatal) for editing via Sanger sequencing. Figure 6B demonstrates that ADP-LNPs successfully delivered Cas9 mRNA and Angelman gRNA and edited approximately 21.00 ± 1.86% of the Angelman gene in the entire brain tissue. We measured the relative expression levels of Ube3a and Ube3a-ats in LNP/Cas9mRNA/gRNA-treated and sham groups. The results show a significant reduction in Ube3a-ats levels in the treated group compared to that in the sham group (Figure 6C). However, we did not observe an upregulation of Ube3a expression levels (Figure S13). This may be because the experiment was conducted in wild-type mice, which have normal Ube3a expression levels compared to those of the maternal copy. In studies using AAVs to edit Ube3a-ats, it has been shown that achieving similar levels of editing can effectively improve the disease phenotype.50,51 These experiments suggest that in utero ICV injection of ADP-LNPs has the potential to efficiently deliver CRISPR enzymes and make therapeutically relevant edits.
Figure 6.
ADP-LNPs containing Cas9 mRNA/gRNA can efficiently edit the Ube3a-ats gene and can potentially treat Angelman syndrome. (A) Schematic diagram describing the editing strategy used to upregulate the expression of Ube3a. Cas9/sgRNA/LNP complexes target the paternal allele of Ube3a-ats, which silences Ube3a. Successful editing downregulates the Ube3a-ats expression, allowing the paternal Ube3a allele to be expressed. (B) The bar graph depicts the indel frequencies observed at 10 days postinjection of ADP-LNPs, demonstrating efficient editing (data are represented as mean ± SEM, n = 3 for sham and n = 4 for LNP/Cas9/gRNA treated (***p < 0.001)). (C) Relative expression levels of Ube3a-ats in sham and treated groups, showing a significant reduction in the treated group (*p < 0.05). Data are represented as mean ± SEM, n = 3 for sham and n = 6 for ADP-LNP/Cas9/gRNA treated.
Conclusions
In this report, we demonstrate that ADP-LNPs can efficiently transfect the mouse fetal brain tissue after an ICV injection. In utero ICV injections of ADP-LNPs resulted in the transfection of NPSCs, which proliferated and populated the entire brain by the time mice were fully developed. ADP-LNPs were also well tolerated after an in utero ICV injection, presumably because of their combination of dense PEGylation and acid degradability. In contrast, traditional MC3-LNPs with 1.0% PEG caused very high levels of toxicity. The injection of ADP-LNPs during the fetal period can transfect a large number of NSPCs. These transfected cells further proliferate, migrate, and develop into mature neurons, resulting in over 40% of neurons in the cerebral cortex, hippocampus, and cerebellum in adult brains being transfected. Finally, ADP-LNPs were able to deliver Cas9 mRNA/gRNA targeting the Angelman Ube3a-ats locus and generated indels in 21% of the brain tissue, 7 days after birth, and also efficiently edited neural stem cells in the Ai9 mouse model. The treatment of neurodevelopmental disorders with gene editing therapeutics requires the development of delivery vectors that can transfect the brain tissue globally. In utero ICV injection with ADP-LNPs is so far the only methodology available for nonvirally transfecting large volumes of the brain tissue and is a promising platform for developing treatments for neurodevelopmental disorders.
Methods
LNP Preparation
DLin-MC3-DMA was purchased from MedKoo Biosciences. DOTAP, DOPE, and DMG-PEG2K were purchased from Avanti Polar Lipids. Cholesterol was purchased from Sigma-Aldrich. Pur-A-Lyzer Midi Dialysis Kits (WMCO, 3.5 kDa) were purchased from Sigma. The CRE and Cas9 mRNA were obtained from Trilink (San Diego, CA, USA). The gRNA for the Ai9 mouse model was obtained from IDT (San Diego, CA, USA) with the targeting sequence AAGTAAAACCTCTACAAATG. Lipid stock solutions of DOTAP, D-Lin, DOPE, Cholesterol, and DMG-PEG2K at a concentration of 10 mg/mL were separately dissolved in ethanol. Commercially available DOTAP is in a solution of chloroform (25 mg/mL), and the solvent was removed under reduced pressure before use. It was then dissolved in ethanol for storage. The acid-degradable PEG lipid (ADP) was synthesized according to our previously described method,25 and the synthetic scheme used to synthesize ADP is described in the Supporting Information (Figure S1). A stock solution of ADP at 40 mg/mL was dissolved in DMSO. All stock solutions were stored at −30 °C. Before LNP formation, lipids were allowed to warm to room temperature or heated up to 37 °C to ensure complete dissolution and vortexed whenever necessary. For the synthesis of ADP-LNPs, 1 μL of a mixed lipid solution was generated, which had the mole ratios described in Table S1, 1 μL of mRNA (1 mg/mL in the original buffer from TriLink) was mixed with 2 μL of DTT (20 mM in PBS pH 7.4), and the solution was thoroughly mixed by pipetting. Subsequently, 1 μL of the mixed lipid solution was mixed with the 3 μL mRNA solution, by pipetting to generate a final 10 mM DTT concentration, and the resulting solution was incubated at room temperature for 15 min. The reduced ADP-LNPs were allowed to store on ice between 30 min and 2 h before administration. For the synthesis of Std-LNPs, 1 μL of a mixed lipid solution was generated, which had the mole ratios described in Table S2, and 1 μL of mRNA (1 mg/mL in citrate buffer pH 4.0) was added to it and mixed by pipetting. The mixed solution was incubated at room temperature for 15 min. The LNPs were stored on ice between 30 min and 2 h before administration.
