Abstract
Background
Fetal growth restriction (FGR) is typically characterised as the fetus’ inability to reach its inherent growth potential. A growing body of evidence points to the important role of the maternal gut microbiota in FGR development. However, comprehensive research on changes in maternal–fetal gut and intrauterine microbiota related to FGR is lacking.
Methods
In this case–control study, we sequenced bacterial 16S rRNA from 35 maternal faecal, 35 meconium, and 31 amniotic fluid samples collected from 19 pregnant women diagnosed with FGR and 16 healthy controls. We identified putative bacterial taxonomic and functional characteristics associated with FGR by comparing these to control samples.
Results
We identified 34 differential operational taxonomic units (OTUs) in amniotic fluid, seven differential OTUs in maternal faecal matter, and two differential OTUs in meconium. Compared to controls, FGR subjects exhibited enriched bacterial OTUs of the genus Bacteroides in the maternal gut. They also had depleted OTUs of the order Enterobacterales and genus Pseudomonas in the amniotic fluid and genus Stenotrophomonas in the fetal gut. These altered bacterial OTUs showed a significant correlation with neonatal weight and fetal ultrasonographic indexes. Additionally, we identified differential microbial functional pathways related to glycan and lipid metabolism in the maternal gut. We developed diagnostic biomarkers for FGR based on the maternal–fetal gut and amniotic fluid microbiota.
Conclusions
This study offers a comprehensive overview of the shifts in microbial composition and functional pathways in the maternal–fetal gut and amniotic fluid microbiota related to FGR, and present novel insights into the development and screening of FGR. However, the assessment of contamination’s impact on meconium and amniotic fluid remains inconclusive, necessitating further rigorous experimentation to address this scientific inquiry in future studies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12884-024-06930-0.
Keywords: Fetal growth restriction, 16S rRNA sequencing, Gut microbiota, Amniotic fluid microbiota
Introduction
Fetal growth restriction (FGR) is defined as the failure of the fetus to reach its genetic growth potential, often evaluated as an estimated fetal weight or abdominal circumference that falls below the 10th percentile for the gestational age, as determined by ultrasound [1]. Globally, the incidence rate of FGR ranges from 3 to 9%, with 20 ~ 50% of stillbirths associated with FGR [2, 3]. This condition increases the risk of perinatal morbidity and mortality, and also heightens the risk of cardiovascular and metabolic diseases such as obesity and impaired glucose tolerance in children or adults post birth [4–6]. Given the adverse effects of FGR on perinatal and long-term postnatal outcomes, there is an urgent need for additional studies to explore novel molecular mechanisms of FGR and identify potential biomarkers for its diagnosis.
Microbial colonisation plays a vital role in immunological and physiological development [7]. The human gut microbiota significantly influences various diseases and is crucial for immunity, metabolism, and nutrient absorption. A growing body of research suggests that dysbiosis in maternal gut microbiota could contribute to maternal and fetal complications, such as gestational diabetes mellitus (GDM), preeclampsia, and preterm birth [8–12]. Recent studies suggest that FGR may be associated with distinct characteristics of maternal gut, placental and intrauterine microbiota [13–16]. Disorders in maternal intestinal flora can incite systemic and placental inflammatory responses and insulin resistance, leading to placental chorioamnionitis, which in turn damages placental vascular endothelial cells and impairs placental function, thereby affecting fetal development and resulting in low fetal weight [17]. Experiments with pregnant mice transplanted with microbiota from FGR mothers have resulted in FGR and placental dysfunction [13]. However, considerable knowledge gaps persist regarding the differences in maternal-fetal gut and amniotic fluid microbiota composition between FGR and normal pregnancies, and concrete data on the relationship between fetal gut microbiota and FGR remain scarce.
In this study, we conducted a case–control study using 16S rRNA gene sequencing to identify changes in maternal–fetal gut and amniotic fluid microbiota in pregnant women with FGR compared with those in healthy pregnancies. Our aim was to explore the role of these alterations in the development of FGR.
Methods
Study design and participants
This study received approval from the Internal Ethics Committee of Zhujiang Hospital, Southern Medical University, Guangzhou, China (No. 2022-KY-186-01). All participants provided written informed consent before enrolment. Samples were collected from singleton pregnant women who underwent elective caesarean sections at Zhujiang Hospital of Southern Medical University, the Seventh Affiliated Hospital of Southern Medical University, and the Second People’s Hospital of Qingyuan City. The caesarean section indications were restricted to advanced maternal age, abnormal presentation, repeat caesarean section, and social factors. For the FGR group, indications also included fetal abnormalities such as abnormal fetal heart rate and umbilical blood flow abnormalities.
Exclusion criteria encompassed pregnant women who were carrying twins or multiple pregnancies; had pregnancy complications; had fetal or neonatal abnormalities; were exposed to tobacco or alcohol during pregnancy; had premature rupture of membranes; or had taken antibiotics or other drugs that could affect microbiome diversity before sample collection. The FGR group inclusion criteria, referencing expert consensus and Chinese birth weight references [4, 18], were as follows: (1) birth weight < 10th percentile for gestational age, with prenatal evidence of uterine placental insufficiency defined as an umbilical or uterine artery pulsatility index > 95th percentile or absent end-diastolic flow in the umbilical artery (< 32 weeks gestation) or cerebroplacental ratio < 5th percentile (≥ 32 weeks gestation); and/or with abdominal circumference (AC) < 10th percentile; or (2) birth weight < 3rd percentile. Healthy controls were those with fetal growth appropriate for the gestational age (birth weight between the 10th and 90th percentile). A total of 35 mother–infant pairs (19 FGR and 16 controls) were included for the final analysis.
