Abstract
Circulating tumor DNA (ctDNA) detection has multiple promising applications in oncology, but the road toward implementation in clinical practice is unclear. We aimed to support the implementation process by exploring potential future pathways of ctDNA testing. To do so, we studied four ctDNA‐testing applications in two cancer types and elicited opinions from 30 ctDNA experts in the Netherlands. Our results showed that the current available evidence differed per application and cancer type. Tumor profiling and monitoring treatment response were found most likely to be implemented in non‐small cell lung cancer (NSCLC) within 5 years. For colorectal cancer, applications of ctDNA testing were found to be at an early stage in the implementation process. Demonstrating clinical utility was found a key aspect for successful implementation, but there was no consensus regarding the evidence requirements. The next step toward implementation is to define how clinical utility of biomarkers should be evaluated. Finally, these data indicate that specific challenges for each clinical application and tumor type should be appropriately addressed in a deliberative process involving all stakeholders to ensure implementation of ctDNA testing and timely access for patients.
Keywords: clinical utility, ctDNA, HTA, implementation, oncology, scenarios
Analysis of circulating tumor DNA (ctDNA) is a promising technology with several applications in oncology. We identified the main challenges and opportunities to inform the implementation process in the Netherlands by eliciting opinions of experts. Next steps toward implementation of ctDNA is to define the evidence to prove clinical utility and to address the specific challenges for each application.
Abbreviations
- cieBOD
Commissie Beoordeling Diagnostiek (Dutch advisory committee for diagnostics)
- cieBOM
Commissie Beoordeling Oncologische Middelen (Dutch advisory committee for oncological agents)
- COIN
ctDNA on the road to implementation in the Netherlands
- CRC
colorectal cancer
- ctDNA
circulating tumor DNA
- DCCG
Dutch Colorectal Cancer Group
- DORP
Dutch Oncology Research Platform
- eHTA
early health technology assessment
- HTA
health technology assessment
- MRD
minimal residual disease
- NSCLC
non‐small cell lung cancer
- PLCRC
Prospective Dutch Colorectal Cancer Cohort
- RCT
randomized clinical trial
1. Introduction
The use of biomarkers to detect and characterize cancer evolution over time is important to improve treatment decision‐making and intervene in the progression of disease [1, 2]. While microscopic evaluation of tumor tissue biopsies is the gold standard for cancer diagnostics, detection of cell‐free circulating tumor DNA (ctDNA) in liquid biopsies is a promising new technology that enables biomarker identification in a minimally invasive way [3]. CtDNA is composed of fragments of tumoral DNA present in blood or other body fluids, and it provides quantitative and qualitative information about a patient's tumor [4, 5]. This makes ctDNA testing useful in many different applications for a wide range of cancer types. For example, for identification of mutations to guide treatment decisions, real‐time monitoring of tumor evolution, minimal residual disease (MRD) detection, or even for screening purposes [6, 7, 8, 9, 10, 11, 12, 13]. The increasing interest in this technology is further reflected in the number of clinical studies currently including or investigating ctDNA testing, the establishment of numerous companies focused on ctDNA testing over the recent years, and the wide variety of ctDNA tests under development [14, 15]. The road for a promising technology such as ctDNA testing to reach implementation in clinical practice is long and complex [16]. Currently, the only ctDNA application included so far in clinical guidelines is the detection of resistance mechanisms in metastatic non‐small cell lung cancer (NSCLC), when tumor tissue is not available [17]. This application is currently used in routine diagnostics, albeit in a non‐coordinated way, leading to diversity in the analytical procedures amongst laboratories, and unequal access for patients to the latest diagnostic developments [18]. Therefore, elucidating the steps toward ctDNA implementation is an emerging need to bring ctDNA testing to patients in a structured way [19]. The Dutch multi‐disciplinary “CtDNA on the road to implementation in the Netherlands” (COIN)‐consortium (www.cfdna.nl/coin) is working to enable controlled, evidence‐based introduction of ctDNA testing in the Dutch healthcare system. An important task of COIN is to perform an early health technology assessment (eHTA) of ctDNA testing to comprehensively evaluate the expected impact of ctDNA testing in clinical practice [20]. HTA is often initiated in the last stages of implementation after the association between use of the test and change in health outcome (i.e. clinical utility) has been proven to inform policy and reimbursement decisions. Starting HTA earlier in the process (eHTA) helps to make evidence‐informed decisions to guide the development of the technology, design future clinical studies, and inform the implementation process [20, 21, 22]. As a result, the road toward implementation can be anticipated and steered toward timely access for patients.
However, the degree of uncertainty is higher when HTA is performed in an earlier stage, as fewer data are available [22]. Therefore, in this study, we aimed to explore potential future pathways to better understand the uncertainties, expectations, and potential barriers to plasma‐based ctDNA testing implementation. To do so, we elicited experts' opinions on developments in the field within 5 years in the Netherlands. We focused on two cancer types, NSCLC and colorectal cancer (CRC), and four different applications of ctDNA testing: early detection of cancer for screening, MRD detection, tumor profiling, and monitoring treatment evaluation. The results of this study can set the basis for future HTAs in the ctDNA field and can help elucidate the specific steps toward implementation needed in the near future.
2. Materials and methods
A scenario‐drafting study was conducted to explore future pathways (i.e. scenarios) of ctDNA testing. This methodology is based on environmental scenario analysis, which is a common practice in environmental science and policy and can also be valuable in health sciences [23, 24, 25, 26]. Scenario drafting can be used to understand and reflect upon the uncertainties about future developments in multiple dimensions, by building potential scenarios, comparing them, and evaluating their impact [23]. This study focuses on the first two features; building and comparing scenarios. It has a three‐step design (Fig. 1): First, relevant aspects that influence the implementation of ctDNA testing were identified. Second, scenarios were drafted based on these aspects. Third, the likelihood of the scenarios was elicited with an online questionnaire among ctDNA experts.
Fig. 1.
