Simple Summary
The rapid growth of cancer cells is fuelled by excessive sugar uptake and utilisation. While laboratory experiments modelling cancer cell metabolism primarily focus on glucose, in this paper, we address the unexplored roles of other sugars (fructose, galactose, mannose and xylose) that are heavily abundant in the diet. In this study, we feed cancer cells with stable isotopes of those sugars and, using nuclear magnetic resonance spectroscopy, we assess how they are being metabolised by acute myeloid leukaemia cells. We provide a metabolic map detailing the unique metabolism of each of those sugars in cancer cells and we identify a novel role of galactose in supporting their building blocks’ biosynthesis. We also demonstrate the ability of galactose to modulate cancer cells’ responses to various drugs and chemotherapies. This study highlights the importance of mimicking human dietary compositions when studying cancer cell metabolism in the lab.
Keywords: AML, sugar metabolism, isotope tracing, NMR spectroscopy
Abstract
Metabolism plays a central role in cancer progression. Rewiring glucose metabolism is essential for fulfilling the high energy and biosynthetic demands as well as for the development of drug resistance. Nevertheless, the role of other diet-abundant natural sugars is not fully understood. In this study, we performed a comprehensive 2D NMR spectroscopy tracer-based assay with a panel of 13C-labelled sugars (glucose, fructose, galactose, mannose and xylose). We assigned over 100 NMR signals from metabolites derived from each sugar and mapped them to metabolic pathways, uncovering two novel findings. First, we demonstrated that mannose has a semi-identical metabolic profile to that of glucose with similar label incorporation patterns. Second, next to the known role of fructose in driving one-carbon metabolism, we explained the equally important contribution of galactose to this pathway. Interestingly, we demonstrated that cells growing with either fructose or galactose became less sensitive to certain one-carbon metabolism inhibitors such as 5-Flurouracil and SHIN1. In summary, this study presents the differential metabolism of natural sugars, demonstrating that mannose has a comparable profile to that of glucose. Conversely, galactose and fructose contribute to a greater extent to one-carbon metabolism, which makes them important modulators for inhibitors targeting this pathway. To our knowledge, this is the first NMR study to comprehensively investigate the metabolism of key natural sugars in AML and cancer.
1. Introduction
Cancer metabolism is dysregulated and reprogrammed to fulfil the high energy demands and synthetic needs of cancer cells [1,2,3]. Altered glucose metabolism has been well studied in cancer cells, which convert their glucose to lactate regardless of oxygen levels, a phenomenon known as the Warburg effect [4,5]. These findings led to several preclinical and clinical studies testing new therapies against glucose metabolism [6,7]. Nevertheless, many of those studies did not yield satisfactory clinical results, in part due to the metabolic adaptation of the cancer cells and their ability to utilise alternative nutrients [8].
While glucose is the main sugar used in energy generation, human dietary intake entails several other sugars, including fructose, galactose, xylose, mannose and sucrose [9,10,11]. Several studies have addressed the role of natural sugars in cancer, such as fructose [12,13] and mannose [14,15] in acute myeloid leukaemia (AML). In fact, AML cells were shown to upregulate fructose transporters to enhance their one-carbon metabolism and supply for nucleotide synthesis. Nevertheless, the metabolism of most natural sugars has yet to be fully explored. Understanding these metabolic drivers is particularly important in cancer considering the high adaptability of cancer cells to stress conditions and nutrient scarcity.
In this study, we present a comprehensive NMR-based metabolic map of natural sugars in AML using tracer-based assays. We demonstrate the ability of certain sugars to fully substitute glucose, and we determine novel roles of other sugars in supplementing certain metabolic pathways and modulating drug responses.
2. Materials and Methods
2.1. Cell Culture and Maintenance
Molm13, HEL, MV4-11, THP1 and Kasumi cell lines were obtained from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkultur GmbH, Braunschweig, Germany) and were maintained at 37 °C with 5% CO2. RPMI 1640 (Gibco/ThermoFisher, Waltham, MA, USA) standard medium was supplemented with 10% heat-inactivated foetal bovine serum (FBS) (Sigma Aldrich, Burlington, MA, USA) and 1% P/S (Penicillin-Streptomycin 10.000 U/mL). Cells were grown for at least 14 days before the experiments were performed. Glucose-free RPMI 1640 medium (Gibco) was supplemented with 10% heat-inactivated FBS (foetal bovine serum from Sigma Aldrich) and 1% PS (Penicillin-Streptomycin 10.000 U/mL) as well as 10 mM of the corresponding sugar (glucose, fructose, mannose, galactose, xylose and sucrose obtained from Sigma Aldrich). The cell density was adjusted to between 5 and 8 × 105 cell/mL. The cells were split every two to three days and supplied with fresh medium. No ethics approval was needed for the cell lines experiment (no primary patient materials were used, and no animal experiments were performed).
2.2. Proliferation Assays
AML cell lines were seeded at a density of 4 × 105 cell/mL and grown in the presence of the different sugars (unlabelled). Cells were counted every two days and replated back to the original density in fresh medium for a period of eight days. Cells were counted using Trypan blue on a Countness 3 cell counter (Invitrogen).
