Abstract
Background
The tumor immune microenvironment in cancer treatment response and resistance is of increasing interest. This retrospective study characterized and investigated programmed death-ligand 1 (PD-L1), PD-L2, and the immune gene expression signature and their association with clinical outcomes in locoregionally advanced head and neck squamous cell carcinoma (LA HNSCC).
Patients and methods
PD-L1 and PD-L2 expression on tumor and immune-infiltrating cells (positivity defined as combined positive score or immunohistochemistry proportion score >1) and T-cell-inflamed gene expression profile (TcellinfGEP) were evaluated in patients with LA HNSCC treated in South Korea from 2000 to 2015. Correlations among the three biomarkers and their associations with overall survival and recurrence-free survival were assessed.
Results
Among 366 patients, 38.8% had human papillomavirus-positive disease. PD-L1-positive, PD-L2-positive, and high TcellinfGEP (≤−0.162) status were observed in 83.6%, 85.4%, and 73.2% of patients, respectively; 4.1% were posttreatment samples. Correlation between PD-L1 and PD-L2 scores was moderate (rSpearman = 0.50), and each biomarker was slightly less correlated with TcellinfGEP (0.41-0.45). PD-L1 expression and high TcellinfGEP status were associated with human papillomavirus positivity. Higher levels of all biomarkers were observed in oral cavity and oropharyngeal cancers compared with other HNSCC sites. In a multivariable analysis that simultaneously adjusted for all three biomarkers, only high TcellinfGEP was significantly associated with longer overall survival (adjusted hazard ratio, 0.57; 95% confidence interval 0.33-0.98) and recurrence-free survival (adjusted hazard ratio, 0.41; 95% confidence interval 0.23-0.74).
Conclusion
High TcellinfGEP status, but not PD-L1 or PD-L2 expression, was independently associated with longer survival in patients with LA HNSCC. Results may have implications for evaluating therapies targeting programmed cell death protein 1 (PD-1)/programmed death-ligand 1 (PD-L1) in HNSCC.
Key words: head and neck squamous cell carcinoma, PD-L1, PD-L2, gene expression profile, programmed cell death protein 1
Highlights
-
•
PD-L1, PD-L2, and TcellinfGEP were expressed at high levels in locally advanced head and neck squamous cell carcinoma.
-
•
PD-L1 and PD-L2 were moderately correlated, and each was less correlated with TcellinfGEP.
-
•
High TcellinfGEP status was independently associated with longer overall survival and recurrence-free survival.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the seventh most common malignancy worldwide, with >740 000 cases of lip/oral cavity, oropharynx, hypopharynx, and larynx cancers diagnosed annually, and a corresponding 365 000 deaths.1,2 Although alcohol and tobacco consumption are the main risk factors, human papillomavirus (HPV) accounts for ∼25% of HNSCC, mostly in the oropharynx.3,4 Approximately 60% of patients with HNSCC are diagnosed with locoregionally advanced (LA) disease,5 and ≥50% will develop recurrence or distant metastasis.3 Historically, LA HNSCC was managed through a multidisciplinary approach, including combinations of radiation, surgery, and chemotherapy or cetuximab.3
Recent therapeutics have targeted the interaction between programmed cell death protein 1 (PD-1) receptor and its ligands, programmed death-ligand 1 (PD-L1) and PD-L2, a key signaling pathway wherein cancer cells evade immune surveillance through inhibition of T-cell activation, cytokine production, and T-cell apoptosis.6,7 PD-1 inhibitors have shown efficacy in combination with chemoradiation in studies of LA HNSCC.8 PD-1 inhibitors pembrolizumab and nivolumab are approved for the treatment of recurrent or metastatic (R/M) HNSCC,9,10 with companion diagnostic PD-L1 IHC 22C3 pharmDx identifying patients with HNSCC eligible for treatment with pembrolizumab.11
Aside from PD-L1, other features of the tumor-immune interface have been explored to better predict clinical benefit. PD-L2 expression was reported to have additional effects on response and survival to PD-1 inhibition in R/M HNSCC.12 Recent studies show PD-L1 and PD-L2 expression are upregulated by interferon-γ (IFN-γ), and the T-cell-inflamed gene expression profile (TcellinfGEP), comprising IFN-γ-responsive genes related to antigen presentation, chemokine expression, cytotoxic activity, and adaptive immune resistance, was predictive of clinical response to PD-1 blockade in multiple cancer subtypes.13
Real-world data characterizing and evaluating, in an integrative manner, the relationship of these biomarkers with clinical outcomes under conventional therapies, however, is scant in LA HNSCC. Prior studies of singular analytes in LA HNSCC have yielded conflicting results, with some showing an association between PD-L1 or PD-L2 expression and shortened14 or prolonged survival,15 whereas others showed no clinical impact.16,17 Discrepant results may be attributed to intertumoral/intratumoral heterogeneity and differences in immunohistochemical detection methods or scoring.18 Evidence also suggests that PD-L1 and PD-L2 scoring in both tumor and immune-infiltrating cells is more clinically relevant than scoring in tumor cells alone.12,19
This study is intended to characterize the immune biomarker profile of the tumor microenvironment, including expression of PD-L1 and PD-L2 in tumor and immune cells and TcellinfGEP signature, and the association of these biomarkers with outcomes in Korean patients receiving standard-of-care primary treatment of LA HNSCC.
Materials and methods
Study design and patient population
Adults aged ≥18 years at diagnosis with histologically confirmed American Joint Committee on Cancer (AJCC) seventh edition stage III-IVB20 squamous cell carcinoma of the oral cavity, oropharynx, hypopharynx, or larynx between 2000 and 2015 and who received primary curative treatment were retrospectively identified from Samsung Medical Center and Severance Hospital of the Yonsei University Health System. Patients with sufficient and evaluable tissue for PD-L1, PD-L2, and TcellinfGEP assessment were included. Patients with a history of other primary cancers within 5 years of HNSCC diagnosis were excluded, except for those with basal cell carcinoma of the skin, superficial bladder cancer, squamous cell carcinoma of the skin, and in situ cervical cancer.
The study was conducted in accordance with Good Clinical and Pharmacoepidemiology Practice according to a protocol approved by the ethics committees at both institutions. Given this analysis was retrospective, informed consent was not required.
