Abstract
Background
Obesity is a complex, diverse and multifactorial disease that has become a major public health concern in the last decades. The current classification systems relies on anthropometric measurements, such as BMI, that are unable to capture the physiopathological diversity of this disease. The aim of this study was to redefine the classification of obesity based on the different H-NMR metabolomics profiles found in individuals with obesity to better assess the risk of future development of cardiometabolic disease.
Materials and methods
Serum samples of a subset of the Di@bet.es cohort consisting of 1387 individuals with obesity were analyzed by H-NMR. A K-means algorithm was deployed to define different H-NMR metabolomics-based clusters. Then, the association of these clusters with future development of cardiometabolic disease was evaluated using different univariate and multivariate statistical approaches. Moreover, machine learning-based models were built to predict the development of future cardiometabolic disease using BMI and waist-to-hip circumference ratio measures in combination with H-NMR metabolomics.
Results
Three clusters with no differences in BMI nor in waist-to-hip circumference ratio but with very different metabolomics profiles were obtained. The first cluster showed a metabolically healthy profile, whereas atherogenic dyslipidemia and hypercholesterolemia were predominant in the second and third clusters, respectively. Individuals within the cluster of atherogenic dyslipidemia were found to be at a higher risk of developing type 2 DM in a 8 years follow-up. On the other hand, individuals within the cluster of hypercholesterolemia showed a higher risk of suffering a cardiovascular event in the follow-up. The individuals with a metabolically healthy profile displayed a lower association with future cardiometabolic disease, even though some association with future development of type 2 DM was still observed. In addition, H-NMR metabolomics improved the prediction of future cardiometabolic disease in comparison with models relying on just anthropometric measures.
Conclusions
This study demonstrated the benefits of using precision techniques like H-NMR to better assess the risk of obesity-derived cardiometabolic disease.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12933-024-02488-5.
Keywords: Obesity, Metabolomics , Type 2 diabetes mellitus, Cardiovascular disease, Atherogenic dyslipidemia, Hypercholesterolemia, Metabolically healthy obesity
Introduction
Obesity is a chronic disease, characterized by abnormal and/or excess of body fat accumulation [1]. This disease has been linked with several comorbidities such as type 2 diabetes mellitus (type 2 DM), cardiovascular disease (CVD), sleep apnea and several types of cancer [1–4]. During the last decades, this disease has become a major public health concern, reaching epidemic proportions. According to the world health statistics report developed by the World Health Organization (WHO) in 2023, 13.1% of adults globally had obesity in 2016, up from 8.7% in 2000 [5]. Currently, obesity is classified based on the body mass index (BMI), which is calculated as measured body weight (kg) divided by measured height squared (m2). Obesity is defined by a BMI higher or equal to 30 kg/m2 [3]. Moreover, the National Institute of Health (NIH) and the WHO distinguish between three classes of obesity: (1) obesity class I comprising BMI values from 30 to 34.9 kg/m2, (2) obesity class II comprising BMI values from 35 to 39.9 kg/m2 and (3) obesity class III when BMI is greater or equal to 40 kg/m2 [6–7]. However, the cause of obesity is complex and multifactorial [1–2, 7–8]. A notable heterogeneity of phenotypes has been addressed among individuals with obesity due to the impact of different environmental, social, genetic and economic factors on this disease [8]. In fact, the observational data from several studies has allowed the definition of the so-called metabolically healthy obesity (MHO) subphenotype [8–11]. These individuals are characterized by the absence of cardiometabolic abnormalities, such as insulin resistance (IR), impaired glucose tolerance (IGT), dyslipidemia and arterial hypertension (AHT), in spite of the excess of fat accumulated [9]. Therefore, the current BMI-based classification of obesity has limitations in explaining the relationship between obesity and morbidity or mortality [8, 12]. In response to this situation, abdominal obesity based on the waist-to-hip circumference ratio (WHR) has emerged as a more accurate risk assessment measure [13]. The WHR allows the definition of abdominal obesity, using the thresholds of WHR > 1 for men and > 0.85 for women [14, 15]. Nevertheless, WHR has some limitations to distinguish between subcutaneous and visceral abdominal fat and therefore, may not account for large variations in the level of total fat and abdominal visceral adipose tissues [16]. This highlights the necessity of a better characterization of obesity, based on its underlying pathophysiological mechanisms, to better assess the pathological risk derived from this complex condition. Of special interest is the risk of CVD associated with obesity, since this is the leading cause of death among individuals with obesity [17]. Dyslipidemia, which is present in obesity, is closely associated with CVD [2, 6]. Indeed, two main lipid profiles have been often described in patients with metabolically unhealthy obesity (MUO): (1) a profile characterized by hypercholesterolemia (HC) and (2) a profile characterized by the presence of atherogenic dyslipidemia, which consists of hypertriglyceridemia (HTG), low HDL cholesterol (HDLC) and high concentrations of small dense LDL particles [18]. The presence of different profiles in MUO in addition to the previously mentioned MHO phenotype suggests that the metabolic diversity of obesity may play a key role in the development of comorbidities derived from it.
Metabolomics is an emerging approach capable of measuring a large number of metabolites in several biological matrices, thereby providing the integration of genomic, transcriptomic and proteomic variation, as well as the impact of environmental factors [19–20]. Its closeness to the phenotype makes metabolomics a powerful tool to study complex diseases, like obesity. Although metabolomics approaches have already been applied to obesity, most of them have focused on the metabolic differences between obese and non-obese individuals [8, 21, 22]. Other approaches have developed machine learning (ML)-based models to predict BMI using metabolomics data, in order to evaluate its concordance with traditional BMI [23–25]. However, none of these approaches has focused on exploring differential metabolomics profiles within obesity and associating them with the future risk of developing cardiometabolic disease. The aim of this study was to find distinct proton nuclear magnetic resonance (H-NMR) metabolomics-based profiles of obesity to better assess the risk of the future development of comorbidities associated with this disease.
Materials and methods
Study population
A subset of the Di@bet.es cohort, consisting of 1387 individuals from the general population with a BMI higher or equal to 30, was used in this study (Supplementary Fig. 1). The Di@bet.es Study, the first national study in Spain to examine the prevalence of diabetes and impaired glucose regulation, consists of 4538 individuals with BMI and metabolomics data available (43% men), with ages ranging from 18 to 93 years old, of whom 2181 participated in the follow-up 8 years later. This research was carried out in accordance with the Declaration of Helsinki of the World Medical Association [26]. Written informed consent was obtained from all the participants. The study was approved by the Ethics and Clinical Investigation Committee of the Hospital Regional Universitario de Málaga (Málaga, Spain) in addition to other regional ethics and clinical investigation committees all over Spain.
Clinical data
Participants were invited to attend an examination at their health center, at each visit. Information was collected by means of an interviewer-administered structured questionnaire, followed by a physical examination and blood sampling. Medical history and medications were also recorded. Weight, height and waist and hip circumferences were measured by standardized methods. The BMI and WHR were calculated, and categorized according to WHO criteria [27–28]. Blood pressure was measured using a blood pressure monitor (Hem-703 C, Omron, Barcelona, Spain) after several minutes in a seated position and the mean of two measurements taken 1–2 min apart was used for analysis. AHT was considered if there was a previously physician-diagnosed hypertension and/or if the mean systolic blood pressure was ≥ 140 mmHg and/or the mean diastolic blood pressure was ≥ 90 mmHg [29]. Blood samples were obtained in fasting conditions. Additionally, a standard oral glucose tolerance test (OGTT) was performed in all individuals not receiving treatment for diabetes, using 2-h venous samples. Samples were immediately centrifuged and the serum was frozen until analysis. Serum glucose was measured enzymatically. Diabetes and pre-diabetes were diagnosed and classified according to WHO criteria [30]. Incident cardiovascular events (CV event) during follow-up, including coronary heart disease (CHD), stroke and peripheral artery disease (PAD), were recorded by questionnaire at the follow-up examination. Incident diabetes was considered if fasting plasma glucose at follow-up examination was ≥ 126 mg/dl, if 2 h post OGTT plasma glucose was ≥ 200 mg/dl [30] or if a clinical diagnosis of diabetes already existed and the treatment was ongoing. Samples were managed by the biochemistry laboratory of the Hospital Regional Universitario de Málaga, the IBIMA Biobank and the CIBERDEM Biorepository (IDIBAPS Biobank).