2.3. Cell Viability Assays
After cells were grown with the corresponding sugar for a given time, an equal number of cells were plated in 96-well plates (Greiner, Kremsmünster, Austria, (chimney 96-well white plates)). Next, 20 μL Cell Titer-Glo (Promega, Madison, Wisconsin, USA) was added to the cells, briefly mixed then incubated at 37 °C for 10 min before reading luminescence with a Tecan Reader (Infinite M200 Pro, TECAN, Männedorf, Switzerland).
2.4. Consumption Rates
To measure sugar consumption, AML cells were grown with unlabelled sugars at a density of 8 × 105 cells/mL. Twenty-four hours later, media samples were harvested and analysed by NMR as described in the NMR section below.
2.5. 13C Tracer-Based Assay
Glucose-free media (RPMI 1640 from Gibco) were supplemented with 10 mM 13C-labelled sugars (mannose, galactose, xylose, fructose, and glucose) (obtained from Eurisotop), 10% heat-inactivated FBS and 1% PS. Cells were washed then resuspended with the corresponding media at a density of around 8 × 105 cell/mL (an approximate total of 40 million cells by the day of extraction). Cells were grown at 37 °C and 5% CO2. Twenty-four hours later, cells were extracted as described below.
2.6. Metabolites Extraction
On the day of extraction (24 h post-labelling), cells were counted, and 40 million cells were centrifugated at 2000 rpm for 4 min. After discarding the supernatant, the pellet was washed with 1 mL PBS and centrifugated again at 13.000 rpm for 20 s and 4 °C. The supernatant was discarded, and the pellet was lysed with 400 μL methanol and vortexed for 10 s. Samples were then transferred to glass vials and 375 μL distilled water and 400 μL chloroform were added. The samples were vortexed for 40 s, then placed on ice for 10 min. The samples were then centrifugated at 4000 rpm for 10 min at 4 °C in a swingout rotor and samples were placed at room temperature for 10 minutes. The upper polar phase was obtained using a Hamilton syringe and dried in a speed vac at 4 °C.
2.7. Sample Preparation and NMR Measurements
Dried metabolites extracts were resuspended in sodium phosphate buffer (pH 7, 100% D2O and 0.5 mM TMSP. Samples were loaded in the NMR tubes (either 40 µL in 1.7 mm or 190 µL in 3 mm tubes, Bruker LabScape Scream, Billerica, MA, USA). For media samples (consumption assays), 190 µL media were transferred into 3mm NMR tubes. Samples were measured on a 600MHz NEO Bruker spectrometer equipped with a Cryo TCI probe at 298 K. We recorded Spectra with 57,142 time-domain points, 512 scans using the noesygppr1d pulse programme. 1H-13C- Heteronuclear Single Quantum Coherence (HSQC) spectra were recorded using the phase sensitive standard Bruker pulse programme hsqcctetgpsp with 1426 complex points in the direct dimension and 1328 increments in the indirect dimension, 16–64 scans and 50% NUS (non-uniform sampling) at 298 K. For the purpose of validating the chemical shift assignments, HMQC-COSY spectra were recorded using the standard Bruker pulse programme h2bcetgpl3 with 2048 complex points in the direct dimension, 256 increments in the indirect dimension and 128 scans. Spectral assignment was performed with the help of online available metabolites reference spectra from the Human Metabolome Data Base (HMDB) and the Biological Magnetic Resonance Bank (BMRB). All spectra were processed in a Bruker Topspin. The 1D Spectra were analysed using Mestrenova and the 1H-13C-HSQC-Spectra were analysed using nmrfam Sparky 1.470.
2.8. Drug Treatment
AML cell lines were grown with the different sugars (unlabelled) for 6 days to adapt them prior to drug treatments. Cells were then seeded at a density of 2.2 × 105 cell/mL in 96-well plates with the corresponding sugars and treated with different concentrations of the inhibitors (5-Flurouracil and SHIN1 (obtained from Selleckchem, Houston, TA, USA). Cell viability was recorded 72 h later using Cell Titer-Glo as described above. The data analysis and statistical analysis were performed in GraphPad Prism 10.3.1. The experiment was performed in technical triplicates and repeated twice.