Biomarker assessment
Immunohistochemistry (IHC) was carried out at a single central laboratory or sponsor-approved laboratory vendor per assay, on 4-μm thick, formalin-fixed paraffin-embedded tumor tissue to determine PD-L1 and PD-L2 expression, using PD-L1 IHC 22C3 pharmDx (Agilent Technologies, Carpinteria, CA) and PD-L2 clone MEB123.3G2.038.12 PD-L1 expression levels were presented as combined positive score (CPS), defined as the number of PD-L1-positive cells (tumor cells, lymphocytes, and macrophages) divided by the total number of tumor cells × 100. CPS ≥1 was considered PD-L1-positive. PD-L2 expression levels were reported as the proportion of PD-L2-positive cells (tumor and immune cells) out of all cells. A proportion score ≥1 was considered PD-L2-positive. All samples were considered adequate for PD-L1 and PD-L2 quantification and interpretation if at least 100 viable tumor cells were present on non-decalcified tissue.21,22
A TcellinfGEP signature comprising 18 genes indicative of a T-cell-activated tumor microenvironment was previously derived across multiple solid tumors and was associated with response to pembrolizumab.23,24 Tumor RNA, extracted from formalin-fixed paraffin-embedded slides, was analyzed on the NanoString nCounter system (Seattle, WA) using a research version of the TcellinfGEP assay. TcellinfGEP scores were calculated as a weighted sum of normalized expression values for the 18 genes.13 TcellinfGEP status was defined as low (<−0.318), intermediate (−0.318 to <−0.162), or high (≥0.162).13
HPV status was determined using p16 IHC clone E6H4 (Roche, Basel, Switzerland). HPV oncogene expression was assessed using a >70% cut point for tumor cells stained for p16 IHC.25
Outcomes and statistical analysis
The primary endpoints were overall survival (OS), defined as the time from primary treatment initiation to date of death from any cause, and recurrence-free survival (RFS), calculated as the time from primary treatment initiation to date of locoregional recurrence, distant metastasis, or death from any cause, whichever came first. Data were censored when date of death was unknown at last follow-up.
Differences in patient and clinicopathologic characteristics between biomarker categories were evaluated using chi-square (χ2) statistics. Statistical significance was based on P < 0.05. Correlations between biomarkers were evaluated using the Spearman correlation coefficient. PD-L1 and PD-L2 scores were square-root transformed to minimize the outsized influence of large values.
The relationship between survival endpoints and biomarkers was analyzed using the Kaplan–Meier method with log-rank test. Crude and adjusted hazard ratios (aHRs) with 95% confidence intervals (CIs) were generated using multivariable Cox proportional hazards models. Model parsimony was established through stepwise variable selection, considering various demographic, clinicopathologic, tumor and treatment characteristics, and clinical site (Table 1).
Table 1.
Baseline characteristics by clinic site
| Characteristics | Total (N = 366) |
Samsung hospital (n = 193) |
Severance hospital (n = 173) |
2 test P valuec |
|---|---|---|---|---|
| n (%) | n (%) | n (%) | ||
| Age at LA HNSCC diagnosis, years | ||||
| Median | 59 | 60 | 58 | |
| ≤65 | 273 (74.6) | 140 (72.5) | 133 (76.9) | 0.3410 |
| >65 | 93 (25.4) | 53 (27.5) | 40 (23.1) | |
| Sex | ||||
| Female | 62 (16.9) | 32 (27.5) | 30 (17.3) | 0.8464 |
| Male | 304 (83.1) | 161 (72.5) | 143 (82.7) | |
| Smoking status (All tobacco)a | ||||
| Current | 131 (35.8) | 61 (34.5) | 70 (44.6) | 0.1129 |
| Former | 93 (25.4) | 50 (28.2) | 43 (27.4) | |
| Never | 110 (30.1) | 66 (37.3) | 44 (28.0) | |
| Unknown | 32 (8.7) | 16 | 16 | |
| Smoking volume | ||||
| Never smokers | 110 (30.1) | 66 (34.2) | 44 (25.4) | 0.2280 |
| <10 pack-years | 13 (3.6) | 5 (2.9) | 8 (5.6) | |
| ≥10 pack-years | 190 (51.9) | 100 (58.5) | 90 (63.4) | |
| Unknown (among smokers) | 21 (5.7) | 6 | 15 | |
| HPV status | ||||
| Negative | 221 (60.4) | 125 (65.6) | 96 (55.8) | 0.0605 |
| Positive | 142 (38.8) | 66 (34.4) | 76 (44.2) | |
| Unknown | 3 (0.8) | 2 | 1 | |
| Primary HNSCC site | ||||
| Hypopharynx | 40 (10.9) | 14 (7.3) | 26 (15.0) | <0.0001 |
| Larynx | 72 (19.7) | 56 (29.0) | 16 (9.3) | |
| Lip and oral cavity | 88 (24.0) | 41 (21.2) | 47 (27.2) | |
| Oropharynx | 132 (36.1) | 49 (25.4) | 83 (48.0) | |
| Other ill-defined sites in lip, oral cavity, and pharynx | 34 (9.3) | 33 (17.1) | 1 (0.6) | |
| Stage at diagnosisb | ||||
| III | 90 (24.6) | 64 (33.2) | 26 (15.0) | 0.0001 |
| IV A/B | 276 (75.4) | 129 (66.8) | 147 (85.0) | |
| Histologic grade | ||||
| Well differentiated | 87 (23.8) | 53 (31.7) | 34 (20.9) | 0.0700 |
| Moderately differentiated | 185 (50.5) | 85 (50.9) | 100 (61.3) | |
| Poorly differentiated | 58 (15.8) | 29 (17.4) | 29 (17.8) | |
| Unknown | 36 (9.8) | 26 | 10 | |
| ECOG status | ||||
| 0 | 116 (31.7) | 86 (44.6) | 30 (30.0) | 0.0351 |
| 1 | 160 (43.7) | 95 (49.2) | 65 (65.0) | |
| 2+ | 17 (4.6) | 12 (6.2) | 5 (5.0) | |
| Unknown | 73 (19.9) | 0 | 73 | |
| Primary treatment (planned) | ||||
| Definitive CCRT | 19 (5.2) | 8 (4.1) | 11 (6.4) | <0.0001 |
| Surgery + CCRT | 127 (34.7) | 30 (15.5) | 97 (56.1) | |
| Surgery + RT | 191 (52.2) | 132 (68.4) | 59 (34.1) | |
| RT or surgery alone | 29 (7.9) | 23 (11.9) | 6 (3.5) | |
| Tissue sample collected before chemotherapy | ||||
| No | 15 (4.1) | 1 (0.5) | 14 (8.1) | 0.0003 |
| Yes | 351 (95.9) | 192 (99.5) | 159 (91.9) | |
| Neoadjuvant chemotherapy | ||||
| No | 350 (95.6) | 193 (100.0) | 157 (90.8) | <0.0001 |
| Yes | 16 (4.4) | 0 (0.0) | 16 (9.2) |
AJCC, American Joint Committee on Cancer; CCRT, concurrent chemoradiation; ECOG, Eastern Cooperative Oncology Group; HNSCC, head and neck squamous cell carcinoma; HPV, human papillomavirus; LA, locoregionally advanced; RT, radiotherapy.
Former smoker is one who quit >1 year before diagnosis; current smoker is one who currently smokes or quit <1 year before diagnosis.
According to AJCC seventh edition criteria.20
P value is to test the difference in clinicopathologic characteristics between clinical sites; the unknown category was excluded from this calculation.
