Abstract
Glioblastoma (GBM) is the most lethal and common primary tumor of central nervous system with a poor prognosis. Glioma stem cells (GSCs) are particularly significant in GBM proliferation, invasion, self-renewal and recurrence. Circular RNAs (circRNAs) play important roles in various physiological and pathological processes, including regulating the biological behavior of GBM. Therefore, discovering novel circRNAs related to GSCs may contribute to a promising approach for treatment of GBM. Herein, we find out a novel circRNA termed circZEB1 with a high expression in glioma. Limb-bud and heart (LBH) is a transcription cofactor and promotes glioma stem cell tumorigenicity in our study. Mechanistically, circZEB1 can upregulate the expression of transcription cofactor LBH via sponging miR-128-3p in GSCs. LBH can facilitate the expression of tumor necrosis factor-α (TNF-α), thus activating the NF-κB signaling pathway to promote the glioma progression. Meanwhile, LBH can also upregulate the RNA binding protein Fused in Sarcoma (FUS) expression, which can bind to and maintain the stability of circZEB1. A positive feedback loop is formed among FUS, circZEB1, miR-128-3p and LBH in GSCs. Our study uncovers a critical role of circZEB1 and provides a novel biomarker for treating GBM.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-024-03526-8.
Keywords: Glioblastoma, CircZEB1, Glioma stem cells, Limb-bud and heart, NF-κB signaling pathway
Background
Glioblastoma (GBM) is a common and highly aggressive primary brain tumor [1]. GBM patients have a poor prognosis with the median survival time of less than 15 months, even though surgical resection, radiotherapy and chemotherapy are applied for patient outcomes [2, 3]. Glioma stem cells (GSCs) are a vital kind of tumor cells in GBM with various functions, including tumor proliferation, invasion, self-renewal and chemoradiotherapy resistance [4, 5]. Therefore, to explore novel oncogenes associated with GSCs may provide potential therapeutic strategies for GBM patients [6].
Circular RNAs (circRNAs) are a kind of noncoding RNAs derived from RNA back splicing, characterized as closed-loop structures [7, 8]. More and more studies reveal that circRNAs participate in the pathology of cancers via sponging micro RNAs (miRNAs) [9–11]. Briefly, circRNAs can bind to miRNAs response elements and regulate corresponding gene expression. Therefore, finding out novel circRNAs related to GSCs is of great significance to provide reliable biomarkers and alternative targets for GBM [12]. RNA-binding proteins (RBPs) can bind to and maintain the stability of RNA, thus regulating various processes in cancers [13, 14]. Fused in Sarcoma (FUS), one of the members of the FET (FUS/EWS/TAF15) protein family, has been reported to take part in regulating oncogenes in a series of cancers [15, 16]. A recent study has proved that FUS could bind to and maintain the stability of circEZH2, thus facilitating the epithelial and mesenchymal transition of breast cancer [17]. Another study revealed FUS could promote FGFR1 transcription and facilitate prostate cancer progression [18]. Therefore, the functions of FUS in GSCs are worthy of further exploration.
Limb-bud and heart (LBH), a transcription cofactor, participates in tumorigenesis and tumor progression in kinds of cancers [19, 20]. Our previous study indicated LBH could promote angiogenesis in glioma under hypoxia [21]. Moreover, studies proved that LBH could suppress progression and angiogenesis [22]. However, very little is known regarding whether LBH contributes to malignant progression in GSCs.
In this work, we uncover a novel circRNA termed circZEB1 that promotes the transcription cofactor LBH expression via sponging miR-128-3p in GSCs. Then, LBH can promote malignant phenotype of GBM by upregulating TNF-α expression and activating NF-κB signaling pathway. Besides, LBH can promote FUS expression, which can bind to and maintain the stability of circZEB1. This study reveals a novel feedback loop and may provide new strategies for GBM therapy.
Materials and methods
Patient sample and ethics approval
A total of 70 glioma tissue samples were collected from glioma patients who underwent surgery in the First Affiliated Hospital of China Medical University from January 2015 to October 2019. Among them, according to the World Health Organization grading guidelines, there were 20 cases of glioma grade II, 25 cases of grade III, and 25 cases of grade IV. In addition, we also collected adjacent normal brain tissue (NBT) samples from 10 GBM patients as a normal control group. More detailed information is presented in Table 1. All participants signed a written informed consent, and this study was approved by the Ethics Committee of the First Affiliated Hospital of China Medical University.
Table 1.
Relationship of circZEB1 expression to clinical features of glioma patients
| Clinical features | Samples (n = 70) |
CircZEB1 expressiona | P-value | |
|---|---|---|---|---|
| Low (n = 35) | High (n = 35) | |||
| Sex | ||||
| Male | 37 | 18 | 19 | P = 0.623 |
| Female | 33 | 17 | 16 | |
| Age | ||||
| ≤ 50 | 22 | 10 | 12 | P = 0.371 |
| > 50 | 48 | 25 | 23 | |
| WHO grade | ||||
| II | 20 | 15 | 5 | P < 0.001 |
| III | 25 | 13 | 12 | |
| IV | 25 | 7 | 18 | |
| IDH status | ||||
| Wild | 52 | 20 | 32 | P = 0.017 |
| Mutant | 18 | 15 | 3 | |
| 1p/19q status | ||||
| Codeletion | 12 | 8 | 4 | P = 0.019 |
| Non-codeletion | 58 | 27 | 31 | |
| H3F3A status | ||||
| Wild | 18 | 13 | 5 | P = 0.033 |
| Mutant | 52 | 22 | 30 | |
| MGMT status | ||||
| Methylation | 43 | 28 | 15 | P = 0.014 |
| Unmethylation | 27 | 7 | 20 | |
The high expression of circZEB1 was defined as the expression level higher than the median expression level of circZEB1
aCircZEB1 expression was detected by qRT-PCR and ranked from low to high
Cell culture and GSCs isolation
Primary glioma stem cells (GSCs) were isolated from 6 patients derived WHO grade IV specimens (GSC103, GSC107, GSC108, GSC109, GSC111 and GSC112), and the clinical data of 6 GBM specimens are detailed in Table S1. Briefly, freshly excised GBM tissue was digested and isolated into single cells using type IV collagenase, and then free cells were cultivated in serum-free DMEM with 2% B27, 20 ng/mL recombinant human (rh) basic fibroblast growth factor (rh-FGF) and 20 ng/mL rh-epidermal growth factor (rh-EGF) (Gibco, Gaithersburg, MD, USA) at 37 °C, 5% CO2 atmosphere for two weeks. All GSCs have passed mycoplasma and short tandem repeat (STR) DNA profiling test. All GSCs used in this study were cultivated for less than 20 passages for experiments.
Construction and transfection of lentiviral vector
The overexpression of circZEB1, LBH and FUS was constructed by the Gene-Chem (Shanghai, China) lentivirus-based vectors, and the silencing of circZEB1, LBH and FUS using RNAi-mediated lentiviral vectors (Gene-Chem). MiR-128-3p mimics, inhibitors, and negative controls were obtained by Thermo Fisher Scientific (Thermo Fisher Scientific, Waltham, MA, USA). The lentivirus transfection efficacy was validated by qPCR or western blotting. All siRNA sequences are shown in Table S2.
qRT-PCR (real-time quantitative reverse transcription PCR)
Total RNAs from GSCs or tissues were extracted using the MiniBEST Universal RNA Extraction Kit (TaKaRa, Kyoto, Japan). The amount and quality of isolated RNAs were tested with Nanodrop (Thermo Fisher Scientific). Construct cDNA libraries with Prime-Scrip RT reagent kit (TaKaRa, Shiga, Japan). The circRNA and mRNA were reverse transcribed, and then the SYBR Green Master Mix (TaKaRa) was performed using PCR LightCycler480 (Roche Diagnostics, Basel, Switzerland). MiR-128-3p expression levels were measured using TaqMan Universal Master Mix II (Applied Biosystems, Foster City, CA, USA). The β-actin was used as an endogenous control. The primers used here are shown in Table S3.
RNase R assay
RNase R can degrade the activity of linear RNA but is ineffective against circular RNA. RNase R assay was used to determine the circular structure of circular RNA. Incubate 2 μg total RNA with 5U/μg RNase R (Epicentre Technologies, Madison, WI, USA) for 30 min at 37 °C. The expression of Linear RNAs and circular RNAs were then detected by qRT-PCR.
