Abstract
Even before the first vertebrates appeared on our planet, the aryl hydrocarbon receptor (AHR) gene was present to carry out one or more critical life functions. The vertebrate AHR then evolved to take on functions of detecting and responding to certain classes of environmental toxicants. These environmental pollutants include polycyclic aromatic hydrocarbons (e.g., benzo[a]pyrene), polyhalogenated hydrocarbons, dibenzofurans, and the most potent small-molecular-weight toxicant known, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD or dioxin). After binding of these ligands, the activated AHR translocates rapidly from the cytosol to the nucleus, where it forms a heterodimer with aryl hydrocarbon nuclear translocator, causing cellular responses that lead to toxicity, carcinogenesis, and teratogenesis. The nuclear form of the activated AHR/aryl hydrocarbon nuclear translocator complex is responsible for alterations in immune, endocrine, reproductive, developmental, cardiovascular, and central nervous system functions whose mechanisms remain poorly understood. Here, we show that the second messenger, cAMP (an endogenous mediator of hormones, neurotransmitters, and prostaglandins), activates the AHR, moving the receptor to the nucleus in some ways that are similar to and in other ways fundamentally different from AHR activation by dioxin. We suggest that this cAMP-mediated activation may reflect the true endogenous function of AHR; disruption of the cAMP-mediated activation by dioxin, binding chronically to the AHR for days, weeks, or months, might be pivotal in the mechanism of dioxin toxicity. Understanding this endogenous activation of the AHR by cAMP may help in developing methods to counteract the toxicity caused by numerous environmental and food-borne toxic chemicals that act via the AHR.
Keywords: protein kinase A, nuclear translocation, dioxin toxicity, cAMP signaling, aryl hydrocarbon receptor physiology
The aryl hydrocarbon receptor (AHR) is an ancient protein that evolved >550 million years ago and is conserved in vertebrates and invertebrates, indicating its important function through evolution (1). AHR is a member of the basic helix-loop-helix (bHLH)/Per-ARNT-Sim (PAS) homology domain family of transcriptional regulators involved in homeostatic response to hypoxia, circadian rhythm, and cellular differentiation (2-4). The gene product of the Drosophila spineless (ss) gene, the closest known relative to the mammalian AHR gene, controls development of the antennae, bristles, and tarsal regions of the legs (5). Unlike the invertebrate family members, AHR conditionally binds exogenous ligands, including carcinogenic and/or teratogenic aromatic and halogenated hydrocarbons found in air pollution, cigarette smoke, or foods (e.g., dioxin or benzo[a]pyrene) (6). Ligand-free AHR is predominantly cytoplasmic or nucleocytoplasmic, depending on the cell type (7-9). Exposure to dioxin leads to abundant nuclear translocation of AHR, heterodimerization with ARNT (AHR-nuclear translocator), and activation of many genes, including several that encode xenobiotic/carcinogen metabolizing enzymes such as cytochrome P450 (CYP) 1A1, CYP1A2, and CYP1B1 (10, 11). Participation of AHR in liver development, nephrogenesis, function of the immune system, cell proliferation, and differentiation as well as retinoic acid metabolism (12-16) suggests that the AHR may be translocated to the nucleus to regulate those processes also in the absence of exogenous ligands. In fact, in the developing mouse embryo, nuclear localization of AHR and activation of an AHR target gene (CYP1A1) during defined stages of embryonic development has been reported (17, 18). However, it is an open question what signal renders AHR nuclear in absence of exogenous ligands.
We investigated whether such a signal could be given by cAMP, a universal intracellular second messenger and mediator of many hormones, neurotransmitters, and prostaglandins action, executing their cellular responses by activating protein kinase A (PKA).
Materials and Methods
Cell Culture. The mouse hepatoma cell line, Hepa1c1c7 (Hepa1) cells, a kind gift from Oliver Hankinson (University of California, Los Angeles), was propagated in α-MEM (Invitrogen) and supplemented in all experiments with 10% FCS (Greiner, Nurtingen, Germany). Cells were maintained as monolayers in an atmosphere of 5% CO2 and 95% air under saturating humidity at 37°C.
Expression Vectors. Plasmids pcDNAI/B6AhR-GFP (GFP-tagged murine AhR), p1646P1Luc3 [encoding murine Cyp1a1 5′ regulatory sequences from -1646 to +57, including six dioxin responsive element (DRE) (also called XRE, AHRE) motifs and NRE], and pAhRDtkLuc3 (driven by Cyp1a1 AhRD enhancer from -1100 to -896, containing three DRE motifs) cloned (19) into the pGL3-basic vector were a kind gift from Alvaro Puga (University of Cincinnati, Cincinnati). The pCMX-ARNT was created by the recloning of full-length cDNA of ARNT from pcDNAI/Neo/mARNT, a kind gift from Oliver Hankinson.
