Abstract
Background & Aims
XBP1, most conserved transcription factor of endoplasmic reticulum stress, plays important roles in physiological and pathologic settings and has profound effects on disease progression and prognosis, so it is necessary to investigate XBP1 in macrophage-originated sterile inflammation during liver ischemia/reperfusion injury (IRI). Macrophage XBP1 expression and liver injury are analyzed in patients undergoing ischemia-related hepatectomy.
Methods
A myeloid-specific male XBP1-knockout (XBP1M-KO) strain is created for function and mechanism of XBP1 on macrophage-derived sterile inflammation in murine liver IRI with in vitro parallel research. Macrophages cocultured with hypoxia-treated hepatocytes are applied to investigate impact of XBP1 in vitro, with analysis of RNA sequencing and databases.
Results
Clinically, macrophage XBP1 expression significantly increases in ischemic liver tissues and positively correlates with liver injury after hepatectomy. Less hepatocellular damage is presented in XBP1M-KO mice than in XBP1-proficient (XBP1FL/FL) control animals. In vitro, XBP1 deficiency inhibits sterile inflammation and migration in macrophages cocultured with hypoxia-treated hepatocytes. Analysis of RNA sequencing and databases determines Metallothionein 2 (MT2) as XBP1 target gene, negatively regulated by binding with its promoter. XBP1 deficiency increases MT2 and IKBα expression, but inhibits nuclear factor-κB-p65 phosphorylation, markedly neutralizing XBP1M-KO-related benefits by promoting sterile inflammation during liver IRI.
Conclusions
XBP1 promotes macrophage-originated sterile inflammation, increases liver IRI by binding to MT2 promoter, and regulates MT2/nuclear factor-κB pathway, potentially therapeutic for clinical liver IRI.
Keywords: Macrophages, XBP1, MT2, NF-κB, Ischemia/Reperfusion Injury
Graphical abstract
Summary.
This is the first study to document the novel role of XBP1 in macrophage-originated sterile inflammation via MT2/NF-κB axis in liver IRI. XBP1 inhibits the target gene MT2, which is required for the activation of the NF-κB pathway.
The liver, a highly vascularized organ, receiving oxygenated blood from the hepatic artery and portal vein, exhibits a substantial ability to withstand ischemic insult. Nevertheless, hepatic ischemia-reperfusion injury (IRI), a local sterile inflammation, is of paramount importance in poor outcomes and is related to multiple injurious factors/pathways, including reactive oxygen species (ROS) overproduction, mitochondrial dysfunction, calcium overload, recruitment of inflammatory cells, and excessive generation of proinflammatory cytokines, inevitably occurring after partial hepatectomy (PH) or liver transplantation.1, 2, 3
Liver resident macrophages (Kupffer cells [KCs]), derived from erythromyeloid progenitors in the fetal liver, promote inflammation during liver injury instead of scavenging circulating antigens and presenting them to T cells in homeostasis. During liver IRI, the KCs can recognize damage-associated molecular pattern molecules, including free fatty acids, high mobility group box-1, and heat shock proteins, which are released from the injury to liver sinusoidal endothelial cells and hepatocytes.4 Activation of Toll-like receptor 4 promotes proinflammatory cytokines to synthesize and secrete in the early stage of reperfusion from KCs. These cytokines upregulate intercellular adhesion molecule-1 expression on liver sinusoidal endothelial cells, promote neutrophil adhesion, and release ROS from neutrophils, resulting in tissue injury.4,5 In addition, infiltrating macrophages are also involved in the liver immune response against ischemia-reperfusion (IR) and play distinctive roles. Accumulating evidence has shown the proinflammatory role of infiltrating macrophages.6, 7, 8 Therefore, regulating macrophage-originated sterile inflammation is key to alleviate the liver IRI.
X-box binding protein-1 (XBP1) is a unique transcription factor with immune function, whose dynamic form is commanded by an alternative splicing response based on disturbance of endoplasmic reticulum (ER) homeostasis and activation of the unfolded protein response (UPR).9, 10, 11 Previous studies have revealed enormous roles of XBP1 in cancer progression and metastasis.11 Splicing of XBP1 can be induced by Toll-like receptor 4, and its deficiency is associated with reduced immune response. XBP1 has been shown to play a key role in hepatocarcinoma. Taking hypoxia as an example, lack of oxygen can induce ER stress, trigger the UPR, and cause enhanced transcription of target genes to drive tumor growth and progression.9 Additionally, XBP1 inhibitors can protect against steatohepatitis.12 Thus far, however, the role of XBP1 in liver IRI remains unclear.
Metallothioneins (MTs) are a family of low-molecular-weight (6–7 kDa) metal-binding and metal-absorbing cysteine-rich cytosolic proteins that, in humans, can bind to heavy metals to protect cells from oxidative stress, and can be categorized into 11 isoforms including MT1, MT2, MT3, and MT4. MT2, which is detected in various organs and tissues,13,14 has been evaluated as an emerging target for inflammation and cancer because of its critical role in oxidative stress and apoptosis.15,16 Previous studies have shown that MT2 is linked to the nuclear factor kappa B (NF-κB) pathway via myeloid zinc finger 1 (MZF1) and their interactions to suppress human gastric carcinogenesis, suggesting that MT2 may be chemosensitive.17,18 In addition, MT2 is slightly upregulated in chronic hepatitis, with the potential in the treatment of fibrosis via activation of human hepatic stellate cells to upregulate the expression of collagenase genes.19 Therefore, a better understanding of the function and mechanism of MT2 in hepatic IRI is required.
We explored the role of XBP1 in macrophage-originated sterile inflammation during liver IRI. First, patients undergoing ischemia-related hepatectomy were assessed for XBP1 expression in liver macrophages and liver injury. Second, a myeloid-specific male XBP1-knockout (XBP1M-KO) strain was generated to evaluate the function and mechanism of macrophage XBP1 in liver IRI. Based on SAGER guidelines, we clarify that male strain only was generated because of possible difference between male and female mice, with the IR model in male mice better than female mice. Third, the roles and molecular mechanisms of XBP1 in macrophage-derived sterile inflammation during liver IRI were analyzed in detail.
Results
XBP1 Expression in Macrophages Was Found to be Upregulated and Positively Correlated with IR-Induced Liver Injury
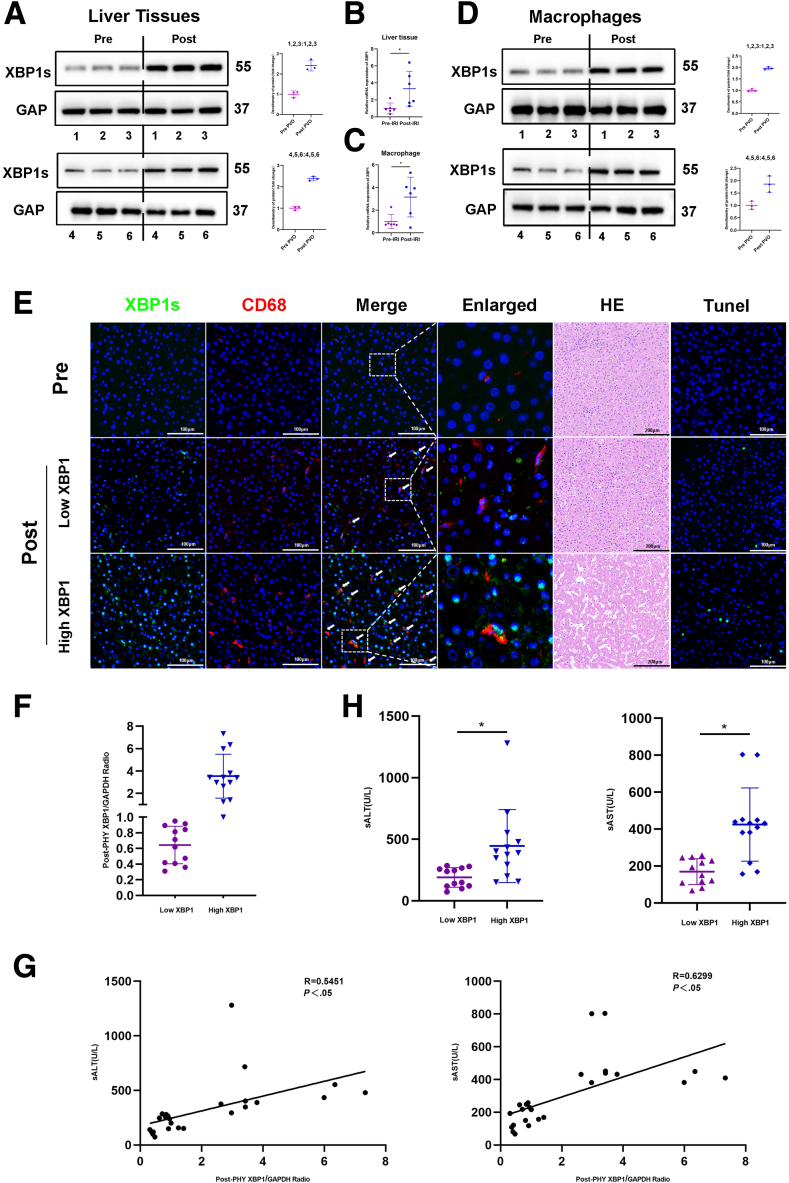
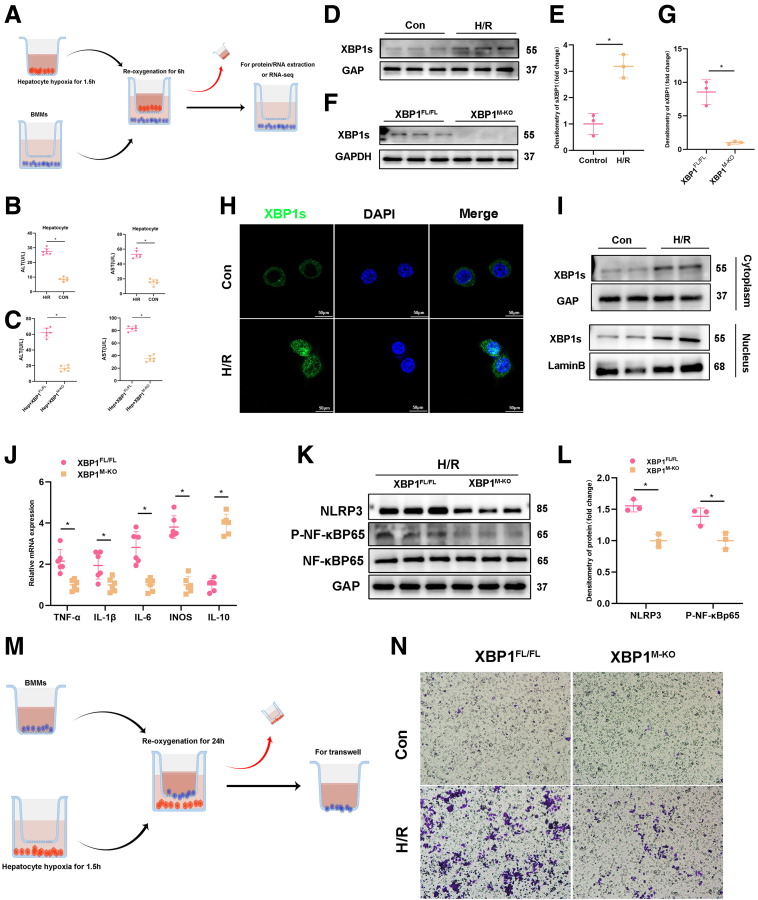
As expected, IR substantially elevated the expression of XBP1 mRNA in liver tissues and macrophages (Figure 1B, C). Next, we examined the expression of XBP1 protein in liver tissues and macrophages from harvested liver specimens. Based on Figure 1A and D, the protein level of XBP1 in liver tissues and isolated macrophages from IR-stressed liver specimens was higher. Dual-immunofluorescence (CD68 and XBP1) staining showed that XBP1 was markedly increased in macrophages of the ischemic liver (Figure 1E). To explore the association between postoperative XBP1 levels and clinical outcomes, the patients were classified into the low-XBP1 and high-XBP1 groups using the postoperative median XBP1/GAPDH ratio as the threshold (Figure 1F). Compared with the low-XBP1 group, the high-XBP1 group exhibited higher alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels at postoperative day 1 (POD1) (Figure 1H). Interestingly, the XBP1 level in postoperative macrophages positively correlated with ALT (P < .05) and AST (P < .05) levels at POD1 (Figure 1G). In addition, the number of infiltrating CD68+XBP1+ cells seemed to be positively correlated with the damage of liver architecture based on hematoxylin and eosin (HE) and TUNEL staining (Figure 1E). All findings indicate an increase in XBP1 expression within macrophages in ischemic livers, presenting a positive correlation with liver injury induced by ischemia reperfusion in patients who went through PH.