Western blotting
Total cell protein extraction kit (KeyGen Biotechnology, Nanjing, China) was used to extract total GSCs protein. Protein quantification and denaturation, SDS-PAGE electrophoresis, followed by transfer of protein to PVDF membrane, blocked with 2% bovine serum albumin (Beyotime Biotechnology, Beijing, China), with a primary antibody against the protein of LBH, TNFα or stem markers (Abcam Technology, Cambridge, UK) at 4 °C overnight. The bands were then incubated with the secondary antibody for 1 h at room temperature, and the bands were detected with a chemiluminescent ECL kit (ProteinTech, Chicago, Illinois, USA). All results were quantified by ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Immunohistochemistry (IHC)
Paraffin-embedded sections of tumor specimens were treated with primary antibody LBH (1:100; Abcam), FUS(1:100; Abcam) and Ki-67 (1:100; Abcam). The sections are then processed with an IHC labeling kit (MaxVision Biotechnology, Fuzhou, China) and photographed with a light microscope (Olympus, Tokyo, Japan). German immunohistochemistry score (GIS) was used to evaluate staining intensity and expression level.
Immunofluorescence (IF)
Cells are incubated with the primary antibody overnight at 4 °C. Fluorescent probe-conjugated secondary antibodies were then added, incubated for 1 h at room temperature, and DAPI solution was used for nuclear counterstaining (SigmaAldrich, St. Louis, MO, USA). Finally, the results of the experiment were visualized with a confocal laser scanning microscope (Olympus).
Cell viability assays
GSCs were seeded into 96-well plates at a density of 1000 cells/well and cultured for 0, 24, 48, 72, 96, and 120 h, respectively. Cell viability was measured with the MTS assay kit (Promega, Madison, WI, USA) according to the manufacturer’s recommendations.
Transwell assay
Due to the suspension neurospheres of GSCs, we firstly digested spherical GSCs to individual cells before performing invasion assays. The 3 × 104 GSCs were seeded into the upper chamber (Corning, Corning, NY, USA) and treated with a Matrigel filter (BD Biosciences, San Jose, CA, USA). The medium in the lower chamber was treated with 20% fetal bovine serum. Then we used 4% paraformaldehyde to fix the invasion cells after 20 h incubation. Crystal violet (Beyotime, Biotechnology) was used to stain the cells, and visualized by a light microscope (Olympus).
5-Ethynyl-20-deoxyuridine (EDU) assay
Due to the suspension neurospheres of GSCs, we firstly digested spherical GSCs to individual cells before performing Edu assays. Then GSCs are seeded into 24-well plates at 1 × 105 cells/well for 20 h. Perform EDU staining with the EDU Assay Kit (KeyGen) according to the manufacturer’s recommendations. GSCs were photographed by laser scanning confocal microscopy (Olympus) and the percentage of edu-positive cells was calculated.
Neurosphere formation assay
GSCs were seeded in 24-well plates at 200 cells/well, cultured in a fresh medium for 7 days, and after neurosphere formation, photographed under a light microscope (Olympus) to calculate the relative size of the neurospheres.
In vitro limiting dilution assay
The GSCs were seeded in 96-well plates based on 1, 10, 20, 30, 40, 50 cells/well, and each density was replicated for 10 times. Then we could count the neurospheres number and figure out the neurosphere formation efficiency by the Extreme Limiting Dilution Analysis (http://bioinf.wehi.edu.au/software/elda) after 7 days.
Luciferase reporter assay
GSCs are seeded into 96-well plates at a density of 5000 cells/well per well, and luciferase reporter plasmids (circZEB1-wt and circZEB1-mt, LBH-3′-UTR-wt and LBH-3′-UTR-mt) are constructed with Gene-Chem. GSCs are co-transfected with luciferase reporter plasmids and incubated for 48 h. Cells are then lysed and tested for luciferase and renal luciferase activity with a dual luciferase reporter assay system (Promega, USA) according to the manufacturer’s instructions.
Chromatin immunoprecipitation (ChIP) assays
Perform chip analysis using the chip analysis kit (Beyotime Biotechnology) according to the manufacturer’s instructions. Chromatin complexes are immunoprecipitated by anti-LBH antibody or normal rabbit IgG (Abcam). The immunoprecipitated DNA was then extracted and purified by qPCR. Primers for ChIP are shown in Table S3.
RNA immunoprecipitation (RIP) assay
RIP assays were performed using the EZ-Magna RIP RNA-binding Protein Immunoprecipitation Kit (Millipore, Darmstadt, Germany). GSCs are lysed in RIP buffer and incubated with magnetic beads conjugated to anti-LBH antibody, anti-FUS antibody, or negative control IgG (Abcam). The RNA was isolated by treating the immunoprecipitated protein-RNAs complex with proteinase K and then purified. The expression of circZEB1 in the precipitants was detected by qPCR.
RNA pull-down assay
The interaction between circZEB1 and LBH was examined using the RNA Protein pull-down Kit (Thermo Fisher Scientific) according to the manufacturer’s guidelines. Briefly, biotinylated circZEB1 or antisense RNA is co-incubated with GSCs and magnetic beading extracts. Then, the protein is washed, purified, and examined with Western blotting. β-actin was used as a control.
RNA stability detection
Block the formation of denovo RNA from GSCs with 2 μg/ml actinomycin D (Sigma-Aldrich). Total RNA was extracted at 12, 24, 36, 48 and 60 h, and the expression of circZEB1 was detected by qRT-PCR. After treatment of actinomycin D, the circZEB1 half-life as 50% RNA levels can be calculated.
Xenograft experiments
Xenograft experiments were performed as described before. Female BALB/c nude mice, 6 weeks old, were purchased from Beijing Weishenghe Laboratory Animal Technology Co., Ltd. (Beijing, China), and all mice were housed under specific sterile conditions at the Laboratory Animal Center of China Medical University. All animal experiments were approved by the Animal Care Committee of China Medical University. The ethics number was provided by Institutional Animal Care and Use Committee (IACUC). IACUC Issue No.2022083. The GSCs with stable knockdown or overexpression of circZEB1, LBH or circZEB1 overexpression combined with LBH knockdown were injected (5 × 104 cells per mouse) in situ by a stereotactic apparatus (n = 5, per group) into the mouse skull, at the lateral 2 mm and the anterior 2 mm to the junction of the coronal and sagittal lines of the mouse skull. The survival time of mice in each group was recorded. Finally, mouse brain tissue was removed immediately on the day of death for H&E staining and immunohistochemistry (IHC) staining. Tumor volume was calculated according to the formula: V = (L × W2) (V = tumor volume, L = the longest diameter of tumor, and W = the shortest diameter of tumor). Survival analysis was calculated by Kaplan–Meier curve.
Bioinformatics analysis
The expression data of circRNA in glioma were obtained from the GEO (Gene Expression Omnibus) dataset. Based on the Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov) and the Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn) datasets, the mRNA expression, WHO grade, isocitrate dehydrogenase (IDH) status (IDH 1/2) of LBH, survival time and status of glioma patients of can be obtained. Then, Gene-set enrichment analysis (GSEA, http://www.broadinstitute.org/gsea/index.jsp) and GSVA were used to detect the enrichment of biological processes or signaling pathways with high and low expression of LBH. TargetScan (https://www.targetscan.org), miRwalk (http://mirwalk.umm.uni-heidelberg.de), and miRDB (https://mirdb.org) were used to predict possible miRNAs targeting LBH. Starbase (https://rnasysu.com/encori/) and circBase (www.circbase.org) to predict possible circular RNAs as miRNA sponges. Besides, Starbase and RBPmap (https://rbpmap.technion.ac.il/) databases were used to predict the proteins binding to circular RNAs.
Statistical analysis
All experiments were repeated at least three times, and results were expressed as mean ± SD. The t-test, chi-square test or one-way ANOVA were used for statistical significance comparison between groups. Pearson correlation analysis was used to analyze the correlation between the two groups. Survival differences were assessed using log-rank test and Kaplan–Meier analysis. SPSS 23.0 software (IBM, Armonk, NY, USA) or GraphPad Prism 8.0 software (GraphPad software Inc, San Diego, C.A, USA) were used for statistical analysis, and the difference was statistically significant when the two-tailed P value was < 0.05.