Immunofluorescence Microscopy. In 2 ml of α-MEM, 105 Hepa1 cells were seeded onto coverslips in six-well dishes and allowed to grow for 20 h at 37°C and 5% CO2. Cells were transfected either with 1 μg of pEGFP (EGFP protein) (Clontech) or pcDNAI/B6-AhR-GFP alone or with pCMX-ARNT. Transfections were performed in accordance with the manufacturer's protocol (Roche). At 48 h posttransfection, cells were incubated for 1 h with either 2 mM db-cAMP or with 2 nM 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD or dioxin) (Campro-Scientific, Emmerich, Germany) or with α-MEM. The visualization of cellular localization of the GFP-tagged AHR was performed with fluorescence microscopy.
For indirect immunofluorescence, 105 Hepa1 cells were seeded onto coverslips in six-well dishes in α-MEM, and after 18 h, the cells were stimulated with 2 mM db-cAMP (Sigma) for 30, 60, 90, and 120 min, or for 60 min with either 10 μM forskolin (Sigma) or 10 nM TCDD. Cells were fixed and permeabilized with acetone for 2 min at -20°C and incubated with anti-AHR (M-20) (Santa Cruz Biotechnology) as first antibodies for 2 h and carbocyanine-conjugated (Cy3) goat anti-mouse IgG (Jackson ImmunoResearch) as the second antibodies for 1 h at room temperature. The AHR cellular localization was evaluated by red immunofluorescence.
Nuclear Extract Preparation and EMSA. For nuclear extract preparation, cells were exposed either to 2 nM TCDD for 1 h or 2 mM db-cAMP or 10 μM forskolin for 1 h and 15 min. To investigate possible interference of cAMP or forskolin with TCDD, cells were preexposed for 15 min with 2 mM db-cAMP or 10 μM forskolin, and then exposure was continued for 1 h in the presence of 2 nM TCDD. To show involvement of PKA in forskolin action, cells were preincubated for 10 min with the PKA inhibitor H89 [N-(2-(p-bromocinnamylamino)ethyl)-5-isoquinolinesulfonamide dihydrochloride], and the incubation was continued in presence of 10 μM forskolin for 1 h. Nuclear extracts were prepared as described in ref. 20. A synthetic oligonucleotide of the Cyp1A1 enhancer 5′-GATCCGGAGTTGCGTGAGAAGAGCCA-3′ (consensus motif is boldfaced for AHR/ARNT) or mutated DRE (mDRE) 5′-GATCCGGAGTTGCGCGAGAAGAGCCA-3′ (mutated base is underlined) was radioactively labeled with 32P at the 5′ ends (21). EMSA was performed as described in ref. 22. Antibodies for AHR (C-20)X or ARNT (C-20)X or C/EBP (14AA)X or preimmune serum (Santa Cruz Biotechnology) were added at the same time as the nuclear extract and allowed to react for 15 min at room temperature before incubation with the radiolabeled probe (300,000 cpm/μl) DRE or mDRE. Samples were analyzed on a 4% nondenaturating gel for 4 h at 4°C, and labeled protein-DNA complexes were visualized by using a Storm 840 PhosphorImager (Molecular Dynamics).
Immunoprecipitation and Immunoblotting Analysis. Subconfluent cultures of Hepa1 cells were stimulated with 2 nM TCDD for 1 h, with 2 mM db-cAMP for either 15 min or 1 h and 15 min or with a combination of both for 1 h, or cells were exposed for 15 min to 2 mM db-cAMP before a 1-h stimulation with 2 nM TCDD. Immunoprecipitation was performed in accordance with the manufacturer's protocol (Santa Cruz Biotechnology) on nuclear extracts by using the anti-AHR antibodies (N19, Santa Cruz Biotechnology), followed by binding to agarose-plus beads. Bound proteins were fractionated by SDS/PAGE and transferred to a poly(vinylidene difluoride) membrane (Millipore). Filters were probed with the anti-ARNT antibody (H-172, Santa Cruz Biotechnology) for 4 h at 6°C, followed by an alkaline phosphatase-conjugated secondary antibody (Calbiochem) for 1 h at room temperature.