LNP Characterization
The size and zeta potential of the ADP-LNPs and Std-LNPs were characterized by a MONO ZetaView (PMX-120, Particle Metrix, Meerbusch, Germany) instrument following the manufacturing manual.
cryo-TEM was used for the direct visualization of LNP suspensions. To prepare LNP samples for cryo-TEM, copper grids (200 mesh, 1.2 μm holes; Ted Pella Inc.) were glow discharged for 30 s at 25 mA using a PELCO EasiGlow system (Ted Pella Inc.). A 4 μL aliquot of LNPs was applied to the carbon side of the grid, blotted for 5 s in the humidity chamber, incubated for 30 s, and plunge-frozen into precooled liquid ethane in an EM GP2 Automatic Plunge Freezer (Leica Microsystems). This process results in the vitrification of LNP samples in a thin layer of amorphous ice to preserve them in their native state and protect them against radiation damage during imaging. The samples were then imaged using a Glacios 2 electron microscope (Thermo Fisher Scientific) equipped with an autoloader with an accelerating voltage of 200 kV for high-resolution grid screening and data collection. Low dose mode was used during image acquisition to minimize radiation damage to the LNP samples. Images were obtained with a GATAN K3 direct electron detector at 45,000× magnification with a defocus value between −2 and −5 μm. The total accumulated dose per image did not exceed 40 e/Å2.
Animal Studies
All animal procedures were approved by The University of California, Davis (UCD) institutional animal care and use committee. All facilities used during the study period were accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International (AAALAC). Ai9 transgenic mice were purchased from The Jackson Laboratory (Jackson stock No. 007909). Ai9 mice were time-mated and bred on-site at UCD. The in utero injections were performed as previously described.15,22,52 E15.5 mice were anesthetized with 5% isoflurane, followed by 2% isoflurane for maintenance, and placed in the supine position on top of a heating pad. After a 2 cm midline abdominal incision was made, the uterus was gently exteriorized using cotton tipped applicators. The number of fetuses on either side of the uterine bifurcation was noted, along with the viability status of each fetus (Figure 2B-a). A glass pipet, which had been pulled to an opening diameter of 50–70 μm, was loaded with 2 μL of Cre mRNA- or Cas9 mRNA-containing LNPs resuspended in PBS and blue food dye in a 10:1 ratio. PBS and blue food dye in a 10:1 ratio were injected as a control (Figure 2B-b). The glass pipet was inserted through the fetal skull and brain tissue into one of the lateral cerebral ventricular spaces. Successful injection into the ventricular system was determined by the spreading of the blue food dye into the VZ as shown in Figure 2B(c). The uterus was then gently interiorized back into the abdomen using cotton tipped applicators and the abdomen was filled with approximately 1 mL 37 °C saline to avoid dehydration. The maternal laparotomy was then sutured closed in 2 layers using absorbable 4–0 Vicryl. The mouse was given a subcutaneous injection of 0.05 mg/kg of buprenorphine for pain management per 12 h in the next 48 h. After the in utero injection, the pregnant dams continued to gestate until the offsprings were delivered via cesarean section either at embryonic day 17.5 or 19.5 (E17.5 or E19.5) or until natural and full-term delivery occurred. Fetal survival was assessed based on size, coloration, and spontaneous movement. Either the whole fetus or dissected brain were imaged by a ChemiDocTM MP imaging system (Bio-Rad, Hercules). The survival study was carried out using three pregnant dams for each group, with std-LNPs and ADP-LNPs injected into the fetuses. Specifically, the fetuses were obtained from three separate dams for each LNP type and pooled into one experiment. Each dam received the same treatment to ensure consistency across the groups. For clarity, we used nested statistical analysis by dividing the fetuses into subgroups based on their birth mothers. This approach allows us to account for any potential variability between different litters and provides a more accurate representation of the treatment effects.
