Abstract
Background
The physiological relevance of cell-to-cell communication mediated by small extracellular vesicle-encapsulated microRNAs (sEV-miRNAs) remains debated because of the limiting representativity of specific miRNAs within the extracellular pool. We hypothesize that sEV-miRNA non-canonical function consisting of the stimulation of Toll-like receptor 7 (TLR7) may rely on a global shift of the sEV cargo rather than on the induction of one or few specific miRNAs. Psoriasis represents an ideal model to test such hypothesis as it is driven by overt activation of TLR7-expressing plasmacytoid dendritic cells (pDCs) following keratinocyte damage.
Methods
To mimic the onset of psoriasis, keratinocytes were treated with a cocktail of psoriatic cytokines or UV-irradiated. SmallRNA sequencing was performed on sEVs released by healthy and UV-treated keratinocytes. sEV-miRNAs were analyzed for nucleotide composition as well as for the presence of putative TLR7-binding triplets. Primary human pDCs where stimulated with sEVs +/- inhibitors of TLR7 (Enpatoran), of sEV release (GW4869 + manumycin) and of TLR7-mediated pDC activation (anti-BDCA-2 antibody). Secretion of type I IFNs and activation of CD8+T cells were used as readouts. qPCR on psoriatic and healthy skin biopsies was conducted to identify induced miRNAs.
Results
sEV-miRNAs released by damaged keratinocytes revealed a significantly higher content of TLR7-activating sequences than healthy cells. As expected, differential expression analysis confirmed the presence of miRNAs upregulated in psoriatic skin, including miR203a. More importantly, 76.5% of induced miRNAs possessed TLR7-binding features and among these we could detect several previously demonstrated TLR7 ligands. In accordance with this in silico analysis, sEVs from damaged keratinocytes recapitulated key events of psoriatic pathogenesis by triggering pDCs to release type I interferon and activate cytotoxic CD8+T cells in a TLR7- and sEV-dependent manner.
Discussion
Our results demonstrate that miR203a is just one paradigmatic TLR7-activating miRNA among the hundreds released by UV-irradiated keratinocytes, which altogether trigger pDC activation in psoriatic conditions. This represents the first evidence that cell damage shifts the miRNA content of sEVs towards a TLR7-activating cargo capable to propagate inflammation and immunity, offering strong support to the physiological role of systemic miRNA-based cell-to-cell communication.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-024-01924-z.
Keywords: Toll-like receptor 7 (TLR7), Plasmacytoid dendritic cells (pDCs), IFN-α, hsa-miR203a-3p, Exosomes, TNF-α, IL-6, IFN-γ, Psoriasis, Extracellular vesicles
Background
Toll-like receptors (TLRs) are innate immune receptors that, upon recognition of conserved molecular components of pathogens, activate inflammation to alert the immune system about infection [1]. Notably, TLRs can also interact with endogenous ligands released under sterile conditions by stressed or damaged tissues to initiate immune cell recruitment and tissue repair. However, sustained TLR activation leads to exacerbated inflammatory responses that pave the way to hyperinflammation and/or autoimmunity, as well exemplified by the pathogenesis of immune-mediated inflammatory diseases (IMIDs) (reviewed in [2]).
One TLR subfamily comprising TLR3, TLR7, TLR8 and TLR9 is characterized by endosomal localization and by recognition of different species of nucleic acids derived from pathogen digestion. Within this family, TLR7 and TLR8 are both activated by single-stranded RNA (ssRNA) ligands but display reciprocal expression and functional significance [3]. Indeed, TLR7 is expressed by pDCs and masters the production of IFN-α, while TLR8 is expressed by conventional DCs, which specialize in the release of inflammatory cytokines [4]. The sequence specificity of ssRNA ligands of TLR7 and TLR8 remained long debated, until it was demonstrated that both recognize short ssRNA sequences derived from endosomal digestion of longer ssRNAs [5]. These studies allowed to bring all previous findings together determining that TLR7 requires guanosine (G) residues to activate a first binding site and a 3-mer containing a nonterminal uridine (U) for the activation of a second binding site, although for synergistic effects sequences with at least two Us are required [3, 5–10]. Since overwhelming evidence exists that endosomal TLRs, and TLR7 in particular, play a central role in the induction and progression of IMIDs [11–14], therapeutic strategies aimed at blocking TLR activation by self-nucleic acids are currently in the limelight of both preclinical and clinical research [13, 15, 16].
MicroRNAs (miRNAs) are a class of short, noncoding, ssRNAs originally discovered as regulators of mRNA expression in the cell cytoplasm of producing cells [17]. However, miRNAs bound to carrier proteins such as Argonaute 2 (Ago2), that preserves them from degradation, can also be transferred from cell to cell, generally through mechanisms involving the shedding of extracellular vesicles (EVs) [18–20]. In recipient cells, extracellular miRNAs can either perform canonical RNA interference or activate pro-inflammatory signaling pathways by direct interaction with TLR7/8 in the endosomes of innate immune cells [6, 21, 22]. Our group and others previously demonstrated that selected extracellular miRNAs, upregulated in settings of cancer, sepsis, neurodegeneration and autoimmunity, could activate the secretion of IFN-α by pDCs via human TLR7 [23] or inflammation via TLR8 [24–26]. However, the physiological relevance of cell-to-cell miRNA-based communication remains uncertain, mainly because of the limiting representativity of specific miRNAs within the extracellular pool.
IMIDs include, among others, common diseases such as psoriasis, systemic lupus erythematosus, rheumatoid arthritis, type 1 diabetes and multiple sclerosis, thus accounting for a high disease burden in the population [27]. These diseases share common pathogenetic mechanism involving a complex combination of environmental factors, which trigger unwanted inflammation, and genetic susceptibility, fostering an undue immune response [27, 28]. In psoriasis, a systemic condition that manifests with skin involvement [29], the development of lesions is long known to follow skin-damaging events such as trauma and ultraviolet (UV) ray exposure (a.k.a Koebner phenomenon [30]), , but the link between such environmental triggers and immuno-mediated tissue destruction was only recently unveiled. Indeed, the initiation of the autoreactive adaptive immune response is now known to depend on the aberrant production of type I interferons (IFNs) by plasmacytoid dendritic cells (pDCs) [31, 32], a small subset of circulating immune cells that are typically absent in peripheral tissues under homeostatic conditions but accumulate within psoriatic skin [14, 33]. Seminal work indicated that pDC activation depended on the accumulation of self-nucleic acids massively released by damaged keratinocytes, which induce unnecessary IFN-α secretion by engaging endosomal TLR7 and TLR9 [34–36]. Of note, several miRNAs were found dysregulated in psoriasis, both at the cellular and extracellular levels [37–39].
By taking advantage of the well characterized pathogenetic mechanisms of psoriasis, this study was designed to explore the hypothesis that the TLR7-activating function of extracellular miRNAs may depend on a global shift of the miRNA cargo of sEVs rather than on the induction of one or few specific miRNAs. In addition to identify so far neglected triggers of pDC activation in psoriasis, and possibly other IMIDs, this scenario would help overcome the limitations of miRNA-based cell-to-cell communication theory.
Methods
Cell preparation and culture
Human primary keratinocytes (KCs) were kindly provided from Banca delle Cute, AOU Città della Salute e della Scienza Torino, Italy. A written informed consent was obtained from all donors of skin grafts. KCs were grown in CnT-Prime Epithelial Cell Culture Medium (CELLnTEC Advanced Cell System). The immortalized human epidermal keratinocyte cell line HaCaT was purchased from American Type Culture Collection (ATCC), and the cells were cultured in DMEM supplemented with 10% FBS, 2 mM L-Glutamine, penicillin and streptomycin (all from Gibco, Thermo Fisher Scientific). pDCs and CD8+ T cells were obtained from buffy coats (through the courtesy of Centro Trasfusionale, Spedali Civili, Brescia, Italy) after immunomagnetic separation with the Plasmacytoid Dendritic Cell Isolation kit II and CD8+ T cell isolation kit (Miltenyi Biotec), respectively. pDCs (1 × 106 cells/ml) were cultured in RPMI 1640 supplemented with 10% heat-inactivated FBS, 2 mM L-Glutamine, penicillin and streptomycin, and 20 ng/ml IL-3 (Miltenyi Biotec).
Cell treatment
KCs and HaCaT cells were stimulated with a mix of proinflammatory cytokines namely IL-1α (Biomart), IL-6 (Peprotech), TNF-α (Prospec), IL-17 (Biomart), IL-22 (Peprotech) and IFN-γ (Peprotech), each at 50 ng/ml, for 24 h. Conditioned media were concentrated 10 times by using centrifugal concentrator with 10,000 MW cutoff (Vivaspin, Sartorius).
Alternatively, KCs and HaCaT cells were exposed for 30 s to UVC irradiation using a germicidal lamp G30T8 (Sankyo Denki) in 10-cm uncovered dishes with a thin layer of PBS at a distance of 40 cm [40]. PBS was removed immediately after UV irradiation, and cells were replenished with serum-free fresh media for 24 h. Where indicated, 10 µM GW4869 and 1 µM manumycin A were added after UV irradiation.
Human samples
Human lesional and nonlesional skin as well as plasma from patients with psoriasis and healthy donors were obtained after informed consent and approval of the Ethics Committee of the Humanitas University (Milan, Italy) and in accordance with the Declaration of Helsinki. Skin samples were immediately stored in RNAlater (Thermo Fisher Scientific) until further processing.
pDC stimulation and T cell coculture
Purified pDCs were stimulated with 10 µg/ml synthetic hsa-miR203a-3p (miR203a) stabilized with a phosphorothioate linkage between each base (Integrated DNA Technologies). Complexation of miR203a with DOTAP (Roche Diagnostics) was performed as previously described [23]. Whenever indicated, pDCs were pretreated for 1 h with 1 µM Enpatoran (Enp, MedChemExpress) or 1 µg/ml of an anti-BDCA2 monoclonal antibody (clone AC144, Miltenyi Biotec). miR203a-activated pDCs were cocultured with CFSE-stained allogenic CD8+ T cells (Miltenyi Biotec) as previously described [41]. After 6 days, alloreactive T cell proliferation was assessed by measuring the loss of the dye CellTrace-CFSE upon cell division using flow cytometry. Positive controls of T cell proliferation were routinely performed using IL-2 plus phytohemagglutinin (PHA, Merck Millipore).