3. Results
3.1. AML Cells Grow Differentially When Grown with Different Natural Sugars
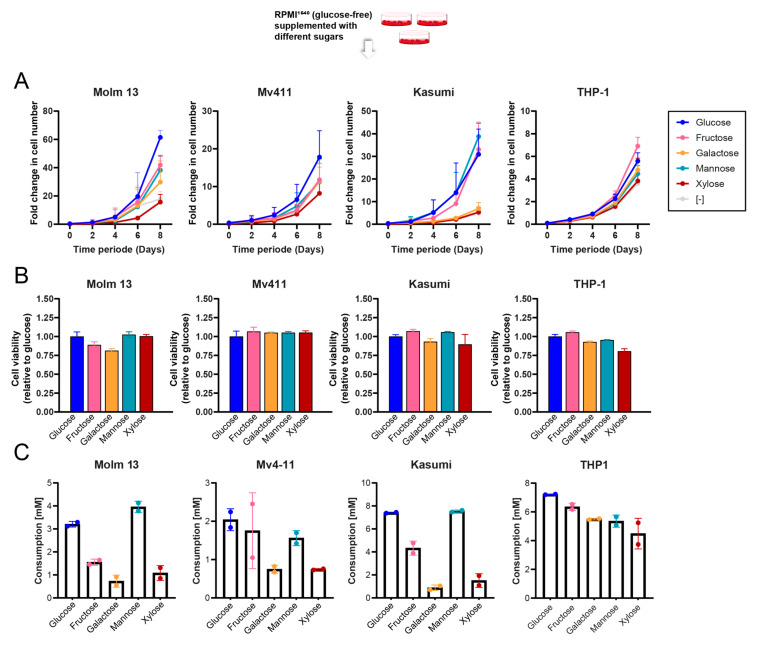
We started by assessing the ability of AML cells to survive on different sugar sources. We grew four AML cell lines (Molm13, Kasumi, MV4-11 and THP1) over a period of 8 days in the presence of either glucose, mannose, fructose, galactose or xylose. Cell lines varied in their ability to grow in the absence of glucose. Generally, most cell lines could grow with mannose and fructose at a comparable rate to that of glucose (Figure 1A). However, when growing under galactose-rich media, the proliferation rates were slower for most cell lines, which is in line with the associated slow metabolism rate of galactose [16]. Finally, cells grown with xylose had the slowest growth pattern observed across all cell lines and cells took longer to adapt and grow after the sugar switch. Nevertheless, when we looked at the ATP levels in the cells eight days after sugar switch, albeit with slight cell line variations, there were only minor differences in ATP content per cell under different sugars (Figure 1B).
Figure 1.
Proliferation and metabolic adaptability of AML cells to natural sugars. (A) Comparisons of growth rates of 4 AML cell lines growing with different sugars over a period of 8 days. (B) Viability of 4 different AML cell lines on day 8 after growing in different sugars relative to day 0. (C) Sugar consumption in 4 different AML cell lines after 24 h of sugar switch (n = 2–3 technical replicates).
Next, we assessed the consumption rates for each of those sugars within 24 h of the sugar switch. This was in line with the proliferation patterns for most cell lines, whereby cells consumed similar amounts of glucose and mannose and slightly less fructose, galactose and xylose (Figure 1C).
3.2. Mapping Sugar Metabolism by 2D NMR Spectroscopy
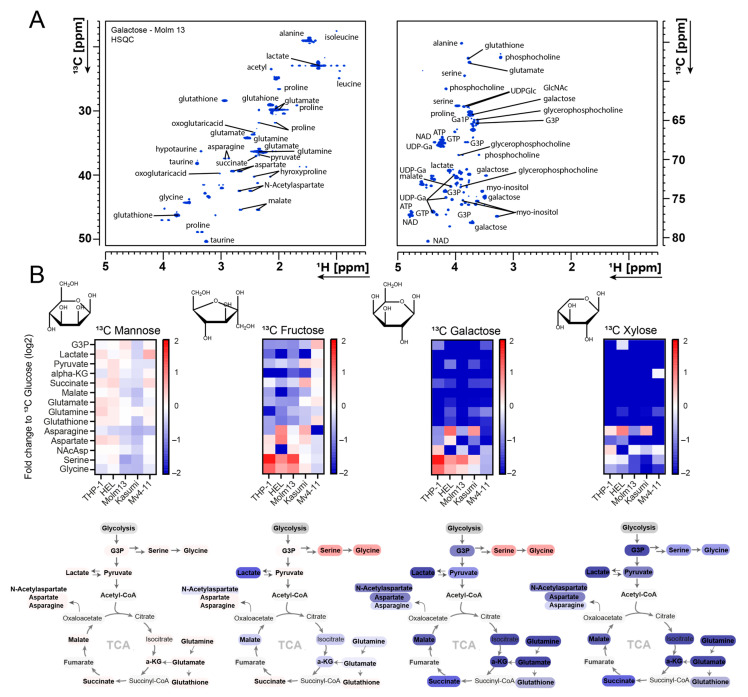
Next, we aimed at assessing how leukaemic cells metabolise different sugars and whether these sugars are substitutable or have different metabolic utilisations. We performed an NMR tracer-based assay using different 13C-labelled sugars in five AML cell lines. Cells were fed with the correspondent 13C-labelled sugar for 24 h, then metabolites were extracted and analysed by NMR spectroscopy. The analysis of the two-dimensional 1H-13C-HSQC spectra using an untargeted approach enabled the assignment and quantification of over 100 signals per spectrum. Figure 2A shows a sample assigned spectrum from Molm13 cells that were fed with 13C galactose.
Figure 2.