Figure 1.
XBP1 expression in macrophages is increased and positively correlates with IR-induced liver injury in patients. Liver tissues and macrophages were harvested from 25 patients undergoing PH. (A-D) Prehepatectomy/posthepatectomy XBP1s expression profiles in liver tissues and macrophages (representative 6 cases) through Western blotting assay. (B, C) Expressions of XBP1 mRNA in liver tissues and in liver macrophages. (E) Dual-immunofluorescence staining for XBP1 (green) and CD68 (red) colocalization in human liver tissues, which were also subjected to HE and TUNEL analysis. (F) Twenty-five patients were divided into the low (n = 12) and high (n = 13) groups using the postoperative median XBP1/GAPDH ratio as the threshold. (G) The ratio of posthepatectomy XBP1/GAPDH positively correlated with serum ALT and AST levels at POD1. (H) Serum ALT and AST levels were measured at POD1 in both low and high groups. ∗P < .05 by Student t test.
Myeloid-Specific XBP1 Deficiency Attenuates IR-Induced Hepatocellular Damage
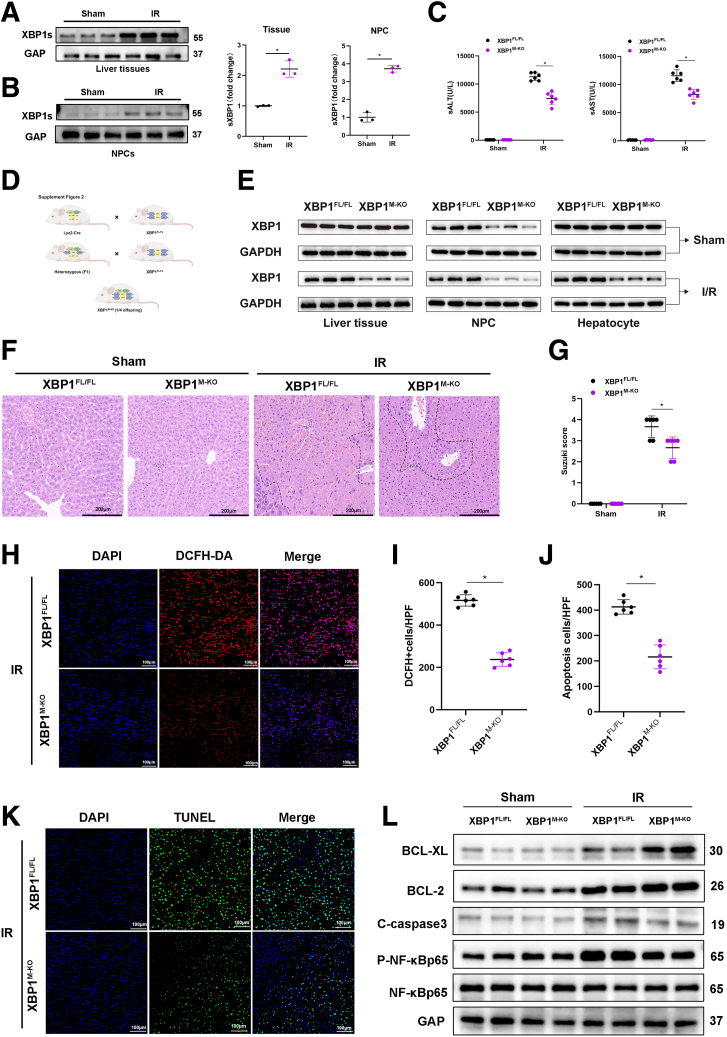
XBP1 expression was analyzed in the mouse hepatic IRI model, which showed that IR increased the XBP1 level in liver tissues and macrophages (nonparenchymal cells) (Figure 2A, B). To ascertain the impact of macrophage XBP1 on hepatocellular harm, ROS production, and inflammation during liver IR, we implemented the Cre-LoxP system to establish myeloid-specific XBP1-KO (XBP1M-KO) mice (Figure 2D). There were no significant differences observed in the overall expression of XBP1 in the livers or hepatocytes of XBP1FL/FL and XBP1M-KO mice before I/R, whereas after I/R, XBP1s reduced in the XBPM-KO mice possibly because of the alleviating of injury and ER stress (Figure 2E). Liver nonparenchymal cells and bone marrow macrophages (BMMs) isolated from XBP1M-KO mice showed significantly lower levels of XBP1 than those isolated from XBP1FL/FL mice. Following 90 minutes of warm ischemia ensued by 6 hours of reperfusion, XBP1M-KO mice exhibited lower levels of ALT and AST in comparison with XBP1FL/FL mice (Figure 2C). Meanwhile, less severe edema, sinusoidal congestion, vacuolization, and extensive hepatocellular necrosis, which were associated with lower Suzuki’s histologic grading of hepatic IRI, were observed in XBP1M-KO mice (Figure 2F, G), implying gentle hepatocellular injury in XBP1M-KO mice. Moreover, DEFH-DA staining showed that myeloid-specific XBP1 deficiency significantly alleviated IR-induced ROS (Figure 2H, I). TUNEL staining showed a reduced number of TUNEL-positive cells in the livers of XBP1M-KO mice (Figure 2J, K). Western blot analysis revealed increased levels of Bcl-2 and Bcl-xl and decreased levels of cleaved-caspase 3 in ischemic livers of XBP1M-KO mice (Figure 2L).
Figure 2.
Myeloid-specific XBP1 deficiency attenuates IR-induced hepatocellular injury. The IRI model mice were subjected to 90-minute ischemia, followed by 6-hour reperfusion. (A, B) XBP1s expression profiles in IR liver tissues and nonparenchymal cells, and their relative quantitation (fold change) via Western blotting assay. (C) Serum ALT and AST levels in the sham and IR groups were measured. (D) Generation flow chart of XBP1M-KO mice. (E) Comparison of XBP1s expression in XBP1FL/FL and XBPM-KO mice under normal conditions (Sham) and after I/R injury. (F) Representative HE staining of ischemic liver tissues, with the scale of 200 μm. (G) Liver damage was evaluated by Suzuki’s histologic scores. (H, I) ROS-sensing dye DCFH-DA staining and quantitation. (J, K) TUNEL staining of liver tissues and positive cell percentage. (L) Bcl-xl, Bcl-2, C-caspase3, P-NF-kB-p65, and NF-kB-p65 profiles in liver tissues based on Western blotting assay. n = 6, ∗P < .05 by Student t test.