H-NMR analysis
Before H-NMR analysis, 200 µl of fasting serum collected at the baseline study were diluted with 50 µl of deuterated water and 300 µl of 50 mM phosphate buffer solution (PBS) at a pH of 7.4. H-NMR spectra were recorded at 306 K on a Bruker Avance III 600 spectrometer operating at a proton frequency of 600.20 MHz.
Lipoprotein analysis
Lipoprotein analysis was performed using the Liposcale® Test, a novel advanced lipoprotein test based on 2D diffusion-ordered H-NMR spectroscopy [31]. The methyl signal was deconvoluted using 9 Lorentzian functions to determine the lipid concentration of the large (l), medium (m) and small (s) subclasses of the main lipoprotein classes (VLDL, LDL and HDL), and their size (Z) associated diffusion coefficients. Then, lipid concentrations were combined with the associated particle volume to quantify the number of particles required to transport the measured lipid concentration of each lipoprotein subclass. Finally, weighted average VLDL, LDL and HDL particle sizes were calculated from various subclass concentrations by totaling the known diameter of each subclass multiplied by its relative percentage of subclass particle number. The variation coefficients for the particle number were between 2% and 4%, and for the particle sizes, they were less than 0.3%.
Glycoprotein analysis
The region of the H-NMR spectrum where the glycoproteins resonate (2.15–1.90 ppm) was analyzed using several analytical functions according to a previously published procedure [32]. For each function, the total area (proportional to concentration) and signal shape (height to bandwidth H/W ratio) were determined. The area of GlycA provided the concentration of protein-bound N- acetylneuraminic acid, and the area of GlycB provided those of N-acetylglucosamine [33]. The GlycF area arises from the concentration of the acetyl groups of N-acetylglucosamine, N-acetylgalactosamine and N-acetylneuraminic acid unbound to proteins (i.e., free fraction) [34]. H/W ratios of GlycA and GlycB, a parameter associated with the aggregation state of the sugar-protein bonds, were also reported. The variation coefficients for the glycoproteins were lower than 3%.
Low molecular weight metabolite (LMWM) analysis
Fully-automated software developed by the company Biosfer Teslab- was used to perform the deconvolution of the signals associated with 15 different LMWMs as previously reported [35]. For each metabolite, the total area was quantified and normalized to obtain the concentration. The variation coefficients obtained for the LMWMs were between 6% and 18%.
Statistical analysis
A non-hierarchical clustering approach was performed deploying K-means and K-medoids algorithms. Three clusters were defined using the following informative 12 variables as predictors: GlycA, LDL cholesterol (LDLC), HDLC, triglycerides (TG), triglyceride to cholesterol ratios of VLDL (TG/C-VLDL), IDL(TG/C-IDL), LDL (TG/C-LDL), HDL (TG/C-HDL), the small to total particle number ratios of VLDL (s/t-VLDLp), LDL (s/t-LDLp) and HDL (s/t-HDLp) and glucose. The optimal number of clusters was selected based on the sum of squared error (SSE) and the average silhouette coefficient. This coefficient is a measure of how similar a sample is to its own cluster compared to other clusters. High values indicate that the sample is well matched to its own cluster and poorly matched to neighboring ones. The silhouette coefficient was also computed for each sample. This showed that the clusters defined by the K-means algorithm had a higher robustness than the ones relying on the K-medoids one (Supplementary Fig. 2). Thus, these clusters were considered for further analysis. A principal component analysis (PCA) was computed using the above mentioned variables as predictors, to display the distribution of the clusters in a reduced dimensional space delimited by the first two principal components (PC). The distribution of obesity classes based on BMI and WHR were also displayed in the same reduced space. The loadings of the PCA were computed to know the contribution of each original variable to the PC. Differences in traditional variables between clusters were evaluated using the Chi-squared test for categorical predictors (i.e., sex, glycemia, AHT, WHR and treatment) and the Kruskal-Wallis test for numerical predictors (i.e., age and BMI). The post-hoc comparisons were computed using the Chi-squared and the Mann-Whitney tests accordingly, and all the resulting p-values were adjusted for multiple comparisons using Benjamini and Hochberg’s method. In addition, to better understand the metabolic differences between clusters, a quantile regression including the age, sex and treatment as covariates was created. The 50% quantile was chosen in order to compare the median values of each metabolite between clusters, instead of the means as in the classical linear regression. The resulting p-values were adjusted using the method developed by Benjamini and Hochberg and the false discovery rate threshold was set to a 5%. After this analysis, associations between clusters and CV event, stroke, peripheral vasculopathy (PV) and DM happening during the 8-years follow up were evaluated using a correspondence analysis (CA). To do that, the corresponding contingency tables were created, displaying the cluster defined in the columns and all the previously mentioned conditions in the rows. Finally, five logistic regression models were computed to predict the future development of cardiometabolic disease in the follow-up. Individuals were considered to have developed cardiometabolic disease if they developed, in the follow-up, any of the conditions listed previously (i.e. cardiovascular event, stroke, PV and DM). The predictors included in each model were: (1) BMI, (2) WHR, (3) PC1 and PC2, (4) BMI, PC1 and PC2 and (5) WHR, PC1 and PC2. A training set comprising the 70% of the individuals was used to train all the models, whose performance was later evaluated in a test set comprising the remaining 30% individuals. The receiver operating curves (ROC) and their corresponding area under the curve (AUC) were computed for all of the models. In addition, the confusion matrix, the sensitivity and specificity were also calculated for each model. The statistical analysis was performed using the scikit-learn package and python software, version 3.6.
Results
H-NMR metabolomics-based clustering of individuals with obesity
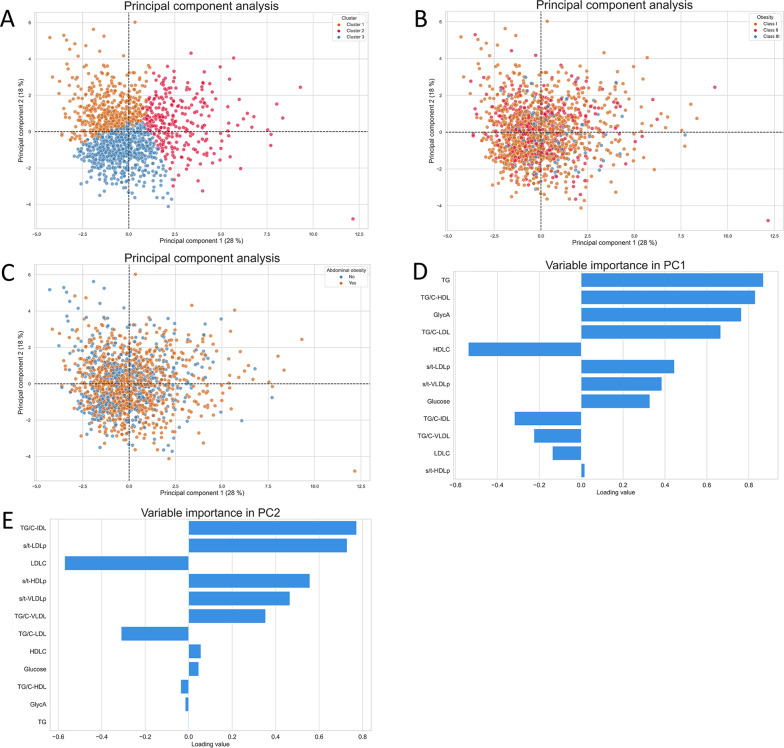
The original Di@bet.es population was filtered by BMI to keep only individuals with obesity, i.e., those having a BMI higher or equal to 30. A K-means algorithm was deployed to define three different clusters. The clusters defined, as well as the current obesity classes defined by WHO and the abdominal obesity based on WHR were displayed in a reduced dimensional space (Fig. 1, A-C). A separation between groups was not observed neither for the WHO obesity classification, nor for abdominal obesity. Therefore, no evident relationship was observed between the metabolomics-based classification of obesity and the classification based on anthropometric traits. The loadings computed for each principal component were also computed (Fig. 1, D-E). Loading values can be interpreted as the weights of each metabolomics feature within the principal component. The most relevant features in the PC1 were TG, TG/C-HDL, GlycA, TG/C-LDL and HDLC. They mainly contributed to the separation of the second cluster from the other two. On the other hand, TG/C-IDL, s/t-LDLp, LDLC, s/t-HDLP and s/t-VLDLP were the features with the highest importance in the PC2, which contributed to the separation of the third cluster from the first one. The two additional obesity classifications were not able to capture the metabolic implications of these features.