Reporter Assay. Transfection of 4 × 104 Hepa1 cells/well was performed in accordance with the manufacturer's protocol (Roche) with 0.2 μg of p1646p1Luc3 or pAhRDtkLuc3 or pGL3-basic (firefly luciferase), 0.01 μg of Renilla gene, and pRL-SV40 (Renilla luciferase), and the cells were incubated at 37°C and 5% CO2. At 24 h posttransfection, the cells were stimulated for 4.5 h with either 10 nM TCDD or 2 mM db-cAMP or 10 μM forskolin or stimulation was performed with 2mM db-cAMP or 0 μM forskolin for 15 min before a 4.5-h exposure to TCDD. The luminescence was measured 16-24 h after transfection in accordance with the manufacturer's instructions (Promega). The relative light units were normalized to expression of the Renilla luciferase.
Results and Discussion
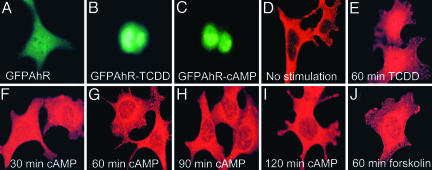
A fusion of AHR and GFP was expressed in Hepa1 cells, which were then stimulated with TCDD or N6-O2′-dibutyryl-cAMP (db-cAMP), a membrane-permeating derivative of cAMP. In the nonstimulated cells, the GFP-tagged AHR was diffusibly distributed in the cytoplasm in the great majority of transfected cells, and in some cells, a weak nuclear staining also was observed (Fig. 1A). As expected, TCDD shifted GFP-tagged AHR to the nucleus (Fig. 1B). Interestingly, db-cAMP caused the AHR-GFP fusion protein to translocate to the nucleus (Fig. 1C). Higher expression of the AHR dimerization partner ARNT had no obvious effect on the localization pattern of AHR-GFP (data not shown). Taking the advantage that Hepa1 cells constitutively express AHR, we have used indirect immunofluorescence as a second method to verify the cAMP-dependent relocation of native AHR. We observed a bright, cytoplasmic, and, in some cells, weak nuclear staining in nonstimulated Hepa1 cells (Fig. 1D and Table 1). As expected, TCDD resulted primarily in either abundant nuclear accumulation of fluorescence (Fig. 1E and Table 1), which in many cases showed a diffuse distribution throughout the cytoplasm and the nucleus, so that the border between these compartments was difficult to distinguish (data not shown).
Fig. 1.
Intracellular elevation of cAMP renders a GFP-tagged and native AHR nuclear. (A-C) Hepa1 cells transfected with GFP-AHR, stimulated with the indicated modulators, immunostained for AHR, and examined for GFP-positive nuclear staining. (D-J) Untransfected Hepa1 cells stimulated as indicated before fixation, stained with the anti-AHR antibodies and examined for AHR nuclear localization. Cells were visualized by immunofluorescence microscopy.
Table 1. A comparison of AHR cellular distribution in the presence of TCDD (dioxin) db-cAMP, and forskolin: Indirect immunostaining of Hepal cells with anti-aryl hydrocarbon receptor antibody.
| Nuclear distribution pattern of AHR
|
||||
|---|---|---|---|---|
| Stimulation | None | Weakly | Strongly | Exclusively |
| No stimulation | 141/200 | 59/200 | 0/200 | 0/200 |
| TCDD (60 min) | 4/200 | 26/200 | 51/200 | 119/200 |
| Db-cAMP | ||||
| 30 min | 7/200 | 59/200 | 134/200 | 0/200 |
| 60 min | 19/200 | 126/200 | 55/200 | 0/200 |
| 90 min | 30/200 | 138/200 | 32/200 | 0/200 |
| 120 min | 36/200 | 140/200 | 24/200 | 0/200 |
| Forskolin (60 min) | 23/200 | 64/200 | 113/200 | 0/200 |
Db-cAMP/forskolin altered the subcellular distribution pattern of AHR qualitatively similarly to, but quantitatively less overwhelming than TCDD. Nonstimulated or stimulated Hepal cells were stained with AHR antibodies and fluorescein secondary antibodies and photographed under a fluorescence microscope. Two hundred cells from at least 12 independent fields were counted, and the AHR subcellular distribution pattern was classified as shown in Table 1. The majority of nonstimulated cells showed very bright cytoplasmic staining, and ~30% of the cells displayed a weak nuclear fluorescence. When the cells were stimulated with 2 nM TCDD, 25% cells showed strong nuclear staining, and 60% of cells showed exclusive nuclear staining. Interestingly, stimulation of cells for 30 min with 2mM db-cAMP resulted in strong nuclear localization of AHR(67% of cells). Even though stimulation with db-cAMP was extended to 120 min, the AHR was never localized entirely to the nucleus. Quite to the contrary, a prolonged exposure to db-cAMP resulted in marked relocation to the cytoplasm with some residual staining in the nucleus. In response to forskolin (a potent activator of adenylate cyclase, leading to a rapid increase of cAMP), the AHR also was targeted to the nucleus.