Results
Patient characteristics
Of 462 patients identified from Samsung Hospital and Severance Hospital with a diagnosis of HNSCC from 2000 to 2015, 96 (20.8%) were excluded or not assessable (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2024.103961). Of the 366 patients with samples evaluated for ≥1 biomarker (193 from Samsung Hospital; 173 from Severance Hospital), 365, 364, and 302 had a PD-L1, PD-L2, and/or TcellinfGEP score, respectively.
Median age at diagnosis was 59 years (range, 23-86 years), 83.1% were male, 61.2% were current or former smokers (All tobacco), and 38.8% had HPV-positive tumors (Table 1). Most tumors were stage IVA/B (75.4%), and most arose in the oropharynx (36.1%) and lip/oral cavity (24.0%). Compared with Severance Hospital, Samsung Medical Center had proportionately more cancers of the larynx (29.0% versus 9.2%) and other ill-defined sites (17.1% versus 0.6%), but fewer oropharyngeal cancers (25.4% versus 48.0%).
Planned primary treatment most often included postsurgical radiation (52.2%) and concurrent chemoradiation (CCRT), either as a definitive therapy (5.2%) or following surgery (34.7%). Postsurgical chemoradiation was more prevalent (56.1% versus 15.5%, respectively) and postsurgical radiation less prevalent (34.1% versus 68.4%, respectively) at Severance Hospital than at Samsung Medical Center. Few patients (4.1%) had samples obtained following chemotherapy treatment.
Biomarker expression
Most patients expressed PD-L1 (83.6%), PD-L2 (85.4%), and high TcellinfGEP status (73.2%; Table 2). Compared with HPV-negative patients, HPV-positive patients had higher proportions of PD-L1 positivity (88.7% versus 80.0%, respectively) and high TcellinfGEP status (85.2% versus 65.5%, respectively). PD-L1, PD-L2, and high TcellinfGEP expression were found in greater proportions in oral cavity and oropharyngeal cancers (range, 72.2%-92.0%) than in laryngeal or hypopharyngeal cancers (range, 54.9%-77.5%). Likewise, overexpression of each biomarker was higher in patients with surgery followed by adjuvant radiation, with or without chemotherapy (range, 73.9%-94.5%), than in those with other treatments (range, 35.7%-75.9%). PD-L2 positivity was slightly higher in patients treated at Samsung Medical Center (89.1%) compared with Severance Hospital (81.3%; Table 2).
Table 2.
Baseline characteristics by biomarker status
| Characteristics | PD-L1 expressionc N = 365 |
2 test P valuef |
PD-L2 expressiond N = 364 |
2 test P valuef |
TcellinfGEP expressione N = 302 |
2 test P valuef |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Negative |
Positive |
Negative |
Positive |
Low |
Intermediate |
High |
||||
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | ||||
| Overall | 60 (16.4) | 305 (83.6) | NA | 53 (14.6) | 311 (85.4) | NA | 49 (16.2) | 32 (10.6) | 221 (73.2) | NA |
| Age, years | ||||||||||
| ≤65 | 44 (16.1) | 229 (83.9) | 0.7755 | 35 (12.9) | 237 (87.1) | 0.1154 | 33 (14.7) | 31 (13.8) | 160 (71.4) | 0.0062 |
| >65 | 16 (17.4) | 76 (82.6) | 18 (19.6) | 74 (80.4) | 16 (20.5) | 1 (1.3) | 61 (78.2) | |||
| Sex | ||||||||||
| Male | 49 (16.2) | 254 (83.8) | 0.7612 | 44 (14.6) | 258 (85.4) | 0.9913 | 43 (17.1) | 23 (9.2) | 185 (73.7) | 0.1582 |
| Female | 11 (17.7) | 51 (82.3) | 9 (14.5) | 53 (85.5) | 6 (11.8) | 9 (17.6) | 36 (70.6) | |||
| HPV status | ||||||||||
| Negative | 44 (20.0) | 176 (80.0) | 0.0291 | 35 (15.9) | 185 (84.1) | 0.2247 | 35 (19.8) | 26 (14.7) | 116 (65.5) | 0.0005 |
| Positive | 16 (11.3) | 126 (88.7) | 16 (11.3) | 125 (88.7) | 13 (10.7) | 5 (4.1) | 104 (85.2) | |||
| Unknown | 0 | 3 | 2 | 1 | 1 | 1 | 1 | |||
| Smoking status (All tobacco) | ||||||||||
| Current | 22 (16.8) | 109 (83.2) | 0.9285 | 18 (13.7) | 113 (86.3) | 0.3481 | 19 (17.9) | 10 (9.4) | 77 (72.6) | 0.0203 |
| Former | 16 (17.4) | 76 (82.6) | 18 (19.6) | 74 (80.4) | 20 (26.3) | 10 (13.2) | 46 (60.5) | |||
| Never | 17 (15.5) | 93 (84.5) | 14 (12.7) | 96 (87.3) | 9 (9.1) | 8 (8.1) | 82 (82.8) | |||
| Unknown | 5 | 27 | 3 | 28 | 1 | 4 | 16 | |||
| Smoking volumea | ||||||||||
| Never | 17 (15.5) | 93 (84.5) | 14 (12.7) | 96 (87.3) | 9 (9.1) | 8 (8.1) | 82 (82.8) | |||
| <10 pack-years | 5 (38.5) | 8 (61.5) | 0.0979 | 1 (7.7) | 12 (92.3) | 0.5263 | 3 (27.3) | 1 (9.1) | 7 (63.6) | 0.0689 |
| ≥10 pack-years | 30 (15.9) | 159 (84.1) | 31 (16.4) | 158 (83.6) | 34 (22.2) | 15 (9.8) | 104 (68.0) | |||
| Unknown (among smokers) | 3 | 18 | 4 | 17 | 2 | 4 | 12 | |||
| Primary HNSCC location | ||||||||||