Fig. 1.
Principal component analysis. Sample distribution within the first two principal components. A Samples are colored according to the clusters defined by the K-means algorithm. B Samples are colored according to the corresponding BMI-based obesity class. C Samples are colored according to abdominal obesity. D Loading values of the first principal component. (E) Loading values of the second principal component
Comparison of the different metabolomics profiles
In order to better understand these implications, a univariate comparison between clusters was conducted. First of all, non-metabolomics variables were compared (Table 1). In regard to age, the individuals within the third cluster displayed a statistically significant higher median age (59 years) than the one observed in the remaining two clusters (57 and 55 years in clusters 2 and 3, respectively). However, statistically significant differences (p = 0.004) were only found between the third and the first cluster. On the other hand, no statistically significant differences in BMI were observed between clusters. In respect to sex, a higher proportion of men was observed in cluster 2 (59%) in comparison to the other two clusters, which displayed a very similar percentage of men between them (41% in cluster 1 and 42% in cluster 3). AHT was also compared between clusters and statistically significant differences were found among all of them. The second cluster displayed the highest prevalence of AHT (76%), followed by the third (67%) and the first one (59%), respectively. Moreover, the highest prevalence of diabetes mellitus was also found in the second cluster (48%), while no statistically significant differences were found between the remaining two clusters (20% and 17% in the third and first clusters, respectively). Abdominal obesity was defined based on WHR. For men, a threshold of 1 was used to define abdominal obesity, while for women a threshold of 0.85 was used. As BMI, abdominal obesity did not show statistically significant differences between clusters. Due to the fact that the cohort used in this study is a representation of the Spanish general population, some individuals were under different treatments, that were not always related with obesity.
Table 1.
Univariate comparison between clusters. The median and the interquartile range are displayed for continuous variables and the observed frequency for the categorical variables. The comparisons of all metabolomics features were adjusted by age, sex and treatment. The post-hoc p-values were adjusted for multiple comparisons using Benjamini– Hochberg’s method
| Variable | Cluster 1 (N = 459) | Cluster 2 (N = 274) | Cluster 3 (N = 654) | Cluster 2 vs. 1 | Cluster 3 vs. 1 | Cluster 3 vs. 2 |
|---|---|---|---|---|---|---|
| Age | 55 [41.0;67.5] | 57 [46.0;67.0] | 59 [46.0;69.0] | 0.147 | 0.004 | 0.361 |
| BMI | 33.0 [31.3;35.8] | 33.0 [31.4;35.8] | 32.5 [31.2;35.6] | 0.884 | 0.162 | 0.173 |
| Sex: | 1.19E-05 | 0.939 | 7.50E-06 | |||
| Male | 189 (41%) | 162 (59%) | 272 (42%) | |||
| Female | 270 (59%) | 112 (41%) | 382 (58%) | |||
| Glycemia: | 2.52E-18 | 0.197 | 9.46E-16 | |||
| Normoglycemia | 299 (65%) | 97 (35%) | 389 (60%) | |||
| Pre-diabetes | 84 (18%) | 47 (17%) | 133(20%) | |||
| Diabetes | 76 (17%) | 130 (48%) | 132 (20%) | |||
| AHT: | 1.19E-05 | 0.034 | 0.010 | |||
| No | 187 (41%) | 65 (24%) | 219 (33%) | |||
| Yes | 272 (59%) | 209 (76%) | 435 (67%) | |||
| Abdominal obesity: | 0.116 | 0.372 | 0.392 | |||
| No | 187 (40%) | 90 (33%) | 219 (36%) | |||
| Yes | 277 (60%) | 274 (67%) | 435 (64%) | |||
| Treatment: | 0.041 | 0.933 | 0.041 | |||
| No | 274 (60%) | 139 (51%) | 386 (59%) | |||
| Yes | 185 (40%) | 135 (49%) | 268 (41%) | |||
| GlycA (µmol/L) | 674 [613;750] | 979 [864;1146] | 730 [652;808] | 2.07E-118 | 3.27E-09 | 3.33E-92 |
| GlycB (µmol/L) | 256 [223;288] | 310 [263;356] | 257 [228;291] | 8.08E-25 | 0.635 | 9.92E-26 |
| GlycF (µmol/L) | 271 [245;301] | 359 [319;399] | 285 [259;313] | 1.46E-87 | 7.84E-06 | 6.80E-70 |
| HWGlycA | 15.8 [14.3;17.4] | 19.8 [18.0;21.6] | 16.4 [15.0;18.1] | 1.93E-62 | 2.99E-05 | 5.20E-47 |
| HWGlycB | 4.2 [3.8;4.7] | 4.8 [4.3;5.3] | 4.3 [3.9;4.7] | 2.28E-19 | 0.082 | 1.12E-15 |
| VLDLTG (mg/dL) | 57.6 [41.8;76.5] | 148 [117;195] | 63.8 [48.7;84.2] | 3.25E-161 | 0.003 | 2.03E-155 |
| IDLTG (mg/dL) | 10.8 [9.1;12.7] | 17.1 [14.7;20.3] | 13.5 [11.1;16.1] | 1.69E-81 | 1.10E-25 | 1.24E-34 |
| LDLTG (mg/dL) | 13.7 [11.9;16.1] | 20.5 [17.1;24.0] | 19.9 [17.191;22.867] | 1.08E-64 | 5.17E-76 | 0.019 |
| HDLTG (mg/dL) | 15.0 [12.8;17.9] | 21.6 [18.2;26.6] | 16.1 [13.2;19.7] | 2.76E-46 | 0.068 | 4.46E-42 |
| TG (mg/dL) | 98.2 [79.8;120] | 207 [174;263] | 115 [96.2;141] | 2.82E-159 | 1.47E-10 | 1.77E-130 |
| VLDLC (mg/dL) | 13.3 [8.8;18.6] | 37.8 [31.1;47.9] | 17.8 [12.9;23.4] | 7.85E-147 | 1.08E-10 | 2.28E-117 |
| IDLC (mg/dL) | 9.6 [7.6;11.4] | 17.0 [14.2;20.9] | 14.1 [11.3;16.9] | 7.35E-80 | 6.87E-45 | 4.25E-19 |
| LDLC (mg/dL) | 126 [112;140] | 127 [107;150] | 152 [136;167] | 0.230 | 1.45E-42 | 3.14E-25 |
| HDLC (mg/dL) | 54.7 [48.2;61.9] | 44.4 [39.3;50.0] | 50.1 [45.5;57.2] | 3.35E-34 | 6.59E-11 | 3.28E-14 |
| TC (mg/dL) | 205 [187;224] | 231 [206;257] | 236 [219;256] | 7.88E-22 | 3.79E-41 | 0.196 |
| VLDLp (nmol/L) | 42.5 [30.8;58.3] | 117 [93.0;155] | 49.2 [37.3;65.5] | 3.93E-169 | 2.23E-04 | 1.03E-158 |
| lVLDLp (nmol/L) | 1.3 [0.9;1.7] | 2.7 [2.24;3.35] | 1.3 [1.0;1.6] | 4.63E-121 | 0.617 | 1.73E-136 |
| mVLDLp (nmol/L) | 3.6 [2.5;4.8] | 7.1 [4.3;11.4] | 4.8 [3.7;6.1] | 1.19E-57 | 3.46E-14 | 4.53E-28 |
| sVLDLp (nmol/L) | 36.8 [26.7;52.3] | 107 [84.6;140] | 43.0 [32.2;58.6] | 8.69E-172 | 0.001 | 1.36E-164 |
| VLDLZ (nm) | 41.9 [41.7;42.2] | 41.5 [41.3;41.8] | 42.0 [41.8;42.2] | 2.48E-21 | 0.004 | 2.45E-35 |
| LDLp (nmol/L) | 1279 [1151;1443] | 1442 [1231;1704] | 1540 [1388;1700] | 1.48E-10 | 5.91E-40 | 4.12E-06 |
| lLDLp (nmol/L) | 171 [155;187] | 173 [142;196] | 213 [195;230] | 0.367 | 2.73E-72 | 1.48E-47 |
| mLDLp (nmol/L) | 336 [278;393] | 327 [262;404] | 487 [420;561] | 0.949 | 6.61E-74 | 2.56E-54 |
| sLDLp (nmol/L) | 773 [693;871] | 925 [771;1114] | 828 [739;927] | 4.28E-24 | 4.14E-05 | 2.26E-13 |
| LDLZ (nm) | 20.8 [20.7;21.0] | 20.6 [20.27;20.9] | 21.1 [21.0;21.3] | 3.01E-22 | 1.25E-42 | 1.55E-95 |
| HDLp (µmol/L) | 28.9 [25.8;32.2] | 26.5 [23.6;29.5] | 26.7 [24.0;29.7] | 3.39E-10 | 1.01E-15 | 0.848 |
| lHDLp (µmol/L) | 0.254 [0.237;0.273] | 0.282 [0.252;0.323] | 0.294 [0.274;0.319] | 1.59E-18 | 8.37E-45 | 0.005 |
| mHDLp (µmol/L) | 8.8 [8.0;9.7] | 8.0 [7.4;9.2] | 9.2 [8.5;10.1] | 1.11E-07 | 2.26E-04 | 2.57E-18 |
| sHDLp (µmol/L) | 20.0 [17.3;22.6] | 18.0 [15.4;20.4] | 17.2 [15.2;19.6] | 9.30E-08 | 2.38E-25 | 0.001 |