Results
CircZEB1 was upregulated in glioblastoma tissues and correlated with the progression and poor prognosis
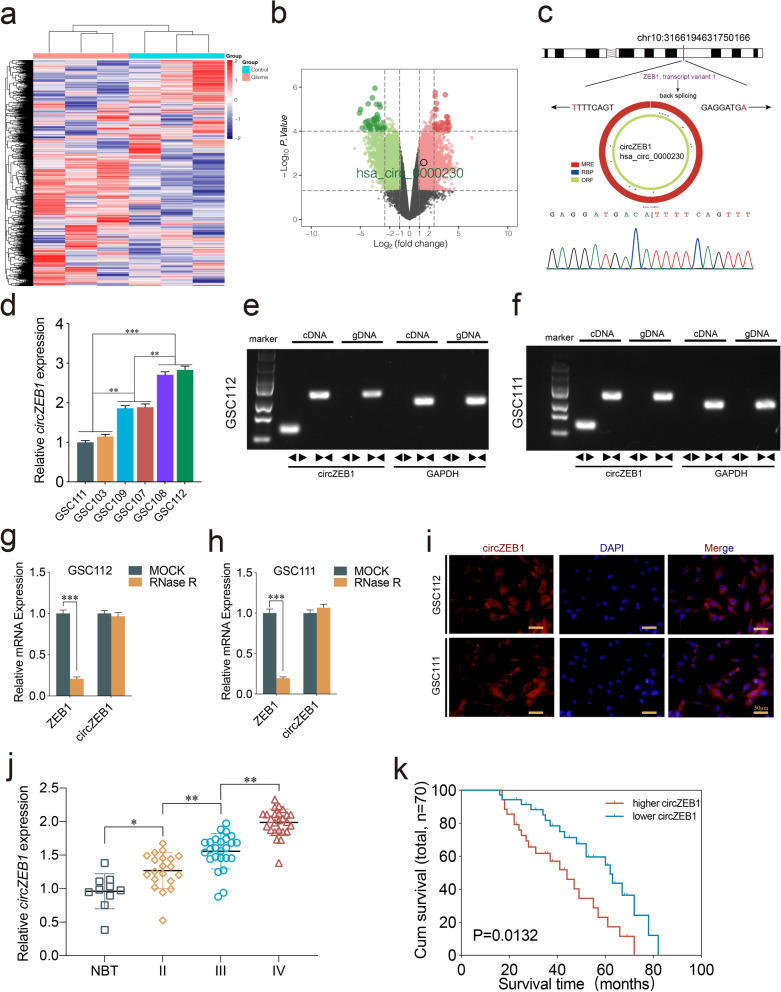
To identify novel and differentially expressed circRNAs in GBM, we conducted analysis about circRNA gene chip GSE109569 from GEO database. Utilizing limma for differential gene analysis, we identified 2871 downregulated circRNAs and 3377 upregulated circRNAs. All upregulated and downregulated circRNAs have been organized in Supplementary Fig. 4. The revised heatmap was presented in Fig. 1a. In our previous research, ZEB1 has emerged as a pivotal transcription factor in gliomas, playing a crucial role in promoting glioma cell invasion and metastasis, glioma oncogenesis and progression, as well as regulating cell differentiation and migration. Notably, there have been no reported studies investigating the role of circRNAs derived from the ZEB1 gene in gliomas. Consequently, we delved deeper into exploring ZEB1-derived circRNAs. Among this vast array of circRNAs, we focused on 27 ZEB1-derived circRNAs for detection and prioritization. We have compiled all circRNAs originating from ZEB1 in Table S5. Ultimately, we uncovered the top five most significantly upregulated circRNAs, including hsa_circ_0048099, hsa_circ_0000230, hsa_circ_0048096, hsa_circ_0051866 and hsa_circ_0044459. Then we performed qRT-PCR using clinical specimen tissues and found that hsa_circ_0000230 (circZEB1) was most upregulated in GBM (Fig. S1a). The volcano plots showed circZEB1 was significantly upregulated in GBM (Fig. 1b). Derived from the ZEB1 gene locus and generated from its exon 9, the sequence of circZEB1 was further validated by Sanger sequencing (Fig. 1c). We cultured six GBM patient derived GSCs (GSC103, GSC107, GSC108, GSC109, GSC111, GSC112) and detected the circZEB1 expression using qPCR. Our results showed the highest circZEB1 expression was in GSC112 and GSC108, while it was in GSC111 and GSC103 with lowest circZEB1 expression (Fig. 1d). Therefore, we performed functional assays using GSC108 and GSC112 as circZEB1 knockdown group, while GSC111 and GSC103 were used as circZEB1 overexpression group. Then, we performed agarose gel electrophoresis assays and found that circZEB1 was only amplified from cDNA in GSC112 and GSC111 (Fig. 1e, f). The RNase R assays revealed that circZEB1 was resistant to RNase R treatment, while the linear RNAs ZEB1 were dramatically decreased (Fig. 1g, h). Moreover, FISH assays proved that circZEB1 was primarily localized in the cytoplasm in GSCs (Fig. 1i). The qPCR results demonstrated that circZEB1 expression was much higher in GBM than normal brain tissues, and the expression increased with higher tumor grade (Fig. 1j). Besides, the Kaplan–Meier survival analysis proved that patients with higher circZBE1 expression had shorter survival time than those with lower circZEB1 expression (Fig. 1k). Taken together, circZEB1 was a significant upregulated circRNA in GBM with the poor prognosis in GBM patients.
Fig. 1.
CircZEB1 was upregulated in glioblastoma tissues and correlated with the progression and poor prognosis. a, b Heatmap (a) and Volcano plots (b) displaying the differentially expressed circRNAs between normal brain tissue and GBM. c Schematic diagram depicting circZEB1 genomics information and results of Sanger sequencing. d qPCR showing expression of circZEB1 in GSCs derived from six patients. e, f Agarose gel electropphoresis of qPCR on circZEB1 expression in GSC112 and GSC111. GAPDH was used as a linear control. g, h The expression of ZEB1 and circZEB1 after RNase R treatment. i FISH assays showing the localization of circZEB1 in GSCs. Scale bar = 50 μm. j circZEB1 expression in different glioma grades and normal brain tissues. k Kaplan–Meier survival curve showing survival rates of patients with high or low circZEB1 expression. All data were expressed as mean ± SD, and each experiment was performed in triplicate. *p < 0.05; **p < 0.01; ***p < 0.001
CircZEB1 promoted malignant phenotype and stemness of GSCs in vitro
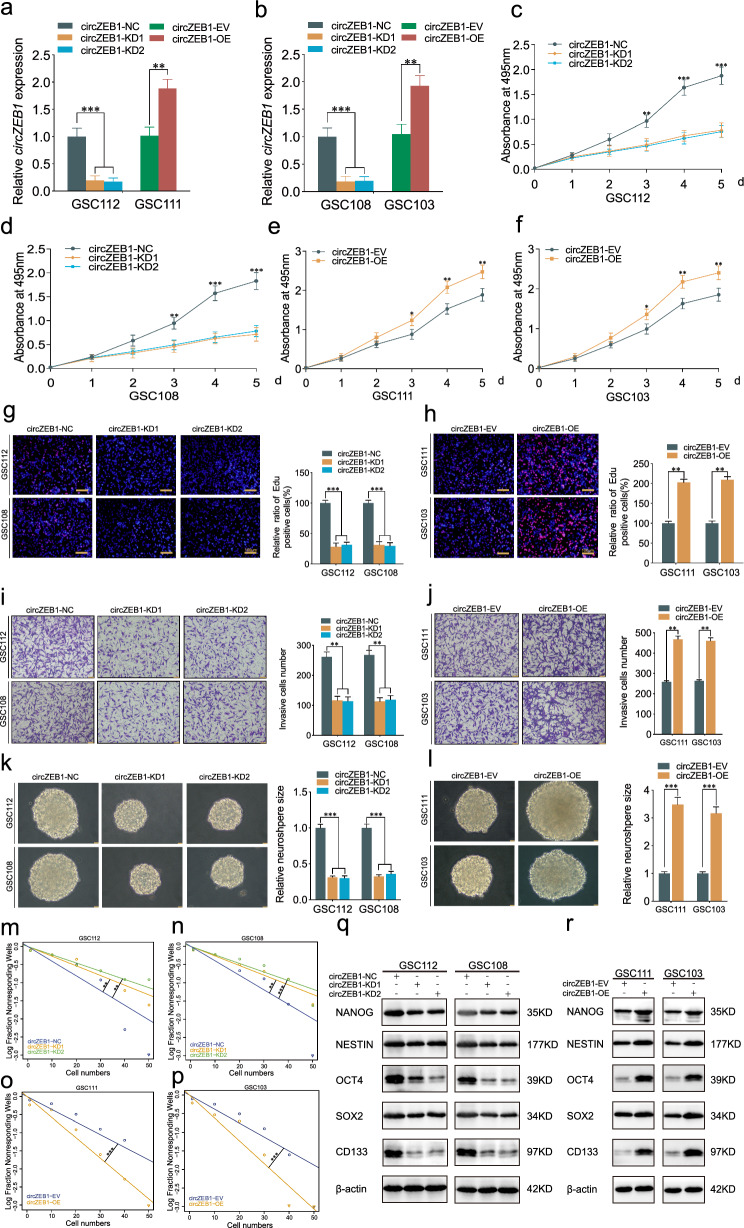
We verified the efficiency of circZEB1 knockdown and overexpression by qPCR detection (Fig. 2a, b). Then, MTS assays revealed the GSCs viabilities were decreased after circZEB1 knockdown, while increased after circZEB1 overexpression (Fig. 2c–f). Edu assays showed circZEB1 knockdown inhibited the ratio of Edu positive cells, which was promoted by circZEB1 overexpression in GSCs (Fig. 2g, h). Moreover, transwell assays proved the GSCs invasion capacity was restrained after circZEB1 knockdown but strengthened after circZEB1 overexpression (Fig. 2i, j). Besides, we performed neurosphere formation assays and ELDA assays to detect the self-renewing ability of GSCs. The neurosphere formation and self-renewing capacities were inhibited after circZEB1 knockdown in GSC108 and GSC112, while obviously promoted after circZEB1 overexpression in GSC103 and GSC111 (Fig. 2k–p). In order to better demonstrate the capability of neurosphere formation, we have provided multiple fields of sphere in supplementary Fig. 4. Our results revealed the number of neurospheres formed under multiple fields of view was significantly reduced after circZEB1 knockdown, while when circZEB1 was overexpressed, the number of neurospheres formed under multiple fields of view is significantly increased. We further conducted western blotting assays to detect the stemness of GSCs through stemness markers, including NANOG, NESTIN, OCT4, SOX2 and CD133. Our results demostrated these stemness markers observably decreased after circZEB1 knockdown, while increased after circZEB1 overexpression (Fig. 2q, r). Taken together, we concluded that circZEB1 promoted malignant phenotype and stemness of GSCs.