| Hypopharynx | 12 (30.8) | 27 (69.2) | 0.0001 | 11 (27.5) | 29 (72.5) | 0.0084 | 7 (22.6) | 5 (16.1) | 19 (61.3) | 0.0012 |
| Larynx | 22 (30.6) | 50 (69.4) | 16 (22.5) | 55 (77.5) | 13 (25.5) | 10 (19.6) | 28 (54.9) | |||
| Lip and oral cavity | 12 (13.6) | 76 (86.4) | 7 (8.0) | 81 (92.0) | 11 (13.9) | 11 (13.9) | 57 (72.2) | |||
| Oropharynx | 11 (8.3) | 121 (91.7) | 14 (10.7) | 117 (89.3) | 13 (10.9) | 3 (2.5) | 103 (86.6) | |||
| Other and ill-defined sites in lip, oral cavity and pharynx | 3 (8.8) | 31 (91.2) | 5 (14.7) | 29 (85.3) | 5 (22.7) | 3 (13.6) | 14 (63.6) | |||
| Histologic grade | ||||||||||
| Well differentiated | 17 (19.5) | 70 (80.5) | 0.1980 | 9 (10.3) | 78 (89.7) | 0.2493 | 15 (20.8) | 8 (11.1) | 49 (68.1) | 0.6325 |
| Moderately differentiated | 31 (16.8) | 153 (83.2) | 32 (17.5) | 151 (82.5) | 23 (15.2) | 17 (11.3) | 111 (73.5) | |||
| Poorly differentiated | 5 (8.6) | 53 (91.4) | 7 (12.1) | 51 (87.9) | 6 (12.0) | 4 (8.0) | 40 (80.0) | |||
| Unknown | 7 | 29 | 5 | 31 | 5 | 3 | 21 | |||
| Stage at diagnosisb | ||||||||||
| III | 13 (14.6) | 76 (85.4) | 0.5919 | 12 (13.5) | 77 (86.5) | 0.7403 | 11 (14.9) | 13 (17.6) | 50 (67.6) | 0.0809 |
| IV A/B | 47 (17.0) | 229 (83.0) | 41 (14.9) | 234 (85.1) | 38 (16.7) | 19 (8.3) | 171 (75.0) | |||
| ECOG status | ||||||||||
| 0 | 13 (11.2) | 103 (88.8) | 0.2152 | 10 (8.6) | 106 (91.4) | 0.1508 | 14 (15.2) | 9 (9.8) | 69 (75.0) | 0.9801 |
| 1 | 28 (17.5) | 132 (82.5) | 22 (13.8) | 137 (86.2) | 21 (16.3) | 11 (8.5) | 97 (75.2) | |||
| 2+ | 1 (6.3) | 15 (93.8) | 4 (23.5) | 13 (76.5) | 2 (12.5) | 2 (12.5) | 12 (75.0) | |||
| Unknown | 18 | 55 | 17 | 55 | 12 | 10 | 43 | |||
| Primary treatment regimen for locally advanced HNSCC | ||||||||||
| Definitive CCRT | 11 (57.9) | 8 (42.1) | <0.0001 | 9 (47.4) | 10 (52.6) | 0.0001 | 6 (42.9) | 3 (21.4) | 5 (35.7) | 0.0070 |
| Surgery + CCRT | 7 (5.5) | 120 (94.5) | 18 (14.3) | 108 (85.7) | 16 (13.9) | 7 (6.1) | 92 (80.0) | |||
| Surgery + RT | 34 (17.8) | 157 (82.2) | 19 (10.0) | 171 (90.0) | 22 (14.4) | 18 (11.8) | 113 (73.9) | |||
| RT alone or surgery alone | 8 (28.6) | 20 (71.4) | 7 (24.1) | 22 (75.9) | 5 (25.0) | 4 (20.0) | 11 (55.0) | |||
| Neoadjuvant therapy | ||||||||||
| No | 57 (16.3) | 292 (83.7) | 0.7986 | 47 (13.5) | 301 (86.5) | 0.0078 | 45 (15.7) | 31 (10.8) | 211 (73.5) | 0.5011 |
| Yes | 3 (18.8) | 13 (81.3) | 6 (37.5) | 10 (62.5) | 4 (26.7) | 1 (6.7) | 10 (66.7) | |||
| Tissue sample collected before chemotherapy | ||||||||||
| No | 4 (26.7) | 11 (73.3) | 0.2750 | 5 (33.3) | 10 (66.7) | 0.0353 | 5 (38.5) | 2 (15.4) | 6 (46.2) | 0.0553 |
| Yes | 56 (16.0) | 294 (84.0) | 48 (13.8) | 301 (86.2) | 44 (15.2) | 30 (10.4) | 215 (74.4) | |||
| Clinic site | ||||||||||
| Samsung Hospital | 32 (16.6) | 161 (83.4) | 0.9382 | 21 (10.9) | 172 (89.1) | 0.0345 | 24 (16.6) | 13 (9.0) | 108 (74.5) | 0.6761 |
| Severance Hospital | 28 (16.3) | 144 (83.7) | 32 (18.7) | 139 (81.3) | 25 (15.9) | 19 (12.1) | 113 (72.0) | |||
AJCC, American Joint Committee on Cancer; CCRT, concurrent chemoradiation; CPS, combined positive score; ECOG, Eastern Cooperative Oncology Group; HNSCC, head and neck squamous cell carcinoma; HPV, human papillomavirus; NA, not applicable; PD-L1, programmed death-ligand 1; PD-L2, programmed death-ligand 2; RT, radiotherapy; TcellinfGEP, T-cell-inflamed gene expression profile.
Former smoker is one who quit >1 year before diagnosis; current smoker is one who currently smokes or quit <1 year before diagnosis.
According to AJCC seventh edition criteria.20
PD-L1 status: positive (CPS ≥1) and negative (CPS <1).
PD-L2 status: positive (proportion score ≥1) and negative (proportion score <1).
TcellinfGEP status: low (score of <−0.318), intermediate (score −0.318 to <−0.162), or high (score ≤−0.162).
P value is based on comparison between subgroups of patient/tumor characteristics within the total population; the unknown category was excluded from this calculation.
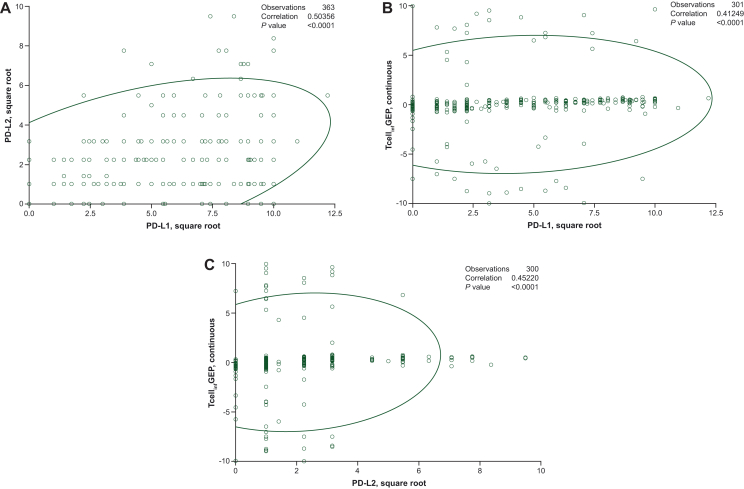
PD-L1 and PD-L2 were moderately correlated with each other (rSpearman = 0.50) but were slightly less correlated with TcellinfGEP (rSpearman = 0.41 for PD-L1, rSpearman = 0.45 for PD-L2; Figure 1A-C).
Figure 1.
Correlation between (A) PD-L1 and PD-L2, (B) PD-L1 and TcellinfGEP, and (C) PD-L2 and TcellinfGEP scores. Both PD-L1 and PD-L2 scores have been square-root transformed to minimize outsized influence of large values. PD-L1, programmed death-ligand 1; PD-L2, programmed death-ligand 2; TcellinfGEP, T-cell- inflamed gene expression profile.