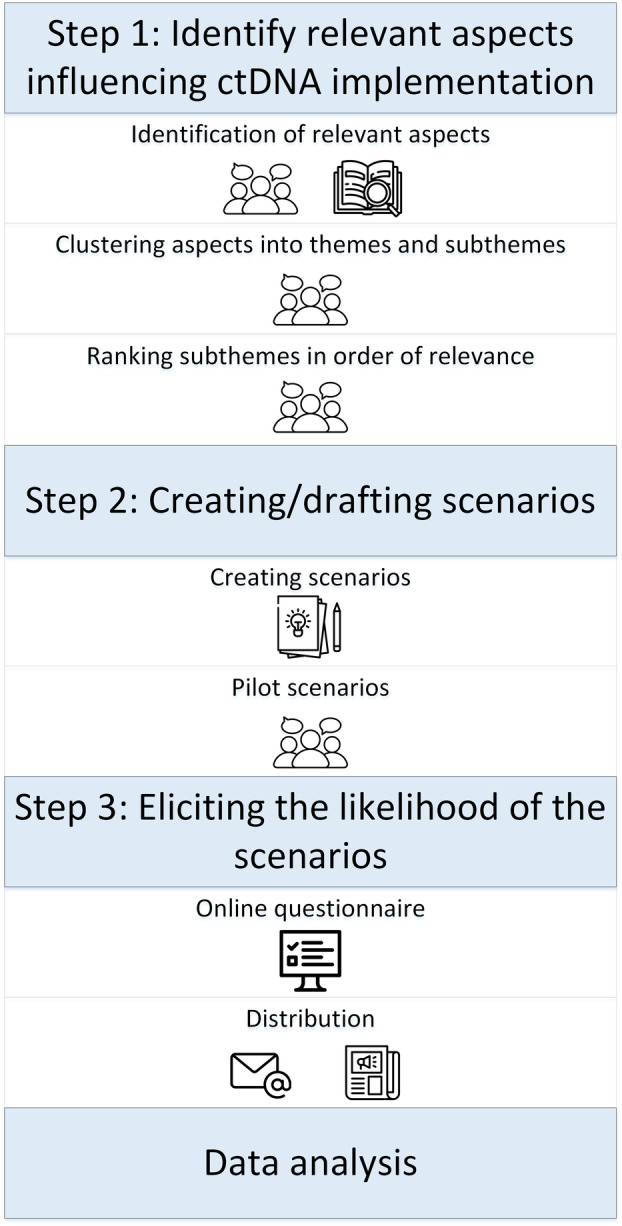
Overview of three‐step study design. Schematic visualization of the three‐step methodology for building and comparing the scenarios.
2.1. Step 1: Identification of relevant aspects and themes influencing ctDNA implementation
Focus groups were organized for the first step and included experts within the field of translational oncology, laboratory medicine, and health technology assessment who were all actively involved in ctDNA research projects. Background information regarding ctDNA implementation was gathered. First, two researchers (AK, CR) performed a scoping literature review to identify key aspects influencing ctDNA testing implementation (both barriers and facilitators), according to the methodology of Peters et al. [27, 28] (Fig. S1). Secondly, the experts verified that no important aspects were missing during the first focus group. The identified aspects were clustered into themes and subthemes and verified with the experts in a second focus group (Fig. S2). The identified main themes were: clinical utility, economic aspects, organizational aspects, technical aspects, and social aspects. Lastly, the experts ranked the subthemes in order of relevance in a final focus group.
2.2. Step 2: Scenario drafting and questionnaire development
The first list of potential scenarios regarding ctDNA testing implementation was drafted: one main scenario about successful implementation, and several scenarios per theme influencing implementation based on the most relevant subthemes found in step 1. To illustrate, an example of a scenario was: “ctDNA testing will be reimbursed within the next five years” (economic subtheme). The scenarios were complemented with additional questions to obtain more context and detailed information about each theme. This first list of scenarios was piloted among eight ctDNA‐experts, and three experts in the field of policy‐making and HTA to evaluate if all scenarios and additional questions included were relevant and uniformly interpreted. The scenarios were adapted according to their feedback.
2.3. Step 3: Eliciting the likelihood of the scenarios
2.3.1. Questionnaire
The final questionnaire was incorporated into an online survey tool (Survalyzer Next Generation). The questionnaire included 12 scenarios and 16 additional questions, divided into three parts: (1) evidence generation, (2) successful implementation, and (3) exploring different scenarios per theme (see Table 1). The first part of the questionnaire consisted of two questions to investigate the awareness about the current stage of evidence for the different ctDNA applications and evidence needed to prove clinical utility in the Netherlands, in order to create context for the scenarios proposed in the second and third parts of the questionnaire. The second part of the questionnaire included the main scenario about successful implementation and investigated the general challenges of successful implementation. “Successful implementation” was defined as the ideal situation in which five theme‐specific scenarios are achieved (representing the five themes found in step 1): (1) The test is included in the clinical guidelines (clinical utility), (2) The costs of the test are reimbursed (economical), (3) Analytical procedures for ctDNA analysis are harmonized (pre‐analytical, analytical, reporting) (technical), (4) All logistics are in place so all patients have access to the test (organizational), (5) The test is offered to all patients who can benefit from ctDNA testing (social). The third part included theme‐specific scenarios and questions to explore specific challenges regarding ctDNA implementation. The complete questionnaire can be found in Appendix S1. The likelihood of the scenarios occurring within 5 years was elicited by using a sliding scale from 0% to 100% (0% = will definitely not occur, 100% = will definitely occur). Additional questions consisted of different types of questions (open, dichotomous, multiple choice, rank‐order scaling, and Likert‐scale). Respondents filled in the questionnaire for NSCLC or CRC only, depending on their expertise, and they could skip questions if they did not feel comfortable answering them. When considered necessary, answers were requested per clinical application of ctDNA testing (monitoring treatment response, target profiling, MRD detection, and early detection/screening). The definitions of the applications included in the questionnaire can be found in Table 2.
Table 1.
Online questionnaire: outline and included themes.
| 1. Evidence generation |
| Awareness about the current stage of evidence |
| Future evidence: Exploring type of evidence needed to prove clinical utility |
| 2. Main implementation scenario |
| Successful implementation of ctDNA |
| Challenges regarding successful implementation |
| 3. Theme‐specific scenarios |
| Clinical utility |
| Inclusion in clinical guidelines |
| ctDNA vs current practice |
| Economical aspects |
| Cost of test |
| Reimbursement/funding systems |
| Organizational aspects |
| Hospital level |
| National level |
| Technical aspects |
| Analytical aspects |
| Competition other biomarkers |
| Social aspects |
| Health care professionals perspective |
| Patient perspective |
Table 2.