NMR metabolic profiling of natural sugars metabolism in AML. (A) 1H-13C 2D-HSQC example NMR spectrum of 13C galactose labelling and spectral assignment in Molm13 cells. Full 1H-13C 2D-HSQC of all sugars for Molm 13 is shown in Supplementary Figure S4. (B) Heat maps displaying fold change in 13C intensity of selected metabolites labelled with different 13C sugars (relative to glucose) in 5 AML cell lines. Full heat map is shown in Supplementary Figure S1. G3P (glycerol-3-phosphate), Ga1P (galactose-1-phosphate), UDP-Glc (UDP-glucose), alpha-KG (alpha-ketoglutaric acid), NAcAsp (N-acetyl aspartate), ATP (Adenosine triphosphate), NAD (nicotinamide adenine dinucleotide), UDP-Ga (uridine diphosphate galactose), GTP (guanosine triphosphate).
Interestingly, mannose-fed cells had an almost identical label incorporation profile to that of glucose with very minor differences between the cell lines (Figure 2B). For instance, we observed similar 13C label incorporation rates into lactate, glutamate, phosphocholine, serine, glutathione and numerous other metabolites.
Overall, 13C intensities arising from other sugars were slightly lower, in line with the slower proliferation patterns observed with the corresponding sugars (fructose flowed by galactose then xylose). We observed relatively low label incorporation from xylose into downstream metabolites compared to other sugars. We could not detect any strong label enrichment pattern in any downstream metabolic pathway in any of the cell lines. While 13C intensities in certain metabolites, such as taurine, myo-inositol and phosphocholine, were comparable to those observed in 13C glucose samples, they were most likely not derived from 13C xylose, and instead were derived from other sources. This is in line with the low proliferative capacity of cells when grown with xylose. Therefore, we did not follow up with xylose and instead focused on fructose and galactose.
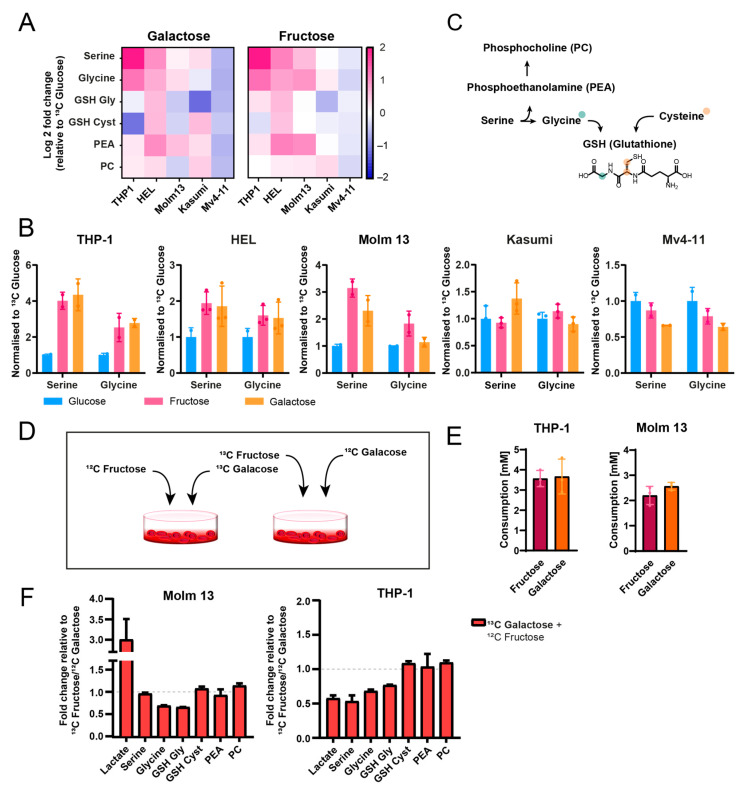
Interestingly, fructose and galactose exhibited similar labelling patterns (except for labelling in their direct metabolic derivatives (i.e., Ga1P and UDP-galactose) but their patterns differed from those of glucose. They both led to strong 13C label enrichment in serine and glycine (Figure 3A,B). While this has been reported previously for fructose [12], we explain here the metabolic utilisation of galactose for feeding the one-carbon metabolism. We only observed this enrichment in Molm13, THP1 and HEL cells, but not in MV4-11 and Kasumi cells, and this was consistent for both fructose and galactose. Interestingly, the fate of this enrichment in glycine and serine varied between the cell lines, whereby HEL cells had a fraction of this label enrichment in serine/glycine transferred to glutathione (originating from serine-derived cysteine, which could not be quantified due to sensitivity limitations), and to phosphocholine synthesis (the latter was also observed for THP1 cells). Nonetheless, Molm13 cells did not show any of this label transfer, suggesting stronger shuttling of the 13C label from serine and glycine towards nucleotide biosynthesis.
Figure 3.