Flow cytometry
pDCs or HaCaT cells were stained with the following antibodies from Miltenyi Biotec or as indicated: APC-Vio770-conjugated anti-human HLA-ABC (clone REA230), Alexa Fluor 488-conjugated anti-human HLA-DR (clone L243, BioLegend), PE-conjugated anti-human CD80 (L307.4, BD Pharmingen), Vioblue-conjugated anti-human CD86 (clone FM95), PE-Vio770-conjugated anti-human CD69 (clone FN50), APC-conjugated anti-human BDCA2 (clone AC144) and APC-conjugated anti-human CD29 (clone TS2/16). KC and HaCaT cell death induced by UV treatment was assessed by Pacific Blue-Annexin V and 7-AAD kit (BioLegend) following the manufacturer’s instructions. To analyze sEV surface proteins, EVs were subjected to bead-based multiplex analysis by using MACSPlex Exosome kit (Miltenyi Biotec). Briefly, 109 EVs were incubated overnight on an orbital shaker with MACSPlex Exosome Capture Beads containing 39 different antibody-coated beads. For counterstaining of EVs bound by capture beads with detection antibodies, APC-conjugated anti-CD9, anti-CD63 and anti-CD81 detection antibodies were added to each sample and incubated at room temperature for 1 h on an orbital shaker. Samples were read on a MACSQuant Analyzer (Miltenyi Biotec) and analyzed with FlowJo (Tree Star Inc.). Median fluorescence intensity (MFI) for all 39 capture beads were background corrected by subtracting respective MFI values from matched media control.
Cytokine detection
IFN-α was detected using a specific Module Set ELISA kit (Invitrogen, Thermo Fisher Scientific). TNF-α, IL-6 and IFN-γ were measured using specific Duo-Set kits (R&D Systems). All assays were performed on cell-free supernatants according to the manufacturer’s protocol.
NF-κB luciferase reporter assay
25,000 human HEK293 cells expressing luciferase under control of the NF-κB promoter and stably transfected with human TLR7 or luciferase alone [23] were seeded in complete DMEM without antibiotics in 96-well plates, as previously described [26, 41]. After 24 h, cells were stimulated with 10 µg/ml miR203a or RNA40 (a TLR7-ligand positive control, Integrated DNA Technologies) for an additional 24 h. After stimulation, cells were lysed using ONE-Glo EX Luciferase Assay System (Promega) and assayed for luciferase activity using the EnSightMultimode Plate Reader (PerkinElmer).
sEV isolation from HaCaT conditioned media
24 h after UV irradiation, HaCaT conditioned media were collected and processed with a serial centrifugation protocol to isolate sEVs [42, 43]. Briefly, conditioned media were firstly centrifuged at 300 g for 5 minutes to discard cells and debris and then centrifuged at 800 g for 30 min at 4 °C to pellet and discard large vesicles and cell debris. Supernatants were centrifuged at 16,000 g for 45 min at 4 °C to pellet medium EVs, and finally ultracentrifuged at 100,000 g for 4 h at 4 °C to collect sEVs (Type 45 Ti rotor, Beckman Optima XPN 80, Beckman Coulter). sEVs were washed with 1 ml of sterile PBS to remove contaminants and pelleted again at 100,000 g for 2 h at 4 °C (rotor TLA-55, Beckman Optima MAX). Finally, sEVs were resuspended in PBS for further analyses. EV protein concentration was measured by Bicinchoninic Acid (BCA) assay (Thermo Fischer Scientific). 1 × 105 pDCs (in 50 µl of RPMI 20% FBS) were stimulated by adding sEVs derived from 12.5 ml HaCaT supernatant resuspended in 50 µl serum free RPMI medium (approximately 1010 sEVs). Throughout the text, sEVs isolated from conditioned media of UV-treated HaCaT cells are indicated as UV-sEVs, while sEVs isolated from conditioned media of resting HaCaT cells are indicated as NT-sEVs.
Atomic force microscopy (AFM) imaging
AFM imaging was performed as previously described [44]. Briefly, sEVs isolated from HaCaT conditioned media were resuspended in 50 µl of PBS and diluted 1:100 v/v with Milli-Q water. The diluted samples (5–10 µl) were then spotted onto freshly cleaved mica sheets (PELCO® Mica discs Grade V-1, thickness 0.15 mm, 10 mm diameter from Ted Pella, Inc). All mica substrates were air dried over a plate heated at 37–40 °C and analyzed using a Nanosurf NaioAFM equipped with Multi75AI-G probes (Budget sensors). Images were acquired in dynamic mode, scan size ranged from 1 to 20 μm and scan speed ranged from 0.8 to 1.5 s per scanning line. AFM images were processed using Gwyddion ver. 2.58. The size of particles was extrapolated using built-in modules from topography images. A minimum of 300 round-shaped vesicles per sample were analyzed. Particle size distribution was then calculated on GraphPad PRISM ver. 8, by plotting particle size against relative abundance.
Purity assessment and titration with CONAN assay
The EV samples were checked for purity (meaning the absence of significant presence of single or aggregated protein contaminants) and subsequently titrated by using the COlorimetric NANoplasmonic (CONAN) assay, an effective method to determine the purity and concentration of EV preparations [45] that is based on the clustering of gold nanoparticles (AuNPs) onto lipid membranes [46]. The CONAN assay was performed as previously described [47]. Briefly, sEVs were resuspended in 50 µL of PBS. 2 µl of EVs were resuspended in 23 µl of water, mixed with 50 µl of 6 nM AuNPs and 25 µl of PBS. When mixed with pure EV formulations, the AuNPs cluster on the EV membrane, whereas in EV formulations that contain soluble protein contaminants the AuNPs are preferentially cloaked by such proteins, which prevents AuNPs from clustering to the EV membrane. When AuNPs cluster, their localized surface plasmon resonance (LSPR) peak shifts and broadens, resulting in a color change of the AuNP solution from red to blue, which can be acquired by Ensight MultiMode Reader (Perkin Elmer). AuNP aggregation is described by the aggregation index ratio (AI%), a numerical value indicating the sample purity and the EV concentration, extrapolated from the UV-VIS spectra of the tested samples. The particle numbers were evaluated comparing the sample AI%, resulting from the assay, with a calibration line built with standard liposome solutions (concentrations ranging from 0.012 to 0.75 mg/mL) quantified with the CONAN assay.
Isolation of miRNAs and qPCR
Skin biopsies were homogenized in Qiazol reagent (Qiagen) by using Ultra-Turrax and centrifuged at 12,000 g for 10 min at 4 °C. Then, miRNAs were purified with miRNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. To purify miRNAs from plasma samples, the miRNeasy Serum/Plasma Kit (Qiagen) was used. miRNAs contained in sEVs were purified with miRNeasy Mini Kit, together with RNeasy Mini Elute Cleanup Kit (Qiagen). To isolate miRNAs from conditioned media, the protocol set up by Turchinovich was followed [48]. Briefly, 1.2 ml of Qiazol were added to 0.4 ml of conditioned media, mixed by vigorous shaking for 10 s and then incubated for 10 min at RT. After supplying 320 µl of chloroform, the mixture was vortexed and allowed to stand for 5 min at RT. Following the centrifugation at 14,000 g for 20 min total RNA was precipitated from the upper (aqueous) phase by adding 1.5 V of 100% ethanol. Purification of extracted total RNA, including miRNAs, was performed with miRNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. To monitor the miRNA extraction efficacy, RNA spike-in (comprising UniSp2 RNA, UniSp4 RNA and UniSp5 RNA or cel-miR39) was added to Qiazol reagent before mixing with the sample. Reverse transcription was performed using miRCURY LNA RT Kit (Qiagen). The levels of indicated miRNAs and RNA spike-in were measured using specific miRCURY LNA miRNA Probe PCR assay in association with miRCURY SYBR Green PCR Kit (Qiagen). Reactions were run in triplicate on a StepOne Plus Real-time PCR System (Applied Biosystems), and the generated products were analyzed by the StepOne Plus Software (Version 2.3, Applied Biosistems). UniSp4 RNA or cel-miR39 was used for the normalization of miRNA expression as indicated.
SDS-PAGE and Western blot
sEVs from HaCaT conditioned media or HaCaT cells were lysed in NP-40/Triton lysis buffer. Equal volumes of extracts (corresponding to 20 µg in UV sample) were analyzed through SDS-PAGE. Western blot was performed with the following antibodies against GM130 (610823, BD Biosciences), TSG101 (sc-7964, Santa Cruz Biotechnology Inc.), CD81 (sc-7637, Santa Cruz Biotechnology Inc.) and Ago2 (MABE253, Merck Millipore). Protein bands were detected with SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Small RNA sequencing and analysis
RNA extraction and next generation sequencing was performed by Genomix4life S.R.L. (Baronissi, Salerno, Italy), using their standard protocols. SmallRNA sequencing libraries were prepared from 700 ng RNA using the TruSeq SmallRNA Sample Prep Kit (Illumina) according to the manufacturer’s instructions. Libraries were pooled at equimolar concentrations and sequenced on the Illumina NovaSeq 6000 System (Illumina) to obtain 75 bp single-end reads. Adapters were trimmed from raw reads using cutadapt [49], and small RNA expression analysis was performed using COMPSRA (v 1.0.3 [50]), , a comprehensive platform to identify and quantify different small RNA types. First of all reads were aligned to the human genome (UCSC, hg38) using the COMPSRA alignment module, a wrapper for STAR aligner (v 2.5.3a [51]), , with default parameters. Mapped reads were annotated to smallRNAs genes (miRNAs, piRNAs, tRNAs, snRNAs, snoRNAs, and circRNAs) using the COMPSRA annotation module. Annotation was performed using, as reference, the following databases: miRbase for miRNAs; piRNABank, piRBase and piRNACluster for piRNA; gtRNA for tRNA; GENECODE release 27 for snRNAs and snoRNAs; circBase for circRNAs. The function module was used to estimate the expression of each smallRNA. smallRNA counts were normalized among different samples using DESeq2 (v 1.36.0 [52]), , and differentially expressed smallRNAs between UV-sEVs and NT-sEVs were identified using the Wald test implemented in DESeq2. smallRNAs with absolute log2(fold change) > 1.5 and adjusted p-value < 0.05 were considered differentially expressed. Only mature miRNAs were used for downstream analysis. Variance stabilized counts (vsd) and fragment per million mapped reads (FPM) expression value were calculated using the varianceStabilizingTransformation and the fpm functions implemented in DESeq2. vsd values of mature miRNAs were used to performed principal component analysis (PCA) using the pca function implemented in the Bioconductor/R package PCAtools (v 2.8.0) with removeVar = 0.2. For heatmap representation FPM were z-score transformed. For specific analysis, mature microRNAs with less than 20 mapped reads per sample were considered not expressed.