Fig. 2.
CircZEB1 promoted malignant phenotype and stemness of GSCs in vitro. a, b The expression of circZEB1 in GSCs after lentiviral-based transfection. c–f MTS assays showing cell viabilities in GSCs after circZEB1 knockdown or overexpression. g, h Edu assyas displaying the proliferation of GSCs after circZEB1 knockdown or overexpression. i, j Transwell assays depicting the invasion abilities of GSCs after circZEB1 knockdown or overexpression. k–p Neurosphere formation and ELDA assays showing the neurosphere formation abilities (k, l) or the GSCs self-renewing capacities (m–p) after circZEB1 knockdown or overexpression. q, r Western blotting detecting the stemness markers expression after circZEB1 knockdown or overexpression. All data were expressed as mean ± SD, and each experiment was performed in triplicate. *p < 0.05; **p < 0.01; ***p < 0.001
LBH promoted malignant phenotype and stemness of GSCs in vitro
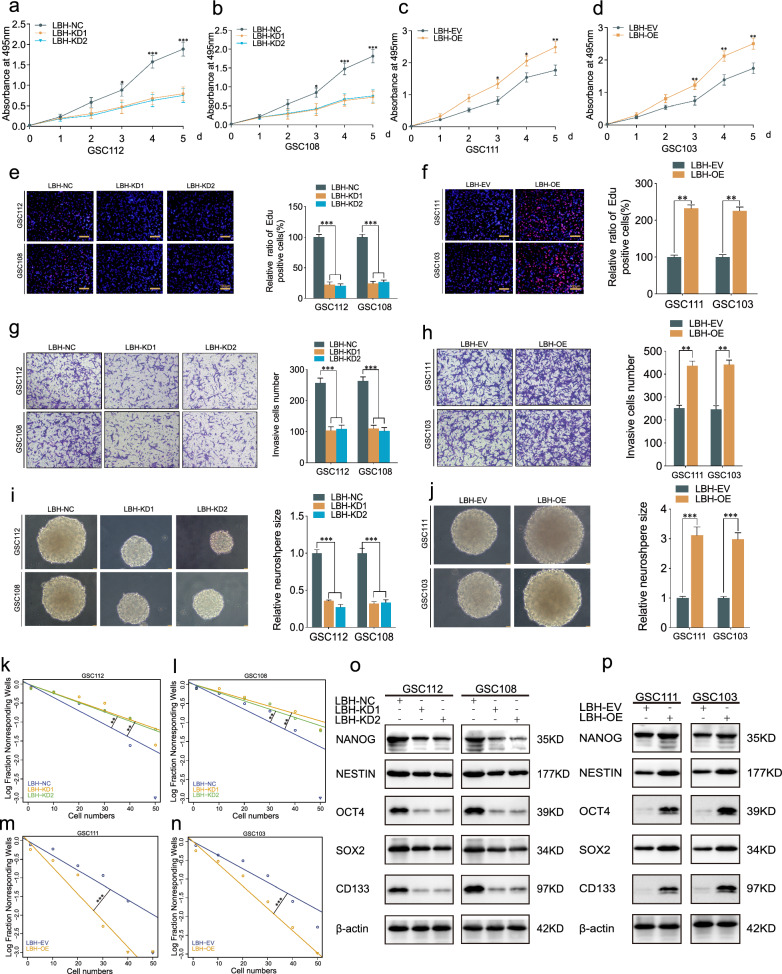
Our previous study proved that LBH promoted angiogenesis in glioma, but the role of LBH in GSCs malignant phenotype and stemness needed to be further explored [21]. QPCR and western blotting assays were carried out to detect the lentiviral-based transfection efficiency of LBH (Fig. S1b–d). Our results of MTS (Fig. 3a–d), Edu (Fig. 3e, f) and transwell assays (Fig. 3g, h) revealed that the cell viability, proliferation ability and invasion capacity were decreased after LBH knockdown in GSC112 and GSC108, while increased after LBH overexpression in GSC111 and GSC103. Besides, we conducted neurosphere formation and ELDA assays to detect GSCs self-renewing abilities. Our results demonstrated that the size of neurosphere was obviously restrained after LBH knockdown, while promoted after LBH overexpression (Fig. 3i–n). Moreover, western blotting assays showed that the GSCs stemness markers were inhibited after LBH knockdown, but facilitated after LBH overexpression (Fig. 3o, p). Therefore, we concluded that LBH significantly promoted malignant phenotype and stemness of GSCs.
Fig. 3.
LBH promoted malignant phenotype and stemness of GSCs in vitro a-d MTS assays displaying GSCs viabilities after LBH knockdown (a, b) or overexpression (c, d). e–h Edu and transwell assays showing the GSCs proliferation (e, f) and invasion (g, h) capacities after LBH knockdown or overexpression. i–n Neurosphere formation and ELDA assays indicating the GSCs neurosphere formation (i, j) and self-renewing (k–n) capacities after LBH knockdown or overexpression. o, p Western blotting detecting the stemness markers expression after LBH knockdown or expression. All data were expressed as mean ± SD, and each experiment was performed in triplicate. *p < 0.05; **p < 0.01; ***p < 0.001
LBH promoted TNF-α transcription and led to NF-κB signaling pathway activation
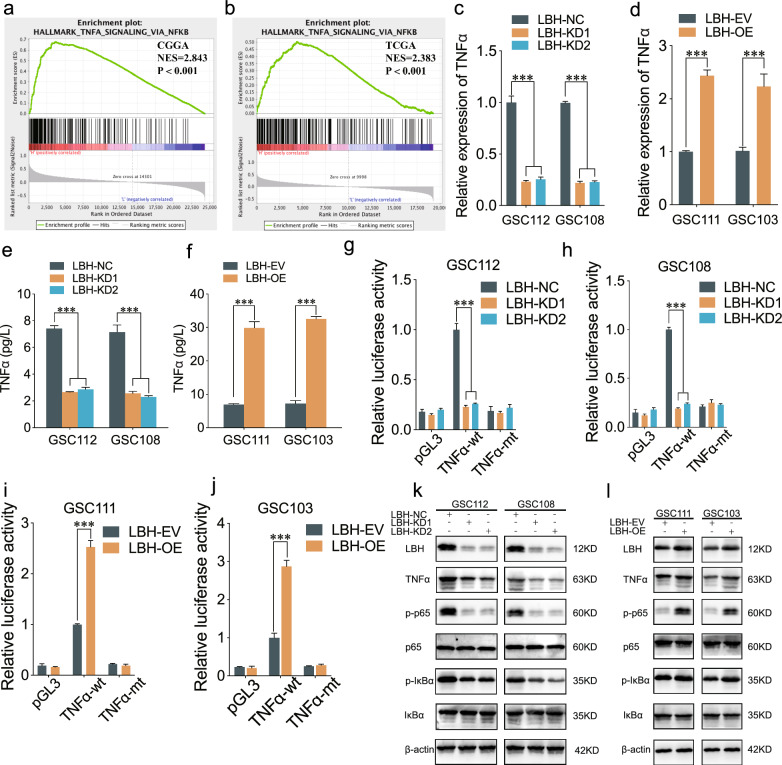
To further explore the exact underlying mechanism of LBH in GSCs, we performed GSEA analysis based on CGGA and TCGA datasets. We found TNF-α mediated NF-κB signaling was significantly enriched in the LBH high expression groups (Fig. 4a, b). Both the qPCR and ELISA assays showed that the expression and secretion of TNF-α were obviously decreased after LBH knockdown, while increased after LBH overexpression (Fig. 4c–f). The luciferase reporter assays revealed that the luciferase activity of pGL3-TNF-α-wt was obviously attenuated after LBH knockdown, while enhanced after LBH overexpression. However, the luciferase activity of pGL3-TNF-α-mt showed no change (Fig. 4g–j). Moreover, we conducted western blotting assays to detect the downstream markers of NF-κB signaling, including p-p65 and p-IκBα. Our results revealed these markers were obviously downregulated after LBH knockdown, while upregulated after LBH overexpression in GSCs (Fig. 4k, l). To investigate whether LBH influenced the malignant phenotype of GSCs by promoting TNF-α expression, we conducted rescue experiments using anti-TNF-α treatment. MTS assays indicated that anti-TNF-α treatment could rescue the cell viability enhanced by LBH overexpression (Fig. S2a, b). Furthermore, EdU and transwell assays demonstrated that anti-TNF-α treatment inhibited the proliferation and invasion abilities facilitated by LBH overexpression in GSCs (Fig. S2c–f). Neurosphere formation and ELDA assays revealed that anti-TNF-α treatment restrained the increased neurosphere size and self-renewing capacity induced by LBH overexpression in GSCs (Fig. S2g–j). Western blotting assays showed that anti-TNF-α treatment could mitigate the expression of stemness markers promoted by LBH overexpression in GSCs (Fig. S2k). Collectively, these findings suggest that anti-TNF-α treatment can counteract the malignant phenotype of GSCs induced by LBH overexpression, and that LBH activates NF-κB signaling by promoting TNF-α transcription. Collectively, LBH could active NF-κB signaling through promoting TNF-α transcription.