| HDLZ (nm) | 8.2 [8.2;8.3] | 8.2 [8.2;8.3] | 8.3 [8.2;8.3] | 0.008 | 1.69E-41 | 1.49E-18 |
| Glucose (mmol/L) | 4.6 [4.1;5.2] | 5.5 [4.7;6.8] | 4.7 [4.2;5.4] | 2.48E-20 | 0.539 | 4.37E-20 |
| Lactate (mmol/L) | 0.82 [0.65;1.05] | 1.05 [0.84;1.42] | 0.82 [0.66;1.09] | 8.02E-11 | 0.705 | 5.07E-13 |
| Creatine (mmol/L) | 0.057 [0.042;0.073] | 0.054 [0.038;0.07] | 0.056 [0.044;0.072] | 0.674 | 0.272 | 0.607 |
| Creatinine (mmol/L) | 0.042 [0.032;0.054] | 0.051 [0.04;0.064] | 0.042 [0.034;0.055] | 1.91E-06 | 0.226 | 9.05E-05 |
| Alanine (mmol/L) | 0.42 [0.37;0.48] | 0.49 [0.43;0.56] | 0.43 [0.37;0.49] | 6.17E-12 | 0.569 | 4.39E-15 |
| Tyrosine (mmol/L) | 0.060 [0.051;0.068] | 0.064 [0.055;0.075] | 0.061 [0.051;0.071] | 0.002 | 0.496 | 0.011 |
| Valine (mmol/L) | 0.22 [0.20;0.25] | 0.26 [0.23;0.29] | 0.22 [0.20;0.26] | 7.52E-12 | 0.055 | 2.59E-08 |
| Leucine (mmol/L) | 0.12 [0.10;0.15] | 0.15 [0.12;0.19] | 0.12 [0.10;0.15] | 2.04E-21 | 0.087 | 7.87E-18 |
| Isoleucine (mmol/L) | 0.057 [0.045;0.069] | 0.075 [0.061;0.089] | 0.056 [0.045;0.071] | 1.70E-19 | 0.617 | 9.75E-20 |
| Acetone (mmol/L) | 0.016 [0.012;0.023] | 0.021 [0.015;0.028] | 0.016 [0.012;0.024] | 1.33E-11 | 0.958 | 9.32E-13 |
The comparison between clusters also included the H-NMR metabolomics-based glycoprotein, lipoprotein and LMWM profiles (Table 1). These comparisons were adjusted by age, sex and treatment to avoid confounding effects. In regard to glycoprotein profile, the individuals within the second cluster displayed the highest glycosylation levels. In the case of GlycA and GlycF, statistically significant differences were also observed between the first and the third cluster (p < 0.01 for GlycA and GlycF), displaying this last one higher concentrations of these two variables. On the other hand, these two clusters had similar values of GlycB (256 µmol/L and 257 µmol/L in clusters 1 and 3, respectively). A similar behavior as the one described for H-NMR-derived glycosylation profile was observed in TG metabolism. Once again, the individuals of the second cluster displayed the highest TG concentrations (207 mg/dL), followed by the third (115 mg/dL) and the first (98.2 mg/dL) cluster, respectively. Moreover, the concentration of TG transported in each of the main lipoprotein classes (i.e. VLDL, IDL, LDL and HDL) was also significantly higher in cluster 2 than in the remaining two clusters (all p < 0.05). In this regard, the third cluster also displayed higher concentrations of TG transported in VLDL, IDL and LDL than the first cluster, but no statistically significant differences between these two clusters were found for TG transported in HDL. The comparison of the total cholesterol (TC), as well as, the cholesterol transported in each lipoprotein class also displayed statistically significant differences between the metabolomics clusters. In this case, the individuals within the third cluster displayed the highest TC concentrations (236 mg/dL). However, the comparison between the third and the second cluster was not statistically significant (p = 0.196). Contrary to what was observed for TG, different tendencies were observed when the cholesterol transported in each lipoprotein class was compared among clusters. For the cholesterol transported in VLDL and IDL, the second cluster displayed the highest concentration (37.8 mg/dL and 17.0 mg/dL, respectively), followed by the third (17.8 mg/dL and 14.1 mg/dL) and the first one (13.3 mg/dL and 9.6 mg/dL). However, the highest levels of LDLC were found in the third cluster (151 mg/dL). Furthermore, no statistically significant differences in LDLC concentrations were observed between the second and the first cluster (p = 0.230). On the other hand, the highest and the lowest HDLC concentrations were found in the first (54.6 mg/dL) and the second (44.4 mg/dL) clusters, respectively. After the comparison of lipid profiles, lipoprotein determinations (VLDLP, LDLP, HDLP and their corresponding large, medium and small subclasses) were compared between clusters. The comparisons of VLDLp, mVLDLp and sVLDLp were statistically significant between all the clusters. For the three determinations, the highest values were found in the second cluster (117 nmol/L, 7.1 nmol/L and 107 nmol/L, respectively) and the lowest in the first one (42.5 nmol/L, 3.6 nmol/L and 36.7 nmol/L). The same trend was observed for lVLDLp, but no statistically significant differences were found between the third and the first cluster (p = 0.617). In regard to the average VLDLZ, the third cluster displayed the largest particle size (42.0 nm) and the first one the smallest (41.9 nm). In relation to LDLp and its subclasses, individuals included in the third cluster displayed statistically significant concentrations of LDLp, lLDLp and mLDLp (all p < 0.01) than the remaining two clusters. In addition, the second cluster had a significantly higher amount of LDLp (1442 nmol/L) than the first cluster (1279 nmol/L), although no statistically significant differences were found for these two clusters with respect to lLDLp and mLDLp (p = 0.367 and 0.949). Interestingly, sLDLp did not follow the trend observed for the rest of the LDL subclasses. In this case, the second cluster displayed the highest concentration of sLDLp (925 nmol/L), followed first by the third one (828 nmol/L) and then by the first (773 nmol/L) cluster. Furthermore, the smallest LDLZ was found in the second cluster (20.6 nm), whereas the highest was observed in the third one (21.1 nm). In respect to HDLp, the first cluster displayed the highest concentration (28.9 µmol/L) but no statistically significant differences were found between the other two clusters (p = 0.848). Individuals within cluster 1 also showed the highest concentration of sHDLp (20.0 µmol/L), followed by the second (18.0 µmol/L) and the third (17.2 µmol/L) clusters. However, the highest concentrations of lHDLP and mHDLp were found in the third cluster (0.29 µmol/L and 9.2 µmol/L, respectively). In the case of lHDLp, the individuals included in the second cluster displayed a higher amount of them (0.28 µmol/L) than individuals within the first cluster (0.25 µmol/L). However, the opposite effect was observed for mHDLp. The LMWM consisting of glucose, lactate, creatine, creatinine, alanine, tyrosine, valine, leucine, isoleucine and acetone was also compared between clusters. All of them, except creatinine, which did not show statistically significant differences between groups, were found to be increased in the second cluster in comparison with the remaining two (all p < 0.01). No statistically significant differences were observed for any of them between the first and the third clusters.