Definition of the clinical applications of ctDNA testing.
| Clinical application ctDNA testing | Definition |
|---|---|
| Monitoring treatment response | Evaluating the response to treatment over time with serial liquid biopsies to detect disease progression during systemic treatment (chemotherapy, targeted therapy, etc.) |
| Tumor profiling | Detect specific mutations in clinically‐relevant targets in liquid biopsies with a single test to guide treatment decisions |
| Minimal residual disease (MRD) detection | Detect presence of ctDNA in liquid biopsies to improve risk stratification and guide adjuvant treatment decisions after treatment with curative intent (e.g. surgery) |
| Early detection/screening | Detect cancer in liquid biopsies at the earliest possible stage to have the best chance for a successful treatment |
2.3.2. Distributing the questionnaire
The target population was experts working in the field of ctDNA testing. The complete questionnaire was distributed via e‐mail and online newsletters from research groups in oncology (Dutch Colorectal Cancer Group; DCCG, and Dutch Oncology Research Platform; DORP) between July and October 2021. Prior to distribution, this study was presented during a COIN consortium meeting in June 2021 to attract respondents. Email recipients included members of the COIN network, members of the researchers' network, and authors of relevant work in the field (127 recipients from 23 Dutch institutes). Recipients were encouraged to forward the questionnaire to their network of ctDNA experts. Reminders were sent to all the recipients of the e‐mail after 2 weeks. All respondents were asked for informed consent before filling in the questionnaire. Respondents who indicated to have no experience in the field of ctDNA testing were excluded from further analysis.
2.3.3. Ethical approval and consent to participate
This study was reviewed and approved by the Institutional Review Board (IRB) from the Netherlands Cancer Institute under number IRBd23‐063. Informed consent was inferred upon completion of the survey and approved by the IRB from the Netherlands Cancer Institute. All methods were carried out in accordance with relevant guidelines.
2.4. Step 4: Data analysis
All responses were anonymized. The data were analyzed in Microsoft Excel (version 2016). Descriptive statistics were used for most questions (frequencies and percentages). Median, minimum, and maximum were estimated from the pooled likelihood estimates of the scenarios. Open additional questions were analyzed by inductive coding of the responses [29]. For rank‐order scaling questions, the ranking positions were scored as follows: rank 1 = 5 points, rank 2 = 4 points, rank 3 = 3 points, rank 4 = 2 points, rank 5 = 1 point. Then, weighted averages were calculated by dividing the total sum of the scores of all respondents by the number of respondents, for each answer. Visualization of the data was performed in Microsoft Excel (version 2016) and using r package “ggplot” in rstudio (https://cran.r‐project.org/web/packages/ggplot2/index.html, r version 4.1.2). Missing answers were excluded from the analysis.
3. Results
3.1. Participant characteristics
Thirty ctDNA‐experts completed the online questionnaire (18 NSCLC‐experts, and 12 CRC‐experts, see Table 3). Most experts had a clinical or technical background and worked in an academic hospital or specialized cancer center, with an average experience with ctDNA testing of 4.9 years. More information about the respondents can be found in Table S1.
Table 3.
Respondent characteristics.
| N | |
|---|---|
| Number of respondents | |
| NSCLC | 18 |
| CRC | 12 |
| Profession | |
| Laboratory specialist | 11 |
| Clinical researcher | 10 |
| Fundamental researcher | 4 |
| Clinical scientist in molecular pathology | 3 |
| Policy maker | 1 |
| HTA‐researcher/Health economist | 1 |
| Place of employment | |
| Academic hospital | 17 |
| Specialized cancer center | 9 |
| General hospital | 2 |
| Healthcare insurance company | 1 |
| University | 1 |
| Years of experience with ctDNA | |
| Mean average | 4.9 years |
3.2. Evidence generation
The awareness about the current stage of evidence in the implementation process in the Netherlands differs among clinical applications of ctDNA testing and per tumor type (see Fig. 2). Of the four clinical applications, tumor profiling was considered to be at the highest stage of evidence generation in both tumor types, with clinical utility proven for both cancer types, and considered ready to use in the clinic for NSCLC. Monitoring response to treatment was also considered ready for use in the clinic by NSCLC experts. For, MRD detection was at the evidence stage of clinical validity in both cancer types. Early detection/screening was considered to be at the lowest stage of evidence generation for both cancer types, with strong consensus.
Fig. 2.
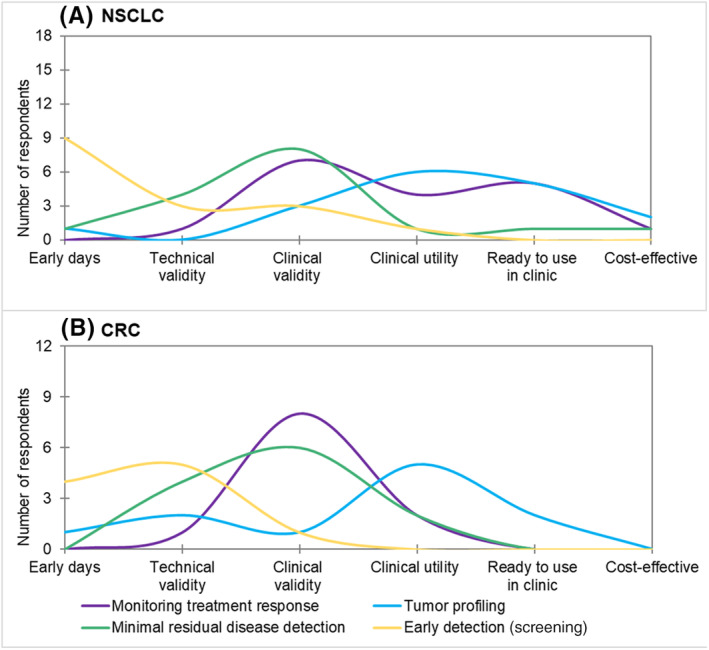
Awareness about the current stage of evidence of the different applications for ctDNA testing in the Netherlands. The y‐axis shows the total number of answers is shown, the upper limit of the y axis is the total number of respondents per tumor type. In the x‐axis shows the stages of evidence [30]. (A) Results for NSCLC. (B) Results for CRC. (1) Early days: new liquid biopsy test is developed. (2) Technical validity: ability to detect and quantify a molecular aberration. (3) Clinical validity: correlation with a clinical outcome such as prognostic value for overall survival. (4) Clinical utility: ability of the liquid biopsy to actually guide treatment decisions that improve clinical outcomes. (5) Ready to use in clinic: level of evidence where clinicians feel the test is ready for use. (6) Cost‐effective: demonstration of an economically viable test relative to the clinical benefit.