Fig. 4.
LBH promoted TNF-α transcription and led to NF-κB signaling pathway activation. a, b CGGA and TCGA analysis showing TNF-α mediated NF-κB signaling in LBH higher expression group. c–f qPCR (c, d) and ELISA (e, f) assays detecting the TNF-α expression after LBH knockdown or overexpression in GSCs. g–j Luciferase report assays showing the luciferase promoter activities of TNF-α after LBH changes in GSCs. k, l Western blotting detecting the downstream markers of NF-κB signaling in GSCs. All data were expressed as mean ± SD, and each experiment was performed in triplicate. *p < 0.05; **p < 0.01; ***p < 0.001
CircZEB1 acted as a miRNA sponge of miR-128-3p and upregulated LBH expression
We analyzed the miRDB, Targetscan, Starbase and Tarbase to explore the potential miRNA sponged by circZEB1. We found two miRNAs (hsa-let-7b-5p, hsa-miR-128-3p) after taking the intersection (Fig. 5a). Then we performed qPCR to detect the expression of LBH after miRNAs knockdown or overexpression. Our results showed LBH expression increased after miR-128-3p inhibitor treatment, while decreased after miR-128-3p mimic treatment (Fig. 5b). However, there were no changes after let-7b-5p knockdown or overexpression (Fig. 5c). Western blotting revealed the LBH expression were promoted after miR-128-3p-inhibitor treatment, while inhibited after miR-128-3p-mimic treatment (Fig. 5d, e). The luciferase reporter assays demonstrated the luciferase activity of LBH-wt was obviously increased after miR-128-3p-inhibitor treatment, while decreased after miR-128-3p-mimics treatment. However, there were no changes in LBH-mt group (Fig. 5f–h). Besides, all the qPCR, luciferase reporter and anti-AgO2 RIP assays proved miR-128-3p was sponged by circZEB1 (Fig. 5i–o). Moreover, western blotting showed that the LBH expression was inhibited after circZEB1 knockdown, while reversed after miR-128-3p-inhibitor treatment in GSC112 (Fig. 5p). The opposite results could be obtained after miR-128-3p-mimic treatment in GSC111 (Fig. 5q). Besides, all the MTS, Edu, transwell and neurosphere formation assays indicated that miR-128-3p-mimic could inhibited the cell viability, proliferation, invasion and neurosphere size promoted by circZEB1 overexpression (Fig. S3a–h). Western blotting assays showed stemness markers could be rescued by miR-128-3p-mimic after circZEB1 overexpression treatment (Fig. S3i). Therefore, we concluded that circZEB1 acted as a miR-128-3p sponge and upregulated LBH expression.
Fig. 5.
CircZEB1 acted as a miRNA sponge of miR-128-3p and upregulated LBH expression. a Identification of miRNAs potentially regulating LBH based on miRDB, Targetscan, Starbase and Tarbase databases. b, c qPCR detecting the LBH expression after miR-128-3p or let-7b-5p changes. d, e Western blotting showing the LBH expression after miR-128-3p inhibitor or mimic treatment. f–h Luciferase report assays showing the luciferase promoter activities of LBH after miR-128-3p inhibitor or mimic treatment. i qPCR detecting miR-128-3p expression after circZEB1 changes. j qPCR detecting circZEB1 expression after miR-128-3p changes. k–m Luciferase report assays showing the luciferase promoter activities of circZEB1 after miR-128-3p changes. n, o RIP assays showing the integration of circZEB1 and miR-128-3p. p, q Western blotting showing the LBH expression after miR-128-3p inhibitor or mimic treatment. All data were expressed as mean ± SD, and each experiment was performed in triplicate. *p < 0.05; **p < 0.01; ***p < 0.001
CircZEB1 promoted the proliferation of GSCs via LBH
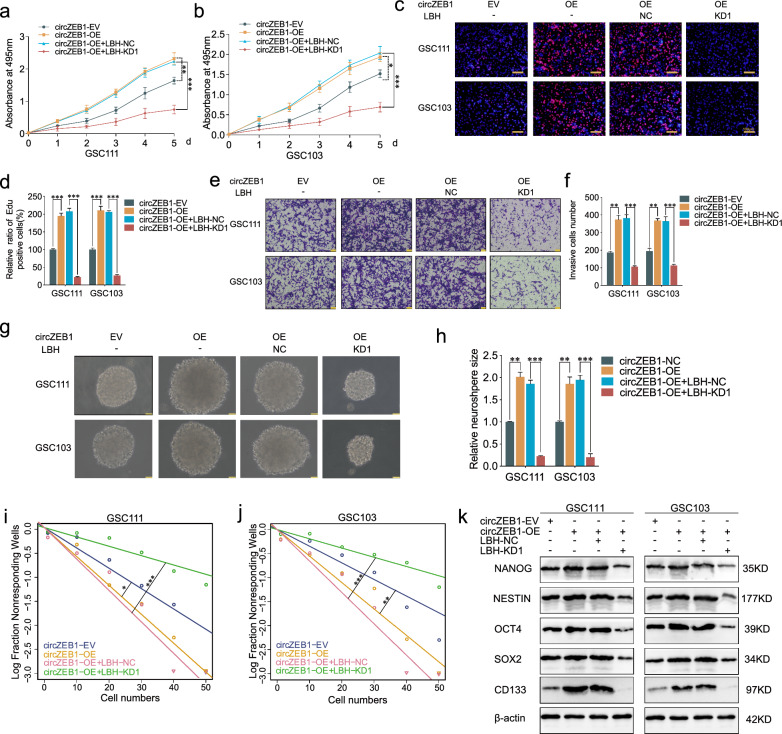
Subsequently, we performed relative rescue experiments to verify whether circZEB1 promoted GSCs proliferation via LBH. All the MTS, Edu, transwell, neurosphere formation and ELDA assays revealed that LBH knockdown could reverse the cell viability, proliferation capacity, invasion ability and self-renewing ability strengthened by circZEB1 overexpression (Fig. 6a–j). Western blotting showed that LBH knockdown could counteract the expression of stemness markers promoted by circZEB1 overexpression (Fig. 6k). Taken together, circZEB1 could promoted the malignant phenotypes and stemness of GSCs via LBH.
Fig. 6.