Sample collection and DNA extraction
Maternal faecal samples were collected within 24 h before delivery. Amniotic fluid samples (~ 10 mL) were collected using sterile syringes before the complete rupture of the amniotic cavity during caesarean section. First-pass meconium samples (~ 20 g) were collected by doctors from nappies (diapers) within the first few hours to 24 h of birth. All samples were immediately stored at − 80℃ until DNA extraction. DNA from maternal and neonatal faecal samples was extracted using the MagaBio Soil and Feces Genomic DNA Purification Kit (Bioer Technology, Huangzhou, China), and from amniotic fluid samples using the Bacterial DNA Extraction Mini Kit (Mabio Biotechnology Co., Ltd., Guangzhou, China). DNA quality and quantity were assessed using a NanoDrop™ One spectrophotometer (Thermo Fisher Scientific, MA, USA), according to the manufacturer’s instructions.
16S rRNA amplification sequencing
The hypervariable region V4 of the bacterial 16S rRNA gene was amplified using specific primers (forward primer 515F: 5′-GTGCCAGCMGCCGCGGTAA-3′ and reverse primer 806R: 5′-GGACTACHVGGGTWTCTAAT-3′) with unique 12-bp barcodes. Amplification was performed with TaKaRa Premix Taq® Version 2.0 (TaKaRa Biotechnology Co. Ltd., Dalian, China) on a BioRad S1000 PCR thermocycler (Bio-Rad Laboratory, Hercules, CA, USA) according to the manufacturer’s instructions. The PCR product’s length and concentration were assessed using 1% agarose gel electrophoresis. The PCR products were mixed equidensity-wise using GeneTools Analysis Software (ver. 4.03.05.0; SynGene, Bengaluru, India) and purified with E.Z.N.A. Gel Extraction Kit (Omega Bio-tek, Norcross, GA, USA). Sequencing libraries were generated using the NEBNext® Ultra™ II DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) and sequenced on an Nova 6000 platform (Illumina, San Diego, CA, USA). The blank controls underwent the same operational procedures as the experimental group, including sample acquisition, use of sterile container, DNA extraction, and PCR amplification, to detect possible contaminating microorganisms.
Sequencing data processing analysis
Fastp (ver. 0.14.1; https://github.com/OpenGene/fastp) was used for raw data quality control, and Cutadapt software (https://github.com/marcelm/cutadapt/) was used to remove primers to obtain paired-end clean reads. The USEARCH fastq_mergepairs command (ver. 10; http://www.drive5.com/usearch/) was employed to obtain the original spliced sequence (raw tags). Fastp was then used again for raw data quality control to yield paired-end clean tags. Operational taxonomic units (OTUs) were clustered using UPARSE (ver. 7.1; http://drive5.com/uparse/), and chimeric sequences and singleton OTUs were removed. The taxonomy of each 16S rRNA gene sequence was analysed with the USEARCH sintax command against the 16S rRNA database (SILVA 16S) using a confidence threshold of 0.8. OTUs annotated as chloroplasts or mitochondria (16S amplicons), or those that could not be annotated to the kingdom level, were removed.
Microbiome source tracking
We conducted microbial source prediction at the OTU level using the FEAST package [19]. Amniotic fluid and maternal gut samples served as bacterial sources, and neonatal gut samples as sinks. We finally selected 15 FGR groups and 16 control groups for analysis, as four amniotic fluid samples were missing from the initial 19 FGR samples. We used the default parameters for the analysis to study transfer relationships at the microbial level. The source tracking results were visualised using the R ‘ggplot2’ package.
Statistical analysis
Continuous variables for participant characteristics in this study were reported as the mean (standard deviation) or median (interquartile range), and categorical variables were expressed as the count (%). Comparisons of participants’ characteristics were conducted using Student’s t-test or the Mann–Whitney U test for continuous variables, and the Chi-squared test for categorical variables.
The α-diversity was assessed using the Shannon diversity index and Chao1 richness estimator to estimate the microbial diversity within individual samples. Significant differences in α-diversities were analysed with the Wilcoxon test using R software (ver. 4.0.2). We also performed β-diversity distance measurements based on the Bray–Curtis distance to investigate the structural variation of microbial communities (between-subject diversity), visualised via principal coordinate analysis (PCoA) using the R ‘vegan’ package. ANOSIM and ADONIS analyses were further applied to reveal the significance of the β-diversity between the groups.
Analysis of the differences in taxa between two groups was conducted based on OTUs with an abundance > 0.01% using the Wilcoxon test with the ‘ggsignif’ package for statistical testing, and the ‘ggplot2’ package for graphic visualisation. A p-value < 0.05 was considered significant. Spearman’s correlation analysis was conducted on significantly differential OTUs and FGR clinical characteristics to assess their putative correlations (significance threshold: p-value < 0.05, r ≥ 0.3). The potential Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology functional categories of microbial communities were predicted using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) method. The disease prediction model was trained using the ‘random Forest’ package, with a 7:3 ratio of training and testing sets, optimising the model according to the importance of each species’ accuracy, thereby maximising the accuracy rate of the three types of samples. Results were evaluated using the 10-fold cross-validation method, and model performance was assessed based on the area under the curve (AUC).
Results
participant characteristics
The final analysis included 19 maternal–fetal pairs with FGR and 16 normal controls. The clinical characteristics of these maternal–fetal pairs are detailed in Table 1. Of the 19 FGR cases, four amniotic fluid samples were missing, so the analysis of the amniotic fluid microbiota encompassed 15 FGR cases and 16 normal controls. Table S1 in Additional file 1 illustrates the clinical features of the cohort. There were no significant differences in maternal age, height, maternal pre-pregnancy body mass index (BMI), maternal weight gain and maternal pre-delivery BMI. However, parity was not balanced between the two groups. As anticipated, the gestational age at birth, neonatal weight, neonatal length, neonatal BMI, and various pre-birth ultrasound measurement indicators, including biparietal diameter (BPD), head circumference (HC), AC, and femur length (FL), were significantly decreased in the FGR group.