Regarding which evidence is necessary to prove clinical utility, the answers differed per application. Notably, the need for large cohort studies was a common answer, while randomized clinical trials (RCTs) were not always considered necessary to prove clinical utility. The suggested endpoints of these large‐cohort studies were both long‐term measures; such as survival benefit for monitoring treatment response and MRD detection, and short‐term measures; such as technical performance and concordance of the test compared to current diagnostic tests for monitoring response to treatment and tumor profiling. For early detection/screening, there was consensus about the need to compare the ctDNA testing approach to the current screening method (e.g. CRC screening program).
3.3. Main scenario: successful implementation
Regarding the main scenario concerning “successful implementation of ctDNA”, NSCLC experts considered tumor profiling and monitoring response to treatment as the respective applications most likely to be successfully implemented in 5 years, with 74% and 82% median likelihoods of achieving the scenario. For all the applications in CRC, the median likelihoods were lower compared to NSCLC and the range of answers was larger. Early detection/screening was seen as the clinical application most distant from successful implementation in both tumor types (NSCLC: 25%, CRC: 4.5% median likelihood) (Fig. 3A and Table S2).
Fig. 3.
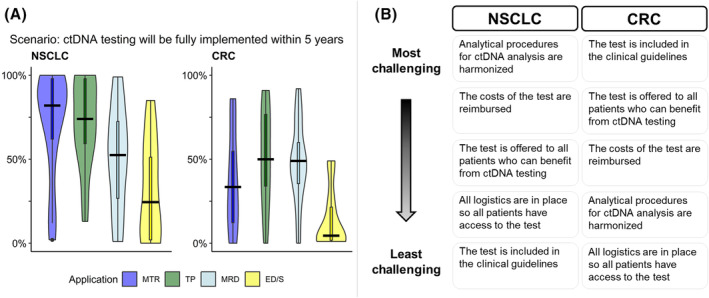
Successful implementation of ctDNA testing: likelihood and main challenges. (A) Results of the scenario per cancer type. Y‐axis shows the 5‐year likelihood between 0% and 100% according to the experts. Black line shows the median. (B) Ranking of the challenges to achieve successful implementation. ED/S, early detection/screening; MTR, monitoring treatment response; TP, tumor profiling.
The order of the ranking from most to least challenging to achieve of the five theme‐specific scenarios differed between CRC and NSCLC (Fig. 3B and Fig. S3). While inclusion in the clinical guidelines was seen as the most challenging scenario by CRC experts, it was seen as the least challenging by NSCLC experts. Having the logistics in place was not considered challenging according to both NSCLC and CRC experts (CRC: ranked 5th, NSCLC: ranked 4th).
3.4. Theme‐specific scenario's
Estimations of the likelihood of in total 11 theme‐specific scenarios were elicited. An overview of elicited likelihoods and median likelihood per scenario and per cancer type can be found in Fig. 4 and Table S2.
Fig. 4.
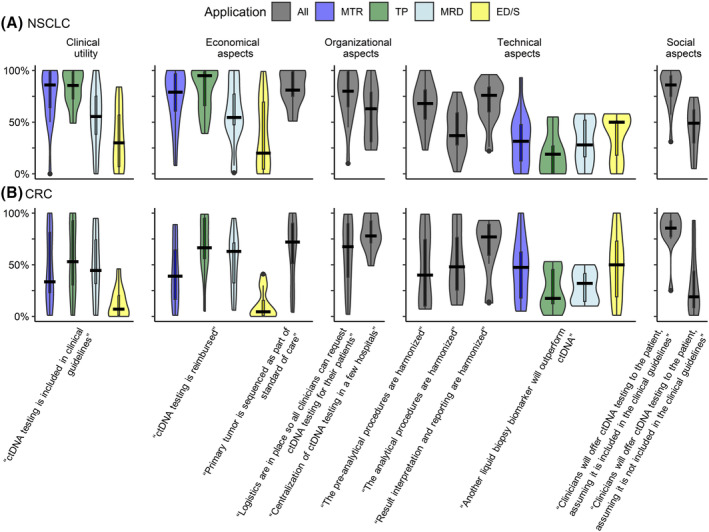
Overview results of 5‐year likelihood per scenario. (A, B) 5‐year likelihood of scenarios regarding ctDNA testing implementation per clinical application. (A) Result for NSCLC. (B) Results for CRC. X‐axis shows the scenarios per theme, y‐axis shows the likelihood between 0 and 100%. Black line shows the median in each scenario. All, all applications; ED/S, early detection/screening; MTR, monitoring treatment response; TP, tumor profiling.
3.4.1. Clinical utility
The median likelihood of the scenario “ctDNA testing will be included in clinical guidelines in the next five years” showed that the respondents considered this scenario most likely to occur for monitoring treatment response and tumor profiling in NSCLC (both 86% median likelihood). In CRC, the answers were widely distributed. In both cancer types, there was consensus about ctDNA testing not being included in guidelines within 5 years for early detection/screening. Additionally, we explored the added value and positioning of ctDNA testing with respect to current standard diagnostic procedures. According to the respondents, ctDNA testing will be used in clinical practice within 5 years along with current diagnostic procedures, and the results will be used in clinical decision‐making (Figs S4 and S5). NSCLC experts deemed that ctDNA testing would significantly improve survival in all applications except early detection/screening, and in CRC this was only indicated for MRD detection (Table S3). Respondents also indicated that the minimally invasive character of liquid biopsies and the possibility of solving an unmet clinical need could also lead to the inclusion of ctDNA testing in guidelines if survival benefit is not yet proven (Table S4).
3.4.2. Economical aspects
The median likelihood of the scenario “ctDNA testing will be reimbursed in the next five years” was highest for tumor profiling and monitoring response to treatment in NSCLC (95% and 79% median likelihood, respectively) and for monitoring response to treatment and MRD detection in CRC (66.5% and 63% median likelihood, respectively). We explored the scenario: “The primary tumor will be sequenced as part of standard of care within five years”, which would limit the costs of ctDNA testing to plasma testing only. Respondents found this scenario likely to occur (NSCLC: 81%, CRC: 72% median likelihood). In addition, most respondents considered it likely that costs of ctDNA testing will decrease or remain stable (63% and 27% of the respondents, respectively) (Fig. S6), and 90% of the respondents with knowledge about the budget for diagnostic procedures indicated that there were currently budget restrictions for ctDNA testing (Fig. S7).