Myeloid-Specific XBP1 Deficiency Decreases Inflammatory Cell Infiltration and Proinflammatory Mediators in Ischemic Livers
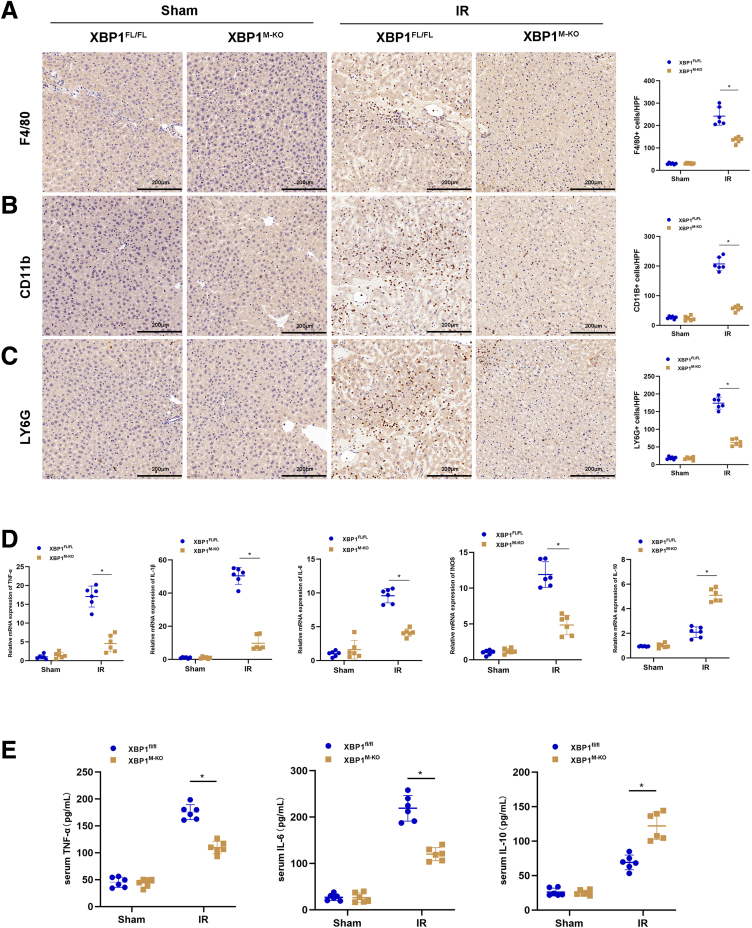
Because inflammatory cells play an essential role in the aggravation of hepatocellular apoptosis and necrosis during reperfusion, we investigated the infiltration of inflammatory cells in the liver after IRI using F4/80, CD11b, and Ly6G staining. Myeloid-specific XBP1 deficiency significantly reduced macrophage/neutrophil infiltration in IR-induced livers compared with XBP1FL/FL mice (Figure 3A-C). Consistent with these results, the mRNA levels of proinflammatory cytokines tumor necrosis factor-α (TNF-α), interleukin-1β (IL1β), interleukin-6 (IL6), and inducible nitric oxide synthase (iNOS) decreased, whereas that of anti-inflammatory cytokine IL10 increased in IR-induced livers of XBP1M-KO mice (Figure 3D). Additionally, decreased levels of serum TNF-α and IL6 and increased levels of serum IL10 were also detected in XBP1M-KO mice after IRI (Figure 3E).
Figure 3.
Myeloid-specific XBP1 deficiency decreases inflammatory cell infiltration and proinflammatory mediators in ischemic livers. The IRI model mice were subjected to 90-minute ischemia, followed by 6-hour reperfusion. (A, B) Macrophage infiltration was analyzed by immunohistologic staining with antibodies against F4/80 or CD11b; F4/80+ or CD11b+ cells were quantitated by counting numbers of positive cells/area, with the scale of 200 μm. (C) Neutrophil infiltration was analyzed by immunohistologic staining with antibodies against Ly6G; Ly6G+ cells were quantitated by counting numbers of positive cells/area, with the scale of 200 μm. (D) Detection of TNF-α, IL1β, IL6, iNOS, and IL10 in ischemic livers using qRT-PCR. (E) Detection of TNF-α, IL6, and IL10 in serum by enzyme-linked immunosorbent assay. n = 6, ∗P < .05 by Student t test.
Myeloid-Specific XBP1 Deficiency Alleviates Sterile Inflammation
Next, we explore the effects of XBP1 on macrophage function by simulating liver IRI models in vitro. Macrophages (BMMs) were cocultured with primary hepatocytes rendered anoxic using an anaerobic cell incubator (Figure 4A). First, Hypoxia/reoxygenation (H/R) significantly increased ALT and AST released by hepatocytes in the supernatant, indicating H/R led to hepatocellular damage, and increased HMGB1 and damage-associated molecular pattern molecules released by damaged cells (Figure 4B). Because the XBP1s expression was detected in macrophages from the XBP1FL/FL and XBP1M-KO mice after cocultured with primary hepatocytes under H/R stress conditions, we also measure the ALT and AST levels in these groups (Figure 4C, F). Figure 4D and E illustrated that the H/R model can stimulate XBP1 expression of BMMs in XBP1FL/FL mice, and we verified the XBP1-knockout model using BMMs of the XBP1FL/FL and XBP1M-KO mice in the cocultured H/R model. As expected, expression of XBP1 was significantly elevated in BMMs isolated from XBP1FL/FL mice, but negative in BMMs isolated from XBP1M-KO mice in the previously mentioned cocultured system (Figure 4D-G). In addition, XBP1 expression was also observed in cocultured XBP1FL/FL macrophages by immunofluorescence staining, displaying more nuclear translocation of XBP1 (Figure 4H). Interestingly, a similar phenotype was identified in the cytoplasm and nucleus of cocultured XBP1FL/FL macrophages through Western blotting assay (Figure 4I). By analyzing inflammatory cytokines in macrophages after coculture for 6 hours, we found that TNF-α, IL1β, IL6, and iNOS expressions decreased, whereas IL10 expression increased in XBP1-deficient BMMs (Figure 4J). NLRP3 inflammasome and NF-κB activation are essential in triggering local macrophage sterile inflammation during the occurrence and progression of immune diseases.20,21 XBP1 deficiency effectively inhibited NLRP3 inflammasome and NF-κB activation in cocultured macrophages (Figure 4K, L). In addition, the migration of macrophages was analyzed by coculture with primary hepatocytes rendered anoxic using an anaerobic cell incubator, displaying that XBP1 deficiency effectively prevented the migration of BMMs in the cocultured system (Figure 4M, N), which consisted of macrophage infiltration in IR stress-induced livers (Figure 3A, B).
Figure 4.
Myeloid-specific XBP1 deficiency alleviates sterile inflammation. (A) BMMs were cocultured with primary hepatocytes rendered anoxic using an anaerobic cell incubator. (B) ALT and AST levels were measured in the supernatant from hepatocytes in H/R system. (C) The ALT and AST levels detected in the cocultured H/R model from XBP1FL/FL and XBP1M-KO mice groups. (D, E) XBP1s expression profiles in XBP1FL/FL BMMs in the control and H/R cocultured system, and their quantitation (fold change) based on Western blotting assay. (F, G) XBP1s expression profiles in XBP1FL/FL and XBP1M-KO BMMs in H/R cocultured system using Western blotting assay. (H) Immunofluorescence staining for XBP1s (green) in cocultured XBP1FL/FL BMMs. (I) XBP1s expression in cytoplasm and nucleus of cocultured XBP1FL/FL BMMs using Western blotting assay. (J) Detection of TNF-α, IL1β, IL6, iNOS, and IL10 in H/R cocultured BMMs by qRT-PCR. (K, L) NLRP3, P-NF-κBp65, and NF-κBp65 protein expression levels in H/R cocultured BMMs and their quantitation (fold change) using Western blotting assay. (M, N) Migratory abilities of BMMs cocultured for 24 hours based on the Transwell assay. n = 6, ∗P < .05 by Student t test.
XBP1 Directly Regulates MT2 to Inhibit the NF-κB Pathway in Macrophage
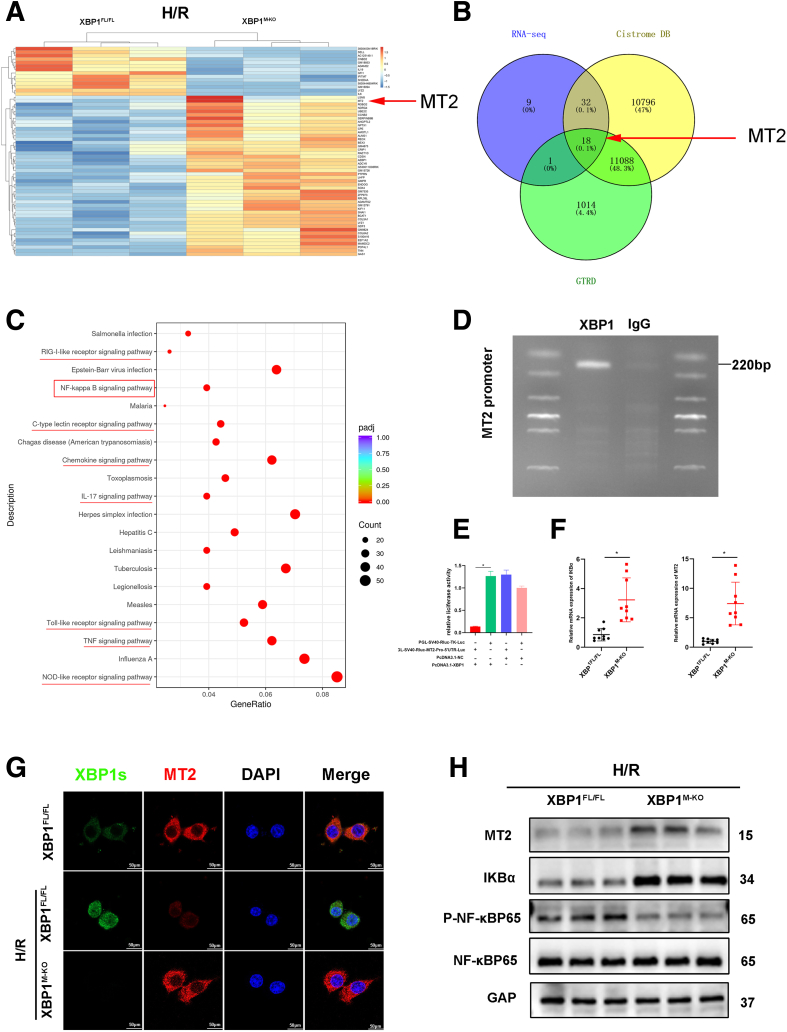
To investigate the underlying mechanism by which XBP1 regulated the sterile inflammation in macrophages, RNA sequencing was performed to identify the genes that were differentially expressed in cocultured macrophages of XBP1FL/FL and XBP1M-KO mice. We screened out a total of 60 mRNAs with the P value of less than .01 and absolute value of more than 1 (Figure 5A). As a unique base leucine zip (bZIP) transcription factor, XBP1 acts by specifically binding to a specific sequence upstream of the 5' terminal of the gene. Therefore, we searched for the downstream target gene of XBP1 through transcription factor databases (Cistrome DB and GTRD), and determined the downstream target gene MT2 of XBP1 and its binding site (Figure 5A, B). Furthermore, based on the Kyoto Encyclopedia of Genes and Genomes analysis, it seems probable that the NF-κB pathway plays a role in regulating XBP1 in BMMs (Figure 5C). Subsequently, we verified the relationship between XBP1 and MT2 through CUT&RUN. CUT&RUN polymerase chain reaction (PCR) in cocultured BMMs was performed to determine the MT2 promoter region using an anti-XBP1 antibody. We designed primers to identify the XBP1/TCF DNA-binding site in the MT2 promoter by PCR after performing CUT&RUN with the XBP1 antibody, showing that XBP1 was situated on the MT2 promoter (Figure 5D). A luciferase reporter assay was used to ascertain whether XBP1 was bound to the MT2 promoter for transcription regulation. The relative luciferase activity was the weakest when 293T cells were transfected with plasmid-XBP1 and plasmid-MT2-promoter (Figure 5E). All the previously mentioned results suggest that MT2 may be a target gene negatively regulated by XBP1. Moreover, MT2 expression was confirmed in XBP1M-KO macrophages by immunofluorescence staining, displaying more nuclear translocation of MT2 in cocultured macrophages (Figure 5G). It is reported that MT2 effectively suppresses NF-kB activation by directly enhancing IKBα transcription.17 Consistent with the previously mentioned reports, our results showed that XBP1 deficiency increased MT2 mRNA expression, thus reducing IKBα mRNA detected in cocultured macrophages (Figure 5F). Notably, XBP1 deficiency markedly increased the protein level of MT2, which contributed to the expression of IKBα protein and inhibited phosphorylation of NF-κB-p65 in cocultured macrophages (Figure 5H). Meanwhile, phosphorylation of NF-κB-p65 was also suppressed in ischemic liver tissues from XBP1M-KO mice (Figure 2L).