CircZEB1 promoted the proliferation of GSCs via LBH. a–j All the MTS (a, b), Edu (c, d), transwell (e, f), neurosphere formation (g, h) and ELDA (i, j) assays showing the cell viabilities, proliferation, invasion and GSCs self-renewing capacities were rescued by LBH knockdown treatment. k, Western blotting showing LBH knockdown could rescue the stemness markers expression promoted by circZEB1 overexpression. All data were expressed as mean ± SD, and each experiment was performed in triplicate. *p < 0.05; **p < 0.01; ***p < 0.001
FUS bound to and upregulated circZEB1 expression in GSCs, while LBH transcriptionally regulated FUS expression
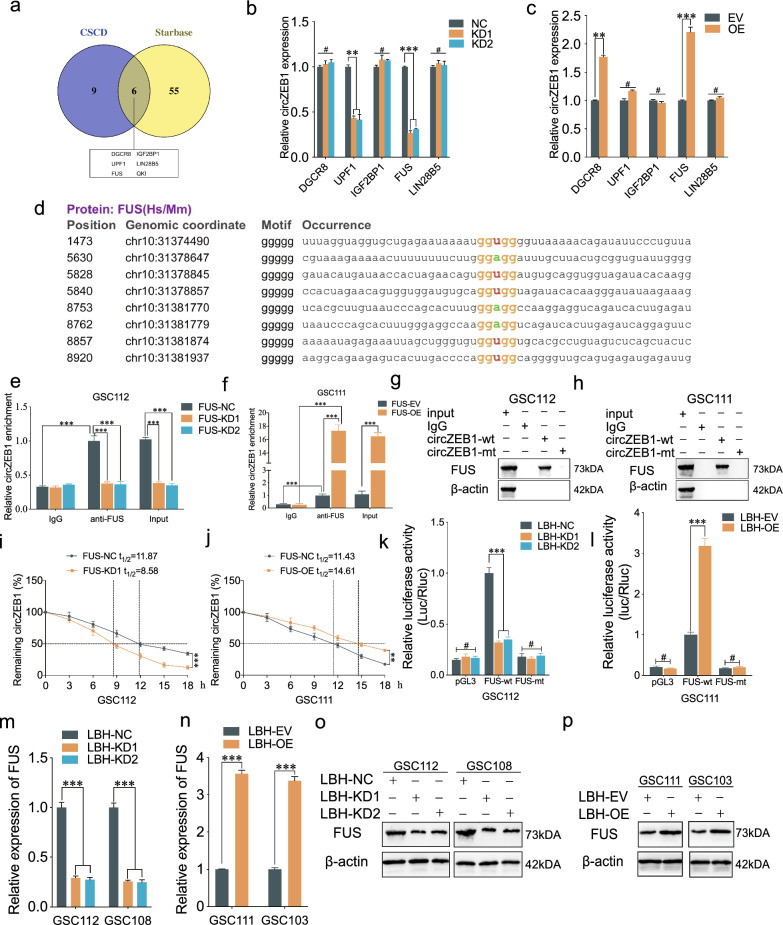
To further explore the potential RBPs interacting with circZEB1, we used CSCD and Starbase to predict and found six candidate RBPs, including DGCR8, UPF1, IGF2BP1, FUS, LIN28B5 and QKI (Fig. 7a). We searched related studies and found that QKI could inhibit GSCs proliferation, which was not suitable for our work [23]. Therefore, we performed qPCR assays to detect circZEB1 expression using DGCR8, UPF1, IGF2BP1, FUS and LIN28B5 knockdown or overexpression treatment. Our results demonstrated that FUS could regulate the expression of circZEB1 in both knockdown and overexpression groups (Fig. 7b, c). There were eight binding sites for FUS on circZEB1 according to the prediction of the RBPmap database (Fig. 7d). We detected the expression of FUS after lentiviral-based transfection using qPCR and western blotting (Fig. S1e–g). Then the results of RIP assays revealed the circZEB1 enrichment was decreased after FUS knockdown, while increased after FUS overexpression (Fig. 7e, f). Besides, the RNA pull-down assays showed the FUS immunoprecipitation with circZEB1 through western blotting in GSCs (Fig. 7g, h). The RNA stability assays demonstrated that FUS knockdown obviously shortened the half-life of circZEB1 in GSC112, and FUS overexpression could prolong the circZEB1 half-life in GSC111 (Fig. 7i, j). All the luciferase reporter, qPCR and western blotting assays proved that LBH knockdown could inhibit the luciferase activity and expression of FUS, while the opposite results could be obtained in LBH overexpression group (Fig. 7k–p). Therefore, FUS bound to and maintained the stability of GSCs, while transcriptionally regulated by LBH.
Fig. 7.
FUS bound to and upregulated circZEB1 expression in GSCs, while LBH transcriptionally regulated FUS expression. a Intersection of CSCD and Starbase showing the potential RBPs binding to circZEB1. b, c qPCR detecting the circZEB1 expression after RBPs changes. d RBPmap database indicating binding sites for FUS on circZEB1. e, f RIP assays showing circZEB1 enrichment after FUS changes. g, h RNA pull-down assays displaying the FUS expression under condition of circZEB1-wt or circZEB1-mt. i, j The expression of circZEB1 after actinomycin D treatment in GSCs. k, l Luciferase report assays showing the luciferase activity of FUS after LBH changes. m–p qPCR (m, n) and western blotting assays (o, p) detecting the FUS expression after LBH knockdown or overexpression. All data were expressed as mean ± SD, and each experiment was performed in triplicate. *p < 0.05; **p < 0.01; ***p < 0.001
The FUS/circZEB1/miR-128-3p/LBH feedback loop promoted GSCs tumorigenesis in vivo
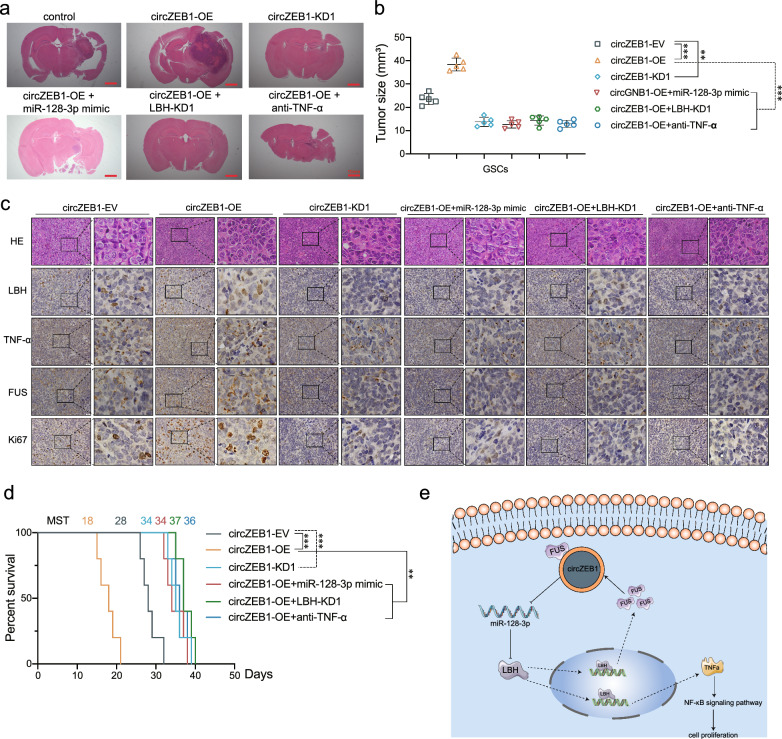
Finally, we carried out orthotopic xenografts to verify the tumorigenesis of circZEB1 and LBH in vivo. Compared with the control group, the tumor volumes were obviously increased in circZEB1 overexpression group, while decreased in circZEB1 knockdown group. Besides, the tumor volumes were significantly decreased in circZEB1 overexpression and miR-128-3p mimic group, circZEB1 overexpression and LBH knockdown group and circZEB1 overexpression and anti-TNF-α group in comparison with the circZEB1 overexpression (Fig. 8a, b). Then we performed IHC to detect LBH, TNF-α, FUS and Ki67 on tumor tissues. The expression of these markers was promoted after circZEB1 overexpression, while decreased after circZEB1 knockdown, miR-128-3p mimic, LBH knockdown and anti-TNF-α treatment (Fig. 8c). The Kaplan Meier survival analysis indicated the MST of circZEB1 overexpression was the shortest, while it was prolonged after circZEB1 knockdown, miR-128-3p mimic, LBH knockdown and anti-TNF-α treatment. The diagram showed that the FUS/circZEB1/miR-128-3p/LBH feedback loop promoted GSCs tumorigenesis in GSCs.
Fig. 8.