Cytokine Analysis
Fetal brain and liver lysates were harvested and assessed for cytokine levels at 48 h post LNP injections. The freshly harvested fetal brain and liver were lysed in Cell Lysis Buffer (Cell Signaling Technology) supplemented with 1% Protease Inhibitor Cocktail (Sigma-Aldrich). Total protein content in each sample was determined with the BCA protein assay kit (Thermo Fisher Scientific) per manufacturer’s instructions. The cytokine levels were assessed using the ProcartaPlex Mouse Cytokine & Chemokine Panel 1, 26plex kit for the Luminex system according to the manufacturer’s instructions. PBS injection served as the control. Cytokine data were normalized to the total protein concentration.
Flow Cytometry
The transfection rate of various fetal organs was measured using flow cytometry based on the percentage of cells positive for td-Tomato following in utero treatment. After organ dissection, single-cell suspensions were obtained as previously described22,53 for staining and flow cytometric analysis. The freshly dissected brains were minced and then digested with 600 μL of 1 mg/mL collagenase type I (Gibco) at 37 °C and 5% CO2 for 20 min in 1.5 mL Eppendorf tubes. Following this, the collected cell suspension was neutralized with 10% fetal bovine serum and kept on ice. Additional 1% collagenase 1 solution was added to the remaining tissue and incubated under the same conditions for an additional 20 min. A single-cell suspension was obtained by filtering the resulting cell suspension solutions through a 70 μm Nylon cell strainer (Falcon). The Attune NxT Flow Cytometer (Thermo Fisher Scientific) was used for performing flow cytometry, and FlowJo software (FlowJo LLC) was used for data analyses.
Histology and Immunofluorescence Staining
The adult mouse brain harvest was followed by PBS and 4% PFA perfusion. Dissected fetal and adult mouse brains were fixed in 4% PFA for 24 h and dehydrated with 15% sucrose for 24 h, followed by 30% sucrose for 24 h, before being embedded in an O.C.T. (Fisher Healthcare; Scigen Scientific Gardena; 4585). Serial 10 μm thickness sections were made using a Cryostat (Leica CM3050S) before the sections were collected onto Matsunami Glass microscope slides. For immunofluorescence staining, tissue sections were washed gently and thoroughly with PBS before undergoing heat antigen retrieval with 10 mM sodium citrate buffer, pH 6.0. Sections were then blocked with 5% Bovine Serum Albumin in PBS at room temperature for 1 h. Sections were stained with primary antibody solutions at 4 °C overnight. The dilutions of primary antibodies were 1:250 for td-Tomato (MyBioSource, MBS448092), 1:100 for NeuN (Cell Signaling Technologies, D4G40, 24307S), 1:250 for GFAP (Invitrogen, PA1-10,004), 1:100 for Iba1 (Cell Signaling Technologies, E404W; 17198T), 1:100 for NG2 (abcam, ab275024), 1:250 for Sox2 (Cell Signaling Technologies, L1D6A2; 4900S), 1:400 for Nestin (abcam, ab6142), and 1:200 for Ki67 (abcam, ab15580). Sections were stained with the corresponding secondary antibody solution at room temperature for 30 min. The dilution of the secondary antibodies was 1:250. Sections were then stained with a 1:5000 dilution of DAPI at room temperature for 5 min. Finally, slides were mounted with Prolong Diamond Antifade Mountant (Invitrogen). The td-Tomato-positive rate of neurons was quantified via fluorescent images, taken at 60× magnification using a Nikon A1 confocal microscope. 3–5 random fields from each tissue section were imaged, and the td-Tomato-positive rate of neurons was quantified in ImageJ based on NeuN and td-Tomato-positive signal.
scRNA-seq Sample Preparation and Sequencing
Single-cell suspension was prepared following the protocol for flow cytometry. The td-Tomato positive cells were sorted in UC Davis Flow Cytometry Shared Resource and performed by BD FACS Aria II. Single-cell sequencing was performed by the DNA Technologies and Expression Analysis Core of UC Davis. The single-cell suspension with 93.9% viability was used to generate single-cell cDNA libraries. The 10× Chromium X using Single Cell 3′ Reagent kit V3.1 was used to create cDNA libraries. Samples were then sequenced on an AVITI sequencer (Element Biosciences).
scRNA-seq Data Analysis
Single nuclei sequencing data obtained from the 10× Genomics Chromium platform were processed using CellRanger 7.2.0 count function, aligning and quantifying against the mm10-2020-A genome assembly.54 Subsequently, the entire data set underwent analysis with Seurat v4.4.0 in R v4.2.3.55,56 Cells with low total UMI counts, few expressed genes, and high mitochondrial proportions were filtered out prior to downstream analysis. The filtered data were normalized using SCTtransform and subjected to PCA using the RunPCA function. Clusters were identified using the FindNeighbors and FindClusters functions in Seurat. Cluster-specific upregulated markers were determined using the FindAllMarkers command. Each cluster was annotated based on previously reported differentially expressed genes. Visualization of the clusters was carried out using PCA, t-distributed Stochastic Neighbor Embedding (t-SNE), or uniform manifold approximation and projection for dimension reduction (UMAP).