Stimulation for 30 min with db-cAMP resulted in a pronounced shift of native AHR protein from the cytoplasm to the nucleus (compare Fig. 1 F and D; Table 1). A similar distribution was seen after 60-, 90-, and 120-min stimulations with db-cAMP, but already a fading of the nuclear fluorescence during these later time points was observed (Fig. 1 G-I and Table 1). In addition, forskolin, a potent activator of adenylate cyclase, known to rapidly increase cAMP level, effectively targeted native AHR to the nucleus (compare Fig. 1 D and J; Table 1). Of note, as seen from Fig. 1 F-J, the db-cAMP- and the forskolin-dependent AHR nuclear shift resulted in a less overwhelming nuclear fluorescence than that of the persistently activating TCDD (Fig. 1E). Moreover, the nucleocytoplasmic border after db-cAMP/forskolin stimulation, in contrast with stimulation with TCDD, always was preserved. It has been reported that dioxin-free AHR (recombinant or native) is present in the nucleus after treatment with geldanamycin (a disruptor of the AHR/HSP90 association), MG132 (an inhibitor of 26S proteasome), or leptomycin (an inhibitor of AHR nuclear export). It is still controversial whether such nuclear form of AHR activates or does not activate AHR/ARNT-dependent transcription of, e.g., CYP1A1/CYP1A2 (19, 23-26).
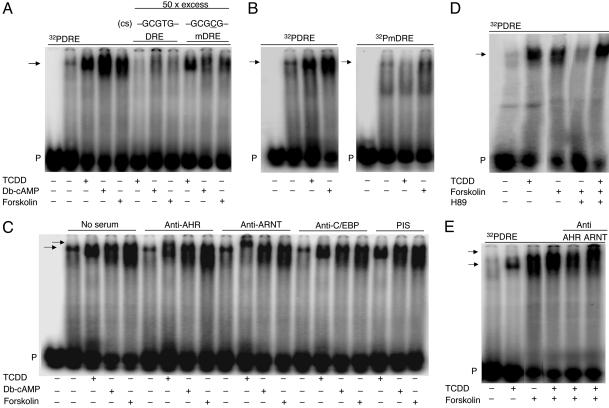
Using DNA-binding analysis, we found that nuclear proteins from either db-cAMP- or forskolin-stimulated cells bound avidly to a 32P-labeled oligonucleotide, representing a fragment of the mouse Cyp1A1 enhancer, called dioxin-responsive element (DRE), containing the AHR/ARNT-binding motif (5′-GCGTG-3′). The db-cAMP/forskolin-generated band had a similar mobility as the TCDD-induced complex (Fig. 2A). The formation of all three complexes was specific, because a 50-fold molar excess of unlabeled DRE completely eliminated their binding to the radioactive probe. However, a 50-fold molar excess of unlabeled oligonucleotide carrying a base mutation T to C (underlined) in the AHR/ARNT consensus motif 5′-GCGCG-3′ (mDRE) only marginally reduced the intensity of the TCDD-dependent signal, but binding of both db-cAMP- and forskolin-dependent complex was substantially reduced (Fig. 2 A). These results were supported by the analysis where equivalent amounts of TCDD- or db-cAMP-induced nuclear proteins were probed with 32P-labeled mDRE. T-to-C mutation fully eliminated binding of the TCDD-dependent complex, whereas the binding of the db-cAMP-induced complex was substantially reduced but clearly present (Fig. 2B) [treatment with forskolin led to similar results (data not shown)]. These findings indicate that the binding site of cAMP-induced nuclear proteins on DRE is not identical but overlaps with the high-affinity binding site of TCDD-activated AHR/ARNT (5′-GCGTG-3′), where the T is supportive but not a strict requirement for the binding of the cAMP-induced nuclear proteins in contrast with the TCDD-activated AHR/ARNT complex, where it is a strict requirement. The mutated base is in the E-box half site (5′-GTG-3′) of the DRE that binds ARNT within the TCDD-activated AHR/ARNT heterodimer. Hence, one possibility is that the treatment with db-cAMP or forskolin leads to a higher affinity of the AHR for an ARNT-related protein than for ARNT itself, and that this partner of AHR has a similar but less stringent sequence specificity within the E-box half site of the DRE.