Survival outcomes
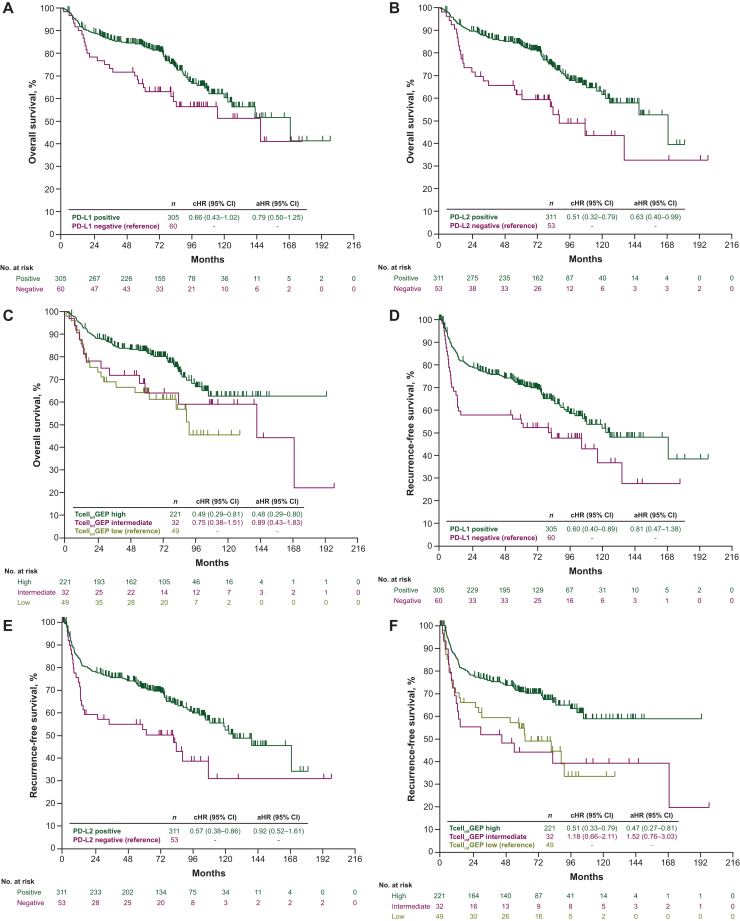
Univariate analyses showed no significant survival with PD-L1 expression (aHR, 0.79; 95% CI, 0.50-1.25) (Figure 2A). OS was significantly longer in patients with high TcellinfGEP status and PD-L2 overexpression versus low or non-expressers (P ≤ 0.01; Figure 2B and C). Similarly, across all three biomarkers, longer RFS was seen in biomarker-high/biomarker-positive patients versus their biomarker-low/biomarker-negative counterparts (P ≤ 0.01; Figure 2D-F).
Figure 2.
Kaplan–Meier curves of (A-C) overall survival and (D-F) recurrence-free survival, by singular biomarker status. Biomarkers were assessed singularly in separate models. aHR, adjusted hazard ratio; cHR, crude hazard ratio; CI, confidence interval; PD-L1, programmed death-ligand 1; PD-L2, programmed death-ligand 2; RFS, recurrence-free survival; TcellinfGEP, T-cell-inflamed gene expression profile.
In multivariable models of singular biomarkers, OS was significantly longer for PD-L2-positive versus PD-L2-negative tumors (aHR, 0.63; 95% CI 0.40-0.99), and for high TcellinfGEP status versus low TcellinfGEP status (aHR, 0.48; 95% CI 0.29-0.80) (Figure 2B and C). When all three biomarkers were simultaneously adjusted along with other explanatory covariates within a single parsimonious model, only high TcellinfGEP status remained significantly and independently associated with longer OS (aHR, 0.57; 95% CI 0.33-0.98; P < 0.05; Table 3).
Table 3.
Multivariable model with simultaneous control of PD-L1, PD-L2, and TcellinfGEP
| aHR (95% CI) for OSa | P value | aHR (95% CI) for RFSb | P value | |
|---|---|---|---|---|
| PD-L1 negative (reference) | — | — | ||
| PD-L1 positive | 0.69 (0.40-1.20) | 0.193 | 0.75 (0.37-1.54) | 0.434 |
| PD-L2 negative (reference) | — | — | ||
| PD-L2 positive | 0.74 (0.43-1.29) | 0.290 | 1.21 (0.61-2.42) | 0.581 |
| TcellinfGEP-low (reference) | — | — | ||
| TcellinfGEP-intermediate | 0.83 (0.40-1.76) | 0.634 | 1.36 (0.66-2.80) | 0.400 |
| TcellinfGEP-high | 0.57 (0.33-0.98) | 0.043 | 0.41 (0.23-0.74) | 0.003 |
aHR, adjusted hazard ratio; CI, confidence interval; ECOG, Eastern Cooperative Oncology Group; OS, overall survival; PD-L1, programmed death-ligand 1; PD-L2, programmed death-ligand 2; RFS, recurrence-free survival; TcellinfGEP, T-cell-inflamed gene expression profile.
Parsimonious model adjusted for all three biomarkers plus age and clinical site.
Parsimonious model adjusted for all three biomarkers plus ECOG, planned treatment, and clinical site.
A significant positive association was observed in multivariate analysis between RFS and high TcellinfGEP status (aHR, 0.47; 95% CI 0.27-0.81); however, no associations were observed for PD-L1 (aHR, 0.81; 95% CI 0.47-1.38) or PD-L2 expression (aHR, 0.92; 95% CI, 0.52-1.61) (Figure 2D-F). In a model that included all three biomarkers and other covariates, only high TcellinfGEP status remained significantly associated with RFS (aHR, 0.41; 95% CI 0.23-0.74; P < 0.01; Table 3).
Discussion
In this retrospective study of Korean patients with LA HNSCC, PD-L1 was common (83.6%) and similar to that reported (84%-90%) in observational studies of patients with varying stages of HNSCC and using the same 22c3 antibody clone and CPS >1 cut point.26, 27, 28 Studies using other PD-L1 antibody clones but similar 1% positivity thresholds in tumor and immune cells in predominantly (≥65%) LA HNSCC have reported varying prevalence of PD-L1 overexpression: 40% with clone EH33,29 71% with E1L3N,30 and 90% with SP142.31
PD-L2 expression was also prevalent (85.4%) and moderately correlated with PD-L1 expression (rSpearman = 0.5). This is comparable to the >90% prevalence and significant correlations reported in a study of archival HNSCC samples using the same PD-L2 antibody clone and 1% cut point in tumor and immune cells12 and a study of LA HNSCC using EPR1161 clones at a 1% cut point in tumor cells.14 In contrast, Kogashiwa et al.32 reported no correlation between PD-L1 and PD-L2 in LA oral cavity cancer; however, any relationship may be challenging to detect given the low PD-L2 prevalence (23.8%) in their study.