AS Gene Editing Rate Quantification
Mouse UBE3A-ATS locus knockout gRNA was obtained from IDT (San Diego, CA, USA) with the targeting sequence GCCCAACCTCTCAAACGTGA. LNPs were formulated as described above using a 1:3 mass ratio of mRNA to gRNA. Brain tissue DNA was extracted following the manufacture recommendation. DNA was used as the template to generate PCR amplicons of the edited region using the primer set: forward primer TGAGCTGCCCAAGCACTTAT and reverse primer TCACAAAGGGCAAACCCTCA. The PCR products were purified and sent for Sanger sequencing. The indel editing efficiency was determined using Synthego ICE Analysis software. The expression level of Ube3A-ATS and Ube3A was evaluated by RT-qRNA. Total RNA was obtained from cells using the RNeasyPlus Mini kit (Qiagen), and cDNA was synthesized from 1 mg of RNA using SuperScript III First-Strand Synthesis SuperMix (Thermo Fisher Scientific). For quantitative analysis, RT-PCR was performed on a StepOnePlus System (Applied Biosystems) using the SYBR green PCR master mix (Applied Biosystems). The primers used were Ube3a-ATS primer: forward-AGTTCTTCCAGGGAAGCAAGGG, reverse-TCTTTGCTGGAATGCCAGGGG, and Ube3a primer: forward-AGACTCTTTCTGCAGTTTACAACAGG, reverse-TGTCCCCAATGAAGAAGGGAGG. GAPDH primer: forward ATTCAACGGCACAGTCAAGG, reverse-TGGATGCAGGGATGATGTTC.
Statistics
Chi-square test was used for comparing the survival rate between ADP-LNP and Std-LNP group using PRISM 7 (GraphPad Software Inc., San Diego, CA, USA). One-way analysis of variance following Tukey’s multiple comparison tests was used to analyze involving the comparison of more than two groups. Kaplan–Meier survival analysis was done by applying the Gehan–Breslow–Wilcoxon test. The student’s t-test was used to analyze body and brain weight. Significance of statistical differences was based on p < 0.05.
Acknowledgments
We would like to acknowledge Shriners Children’s research grant 85400-NCA-24, NIH grants 1R01NS131538-01A1, R01MH125979-01, and 1R21NS133881-01, California Institute for Regenerative Medicine (CIRM) DISC2-14097 and DISC2-14045, the Innovative Genomics Institute, and the CRISPR Cures for Cancer Initiative.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.4c05169.
Comprehensive description of the lipid nanoparticle (LNP) synthesis strategy and detailed LNP characterization data; and full panel of cytokine and chemokine expression profiles, survival analysis data, microscopy images illustrating nanoparticle distribution, flow cytometry results, and single-cell sequencing gene expression profiles (PDF)
Author Contributions
¶ K.G. and H.H. contributed equally to the study.
The authors declare the following competing financial interest(s): K.G., H.H. and N.M. own equity in Opus Biosciences. All the other authors declare no competing interests.