Identification of putative TLR7-activating miRNAs
miRNA sequences were analyzed for nucleotide composition (numbers/percentage of A, C, G, U). miRNA sequences were also analyzed for the presence of putative TLR7-binding triplets as previously described [5, 9, 53]. Specifically, miRNAs were characterized as “high TLR7 binding” if containing at least one UU(U/C) triplet or “medium TLR7 binding” if containing at least one UU(A/G) triplet. miRNAs devoid of these triplets were classified as “no/dampened TLR7 binding”.
Statistics
Normal distribution in each group was always tested using the Shapiro-Wilk test first for the subsequent choice of a parametric (paired or unpaired 2-tailed Student’s t test, 1-way ANOVA with Dunnett’s post-hoc test ANOVA) or non-parametric (Mann-Whitney) test by using GraphPad Prism 9. P < 0.05 was considered significant. Base proportions were modelled using a multinomial regression. Post-hoc contrasts p-values were adjusted using Holm procedure.
Results
Hsa-miR203a-3p is a TLR7-activating ligand upregulated in psoriatic skin and released in plasma and in supernatants of inflamed keratinocytes
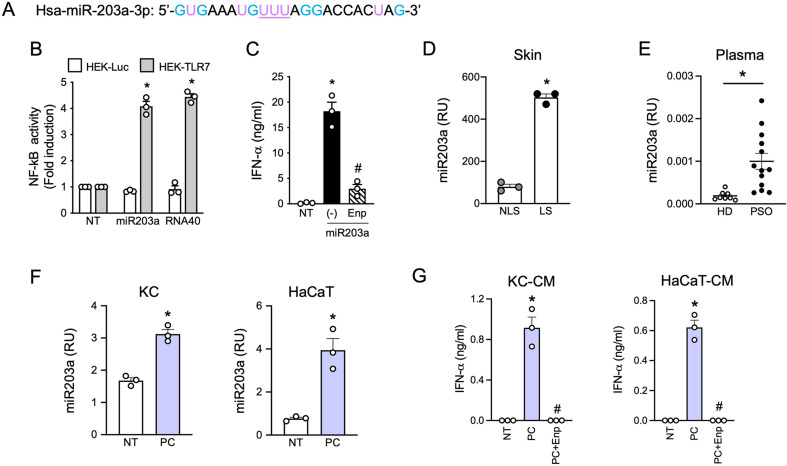
Based on previous work by our group demonstrating the capability of selected extracellular miRNAs to activate human DCs and trigger autoimmunity [23, 26], we observed that hsa-miR203a-3p (miR203a), a skin-specific miRNA that increases in psoriatic keratinocytes [37, 38], displayed six G and a “UUU” motif (Fig. 1A), which identify RNA sequences with full TLR7 binding potential [6, 8, 9]. Indeed, synthetic miR203a activated an NF-κB reporter gene in TLR7 stably transfected cell lines in a way comparable to the control sequence RNA40 [6] (Fig. 1B) and also induced the secretion of IFN-α by human pDCs (Fig. 1C), which express TLR7 as the main ssRNA receptor and do not express TLR8 [54]. Supplemental Fig. 1 shows that pDC activation induced by miR203a was full and functional, including TNF-α and IL-6 secretion (Supplemental Fig. 1A) and phenotypic maturation (Supplemental Fig. 1B), which allowed pDCs to activate allogeneic T cells (Supplemental Fig. 1C and 1D). To demonstrate the involvement of TLR7 in pDC activation by miR203a, pDCs were stimulated in the presence of Enpatoran, a selective TLR7/8 antagonist, which significantly reduced both cytokine secretion (Fig. 1C and Supplemental Fig. 1A) and T cell activation (Supplemental Fig. 1C-D) without affecting pDC viability (Supplemental Fig. 1E).
Fig. 1.
miR203a is upregulated in psoriatic patients and released by inflamed keratinocytes. (A) Sequence of synthetic hsa-miR203a-3p. G and U are highlighted in light blue and purple respectively while the TLR7 full binding motif “UUU” is underlined. (B) HEK293 cells stably transfected with a NF-kB reporter gene and human TLR7 or luciferase alone were stimulated with 10 µg/ml of miR203a or RNA40 for 24 h. NF-κB activation was evaluated in terms of luciferase activity (fold of induction) over unstimulated cells (NT). Data are expressed as mean ± SEM (n = 3); *P < 0.05 vs. “NT” by 1-way ANOVA with Dunnett’s post hoc test. (C) pDCs were pretreated or not with Enpatoran (Enp, 1 µM) for 1 h and then stimulated with miR203a for 24 h. IFN-α secretion was evaluated by ELISA (mean ± SEM, n = 3); *P < 0.05 vs. “NT” or #P < 0.05 vs. (-) by paired Student’s t test. (D) miR203a expression from paired non lesional (NL) and lesional (L) skin biopsies from 3 psoriatic patients was investigated by qPCR. Data are expressed as mean ± SEM of Relative Units (RU) compared to cel-miR39; *P < 0.05 vs. “NLS” by paired Student’s t test. (E) miR203a expression in the plasma of healthy donors (HD, n = 8) and psoriatic patients (PSO, n = 13) by qPCR; *P < 0.05 vs. “HD” by Mann-Whitney test. (F) Primary keratinocytes (KC) and HaCaT cells were stimulated with a mix of psoriatic cytokines (PC, each cytokine at 50 ng/ml) for 24 h and the expression of miR203a in the supernatant was assessed by qPCR (mean ± SEM, n = 3); *P < 0.05 vs. “NT” by paired Student’s t test. (G) pDCs were pretreated or not with Enpatoran for 1 h and then stimulated with conditioned media of KC or HaCaT (CM, 50% vol/vol). IFN-α production was evaluated by ELISA (mean ± SEM, n = 3); *P < 0.05 vs. “NT” or #P < 0.05 vs. (-) by paired Student’s t test
In accordance with previous literature [37, 38, 55], miR203a was significantly upregulated in lesional skin of three psoriatic patients as compared to non-lesional counterparts (Fig. 1D) and also increased in the plasma of psoriatic patients as compared to healthy donors (Fig. 1E). These findings indicate that miR203a upregulation correlates with psoriatic activation and that it is released and accumulate extracellularly upon skin damage, thus potentially representing a pDC-activating trigger in psoriasis. In the attempt to reproduce in vitro this phenomenon, primary (KC) and immortalized (HaCaT cells) keratinocytes were stimulated with a mix of cytokines mimicking the psoriatic inflammatory milieu (IL-1α, IL-6, TNF-α, IL-17, IL-22 and IFN-γ) [56]. Figure 1F demonstrates that miR203a could be detected in the supernatants of both cells and that it was increased by the psoriatic cytokine mix. We then asked whether miR203a-enriched supernatants could induce pDC activation. As shown by Fig. 1G, conditioned media of cells treated with psoriatic cytokines stimulated the production of IFN-α by pDCs in a TLR7 dependent manner, as demonstrated by the lack of production in the presence of Enpatoran. Of note, cytokines alone (unconditioned medium) did not induce IFN-α production by pDCs (Supplemental Fig. 1F).
This first set of experiments demonstrates that miR203a can be released by activated/psoriatic keratinocytes and, in turn, stimulate the production of the pathogenetic mediator IFN-α by pDCs.
miR203a is encapsulated in small EVs (sEVs) released by damaged keratinocytes
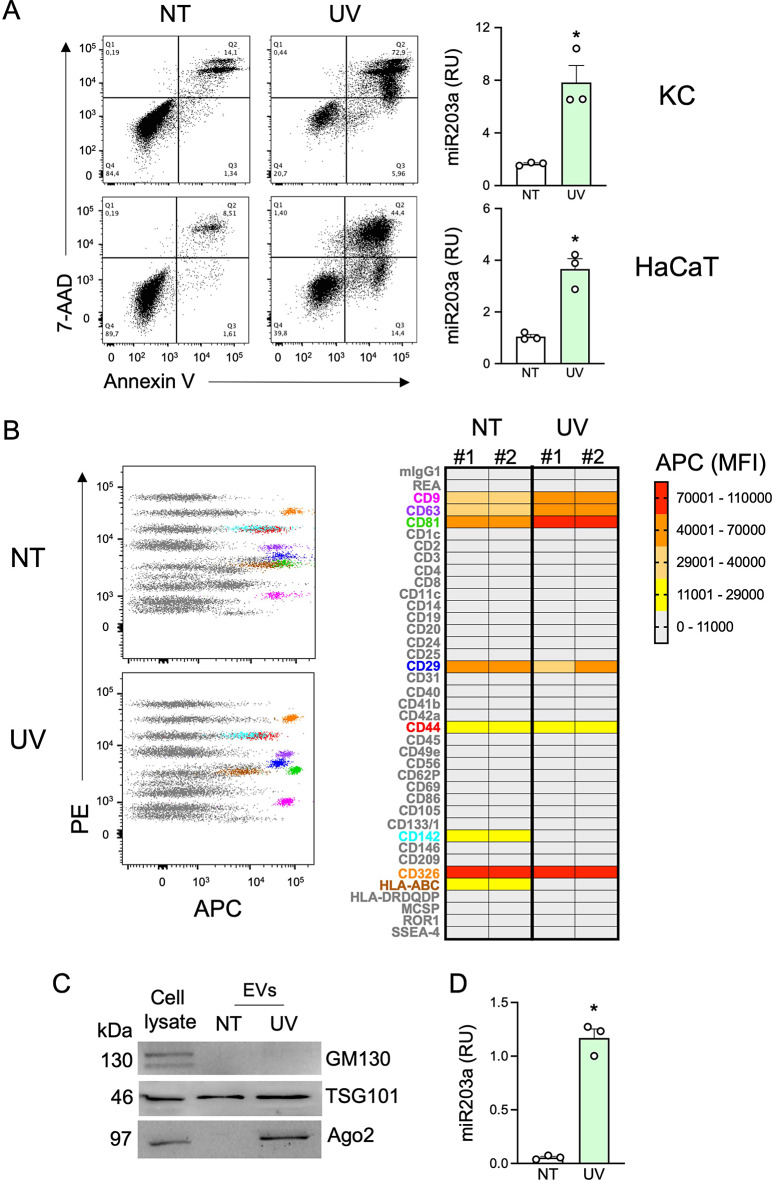
Next, we set up a model to investigate if extracellular miR203a is encapsulated in EVs. This task required the availability of large amounts of supernatants possibly devoid of exogenous molecules such as cytokines used in previous experiments. Since the initiation phase of psoriasis is closely linked with environmental triggers such as skin-damaging events and trauma (a.k.a Koebner phenomenon), keratinocytes and HaCaT cells were exposed to ultraviolet (UV) irradiation damage. As expected, UV exposure induced cell death as assessed by the double positivity for Annexin V and 7-AAD staining (Fig. 2A, left panels). In supernatants, we confirmed the accumulation of miR203a in both cell types (Fig. 2A, right panels). HaCaT cells were used for further experiments because of the limited growth capability of primary keratinocytes that hampered the collection of sufficient amounts of supernatants for EV separation and analysis. sEVs were separated by differential ultra-centrifugations from conditioned media of untreated- and UV-treated HaCaT cells (from now on, indicated as NT-sEVs and UV-sEVs). Morphology and size distribution of EVs were investigated by atomic force microscopy (AFM) and revealed that both EV populations had typical vesicular morphology with a diameter ranging from 35 to 180 nm, compatible with the size of sEVs (Supplemental Fig. 2A-B). Specifically, NT-sEVs showed a peak around 40 nm whereas UV-sEVs showed higher variability in size distribution with a prevalence of EVs with a diameter around 80 nm (Supplemental Fig. 2B). EVs were evaluated for purity by COlorimetric NANoplasmonic (CONAN) assay (Supplemental Fig. 2C) revealing that both NT- and UV-sEVs contained mainly vesicular particles being devoid of significant presence of single or aggregated proteins and thus of non-vesicular particles of diameter < 35 nm. Despite no significant difference could be found in the number of NT-sEVs versus UV-sEVs as well as in the three different preparations (Supplemental Fig. 2D), NT-sEVs and UV-sEVs differed in protein concentration, with a total protein content almost 10-fold higher in UV-sEVs as compared to NT-sEVs (Supplemental Fig. 2E). The expression of specific sEV markers was evaluated by flow cytometry and western blot to investigate extracellular and intracellular proteins, respectively. Multiplex bead-based flow cytometry showed the expression of the canonical vesicle markers CD9, CD63 and CD81 on sEVs, with a more abundant expression in UV-sEVs as indicated by the increase of the Median Fluorescence Intensity (MFI) (Fig. 2B).