Galactose and fructose support one-carbon metabolism. (A) Heat map of the fold change of 13C label incorporation into a selection of metabolites depicting one-carbon and redox metabolism. (B) Overview of fructose and galactose metabolic fates based on the 13C label incorporation patterns. (C) 13C label incorporation ratio from fructose or galactose into serine and glycine in 5 different AML cell lines. (D) Schematic illustration of the two different settings for the contrast; (i) 12C galactose + 13C fructose or (ii) 13C galactose + 12C fructose. (E) Fructose and galactose consumption in a contrasting metabolic assay. (F) Fold change of 13C label enrichment from the contrasting metabolic assay of galactose and fructose. Full bar plot is shown in Supplementary Figure S3.
Finally, for the cell lines that did not show this pattern (Mv4-11 and Kasumi), we did not see an alternative label incorporation fate, suggesting cell line-specific utilisation of fructose and galactose, which might be associated with certain genetic backgrounds. We evaluated whether we could explain this by assessing the expression levels of enzymes and transporters involved in galactose metabolism in publicly available gene expression data [17]. While we did not see differences between those cell lines in their GALK1 or GALM (both mediate galactose incorporation into glycolysis), we saw 4–5-fold higher expression levels of GLUT14 (galactose transporter) in cells that showed galactose enrichment in the one-carbon metabolism (Molm13, HEL and THP1) compared to cell lines that did not (MV4-11 and Kasumi (Supplementary Figure S2)).
Given that these data suggest that galactose might be playing an equally important role in providing for one-carbon metabolism compared to fructose, we sought to further validate this by comparing the relative contribution of both sugars to one-carbon metabolism in a competitive metabolic assay. We grew Molm13 and THP1 cells in a 1:1 mixture of galactose and fructose (in the absence of glucose) in two different settings; (i) 12C galactose + 13C fructose or (ii) 13C galactose + 12C fructose. Interestingly, while both sugars were taken up at similar rates (Figure 3E), there were some differences between the cell lines, with THP1 exhibiting comparable label incorporation from both sugars into serine and glycine (although slightly lower incorporation from galactose) (Figure 3F). Molm13 had higher enrichment from fructose vs. glucose in serine and glycine. With the exception of significantly higher labelling in lactate in Molm13 (a 3-fold increase), labelling did not differ between the two sugars in glutathione, phosphoethanolamine or phosphocholine.
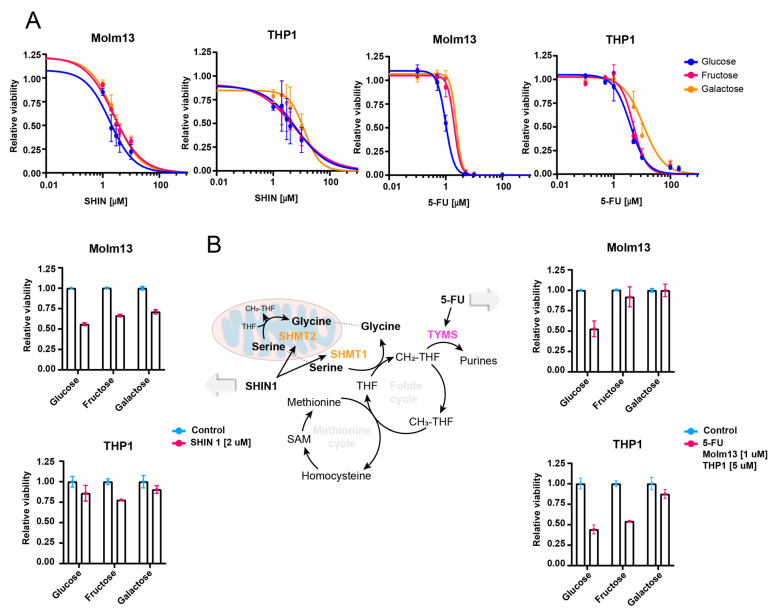
3.3. Galactose Reduces Sensitivity to One-Carbon Metabolism Inhibition
Finally, given the strong shuttling of galactose towards one-carbon metabolism, we aimed at assessing whether cells growing with galactose/fructose would be more resistant to inhibitors targeting the one-carbon metabolism such as 5-Flurouracil (5-FU) and the SHMT1/2 inhibitor SHIN1 [18]. First, we adapted Molm13 and THP1 cells to growing with either glucose, galactose or fructose for 6 days before treatment. Interestingly, we observed lower sensitivity to both inhibitors in cells growing with galactose and fructose compared to those growing with glucose (Figure 4A,B). The pattern was stronger in Molm13 compared to THP1 cells. Although the effect size was not very large, these data further confirm our findings on the role of galactose and fructose in driving the one-carbon metabolism and hence, desensitising cancer cells to inhibitors of this pathway.
Figure 4.
Galactose reduces sensitivity to one-carbon metabolism inhibitors. (A) IC50 curves of Molm13 and THP1 cells treated with SHIN1 and 5-FU. Cells were previously adapted for 6 days’ growth in media containing either glucose, fructose or galactose. Relative survival of Molm13 and THP1 treated with 5-FU or SHIN1 at approximately IC50 concentrations. (B) Schematic displaying the one-carbon metabolism inhibitors [19,20].