High TcellinfGEP status was found in 73.2% of patients, with a modest correlation between TcellinfGEP and both PD-L1 and PD-L2 (rSpearman = 0.41 and 0.45, respectively). The KEYNOTE-012 trial of pembrolizumab in patients with R/M HNSCC used the same TcellinfGEP signature of IFN-γ-responsive genes and also reported a high TcellinfGEP prevalence of 71% and a significant correlation with PD-L1 (rSpearman = 0.51).33 This correlation likely reflects the induction of PD-L1 and PD-L2 expression by IFN-γ, secreted from CD8+ T cells as part of the intratumoral T-cell adaptive immune response,13 rather than induction from an innate, oncogene-driven mechanism, such as activation of the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) pathways.34
Overexpression of all three biomarkers was consistently higher in oral cavity/oropharyngeal cancers versus other HNSCC sites. Anatomic site-based differential in CPS >1 has been reported in other studies and may be a function of a relatively rich inflammatory environment in the tongue and oropharynx.27,35,36 Virus-induced chronic immune cell infiltration in HPV-associated oropharyngeal cancer may also explain why HPV-positive patients had greater levels of PD-L1 and TcellinfGEP in our study.37,38
Biomarker-positive prevalence was also consistently higher in patients treated with postsurgical radiation, with or without chemotherapy, versus other treatment regimens. Cancer treatment could artificially modulate biomarker expression levels39; however, few patients (4.1%) had samples obtained following systemic treatment in our study. Higher expression levels in surgically treated patients may be explained by an underestimation of CPS (and other immune markers) in smaller biopsy samples versus resection specimens, which are more likely to harbor the tumor-invasive front, where immune activity is greatest.26,40
Our results did not show PD-L1 to be associated with clinical outcome. Prior observational studies evaluating 22C3 clone at CPS >1, including one study in Asia, have generally shown no association with survival endpoints in HNSCC with mixed stages of disease.27,36 Another study, however, reported varying results of association with PD-L1 based on the start of OS follow-up; no association was found when OS was defined from time of diagnosis, and a positive association was observed when OS was defined from time of tissue acquisition.26 Two additional studies of patients in Asia, primarily with LA HNSCC, had included immune cells in the PD-L1 scoring methods, using clone SP142 at a 5% positivity threshold. Kim et al.41 reported PD-L1 in immune cells (but not tumor cells) was positively prognostic of OS in surgically treated patients, whereas Ngamphaiboon et al.31 showed no association between PD-L1 (as a composite of tumor- and immune-cell staining) and OS in patients who received definitive treatment.
PD-L2 was not independently associated with survival after adjustment for TcellinfGEP and PD-L1. Prior HNSCC studies using a variety of PD-L2 antibody clones, positivity thresholds, and cell types for scoring reported a negative42 or no association with survival.14,32 Inconsistent results across PD-L1 and PD-L2 studies could be attributed to tumor spatiotemporal heterogeneity [e.g. sample size (biopsy versus resection), tissue origin (primary versus metastatic), or staining pattern (membranous versus cytoplasmic)] or methodologic differences [e.g. variability in the antibodies, positivity cut-offs, or scoring criteria (tumor ± immune cells)].18,26
After adjusting for the other biomarkers, only high TcellinfGEP status was independently associated with longer OS and RFS. This suggests the association of these immune biomarkers of the PD-1/L-1 axis with survival is mainly driven by high TcellinfGEP expression. This relationship may reflect a functionally active T-cell antitumor response, as represented by an elevated IFN-γ signature.35 HNSCC is relatively rich in immune infiltrates; such tumors are likely to have more effective immune surveillance, and therefore, a better prognosis.32
Key strengths of this study include the large sample size, detailed clinical information, and use of a single laboratory to mitigate assay variability. Limitations include the retrospective design, treatment regimen heterogeneity, and inconsistency in patient follow-up schedules, which could impact the timing of outcome assessment. Selection bias may have occurred, given patients were included based on tissue sample availability. We did not account for time-varying changes to therapy following the index primary treatment. Further, HPV DNA is not routinely collected in the clinical setting and therefore no additional HPV analyses were carried out outside of p16 IHC testing. During our study, only two patients received PD-(L)1 inhibitors after the index treatment, which could influence survival, albeit minimally. Additionally, although Korean patients are not representative of other races/ethnicities, no data suggest PD-L1 expression in HNSCC varies by race or ethnicity.
Conclusions
In this retrospective analysis, PD-L1, PD-L2, and TcellinfGEP expression were moderately correlated; however, only high TcellinfGEP status was independently associated with OS and RFS. Although additional studies are needed to confirm these associations, these results could provide a deeper understanding of the relationship between biomarkers of the tumor microenvironment and treatment response in HNSCC. Future studies may benefit from broadening the profile of immune markers under evaluation and replicating analyses across different racial/ethnic populations.
Disclosure
TV, HZ, DC, ALW, BG are employees of Merck Sharp & Dohme (MSD) LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, and hold stock in Merck & Co., Inc., Rahway, NJ. BCC reports funding from MOGAM Institute, LG Chem, Oscotec, Interpark Bio Convergence Corp., GI Innovation, GI-Cell, Abion, AbbVie, AstraZeneca, Bayer, Blueprint Medicines, Boehringer Ingelheim, Champions Oncology, CJ Bioscience, CJ Blossom Park, Cyrus, Dizal Pharma, Genexine, Janssen, Lilly, MSD, Novartis, Nuvalent, Oncternal, Ono, Regeneron, Dong-A ST, Bridge Biotherapeutics, Yuhan, ImmuneOncia, Illumina, Kanaph Therapeutics, Therapex, J INTS Bio, Hanmi, and CHA Bundang Medical Center; royalties from Champions Oncology, Crown Bioscience, and Imagen; has served as a consultant for Abion, BeiGene, Novartis, AstraZeneca, Boehringer Ingelheim, Roche, BMS, CJ, CureLogen, Cyrus Therapeutics, Ono, Onegene Biotechnology, Yuhan, Pfizer, Eli Lilly, GI-Cell, Guardant, HK Inno-N, Imnewrun Biosciences Inc., Janssen, Takeda, MSD, Janssen, MedPacto, Blueprint Medicines, RandBio, and Hanmi; honoraria from ASCO, AstraZeneca, Guardant, Roche, ESMO, IASLC, Korean Cancer Association, Korean Society of Medical Oncology, Korean Society of Thyroid-Head and Neck Surgery, Korean Cancer Study Group, Novartis, MSD, The Chinese Thoracic Oncology Society, and Pfizer; participation on a scientific advisory board for KANAPH Therapeutic Inc., Bridge Biotherapeutics, Cyrus Therapeutics, Guardant Health, and Oscotec Inc.; participates on the Board of Directors for Interpark Bio Convergence Corp., and J INTS Bio; holds stock in TheraCanVac Inc., Gencurix Inc., Bridge Biotherapeutics, KANAPH Therapeutics Inc., Cyrus Therapeutics, Interpark Bio Convergence Corp., and J INTS Bio; is an employee of Yonsei University Health System; and is the founder of DAAN Biotherapeutics. All other authors have declared no conflicts of interest.