Figure 5.
XBP1 directly regulates MT2 to inhibit the NF-κB pathway in macrophages. (A) Differently expressed genes in H/R cocultured BMMs from XBP1FL/FL and XBP1M-KO mice. (B) The Venn diagram for screening differential mRNAs on the basis of RNA sequencing and transcription factor databases Cistrome DB and GTRD. (C) Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of the identified differentially expressed genes. (D) CUT&RUN-PCR analysis of XBP1 binding to the MT2 promoter. (E) Luciferase reporter assay of 293T cells transfected with plasmid-XBP1 or plasmid-NC and SV40-Rluc-TK-Promoteror SV40-Rluc-MT2-Promoter. (F) Detection of IKBα and MT2 by qRT-PCR after RNA-seq. BMMs were cocultured with primary hepatocytes rendered anoxic using an anaerobic cell (hypoxia for 1.5 hours, reoxygenation 6 hours). (G) Immunofluorescence staining of XBP1 (green), MT2 (red), and DAPI in cocultured BMMs. (H) Protein expressions of MT2, IKBα, P-NF-κBp65, and NF-κBp65 in cocultured BMMs based on Western blotting analysis.
MT2/NF-κB Axis Is Critical in XBP1-Induced Inflammatory Responses of Macrophages During Liver IRI
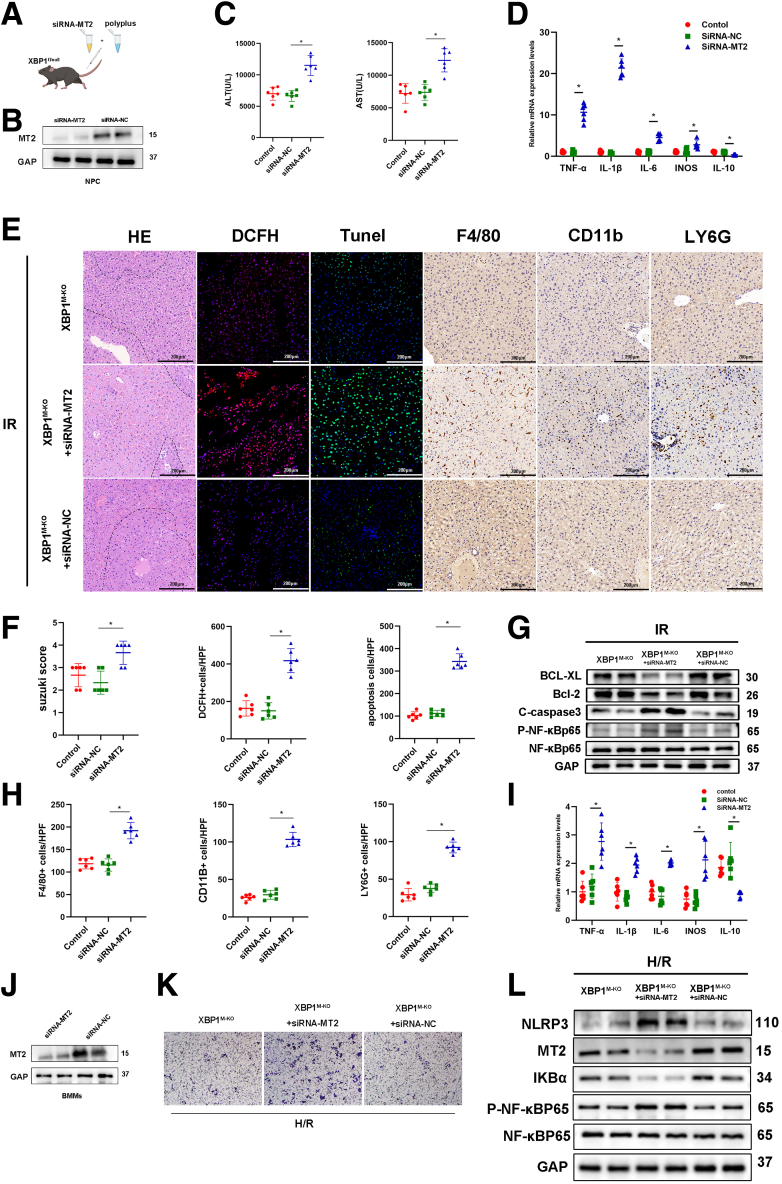
To elucidate whether XBP1-mediated effects of macrophages depended on the MT2 signaling pathway in IR-stressed liver injury, XBP1M-KO mice were injected with siRNA-MT2 or siRNA-NC mixed with mannose-conjugated polymers via the tail vein (Figure 6A). It was observed that siRNA-MT2 effectively inhibited MT2 expression in macrophages in vitro and in vivo (Figure 6B, J). In the liver IRI model, ALT and AST levels were significantly increased in XBP1M-KO mice after siRNA-MT2 treatment compared with siRNA-NC (Figure 6C). As shown in Figure 6D, siRNA-MT2 treatment enhanced TNF-α, IL1β, IL6, and iNOS expressions and decreased IL10 expression in ischemic livers. Histologically, siRNA-MT2 treatment promoted liver injury and inflammatory responses because of more severe necrosis and apoptosis (HE and TUNEL staining), higher ROS levels (DCFH staining), and more inflammatory cell infiltration (F4/80, CD11b, and Ly6G staining) (Figure 6E, F, H). Consistently, siRNA-MT2 treatment inhibited the expression of Bcl-xl and Bcl-2, and increased expression of cleaved-caspase 3 and p-NF-κB-p65 (Figure 6G). In vitro parallel research, XBP1M-KO BMMs treated with siRNA-MT2 or siRNA-NC were cocultured with primary hepatocytes rendered anoxic using an anaerobic cell incubator. By analyzing inflammatory cytokines in the cocultured system, we found increased TNF-α, IL1β, IL6, and iNOS expressions and decreased IL10 expression in the siRNA-MT2 group (Figure 6I). Moreover, siRNA-MT2 administration significantly suppressed the migration of BMMs in the cocultured system (Figure 6K), which consisted of infiltration of F4/80 or CD11b cells in ischemic livers (Figure 6E). Western blotting analysis further showed that siRNA-MT2 treatment inhibited IKBα protein, enhanced NF-κB-p65 phosphorylation, and promoted NLRP3 expression in cocultured macrophages (Figure 6L). All these data demonstrate that MT2 is critical for the XBP1-decorated macrophage function during liver IRI.
Figure 6.
MT2 inhibition abolishes XBP1M-KO-related anti-inflammatory features in vivo and in vitro. The IRI model mice were subjected to 90-minute ischemia, followed by 6-hour reperfusion. (A) XBP1M-KO mice were injected with siRNA-MT2 by caudal vein before establishing hepatic IRI model. (B) Western blotting analysis for the expressions of MT2 in nonparenchymal cells (NPC) after siRNA-MT2/siRNA-NC was injected into mice tail vein. (C) Serum ALT and AST levels were measured. (D) Detection of TNF-α, IL1β, IL6, iNOS, and IL10 in the ischemic liver of mice by qRT-PCR. (E) HE DCFH, TUNEL, F4/80, CD11b, and Ly6G staining, and (F, H) their quantitations. (G) Expressions of Bcl-2, Bcl-xl, C-caspase 3, P-NF-κBp65, and NF-κBp65 in IR liver tissues based on Western blotting analysis. (I) Detection of TNF-α, IL1β, IL6, iNOS, and IL10 in H/R cocultured XBP1M-KO BMMs through qRT-PCR. (J) Western blotting analysis for the expressions of MT2 in BMMs after transfected with siRNA-MT2/siRNA-NC. (K) Migratory abilities of XBP1M-KO BMMs cocultured for 24 hours based on the Transwell assay. (L) Expressions of NLRP3, MT2, IKBα, P-NF-κBp65, and NF-κBp65 in H/R cocultured XBP1M-KO BMMs using Western blotting assay. n = 6, ∗P < .05 by Student t test.