Table 1.
Maternal and fetal clinical characteristics
| Features | Control (n = 16) | FGR (n = 19) | P values |
|---|---|---|---|
| Maternal features | |||
| Maternal age (years) | 30.13 ± 5.21 | 27.84 ± 6.70 | 0.276 |
| Maternal pre-pregnancy BMI (kg/m2) | 20.61 ± 1.87 | 20.01 ± 2.80 | 0.123 |
| Maternal BMI before delivery (kg/m2) | 26.24 ± 3.59 | 24.68 ± 3.65 | 0.959 |
| Maternal weight gain | 14.03 ± 7.11 | 11.18 ± 4.86 | 0.171 |
| Maternal height (m) | 1.57 ± 0.05 | 1.55 ± 0.07 | 0.273 |
| Parity (n (%)) | <0.001 | ||
| Primigravida | 1 (6.25) | 12 (63.16) | |
| Multigravida | 15 (93.75) | 7 (36.84) | |
| Gestational age (weeks) | 38.88 (38.36–39.14) | 38.14 (37.43–38.43) | <0.01 |
| Fetal features | |||
| Gender (n (%)) | 0.641 | ||
| Female | 8 (50.00) | 11 (57.89) | |
| Male | 8 (50.00) | 8 (42.11) | |
| Neonatal weight (kg) | 3.23 (3.07–3.73) | 2.38 (2.07–2.46) | <0.0001 |
| Neonatal BMI (kg/m2) | 13.18 (12.28–14.50) | 10.33 (9.38–10.68) | <0.0001 |
| Neonatal length (m) | 0.50 (0.50–0.51) | 0.48 (0.47–0.48) | <0.0001 |
| BPD (mm) | 92.00 (89.50–93.75) | 86.00 (84.00–87.00) | <0.0001 |
| HC (mm) | 329.50 (320.00–334.80) | 309.00 (302.00–314.00) | <0.0001 |
| AC (mm) | 343.00 (332.30–359.80) | 304.00 (291.00–312.00) | <0.0001 |
| FL (mm) | 70.00 (68.00–71.00) | 65.00 (64.00–68.00) | <0.0001 |
| AFI (mm) | 133.8 ± 41.80 | 108.7 ± 38.77 | 0.10 |
Data were assessed for normality using the Shapiro–Wilk test and expressed as the mean ± standard deviation, median (interquartile range), frequency, and percentage. FGR: fetal growth restriction; BMI: body mass index; F: female; M: male; m: metre; mm: millimetre; kg: kilogram; BPD: biparietal diameter; HC: head circumference; AC: abdominal circumference; FL: femur length; AFI: amniotic fluid index. BMI is defined as weight/height2 (kg/m2)
Overall microbiota diversity in each group
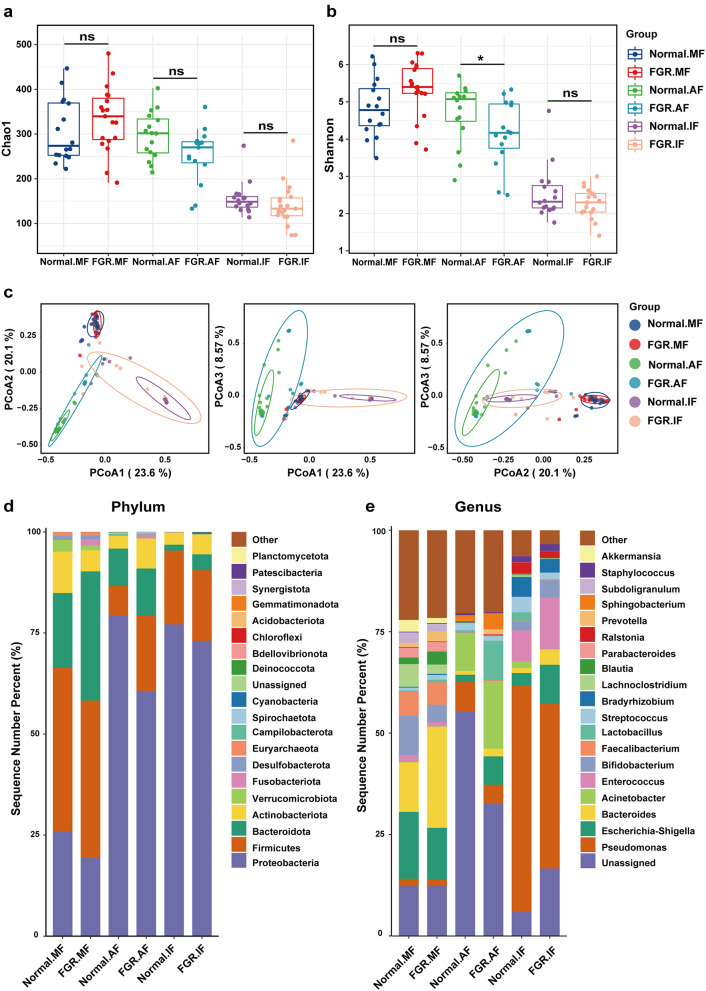
A total of 8,751 different OTUs were identified from 70 faecal samples and 31 amniotic fluid samples using UPARSE. The Shannon diversity and Chao1 richness curves both plateaued, indicating that the sequencing data volume was ample to represent the majority of microbial diversity (see Figure S1 in Additional file 2). The Shannon diversity index and Chao1 richness index were used to measure the α-diversity of each sample’s microbial community. Comparison of the Shannon diversity index showed that the microbial diversity of amniotic fluid in the FGR group was significantly lower than that in the control group (p = 0.046, Wilcoxon test). However, there were no significant differences between the maternal faecal samples from the control group and FGR group (p = 0.066), or between the meconium samples from the control group and FGR group (p = 0.328) (Fig. 1a). The Chao1 richness index showed no significant difference between the FGR and control groups, irrespective of the sample type (Fig. 1b). Additionally, there were significant differences in the Shannon diversity index and Chao1 richness index among different sample types in both the control group and the FGR group (p < 0.001, except between maternal faeces and amniotic fluid in the control group; p = 0.955 for the Shannon index; p = 0.895 for Chao1 index) (Table S2 in Additional file 1).