Fig. 2.
sEVs derived from conditioned media of UV-treated HaCaT cells are enriched in Ago2 protein. (A) KC and HaCaT cells were UV irradiated and cell death was investigated by flow cytometry (left panels). miR203a expression in cell-free supernatants was assessed by qPCR (right panels). Data are expressed as mean ± SEM of Relative Units (RU) compared to UniSp4 RNA; *P < 0.05 vs. “NT” by paired Student’s t test. (B) sEV surface proteins were analysed by multiplex bead-based flow cytometry. PE channel vs. APC-stained channel dot plots (B, left panels) display the signal of single bead types. The bead populations with the brightest staining are colored. (B, right panel) Heatmap of the Median Fluorescence Intensity (MFI) of the APC channel for all 39 bead populations after subtraction of background signal (medium alone) in 2 independent sEV preparations. Markers called present are highlighted with the same colors as in dot plots. (C) Western blot analysis was performed on HaCaT cell lysates (10 µg) or on equal volumes of NT-sEVs and UV-sEVs. One representative experiment out of 3 is shown. (D) miR203a expression in NT-sEVs or UV-sEVs was evaluated by qPCR. Data are expressed as mean ± SEM of Relative Units (RU) compared to UniSp4 RNA; *P < 0.05 vs. “NT” by paired Student’s t test
By contrast, both NT-sEVs and UV-sEVs expressed similar levels of the integrin CD29, the epithelial marker CD326 (also known as EpCAM) and the glycoprotein CD44 (Fig. 2B). Interestingly, the surface expression of MHC-I (a.k.a. HLA-ABC) was downregulated on UV-sEVs (Fig. 2B). The expression of CD29 and MHC-I on sEVs was in line with what observed on the HaCaT cell surface (Supplemental Fig. 2F). sEVs were negative for most of the other surface markers investigated, such as hematopoietic markers, confirming the high specificity of the assay and the quality of sEV preparations (Fig. 2B and Supplemental Fig. 2F). Western blot analysis confirmed the presence of the typical sEV marker TSG101, a component of the ESCRT-I complex, and the absence of GM130, a cis-Golgi matrix protein, thus confirming EV enrichment and the absence of cell contaminants (Fig. 2C, upper panels). While using these protein extracts as controls in unrelated experiments, we unexpectedly noticed an enrichment in Ago2 protein, a common transporter of extracellular miRNAs, in UV-sEVs (Fig. 2C, lower panels). Finally, we confirmed that UV-sEVs were enriched in miR203a as compared to NT-sEVs (Fig. 2D).
These experiments, designed to localize miR203a within sEVs, a population of EVs with the appropriate size for pDCs uptake [57] and content delivery to TLR7, also provide initial evidence that sEVs from untreated and UV-treated keratinocyte differ in several quali-quantitative features such as diameter, protein content and markers that may deserve thorough attention in future studies.
UV-sEVs contain more miRNAs than NT-sEVs and display a higher TLR7-activating potential
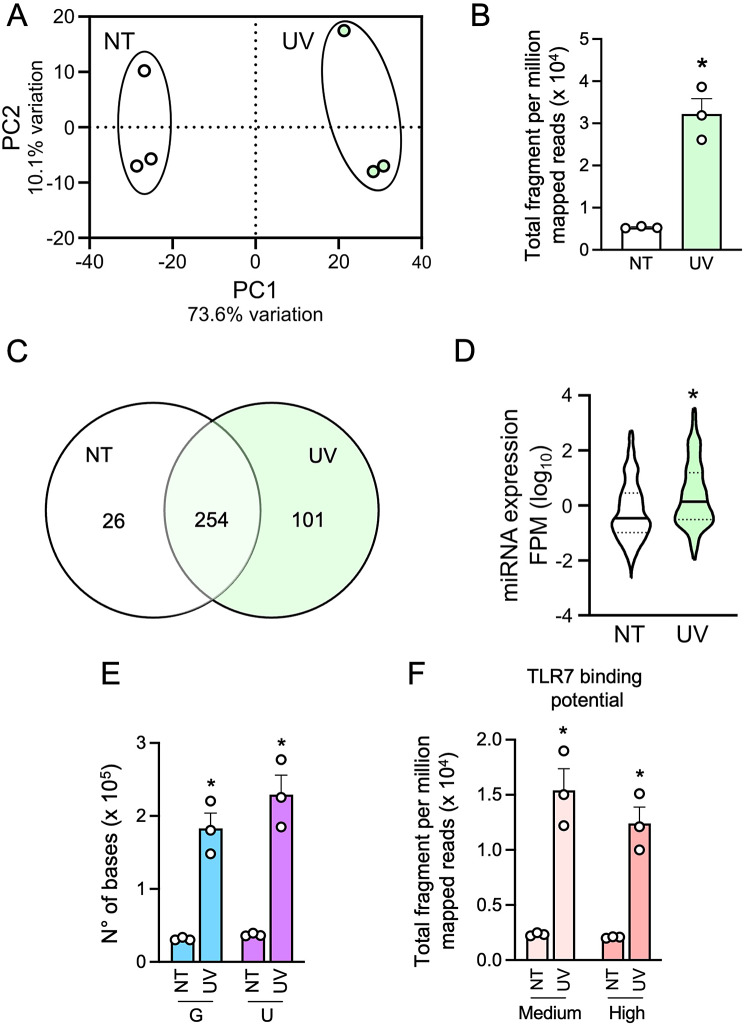
Based on the observed enrichment of UV-sEVs in the miRNA transporter Ago2 protein (Fig. 2C), we hypothesized an enrichment in other miRNAs besides miR203a. Therefore, smallRNA sequencing was performed to thoroughly characterize the vesicle miRNome. Principal component analysis (PCA) of miRNAs from sEVs revealed that UV-sEVs clearly segregate from NT-sEVs (Fig. 3A). According to our hypothesis, reads (FPM) attributed to miRNAs were significantly higher in UV-sEVs as compared to NT-sEVs (fold change UV-sEVs vs. NT-sEVs = 6.05, Fig. 3B). 280 miRNAs could be identified in NT-sEVs and 355 in UV-sEVs: of these, 101 were present only in UV-sEVs, 26 only in NT-sEVs and 254 in both populations (Fig. 3C). Despite this large overlap, miRNAs detected in UV-sEVs were present at significantly higher levels (FPM) as compared to NT-sEVs (Fig. 3D), in accordance with the highest number of global reads shown in Fig. 3B.
Fig. 3.
UV-sEVs contain more miRNAs as compared to NT-sEVs and display a higher TLR7-activating potential. (A) PCA scatterplot based on the variance stabilized counts of mature miRNAs identified in smallRNA-seq data of NT-sEVs (“NT”) and UV-sEVs (“UV”) (n = 3). (B) Total fragment per million mapped reads (FPM) in NT-sEVs and UV-sEVs. Data are expressed as mean ± SEM (n = 3); *P < 0.05 vs. “NT” by paired Student’s t test. (C) Venn diagram of number of miRNAs expressed in NT-sEVs and UV-sEVs as obtained from smallRNA-seq analysis. (D) Violin plot of the expression levels of miRNAs present either in NT-sEVs and UV-sEVs. For each miRNA the FPM value of 3 experiments was plotted. Data are shown as log10 (FPM). (E) G and U content was determined for each miRNA in NT-sEVs and UV-sEVs and adjusted for its expression (FPM). Data are expressed as mean ± SEM (n = 3); *P < 0.05 vs. “NT” by paired Student’s t test. (F) miRNAs were characterized as “high TLR7 binding” if containing at least one UUU/UUC triplet or “medium TLR7 binding” if containing at least one UUA/UUG triplet (medium binding potential) and represented with their respective levels of expression (FPM). Data are expressed as mean ± SEM (n = 3); *P < 0.05 vs. “NT” by paired Student’s t test
Next, we investigated the TLR7-binding properties of miRNAs contained in the two sEV populations. To this extent, we considered two approaches: GU content and content of TLR7-binding triplets [6, 8, 9]. Supplemental Fig. 3A shows that the 280 miRNAs in NT-sEVs and the 355 miRNAs in UV-sEVs displayed an overlapping base composition, as expected based on the high number of shared miRNAs. The quantification of these diagrams (Supplemental Fig. 3A, right panel) highlighted a prevalence of G and U over A and C in both conditions: this prevalence was previously identified as typical of human miRNAs [58, 59] and even more distinctive of extracellular miRNAs [60]. In addition, we noticed a statistically significant decrease in the G content in UV-sEVs, that was compensated by an increase (although in both cases not statistically significant) in U and A. As expected, we also found a similar composition in high- medium- and no-TLR7-binding miRNAs (Supplemental Fig. 3B).