Supplementary Material
References
- Gidziela A.; Ahmadzadeh Y. I.; Michelini G.; Allegrini A. G.; Agnew-Blais J.; Lau L. Y.; Duret M.; Procopio F.; Daly E.; Ronald A.; Rimfeld K.; Malanchini M. A Meta-Analysis of Genetic Effects Associated with Neurodevelopmental Disorders and Co-Occurring Conditions. Nat. Human Behav. 2023, 7 (4), 642–656. 10.1038/s41562-023-01530-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadders-Algra M. Early Diagnostics and Early Intervention in Neurodevelopmental Disorders—Age-Dependent Challenges and Opportunities. J. Clin. Med. 2021, 10 (4), 861. 10.3390/jcm10040861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeron T.What Do We Know about Early Onset Neurodevelopmental Disorders? In Translational Neuroscience: Toward New Therapies; Nikolich K., Hyman S. E., Eds.; MIT Press: Cambridge (MA), 2015. [PubMed] [Google Scholar]
- Palanki R.; Peranteau W. H.; Mitchell M. J. Delivery Technologies for in Utero Gene Therapy. Adv. Drug Delivery Rev. 2021, 169, 51–62. 10.1016/j.addr.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida-Porada G.; Atala A.; Porada C. D. In utero stem cell transplantation and gene therapy: rationale, history, and recent advances toward clinical application. Mol. Ther.--Methods Clin. Dev. 2016, 3, 16020. 10.1038/mtm.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Caneja C.; State M.; Hagerman R.; Jacquemont S.; Marín O.; Bagni C.; Umbricht D.; Simonoff E.; de Andrés-Trelles F.; Kaale A.; Pandina G.; Gómez-Mancilla B.; Wang P.; Cusak J.; Siafis S.; Leucht S.; Parellada M.; Loth E.; Charman T.; Buitelaar J.; Murphy D.; Arango C. A White Paper on a Neurodevelopmental Framework for Drug Discovery in Autism and Other Neurodevelopmental Disorders. Eur. Neuropsychopharmacol. 2021, 48, 49–88. 10.1016/j.euroneuro.2021.02.020. [DOI] [PubMed] [Google Scholar]
- Gonzaludo N.; Belmont J. W.; Gainullin V. G.; Taft R. J. Estimating the Burden and Economic Impact of Pediatric Genetic Disease. Genet. Med. 2019, 21 (8), 1781–1789. 10.1038/s41436-018-0398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.; Duan R.; Li C.; Xie Z.; Wang A.; Xiong L.; Wei J.; Xi H.; Fang J.; Yan H.; Wang J.; Zhang Y.; Mao X.; Wang J.; Wang H. Study on the Economic Burden of Neurodevelopmental Diseases on Patients With Genetic Diagnosis. Front. Public Health 2022, 10, 887796. 10.3389/fpubh.2022.887796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. A.; Mueller-Mathews J. M.. Angelman Syndrome. In Cassidy and Allanson’s Management of Genetic Syndromes; Carey J. C., Battaglia A., Viskochil D., Cassidy S. B., Eds.; Wiley, 2021; pp 61–73. [Google Scholar]
- Madaan M.; Mendez M. D.. Angelman Syndrome. StatPearls; StatPearls Publishing: Treasure Island (FL), 2024. [PubMed] [Google Scholar]
- Atkinson A. J. Intracerebroventricular Drug Administration. Transl. Clin. Pharmacol. 2017, 25 (3), 117. 10.12793/tcp.2017.25.3.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashnonejad A.; Amini Chermahini G.; Gündüz C.; Onay H.; Aykut A.; Durmaz B.; Baka M.; Su Q.; Gao G.; Özkınay F. Fetal Gene Therapy Using a Single Injection of Recombinant AAV9 Rescued SMA Phenotype in Mice. Mol. Ther. 2019, 27 (12), 2123–2133. 10.1016/j.ymthe.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad M. R.; Donsante A.; Zerfas P.; Kaler S. G. Fetal Brain-Directed AAV Gene Therapy Results in Rapid, Robust, and Persistent Transduction of Mouse Choroid Plexus Epithelia. Mol. Ther.--Nucleic Acids 2013, 2, e101 10.1038/mtna.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamodi A. S.; Martinez Sabino A.; Fitzgerald N. D.; Moschou D.; Crair M. C. Transverse Sinus Injections Drive Robust Whole-Brain Expression of Transgenes. eLife 2020, 9, e53639 10.7554/eLife.53639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glascock J. J.; Osman E. Y.; Coady T. H.; Rose F. F.; Shababi M.; Lorson C. L. Delivery of Therapeutic Agents Through Intracerebroventricular (ICV) and Intravenous (IV) Injection in Mice. J. Vis. Exp. 2011, (56), e2968 10.3791/2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.; Lee K.; Panda S.; Gonzales-Rojas R.; Chong A.; Bugay V.; Park H. M.; Brenner R.; Murthy N.; Lee H. Y. Nanoparticle Delivery of CRISPR into the Brain Rescues a Mouse Model of Fragile X Syndrome from Exaggerated Repetitive Behaviours. Nat. Biomed. Eng. 2018, 2 (7), 497–507. 10.1038/s41551-018-0252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L.; Zhang Z.; Lu X.; Wang G.; Meng D.; Liu C.; Yun J.; Xu T.; Zhao C.; Lu J. Elimination of Serotonergic Neurons by Stereotaxic Injection of 5,7-Dihydroxytryptamine in the Dorsal Raphe Nuclei of Mice. J. Vis. Exp. 2020, (159), e60968 10.3791/60968. [DOI] [PubMed] [Google Scholar]