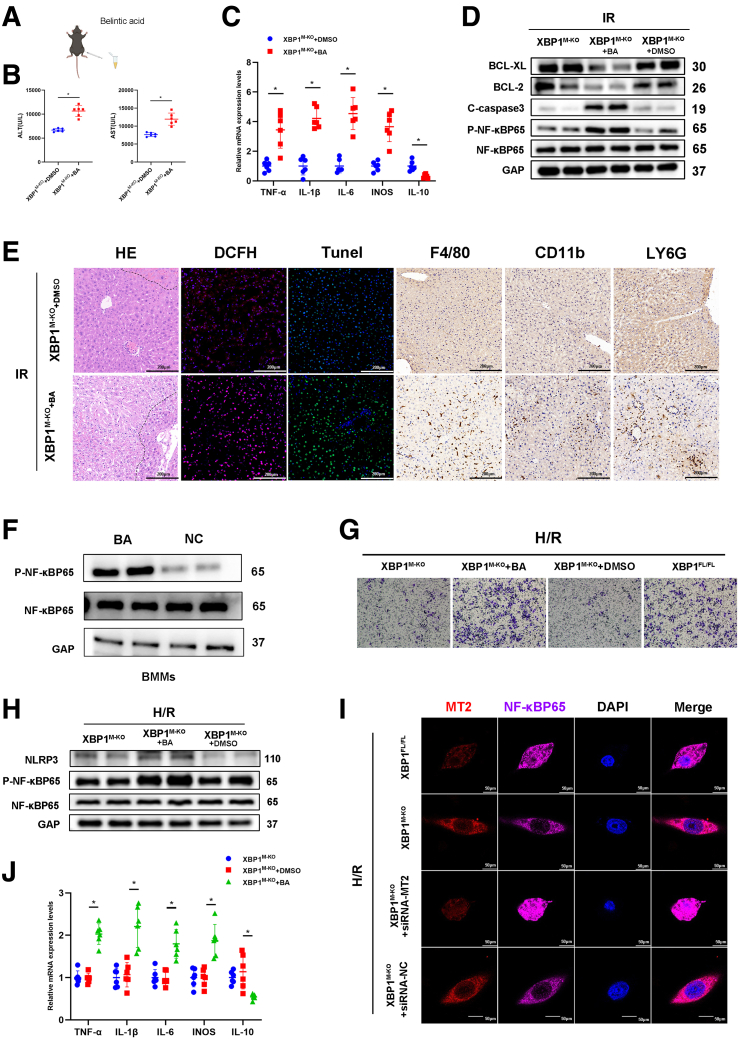
Next, we determined whether XBP1-mediated MT2 decorated the macrophage function depended on NF-κB activity during liver IRI. Betulinic acid (BA), a NF-κB activator, was used to sensitize macrophage NF-κB activity in XBP1M-KO mice (Figure 7A, F). After 90-minute ischemia and 6-hour reperfusion, BA treatment significantly increased ALT and AST levels compared with the control group (Figure 7B). In addition, BA treatment significantly elevated mRNA expressions of TNF-α, IL1β, IL6, and iNOS but inhibited IL10 mRNA expression in the ischemic livers of XBP1M-KO mice (Figure 7C). Pathologic analysis showed that BA treatment promoted hepatic damage and inflammatory responses based on more severe necrosis and apoptosis (HE and TUNEL staining), higher ROS levels (DCFH staining), and more inflammatory cell infiltration (F4/80, CD11b, and Ly6G staining) (Figure 7E). BA treatment reduced the expression of Bcl-xl and Bcl-2 but increased the expression of cleaved-caspase 3 and p-NF-κB-p65 (Figure 7D). BMMs isolated from XBP1M-KO mice were cultured for 7 days. After stimulation with BA, they were cocultured with anoxic primary hepatocytes. It was observed that BA treatment not only significantly promoted the migration of BMMs (Figure 7G), NF-κB-p65 phosphorylation, and NLRP3 expression in the cocultured macrophages (Figure 7H), but also significantly increased mRNA expressions of TNF-α, IL1β, IL6, and iNOS in cocultured BMMs except for IL10 mRNA expression (Figure 7G, H, J). To visually present the relationship between XBP1 and MT2/NF-κB axis in macrophages, XBP1M-KO BMMs treated with siRNA-MT2 were stained by immunofluorescence in a cocultured system. As shown in Figure 7I, XBP1 deficiency increased MT2 expression but inhibited cytoplasm-to-nuclear translocation of NF-κB-p65; siRNA-MT2 effectively reversed this phenomenon. Thus, MT2/NF-κB axis is critical for XBP1-related inflammatory response in macrophages.
Figure 7.
Sensitizing the NF-kB pathway neutralizes XBP1M-KO-related benefits in vivo and in vitro. The IRI model mice were subjected to 90-minute ischemia, followed by 6-hour reperfusion. (A) XBP1M-KO mice were treated with BA (20 mg/kg) before establishing hepatic IRI model. (B) Serum ALT and AST levels were measured. (C) Detection of TNF-α, IL1β, IL6, iNOS, and IL10 in the ischemic liver of mice by qRT-PCR. (D) Expressions of Bcl-2, Bcl-xl, C-caspase 3, P-NF-κBp65, and NF-κBp65 in IR liver tissues based on Western blotting analysis. (E) HE, DCFH, TUNEL, F4/80, CD11b, and Ly6G staining. (F) Western blotting analysis for the expressions of P-NF-κBP65, NF-κBP65 in BMMs after incubated with BA. (G) Migratory abilities of BMMs cocultured for 24 hours based on the Transwell assay. (H) Expressions of NLRP3, P-NF-κBp65, and NF-κBp65 in H/R cocultured XBP1M-KO BMMs using Western blotting assay. (I) Immunofluorescence staining of MT2 (red), NF-κBp65 (purple), and DAPI (blue) in H/R cocultured XBP1M-KO BMMs. (J) Detection of TNF-α, IL1β, IL6, iNOS, and IL10 in H/R cocultured XBP1M-KO BMMs through qRT-PCR. n = 6, ∗P < .05 by Student t test.
Discussion
As far as we know, this study is the first documenting the innovative role of XBP1 in macrophage-induced sterile inflammation via the MT2/NF-κB axis in liver IRI. The principal findings are as follows: (1) XBP1 expression in macrophages is upregulated and positively correlated with IR-induced liver injury in patients undergoing PH; (2) XBP1 deficiency in macrophages attenuates the ROS, inflammation, and injury in the IR liver; (3) XBP1 suppresses the MT2 gene, necessary for the initiation of the NF-κB pathway; and (4) MT2/NF-κB axis is crucial for XBP1-mediated inflammatory response in macrophages during liver IRI. All these findings highlight that the XBP1-mediated MT2/NF-κB axis contributes to macrophage-originated sterile inflammation during liver IRI (Figure 8).
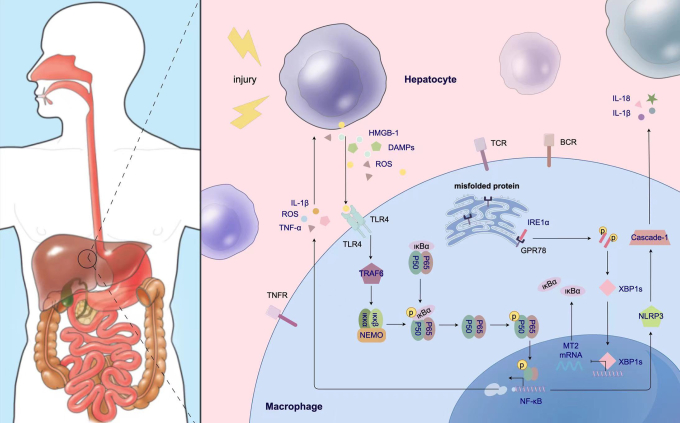
Figure 8.
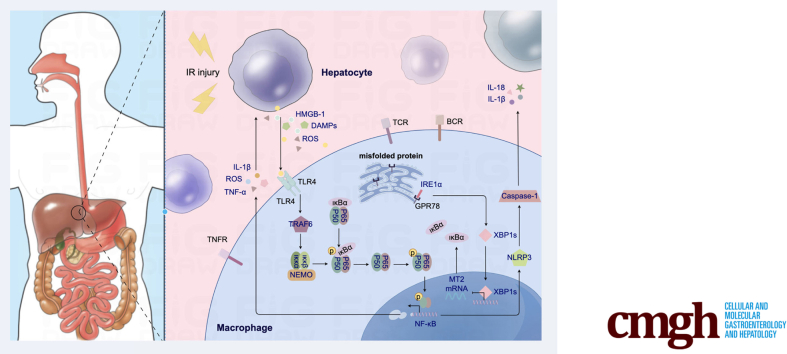
Schematic illustration how XBP1 facilitates NF-κB-p65 nuclear translocation via regulating MT2 transcription in macrophages during liver IRI. XBP1 is increased in macrophages of the IR-stressed liver, and then is bound to MT2 promoter to inhibit its transcription. However, MT2 enhances IKBα transcription and represses NF-κB-p65 phosphorylation and nuclear translocation. Thus, XBP1 facilitates NF-κB-p65 nuclear translocation and contributes to macrophage-originated sterile inflammation in the IR liver.
Under IR conditions, redox alteration, calcium overload, and ATP deficiency leads to dysfunction of key intracellular organelles and trigger cellular stress. Three main signaling cascades, activated by ER transmembrane protein sensors, are involved in mammalian UPR pathways: IRE1α/XBP1, PERK/ATF4, and ATF6. Impairment or excessive/persistent activation of ER stress has been confirmed to cause or aggravate various diseases, including IRI, diabetes, and cancer. Our previous studies have reported that ER stress is closely related to liver IRI: (1) ATF4-CHOP pathway promotes hepatocellular apoptosis and innate inflammation during liver IRI.22 (2) A critical intracellular mechanism in the regulation of innate immune cells during ischemia is ER stress and its specific signaling pathways regulated by ATF6.22 (3) Hyperglycemia triggers ER stress-ATF6-CHOP axis, accelerates innate inflammation, and deteriorates diabetes-related liver IRI. In addition, liver IRI can induce CHOP expression, whereas CHOP deficiency reduces liver IRI by inhibiting apoptosis and reducing the cleavage of caspase 3 protein.23
Accumulating evidence has indicated that macrophage-originated sterile inflammation is a crucial factor in liver IRI. Damage-associated molecular patterns, including high-mobility group box 1 (HMGB1), S100, and heat shock proteins, act via potent pattern-recognition receptors that are also used by pathogen-associated molecular patterns, such as Toll-like receptors, formyl peptide receptor, C-type lectins, and the receptor for advanced glycation endproducts. These receptors activate a shared set of inflammatory pathways, including NF-κB, p38, ERK, inflammasome assembly, and IL1β and IL18 release, and secretion of IL6, TNF, LT-β, IFNγ, and TGF-β, and promote the recruitment of inflammatory cells.24 Macrophage activation can directly result in the release of proinflammatory mediators, which directly or indirectly contribute to hepatocyte death/apoptosis to promote hepatic macrophage activation after reperfusion.25 However, Nrf2 and Notch1 signalings can modulate macrophage function, reduce sterile inflammation, and alleviate hepatic IRI.21,26 In a previous study, we also found that macrophage Nrf2 negatively mediated innate inflammatory responses and attenuated liver IRI by Timp3-suppressed RhoA/ROCK pathway.6 Indeed, UPR plays a crucial role in modulating macrophage phenotypes and functions in various diseases.27, 28, 29 Our previous study demonstrated that Nogo-B, a protein residing in the ER, could trigger macrophage-related innate inflammation and contributed to IR-induced liver injury by activating the MST-mediated Hippo/YAP pathway.7 In an infection model, activation of IRE1 in macrophages promotes inflammasome activation, which is crucial for inducing IL1β production and clearing bacterial infections.30 In obesity induced by a high-fat diet, IRE1-depleted macrophages alleviate pathologic symptoms by supporting the transformation of proinflammatory macrophages into the anti-inflammatory type.27 The activated IRE1 excises an intron from XBP1 and transforms it into spliced XBP1 (XBP1s). XBP1s is transported to the nucleus, where it facilitates gene translation. In the event of ER stress, ATF6 is transported to the Golgi apparatus and cleaved by Site-1 and Site-2 proteases. Following cleavage, ATF6 releases a cytosolic fragment, which directly controls the genes encoding ERAD components, such as the basic transcription factor leucine zipper family and XBP1. The IRE1-XBP1 UPR pathway is responsible for the efficient folding, maturation, and degradation of proteins in the ER.31 Based on our knowledge, XBP1-dependent UPR target genes take part in hepatic ferroptosis through the MT2-cAMP-PKA-IRE1 axis-mediated ER stress.32 XBP1, as a major IRE1 downstream effector, has also shown an excellent orchestrating role in macrophage function. For example, macrophage RIPK3 activates the IRE1α-XBP1 pathway and Foxo1 signaling in IR-stress livers.33 In mice with diet-induced steatohepatitis, EVs-containing XBP1 can recruit monocyte-derived macrophages to the liver, leading to inflammation and injury.29 Our findings demonstrate a significant increase in XBP1 expression within macrophages of ischemic liver, regardless of whether the samples were obtained from humans or mice. To determine the role of XBP1 in macrophages, we assessed the degree of IRI by the Cre-LoxP system to create a myeloid-specific XBP1-deficient strain. As expected, XBP1 deficiency in macrophages significantly improved macrophage-originated sterile inflammation and attenuated liver IRI.