Fig. 1.
Overall microbiota diversity and microbial composition in each group. Diversity and composition of the overall microbiota between the FGR and control groups. Diversity of the overall microbiota between the FGR and control groups based on the (a) Chao1 richness and (b) Shannon diversity index. (c) Separation of FGR and control samples based on PCoA following the Bray–Curtis distance. The microbiota composition of the of FGR and control groups at the (d) phylum and (e) genus level. AF, amniotic fluid; IF, meconium; MF, maternal faeces. *p < 0.05
PCoA based on the Bray–Curtis distance was performed to assess the overall microbial composition diversity in each group. In the PCoA plots, the maternal faecal samples, amniotic fluid, and meconium samples formed distinct clusters, implying significant differences in microbial community structures among the three sample types (Figure S1 in Additional file 2). There was a significant difference in the microbial composition of amniotic fluid between the control and FGR groups (p < 0.05), even though the FGR group’s clustering was not completely separate from the control group (Fig. 1c). No significant differences were observed in the microbial composition of maternal faeces and meconium between the control and FGR groups (Table S3 in Additional file 1).
Microbial composition in each group at the phylum and genus levels
A statistical taxonomic analysis was conducted at the phylum and genus levels. At the phylum level, Proteobacteria, Firmicutes, Bacteroidota, and Actinobacteria constituted the most prevalent taxonomic groups across the samples, representing over 95% of the taxonomy. Firmicutes predominated in maternal faecal samples, whereas Proteobacteria predominated in amniotic fluid and meconium samples. Compared to the control group, Bacteroidota was more abundant in all three sample types in the FGR group, whereas the proportion of Proteobacteria decreased (Fig. 1d).
At the genus level, the top 20 most abundant genera were distinguished, while the remainder were grouped as “Others” (Fig. 1e). In maternal faeces, the dominant genus Bacteroides was significantly abundant in the FGR group, while Escherichia-Shigella and Bifidobacterium were less abundant. In amniotic fluid, the dominant genera Acinetobacter and Lactobacillus were more abundant in the FGR group, while the abundance of Pseudomonas decreased. In meconium, the dominant genus Pseudomonas decreased in the FGR group, whereas Enterococcus and Escherichia-Shigella increased.
Source tracking of meconium microbiota communities
FEAST data can provide valuable insights for tracking the formation of microbial communities, and distinguishing and characterising health conditions related to bacteria. By source tracking the meconium microbial community, we can better understand the origins of its microbiota and their similarities to the maternal microbiota. In this study, FEAST was used at the OTU level to predict the probable source of the meconium microbiota and further discern potential differences between the FGR and control groups, using maternal faeces and amniotic fluid microbiota as potential sources. The analysis revealed that meconium and maternal faeces samples shared a higher level of common OTUs (FGR: 31.53%; control: 23.85%) compared to the meconium–amniotic fluid sample pair (FGR: 3.68%; control: 2.06%) (Fig. 2). When compared with the control group, the proportion of OTUs shared by meconium and maternal faeces, and amniotic fluid samples in the FGR group significantly increased.
Fig. 2.
Source tracking of meconium microbiota communities. The percentage of shared OTUs between the meconium microbiota and the microbiota of maternal faeces and amniotic fluid in the (a) control and (b) FGR groups. The notations MF, IF, and AF are the same as in Fig. 1
Differences in microbiota between the FGR and control groups
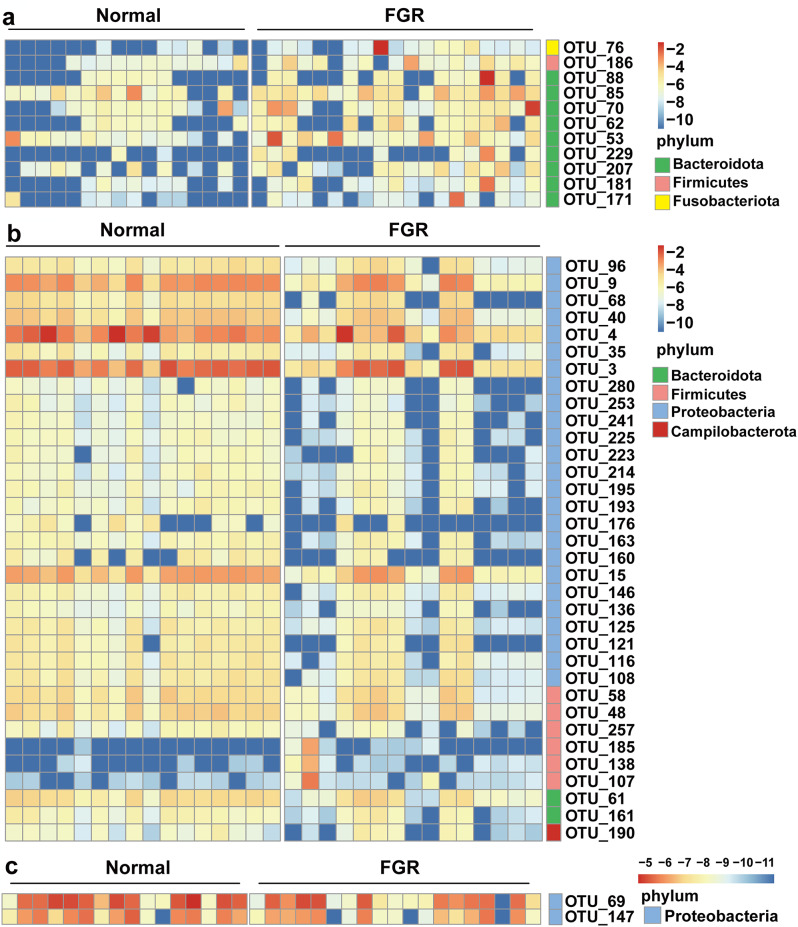
We used the Wilcoxon test on the microbiota composition at the OTU level for various sample types to identify differentially abundant taxa between the FGR and control groups. In total, we identified 34 differential OTUs in amniotic fluid samples, 11 OTUs in maternal faecal samples, and two OTUs in meconium samples. A heatmap composed of these differential OTUs revealed distinct clustering patterns between the FGR and control groups (Fig. 3). Compared to the control group, FGR subjects were characterised by 11 enriched OTUs primarily belonging to Bacteroidota (six OTUs from genus Bacteroides: OTU_62, 70, 85, 88, 181, and 229) in maternal faecal samples (Fig. 3a); 31 depleted OTUs mainly from Proteobacteria (11 OTUs from order Enterobacterales: OTU_3, 4, 9, 68, 96, 121, 136, 163, 176, 193, and 195; and five OTUs from genus Pseudomonas: OTU_15, 116, 146, 253, and 280) and three enriched OTUs from Firmicutes (OTU_107, 138, and 185) in amniotic fluid (Fig. 3b); and two depleted OTUs of Proteobacteria in meconium (Fig. 3c). Table S4 in Additional file 1 provides detailed information on the differential OTUs. These results indicated that there were more differentially abundant taxa in amniotic fluid between the FGR and control groups.