However, when the contents of G and U and TLR7-binding triplets were adjusted for the level of expression of each specific miRNAs (FPM), the neat content of TLR7-activating bases (Fig. 3E) and miRNAs (as evaluated by the content of at least one TLR7-binding triplet, Fig. 3F) appeared significantly higher in UV-sEVs.
These results indicate that, despite NT-sEVs and UV-sEVs largely overlap for the panel of miRNA that they vehiculate, UV treatment increases the miRNA load of sEVs, thus also increasing the TLR7-activating potential of UV-sEVs.
Most miRNAs are upregulated in UV-sEVs and display marked TLR7-activating features
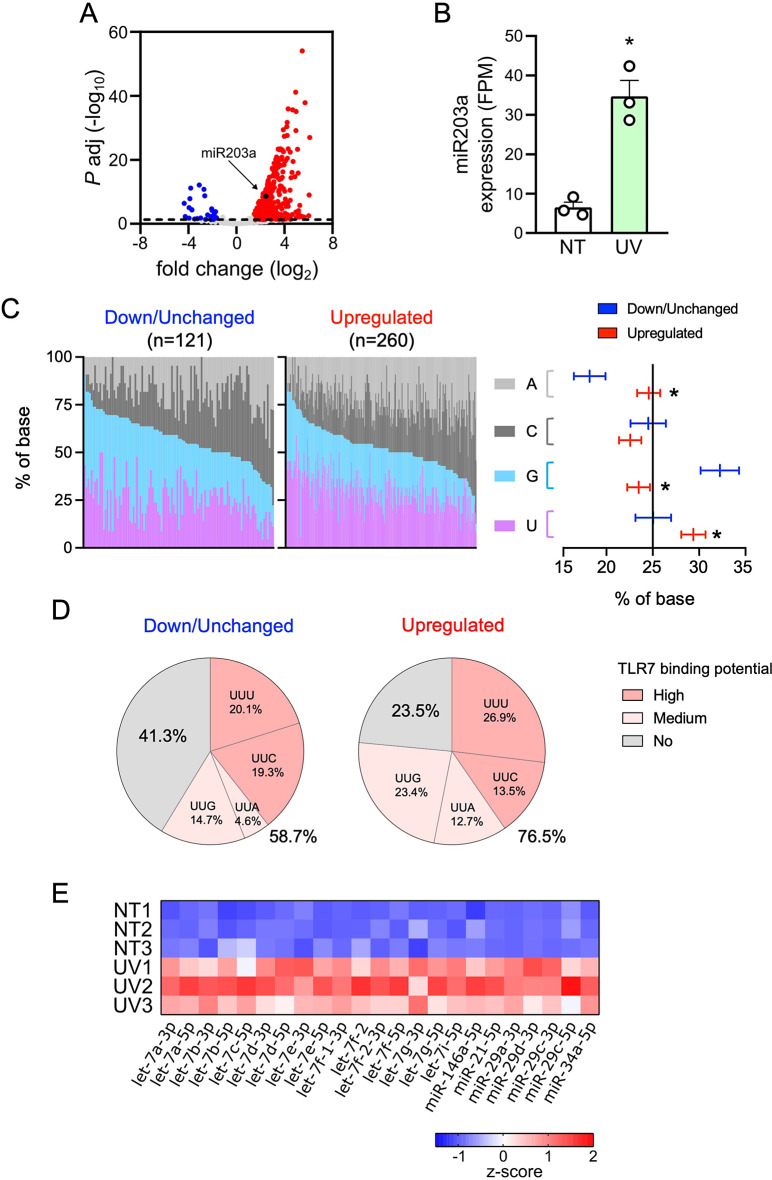
Next, we performed a differential miRNA expression analysis to compare UV-sEVs and NT-sEVs. A total of 281 miRNAs were significantly modulated in UV-sEVs as compared to NT-sEVs; notably, 260 of these miRNAs (92.5%) were upregulated in UV-sEVs (Fig. 4A and Supplemental 4A). Upregulation of miR203a in UV-sEVs was confirmed also in sequencing data (fold change = 5.51, adjusted p-value = 2.53 × 10− 9, Fig. 4B, upregulation assessed by real-time PCR was shown in Fig. 2D). Interestingly, among upregulated miRNAs we noticed miR21 and miR146a, previously described as elevated in psoriasis [37] and also found here to be upregulated in our psoriatic skin biopsies (Supplemental Fig. 4B upper panels), thus further validating that our in vitro model faithfully mimic psoriatic conditions. As an internal control of the accuracy of our bioinformatic analysis, these two miRNAs were confirmed as upregulated also in sequencing data (Supplemental Fig. 4B middle panels) and by real-time PCR in sEVs (Supplemental Fig. 4B lower panels). Additionally, we confirmed by real-time PCR that NT-sEVs and UV-sEVs contained equal levels of some miRNAs that were found unchanged by bioinformatic analysis (Supplemental Fig. 4C, and Methods section).
Fig. 4.
Most miRNAs are upregulated in UV-sEVs and are enriched in TLR7-activating moieties. (A) Volcano plot of differentially expressed miRNAs in NT-sEVs and UV-sEVs. For each miRNA, the log2 (fold change, x-axis) and the -log10 (adjusted p-value, y-axis) are shown. Dashed line shows the 0.05 boundary for adjusted p-value. Red dots: upregulated miRNAs (log2(fold change) > 1.5, adjusted p-value < 0.05; blue dots: downregulated miRNAs (log2(fold change) < -1.5, adjusted p-value < 0.05; grey dots: not differentially expressed miRNAs. Black dot highlights miR203a (B) miR203a expression levels in NT-sEVs (“NT”) and UV-sEVs (“UV”) as obtained from smallRNA-seq data. miR203a expression is reported as FPM (mean ± SEM, n = 3); *P < 0.05 vs. “NT” according to the Wald test implemented in DESeq2. (C, left panel) Base composition (%, colors as indicated in figure) of each down/unchanged or upregulated UV-sEV miRNA is represented. miRNAs were ordered based on decreasing contents of G + U. (C, right panel) Confidence level of the percentage of the base composition of down/unchanged or upregulated miRNAs is represented. Base proportions were modelled using a multinomial regression. Post-hoc contrasts p-values were adjusted using Holm procedure; *P < 0.05 vs. “NT”. (D) Pie charts representing the significantly different mean proportions of TLR7-binding miRNAs in down/unchanged or upregulated UV-sEVs. The distribution of TLR7 high, medium and no binding sequences between down/unchanged and upregulated was compared sing a multinomial logistic regression model and tested using a likelihood ratio test (LRT, p = 0.00055). (E) Heatmap of the modulation of miRNAs previously demonstrated to activate TLRs in UV-sEVs as compared to NT-sEVs. Data are shown as z-score transformed FPM
Differentially expressed miRNAs and unchanged miRNAs were analyzed for their TLR7-binding capability with the same approach of previous Sects. [6, 8, 9]. Base composition analysis (Fig. 4C) clearly showed that upregulated miRNAs were significantly enriched in U and A, while G was decreased compared to downregulated and unchanged miRNAs. In addition, 76.5% upregulated miRNAs contained high- or medium-TLR7-binding triplets, while only 58.7% of down/unchanged did (Fig. 4D). Finally, we found that the expression of a set of GU-rich miRNAs known to be responsible for TLR7/8 activation, namely let7 and miR29 family members, miR21, miR34a and miR146a [21–23, 26, 61–71], was significantly upregulated in UV-sEVs as compared to NT-sEVs (Fig. 4E).
These results reinforce our previous findings, indicating that the enrichment in TLR7-binding sequences in UV-sEVs is striking when considering upregulated miRNAs. Here, indeed, the skewing towards an increased TLR7-activating potential is independent of miRNA abundance and rather suggests the existence of mechanisms of “miRNA sorting” to enrich sEVs of damaged keratinocytes in TLR7-binding species. Further work is granted to confirm this hypothesis by studying the TLR7-binding miRNA cargo in other pathological settings.
UV-sEVs, but not NT-sEVs, activate pDCs via TLR7
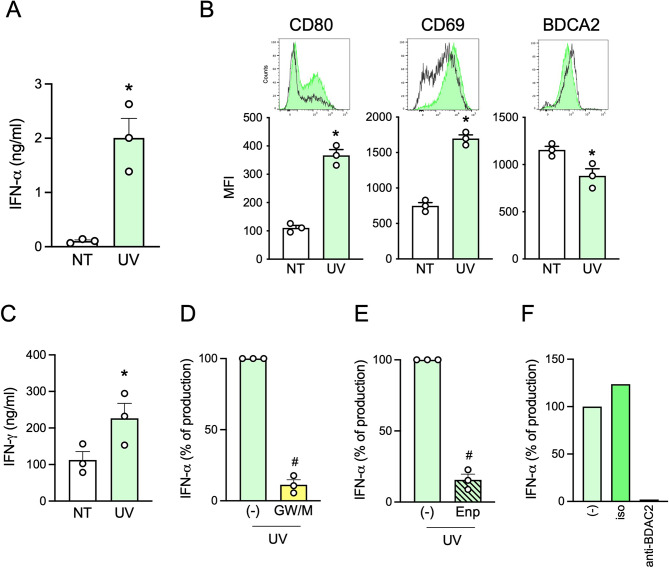
Next, we wanted to demonstrate the TLR7-activating capability of UV-sEVs (and their carried miRNome) in a biological setting. pDCs were stimulated with NT-sEVs and UV-sEVs, showing that UV-sEVs could induce IFN-α secretion (Fig. 5A), phenotypical maturation (Fig. 5B) and CD8+ T cell activation as shown by the increased release of IFN-γ (Fig. 5C). To better address the role of sEVs in pDC activation, HaCaT cells were treated with UVs in the presence of GW4869 and manumycin A (GW/M), which inhibit the ESCRT-independent and ESCRT-dependent EV secretory pathway respectively, prior to supernatant collection and sEV isolation. The inhibitor combination reduced the number (Supplemental Fig. 5A), the total protein content (Supplemental Fig. 5B), the expression of the sEV markers TSG101 and CD81 and of Ago2 protein (Supplemental Fig. 5C) as well as the content of miR203a (Supplemental Fig. 5D) of UV-sEVs. Accordingly, these sEVs induced low or no IFN-α when used to stimulate pDCs (Fig. 5D). Also, the block of IFN-α release in the presence of Enpatoran demonstrated that pDC activation almost completely depended on TLR7 triggering by sEV-vehiculated ligands (Fig. 5E). Finally, we show that pDC activation by sEVs can be abolished in the presence of an antibody directed against the inhibitory pDC molecule, BDCA2 [72](Fig. 5F).