Another distinguishing finding in our study was that MT2 expression was significantly enhanced in XBP1-deficient macrophages. MT2 detected in various organs and tissues has been considered an emerging target for inflammation and cancer. It can effectively inhibit NF-κB activation by enhancing IκB-α transcription directly in cancer cells. NF-κB activation is crucial in triggering local macrophage sterile inflammation in various diseases.25 Here, we found that XBP1 binded to the promoter of MT2 to inhibit MT2 expression, reduce IκB-α expression, and promote NF-κB-p65 phosphorylation in macrophages. In addition, KEGG analysis also suggested other important signaling pathways involved in macrophage-originated sterile inflammation during various diseases. (1) RIG-I-like receptor signaling pathway: plays a major role in triggering innate immunity and inflammation and imparting gene expression involved in many autoimmune diseases.34 (2) IL17 signaling pathway: IL17 neutralization can effectively prevent the enhanced hepatocellular damage and liver inflammation during liver IRI.35 (3) TNF signaling pathway: this pathway is crucial to multiple physiological and pathologic processes, such as the regulation of immune responses and the initiation of inflammation.36 Indeed, NF-κB is involved in the transcription regulation of a large number of “inflammation activation” genes, including the previously mentioned signaling pathways.
To validate the role of MT2/NF-κB axis in XBP1-induced inflammatory response in macrophages during liver IRI, siRNA-MT2 and NF-κB activator were applied to the animal model. In vivo data showed that siRNA-MT2 and NF-κB activator abolished the benefits of macrophage XBP1 deficiency in IR-induced liver inflammation and injury. In vitro, siRNA-MT2 inhibited MT2 expression, reduced IκB-α expression, promoted NF-κB-p65 phosphorylation and nuclear translocation, thus fostering migration and production of inflammatory factors in cocultured XBP1-deficient macrophages. Additionally, NF-κB activator could also abrogate anti-inflammatory functions of XBP1 deficiency in the cocultured system. In a previous study, researchers examined the expression of several crucial hepatocyte ER stress markers, including XBP1s, and representative XBP1-dependent UPR target genes, including Dnajb9, Edem1, and Sec61a1, with the aim of further investigating the potential roles of melatonin in ferroptosis. The results indicated that melatonin could alleviate ER stress in NAFLD via the MT2/IRE1 signaling pathway, with XBP1s decreased by.32 However, further investigation is required to elucidate the interaction between XBP1s and MT2.
Hepatocytes XBP1 also play an important role in liver injury and repair. Previous studies have demonstrated that XBP1 expression is markedly upregulated in liver samples from patients with MASH, and XBP1M-KO knockout promotes M2 polarization, and inhibits hepatic stellate cell activation by reducing TGF-β1 expression during liver fibrosis.12 These results indicated that macrophage XBP1 matters a lot in liver injury and repair. However, our study focuses on the role of XBP1 in macrophages, with no further consideration of hepatocyte XBP1. To investigate the impact of hepatocyte XBP1 in liver IRI, we will also measure XBP1 expression in hepatocytes for IRI liver from patients and mice model. Then, hepatocyte XBP1 knockout mouse will be used to analyze underlying mechanisms by molecular biology methods.
Conclusions
Macrophage-specific XBP1 deficiency suppresses the NF-κB signaling pathway, mitigating liver damage via upregulation of MT2 transcription as a target gene during liver IRI. By identifying the molecular pathways through which the macrophage XBP1 regulates MT2/NF-κB-mediated innate immunity, our findings present a rationale for novel therapeutic strategies to alleviate sterile inflammatory liver injury.
Material and Methods
All authors had access to the all data and have reviewed and approved the final manuscript.
Patients and Clinical Samples
Biopsy samples were obtained from 25 patients (Table 1) with hepatic hemangioma who underwent PH with the Pringle maneuver between December 2021 and December 2022. Prehepatectomy biopsies were obtained following laparotomy before hepatic portal occlusion, and posthepatectomy biopsies were collected 1.5–2 hours after reperfusion (before abdominal closure). The ischemia time was 15–30 minutes. To monitor the degree of hepatocellular injury, serum ALT and serum AST levels were both assessed on POD1. Each patient gave written informed consent. The trial was approved by the Institutional Review Board (approval number: 20220831), and was performed according to the ethical guidelines of the 1975 Declaration of Helsinki.
Table 1.
Patient Characteristics of Hepatectomy
| Variables | Patients |
|---|---|
| N | 25 |
| Sex (male/female) | 5/20 |
| Age, Y | 48.88 ± 9.09 |
| Ischemia time, MIN | 15–30 |
| Reperfusion time, H | 1.5–2 |
| ALT of POD1, U/L | 322.652 ± 247.31 |
| AST of POD1, U/L | 302.408 ± 193.03 |
ALT, alanine aminotransferase; AST, aspartate aminotransferase; POD1, postoperative day 1.
Hepatic IRI Model and Treatments
Our work in animal experiments has been reported in accordance with the ARRIVE guidelines (Animals in Research: Reporting In Vivo Experiments).37 XBP1-LoxP mice (XBP1FL/FL) were crossed with myeloid-specific Cre mice (Lyz2-Cre) to generate myeloid-specific male XBP1-knockout (XBP1M-KO) mice. Male XBP1FL/FL and XBP1M-KO mice, aged between 6 and 8 weeks and on a C57BL/6 background, were obtained. Based on knowledge, several previous studies have proven that one could induce the IR model in male mice better than female mice, and sex both in homo sapiens and mice can make a difference in liver IRI.38, 39, 40 The mouse hepatic IRI model was established according to previously described methods.6,7 Clamping for 90 minutes was performed on the arterial and portal vessels leading to the cephalic lobes. However, the sham-controlled mice did not exhibit any vascular occlusion. The pathologic injury's severity was assessed blindly on a scale ranging from 0 to 4, using Suzuki's criteria. Additionally, 4 hours before the establishment of the hepatic IRI model, some mice were injected with MT2-small interfering RNA (siRNA-MT2) or nonspecific control (siRNA-NC) mixing with mannose-conjugated polymers at a ratio through the tail vein. Saline/BA (30 mg/kg) was intraperitoneally injected for 24 hours before the mouse hepatic IRI.6,7,41
Cell Isolation and Culture
BMMs were obtained from the bone marrow of 6- to 8-week-old male mice by culture in 1 × DMEM containing 10% fetal bovine serum and 20% L929 conditioned medium for 7 days as previously described.7,26,42 BMMs (5 × 106/well) were then replated and cultured overnight in new culture dishes. BMMs were incubated with siRNA-MT2 or siRNA-NC (corues Biotechnology, China) for 48 hours. After that, the BMMs were harvested for further experiments. Primary hepatocytes isolated from mice were cocultured with primary macrophages isolated from the BMMs mice after rendered anoxic using an anaerobic cell incubator (6 hours for PCR and Western blotting; 24 hours for Transwell). Human hepatic macrophages were isolated from liver tissue specimens that were cut into small pieces and digested in collagenase IV (50 IU/mL). The dissociated cells were filtered through a 75-μm cell strainer and separated by Ficoll centrifugation, and the mononuclear cells were cultured in RPMI 1640 containing 10% fetal bovine serum. Human macrophages were subsequently isolated using CD68 magnetic beads (Miltenyi Biotec). The detailed process by which the hepatocytes, KCs and nonparenchymal cells were isolated from mice was described in our previous study.6,26,42
Generation of M-KO Mice and Breeding Information
Wild-type, male, 6- to 8-week-old, FloxP-Xbp1 (Xbp1FL/FL), Lyz2-Cre Xbp1-knockout (XBP1M-KO) mice on a C57BL/6 background (Nanjing Biomedical Research Institute of Nanjing University, Nanjing, China) were used in the experiments. They were fed sterilized water and food under specific pathogen-free conditions.
XBP1FL/FL (carrying the a floxed allele) and Lyz2-Cre (expressing the Cre recombinase specifically in myeloid cells) mice were both acquired from GemPharmatech Co Ltd (Nanjing, China). XBP1 conditional knockout mice were made via CRISPR/Cas9 system. First, 2 sgRNAs-targeting the introns on both sides of the floxed region of XBP1 were respectively constructed and transcribed in vitro. The donor vector with the loxp fragment was designed and constructed in vitro. Then Cas9 mRNA, sgRNA, and donor were coinjected into zygotes. Thereafter, the zygotes were transferred into the oviduct of pseudopregnant ICR females at 0.5 dpc. F0 mice were birthed after 19∼21 days of transplantation; all the offsprings of ICR females (F0 mice) were identified by PCR and sequencing of tail DNA. Positive F0 mice were genetyped by the methods. Finally, F0 mice were crossed with C57BL/6J mouse to build up heterozygous mice. A stable F1 generation mouse model was obtained by mating positive F0 generation mice with C57BL/6J mice.