Fig. 3.
Heat map consisting of differential OTUs between the FGR and control groups. Heat map illustrating the differential OTUs in (a) maternal faeces, (b) amniotic fluid, and (c) meconium between the FGR and control groups. Red shading represents higher abundance; blue shading represents lower abundance
Association between altered microbiota and clinical parameters
Subsequently, we conducted Spearman’s correlation analysis between differential OTUs and FGR-associated clinical measures, including neonatal weight, neonatal length, neonatal BMI, BPD, HC, AC, FL, and amniotic fluid index (AFI). In maternal faeces, the six enriched OTUs of the genus Bacteroides (OTU_62, 70, 85, 88, 181, and 229) in the FGR group were negatively correlated with neonatal weight, and two enriched OTUs (OTU_76 and 229) were negatively correlated with neonatal weight, neonatal length, neonatal BMI, HC, AC, and FL (Figure S2a in Additional file 2). In the amniotic fluid, 29 depleted OTUs were positively correlated with neonatal weight, among which were ten OTUs belonging to the order Enterobacterales (OTU_3, 4, 9, 68, 96, 121, 136, 163, 193, and 195) and five OTUs belonging to the genus Pseudomonas (OTU_15, 116, 146, 253, and 280). Notably, three depleted OTUs (OTU_4, 136, and 253) were positively correlated with neonatal weight, neonatal length, neonatal BMI, BPD, HC, and AC, while the enriched OTU_138 (g_Agathobacter) was negatively correlated with them (Figure S2b in Additional file 2). In meconium, the depleted OTU_69 (g_Stenotrophomonas) was positively correlated with neonatal weight, neonatal BMI, and HC (Figure S2c in Additional file 2).
Differential functions of bacteria between the FGR and control groups
To gain a deeper understanding of the relationship between FGR and the maternal–fetal gut microbiome functions, we mapped the 16s sequences to the genes and pathways that these bacterial populations may harbour using the PICRUSt method. The functional category enrichment results revealed 16 pathways that differed significantly in maternal faeces (P < 0.05) between the FGR and control groups; no significant differences were found in pathways between the two groups of meconium (Fig. 4).
Fig. 4.
The differential KEGG pathways of microbiota of maternal faeces between the FGR and control groups. The notations MF is the same as in Fig. 1
The functional categories differentially enriched in maternal faeces between the two groups were mainly involved in glycan and lipid metabolism, including “Galactose metabolism”, “Other glycan degradation”, “Glycerophospholipid metabolism”, “Sphingolipid metabolism”, “Linoleic acid metabolism”, and “Steroid hormone biosynthesis”. Table S5 in Additional file 1 provides detailed information on these functional pathways. These results suggest that the maternal gut microbiota may influence the occurrence and development of FGR by affecting specific metabolic pathways.
Prediction of FGR based on maternal faeces, amniotic fluid, and meconium microbiota signature
Finally, we used the random forest algorithm to assess the potential value of maternal faeces, amniotic fluid, and meconium microbiota as biomarkers to predict FGR risk. A random forest model based on a 23-OTU combination achieved the highest prediction performance (AUC: 0.88, 95% CI: 75.67–100.00, accuracy: 83.5%) in the amniotic fluid (Fig. 5a). Several depleted OTUs in FGR amniotic fluid, including OTU_4 (o_Enterobacterales), OTU_9 (f_Enterobacteriaceae), OTU_193 (g_Pectobacterium), OTU_136 (g_Morganella), all belonging to Enterobacterales, as well as OTU_253 (g_Pseudomonas), featured high scores for the discrimination between FGR and healthy controls (Fig. 5b). Furthermore, FGR could be differentiated from controls using models based on a 10-OTU combination in maternal faeces (AUC: 86.23%, 95% CI: 0.74–0.98, accuracy: 77.0%) and a 13-OTU combination in meconium (AUC: 84.9%, 95% CI: 0.72–0.98, accuracy: 74.3%) (Fig. 5c and e). Several increased OTUs in FGR maternal faeces, including OTU_76 (g_Fusobacterium), OTU_62 (g_Bacteroides), and OTU_53 (g_Prevotella), featured high scores for the discrimination between FGR and healthy controls (Fig. 5d). The depleted OTU_69 (g_Stenotrophomonas) in FGR meconium was also present in the random forest model of meconium, indicating its importance (Fig. 5f).