The FUS/circZEB1/miR-128-3p/LBH feedback loop promoted GSCs tumorigenesis in vivo. a Representative photograph showing the intracranial tumors in different groups. Scale bar = 10 mm. b The tumor sizes of these groups. c Representative immunohistochemical images showing the expression of LBH, TNF-α, FUS and Ki67. d Kaplan–Meier survival curves showing the survival time of these groups. e Schematic diagram displaying the FUS/circZEB1/miR-128-3p/LBH feedback loop promoted tumorigenesis in GSCs. All data were expressed as mean ± SD, and each experiment was performed in triplicate. *p < 0.05; **p < 0.01; ***p < 0.001
Discussion
Glioma stem cells play vital roles in GBM due to their tumorigenesis, chemotherapy and radiotherapy resistance and recurrence [24]. Recently, more and more studies have found that circRNAs participate in a series of malignant tumors [25, 26]. Therefore, exploring the underlying mechanism between circRNAs and cancer stem cells warrants further investigation and may provide promising targets for cancers [27]. For instance, the HIF-1α/circ-CDYL-COL14A1 axis could promote lung metastasis of cancer stem cells from hepatocellular carcinoma [28]. CircSLC4A7 could promote gastric cancer stem cells progression by interacting with HSP90 and activating NOTCH1 signaling pathway [29]. CircRNF10 facilitated glioma stem cells progression by the circRNF10/ZBTB48/IGF2BP3 feedback loop [30]. Our study identifies a novel circRNA termed circZEB1 highly expressed in GSCs with a poor prognosis. We describe that circZEB1 upregulates TNF-α expression and actives NF-κB signaling, which may provide an effective target for anticancer therapy.
Additionally, there are also many other biological mechanisms of circRNAs affecting the GBM progress, including functioning as a scaffold, promoting angiogenesis, regulating the tumor microenvironment and encoding proteins [31, 32]. For instance, circLRFN5 could bind to PRRX2 protein and promoted its degradation, which inhibited the progression of GBM [33]. CircNEIL3 could promote glioma progression and macrophage immunosuppressive polarization through stabilizing IGF2BP3 [34]. Moreover, circMET could encode a 404-amino-acid MET variant to promote glioblastoma tumorigenesis [35]. Therefore, there may be other downstream factors regulated by circZEB1 and warrant further investigation in the future.
As a transcription cofactor, LBH is involved in a series of pathogenesis and pathological process [36]. For instance, LBH could be inhibited by miR-31 host gene to drive oral carcinoma oncogenicity [37]. Our previous study has reported that LBH promotes glioma angiogenesis via VEGFA-mediated ERK signaling pathway [21]. In this work, we discover that LBH can facilitate GSCs proliferation and stemness via upregulating TNF-α and FUS. Herein, it is quite crucial to furtherly uncover the mechanism of LBH in regulating GSCs. TNF-a mediated NF-κB signaling is also involved in physiological process and cancers. A recent study reported TNF-a/ NF-κB could be repressed by khasianine to ameliorate psoriasis-like skin inflammation [38]. And TNF-a/NF-κB signaling could be inhibited by OTUD1 to sensitize ccRCC to TKIs [39]. Furthermore, several studies have reported that FUS could bind to circRNAs and promote cancer progression. FUS could interact with circTBC1D14 to promote autophagy in triple-negative breast cancer [40]. Our results in vivo tumorigenesis assays indicated that LBH could promote TNF-a expression and increase brain tumor growth in mice.
In summary, we report a novel mechanism of circZEB1 in the process of GSCs and provide a potential target for therapy.
Supplementary Information
Supplementary Figure 1. Validation of the top 5 upregulated circRNAs and efficiency of lentiviral vectors transfection. a qPCR detecting the top 5 upregulated circRNAs between NBT and GBM. b-d qPCR and western blotting showing the LBH expression after transfection. e-g qPCR and western blotting displaying the FUS expression after transfection. All data were expressed as mean ± SD, and each experiment was performed in triplicate. *p < 0.05; **p < 0.01; ***p < 0.001.
Supplementary Figure 2. LBH promotes the malignant phenotype of GSCs via transcriptional regulating TNF-α. a-j All the MTS, Edu, transwell, neurosphere formationand ELDAassays indicating anti-TNF-α could rescue the cell viabilities, proliferation, invasion and self-renewing capacities facilitated by LBH overexpression. k Western blotting assays showing the stemness markers expression was rescued by anti-TNF-α. All data were expressed as mean ± SD, and each experiment was performed in triplicate. *p < 0.05; **p < 0.01; ***p < 0.001.
Supplementary Figure 3. CircZEB1 promotes the malignant phenotype of GSCs via miR-128-3p. a-h All the MTS, Edu, transwelland neurosphere formationassays showing miR-128-3p-mimic could rescue the cell viabilities, proliferation, invasion and self-renewing capacities promoted by circZEB1 overexpression. i Western blotting assays showing the stemness markers expression was rescued by miR-128-3p-mimic. All data were expressed as mean ± SD, and each experiment was performed in triplicate. *p < 0.05; **p < 0.01; ***p < 0.001.
Supplementary Figure 4. Multiple fields of neurospheres
Acknowledgements
We appreciated all the lab colleagues for their support for this work.
Abbreviations
- GBM
Glioblastoma
- GSCs
Glioma stem cells
- CircRNAs
Circular RNAs
- LBH
Limb-bud and heart
- TNF-α
Tumor necrosis factor-α
- FUS
Fused in sarcoma
- RBPs
RNA-binding proteins
- NBT
Normal brain tissue
- qRT-PCR
Real-time quantitative reverse transcription PCR
- IHC
Immunohistochemistry
- IF
Immunofluorescence
- CHIP
Chromatin immunoprecipitation
- ELDA
Extreme limiting dilution analysis
- RIP
RNA immunoprecipitation
- TCGA
The Cancer Genome Atlas
- CGGA
Chinese Glioma Genome Atlas
Author contributions
ZTJ and YJ conceived and designed the study; GQZ and ZCW performed the experiments, interpreted the results and wrote the manuscript.; JPH and YFW performed bioinformatics analysis; ZTG and XQL collected and analyzed the data. All authors read and approved the final version of the manuscript.
Funding
The research was supported by the National Natural Science Foundation of China (Nos. 82072794) and the Shanghai Sailing Program (No. 21YF1449900).
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Patients and controls were acquired with informed consent approved by the First Hospital of China Medical University research ethics committee, and animals have been approved by ethics committee of China Medical University.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Guoqing Zhang, Yang Jiang and Zhichao Wang contributed equally to this work.
Contributor Information
Yongfeng Wang, Email: wyf1208@163.com.
Zhitao Jing, Email: jingzhitao@hotmail.com.
References
- 1.Lee JH, Lee JE, Kahng JY, Kim SH, Park JS, Yoon SJ, et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature. 2018;560(7717):243–7. [DOI] [PubMed] [Google Scholar]
- 2.Eltoukhy M, Kandula V, Joseph S, Albanese E, Giridharan S. Should redo surgery be offered to patients with relapsed glioblastoma?-outcome analyses of a single institution comparative cohort study. World Neurosurg. 2023;176:e543–7. [DOI] [PubMed] [Google Scholar]
- 3.Smith K, Nakaji P, Thomas T, Pinnaduwage D, Wallstrom G, Choi M, et al. Safety and patterns of survivorship in recurrent GBM following resection and surgically targeted radiation therapy: results from a prospective trial. Neuro Oncol. 2022;24(Suppl 6):S4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jhaveri N, Chen TC, Hofman FM. Tumor vasculature and glioma stem cells: contributions to glioma progression. Cancer Lett. 2016;380(2):545–51. [DOI] [PubMed] [Google Scholar]
- 5.Wakimoto H, Kesari S, Farrell CJ, Curry WT Jr, Zaupa C, Aghi M, et al. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009;69(8):3472–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Liu G, Wang X, Hong H, Li T, Li L, et al. Glioblastoma stem cell-specific histamine secretion drives pro-angiogenic tumor microenvironment remodeling. Cell Stem Cell. 2022;29(11):1531-46 e7. [DOI] [PubMed] [Google Scholar]