4. Discussion
Glucose metabolism has been well studied in cancer. Although some studies have explored fructose [12] and mannose metabolism [14,15], a comprehensive understanding of natural sugars is still lacking. In this study, we performed an NMR tracer-based metabolic analysis and mapped the metabolic fates of most abundant dietary sugars in AML cell lines. To our knowledge, this is the first NMR (or mass spectrometry) study that provides an overall picture of sugars metabolism in cancer.
Our untargeted NMR approach is significantly more comprehensive than most published NMR metabolic screenings in terms of spectral assignment and the number of signals quantified. We achieved a spectral assignment of over 72%. Given the significantly higher reproducibility of NMR compared to mass spectrometry and its non-destructive nature, our study gives a strong boost to NMR spectroscopy [21,22,23] in steady-state stable isotope tracing.
We highlight two main findings in this study: first, the role of galactose (next to fructose) in driving the one-carbon metabolism in the absence of glucose. One-carbon metabolism plays a pivotal role in nucleotide synthesis, methylation, and redox balance, and this is particularly important in cancer cells given their high biosynthetic and redox control demands [24]. Previously, fructose has been reported to be essential for feeding the one-carbon metabolism at a rate that is higher than that of glucose [12]. Here, we reveal an equally important role of galactose in supplying the one-carbon metabolism. We also show in a competitive metabolic assay between galactose and fructose that AML cells comparably rely on and consume galactose to supply one-carbon metabolism, albeit at slightly lower rates in some cell lines. This is presumably due to the slow metabolism of galactose [16], which, in contrast to glucose, does not yield any net ATP when metabolised through glycolysis. This slower metabolism of galactose is also reflected in the slower proliferation of cells grown with galactose. Given all of this (and in spite of the high label incorporation observed in lactate), galactose is unlikely to be essential for energy generation but rather for nucleotide synthesis. This is illustrated in its desensitisation effect to one-carbon metabolism inhibitors. While we did not assess its importance beyond that, labelling in glutathione suggests the importance of galactose for redox metabolites [25] and labelling in phosphocholine suggests a role in lipid metabolism [26].
It is worthy of note that this role of galactose in supplying the one-carbon metabolism is not observed in all AML genetic backgrounds. In fact, our analysis of publicly available gene expression data suggests that this only happens in cell lines with higher expression of the galactose transporter GLUT-14. Indeed, a newly emerging report has demonstrated a correlation between poor disease-free survival and galactose metabolism in breast cancer patients [27]. Moreover, the expression level of GALK1 (the enzyme that converts galactose to galactose-1-phosphate) was shown to correlate with poor prognosis in glioblastoma [28] as well as in colorectal cancer [29].
Importantly, we show in this study that a galactose-rich medium reduces sensitivity to 5-flurouracil and SHMT1 inhibition. This is highly relevant when studying drug sensitivities in vitro where cells are grown in media that lack many molecules that are present in a physiological context (including galactose, which is amongst the most abundant dietary sugars (monomer of lactose)). An example of this is the role of uric acid in driving resistance towards 5-flurouracil, which was only observed when Cantor et al. designed a physiological cell culture that contains uric acid [30].
The second finding of this study was the ability of mannose to substitute glucose as a source for glycolysis and most other metabolic pathways. This potential for substitution was observed not only in metabolic labelling patterns but also in sugar consumption rates, ATP levels and proliferation ability. These results are in line with a recent report on mannose metabolism and its role in driving glycolysis in the absence of glucose [14]. Another report [15] revealed a slightly different role of mannose in supplying fatty acid metabolism via the high expression of the mannose-6-phosphate isomerase (MPI). Inhibition of MPI increased AML cells’ susceptibility to FLT3-inhibitors by increasing ferroptosis. While our study does not address fatty acid metabolism (only polar metabolites were analysed), our data do not contradict that report, given the role of glucose (and presumably mannose) in feeding multiple metabolic pathways, including fatty acid metabolism.
Furthermore, we did not observe any particular labelling pattern for xylose in any downstream metabolic network, which, together with its insufficiency for driving cell proliferation, suggests its insignificance in supporting metabolic networks.
Finally, our results need to be further validated in vivo due to the limitation of cell culture models, particularly in the context of metabolism (the lack of complete physiological nutrient composition and its implications for drug responses and metabolic adaptions).
5. Conclusions
In summary, our study is the first comprehensive NMR metabolic study that maps the metabolism of natural sugars in AML. Our untargeted approach and the nearly full spectral assignment provided useful insights into the differential metabolism of those sugars. We have revealed an essential role of galactose in feeding the one-carbon metabolism and demonstrated its implications for one-carbon metabolism inhibitors. Further work is needed to investigate less abundant dietary sugars and also to explore lipid metabolism.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/cancers16213576/s1. Supplementary Figure S1. Full heatmap showing the fold change of 13C intensity (relative to 13C glucose) in a large panel of metabolites labelled with different 13C sugars in 5 AML cell lines. Supplementary Figure S2. Expression levels of galactose metabolism-related genes. Gene expression data for enzymes and nutrient transporters involved in galactose metabolism (GALM, GALK1, GLUT3 and GLUT14) in 5 AML cell lines. Supplementary Figure S3. Metabolic competition assay of fructose and galactose. Bar plots displaying the 13C intensity of a panel of metabolites labelled with 13C galactose (mixed 1:1 with 12C Fructose) in Molm13 and THP1 cells relative to those labelled with 13C fructose (mixed 1:1 with 12C Galactose. Supplementary Figure S4. 1H-13C 2D-HSQC NMR spectra of 13C labelled sugars (glucose, fructose, mannose, galactose and xylose. An overlaid spectrum of all sugars is also shown.