Associations of different clusters with future development of cardiometabolic disease
Once the clusters had been described, a CA was computed to evaluate the association of the clusters with the development of several cardiometabolic conditions in the follow-up (Fig. 2 ). It was observed that the third cluster was located closer to conditions associated with CVD, especially to the development of a CV event, in the reduced dimensions space. On the other hand, DM was separated from the other three CVD-related conditions. In this case, the second cluster was found to be associated with the development of this disease in the follow-up. Finally, the first cluster was the most isolated one. Thus, it was the one showing the weakest association with any of the conditions considered, even though some association with DM development was observed.
Fig. 2.
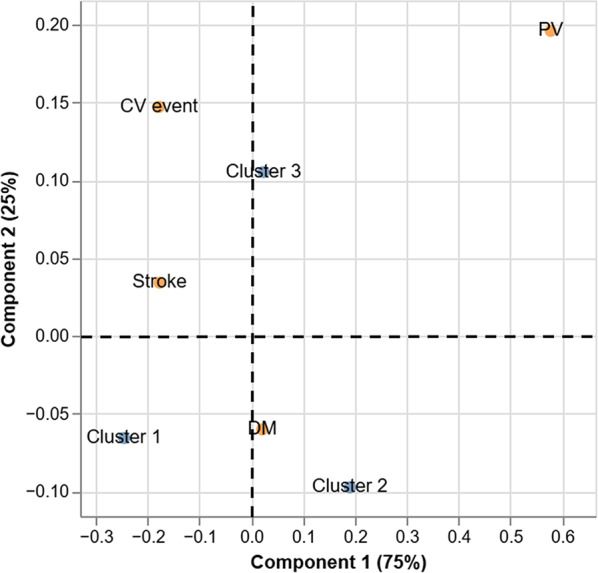
Corresponding analysis. Associations of each cluster with future events of cardiometabolic disease
Prediction of future cardiometabolic disease in individuals with obesity
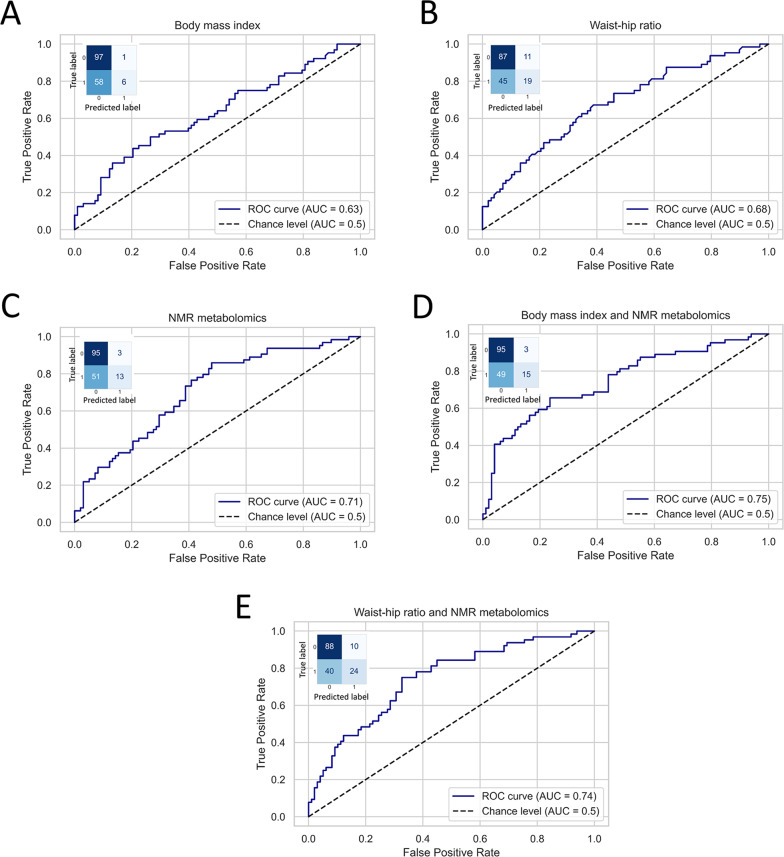
The last analysis of this study consisted of building several multivariate statistical models to predict the future development of cardiometabolic disease in the follow-up (Fig. 3 ). The aim of this analysis was to determine whether the presented H-NMR-metabolomics approach could improve the assessment of the obesity-derived risk of developing any cardiometabolic disease in the future. At first, a total of two models were trained and tested, using BMI and WHR respectively to predict the future development of cardiometabolic disease. The models displayed AUC values of 0.63 and 0.68, respectively (Fig. 3, A-B). Although these models displayed AUC far away from the chance level (AUC of 0.5) and showed to be very specific, their sensitivity was low (Supplementary Table 2). Then, a third model based on NMR metabolomics, i.e., including PC1 and PC2 as predictors was trained. The AUC obtained was 0.71, which was slightly higher than the AUC values showed by the previous models (Fig. 3, C). Two additional models were trained and tested combining each anthropometric measure with the information resulting from the classification based on H-NMR metabolomics presented in this study. In both cases, the incorporation of H-NMR metabolomics to the models notably improved the predictive performance of the models. The AUC increased from 0.63 to 0.75 when BMI and NMR-metabolomics were combined and from 0.68 to 0.74 when WHR and BMI were used together as predictors. Moreover, the improvement of the predictive performance of the models was due to an increase of their sensitivity without losing specificity (Fig. 3, D-E and Supplementary Table 2 ).
Fig. 3.
Logistic regression analysis. Several logistic regressions were fitted to predict future development of obesity-associated cardiometabolic disease in the follow-up. The confusion matrix computed in the testing of each model is also displayed. The y-axis shows the true classes, with 0 corresponding to no development of cardiometabolic disease in the follow-up and 1 corresponding to the development of cardiometabolic disease in the follow-up. The x-axis displays the label predicted by the model
Discussion
The current study aimed to define obesity beyond BMI to better capture the metabolic diversity behind this complex condition and to better assess the risk of developing future cardiometabolic disease. In order to do that, the K-means clustering algorithm was deployed and three metabolomics-based clusters were defined within a subset of the general population cohort Di@bet.es, that only included individuals with obesity. These clusters were designed to maximize the differences of several well-known obesity associated predictors among individuals. On the one hand, the traditional lipid profiling consisting of TG, LDLC and HDLC was considered for clustering, since its association with obesity has been clearly described in several clinical guidelines [6–36]. On the other hand, glucose was added to the model to account for the association between obesity and IR [2, 3, 8]. In addition, novel H-NMR metabolomics-based predictors were also considered. First, ratios of small lipoproteins to the total circulating particle number (s/t-VLDLp, s/t-LDLp and s/t-HDLp) were computed to incorporate information about lipoprotein number and size into the clustering algorithm, since both have been associated with obesity [2, 37–38]. Furthermore, the functionality of the main lipoprotein classes (VLDL, IDL, LDL and HDL) was also considered, by including the TG/C ratios for each class in the clustering algorithm. The presence of dysfunctional TG-enriched HDL lipoproteins, in combination with high concentrations of TG-rich lipoproteins (TRL) have been found to be associated with a higher risk of CVD [24, 37–38]. GlycA was the last predictor incorporated into the clustering algorithm, since this parameter is a robust biomarker of systemic inflammation [32–34, 39]. Based on all the parameters described, three different clusters were defined. Even though statistically significant differences were not observed for BMI and abdominal obesity between clusters, several metabolic differences were found among them. Each of them seemed to be describing different subtypes of obesity, that had already been mentioned in the first section of this document: (1) MUO with predominant atherogenic dyslipidemia, (2) MUO with predominant HC and (3) MHO. However, the current analysis enabled a detailed description of several pathogenic mechanisms that were taking place in addition to dyslipidemia and evaluated the differential cardiometabolic risk derived from each of the profiles found.