Acknowledgements
The authors thank the following individuals, who were employees of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, when the study was conducted: Shawna Calhoun (project management); Adriane Zernhelt, Nena DiMaano, Diane Levitan, and Jennifer Stouffer (biomarker/laboratory consultants); Bo Zheng (statistical programming); and Lingkang Huang (biostatistics) for contributions in the design, implementation, or interpretation of the study. Medical writing and editorial assistance were provided by Mallory Campbell, PhD, and Matthew Grzywacz, PhD, of ApotheCom (Yardley, PA). This assistance was funded by Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ.
Funding
This work was supported by Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ (MSD) (no grant number). The funders contributed to the study design, data collection, data analysis, and data interpretation in collaboration with the authors; all authors had full access to the data. Investigators and site personnel collected data, which were housed in an MSD database. The corresponding authors had full access to all the data and had final responsibility for the decision to submit for publication.
Data sharing
Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ (MSD), is committed to providing qualified scientific researchers access to anonymized data and clinical study reports from the company’s clinical trials for the purpose of conducting legitimate scientific research. MSD is also obligated to protect the rights and privacy of trial participants and, as such, has a procedure in place for evaluating and fulfilling requests for sharing company clinical trial data with qualified external scientific researchers. The MSD data sharing website (available at: http://engagezone.msd.com/ds_documentation.php) outlines the process and requirements for submitting a data request. Applications will be promptly assessed for completeness and policy compliance. Feasible requests will be reviewed by a committee of MSD subject matter experts to assess the scientific validity of the request and the qualifications of the requestors. In line with data privacy legislation, submitters of approved requests must enter into a standard data-sharing agreement with MSD before data access is granted. Data will be made available for request after product approval in the USA and European Union or after product development is discontinued. There are circumstances that may prevent MSD from sharing requested data, including country- or region-specific regulations. If the request is declined, it will be communicated to the investigator. Access to genetic or exploratory biomarker data requires a detailed, hypothesis-driven statistical analysis plan that is collaboratively developed by the requestor and MSD subject matter experts; after approval of the statistical analysis plan and execution of a data-sharing agreement, MSD will either perform the proposed analyses and share the results with the requestor or will construct biomarker covariates and add them to a file with clinical data that is uploaded to an analysis portal so that the requestor can perform the proposed analyses.
Contributor Information
B.C. Cho, Email: CBC1971@yuhs.ac.
M.-J. Ahn, Email: silkahn@skku.edu, silk.ahn@samsung.com.
Supplementary data
References
- 1.International Agency for Research on Cancer (IARC) Estimated number of new cases in 2020, worldwide, both sexes, all ages (excl. NMSC). IARC. https://gco.iarc.fr/today/online-analysis-table?v=2020&mode=cancer&mode_population=continents&population=900&populations=900&key=asr&sex=0&cancer=39&type=0&statistic=5&prevalence=0&population_group=0&ages_group%5B%5D=0&ages_group%5B%5D=17&group_cancer=1&i Available at.
- 2.International Agency for Research on Cancer (IARC) Estimated number of deaths in 2020, World, both sexes, all ages (excl. NMSC). World Health Organization. https://gco.iarc.fr/today/online-analysis-table?v=2020&mode=cancer&mode_population=regions&population=900&populations=900&key=asr&sex=0&cancer=39&type=1&statistic=5&prevalence=0&population_group=0&ages_group%5B%5D=0&ages_group%5B%5D=17&group_cancer=1&incl Available at.
- 3.Argiris A., Karamouzis M.V., Raben D., Ferris R.L. Head and neck cancer. Lancet. 2008;371(9625):1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillison M.L., Koch W.M., Capone R.B., et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92(9):709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y.-G., Kang E.J., Keam B., et al. Treatment strategy and outcomes in locally advanced head and neck squamous cell carcinoma: a nationwide retrospective cohort study (KCSG HN13–01) BMC Cancer. 2020;20(1):813. doi: 10.1186/s12885-020-07297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou W., Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol. 2008;8(6):467–477. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
- 7.Latchman Y., Wood C.R., Chernova T., et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2(3):261–268. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 8.Cohen E.E.W., Bell R.B., Bifulco C.B., et al. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC) J Immunother Cancer. 2019;7(1):184. doi: 10.1186/s40425-019-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bristol Myers Squibb Company . Bristol Myers Squibb Company; Princeton, NJ: 2022. OPDIVO (Nivolumab) Injection, for Intravenous Use. [Google Scholar]
- 10.Merck Sharp & Dohme LLC KEYTRUDA® (Pembrolizumab) Injection, for Intravenous Use. LLC: Rahway, NJ: Merck Sharp & Dohme. 2023 [Google Scholar]
- 11.Agilent Technologies I. PD-L1 IHC 22C3 pharmDx Agilent Technologies, Inc. https://www.accessdata.fda.gov/cdrh_docs/pdf15/P150013S016C.pdf Available at. Updated July 29, 2019.
- 12.Yearley J.H., Gibson C., Yu N., et al. PD-L2 expression in human tumors: relevance to anti-PD-1 therapy in cancer. Clin Cancer Res. 2017;23(12):3158–3167. doi: 10.1158/1078-0432.CCR-16-1761. [DOI] [PubMed] [Google Scholar]
- 13.Ayers M., Lunceford J., Nebozhyn M., et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller T., Braun M., Dietrich D., et al. PD-L1: a novel prognostic biomarker in head and neck squamous cell carcinoma. Oncotarget. 2017;8(32):52889–52900. doi: 10.18632/oncotarget.17547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balermpas P., Rödel F., Krause M., et al. The PD-1/PD-L1 axis and human papilloma virus in patients with head and neck cancer after adjuvant chemoradiotherapy: a multicentre study of the German Cancer Consortium Radiation Oncology Group (DKTK-ROG) Int J Cancer. 2017;141(3):594–603. doi: 10.1002/ijc.30770. [DOI] [PubMed] [Google Scholar]
- 16.Ock C.Y., Kim S., Keam B., et al. PD-L1 expression is associated with epithelial-mesenchymal transition in head and neck squamous cell carcinoma. Oncotarget. 2016;7(13):15901–15914. doi: 10.18632/oncotarget.7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doescher J., Minkenberg P., Laban S., et al. Immune checkpoint expression in HNSCC patients before and after definitive chemoradiotherapy. Head Neck. 2021;43(3):778–787. doi: 10.1002/hed.26534. [DOI] [PubMed] [Google Scholar]