Author Contributions
C.M. performed the experiments. C.M., I.A. and C.R. helped with experimental design and data analysis. All authors (C.M., I.A., C.R., H.S. (Hubert Serve) and H.S. (Harald Schalbe)) contributed to discussing the project and writing of the manuscript. C.M., I.A. and H.S. (Harald Schwalbe) conceptualised and initiated the project. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable. This study did not involve humans or animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
All supplementary data generated for this work are available online with this manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was supported in part by LOEWE FCI (Frankfurt Cancer Institute) funded by the Hessian Ministry of Higher Education, Research and the Arts [III L 5-519/03/03.001-(0015)] and DKTK (German Consortium for Translational Cancer Research). The work at BMRZ was supported by the state of Hesse.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Finley L.W.S. What Is Cancer Metabolism? Cell. 2023;186:1670–1688. doi: 10.1016/j.cell.2023.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alshamleh I., Kurrle N., Makowka P., Bhayadia R., Kumar R., Süsser S., Seibert M., Ludig D., Wolf S., Koschade S.E., et al. PDP1 Is a Key Metabolic Gatekeeper and Modulator of Drug Resistance in FLT3-ITD-Positive Acute Myeloid Leukemia. Leukemia. 2023;37:2367–2382. doi: 10.1038/s41375-023-02041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker F., Polat I.H., Abou-el-ardat K., Alshamleh I., Thoelken M., Hymon D., Gubas A., Koschade S.E., Vischedyk J.B., Kaulich M., et al. Metabolic Rewiring Is Essential for AML Cell Survival to Overcome Autophagy Inhibition by Loss of ATG3. Cancers. 2021;13:6142. doi: 10.3390/cancers13236142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martínez-Reyes I., Chandel N.S. Cancer Metabolism: Looking Forward. Nat. Rev. Cancer. 2021;21:669–680. doi: 10.1038/s41568-021-00378-6. [DOI] [PubMed] [Google Scholar]
- 5.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stine Z.E., Schug Z.T., Salvino J.M., Dang C.V. Targeting Cancer Metabolism in the Era of Precision Oncology. Nat. Rev. Drug Discov. 2022;21:141–162. doi: 10.1038/s41573-021-00339-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreitz J., Schönfeld C., Seibert M., Stolp V., Alshamleh I., Oellerich T., Steffen B., Schwalbe H., Schnütgen F., Kurrle N., et al. Metabolic Plasticity of Acute Myeloid Leukemia. Cells. 2019;8:805. doi: 10.3390/cells8080805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mishra S.K., Millman S.E., Zhang L. Metabolism in Acute Myeloid Leukemia: Mechanistic Insights and Therapeutic Targets. Blood. 2023;141:1119–1135. doi: 10.1182/blood.2022018092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alam Y.H., Kim R., Jang C. Metabolism and Health Impacts of Dietary Sugars. J. Lipid Atheroscler. 2022;11:20–38. doi: 10.12997/jla.2022.11.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conte F., Noga M.J., van Scherpenzeel M., Veizaj R., Scharn R., Sam J.E., Palumbo C., van den Brandt F.C.A., Freund C., Soares E., et al. Isotopic Tracing of Nucleotide Sugar Metabolism in Human Pluripotent Stem Cells. Cells. 2023;12:1765. doi: 10.3390/cells12131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Laar A., Grootaert C., Van Nieuwerburgh F., Deforce D., Desmet T., Beerens K., Van Camp J. Metabolism and Health Effects of Rare Sugars in a CACO-2/HepG2 Coculture Model. Nutrients. 2022;14:611. doi: 10.3390/nu14030611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong S., Savino A.M., Chirayil R., Barin E., Cheng Y., Park S.M., Schurer A., Mullarky E., Cantley L.C., Kharas M.G., et al. High Fructose Drives the Serine Synthesis Pathway in Acute Myeloid Leukemic Cells. Cell Metab. 2021;33:145–159.e6. doi: 10.1016/j.cmet.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W.L., Wang Y.Y., Zhao A., Xia L., Xie G., Su M., Zhao L., Liu J., Qu C., Wei R., et al. Enhanced Fructose Utilization Mediated by SLC2A5 Is a Unique Metabolic Feature of Acute Myeloid Leukemia with Therapeutic Potential. Cancer Cell. 2016;30:779–791. doi: 10.1016/j.ccell.