Fig. 5.
Classification of FGR status based on the relative abundances of microbial biomarkers. Receiver operating characteristic curve analysis of the classification of preeclampsia patients and healthy controls based on microbiota of the (a) amniotic fluid, (c) maternal faeces, and (e) meconium, as assessed by the AUC. The discriminant OTUs in the model of (b) amniotic fluid, (d) maternal faeces, and (f) meconium classifying the FGR and control groups. Colour-coding indicates enrichment in FGR (red) or the control (blue)
Discussion
To explore the alterations in maternal–fetal gut and amniotic fluid microbiota associated with FGR, we analysed a total of 101 samples from 19 FGR cases and 16 controls. The results demonstrated significant differences in maternal and fetal gut and amniotic fluid microbiota between FGR and normal pregnancies. Compared to the control group, the FGR subjects were characterised by an enrichment of bacterial OTUs from the genus Bacteroides in the maternal gut, depletion of OTUs from the order Enterobacterales and genus Pseudomonas in the amniotic fluid, as well as a depletion of the genus Stenotrophomonas in the fetal gut. These characteristic bacterial OTU alterations significantly correlated with neonatal weight and fetal ultrasonography indices. Furthermore, the functional pathway disorders of the maternal gut microbiota may be associated with the occurrence of FGR. These well-defined maternal–fetal gut and amniotic fluid microbial biomarkers could contribute to the early warning of FGR status.
Initially, the maternal, fetal gut, and amniotic fluid microbiota were analysed using the α- and β-diversity indices. We discovered that the richness and diversity of FGR amniotic fluid microbiota were lower than those in normal pregnancies. Similarly, the structural composition of FGR amniotic fluid microbiota differed significantly from normal pregnancies. Concurrently, we observed higher richness and diversity in the maternal gut microbiota of FGR subjects, contrasted by lower richness and higher diversity in fetal gut microbiota, albeit without statistical significance. Alterations in the maternal gut microbiota of FGR subjects align with previous study [14]. Zheng et al. found significantly lower bacterial richness and evenness in the placentas of low-birth-weight newborns compared to normal-weight newborns [20]. Chen et al. similarly highlighted lower richness and diversity in the intrauterine microbiome of FGR subjects [15]. These findings suggest a potential association between decreased bacterial richness and diversity in the intrauterine environment and abnormal fetal development. Individuals with lower bacterial richness exhibited more pronounced obesity, insulin resistance, dyslipidaemia, and a more pronounced inflammatory phenotype compared to those with higher bacterial richness [21].
Existing studies generally consider that the development of gut microbiota ecology begins in the fetal stage and can be influenced by maternal factors during pregnancy, even though the fetal gut microbiome differs from the placental and maternal oral, skin, vaginal, and faecal microbiome [22, 23]. Our study aligns with these findings, supporting the unique characteristics of the fetal gut microbiome. Source tracking results indicated that the fetal gut microbiota shared more common OTUs with the maternal gut microbiota, and the proportion of unique fetal gut microbiota in the FGR group decreased compared to normal pregnancies. This variation in microbiota could potentially contribute to the development of FGR.
The OTUs that increased in the maternal gut of the FGR group predominantly belonged to the genus Bacteroides of the phylum Bacteroidota, aligning with previous research [14]. Bacteroides in pregnant women’s intestines is associated with lipid status and maternal dyslipidaemia during pregnancy, as it interferes with inflammatory pathways [24]. Interestingly, we also found that disturbances of lipid metabolism functions (e.g. sphingolipid metabolism, linoleic acid metabolism, and steroid hormone biosynthesis) occurred in the FGR maternal gut. Prior research has found that maternal dyslipidaemia is associated with accelerated placental epigenetic aging [25]. Premature placental aging is linked to adverse obstetric complications such as preeclampsia, low birth weight, and preterm birth [26]. Therefore, our findings suggest that an increase in the abundance of Bacteroidota, especially the genus Bacteroides, in the maternal gut may contribute to the occurrence and development of FGR via lipid metabolic pathways. Further metabolomics research is needed to identify specific microbial metabolites and pathways involved in FGR pathogenesis.
Further investigation into the differential microbiome between FGR and normal pregnancies revealed that the main distinguishing microbiota in the amniotic fluid and fetal gut of the FGR group primarily belonged to the Proteobacteria, whereas in the maternal gut, it mainly belonged to the Bacteroidota. Out of the 29 differential OTUs in amniotic fluid positively correlated with fetal weight, ten OTUs belonged to the order Enterobacterales, three of which belonged to the family Enterobacteriaceae, and five OTUs belonged to the genus Pseudomonas in the family Pseudomonadaceae. The differential OTU in meconium positively correlated with fetal weight belonged to the genus Stenotrophomonas. Interestingly, the order Enterobacterales and the genera Pseudomonas and Stenotrophomonas were all gram-negative bacteria. The decrease in the abundance of gram-negative bacteria in amniotic fluid and fetal gut may be associated with FGR. A previous study by Chen et al. also found that gram-negative intrauterine bacteria were linked to a decreased risk of FGR [15]. Kandasamy et al. reported that selected gram-negative probiotics proved more effective than gram-positive probiotics in stimulating protective immunity against pathogens in human and animal models [27]. Impaired immunity and infection resistance in newborns with FGR, alongside the increased risk of infection, may be associated with immune dysfunction mediated by fetal microbial exposure disorders [28].