Fig. 5.
The miRNA cargo of UV-sEVs activates pDCs via TLR7. (A-B) pDCs were stimulated for 24 h with an equal volume (approximately 1010 sEVs) of NT-sEVs (“NT”) or UV-sEVs (“UV”). IFN-α production was evaluated by ELISA (A) while pDC maturation was assessed by flow cytometry (B). (A-B) Data are expressed as mean ± SEM (n = 3); *P < 0.05 vs. “NT” by paired Student’s t test. (C) sEV-activated pDCs were cocultured for 6 days with allogenic CD8+ T cells and the secretion of IFN-γ was evaluated by ELISA in cell-free supernatants. Data are expressed as mean ± SEM (n = 3); *P < 0.05 vs. “NT” by paired Student’s t test. (D) pDCs were stimulated for 24 h with UV-sEVs produced in the presence or absence of 10 µM GW4869 (GW) and 1 µM manumycin A (M). (E) pDCs were stimulated with UV-sEVs in the presence or absence of Enpatoran (Enp, 1 µM) for 24 h. (D-E) IFN-α production was assessed by ELISA. Data are expressed as mean ± SEM (n = 3) of the percentage of IFN-α production; #P < 0.05 vs. (-) by paired Student’s t test. (F) pDCs were pretreated for 1 h with an anti-BDCA2 monoclonal antibody or an isotype control (both at 1 µg/ml) and then stimulated with UV-sEVs. The production of IFN-α was evaluated by ELISA and represented as percentage of IFN-α production. One representative donor out of 2 is shown
Taken together, these results demonstrate that UV-sEVs induce pDCs activation through the interaction between the miRNome carried by sEVs and TLR7 expressed by pDCs.
Discussion
miR203a is a well-known keratinocyte-specific miRNA upregulated in psoriatic skin [37, 38, 55]. Here, we show that miR203a is released by psoriatic keratinocytes encapsulated in sEVs and that it triggers the production of IFN-α by pDCs via TLR7 activation. More importantly, we show that miR203a represents only one paradigmatic TLR7-activating miRNA among the hundreds that are released by damaged keratinocytes, which we propose to work as an alarmin cargo contributing to pDC activation in the earliest phases of psoriatic pathogenesis.
To date, the existence of an unconventional mechanisms of TLR7/8 stimulation by extracellular miRNAs is well documented and linked to the establishment of detrimental inflammation in several pathological settings including cancer, neurodegeneration, sepsis, graft-vs.-host diseases and different forms of autoimmunity [21–23, 25, 26, 61–71, 73]. Most studies identified one or few upregulated GU-rich miRNAs as responsible of TLR7/8 activation, for a total of less than twenty activatory sequences characterized so far (let-7 and miR29 family members, miR-7a, miR-21, miR-34a, miR-122, miR-133a, miR-142, miR-145, miR-146a and miR-208a).
However, the current knowledge about RNA recognition by TLR7 [3, 5, 9] suggests that many more miRNAs than previously thought may work as ligands after endosomal digestion into smaller sequences. In accordance, we found that sEVs released by damaged keratinocytes are strongly enriched in TLR7 binding sequences (in addition to miR203a) as compared to sEVs from resting keratinocytes and that such enrichment correlated with the potential to activate pDCs via TLR7. Thus, unconventional TLR-stimulating functions may need to be rethought as dependent on whole sEV miRNA cargoes shifted towards stronger TLR7-activating potential rather than on a single/few specific upregulated miRNAs. This hypothesis needs further research to demonstrate the existence of such TLR-binding shift and related TLR-dependent immune cell activation in other pathological conditions. In this direction, one bioinformatic study reported that apoptotic endothelial cells release sEVs loaded with immunostimulatory viral-like RNAs strongly enriched in U-rich motives which could represent TLR7/8 ligands, but no functional activation of TLR7/8 expressing cells is shown [60]. To the best of our knowledge, our study shows for the first time a clear functional correlation between increased TLR7-binding sequences and activation of primary human TLR7-expressing immune cells.
Our results also add evidence in support of the physiological relevance of cell-to-cell miRNA-based communication, which remains uncertain because of the low representativity of specific miRNAs within the extracellular pool [48, 74–76]. Indeed, to trigger measurable effects via conventional post-transcriptional regulation, a threshold of about one thousand copies of target-specific miRNAs has to be reached in the cytoplasm of each recipient cell [77], which implies an estimated delivery of tens of thousands of sEVs and also an efficient escape of undigested miRNAs from the endosomes [78], a well-known rate-limiting step for both therapeutic short interference RNA delivery and viral infection [79, 80]. By contrast, unconventional TLR-binding activity does not require the simultaneous entry of hundreds of copies of one target-specific miRNA in the recipient cell. Rather, as indicated by our results, numerous TLR-binding miRNAs would act synergistically to activate TLR7. In addition, receptor activation by miRNA-loaded sEVs is plausible, given that the concentration of a single RNA molecule within sEVs is calculated in the micromolar range, which matches TLR7 affinity for GU-rich sequences [9, 24, 81]. Finally, since inflammation is an auto-amplifying process, it easily transfers even weak local signals to the organism level, as well documented by IFN-α production by pDCs that ignite a systemic autoimmune response [31, 32, 34].
Extracellular miRNAs can be either free or encapsulated into EVs [48, 82, 83]. Our analysis focused on sEV-encapsulated miRNA which could activate pDCs in a TLR7-dependent manner, in accordance with most of the works describing unconventional TLR-binding activity [21, 23, 26, 61, 63, 65–71]. Indeed, EV-encapsulation facilitates the entering in the endocytic pathway where miRNAs may directly engage TLR7/8 following necessary fragmentation [5, 84]. In this regard immune cells, including pDCs, not only represent the major sites of TLR7 or TLR8 expression but also display a marked attitude to particle engulfment correlated to their antimicrobial and/or antigen presenting functions [85, 86]. Interestingly, nanometric particles were shown to be selectively internalized by pDCs leading to the production of large amounts of IFN-α, in accordance the activation by nanometric sEVs demonstrated here [57]. However, the present experimental setting does not allow to exclude that micrometric particles and/or free miRNAs might also reach endosomal TLR7 in pDCs. In this regard, the uptake of free miRNAs by pDCs was previously described to take place following the recognition of Ago2 by the receptor Neuropilin-1 [87].
In line with theories suggesting that EVs derived from healthy tissues are essentially immune-silent [88], sEVs from resting keratinocytes did not activate pDCs. Based on the effects of Enpatoran, which dramatically decreases pDC activation by UV-sEVs, we ascribe this difference to the “healthy” sEV cargo of resting cells, characterized by reduced TLR7-binding potential. However, our EV characterization shows that also protein markers and composition of the sEV populations produced by stressed cells differ from healthy ones, which may also play a role in making vesicles more activatory. Previous studies indicated that sEVs released in acute stress responses concentrate immune active molecules apart from miRNAs, including lipids, prostaglandins, proteins and metabolites: these, delivered as a bundle, were proposed to initiate sterile inflammation and shape the outcome of the immune response in different contexts [88–92]. In such view, it makes sense to hypothesize that acute stress messages are sent more easily and efficiently by increasing the sorting of GU-rich miRNA into sEVs rather than by upregulating one single specific miRNA. We also propose that, while immune cells equipped with a broader/different variety of alarmin receptors may respond to several EV components [91], pDC activation may mostly depend on the miRNA cargo.
Finally, inhibitor experiments shown here indicate that pDC activation largely depends on unconventional TLR7 activation by miRNAs but do not completely rule out the possibility that sEV-delivered miRNAs may exert marginal post-transcriptional regulation in pDCs.
From a translational point of view, the inhibition of IFN-α production by both Enpatoran and anti-BDCA2 antibody has a therapeutical significance, being both drugs in clinical trials for the treatment of IMIDs [13, 15, 16, 93].