- 18.Miranda-Galvis M., Rumayor Piña A., Sales de Sá R., et al. PD-L1 expression patterns in oral cancer as an integrated approach for further prognostic classification. Oral Dis. 2021;27(7):1699–1710. doi: 10.1111/odi.13714. [DOI] [PubMed] [Google Scholar]
- 19.Chow L.Q., Haddad R., Gupta S., et al. Antitumor activity of pembrolizumab in biomarker-unselected patients with recurrent and/or metastatic head and neck squamous cell carcinoma: results from the phase Ib KEYNOTE-012 expansion cohort. J Clin Oncol. 2016;34(32):3838–3845. doi: 10.1200/JCO.2016.68.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.American Joint Committee on Cancer . New York, Dordrecht, Heidelberg, London; Springer: 2010. AJCC Cancer Staging Manual. ISBN: 978-0-387-88440-0. [Google Scholar]
- 21.Naito T., Udagawa H., Sato J., et al. A minimum Of 100 tumor cells in a single biopsy sample is required to assess programmed cell death ligand 1 expression in predicting patient response to nivolumab treatment in nonsquamous non-small cell lung carcinoma. J Thorac Oncol. 2019;14(10):1818–1827. doi: 10.1016/j.jtho.2019.06.019. [DOI] [PubMed] [Google Scholar]
- 22.Strickland A.L., Blacketer S., Molberg K., Markantonis J., Lucas E. Effects of decalcifying agents of variable duration on PD-L1 immunohistochemistry. Am J Clin Pathol. 2020;153(2):258–265. doi: 10.1093/ajcp/aqz161. [DOI] [PubMed] [Google Scholar]
- 23.Danaher P., Warren S., Lu R., et al. Pan-cancer adaptive immune resistance as defined by the Tumor Inflammation Signature (TIS): results from The Cancer Genome Atlas (TCGA) J Immunother Cancer. 2018;6(1):63. doi: 10.1186/s40425-018-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallden B., Pekker I., Popa S., et al. Development and analytical performance of a molecular diagnostic for anti-PD1 response on the nCounter® Dx Analysis System. J Clin Oncol. 2016;34(15_suppl):3034. [Google Scholar]
- 25.Fakhry C., Lacchetti C., Rooper L.M., et al. Human papillomavirus testing in head and neck carcinomas: ASCO Clinical Practice Guideline Endorsement of the College of American Pathologists Guideline. J Clin Oncol. 2018;36(31):3152–3161. doi: 10.1200/JCO.18.00684. [DOI] [PubMed] [Google Scholar]
- 26.De Keukeleire S.J., Vermassen T., Deron P., et al. Concordance, correlation, and clinical impact of standardized PD-L1 and TIL scoring in SCCHN. Cancers (Basel) 2022;14(10):2431. doi: 10.3390/cancers14102431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wusiman D., Guo L., Huang Z., et al. The clinicopathological significance of PD-L1 expression assessed by the combined positive score (CPS) in head and neck squamous cell carcinoma. Pathol Res Pract. 2022;236:153934. doi: 10.1016/j.prp.2022.153934. [DOI] [PubMed] [Google Scholar]
- 28.Dewan A., Sharma S., Bhardwaj N., et al. Evaluation of PD-L1 immunohistochemical expression in the head and neck squamous cell carcinoma and its correlation with various clinicopathologic parameters: a study from a tertiary referral center in India. Lab Invest. 2021;101(suppl 1):767. [Google Scholar]
- 29.Sanchez-Canteli M., Granda-Díaz R., Del Rio-Ibisate N., et al. PD-L1 expression correlates with tumor-infiltrating lymphocytes and better prognosis in patients with HPV-negative head and neck squamous cell carcinomas. Cancer Immunol Immunother. 2020;69(10):2089–2100. doi: 10.1007/s00262-020-02604-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ou D., Adam J., Garberis I., et al. Clinical relevance of tumor infiltrating lymphocytes, PD-L1 expression and correlation with HPV/p16 in head and neck cancer treated with bio- or chemo-radiotherapy. Oncoimmunology. 2017;6(9):e1341030. doi: 10.1080/2162402X.2017.1341030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ngamphaiboon N., Chureemas T., Siripoon T., et al. Characteristics and impact of programmed death-ligand 1 expression, CD8+ tumor-infiltrating lymphocytes, and p16 status in head and neck squamous cell carcinoma. Med Oncol. 2019;36(2):21. doi: 10.1007/s12032-018-1241-1. [DOI] [PubMed] [Google Scholar]
- 32.Kogashiwa Y., Yasuda M., Sakurai H., et al. PD-L1 expression confers better prognosis in locally advanced oral squamous cell carcinoma. Anticancer Res. 2017;37(3):1417–1424. doi: 10.21873/anticanres.11465. [DOI] [PubMed] [Google Scholar]
- 33.Haddad R., Seiwert T.Y., Chow L.Q.M., et al. Influence of tumor mutational burden, inflammatory gene expression profile, and PD-L1 expression on response to pembrolizumab in head and neck squamous cell carcinoma. J Immunother Cancer. 2022;10:e003026. doi: 10.1136/jitc-2021-003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenouvel D., González-Moles M., Talbaoui A., et al. An update of knowledge on PD-L1 in head and neck cancers: physiologic, prognostic and therapeutic perspectives. Oral Dis. 2020;26(3):511–526. doi: 10.1111/odi.13088. [DOI] [PubMed] [Google Scholar]
- 35.De Keukeleire S.J., Vermassen T., Hilgert E., Creytens D., Ferdinande L., Rottey S. Immuno-oncological biomarkers for squamous cell cancer of the head and neck: current state of the art and future perspectives. Cancers (Basel) 2021;13(7):1714. doi: 10.3390/cancers13071714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirshoren N., Al-Kharouf I., Weinberger J.M., et al. Spatial intratumoral heterogeneity expression of PD-L1 antigen in head and neck squamous cell carcinoma. Oncology. 2021;99(7):464–470. doi: 10.1159/000515441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang W.F., Wong M.C.M., Thomson P.J., Li K.Y., Su Y.X. The prognostic role of PD-L1 expression for survival in head and neck squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2018;86:81–90. doi: 10.1016/j.oraloncology.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 38.Hanna G.J., Liu H., Jones R.E., et al. Defining an inflamed tumor immunophenotype in recurrent, metastatic squamous cell carcinoma of the head and neck. Oral Oncol. 2017;67:61–69. doi: 10.1016/j.oraloncology.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 39.De Meulenaere A., Vermassen T., Aspeslagh S., et al. Turning the tide: clinical utility of PD-L1 expression in squamous cell carcinoma of the head and neck. Oral Oncol. 2017;70:34–42. doi: 10.1016/j.oraloncology.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Paolino G., Pantanowitz L., Barresi V., et al. PD-L1 evaluation in head and neck squamous cell carcinoma: insights regarding specimens, heterogeneity and therapy. Pathol Res Pract. 2021;226:153605. doi: 10.1016/j.prp.2021.153605. [DOI] [PubMed] [Google Scholar]
- 41.Kim H.R., Ha S.J., Hong M.H., et al. PD-L1 expression on immune cells, but not on tumor cells, is a favorable prognostic factor for head and neck cancer patients. Sci Rep. 2016;6:36956. doi: 10.1038/srep36956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiao Y., Liu C., Zhang X., et al. PD-L2 based immune signature confers poor prognosis in HNSCC. Oncoimmunology. 2021;10(1):1947569. doi: 10.1080/2162402X.2021.1947569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.