- Amer M. H.; Rose F. R. A. J.; Shakesheff K. M.; Modo M.; White L. J. Translational Considerations in Injectable Cell-Based Therapeutics for Neurological Applications: Concepts, Progress and Challenges. npj Regener. Med. 2017, 2 (1), 23. 10.1038/s41536-017-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest J.; Benavides F.; Padgett K.; Mendez E.; Tovar D. Technical Aspects of Spinal Cord Injections for Cell Transplantation. Clinical and Translational Considerations. Brain Res. Bull. 2011, 84 (4–5), 267–279. 10.1016/j.brainresbull.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Palanki R.; Bose S. K.; Dave A.; White B. M.; Berkowitz C.; Luks V.; Yaqoob F.; Han E.; Swingle K. L.; Menon P.; Hodgson E.; Biswas A.; Billingsley M. M.; Li L.; Yiping F.; Carpenter M.; Trokhan A.; Yeo J.; Johana N.; Wan T. Y.; Alameh M.-G.; Bennett F. C.; Storm P. B.; Jain R.; Chan J.; Weissman D.; Mitchell M. J.; Peranteau W. H. Ionizable Lipid Nanoparticles for Therapeutic Base Editing of Congenital Brain Disease. ACS Nano 2023, 17 (14), 13594–13610. 10.1021/acsnano.3c02268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever D. P.; Scharenberg S. G.; Camarena J.; Kildebeck E. J.; Clark J. T.; Martin R. M.; Bak R. O.; Tang Y.; Dohse M.; Birgmeier J. A.; Jagadeesh K. A.; Bejerano G.; Tsukamoto A.; Gomez-Ospina N.; Uchida N.; Porteus M. H. CRISPR/Cas9 Genome Engineering in Engraftable Human Brain-Derived Neural Stem Cells. iScience 2019, 15, 524–535. 10.1016/j.isci.2019.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K.; Li J.; Song H.; Han H.; Wang Y.; Yin B.; Farmer D. L.; Murthy N.; Wang A. In Utero Delivery of mRNA to the Heart, Diaphragm and Muscle with Lipid Nanoparticles. Bioact. Mater. 2023, 25, 387–398. 10.1016/j.bioactmat.2023.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S. K.; Menon P.; Peranteau W. H. InUtero Gene Therapy: Progress and Challenges. Trends Mol. Med. 2021, 27 (8), 728–730. 10.1016/j.molmed.2021.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiaie S. H.; Majidi Zolbanin N.; Ahmadi A.; Bagherifar R.; Valizadeh H.; Kashanchi F.; Jafari R. Recent Advances in mRNA-LNP Therapeutics: Immunological and Pharmacological Aspects. J. Nanobiotechnol. 2022, 20 (1), 276. 10.1186/s12951-022-01478-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S.; Gao K.; Han H.; Stenzel M.; Yin B.; Song H.; Lawanprasert A.; Nielsen J. E.; Sharma R.; Arogundade O. H.; Pimcharoen S.; Chen Y.-J.; Paul A.; Tuma J.; Collins M. G.; Wyle Y.; Cranick M. G.; Burgstone B. W.; Perez B. S.; Barron A. E.; Smith A. M.; Lee H. Y.; Wang A.; Murthy N. Acid-Degradable Lipid Nanoparticles Enhance the Delivery of mRNA. Nat. Nanotechnol. 2024, 10.1038/s41565-024-01765-4. [DOI] [PubMed] [Google Scholar]
- Chen K.; Han H.; Zhao S.; Xu B.; Yin B.; Lawanprasert A.; Trinidad M.; Burgstone B. W.; Murthy N.; Doudna J. A. Lung and liver editing by lipid nanoparticle delivery of a stable CRISPR–Cas9 ribonucleoprotein. Nat. Biotechnol. 2024, 10.1038/s41587-024-02437-3. [DOI] [PubMed] [Google Scholar]
- Encha-Razavi F.; Sonigo P. Features of the Developing Brain. Childs Nerv. Syst. 2003, 19 (7–8), 426–428. 10.1007/s00381-003-0765-6. [DOI] [PubMed] [Google Scholar]
- Modo M. Bioscaffold-Induced Brain Tissue Regeneration. Front. Neurosci. 2019, 13, 1156. 10.3389/fnins.2019.01156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed K.; Reddy V.; Lui F.. Neuroanatomy, Choroid Plexus. StatPearls; StatPearls Publishing: Treasure Island (FL), 2024. [PubMed] [Google Scholar]