We acknowledge that our study has several limitations. Firstly, cross-sectional nature prevents us from establishing a causal relationship. Secondly, our study only included maternal feces samples taken before delivery, thereby failing to reflect the dynamic changes of the microbiota throughout pregnancy. Thirdly, gestational age at birth of the FGR group were notably lower than those of the control group. This uncertainty leaves us unable to ascertain whether different gestational ages had an impact on the baseline level of the microbiota. Additionally, the limited sample sizes in our study may have constrained the reliability of predictions made through machine learning analysis, so more longitudinal studies with larger samples of pregnant women could lend further support to our hypothesis. Moreover, several studies suggest that the identified microbial signals originating from the fetus and the intrauterine environment are likely attributed to contamination during clinical procedures for fetal sample acquisition, DNA extraction or sequencing processes [29, 30], and our study was also affected by the same contamination issue, potentially resulting in contamination-associated findings such as the presence of genus Pseudomonas and genus Stenotrophomonas [31]. To mitigate the impact of contamination, corresponding blank controls were established for each type of specimen in our study. The blank controls underwent the same operational procedures as the experimental group, including sample acquisition, use of sterile container, DNA extraction, and PCR amplification. However, we didn’t use blank control for library construction and sequencing, which might cause contamination. The implementation of a corresponding blank control at each step is essential to mitigate contamination, thereby enhancing the reliability of the obtained results. In the end, it is important to note that the meconium samples were not obtained with anal swabs during caesarean section but collected several hours after birth. Consequently, the results may be influenced by microbiota of maternal skin, breast milk, or the external environment. Therefore, it would be more appropriate to consider the microbes of the meconium as part of the newborn gut microbiome rather than solely representing the fetal gut microbiome.
Conclusions
In conclusion, we carried out a comprehensive investigation of changes in microbial composition and functional pathways in the maternal–fetal gut and amniotic fluid microbiota associated with FGR in this case–control study. Our results illustrate the differences between the maternal gut, amniotic fluid, and fetal gut microbiota in FGR states and normal pregnancies. We hope to leverage larger sample sizes in future studies to investigate whether microbiota can be used as a biomarker to distinguish FGR. Although the assessment of contamination’s impact on meconium and amniotic fluid remains inconclusive, necessitating further rigorous experimentation to address this scientific inquiry in future studies. These findings may present novel insights into the development of FGR and provide valuable cues for its screening.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
We sincerely thank all doctors, midwives who helped with patient recruitment. We are grateful to all the team members for their contributions to data collection and integrity.
Abbreviations
- FGR
Fetal growth restriction
- OTUs
Operational taxonomic units
- GDM
Gestational diabetes mellitus
- AC
Abdominal circumference
- PCoA
Principal coordinate analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
- AUC
Area under the curve
- BMI
Body mass index
- BPD
Biparietal diameter
- HC
Head circumference
- FL
Femur length
- AFI
Amniotic fluid index
Author contributions
CX, FY and DH conceived and designed the study. Patient recruitment and sample collection were undertaken by XP, YP, PN, GZ and DW. Experiments and data collection were performed by YX and SZ. Data analyses and interpretation were performed by YX, ML and SZ. The manuscript was written by YX, ML, SZ and revised by CX. All authors read and approved the final manuscript.
Funding
This study was supported by The China Postdoctoral Science Foundation(Grant number: 2022M721519); The Foshan Science and Technology Innovation Project (Grant number: 2020001005124); President Foundation of Zhujiang Hospital, Southern Medical University(Grant number: yzjj2022qn09).
Data availability
16S rRNA gene sequencing data is available in the NCBI’ s Sequence Read Archive database (BioProject ID PRJNA1024934).
Declarations
Ethics approval and consent to participate
This study was approved by the Internal Ethics Committee of Zhujiang Hospital, Southern Medical University (IRB No. 2022-KY-186-01). All participants provided written informed consent in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yuanling Xiao, Meng Li and Shaoping Zheng contribute to this work equally. The order of author names was determined based on the contributions to the writing of the manuscript.
Contributor Information
Dongmei Hu, Email: zjyyhdm@163.com.
Fang Yang, Email: 964175870@qq.com.
Cailing Xu, Email: xucailing0@163.com.
References
- 1.Fetal Growth Restriction. <ArticleTitle Language=“En”>ACOG Practice Bulletin, Number 227. Obstet Gynecol. 2021;137(2):e16–28. [DOI] [PubMed] [Google Scholar]
- 2.Nardozza L, Caetano A, Zamarian A, Mazzola J, Silva C, Marçal V, et al. Fetal growth restriction: current knowledge. Arch Gynecol Obstet. 2017;295(5):1061–77. [DOI] [PubMed] [Google Scholar]
- 3.Ego A, Monier I, Skaare K, Zeitlin J. Antenatal detection of fetal growth restriction and risk of stillbirth: population-based case-control study. Ultrasound Obstet gynecology: official J Int Soc Ultrasound Obstet Gynecol. 2020;55(5):613–20. [DOI] [PubMed] [Google Scholar]
- 4.Gordijn S, Beune I, Thilaganathan B, Papageorghiou A, Baschat A, Baker P, et al. Consensus definition of fetal growth restriction: a Delphi procedure. Ultrasound Obstet gynecology: official J Int Soc Ultrasound Obstet Gynecol. 2016;48(3):333–9. [DOI] [PubMed] [Google Scholar]