3.4.3. Organizational aspects
The likelihood of the scenario “Logistics are in place so every patient has access to the test in five years” was considered high according to the respondents (median likelihood NSCLC: 81%, CRC: 67.5%). Other suggested requirements to ensure patient access were: reimbursement, education, use of national existing logistics, and the need for communication with hospitals and patients (Table S5). The second organizational scenario “ctDNA testing will be centralized in a few hospitals in the next five years” was deemed likely for most of the respondents, with high consensus for CRC specialists, but a large variety in the opinions among NSCLC experts (median likelihood NSCLC: 57%, CRC: 78%). Cost reduction, increased expertise, and scale efficiencies were seen as the main advantage of centralization, but complex logistics and a possible increase in the turnaround time were mentioned as the main disadvantages (Table S6). Additionally, the possibility of centralization in a single center was considered not helpful for implementation by 80% of the respondents (Fig. S8).
3.4.4. Technical aspects
The likelihood of the scenarios that “ctDNA analysis will be harmonized on a national level in the next five years” for each part of the analysis (pre‐analytical, analytical, and post‐analytical procedures) was explored. Harmonization of post‐analytical procedures (i.e. results interpretation and reporting) was considered likely to occur (median likelihood NSCLC: 76%, CRC: 77%), and harmonization of analytical procedures was considered least likely (median likelihood NSCLC: 37%, CRC: 48%). The likelihood of the scenario that “another liquid biopsy biomarker will outperform ctDNA within five years” was considered unlikely (median likelihoods: 19–50%).
3.4.5. Social aspects
The likelihood of the scenario “clinicians will offer ctDNA testing to the patient within five years” was considered higher if the test is included in the clinical guidelines (NSCLC: 86%, CRC 85.5%) compared to when ctDNA testing is not included in guidelines (NSCLC: 49%, CRC: 19%). Most ctDNA experts agreed that patients would prefer ctDNA testing over current diagnostic methods (Fig. S9).
4. Discussion
In this study, we investigated the opinions and expectations of 30 ctDNA‐experts on 12 future scenarios to explore the current status, future developments, and requirements for implementation of ctDNA testing for oncology in the Netherlands. The applications which were considered to have the highest stage of evidence in the implementation process in the Netherlands were also considered more likely to be implemented within 5 years. Early detection/screening was considered least likely to be implemented within this time frame. Based on the results, we identified that demonstrating clinical utility is the main facilitator for ctDNA implementation in Dutch clinical practice; as it could accelerate implementation on multiple levels. However, the type of evidence required to demonstrate clinical utility was unclear and differed per indication and tumor type. We also found that the challenges regarding implementation differed amongst clinical applications and tumor types, for example, inclusion of ctDNA testing in clinical guidelines and centralization of ctDNA analysis. Therefore, to achieve optimal implementation of ctDNA testing in clinical practice, it seems that the implementation process of each clinical application of ctDNA testing should be evaluated per tumor type in context of the clinical application.
The evaluation of diagnostic tests for the implementation in clinical practice has been subject of discussion for many years [6]. The present study confirmed that it is critical to demonstrate clinical utility to achieve successful implementation, in line with the conclusions of a similar study in the Australian setting [30]. In addition, we found that the specific evidence required (i.e. type of study, type of evidence, definition of “sufficient” evidence) is not yet clearly defined for diagnostic tests. For new oncological drugs, the evaluation criteria and endpoints are clearly defined by the Dutch advisory committee for oncological agents “Commissie Beoordeling Oncologische Middelen” (cieBOM) [31]. However, it remains ambiguous which criteria a diagnostic test must meet and which endpoints should be evaluated to demonstrate its clinical utility [7]. Moreover, new drugs need to be investigated in RCTs, but successfully completing an RCT for the investigation of biomarkers is arduous [32]. Our results suggest that evidence from traditional RCTs with long follow‐up (level 1 evidence) is not always considered necessary to demonstrate clinical utility of ctDNA testing, which could save time and costs in evidence generation. Alternatives for RCTs that have been suggested in literature for evaluating the clinical utility of a biomarker are real‐world cohorts, prospective‐retrospective studies, or decision models [32, 33, 34, 35]. Recently, the advisory committee “Commissie Beoordeling Diagnostiek” (cieBOD) was established in the Netherlands to advise on the effectiveness and positioning of new biomarkers to accelerate implementation [36]. This advisory committee could be involved in a deliberative process with all stakeholders (e.g. regulatory bodies, patient advocates, clinicians, etc.) to evaluate if any endpoints besides survival benefits can also be considered in their advice. By defining the specific evidence required to prove clinical utility and the evaluation criteria, clinical studies can be designed more optimally to obtain the type of data needed, which will bring new diagnostic developments to the patient faster.
In general, ctDNA testing was considered more likely to be implemented in NSCLC than in CRC in the next 5 years for all the applications. In addition, we found that CRC experts considered inclusion in clinical guidelines the biggest challenge for successful implementation; even more than ensuring reimbursement, while NSCLC experts consider it least challenging. These differences could be explained by the different clinical needs per tumor type. In NSCLC, there is a clinical need for mutational predictive testing as there are several approved targeted treatments whose administration is based on molecular profiling. As obtaining a tumor tissue biopsy is challenging in ~15% of patients and associated with complications, the use of ctDNA has high clinical need [37]. This promoted the execution of relevant clinical studies with clear endpoints and expedited the inclusion of ctDNA testing in clinical guidelines for metastatic NSCLC, and could explain the more optimistic attitude toward ctDNA testing of NSCLC experts [38, 39, 40, 41]. In contrast, there are currently limited targeted treatment options for CRC, and tumor tissue biopsies are more easily available. Arguably, this resulted in fewer large cohort studies focused on ctDNA analysis and fewer data available for CRC. As the field of personalized medicine evolves, the expectation is that more targeted therapies will become available for CRC as well in the coming years [42], and liquid biopsies testing will also become a minimally invasive option for the new clinical need of profiling the tumor of these patients.