In conclusion, our results indicate that the miRNA cargo of sEVs enriched in TLR7-activating sequences may contribute to unwanted inflammation and represent a novel mechanism capable to trigger psoriasis and possibly other IMIDs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- AFM
Atomic force microscopy
- Ago2
Argonaute 2
- CONAN
Colorimetric Nanoplasmonic
- DCs
Dendritic cells
- FPM
Fragment per million mapped reads
- IFN
Interferon
- IMIDs
Immune-mediated inflammatory diseases
- KCs
Keratinocytes
- MFI
Median fluorescence intensity
- miRNAs
MicroRNAs
- NT-sEVs
Small extracellular vesicles from conditioned media of untreated HaCaT cells
- pDCs
Plasmacytoid dendritic cells
- PHA
Phytohemagglutinin
- qPCR
Quantitative polymerase chain reaction
- sEVs
Small extracellular vesicles
- ssRNA
Single-stranded RNA
- TLR
Toll-like receptor
- UV
Ultraviolet
- UV-sEVs
Small extracellular vesicles from conditioned media of UV-treated HaCaT cells
Author contributions
Conceptualization, VS, DB. Methodology, VS, BM, FB, SA, PB, DB. Validation and formal analysis, VS, BM, GG, LT, PB, SC, FB, DB. Investigation, VS, CG, BM, GG, SA, VG, AC, CS. Data curation, VS, CG, BM, GG, SA. Writing—original draft preparation, VS, BM, DB. Writing—review and editing, VS, ADP, SS, FB, CS, DB. Visualization, VS, CG, BM, GG, SA. Supervision, VS, FB, DB. Funding acquisition, VS, LT, ADP, SS, DB. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Italian Ministry of the University and Research (MUR-PRIN 2022PYH73K to VS, MUR-PRIN PNRR P2022L3LJN to DB), PNRR-CN3 (National Center for Gene Therapy and Drugs based on RNA Technology to SS), University of Brescia (Fondi Locali to VS, LT, ADP, DB). These funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Medzhitov R, Preston-Hurlburt P, Janeway CA. A human homologue of the Drosophila toll protein signals activation of adaptive immunity. Nature. 1997;388:394–7. [DOI] [PubMed] [Google Scholar]
- 2.Yu L, Wang L, Chen S. Endogenous toll-like receptor ligands and their biological significance. J Cell Mol Med. 2010;14:2592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimizu T. Structural insights into ligand recognition and regulation of nucleic acid-sensing toll-like receptors. Curr Opin Struct Biol. 2017;47:52–9. [DOI] [PubMed] [Google Scholar]
- 4.Gorden KB, Gorski KS, Gibson SJ, Kedl RM, Kieper WC, Qiu X, Tomai MA, Alkan SS, Vasilakos JP. Synthetic TLR agonists reveal functional differences between human TLR7 and TLR8. J Immunol. 2005;174:1259–68. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z, Ohto U, Shibata T, Krayukhina E, Taoka M, Yamauchi Y, Tanji H, Isobe T, Uchiyama S, Miyake K, Shimizu T. Structural analysis reveals that toll-like receptor 7 is a dual receptor for Guanosine and single-stranded RNA. Immunity. 2016;45:737–48. [DOI] [PubMed] [Google Scholar]
- 6.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–9. [DOI] [PubMed] [Google Scholar]
- 7.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–31. [DOI] [PubMed] [Google Scholar]
- 8.Forsbach A, Nemorin JG, Montino C, Müller C, Samulowitz U, Vicari AP, Jurk M, Mutwiri GK, Krieg AM, Lipford GB, Vollmer J. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol. 2008;180:3729–38. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z, Ohto U, Shibata T, Taoka M, Yamauchi Y, Sato R, Shukla NM, David SA, Isobe T, Miyake K, Shimizu T. Structural analyses of toll-like receptor 7 Reveal detailed RNA sequence specificity and recognition mechanism of Agonistic Ligands. Cell Rep. 2018;25:3371–e33813375. [DOI] [PubMed] [Google Scholar]
- 10.Diebold SS, Massacrier C, Akira S, Paturel C, Morel Y, Reis e Sousa C. Nucleic acid agonists for toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur J Immunol. 2006;36:3256–67. [DOI] [PubMed] [Google Scholar]
- 11.Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–28. [DOI] [PubMed] [Google Scholar]
- 12.Berland R, Fernandez L, Kari E, Han JH, Lomakin I, Akira S, Wortis HH, Kearney JF, Ucci AA, Imanishi-Kari T. Toll-like receptor 7-dependent loss of B cell tolerance in pathogenic autoantibody knockin mice. Immunity. 2006;25:429–40. [DOI] [PubMed] [Google Scholar]
- 13.Farrugia M, Baron B. The Role of Toll-Like receptors in Autoimmune diseases through failure of the self-recognition mechanism. Int J Inflam. 2017;2017:8391230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sozzani S, Del Prete A, Bosisio D. Dendritic cell recruitment and activation in autoimmunity. J Autoimmun. 2017;85:126–40. [DOI] [PubMed] [Google Scholar]
- 15.Klopp-Schulze L, Shaw JV, Dong JQ, Khandelwal A, Vazquez-Mateo C, Goteti K. Applying modeling and simulations for rational dose selection of Novel Toll-Like receptor 7/8 inhibitor enpatoran for indications of High Medical need. Clin Pharmacol Ther. 2022;112:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klopp-Schulze L, Gopalakrishnan S, Yalkinoglu Ö, Kuroki Y, Lu H, Goteti K, Krebs-Brown A, Nogueira Filho M, Gradhand U, Fluck M et al. Asia-inclusive global development of enpatoran: results of an ethno-bridging study, intrinsic/extrinsic factor assessments and disease trajectory modeling to inform design of a phase II multiregional clinical trial. Clin Pharmacol Ther. 2024;115(6):1346–57. [DOI] [PubMed]
- 17.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. [DOI] [PubMed] [Google Scholar]
- 18.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. [DOI] [PubMed] [Google Scholar]
- 19.Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miRNAs: cell-cell communication function? Front Genet. 2013;4:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–32. [DOI] [PubMed] [Google Scholar]
- 21.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, et al. MicroRNAs bind to toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehmann SM, Krüger C, Park B, Derkow K, Rosenberger K, Baumgart J, Trimbuch T, Eom G, Hinz M, Kaul D, et al. An unconventional role for miRNA: let-7 activates toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–35. [DOI] [PubMed] [Google Scholar]
- 23.Salvi V, Gianello V, Busatto S, Bergese P, Andreoli L, D’Oro U, Zingoni A, Tincani A, Sozzani S, Bosisio D. Exosome-delivered microRNAs promote IFN-α secretion by human plasmacytoid DCs via TLR7. JCI Insight 2018, 3. [DOI] [PMC free article] [PubMed]
- 24.Bosch S, Young NA, Mignot G, Bach JM. Epigenetic mechanisms in Immune Disease: the significance of toll-like receptor-binding Extracellular Vesicle-Encapsulated microRNA. Front Genet. 2020;11:578335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosisio D, Gianello V, Salvi V, Sozzani S. Extracellular miRNAs as activators of innate immune receptors. Cancer Lett. 2019;452:59–65. [DOI] [PubMed] [Google Scholar]
- 26.Gaudenzi C, Schioppa T, Passari M, Zucchi G, Tiberio L, Vahidi Y, Scutera S, Musso T, Sozzani S, Del Prete A, et al. Extracellular microRNAs induce dendritic cell-dependent joint inflammation and potentiate osteoclast differentiation via TLR7/8 engagement. J Autoimmun. 2024;145:103189. [DOI] [PubMed] [Google Scholar]
- 27.David T, Ling SF, Barton A. Genetics of immune-mediated inflammatory diseases. Clin Exp Immunol. 2018;193:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surace AEA, Hedrich CM. The role of epigenetics in Autoimmune/Inflammatory Disease. Front Immunol. 2019;10:1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boehncke WH, Schön MP. Psoriasis. Lancet. 2015;386:983–94. [DOI] [PubMed] [Google Scholar]
- 30.Sagi L, Trau H. The Koebner phenomenon. Clin Dermatol. 2011;29:231–6. [DOI] [PubMed] [Google Scholar]
- 31.Nestle FO, Conrad C, Tun-Kyi A, Homey B, Gombert M, Boyman O, Burg G, Liu YJ, Gilliet M. Plasmacytoid predendritic cells initiate psoriasis through interferon-alpha production. J Exp Med. 2005;202:135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conrad C, Boyman O, Tonel G, Tun-Kyi A, Laggner U, de Fougerolles A, Kotelianski V, Gardner H, Nestle FO. Alpha1beta1 integrin is crucial for accumulation of epidermal T cells and the development of psoriasis. Nat Med. 2007;13:836–42. [DOI] [PubMed] [Google Scholar]
- 33.Wollenberg A, Wagner M, Günther S, Towarowski A, Tuma E, Moderer M, Rothenfusser S, Wetzel S, Endres S, Hartmann G. Plasmacytoid dendritic cells: a new cutaneous dendritic cell subset with distinct role in inflammatory skin diseases. J Invest Dermatol. 2002;119:1096–102. [DOI] [PubMed] [Google Scholar]
- 34.Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, Cao W, Su B, Nestle FO, Zal T, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–9. [DOI] [PubMed] [Google Scholar]
- 35.Lande R, Chamilos G, Ganguly D, Demaria O, Frasca L, Durr S, Conrad C, Schröder J, Gilliet M. Cationic antimicrobial peptides in psoriatic skin cooperate to break innate tolerance to self-DNA. Eur J Immunol. 2015;45:203–13. [DOI] [PubMed] [Google Scholar]
- 36.Ganguly D, Chamilos G, Lande R, Gregorio J, Meller S, Facchinetti V, Homey B, Barrat FJ, Zal T, Gilliet M. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J Exp Med. 2009;206:1983–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonkoly E, Wei T, Janson PC, Sääf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS ONE. 2007;2:e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lerman G, Avivi C, Mardoukh C, Barzilai A, Tessone A, Gradus B, Pavlotsky F, Barshack I, Polak-Charcon S, Orenstein A, et al. MiRNA expression in psoriatic skin: reciprocal regulation of hsa-miR-99a and IGF-1R. PLoS ONE. 2011;6:e20916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen JQ, Papp G, Szodoray P, Zeher M. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev. 2016;15:1171–80. [DOI] [PubMed] [Google Scholar]
- 40.Takasawa R, Nakamura H, Mori T, Tanuma S. Differential apoptotic pathways in human keratinocyte HaCaT cells exposed to UVB and UVC. Apoptosis. 2005;10:1121–30. [DOI] [PubMed] [Google Scholar]
- 41.Salvi V, Nguyen HO, Sozio F, Schioppa T, Gaudenzi C, Laffranchi M, Scapini P, Passari M, Barbazza I, Tiberio L et al. SARS-CoV-2-associated ssRNAs activate inflammation and immunity via TLR7/8. JCI Insight. 2021;6(18):e150542. [DOI] [PMC free article] [PubMed]
- 42.Montis C, Zendrini A, Valle F, Busatto S, Paolini L, Radeghieri A, Salvatore A, Berti D, Bergese P. Size distribution of extracellular vesicles by optical correlation techniques. Colloids Surf B Biointerfaces. 2017;158:331–8. [DOI] [PubMed] [Google Scholar]
- 43.Radeghieri A, Savio G, Zendrini A, Di Noto G, Salvi A, Bergese P, Piovani G. Cultured human amniocytes express hTERT, which is distributed between nucleus and cytoplasm and is secreted in extracellular vesicles. Biochem Biophys Res Commun. 2017;483:706–11. [DOI] [PubMed] [Google Scholar]