- 7.Tao M, Zheng M, Xu Y, Ma S, Zhang W, Ju S. CircRNAs and their regulatory roles in cancers. Mol Med. 2021;27(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao X, Zhong Y, Wang X, Shen J, An W. Advances in circular RNA and its applications. Int J Med Sci. 2022;19(6):975–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhenhao Z, Ru C, Xiaofeng C, Heng Y, Gongxian W. A novel circular RNA, circMAML3, promotes tumor progression of prostate cancer by regulating miR-665/MAPK8IP2 axis. Cell Death Discov. 2023;9(1):455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miao Z, Li J, Wang Y, Shi M, Gu X, Zhang X, et al. Hsa_circ_0136666 stimulates gastric cancer progression and tumor immune escape by regulating the miR-375/PRKDC axis and PD-L1 phosphorylation. Mol Cancer. 2023;22(1):205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu S, Lu J, Zhu H, Wu F, Mo Y, Xie L, et al. A novel axis of circKIF4A-miR-637-STAT3 promotes brain metastasis in triple-negative breast cancer. Cancer Lett. 2024;581: 216508. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Y, Zhao J, Liu Y, Hu J, Gao L, Wang H, et al. CircKPNB1 mediates a positive feedback loop and promotes the malignant phenotypes of GSCs via TNF-alpha/NF-kappaB signaling. Cell Death Dis. 2022;13(8):697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehta M, Raguraman R, Ramesh R, Munshi A. RNA binding proteins (RBPs) and their role in DNA damage and radiation response in cancer. Adv Drug Deliv Rev. 2022;191: 114569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao C, Xie W, Zhu H, Zhao M, Liu W, Wu Z, et al. LncRNAs and their RBPs: how to influence the fate of stem cells? Stem Cell Res Ther. 2022;13(1):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Yang L, Wu P, Li X, Tang Y, Ou X, et al. The circROBO1/KLF5/FUS feedback loop regulates the liver metastasis of breast cancer by inhibiting the selective autophagy of afadin. Mol Cancer. 2022;21(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen T, Wang X, Li C, Zhang H, Liu Y, Han D, et al. CircHIF1A regulated by FUS accelerates triple-negative breast cancer progression by modulating NFIB expression and translocation. Oncogene. 2021;40(15):2756–71. [DOI] [PubMed] [Google Scholar]
- 17.Liu P, Wang Z, Ou X, Wu P, Zhang Y, Wu S, et al. The FUS/circEZH2/KLF5/ feedback loop contributes to CXCR4-induced liver metastasis of breast cancer by enhancing epithelial-mesenchymal transition. Mol Cancer. 2022;21(1):198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang R, Zhong J, Pan X, Su Z, Xu Y, Zhang M, et al. A novel intronic circular RNA circFGFR1(int2) up-regulates FGFR1 by recruiting transcriptional activators P65/FUS and suppressing miR-4687-5p to promote prostate cancer progression. J Transl Med. 2023;21(1):840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu R, Li Z, Zhang C, Song H, Deng M, Sun L, et al. Elevated limb-bud and heart development (LBH) expression indicates poor prognosis and promotes gastric cancer cell proliferation and invasion via upregulating Integrin/FAK/Akt pathway. PeerJ. 2019;7: e6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Q, Li E, Huang L, Cheng M, Li L. Limb-bud and heart overexpression inhibits the proliferation and migration of PC3M Cells. J Cancer. 2018;9(2):424–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y, Zhou J, Zou D, Hou D, Zhang H, Zhao J, et al. Overexpression of limb-bud and heart (LBH) promotes angiogenesis in human glioma via VEGFA-mediated ERK signalling under hypoxia. EBioMedicine. 2019;48:36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu A, Luo N, Xu Y, Du N, Li L, Liu Q. Exosomal LBH inhibits epithelial-mesenchymal transition and angiogenesis in nasopharyngeal carcinoma via downregulating VEGFA signaling. Int J Biol Sci. 2022;18(1):242–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han B, Wang R, Chen Y, Meng X, Wu P, Li Z, et al. QKI deficiency maintains glioma stem cell stemness by activating the SHH/GLI1 signaling pathway. Cell Oncol (Dordr). 2019;42(6):801–13. [DOI] [PubMed] [Google Scholar]
- 24.Eckerdt F, Platanias LC. Emerging role of glioma stem cells in mechanisms of therapy resistance. Cancers (Basel). 2023;15(13):3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pisignano G, Michael DC, Visal TH, Pirlog R, Ladomery M, Calin GA. Going circular: history, present, and future of circRNAs in cancer. Oncogene. 2023;42(38):2783–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Z, Ji Y, Zhou J, Li G, Wu Y, Liu W, et al. Exosomal circRNAs in cancer: implications for therapy resistance and biomarkers. Cancer Lett. 2023;566: 216245. [DOI] [PubMed] [Google Scholar]
- 27.Han T, Chen L, Li K, Hu Q, Zhang Y, You X, et al. Significant circRNAs in liver cancer stem cell exosomes: mediator of malignant propagation in liver cancer? Mol Cancer. 2023;22(1):197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong R, Wei W, Man Q, Chen L, Jia Y, Zhang H, et al. Hypoxia-induced circ-CDYL-EEF1A2 transcriptional complex drives lung metastasis of cancer stem cells from hepatocellular carcinoma. Cancer Lett. 2023;578: 216442. [DOI] [PubMed] [Google Scholar]
- 29.Hui Y, Wenguang Y, Wei S, Haoran W, Shanglei N, Ju L. circSLC4A7 accelerates stemness and progression of gastric cancer by interacting with HSP90 to activate NOTCH1 signaling pathway. Cell Death Dis. 2023;14(7):452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Zhang M, Liu Y, Cui D, Gao L, Jiang Y. CircRNF10 triggers a positive feedback loop to facilitate progression of glioblastoma via redeploying the ferroptosis defense in GSCs. J Exp Clin Cancer Res. 2023;42(1):242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed SP, Castresana JS, Shahi MH. Role of circular RNA in brain tumor development. Cells. 2022;11(14):2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balandeh E, Mohammadshafie K, Mahmoudi Y, Hossein Pourhanifeh M, Rajabi A, Bahabadi ZR, et al. Roles of non-coding RNAs and angiogenesis in glioblastoma. Front Cell Dev Biol. 2021;9: 716462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang Y, Zhao J, Li R, Liu Y, Zhou L, Wang C, et al. CircLRFN5 inhibits the progression of glioblastoma via PRRX2/GCH1 mediated ferroptosis. J Exp Clin Cancer Res. 2022;41(1):307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan Z, Zhao R, Li B, Qi Y, Qiu W, Guo Q, et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol Cancer. 2022;21(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong J, Wu X, Gao Y, Chen J, Zhang M, Zhou H, et al. Circular RNA encoded MET variant promotes glioblastoma tumorigenesis. Nat Commun. 2023;14(1):4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young IC, Brabletz T, Lindley LE, Abreu M, Nagathihalli N, Zaika A, et al. Multi-cancer analysis reveals universal association of oncogenic LBH expression with DNA hypomethylation and WNT-Integrin signaling pathways. Cancer Gene Ther. 2023;30(9):1234–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang KW, Hung WW, Chou CH, Tu HF, Chang SR, Liu YC, et al. LncRNA MIR31HG drives oncogenicity by inhibiting the limb-bud and heart development gene (LBH) during oral carcinoma. Int J Mol Sci. 2021;22(16):8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, Zhang Y, Chen X, Su Z, Deng Y, Zhao Q. Khasianine ameliorates psoriasis-like skin inflammation and represses TNF-alpha/NF-kappaB axis mediated transactivation of IL-17A and IL-33 in keratinocytes. J Ethnopharmacol. 2022;292: 115124. [DOI] [PubMed] [Google Scholar]
- 39.Liu W, Yan B, Yu H, Ren J, Peng M, Zhu L, et al. OTUD1 stabilizes PTEN to inhibit the PI3K/AKT and TNF-alpha/NF-kappaB signaling pathways and sensitize ccRCC to TKIs. Int J Biol Sci. 2022;18(4):1401–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Liu Y, He Y, Zhang N, Zhang S, Li Y, et al. Hypoxia-induced FUS-circTBC1D14 stress granules promote autophagy in TNBC. Adv Sci (Weinh). 2023;10(10): e2204988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Validation of the top 5 upregulated circRNAs and efficiency of lentiviral vectors transfection. a qPCR detecting the top 5 upregulated circRNAs between NBT and GBM. b-d qPCR and western blotting showing the LBH expression after transfection. e-g qPCR and western blotting displaying the FUS expression after transfection. All data were expressed as mean ± SD, and each experiment was performed in triplicate. *p < 0.05; **p < 0.01; ***p < 0.001.
Supplementary Figure 2. LBH promotes the malignant phenotype of GSCs via transcriptional regulating TNF-α. a-j All the MTS, Edu, transwell, neurosphere formationand ELDAassays indicating anti-TNF-α could rescue the cell viabilities, proliferation, invasion and self-renewing capacities facilitated by LBH overexpression. k Western blotting assays showing the stemness markers expression was rescued by anti-TNF-α. All data were expressed as mean ± SD, and each experiment was performed in triplicate. *p < 0.05; **p < 0.01; ***p < 0.001.
Supplementary Figure 3. CircZEB1 promotes the malignant phenotype of GSCs via miR-128-3p. a-h All the MTS, Edu, transwelland neurosphere formationassays showing miR-128-3p-mimic could rescue the cell viabilities, proliferation, invasion and self-renewing capacities promoted by circZEB1 overexpression. i Western blotting assays showing the stemness markers expression was rescued by miR-128-3p-mimic. All data were expressed as mean ± SD, and each experiment was performed in triplicate. *p < 0.05; **p < 0.01; ***p < 0.001.
Supplementary Figure 4. Multiple fields of neurospheres
Data Availability Statement
No datasets were generated or analysed during the current study.