One of the clear differences observed in the results was that CRC experts considered centralization more likely than NSCLC experts. Possibly, this can be explained by the fact that in CRC, blood samples are already collected and stored centrally as part of ongoing substudies of a national cohort study (Prospective Dutch Colorectal Cancer Cohort, PLCRC) [35]. Contrarily, in metastatic NSCLC at least 9 hospitals are already analyzing ctDNA in their diagnostic laboratories for monitoring progression while on targeted therapies and the detection of resistant mechanisms [43]. Centralization of ctDNA testing in a few hospitals regionally in the coming years would facilitate harmonization and would render other advantages mainly derived from scale efficiencies (e.g. lower costs, increased expertise). Even though the most commonly mentioned disadvantage of centralization in this study was that the turnaround time would increase, recent research in Dutch setting has shown that a minimum number of samples needs to be analyzed weekly in a hospital to make the cost per sample acceptable [44]. Currently, the volume per laboratory is low, so regional collaboration would enable reaching the minimum number of samples in a shorter time and the suspected increase in turnaround time due to shipment of the sample would be compensated by the possibility of immediate analysis [19]. In case ctDNA testing is centralized, the analysis and result interpretation could be performed in a central (regional) molecular laboratory with a multi‐disciplinary molecular tumor board [19, 45], and knowledge transfer must be ensured so all hospitals remain involved in the advances in the field, and expertise is not limited to the executing laboratories.
This study also has limitations. First of all, the study was designed as a first step in the implementation process of ctDNA testing to better understand the broader uncertainties, expectations, and potential barriers in multiple themes. Considering our extensive and broad questionnaire, some important topics like the choice of ctDNA testing technology could not be investigated thoroughly within this study. However, the importance of this topic was also recognized within the COIN project, so another work package has been evaluating and publishing on the pre‐analytical and analytical aspects of ctDNA testing in the Netherlands [18]. A second limitation is the timing of the survey. The survey was conducted in 2021, and new evidence have become available on the clinical utility of ctDNA testing, for amongst others for MRD detection in CRC [46, 47]. While the answers to some of the scenarios and questions would be different in case the survey was distributed more recently, we are under the impression that the identified challenges and opportunities still exist and remain a relevant for the implementation of ctDNA testing [39]. Finally, two other important limitations of this study are the number of respondents, and the intrinsic variability that eliciting opinions entails; this resulted in a wide range in the likelihood of the scenarios and the answers to the questions, which reflects the need for a multidisciplinary structured approach toward implementation. Nevertheless, we collected valuable information about views on the current and future status of ctDNA testing. Another point to take into consideration is that, while one strength of this study lies in involving all stakeholders in the development of the questionnaire, most respondents had laboratory background and/or in research. For timely implementation and adoption of ctDNA testing, it is important that the perspectives of all stakeholders are actively participating in future studies from an early stage, such as patient representatives, physicians, and policy‐makers.
5. Conclusions
In conclusion, we present a structured description of the opinions of ctDNA experts about potential future pathways for ctDNA testing in the Netherlands. It can be expected that ctDNA testing will continue to be gradually implemented in clinical practice. Challenges related to the rapid advancements in the field, as well as the specific challenges per application and tumor type should be addressed to ensure smooth implementation. The next step toward implementation is to define how clinical utility of biomarkers is demonstrated. This, and other remaining challenges, can be addressed in a deliberative process involving all stakeholders, to ultimately deliver optimal patient care.
Conflict of interest
CR‐A reports nonfinancial support from Personal Genome Diagnostics and Cergentis BV. DB has provided lectures, expert testimony, and advisory board presence, for Roche diagnostics, all outside the submitted work and all financial supports transferred to institute. GAM is co‐founder and board member (CSO) of CRCbioscreen BV, he has a research collaboration with CZ Health Insurances (cash matching to ZonMW grant), he has research collaborations with Exact Sciences, Sysmex, Sentinel Ch. SpA, Personal Genome Diagnostics (PGDX), DELFi and Hartwig Medical Foundation; these companies provide materials, equipment and/or sample/genomic analyses, and he has several patents pending/issued. GRV reported grants and/or nonfinancial support from BMS, Merck, Servier, Personal Genome Diagnostics, Bayer, Sirtex, Pierre Fabre, Lilly, Delfi Diagnostics, all outside the submitted work and all financial supports transferred to institute. ES provided lectures for Bio‐Rad, Seracare, Roche, Biocartis, Lilly, Agena Bioscience, Illumina, received provided consultation for MSD/Merck, AstraZeneca, Roche, Novartis, Bayer, BMS, Lilly, Amgen, Illumina, Agena Bioscience, CC Diagnostics, Janssen Cilag (Johnson&Johnson), Astellas Pharma, and received research grants from Abbott, Biocartis, Invitae‐ArcherDX, AstraZeneca, Bayer, Bio‐Rad, Roche, Agena Bioscience, CC diagnostics, and Boehringer Ingelheim, all outside the submitted work and all financial supports transferred to institute. RJAF reports grants and nonfinancial support from Personal Genome Diagnostics, DELFI Diagnostics and Cergentis BV; grants from MERCK BV; and nonfinancial support from Pacific Biosciences, outside the submitted work. In addition, R.J.A.F. has several patents pending. VPR received in the past 3 years an unrestricted grant from Intuitive BV, outside of the current work. The remaining authors declare no potential competing interests.
Author contributions
AK, CR‐A, DB, DCLV, IE, RJAF, VMHC, and VPR are all experts who participated in the focus groups contributed to the conceptualization. AK and CR‐A were involved in methodology, formal analysis, investigation, data curation, writing main manuscript, and visualization. VPR and VMHC were involved in methodology, supervision, project administration, and funding acquisition. DB, GAM, and RJAF were involved in funding acquisition. AK, CR‐A, DB, DCLV, IE, GAM, GRV, ES, RJAF, VMHC, and VPR were involved in interpretation of the data, reviewing and editing the manuscript, and approval of the final version.
Peer review
The peer review history for this article is available at https://www.webofscience.com/api/gateway/wos/peer‐review/10.1002/1878‐0261.13562.
Supporting information
Appendix S1. Final questionnaire ctDNA testing implementation.
Fig. S1. Flowchart scoping literature review.
Fig. S2. Identified themes.
Fig. S3. Ranking of challenges to achieve successful implementation of ctDNA testing.
Fig. S4. Positioning of ctDNA testing in the diagnostic procedures.
Fig. S5. Role of ctDNA testing in clinical decision making.