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito Y., Kinoshita M., Yamada A., Kawano S., Liu H.S., Kamimura S., Nakagawa M., Nagasawa S., Taguchi T., Yamada S., et al. Mannose and Phosphomannose Isomerase Regulate Energy Metabolism under Glucose Starvation in Leukemia. Cancer Sci. 2021;112:4944–4956. doi: 10.1111/cas.15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodley K., Dillingh L.S., Giotopoulos G., Madrigal P., Rattigan K.M., Philippe C., Dembitz V., Magee A.M.S., Asby R., van de Lagemaat L.N., et al. Mannose Metabolism Inhibition Sensitizes Acute Myeloid Leukaemia Cells to Therapy by Driving Ferroptotic Cell Death. Nat. Commun. 2023;14:2132. doi: 10.1038/s41467-023-37652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conte F., van Buuringen N., Voermans N.C., Lefeber D.J. Galactose in Human Metabolism, Glycosylation and Congenital Metabolic Diseases: Time for a Closer Look. Biochim. Biophys. Acta Gen. Subj. 2021;1865:129898. doi: 10.1016/j.bbagen.2021.129898. [DOI] [PubMed] [Google Scholar]
- 17.DepMap, Broad (2024). DepMap 24Q2 Public. Figshare+. Dataset. [(accessed on 19 September 2024)]. Available online: [DOI]
- 18.Sun W., Liu R., Gao X., Lin Z., Tang H., Cui H., Zhao E. Targeting Serine-Glycine-One-Carbon Metabolism as a Vulnerability in Cancers. Biomark. Res. 2023;11:48. doi: 10.1186/s40364-023-00487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pikman Y., Ocasio-Martinez N., Alexe G., Dimitrov B., Kitara S., Diehl F.F., Robichaud A.L., Conway A.S., Ross L., Su A., et al. Targeting Serine Hydroxymethyltransferases 1 and 2 for T-Cell Acute Lymphoblastic Leukemia Therapy. Leukemia. 2022;36:348–360. doi: 10.1038/s41375-021-01361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahmood K., Emadi A. 1-C Metabolism—Serine, Glycine, Folates—In Acute Myeloid Leukemia. Pharmaceuticals. 2021;14:190. doi: 10.3390/ph14030190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wishart D.S., Cheng L.L., Edison A.S., Eghbalnia H.R., Hoch J.C., Gouveia G.J., Pathmasiri W., Powers R., Schock T.B., Sumner L.W., et al. NMR and Metabolomics—A Roadmap for the Future. Metabolites. 2022;12:678. doi: 10.3390/metabo12080678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrigan J.B., Reed M.A.C., Ludwig C., Khanim F.L., Bunce C.M., Günther U.L. Tracer-Based Metabolic NMR-Based Flux Analysis in a Leukaemia Cell Line. Chempluschem. 2016;81:453–459. doi: 10.1002/cplu.201500549. [DOI] [PubMed] [Google Scholar]
- 23.Alshamleh I., Krause N., Richter C., Kurrle N., Serve H., Günther U.L., Schwalbe H. Real-Time NMR Spectroscopy for Studying Metabolism. Angew. Chem.—Int. Ed. 2020;59:2304–2308. doi: 10.1002/anie.201912919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenzweig A., Blenis J., Gomes A.P. Beyond the Warburg Effect: How Do Cancer Cells Regulate One-Carbon Metabolism? Front. Cell Dev. Biol. 2018;6:402976. doi: 10.3389/fcell.2018.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerksick C., Willoughby D. The Antioxidant Role of Glutathione and N-Acetyl-Cysteine Supplements and Exercise-Induced Oxidative Stress. J. Int. Soc. Sports Nutr. 2005;2:38. doi: 10.1186/1550-2783-2-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Veen J.N., Kennelly J.P., Wan S., Vance J.E., Vance D.E., Jacobs R.L. The Critical Role of Phosphatidylcholine and Phosphatidylethanolamine Metabolism in Health and Disease. Biochim. Biophys. Acta-Biomembr. 2017;1859:1558–1572. doi: 10.1016/j.bbamem.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Han X., Han B., Luo H., Ling H., Hu X. Integrated Multi-Omics Profiling of Young Breast Cancer Patients Reveals a Correlation between Galactose Metabolism Pathway and Poor Disease-Free Survival. Cancers. 2023;15:4637. doi: 10.3390/cancers15184637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharpe M.A., Ijare O.B., Baskin D.S., Baskin A.M., Baskin B.N., Pichumani K. The Leloir Cycle in Glioblastoma: Galactose Scavenging and Metabolic Remodeling. Cancers. 2021;13:1815. doi: 10.3390/cancers13081815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu G., Wu X., Chen J. Identification and Validation of a Glycolysis-Related Gene Signature for Depicting Clinical Characteristics and Its Relationship with Tumor Immunity in Patients with Colon Cancer. Aging. 2022;14:8700. doi: 10.18632/aging.204226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cantor J.R., Abu-Remaileh M., Kanarek N., Freinkman E., Gao X., Louissaint A., Lewis C.A., Sabatini D.M. Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell. 2017;169:258–272.e17. doi: 10.1016/j.cell.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All supplementary data generated for this work are available online with this manuscript.