- Finlay B. L.; Darlington R. B. Linked Regularities in the Development and Evolution of Mammalian Brains. Science 1995, 268 (5217), 1578–1584. 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- Tuma J.; Chen Y.-J.; Collins M. G.; Paul A.; Li J.; Han H.; Sharma R.; Murthy N.; Lee H. Y. Lipid Nanoparticles Deliver mRNA to the Brain after an Intracerebral Injection. Biochemistry 2023, 62 (24), 3533–3547. 10.1021/acs.biochem.3c00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasistha N. A.; Khodosevich K. The Impact of (Ab)Normal Maternal Environment on Cortical Development. Prog. Neurobiol. 2021, 202, 102054. 10.1016/j.pneurobio.2021.102054. [DOI] [PubMed] [Google Scholar]
- White T. A.; Miller S. L.; Sutherland A. E.; Allison B. J.; Camm E. J. Perinatal Compromise Affects Development, Form, and Function of the Hippocampus Part One; Clinical Studies. Pediatr. Res. 2024, 95 (7), 1698–1708. 10.1038/s41390-024-03105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L.; Calcagnotto M. E.; Paredes M. F. Cortical Malformations: Lessons in Human Brain Development. Front. Cell. Neurosci. 2020, 13, 576. 10.3389/fncel.2019.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch T.; Wulff P. The Hippocampus in Aging and Disease: From Plasticity to Vulnerability. Neuroscience 2015, 309, 1–16. 10.1016/j.neuroscience.2015.07.084. [DOI] [PubMed] [Google Scholar]
- Dhikav V.; Anand K. Hippocampus in Health and Disease: An Overview. Ann. Indian Acad. Neurol. 2012, 15 (4), 239. 10.4103/0972-2327.104323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasinghe-Mudiyanselage P. D. E.; Ang M. J.; Kang S.; Kim J.-S.; Moon C. Structural Plasticity of the Hippocampus in Neurodegenerative Diseases. Int. J. Mol. Sci. 2022, 23 (6), 3349. 10.3390/ijms23063349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl E. C.; Sabo J. K.; Kang M. H.; Allen R.; Applegate E.; Kim S. E.; Kwon Y.; Seth A.; Lemus N.; Salinas-Rios V.; Soczek K. M.; Trinidad M.; Vo L. T.; Jeans C.; Wozniak A.; Morris T.; Kimberlin A.; Foti T.; Savage D. F.; Doudna J. A. Genome Editing in the Mouse Brain with Minimally Immunogenic Cas9 RNPs. Mol. Ther. 2023, 31 (8), 2422–2438. 10.1016/j.ymthe.2023.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger J. M.; Wang Y.; Neuman S. S.; Snow K. J.; Murray S. A.; Lutz C. M.; Bondarenko V.; Felton J.; Gimse K.; Xie R.; Li D.; Zhao Y.; Flowers M. T.; Simmons H. A.; Roy S.; Saha K.; Levine J. E.; Emborg M. E.; Gong S. Efficient in Vivo Neuronal Genome Editing in the Mouse Brain Using Nanocapsules Containing CRISPR-Cas9 Ribonucleoproteins. Biomaterials 2023, 293, 121959. 10.1016/j.biomaterials.2022.121959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. J.; Pauler F. M.; McKenna A.; Shendure J.; Hippenmeyer S.; Horwitz M. S. Simultaneous Brain Cell Type and Lineage Determined by scRNA-Seq Reveals Stereotyped Cortical Development. Cell Syst. 2022, 13 (6), 438–453.e5. 10.1016/j.cels.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G.; Siletti K.; Furlan A.; Gyllborg D.; Vinsland E.; Mossi Albiach A.; Mattsson Langseth C.; Khven I.; Lederer A. R.; Dratva L. M.; Johnsson A.; Nilsson M.; Lönnerberg P.; Linnarsson S. Molecular Architecture of the Developing Mouse Brain. Nature 2021, 596 (7870), 92–96. 10.1038/s41586-021-03775-x. [DOI] [PubMed] [Google Scholar]
- Ruan X.; Kang B.; Qi C.; Lin W.; Wang J.; Zhang X. Progenitor Cell Diversity in the Developing Mouse Neocortex. Proc. Natl. Acad. Sci. U.S.A. 2021, 118 (10), e2018866118 10.1073/pnas.2018866118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandler R. C.; Vitali I.; Delgado R. N.; Ho M. C.; Dvoretskova E.; Ibarra Molinas J. S.; Frazel P. W.; Mohammadkhani M.; Machold R.; Maedler S.; Liddelow S. A.; Nowakowski T. J.; Fishell G.; Mayer C. Single-Cell Delineation of Lineage and Genetic Identity in the Mouse Brain. Nature 2022, 601 (7893), 404–409. 10.1038/s41586-021-04237-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A.; Mao C.; Hadjantonakis A.-K.; Klein W. H.; Broccoli V. Tbr2 Directs Conversion of Radial Glia into Basal Precursors and Guides Neuronal Amplification by Indirect Neurogenesis in the Developing Neocortex. Neuron 2008, 60 (1), 56–69. 