On the one hand, atherogenic dyslipidemia was found to be present on the second cluster. All its classical signs, consisting of HTG, low HDLC accompanied by TG-enriched HDLP and a high amount of sLDLp despite having moderate LDLC values [2, 38–39] were observed in this cluster. This lipid profile has been often described in individuals with obesity in whom IR was also present [18]. Coherently, several markers of IR such as glucose, lactate, alanine and branched-chain amino acids (BCAA) [8] reached their highest concentration values in this cluster. In addition, the highest concentrations of remnant cholesterol (i.e. VLDLC and IDLC) were also observed in this cluster. A recent study attributed a fundamental role to remnant cholesterol in the development of type 2 DM, especially in individuals with moderate levels of LDLC as the ones included in this cluster [40]. On top of the described metabolic disturbances, the presence of a high inflammatory state was assessed by the elevated levels of GlycA and GlycB found in this cluster [32–34, 41]. All the pathogenic alterations described pointed in the direction of a predisposition of individuals with this specific obesity profile to develop type 2 DM. The analysis of the follow-up data showed an association between this cluster and the future development of type 2 DM, thereby confirming the previous hypothesis.
On the other hand, predominant HC was found to be present in the third cluster. This cluster displayed the highest TC and LDLC concentrations. Accordingly, this cluster was found to have the largest amount of LDLp. However, the lLDLp and mLDLp were the most increased fractions. It is well known that LDLC-driven HC is one of the major risk factors of cardiovascular disease, atherosclerosis and CHD [42, 43]. Coherently with this, our results showed this cluster to be the most associated one with future CV events, including CHD among other conditions. Another interesting characteristic of this cluster was the GlycA-driven inflammation, in absence of increased GlycB levels. Our previous work showed that increased values of GlycA in combination with moderately increased values of GlycB were associated with a higher risk of developing type 2 DM in hyperglycemic individuals with similar TC and LDLC concentrations [44]. Thus, GlycB-derived glycosylation might be associated with TG metabolism rather than with serum cholesterol levels, and thereby, might be more strongly contributing to IR and type 2 DM than to CV events.
The last cluster defined, the one named as cluster 1, was the closest to MHO. The lowest concentrations of TC and TG in all the lipoprotein classes were found in this cluster, except for HDL. Indeed, the highest HDLC together with the lowest HDLTG concentrations were displayed by this cluster. These findings reflected a healthy HDL function that has been associated with a better cardiovascular health [39]. Accordingly, the individuals within this cluster displayed the lowest amount of ApoB-containing atherogenic lipoproteins (VLDL and LDL) and the highest concentrations of apoA-containing lipoproteins (HDLP). These findings, together with the absence of IR, AHT and dyslipidemia have been described in MHO phenotype [8–11]. Interestingly, our results showed a higher amount sHLDp and the smallest HDLP diameter to be present in this cluster. This contradicts previous literature describing larger HDLP in MHO [38, 41]. It must be noted that the MHO is still an ambiguous phenotype, whose definition is not currently standardized [8–11] and that the particle size thresholds vary between different techniques. Although this cluster showed the lowest association with future cardiometabolic disease, this association was not negligible. Several studies have discussed the fluctuant character of MHO, arguing that the lower association of this phenotype with type 2 DM could just be a matter of time [9, 11]. Moreover, specific associations between MHO and future type 2 DM have been found and attributed to abnormalities in Bromodomain and extra terminal (BET) proteins which promote pancreatic β-cell function and proliferation [8, 45].
All that has been discussed above made it clear that obesity is a complex disease that cannot be explained using just anthropometric measures, such as BMI or WHR. Nevertheless, it is not our aim to state that these traditionally used methods are useless but to demonstrate that complementing them with information derived from H-NMR metabolomics can improve the characterization of this complex disease. Indeed, it was observed, that metabolomics alone was not able to significantly improve the performance of the models relying on BMI and WHR. Nevertheless, it was demonstrated that including H-NMR-metabolomics information together with BMI and WHR improved the prediction of the development of future cardiovascular disease in a 12% and a 6%, respectively. Moreover, the inclusion of metabolomics data increased the specificity of the models by 24% and 8%, when it was used together with BMI and WHR. This showed that the incorporation of H-NMR metabolomics into the predictive models enabled a better identification of those individuals being at risk of developing cardiometabolic disease in the future. Although further validation of the clusters defined in this study is needed, the current results suggested potential benefits of using H-NMR metabolomics as a complementary tool of the currently available clinical measures of obesity.
This study presents several strengths. First of all, a large sample that comprised men and women with obesity in a wide age range, was used for the definition of the clusters. Treated individuals were also included, thereby reflecting in a more accurate way the general population. Although different treatments might have a different impact on metabolism, the exclusion of treated individuals would bias the results since the population obtained would not be representative of the general population. In addition, H-NMR was used to conduct the metabolomics measurements. This is a non-destructive technology that requires a minimum sample processing to quantify the most abundant metabolites and macromolecules present in different biological matrices, even if these analytes have identical molecular masses [46]. In addition, NMR spectroscopy is unbiased, fast, very reproducible and highly automatable [14]. However, some limitations must be also acknowledged in this study. First, the validation of the results in a completely independent large cohort was not possible due to the lack of another cohort equally profiled. In addition, the number of individuals developing different kinds of cardiometabolic diseases in the follow-up was limited. To overcome this issue, the different conditions were grouped under the broader category of cardiometabolic disease, so balanced groups could be obtained for the predictive models.
Conclusion
In this study, three clusters describing three obesity subtypes with very different metabolomics profiles were defined. Each subtype was deeply characterized and found to be associated with a different type of cardiometabolic risk. It was demonstrated that metabolomics added an additional layer of information regarding the development of obesity-derived comorbidities that BMI and WHR are unable to capture. This study is a first step in defining novel endotypes within obesity that should be further evaluated in cohorts including a larger number of cases of the diseases comprised by the term cardiometabolic disease in this study. Indeed, we believe that the use of metabolomics profiling in combination with the currently available anthropometric measures in the clinical definition of such a complex disease like obesity, will improve the assessment of the future cardiometabolic risk and enable an earlier and more precise clinical management of it.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors wish to acknowledge the entire working group of the di@bet.es study.More information about it can be found at https://www.ciberdem.org/en/research-programmes/projects/di-betes-study.
Abbreviations
- AHT
Arterial Hypertension
- AUC
Area Under the Curve
- BCAA
Branched-Chain Amino Acids
- BMI
Body Mass Index
- BET
Bromodomain and extra terminal
- CA
Correspondence Analysis
- CHD
Coronary Heart Disease
- CVD
Cardiovascular Disease
- DM
Diabetes Mellitus
- HC
Hypercholesterolemia
- HDLC
High Denisty Lipoprotein Cholesterol
- HDLZ
HDL Particle Size
- H-NMR
Proton Nuclear Magnetic Resonance
- HTG
Hypertriglyceridemia
- IGT
Impaired Glucose Tolerance
- IR
Insulin Resistance
- LDLC
Low Density Lipoprotein Cholesterol
- LDLZ
LDL Particle Size
- lHDLp
Large HDL Particles
- lLDLp
Large LDL Particles
- LMWM
Low Molecular Weight Metabolites
- lVLDLp
Large VLDL Particles
- mHDLp
Medium HDL Particles
- MHO
Metabolically Healthy Obesity
- mLDLp
Medium LDL Particles
- MUO
Metabolically Unhealthy Obesity
- mVLDLp
Medium VLDL Particles
- NIH
National Institute of Health
- OGTT
Oral Glucose Tolerance Test
- PAD
Peripheral Artery Disease
- PCA
Principal Component Analysis
- ROC
Receiver Operating Curves
- sHDLp
Small HDL Particles
- sLDLp
Small LDL Particles
- s/t-LDLp
small to total Very Low Density Lipoprotein particle ratio
- s/t-HDLp
small to total Very Low Density Lipoprotein particle ratio
- s/t-VLDLp
small to total Very Low Density Lipoprotein particle ratio
- sVLDLp
Small VLDL Particles
- SSE
Sum of Squared Error
- TC
Total Cholesterol
- TG
Triglycerides
- TG/C-HDL
Triglyceride to Cholesterol High Density Lipoprotein ratio
- TG/C-IDL
Triglyceride to Cholesterol Intermediate Density Lipoprotein ratio
- TG/C-LDL
Triglyceride to Cholesterol Low Density Lipoprotein ratio
- TG/C-VLDL
Triglyceride to Cholesterol Very Low Density Lipoprotein ratio
- TRL
Triglyceride Rich Lipoprotein
- VLDLZ
VLDL Particle Size
- WHO
World Health Organization
- WHR
Waist-to-Hip Circumference Ratio
Author contributions
J.R., M.G., E.O. and G.R. contributed to the conception and design of the work. E.O., N.A., S.V., P.R. and W.O. contributed to the acquisition of data. E.O. and M.G. performed the analysis and the interpretation of data. E.O. wrote the draft of the manuscript and all authors revisited it critically and approved and agreed with the final version of the manuscript. E.O. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Funding
This work was supported by the Spanish Ministerio de Economía y Competitividad (PI21/01294; PI16/00507), Fondo Europeo de Desarrollo Regional (FEDER), and CIBERDEM (CIBER de Diabetes y Enfermedades Metabólicas Asociadas), which are initiatives of Instituto de Salud Carlos III (ISCIII); and by the Cerca Programme, Generalitat de Catalunya. P. R. is a recipient of a predoctoral fellowship from the Spanish Ministerio de Universidades (grant number FPU19/04610). GRM belongs to the regional Nicolás Monardes research program of the Consejería de Salud (RC-0006-2016; Junta de Andalucía, Spain). WOB is recipient of a ‘PFIS’ predoctoral research contract (FI2100040, Instituto de Salud Carlos III).