For producing myeloid-specific knockout mice, homozygous XBP1FL/FL mice were bred with homozygous Lyz2-Cre mice, and their heterozygous offspring were back crossed with homozygous XBP1FL/FL mice (Figure 2D).
Blood samples were obtained when the mice were sacrificed and centrifuged at 7000 rpm for 10 minutes at 4°C to collect the serum. The levels of serum ALT and serum AST were measured by an automatic chemical analyzer (Olympus Company, Tokyo, Japan).
Histopathology, Immunohistochemical Staining, ROS Staining, Immunofluorescence Staining, and TUNEL Staining
Hepatic tissue samples were obtained when the mice were sacrificed, fixed in formaldehyde solution for approximately 3 days, and embedded in paraffin based on standard procedures. For analysis of hepatic histopathology, all paraffin blocks were cut into 4-μm slices, dyed with HE, and then observed by light microscopy. The Suzuki score, which consists of 3 aspects (hepatocyte necrosis, sinusoidal congestion, and ballooning degeneration), was rated on a scale of 1 to 4, as defined by Suzuki et al. Hepatic macrophages and neutrophils were analyzed by IHC staining and incubation with primary rat anti-mouse F4/80, CD11b and Ly6G mAbs (Cell Signaling Technology). Positive cells were counted in a blinded manner in at least 3 fields per section (×200). The expression of XBP1, P65 (Cell Signaling Technology), and MT2 (Abclonal, WuHan, China) in BMMs was determined by immunofluorescence staining. The secondary biotinylated goat anti-rat IgG (Vector, Burlingame, CA) was added and incubated with immunoperoxidase (ABC Kit, Vector) according to the manufacturer’s protocols. DAPI was used for the nuclear counterstaining. To detect the DNA fragmentation that is characteristic of apoptosis in formalin-fixed paraffin-embedded tumor slices, we performed TUNEL staining using the Klenow-FragEL DNA Fragmentation Detection Kit (Roche, Basel, Switzerland) according to the manufacturer’s instructions. Ten fields from each slide were selected randomly for examination (×400). ROS accumulation was detected using a fluorescent probe,2,7-dichlorohydrofluorescein diacetate (DCFH-DA) (Servicebio, Wuhan, China), according to the manufacturer’s instructions. ROS levels were imaged by fluorescence microscopy (Olympus, Japan); red fluorescence represented the amount of ROS (×200).
Protein Extraction and Western Blotting
We used a whole protein extraction kit (KeyGEN BioTECH) to extract total protein from tissues and cells, and we used the nuclear and cytoplasmic protein extraction kit (KeyGEN BioTECH) to extract nuclear and cytoplasmic proteins from KCs according to the manufacturer's protocols. All protein lysates were separated by standard sodium dodecyl-sulfate polyacrylamide gel electrophoresis and then transferred onto polyvinylidene fluoride membranes. The antibodies used were as follows: XBP1s, Bcl2, Bcl-XL, C-Caspase3, NF-κB p65, P-NF-κB p65, GAPDH (Cell Signaling Technology), and MT2 (Abclonal, WuHan, China).
RNA Extraction and qRT-PCR
We used the EasyPure RNA Kit (Ruijie Biotech) to extract total RNA from tissue and cell samples according to the manufacturer’s protocols. Briefly, tissues or cells were mixed with lysis buffer. After efficient lysis was confirmed, the lysis solution was collected and vortexed for 10 seconds, after which an equal volume of absolute ethyl alcohol was added and mixed. The lysis solution was transferred into an RNA column, which was placed in a centrifuge tube. The tube was centrifuged at 4000 × g for 1 minute at 4°C and washed with 500 μL of wash buffer, followed by centrifugation at 12,000 × g for 1 minute. Subsequently, the RNA column was transferred to a new and RNase-free tube and air-dried for 2 minutes. We added 20 μL of elution buffer to the center of the RNA column and waited for 2 minutes. Subsequently, the tube was centrifuged at 12,000 × g for 1 minute, and the extracted RNA was stored at -80°C before further experiments. For cDNA synthesis, a 20 μL/well reaction mix composed of 1 μg of total RNA, 4 μL of 5 × HiScript Ⅱ qRT SuperMix (Vazyme Biotech Co, Ltd), and RNase-free ddH2O was prepared.
We performed qRT-PCR with ChamQTM Universal SYBR qPCR master mix (Vazyme Biotech Co, Ltd) based on standard protocols. The primer sequences used in this study for PCR amplification are listed in Table 2.
Table 2.
Primer Sequences for the Amplification
| Gene | Forward Primer (5′ → 3′) | Reverse Primer (5′ → 3′) |
|---|---|---|
| TNF-α | 5’-CAGGCGGTGCCTATGTCTC-3’ | 5’-CGATCACCCCGAAGTTCAGTAG-3’ |
| IL1β | 5’-GAAATGCCACCTTTTGACAGTG-3’ | 5’-TGGATGCTCTCATCAGGACAG-3’ |
| IL6 | 5’-CTGCAAGAGACTTCCATCCAG-3’ | 5’-AGTGGTATAGACAGGTCTGTTGG-3’ |
| INOS | 5’-GTTCTCAGCCCAACAATACAAGA-3’ | 5’-GTGGACGGGTCGATGTCAC-3’ |
| IL10 | 5’-CTTACTGACTGGCATGAGGATCA-3’ | 5’-GCAGCTCTAGGAGCATGTGG-3’ |
| XBP1 (m) | 5’- AGCTTTTACGGGAGAAAACTCAC-3’ | 5’-CCTCTGGAACCTCGTCAGGA-3’ |
| XBP1 (H) | 5’-CCCTCCAGAACATCTCCCCAT-3’ | 5’-ACATGACTGGGTCCAAGTTGT-3’ |
| GAP (m) | 5’-AGGTCGGTGTGAACGGATTTG-3’ | 5’-GGGGTCGTTGATGGCAACA-3’ |
| GAP (H) | 5’-GGAGCGAGATCCCTCCAAAAT-3’ | 5’-GGCTGTTGTCATACTTCTCATGG-3’ |
Enzyme-Linked Immunosorbent Assay
We obtained murine serum and macrophage culture supernatants to analyze the levels of cytokines. The levels of TNF-α, IL6, and IL10 in murine serum were measured by using enzyme-linked immunosorbent assay kits (eBioscience, San Diego, CA) according to the manufacturer’s instructions.
RNA Sequencing, CUT&RUN
BMMs were isolated from XBP1FL/FL and XBP1M-KO mice, and cultured for 7 days. Then primary hepatocytes isolated from mice were cocultured with primary macrophages isolated from the BMMs mice after rendered anoxic (1.5 hours) using an anaerobic cell incubator (6 hours). TRIzol reagent was added to the culture dishes to lyse the BMMs, and the lysate was harvested for further RNA-seq analysis (Novogene, China).
CUT&RUN analysis was carried out using the CUT&RUN Assay Kit HD101 (Vazyme Biotech Co, Ltd). Briefly, BMMs were isolated from XBP1FL/FL and XBP1M-KO mice, and cultured for 7 days. Then primary hepatocytes isolated from mice were cocultured with primary macrophages isolated from the BMMs mice after rendered anoxic (1.5 hours) using an anaerobic cell incubator (6 hours). BMMs were collected at room temperature and counted. Then protein-DNA complexes were washed and eluted, followed by a cross-link reversal step, and the resulting DNA was purified. DNA from each immunoprecipitation reaction was examined by PCR. The primers for the XBP1-responsive region of the MT2 promoter were as follows:
forward:5′- CTAAGAGCTGCGAGGACAGC-3′ and
reverse: 5′- GGTGTTTCCCCACTTGCTTA-3′.
Luciferase Assays
Luciferase reporter plasmid was constructed in the pGL3 luciferase vector according to the manufacturer’s instructions (corues Biotechnology, China). 293T cells were transfected with 0.5 μg of pGL-MT2-luciferase vector and then transfected with the PcDNA3.1-XBP1 (XBP1-vector). Transcriptional activity was measured using a Dual-Luciferase Reporter Assay System according to the manufacturer’s instructions (GeneChem, Shanghai, China).
Migration Assays
BMMs were isolated from XBP1FL/FL and XBP1M-KO mice, cultured, and differentiated for 7 days and then incubated with coculture for 24 hours. The supernatant was harvested for use as conditioned medium in subsequent experiments. Then, 1 × 104 BMMs in 400 μL were seeded in the upper chambers of Transwell units with 8-μm pore-size polycarbonate filters (Millipore, Bedford, MA) in fetal bovine serum–free medium, and 600 μL of the indicated conditioned media was added to the lower chambers. After being incubated for 24 hours, the migratory BMMs were stained with 0.1% crystal violet for 30 minutes and then counted and analyzed by using a digital microscope system (Leica, Wetzlar, Germany).
Statistical Analysis
All findings in this study were illustrated as the mean ± standard deviation. Student t test or analysis of variance was used to assess statistical differences between multiple groups. All experiments were repeated at least 3 times, and discrepancies were statistically significant if P was less than .05. SPSS 20.0 and GraphPad Prism 8.0 software were used to analyze all data.
Acknowledgments
CRediT Authorship Contributions
Jianhua Rao (Conceptualization: Lead; Formal analysis: Lead; Funding acquisition: Lead; Methodology: Lead; Project administration: Lead; Supervision: Lead; Writing – review & editing: Lead)
Zeng Wang (Data curation: Equal; Investigation: Equal; Resources: Equal; Software: Equal; Validation: Equal)
Fei Yu (Formal analysis: Equal; Investigation: Equal; Validation: Equal; Visualization: Equal; Writing – original draft: Equal)
Junda Li (Data curation: Equal; Methodology: Equal; Software: Equal; Visualization: Equal)
Wenzhu Li (Conceptualization: Equal; Investigation: Equal; Methodology: Equal; Software: Equal; Validation: Equal; Visualization: Equal)
Zhengfeng Xuan (Validation: Equal; Visualization: Equal)
Yongquan Chi (Supervision: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Feng Zhang (Formal analysis: Equal; Methodology: Equal; Project administration: Equal)
Liming Tang (Conceptualization: Equal; Supervision: Equal)
Feng Cheng (Methodology: Equal; Project administration: Equal; Supervision: Equal; Writing – review & editing: Equal
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding The project was supported by The National Natural Science Foundation of China (81871259), The Six Talent Peaks Project in Jiangsu Province (2017-WSW-019), and Clinical and Translational Medicine Research Project of Chinese Academy of Medical Sciences (2023-I2M-CT-B-126). The funding body had no role in the design of the study, data collection, analysis, interpretation, or writing the manuscript.