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo L.; Simon J. M.; Xing L.; McCoy E. S.; Niehaus J. K.; Guo J.; Anton E. S.; Zylka M. J. Single-Cell Transcriptomic Analysis of Mouse Neocortical Development. Nat. Commun. 2019, 10 (1), 134. 10.1038/s41467-018-08079-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wizeman J. W.; Guo Q.; Wilion E. M.; Li J. Y. Specification of Diverse Cell Types during Early Neurogenesis of the Mouse Cerebellum. eLife 2019, 8, e42388 10.7554/eLife.42388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratz M.; Von Berlin L.; Larsson L.; Martin M.; Westholm J. O.; La Manno G.; Lundeberg J.; Frisén J. Clonal Relations in the Mouse Brain Revealed by Single-Cell and Spatial Transcriptomics. Nat. Neurosci. 2022, 25 (3), 285–294. 10.1038/s41593-022-01011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer P. L.; Hanayama R.; Bloodgood B. L.; Mardinly A. R.; Lipton D. M.; Flavell S. W.; Kim T.-K.; Griffith E. C.; Waldon Z.; Maehr R.; Ploegh H. L.; Chowdhury S.; Worley P. F.; Steen J.; Greenberg M. E. The Angelman Syndrome Protein Ube3A Regulates Synapse Development by Ubiquitinating Arc. Cell 2010, 140 (5), 704–716. 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y.; Sonzogni M. UBE3A Reinstatement as a Disease-modifying Therapy for Angelman Syndrome. Develop. Med. Child Neuro. 2021, 63 (7), 802–807. 10.1111/dmcn.14831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Santos S.; van Woerden G. M.; Bruinsma C. F.; Mientjes E.; Jolfaei M. A.; Distel B.; Kushner S. A.; Elgersma Y. Ube3a reinstatement identifies distinct developmental windows in a murine Angelman syndrome model. J. Clin. Invest. 2015, 125, 2069. 10.1172/JCI80554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter J. M.; Mao H.; Fragola G.; Simon J. M.; Krantz J. L.; Bazick H. O.; Oztemiz B.; Stein J. L.; Zylka M. J. Cas9 Gene Therapy for Angelman Syndrome Traps Ube3a-ATS Long Non-Coding RNA. Nature 2020, 587 (7833), 281–284. 10.1038/s41586-020-2835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid R. S.; Deng X.; Panikker P.; Msackyi M.; Breton C.; Wilson J. M. CRISPR/Cas9 Directed to the Ube3a Antisense Transcript Improves Angelman Syndrome Phenotype in Mice. J. Clin. Invest. 2021, 131 (5), e142574 10.1172/JCI142574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P.; Gao K.; Wang C.; Pivetti C.; Lankford L.; Farmer D.; Wang A. In Utero Transplantation of Placenta-Derived Mesenchymal Stromal Cells for Potential Fetal Treatment of Hemophilia A. Cell Transplant. 2018, 27 (1), 130–139. 10.1177/0963689717728937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K.; He S.; Kumar P.; Farmer D.; Zhou J.; Wang A. Clonal Isolation of Endothelial Colony-Forming Cells from Early Gestation Chorionic Villi of Human Placenta for Fetal Tissue Regeneration. World J. Stem Cells 2020, 12 (2), 123–138. 10.4252/wjsc.v12.i2.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G. X. Y.; Terry J. M.; Belgrader P.; Ryvkin P.; Bent Z. W.; Wilson R.; Ziraldo S. B.; Wheeler T. D.; McDermott G. P.; Zhu J.; Gregory M. T.; Shuga J.; Montesclaros L.; Underwood J. G.; Masquelier D. A.; Nishimura S. Y.; Schnall-Levin M.; Wyatt P. W.; Hindson C. M.; Bharadwaj R.; Wong A.; Ness K. D.; Beppu L. W.; Deeg H. J.; McFarland C.; Loeb K. R.; Valente W. J.; Ericson N. G.; Stevens E. A.; Radich J. P.; Mikkelsen T. S.; Hindson B. J.; Bielas J. H. Massively Parallel Digital Transcriptional Profiling of Single Cells. Nat. Commun. 2017, 8 (1), 14049. 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A.; Hoffman P.; Smibert P.; Papalexi E.; Satija R. Integrating Single-Cell Transcriptomic Data across Different Conditions, Technologies, and Species. Nat. Biotechnol. 2018, 36 (5), 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E. Z.; Basu A.; Satija R.; Nemesh J.; Shekhar K.; Goldman M.; Tirosh I.; Bialas A. R.; Kamitaki N.; Martersteck E. M.; Trombetta J. J.; Weitz D. A.; Sanes J. R.; Shalek A. K.; Regev A.; McCarroll S. A. Highly Parallel Genome-Wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161 (5), 1202–1214. 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.