Data availability
Raw data for the dataset used in this study are not publicly available to preserve individuals’ privacy under the European General Data Protection Regulation.
Declarations
Ethics approval and consent to participate
Written informed consent was obtained from all the participants. The study was approved by the Ethics and Clinical Investigation Committee of the Hospital Regional Universitario de Málaga (Málaga, Spain) in addition to other regional ethics and clinical investigation committees all over Spain.
Competing interests
N.A. is stock owner of Biosfer Teslab and has a patent on the method for lipoprotein profiling described in the present manuscript. E.O. was employed at Biosfer Teslab. The other authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper interests or personal relationships that could have appeared to influence the work reported in this paper.
Consent of publication
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Equally contributed: Enrique Ozcariz and Montse Guardiola
References
- 1.Sharaiha RZ, Shikora S, White KP, Macedo G, Toouli J, Kow L. <ArticleTitle Language="En">Summarizing Consensus guidelines on obesity management: a Joint, Multidisciplinary Venture of the International Federation for the Surgery of Obesity & Metabolic Disorders (IFSO) and World Gastroenterology Organisation (WGO). J Clin Gastroenterol. 2023. 10.1097/MCG.0000000000001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Piché ME, Tchernof A, Després JP. Obesity phenotypes, diabetes, and cardiovascular diseases. Circ Res. 2020. 10.1161/CIRCRESAHA.120.316101. [DOI] [PubMed] [Google Scholar]
- 3.ElSayed NA, Aleppo G, Aroda VR, Bannuru RR, Brown FM, Bruemmer D, Collins BS, Hilliard ME, Isaacs D, Johnson EL, Kahan S, Khunti K, Leon J, Lyons SK, Perry ML, Prahalad P, Pratley RE, Seley JJ, Stanton RC, Gabbay RA, on behalf of the American Diabetes Association. 8. Obesity and weight management for the prevention and treatment of type 2 diabetes: standards of care in diabetes-2023. Diabetes Care. 2023; 10.2337/dc23-S008. PMID: 36507637. [DOI] [PMC free article] [PubMed]
- 4.Avgerinos KI, Spyrou N, Mantzoros CS, Dalamaga M. Obesity and cancer risk: emerging biological mechanisms and perspectives. Metabolism. 2019. 10.1016/j.metabol.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 5.World health statistics. 2023: Monitoring health for the SDGs, sustainable development goals. Geneva: World Health Organization; 2023. https://www.who.int/publications/i/item/9789240074323. Accessed 4 June 2024.
- 6.Yumuk V, Tsigos C, Fried M, Schindler K, Busetto L, Micic D, Toplak H. Obesity management task force of the european association for the study of obesity. European guidelines for obesity management in adults. Obes Facts. 2015. 10.1159/000442721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weir CB, Jan A, BMI classification percentile and cut off points. 2024. https://www.ncbi.nlm.nih.gov/books/NBK541070/. Accessed 12 March 2024. [PubMed]
- 8.Mayoral LP, Andrade GM, Mayoral EP, Huerta TH, Canseco SP, Rodal Canales FJ, Cabrera-Fuentes HA, Cruz MM, Pérez Santiago AD, Alpuche JJ, Zenteno E, Ruíz HM, Cruz RM, Jeronimo JH, Perez-Campos E. Obesity subtypes, related biomarkers & heterogeneity. Indian J Med Res. 2020. 10.4103/ijmr.IJMR_1768_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthias Blüher MH. Obesity. Endocr Rev. 2020. 10.1210/endrev/bnaa004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho YK, Jung CH. Metabolically healthy obesity: epidemiology, criteria, and implications in chronic kidney disease. J Obes Metab Syndr. 2022. 10.7570/jomes22036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soriguer F, Gutiérrez-Repiso C, Rubio-Martín E, García-Fuentes E, Almaraz MC, Colomo N, Esteva de Antonio I, de Adana MS, Chaves FJ, Morcillo S, Valdés S, Rojo-Martínez G. Metabolically healthy but obese, a matter of time? Findings from the prospective Pizarra study. J Clin Endocrinol Metab. 2013. 10.1210/jc.2012-4253. [DOI] [PubMed] [Google Scholar]
- 12.Kim YH, Kim SM, Han KD, Jung JH, Lee SS, Oh SW, Park HS, Rhee EJ, Lee WY, Yoo SJ. Waist circumference and all-cause mortality independent of body mass index in korean population from the national health insurance health checkup 2009⁻2015. J Clin Med. 2019. 10.3390/jcm8010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Åberg F, Färkkilä M, Salomaa V, Jula A, Männistö S, Perola M, Lundqvist A, Männistö V. Waist-hip ratio is superior to BMI in predicting liver-related outcomes and synergizes with harmful alcohol use. Commun Med (Lond). 2023. 10.1038/s43856-023-00353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steven McGee. Evidence-based physical diagnosis (Fourth Edition). Elsevier. 2018; 10.1016/B978-0-323-39276-1.00013-5.