Contributor Information
Jianhua Rao, Email: raojh@njmu.edu.cn.
Liming Tang, Email: drtangliming@163.com.
Feng Cheng, Email: docchengfeng@njmu.edu.cn.
References
- 1.Mao B., Yuan W., Wu F., et al. Autophagy in hepatic ischemia-reperfusion injury. Cell Death Discov. 2023;9:115. doi: 10.1038/s41420-023-01387-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montalvo-Jave E.E., Escalante-Tattersfield T., Ortega-Salgado J.A., et al. Factors in the pathophysiology of the liver ischemia-reperfusion injury. J Surg Res. 2008;147:153–159. doi: 10.1016/j.jss.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan T.S., Cassim S., Raymond V.A., et al. Upregulation of Krebs cycle and anaerobic glycolysis activity early after onset of liver ischemia. PLoS One. 2018;13 doi: 10.1371/journal.pone.0199177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guillot A., Tacke F. Spatial dimension of macrophage heterogeneity in liver diseases. eGastroenterology. 2023;1 [Google Scholar]
- 5.Dar W.A., Sullivan E., Bynon J.S., et al. Ischaemia reperfusion injury in liver transplantation: cellular and molecular mechanisms. Liver Int. 2019;39:788–801. doi: 10.1111/liv.14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao J., Qiu J., Ni M., et al. Macrophage nuclear factor erythroid 2-related factor 2 deficiency promotes innate immune activation by tissue inhibitor of metalloproteinase 3-mediated RhoA/ROCK pathway in the ischemic liver. Hepatology. 2022;75:1429–1445. doi: 10.1002/hep.32184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao J., Cheng F., Zhou H., et al. Nogo-B is a key mediator of hepatic ischemia and reperfusion injury. Redox Biol. 2020;37 doi: 10.1016/j.redox.2020.101745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehal W. Mechanisms of liver fibrosis in metabolic syndrome. eGastroenterology. 2023;1 doi: 10.1136/egastro-2023-100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hetz C., Zhang K., Kaufman R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol. 2020;21:421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z., Wang M., Wang X., et al. XBP1 deficiency promotes hepatocyte pyroptosis by impairing mitophagy to activate mtDNA-cGAS-STING signaling in macrophages during acute liver injury. Redox Biol. 2022;52 doi: 10.1016/j.redox.2022.102305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S., Chen J., Hua X., et al. The emerging role of XBP1 in cancer. Biomed Pharmacother. 2020;127 doi: 10.1016/j.biopha.2020.110069. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q., Zhou H., Bu Q., et al. Role of XBP1 in regulating the progression of non-alcoholic steatohepatitis. J Hepatol. 2022;77:312–325. doi: 10.1016/j.jhep.2022.02.031. [DOI] [PubMed] [Google Scholar]
- 13.Kim H.G., Kim J.Y., Han E.H., et al. Metallothionein-2A overexpression increases the expression of matrix metalloproteinase-9 and invasion of breast cancer cells. FEBS Lett. 2011;585:421–428. doi: 10.1016/j.febslet.2010.12.030. [DOI] [PubMed] [Google Scholar]
- 14.Gao T., Wang T., Wang Z., et al. Melatonin-mediated MT2 attenuates colitis induced by dextran sodium sulfate via PI3K/AKT/Nrf2/SIRT1/RORalpha/NF-kappaB signaling pathways. Int Immunopharmacol. 2021;96 doi: 10.1016/j.intimp.2021.107779. [DOI] [PubMed] [Google Scholar]
- 15.Takano H., Inoue K., Yanagisawa R., et al. Protective role of metallothionein in acute lung injury induced by bacterial endotoxin. Thorax. 2004;59:1057–1062. doi: 10.1136/thx.2004.024232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Si M., Lang J. The roles of metallothioneins in carcinogenesis. J Hematol Oncol. 2018;11:107. doi: 10.1186/s13045-018-0645-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin S., Wang X., Pan Y., et al. Transcription factor myeloid zinc-finger 1 suppresses human gastric carcinogenesis by interacting with metallothionein 2A. Clin Cancer Res. 2019;25:1050–1062. doi: 10.1158/1078-0432.CCR-18-1281. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009;1 doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X.L., Schnoor M., Yin L.M. Metallothionein-2: an emerging target in inflammatory diseases and cancers. Pharmacol Ther. 2023;244 doi: 10.1016/j.pharmthera.2023.108374. [DOI] [PubMed] [Google Scholar]
- 20.Jin Y., Li C., Xu D., et al. Jagged1-mediated myeloid Notch1 signaling activates HSF1/Snail and controls NLRP3 inflammasome activation in liver inflammatory injury. Cell Mol Immunol. 2020;17:1245–1256. doi: 10.1038/s41423-019-0318-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun S.C. The non-canonical NF-κB pathway in immunity and inflammation. Nat Rev Immunol. 2017;17:545–558. doi: 10.1038/nri.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao J., Qin J., Qian X., et al. Lipopolysaccharide preconditioning protects hepatocytes from ischemia/reperfusion injury (IRI) through inhibiting ATF4-CHOP pathway in mice. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang C., Wang Z., Hu Y., et al. Hyperglycemia-triggered ATF6-CHOP pathway aggravates acute inflammatory liver injury by β-catenin signaling. Cell Death Discov. 2022;8:115. doi: 10.1038/s41420-022-00910-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsung A. The nuclear factor HMGB1 mediates hepatic injury after murine liver ischemia-reperfusion. J Exp Med. 2005;7:1135–1143. doi: 10.1084/jem.20042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhai Y., Petrowsky H., Hong J.C., et al. Ischaemia-reperfusion injury in liver transplantation: from bench to bedside. Nat Rev Gastroenterol Hepatol. 2013;10:79–89. doi: 10.1038/nrgastro.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu L., Yue S., Jiang L., et al. Myeloid Notch1 deficiency activates the RhoA/ROCK pathway and aggravates hepatocellular damage in mouse ischemic livers. Hepatology. 2018;67:1041–1055. doi: 10.1002/hep.29593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shan B., Wang X., Wu Y., et al. The metabolic ER stress sensor IRE1α suppresses alternative activation of macrophages and impairs energy expenditure in obesity. Nat Immunol. 2017;18:519–529. doi: 10.1038/ni.3709. [DOI] [PubMed] [Google Scholar]
- 28.Raines L.N., Zhao H., Wang Y., et al. PERK is a critical metabolic hub for immunosuppressive function in macrophages. Nat Immunol. 2022;23:431–445. doi: 10.1038/s41590-022-01145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dasgupta D., Nakao Y., Mauer A.S., et al. IRE1A stimulates hepatocyte-derived extracellular vesicles that promote inflammation in mice with steatohepatitis. Gastroenterology. 2020;159:1487–1503.e17. doi: 10.1053/j.gastro.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bronner D.N., Abuaita B.H., Chen X., et al. Endoplasmic reticulum stress activates the inflammasome via NLRP3- and Caspase-2-driven mitochondrial damage. Immunity. 2015;43:451–462. doi: 10.1016/j.immuni.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X., Shi C., He M., et al. Endoplasmic reticulum stress: molecular mechanism and therapeutic targets. Sig Transduct Target Ther. 2023;8:352. doi: 10.1038/s41392-023-01570-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guan Q., Wang Z., Hu K., et al. Melatonin ameliorates hepatic ferroptosis in NAFLD by inhibiting ER stress via the MT2/cAMP/PKA/IRE1 signaling pathway. Int J Biol Sci. 2023;19:3937–3950. doi: 10.7150/ijbs.85883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qu X., Yang T., Wang X., et al. Macrophage RIPK3 triggers inflammation and cell death via the XBP1-Foxo1 axis in liver ischaemia-reperfusion injury. JHEP Rep. 2023;5 doi: 10.1016/j.jhepr.2023.100879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loo Y.M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loi P., Yuan Q., Torres D., et al. Interferon regulatory factor 3 deficiency leads to interleukin-17-mediated liver ischemia-reperfusion injury. Hepatology. 2013;57:351–361. doi: 10.1002/hep.26022. [DOI] [PubMed] [Google Scholar]
- 36.Xu G., Shi Y. Apoptosis signaling pathways and lymphocyte homeostasis. Cell Res. 2007;17:759–771. doi: 10.1038/cr.2007.52. [DOI] [PubMed] [Google Scholar]
- 37.Kilkenny C., Browne W.J., Cuthill I.C., et al. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Epps H.V., Astudillo O., Martin Y.D.P., et al. The Sex and Gender Equity in Research (SAGER) guidelines: implementation and checklist development. European Science Editing. 2022;48 [Google Scholar]
- 39.Fang X., Zhang J., Li Y., et al. Malic enzyme 1 as a novel anti-ferroptotic regulator in hepatic ischemia/reperfusion injury. Adv Sci. 2023;10 doi: 10.1002/advs.202205436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong J., Ke M.Y., Wu X.N., et al. SRY is a key mediator of sexual dimorphism in hepatic ischemia/reperfusion injury. Ann Surg. 2022;276:345. doi: 10.1097/SLA.0000000000004422. [DOI] [PubMed] [Google Scholar]
- 41.Rao J., Yue S., Fu Y., et al. ATF6 mediates a pro-inflammatory synergy between ER stress and TLR activation in the pathogenesis of liver ischemia-reperfusion injury. Am J Transplant. 2014;14:1552–1561. doi: 10.1111/ajt.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao J., Qian X., Li G., et al. ATF3-mediated NRF2/HO-1 signaling regulates TLR4 innate immune responses in mouse liver ischemia/reperfusion injury. Am J Transplant. 2015;15:76–87. doi: 10.1111/ajt.12954. [DOI] [PubMed] [Google Scholar]