Fig. S6. Expectations about cost of ctDNA testing.
Fig. S7. Perception about budget restrictions for ctDNA testing.
Fig. S8. Opinion about centralization in one center.
Fig. S9. Opinion about patient preference for ctDNA testing over current diagnostic methods.
Table S1. Detailed respondent characteristics.
Table S2. Detailed overview of the results for all scenarios.
Table S3. Expectation about obtaining survival benefit as a result of ctDNA testing.
Table S4. Aspects different to survival benefit leading to inclusion of ctDNA testing in clinical guidelines.
Table S5. Ensuring patient access.
Table S6. Main advantages and disadvantages of centralization.
Acknowledgements
We thank Zorginstuut Nederland, CZ, and all experts for their contribution to the development of the questionnaire and all respondents for answering the questionnaire. This study was supported by scientific grants for: the COIN project, funded by the ZonMw ‘Personalized Medicine’ program (project number 848101011), PGDX and CZ, and by the Dutch Cancer Society (grant no. 11809), The Netherlands. The collaboration project is co‐funded by the PPP Allowance made available by Health~Holland, Top Sector Life Sciences & Health, to the Dutch Cancer Society to stimulate public‐private partnerships.
Astrid Kramer and Carmen Rubio‐Alarcón contributed equally to this work, and are shared first authors.
Data accessibility
The questionnaire used is provided as Appendix S1. Data from the answers to the questionnaire are available upon reasonable request from the authors.
References
- 1. Kalia M. Biomarkers for personalized oncology: recent advances and future challenges. Metabolism. 2015;64(3):S16–S21. [DOI] [PubMed] [Google Scholar]
- 2. La Thangue NB, Kerr DJ. Predictive biomarkers: a paradigm shift towards personalized cancer medicine. Nat Rev Clin Oncol. 2011;8(10):587–596. [DOI] [PubMed] [Google Scholar]
- 3. Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61(1):112–123. [DOI] [PubMed] [Google Scholar]
- 4. Volckmar AL, Sultmann H, Riediger A, Fioretos T, Schirmacher P, Endris V, et al. A field guide for cancer diagnostics using cell‐free DNA: from principles to practice and clinical applications. Genes Chromosomes Cancer. 2018;57(3):123–139. [DOI] [PubMed] [Google Scholar]
- 5. Wan JCM, Massie C, Garcia‐Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. [DOI] [PubMed] [Google Scholar]
- 6. Corcoran RB, Chabner BA. Application of cell‐free DNA analysis to cancer treatment. N Engl J Med. 2018;379(18):1754–1765. [DOI] [PubMed] [Google Scholar]
- 7. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer‐genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–484. [DOI] [PubMed] [Google Scholar]
- 8. Rolfo C, Mack PC, Scagliotti GV, Baas P, Barlesi F, Bivona TG, et al. Liquid biopsy for advanced non‐small cell lung cancer (NSCLC): a statement paper from the IASLC. J Thorac Oncol. 2018;13(9):1248–1268. [DOI] [PubMed] [Google Scholar]
- 9. Mauri D, Kamposioras K, Matthaios D, Tolia M, Nixon I, Dambrosio M, et al. Next‐generation sequencing of circulating tumor DNA can optimize second‐line treatment in RAS wild‐type metastatic colorectal cancer after progression on anti‐EGFR therapy: time to rethink our approach. Oncol Res Treat. 2022;45(4):216–220. [DOI] [PubMed] [Google Scholar]
- 10. Pantel K, Alix‐Panabières C. Liquid biopsy and minimal residual disease—latest advances and implications for cure. Nat Rev Clin Oncol. 2019;16(7):409–424. [DOI] [PubMed] [Google Scholar]
- 11. Sanz‐Garcia E, Zhao E, Bratman SV, Siu LL. Monitoring and adapting cancer treatment using circulating tumor DNA kinetics: current research, opportunities, and challenges. Sci Adv. 2022;8(4):eabi8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen M, Zhao H. Next‐generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019;13(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome‐wide cell‐free DNA fragmentation in patients with cancer. Nature. 2019;570(7761):385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol Oncol. 2022;15(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. ClinicalTrials.gov [Internet]. U.S. National Library of Medicine; 2022 [cited 2022 Oct 13]. Available from: https://clinicaltrials.gov/search?cond=cancer&intr=ctdna
- 16. Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic—implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18(5):297–312. [DOI] [PubMed] [Google Scholar]
- 17. NVALT . Niet kleincellig longcarcinoom Utrecht, the Netherlands: Federatie Medisch Specialisten. 2020. [updated 2020 Jan 24], [cited 2022 Oct 13]. Available from: https://richtlijnendatabase.nl/richtlijn/niet_kleincellig_longcarcinoom/systemische_behandeling_stadium_iv_nsclc/moleculaire_tumor_board_nsclc.html
- 18. van der Leest P, Ketelaar EM, van Noesel CJM, van den Broek D, van Boerdonk RAA, Deiman B, et al. Dutch National Round Robin Trial on plasma‐derived circulating cell‐free DNA extraction methods routinely used in clinical pathology for molecular tumor profiling. Clin Chem. 2022;68(7):963–972. [DOI] [PubMed] [Google Scholar]
- 19. Advies moleculaire diagnostiek in de oncologie. Diemen: Zorginstituut Nederland; 2021. [Google Scholar]
- 20. O'Rourke B, Oortwijn W, Schuller T. The new definition of health technology assessment: a milestone in international collaboration. Int J Technol Assess Health Care. 2020;36(3):187–190. [DOI] [PubMed] [Google Scholar]
- 21. Retel VP, Joore MA, Linn SC, Rutgers EJ, van Harten WH. Scenario drafting to anticipate future developments in technology assessment. BMC Res Notes. 2012;5:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. IJzerman MJ, Koffijberg H, Fenwick E, Krahn M. Emerging use of early health technology assessment in medical product development: a scoping review of the literature. Pharmacoeconomics. 2017;35(7):727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alcamo J. Environmental futures: the practice of environmental scenario analysis. Amsterdam: Elsevier; 2008. [Google Scholar]