- 15.Soriguer F, Goday A, Bosch-Comas A, Bordiú E, Calle-Pascual A, Carmena R, Casamitjana R, Castaño L, Castell C, Catalá M, Delgado E, Franch J, Gaztambide S, Girbés J, Gomis R, Gutiérrez G, López-Alba A, Martínez-Larrad MT, Menéndez E, Mora-Peces I, Ortega E, Pascual-Manich G, Rojo-Martínez G, Serrano-Rios M, Valdés S, Vázquez JA, Vendrell J. Prevalence of diabetes mellitus and impaired glucose regulation in Spain: the Di@bet.es Study. Diabetologia. 2012. 10.1007/s00125-011-2336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan DC, Watts GF, Barrett PH, Burke V. Waist circumference, waist-to-hip ratio and body mass index as predictors of adipose tissue compartments in men. QJM. 2003. 10.1093/qjmed/hcg069. [DOI] [PubMed] [Google Scholar]
- 17.Afshin A, Forouzanfar MH, Reitsma MB, Sur P, Estep K, Lee A, Marczak L, Mokdad AH, Moradi-Lakeh M, Naghavi M, et al. GBD 2015 obesity collaborators. Health effects of overweight and obesity in 195 countries over 25 years. N Engl J Med. 2017. 10.1056/NEJMoa1614362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bays HE, Kirkpatrick CF, Maki KC, Toth PP, Morgan RT, Tondt J, Christensen SM, Dixon DL, Jacobson TA. Obesity, dyslipidemia, and cardiovascular disease: a joint expert review from the obesity medicine association and the national lipid association 2024. J Clin Lipidol. 2024. 10.1016/j.jacl.2024.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Azad RK, Shulaev V. Metabolomics technology and bioinformatics for precision medicine. Brief Bioinform. 2019. 10.1093/bib/bbx170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satheesh G, Ramachandran S, Jaleel A. Metabolomics-based prospective studies and prediction of Type 2 diabetes mellitus risks. Metab Syndr Relat Disord. 2020. 10.1089/met.2019.0047. [DOI] [PubMed] [Google Scholar]
- 21.Bellot PENR, Braga ES, Omage FB, da Silva Nunes FL, Lima SCVC, Lyra CO, Marchioni DML, Pedrosa LFC, Barbosa F Jr, Tasic L, Sena-Evangelista KCM. Plasma lipid metabolites as potential biomarkers for identifying individuals at risk of obesity-induced metabolic complications. Sci Rep. 2023. 10.1038/s41598-023-38703-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu B, Hu M, Jiang W, Ma Y, Ye J, Wu Q, Guo W, Sun Y, Zhou M, Xu Y, Wu Z, Wang Y, Lam SM, Shui G, Gu J, Li JZ, Fu Z, Gong Y, Zhou H. Ceramide d18:1/24:1 as a potential biomarker to differentiate obesity subtypes with unfavorable health outcomes. Lipids Health Dis. 2023. 10.1186/s12944-023-01921-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ottosson F, Smith E, Ericson U, Brunkwall L, Orho-Melander M, Di Somma S, Antonini P, Nilsson PM, Fernandez C, Melander O. Metabolome-defined obesity and the risk of future Type 2 diabetes and mortality. Diabetes Care. 2022. 10.2337/dc21-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cirulli ET, Guo L, Leon Swisher C, Shah N, Huang L, Napier LA, Kirkness EF, Spector TD, Caskey CT, Thorens B, Venter JC, Telenti A. Profound perturbation of the metabolome in obesity is associated with health risk. Cell Metab. 2019. 10.1016/j.cmet.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong P, Tan S, Zhu Z, Bulloch G, Long E, Huang W, He M, Wang W. Metabolomic phenotyping of obesity for profiling cardiovascular and ocular diseases. J Transl Med. 2023. 10.1186/s12967-023-04244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Medical Association. World Medical Association declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013. 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 27.WHO Consultation on Obesity,World Health Organization. Obesity: preventing and managing the global epidemic: report of a WHO consultation.World Health Organization. 2000. https://apps.who.int/iris/handle/10665/42330. Accessed 8 July 2024. [PubMed]
- 28.World Health Organization. Waist circumference and waist-hip ratio: report of a WHO expert consultation. World Health Organization. 2011 https://www.who.int/publications/i/item/9789241501491. Accessed 11 June 2024.
- 29.Chobanian AV, et al. Seventh report of the Joint National Committee on prevention, detection, evaluation, and treatment of high blood pressure. Hypertension. 2003. 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 30.World Health Organization. Definition, diagnosis and classification of diabetes mellitus and its complications: report of a WHO consultation. Part 1: diagnosis and classification of diabetes mellitus. Diabet Med. 1998; doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed]
- 31.Mallol R, Amigó N, Rodríguez MA, Heras M, Vinaixa M, Plana N, Rock E, Ribalta J, Yanes O, Masana L, Correig X. Liposcale: a novel advanced lipoprotein test based on 2D diffusion-ordered 1H NMR spectroscopy. J Lipid Res. 2015. 10.1194/jlr.D050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuertes-Martín R, Taverner D, Vallvé JC, Paredes S, Masana L, Correig Blanchar X, Amigó Grau N. Characterization of 1H NMR plasma glycoproteins as a new strategy to identify inflammatory patterns in rheumatoid arthritis. J Proteome Res. 2018. 10.1021/acs.jproteome.8b00411. [DOI] [PubMed] [Google Scholar]
- 33.Mallagaray A, Rudolph L, Lindloge M, Mölbitz J, Thomsen H, Schmelter F, Alhabash MW, Abdullah MR, Saraei R, Ehlers M, Graf T, Sina C, Petersmann A, Nauck M, Günther UL. Towards a precise NMR quantification of acute phase inflammation proteins from human serum. Angew Chem Int Ed Engl. 2023. 10.1002/anie.202306154. [DOI] [PubMed] [Google Scholar]
- 34.Fuertes-Martín R, Correig X, Vallvé JC, Amigó N. Human serum/plasma glycoprotein analysis by 1H-NMR, an emerging method of inflammatory assessment. J Clin Med. 2020. 10.3390/jcm9020354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miranda J, Simões RV, Paules C, Cañueto D, Pardo-Cea MA, García-Martín ML, Crovetto F, Fuertes-Martin R, Domenech M, Gómez-Roig MD, Eixarch E, Estruch R, Hansson SR, Amigó N, Cañellas N, Crispi F. Gratacós. Metabolic profiling and targeted lipidomics reveals a disturbed lipid profile in mothers and fetuses with intrauterine growth restriction. Sci Rep. 2018. 10.1038/s41598-018-31832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garvey WT, Mechanick JI, Brett EM, Garber AJ, Hurley DL, Jastreboff AM, Nadolsky K, Pessah-Pollack R, Plodkowski R. Reviewers of the AACE/ACE obesity clinical practice guidelines. American Association of Clinical Endocrinologists and American Association of Endocrinology Comprehensive Clinical Practice Guidelines for Medical Care of Patients with Obesity. Endocr Pract. 2016. 10.4158/EP161365.GL. [DOI] [PubMed] [Google Scholar]
- 37.Vekic J, Zeljkovic A, Stefanovic A, Jelic-Ivanovic Z, Spasojevic-Kalimanovska V. Obesity and dyslipidemia. Metabolism. 2019. 10.1016/j.metabol.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Phillips CM, Perry IJ. Lipoprotein particle subclass profiles among metabolically healthy and unhealthy obese and non-obese adults: does size matter? Atherosclerosis. 2015. 10.1016/j.atherosclerosis.2015.07.040. [DOI] [PubMed] [Google Scholar]
- 39.Stadler JT, Lackner S, Mörkl S, Trakaki A, Scharnagl H, Borenich A, Wonisch W, Mangge H, Zelzer S, Meier-Allard N, Holasek SJ, Marsche G. Obesity affects HDL metabolism, Composition and Subclass Distribution. Biomedicines. 2021. 10.3390/biomedicines9030242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu X, Liu Q, Guo X, Wang W, Yu B, Liang B, Zhou Y, Dong H, Lin J. The role of remnant cholesterol beyond low-density lipoprotein cholesterol in diabetes mellitus. Cardiovasc Diabetol. 2022;2. 10.1186/s12933-022-01554-0. [DOI] [PMC free article] [PubMed]
- 41.Manmadhan A, Lin BX, Zhong J, Parikh M, Berger JS, Fisher EA, Heffron SP. Elevated GlycA in severe obesity is normalized by bariatric surgery. Diabetes Obes Metab. 2019. 10.1111/dom.13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schubert J, Lindahl B, Melhus H, Renlund H, Leosdottir M, Yari A, Ueda P, James S, Reading SR, Dluzniewski PJ, Hamer AW, Jernberg T, Hagström E. Low-density lipoprotein cholesterol reduction and statin intensity in myocardial infarction patients and major adverse outcomes: a Swedish nationwide cohort study. Eur Heart J. 2021. 10.1093/eurheartj/ehaa1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cannon CP, Cariou B, Blom D, McKenney JM, Lorenzato C, Pordy R, Chaudhari U, Colhoun HM. ODYSSEY COMBO II Investigators. Efficacy and safety of alirocumab in high cardiovascular risk patients with inadequately controlled hypercholesterolaemia on maximally tolerated doses of statins: the ODYSSEY COMBO II randomized controlled trial. Eur Heart J. 2015. 10.1093/eurheartj/ehv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozcariz E, Guardiola M, Amigó N, Rojo-Martínez G, Valdés S, Rehues P, Masana L, Ribalta J. NMR-based metabolomic profiling identifies inflammation and muscle-related metabolites as predictors of incident type 2 diabetes mellitus beyond glucose: The Di@bet.es study. Diabetes Res Clin Pract. 2023. 10.1016/j.diabres.2023.110772. [DOI] [PubMed] [Google Scholar]
- 45.Wang F, Liu H, Blanton WP, Belkina A, Lebrasseur NK, Denis GV. Brd2 disruption in mice causes severe obesity without Type 2 diabetes. Biochem J. 2009. 10.1042/BJ20090928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soininen P, Kangas AJ, Würtz P, Suna T, Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet. 2015. 10.1161/CIRCGENETICS.114.000216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data for the dataset used in this study are not publicly available to preserve individuals’ privacy under the European General Data Protection Regulation.