- 5.Miller S, Huppi P, Mallard C. The consequences of fetal growth restriction on brain structure and neurodevelopmental outcome. J Physiol. 2016;594(4):807–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crispi F, Miranda J, Gratacós E. Long-term cardiovascular consequences of fetal growth restriction: biology, clinical implications, and opportunities for prevention of adult disease. Am J Obstet Gynecol. 2018;218:S869–79. [DOI] [PubMed] [Google Scholar]
- 7.Koleva P, Kim J, Scott J, Kozyrskyj A. Microbial programming of health and disease starts during fetal life. Birth defects Res Part C Embryo today: reviews. 2015;105(4):265–77. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Li P, Liu M, Zheng H, He Y, Chen M, et al. Gut dysbiosis induces the development of pre-eclampsia through bacterial translocation. Gut. 2020;69(3):513–22. [DOI] [PubMed] [Google Scholar]
- 9.Gershuni V, Li Y, Elovitz M, Li H, Wu G, Compher C. Maternal gut microbiota reflecting poor diet quality is associated with spontaneous preterm birth in a prospective cohort study. Am J Clin Nutr. 2021;113(3):602–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Simone N, Santamaria Ortiz A, Specchia M, Tersigni C, Villa P, Gasbarrini A, et al. Recent Insights on the Maternal Microbiota: Impact on Pregnancy Outcomes. Front Immunol. 2020;11:528202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Zheng J, Shi W, Du N, Xu X, Zhang Y, et al. Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut. 2018;67(9):1614–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Arango L, Barrett H, McIntyre H, Callaway L, Morrison M, Dekker Nitert M. Increased Systolic and Diastolic Blood Pressure Is Associated With Altered Gut Microbiota Composition and Butyrate Production in Early Pregnancy. Hypertens (Dallas Tex: 1979). 2016;68(4):974–81. [DOI] [PubMed] [Google Scholar]
- 13.Tao Z, Chen Y, He F, Tang J, Zhan L, Hu H, et al. Alterations in the Gut Microbiome and Metabolisms in Pregnancies with Fetal Growth Restriction. Microbiol Spectr. 2023;11(3):e0007623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu X, Duan C, Lin B, Li K, Gao J, Yan H, et al. Characteristics of the gut microbiota in pregnant women with fetal growth restriction. BMC Pregnancy Childbirth. 2022;22(1):297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C, Tang P, Liang J, Huang D, Pan D, Lin M, et al. Association between Intrauterine Microbiome and Risk of Intrauterine Growth Restriction: A Case-Control Study Based on Guangxi Zhuang Birth Cohort in China. Tohoku J Exp Med. 2022;258(1):11–21. [DOI] [PubMed] [Google Scholar]
- 16.Hu J, Benny P, Wang M, Ma Y, Lambertini L, Peter I, et al. Intrauterine Growth Restriction Is Associated with Unique Features of the Reproductive Microbiome. Reproductive Sci (Thousand Oaks Calif). 2021;28(3):828–37. [DOI] [PubMed] [Google Scholar]
- 17.Gorczyca K, Obuchowska A, Kimber-Trojnar Ż, Wierzchowska-Opoka M, Leszczyńska-Gorzelak B. Changes in the Gut Microbiome and Pathologies in Pregnancy. Int J Environ Res Public Health 2022, 19(16). [DOI] [PMC free article] [PubMed]
- 18.Dai L, Deng C, Li Y, Zhu J, Mu Y, Deng Y, et al. Birth weight reference percentiles for Chinese. PLoS ONE. 2014;9(8):e104779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shenhav L, Thompson M, Joseph T, Briscoe L, Furman O, Bogumil D, et al. FEAST: fast expectation-maximization for microbial source tracking. Nat Methods. 2019;16(7):627–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng J, Xiao X, Zhang Q, Mao L, Yu M, Xu J. The Placental Microbiome Varies in Association with Low Birth Weight in Full-Term Neonates. Nutrients. 2015;7(8):6924–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–6. [DOI] [PubMed] [Google Scholar]
- 22.Tapiainen T, Paalanne N, Tejesvi M, Koivusaari P, Korpela K, Pokka T, et al. Maternal influence on the fetal microbiome in a population-based study of the first-pass meconium. Pediatr Res. 2018;84(3):371–9. [DOI] [PubMed] [Google Scholar]
- 23.Williams N, Vella R, Zhou Y, Gao H, Mass K, Townsel C et al. Investigating the origin of the fetal gut and placenta microbiome in twins. The journal of maternal-fetal & neonatal medicine: the official journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies, the International Society of Perinatal Obstetricians 2022, 35(25):7025–35. [DOI] [PubMed]
- 24.Yang X, Zhang M, Zhang Y, Wei H, Guan Q, Dong C, et al. Ecological change of the gut microbiota during pregnancy and progression to dyslipidemia. NPJ biofilms microbiomes. 2023;9(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shrestha D, Workalemahu T, Tekola-Ayele F. Maternal dyslipidemia during early pregnancy and epigenetic ageing of the placenta. Epigenetics. 2019;14(10):1030–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polettini J, Dutta E, Behnia F, Saade G, Torloni M, Menon R. Aging of intrauterine tissues in spontaneous preterm birth and preterm premature rupture of the membranes: A systematic review of the literature. Placenta. 2015;36(9):969–73. [DOI] [PubMed] [Google Scholar]
- 27.Kandasamy S, Vlasova A, Fischer D, Chattha K, Shao L, Kumar A, et al. Unraveling the Differences between Gram-Positive and Gram-Negative Probiotics in Modulating Protective Immunity to Enteric Infections. Front Immunol. 2017;8:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bæk O, Ren S, Brunse A, Sangild P, Nguyen D. Impaired Neonatal Immunity and Infection Resistance Following Fetal Growth Restriction in Preterm Pigs. Front Immunol. 2020;11:1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy KM, de Goffau MC, Perez-Muñoz ME, Arrieta MC, Bäckhed F, Bork P, et al. Questioning the fetal microbiome illustrates pitfalls of low-biomass microbial studies. Nature. 2023;613(7945):639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panzer JJ, Romero R, Greenberg JM, Winters AD, Galaz J, Gomez-Lopez N, et al. Is there a placental microbiota? A critical review and re-analysis of published placental microbiota datasets. BMC Microbiol. 2023;23(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA gene sequencing data is available in the NCBI’ s Sequence Read Archive database (BioProject ID PRJNA1024934).