- 24. Lindenberg M, Retel V, Rohaan M, van den Berg J, Haanen J, van Harten W. Evaluating different adoption scenarios for TIL‐therapy and the influence on its (early) cost‐effectiveness. BMC Cancer. 2020;20(1):712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Joosten SEP, Retel VP, Coupe VMH, van den Heuvel MM, van Harten WH. Scenario drafting for early technology assessment of next generation sequencing in clinical oncology. BMC Cancer. 2016;16:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van de Ven M, Simons MJ, Koffijberg H, Joore MA, IJzerman MJ, Retèl VP, et al. Whole genome sequencing in oncology: using scenario drafting to explore future developments. BMC Cancer. 2021;21(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peters MD, Marnie C, Tricco AC, Pollock D, Munn Z, Alexander L, et al. Updated methodological guidance for the conduct of scoping reviews. JBI Evid Synth. 2020;18(10):2119–2126. [DOI] [PubMed] [Google Scholar]
- 28. Munn Z, Peters MD, Stern C, Tufanaru C, McArthur A, Aromataris E. Systematic review or scoping review? Guidance for authors when choosing between a systematic or scoping review approach. BMC Med Res Methodol. 2018;18(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thomas DR. A general inductive approach for qualitative data analysis. Auckland: School of Population Health UoA; 2003. [Google Scholar]
- 30. IJzerman MJ, de Boer J, Azad A, Degeling K, Geoghegan J, Hewitt C, et al. Towards routine implementation of liquid biopsies in cancer management: it is always too early, until suddenly it is too late. Diagnostics (Basel). 2021;11(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. BOM Utrecht, the Netherlandse . Nederlandse Vereniging voor Medische Oncologie [cited 2022 Oct 13]. Available from: https://www.nvmo.org/over‐de‐adviezen/
- 32. Bossuyt PM, Reitsma JB, Linnet K, Moons KG. Beyond diagnostic accuracy: the clinical utility of diagnostic tests. Clin Chem. 2012;58(12):1636–1643. [DOI] [PubMed] [Google Scholar]
- 33. Cook NR. Quantifying the added value of new biomarkers: how and how not. Diagn Progn Res. 2018;2(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang W, Zhang Z, Krushkal J, Liu A. Group testing can improve the cost‐efficiency of prospective‐retrospective biomarker studies. BMC Med Res Methodol. 2021;21(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Derksen JW, Vink GR, Elferink MA, Roodhart JM, Verkooijen HM, van Grevenstein WM, et al. The Prospective Dutch Colorectal Cancer (PLCRC) cohort: real‐world data facilitating research and clinical care. Sci Rep. 2021;11(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Venrooij T. Behoefte aan overzicht noodzakelijke genetische testen is groot. Amsterdam: Oncologie Vandaag; 2022 [cited 2022 Dec 01]. Available from: https://oncologievandaag.nl/artikel/moleculaire‐oncologie/behoefte‐aan‐overzicht‐noodzakelijke‐genetische‐testen‐is‐groot/ [Google Scholar]
- 37. Otto S, Mensel B, Friedrich N, Schäfer S, Mahlke C, von Bernstorff W, et al. Predictors of technical success and rate of complications of image‐guided percutaneous transthoracic lung needle biopsy of pulmonary tumors. PLoS One. 2015;10(4):e0124947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heitzer E, van den Broek D, Denis M, Hofman P, Hubank M, Mouliere F, et al. Recommendations for a practical implementation of circulating tumor DNA mutation testing in metastatic non‐small‐cell lung cancer. ESMO Open. 2022;7(2):100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pascual J, Attard G, Bidard F‐C, Curigliano G, De Mattos‐Arruda L, Diehn M, et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2022;33:750–768. [DOI] [PubMed] [Google Scholar]
- 40. Planchard D, Popat S, Kerr K, Novello S, Smit E, Faivre‐Finn C, et al. Metastatic non‐small cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow‐up. Ann Oncol. 2018;29:iv192–iv237. [DOI] [PubMed] [Google Scholar]
- 41. Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Mol Diagn. 2018;20(2):129–159. [DOI] [PubMed] [Google Scholar]
- 42. Xie Y‐H, Chen Y‐X, Fang J‐Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther. 2020;5(1):1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fairley JA, Cheetham MH, Patton SJ, Rouleau E, Denis M, Dequeker E, et al. Results of a worldwide external quality assessment of cfDNA testing in lung cancer. BMC Cancer. 2022;22(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kramer A, Schuuring E, Vessies DC, van der Leest P, Geerlings MJ, Rozendal P, et al. A micro‐costing framework for circulating tumor DNA testing in Dutch clinical practice. J Mol Diagn. 2022;25:36–45. [DOI] [PubMed] [Google Scholar]
- 45. Koopman B, Groen HJ, Ligtenberg MJ, Grünberg K, Monkhorst K, Langen AJ, et al. Multicenter comparison of molecular tumor boards in the Netherlands: definition, composition, methods, and targeted therapy recommendations. Oncologist. 2021;26(8):e1347–e1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tie J, Cohen JD, Lahouel K, Lo SN, Wang Y, Kosmider S, et al. Circulating tumor DNA analysis guiding adjuvant therapy in stage II colon cancer. N Engl J Med. 2022;386:2261–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kotani D, Oki E, Nakamura Y, Yukami H, Mishima S, Bando H, et al. Molecular residual disease and efficacy of adjuvant chemotherapy in patients with colorectal cancer. Nat Med. 2023;29(1):127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Final questionnaire ctDNA testing implementation.
Fig. S1. Flowchart scoping literature review.
Fig. S2. Identified themes.
Fig. S3. Ranking of challenges to achieve successful implementation of ctDNA testing.
Fig. S4. Positioning of ctDNA testing in the diagnostic procedures.
Fig. S5. Role of ctDNA testing in clinical decision making.
Fig. S6. Expectations about cost of ctDNA testing.
Fig. S7. Perception about budget restrictions for ctDNA testing.
Fig. S8. Opinion about centralization in one center.
Fig. S9. Opinion about patient preference for ctDNA testing over current diagnostic methods.
Table S1. Detailed respondent characteristics.
Table S2. Detailed overview of the results for all scenarios.
Table S3. Expectation about obtaining survival benefit as a result of ctDNA testing.
Table S4. Aspects different to survival benefit leading to inclusion of ctDNA testing in clinical guidelines.
Table S5. Ensuring patient access.
Table S6. Main advantages and disadvantages of centralization.
Data Availability Statement
The questionnaire used is provided as Appendix S1. Data from the answers to the questionnaire are available upon reasonable request from the authors.