- 44.Busatto S, Giacomini A, Montis C, Ronca R, Bergese P. Uptake profiles of human serum exosomes by murine and human tumor cells through combined use of Colloidal Nanoplasmonics and Flow Cytofluorimetric Analysis. Anal Chem. 2018;90:7855–61. [DOI] [PubMed] [Google Scholar]
- 45.Maiolo D, Paolini L, Di Noto G, Zendrini A, Berti D, Bergese P, Ricotta D. Colorimetric nanoplasmonic assay to determine purity and titrate extracellular vesicles. Anal Chem. 2015;87:4168–76. [DOI] [PubMed] [Google Scholar]
- 46.Montis C, Caselli L, Valle F, Zendrini A, Carlà F, Schweins R, Maccarini M, Bergese P, Berti D. Shedding light on membrane-templated clustering of gold nanoparticles. J Colloid Interface Sci. 2020;573:204–14. [DOI] [PubMed] [Google Scholar]
- 47.Zendrini A, Paolini L, Busatto S, Radeghieri A, Romano M, Wauben MHM, van Herwijnen MJC, Nejsum P, Borup A, Ridolfi A, et al. Corrigendum: augmented COlorimetric NANoplasmonic (CONAN) Method for Grading Purity and Determine Concentration of EV Microliter volume solutions. Front Bioeng Biotechnol. 2021;9:674507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kechin A, Boyarskikh U, Kel A, Filipenko M. cutPrimers: a New Tool for Accurate cutting of primers from reads of targeted next generation sequencing. J Comput Biol. 2017;24:1138–43. [DOI] [PubMed] [Google Scholar]
- 50.Li J, Kho AT, Chase RP, Pantano L, Farnam L, Amr SS, Tantisira KG. COMPSRA: a COMprehensive platform for small RNA-Seq data analysis. Sci Rep. 2020;10:4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love MI, Huber W, Anders S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moreno-Eutimio MA, López-Macías C, Pastelin-Palacios R. Bioinformatic analysis and identification of single-stranded RNA sequences recognized by TLR7/8 in the SARS-CoV-2, SARS-CoV, and MERS-CoV genomes. Microbes Infect. 2020;22:226–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F, Liu YJ. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J Exp Med. 2001;194:863–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mostafa SA, Mohammad MHS, Negm WA, Batiha GES, Alotaibi SS, Albogami SM, Waard M, Tawfik NZ, Abdallah HY. Circulating microRNA203 and its target genes’ role in psoriasis pathogenesis. Front Med (Lausanne). 2022;9:988962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bocheńska K, Smolińska E, Moskot M, Jakóbkiewicz-Banecka J, Gabig-Cimińska M. Models in the research process of psoriasis. Int J Mol Sci. 2017;18(12):2514. [DOI] [PMC free article] [PubMed]
- 57.Rettig L, Haen SP, Bittermann AG, von Boehmer L, Curioni A, Krämer SD, Knuth A, Pascolo S. Particle size and activation threshold: a new dimension of danger signaling. Blood. 2010;115:4533–41. [DOI] [PubMed] [Google Scholar]
- 58.Fang Z, Du R, Edwards A, Flemington EK, Zhang K. The sequence structures of human microRNA molecules and their implications. PLoS ONE. 2013;8:e54215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rolle K, Piwecka M, Belter A, Wawrzyniak D, Jeleniewicz J, Barciszewska MZ, Barciszewski J. The sequence and structure determine the function of mature human miRNAs. PLoS ONE. 2016;11:e0151246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hardy MP, Audemard É, Migneault F, Feghaly A, Brochu S, Gendron P, Boilard É, Major F, Dieudé M, Hébert MJ, Perreault C. Apoptotic endothelial cells release small extracellular vesicles loaded with immunostimulatory viral-like RNAs. Sci Rep. 2019;9:7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He WA, Calore F, Londhe P, Canella A, Guttridge DC, Croce CM. Microvesicles containing miRNAs promote muscle cell death in cancer cachexia via TLR7. Proc Natl Acad Sci U S A. 2014;111:4525–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park CK, Xu ZZ, Berta T, Han Q, Chen G, Liu XJ, Ji RR. Extracellular microRNAs activate nociceptor neurons to elicit pain via TLR7 and TRPA1. Neuron. 2014;82:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salama A, Fichou N, Allard M, Dubreil L, De Beaurepaire L, Viel A, Jégou D, Bösch S, Bach JM. MicroRNA-29b modulates innate and antigen-specific immune responses in mouse models of autoimmunity. PLoS ONE. 2014;9:e106153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu HY, Huang CM, Hung YF, Hsueh YP. The microRNAs Let7c and miR21 are recognized by neuronal toll-like receptor 7 to restrict dendritic growth of neurons. Exp Neurol. 2015;269:202–12. [DOI] [PubMed] [Google Scholar]
- 65.Yelamanchili SV, Lamberty BG, Rennard DA, Morsey BM, Hochfelder CG, Meays BM, Levy E, Fox HS. Correction: MiR-21 in Extracellular vesicles leads to Neurotoxicity via TLR7 Signaling in SIV neurological disease. PLoS Pathog. 2015;11:e1005131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim SJ, Chen Z, Essani AB, Elshabrawy HA, Volin MV, Volkov S, Swedler W, Arami S, Sweiss N, Shahrara S. Identification of a Novel toll-like receptor 7 endogenous ligand in Rheumatoid Arthritis Synovial Fluid that can provoke arthritic joint inflammation. Arthritis Rheumatol. 2016;68:1099–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coleman LG, Zou J, Crews FT. Microglial-derived miRNA let-7 and HMGB1 contribute to ethanol-induced neurotoxicity via TLR7. J Neuroinflammation. 2017;14:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feng Y, Zou L, Yan D, Chen H, Xu G, Jian W, Cui P, Chao W. Extracellular MicroRNAs induce potent Innate Immune responses via TLR7/MyD88-Dependent mechanisms. J Immunol. 2017;199:2106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ranganathan P, Ngankeu A, Zitzer NC, Leoncini P, Yu X, Casadei L, Challagundla K, Reichenbach DK, Garman S, Ruppert AS, et al. Serum miR-29a is upregulated in Acute Graft-versus-host disease and activates dendritic cells through TLR binding. J Immunol. 2017;198:2500–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young NA, Valiente GR, Hampton JM, Wu LC, Burd CJ, Willis WL, Bruss M, Steigelman H, Gotsatsenko M, Amici SA, et al. Estrogen-regulated STAT1 activation promotes TLR8 expression to facilitate signaling via microRNA-21 in systemic lupus erythematosus. Clin Immunol. 2017;176:12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu J, Feng Y, Jeyaram A, Jay SM, Zou L, Chao W. Circulating plasma Extracellular vesicles from septic mice induce inflammation via MicroRNA- and TLR7-Dependent mechanisms. J Immunol. 2018;201:3392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tiberio L, Laffranchi M, Zucchi G, Salvi V, Schioppa T, Sozzani S, Del Prete A, Bosisio D. Inhibitory receptors of plasmacytoid dendritic cells as possible targets for checkpoint blockade in cancer. Front Immunol. 2024;15:1360291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, Liang H, Jin F, Yan X, Xu G, Hu H, Liang G, Zhan S, Hu X, Zhao Q, et al. Injured liver-released miRNA-122 elicits acute pulmonary inflammation via activating alveolar macrophage TLR7 signaling pathway. Proc Natl Acad Sci U S A. 2019;116:6162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, Tuschl T. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc Natl Acad Sci U S A. 2013;110:4255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111:14888–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Albanese M, Chen YA, Hüls C, Gärtner K, Tagawa T, Mejias-Perez E, Keppler OT, Göbel C, Zeidler R, Shein M, et al. MicroRNAs are minor constituents of extracellular vesicles that are rarely delivered to target cells. PLoS Genet. 2021;17:e1009951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, Baccarini A, Lazzari G, Galli C, Naldini L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–67. [DOI] [PubMed] [Google Scholar]
- 78.Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang Z, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johannes L, Lucchino M. Current challenges in Delivery and Cytosolic translocation of therapeutic RNAs. Nucleic Acid Ther. 2018;28:178–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Staring J, Raaben M, Brummelkamp TR. Viral escape from endosomes and host detection at a glance. J Cell Sci. 2018;131(15):jcs216259. [DOI] [PubMed]
- 81.Crozat K, Beutler B. TLR7: a new sensor of viral infection. Proc Natl Acad Sci U S A. 2004;101:6835–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE. 2012;7:e30679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles 2014, 3. [DOI] [PMC free article] [PubMed]
- 85.Lin F, Yin HB, Li XY, Zhu GM, He WY, Gou X. Bladder cancer cell–secreted exosomal miR–21 activates the PI3K/AKT pathway in macrophages to promote cancer progression. Int J Oncol. 2020;56:151–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun LH, Tian D, Yang ZC, Li JL. Exosomal miR-21 promotes proliferation, invasion and therapy resistance of colon adenocarcinoma cells through its target PDCD4. Sci Rep. 2020;10:8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Prud’homme GJ, Glinka Y, Lichner Z, Yousef GM. Neuropilin-1 is a receptor for extracellular miRNA and AGO2/miRNA complexes and mediates the internalization of miRNAs that modulate cell function. Oncotarget. 2016;7:68057–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fleshner M, Crane CR. Exosomes, DAMPs and miRNA: features of stress physiology and Immune Homeostasis. Trends Immunol. 2017;38:768–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dieudé M, Bell C, Turgeon J, Beillevaire D, Pomerleau L, Yang B, Hamelin K, Qi S, Pallet N, Béland C, et al. The 20S proteasome core, active within apoptotic exosome-like vesicles, induces autoantibody production and accelerates rejection. Sci Transl Med. 2015;7:318ra200. [DOI] [PubMed] [Google Scholar]
- 90.Walsh SA, Hoyt BW, Rowe CJ, Dey D, Davis TA. Alarming cargo: the role of exosomes in trauma-induced inflammation. Biomolecules. 2021;11(4):522. [DOI] [PMC free article] [PubMed]
- 91.Vulpis E, Soriani A, Cerboni C, Santoni A, Zingoni A. Cancer exosomes as conveyors of stress-induced molecules: new players in the modulation of NK cell response. Int J Mol Sci. 2019;20(3):611. [DOI] [PMC free article] [PubMed]
- 92.Beninson LA, Brown PN, Loughridge AB, Saludes JP, Maslanik T, Hills AK, Woodworth T, Craig W, Yin H, Fleshner M. Acute stressor exposure modifies plasma exosome-associated heat shock protein 72 (Hsp72) and microRNA (mir-142-5p and miR-203). PLoS ONE. 2014;9:e108748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Furie RA, van Vollenhoven RF, Kalunian K, Navarra S, Romero-Diaz J, Werth VP, Huang X, Clark G, Carroll H, Meyers A, et al. Trial of Anti-BDCA2 antibody litifilimab for systemic Lupus Erythematosus. N Engl J Med. 2022;387:894–904. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.