Fig. 2.
cAMP-dependent formation of a nuclear protein complex on dioxin responsive element (DRE) of Cyp1a1 enhancer in an electrophoretic mobility shift assay. (A) DRE consensus sequence (cs) for TCDD-activated AHR/ARNT and mutated (underlined) mDRE. For the competition studies a 50-fold molar excess of unlabeled DRE or mDRE was used. (B) TCDD-dependent complex failes to bind, and binding of db-cAMP-dependent complex is substantially reduced, when radioactively labeled mDRE is used as a probe. (C) Supershift with the corresponding antisera or preimmune serum (PIS). The top and the bottom arrows indicate supershift (partial supershift of AHR; total supershift of ARNT) and specific complexes, respectively. (D) PKA inhibitor H89 [N-(2-(p-bromocinnamylamino)ethyl)-5-isoquinolinesulfonamide dihydrochloride] abolished formation of forskolin-dependent complex. (E) AHR and ARNT proteins are present in the forskolin/TCDD-induced complex bound to DRE. Arrows in A, B, and D show specific complexes. The lower arrow in C and E shows specific complexes. The upper arrow shows supershift. P, probe (radioactively labeled oligonucleotide). Lane 1 in A-C shows probe alone.
To identify proteins bound to DRE, we used in the binding reaction specific antibodies directed against AHR or ARNT-C-terminal region in order not to interfere with their DNA-binding and protein-interaction domains. Both proteins were recognized in the TCDD-induced complex (indicated by its supershifting) but not in the db-cAMP- or forskolin-induced complexes (Fig. 2C), irrespective of whether incubation took place before or after DNA addition to the binding reaction. This finding suggests that the nuclear proteins of db-cAMP- and forskolin-dependent complexes bound to DRE differ from those present in the TCDD-dependent complex. Alternatively, those proteins may be the same as these present in the TCDD complex but because of possible conformational changes during cAMP modulation may not have an equivalent affinity or free availability of the immunocompetent sites for anti-AHR or anti-ARNT antibodies. Experiments with AHR and with ARNT-deficient Hepa1 cells should be able to delineate the necessity of the presence of AHR and/or ARNT for the observed cAMP-dependent complex to form on DRE. Such experiments are now in progress. Antibodies against C/EBP (CCAAT/enhancer-binding protein), which regulates along with AHR the glutathione S-transferase Ya gene via DRE (27), did not interfere with the formation of any of the investigated complexes (Fig. 2C).
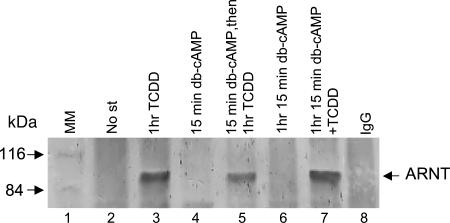
If subcellular distribution of AHR is governed by cAMP-dependent PKA, then PKA inhibition should block cAMP-dependent nuclear effects. Indeed, the PKA inhibitor H89 [N-(2-(p-bromocinnamylamino)ethyl)-5-isoquinolinesulfonamide dihydrochloride] abolished the binding of forskolin-induced complex to DRE (Fig. 2D). Exposure of cells to forskolin for 15 min before TCDD resulted in the formation of a DRE-bound complex containing the immunoreactive AHR and ARNT proteins but migrating with a slightly slower mobility, compared with the complex induced by TCDD alone, therefore implying a higher molecular mass or a different conformation of forskolin/TCDD complex (Fig. 2E). To further distinguish the db-cAMP/forskolin-dependent nuclear form of AHR from the TCDD-dependent nuclear form of AHR, we checked the association of the former with ARNT (heterodimer essential for TCDD-dependent Cyp1A1 induction), and we found in immunoprecipitation experiments no AHR/ARNT interaction in the presence of cAMP (Fig. 3).
Fig. 3.
ARNT does not coprecipitate with the cAMP-dependent nuclear form of AHR. Nuclear extracts prepared from nonstimulated (No st) Hepa1 cells or stimulated as indicated were immunoprecipitated with anti-AHR antibody or with IgG antibody [negative control after stimulation with TCDD; the negative controls after other modulations gave similar results (data not shown)]. The AHR/ARNT complexes were resolved by SDS/PAGE, and ARNT was detected by Western blotting with anti-ARNT antibody. Western blot analysis revealed a 90-kDa band, corresponding to ARNT when cells were exposed to TCDD (lane 3) but not after exposure to db-cAMP, even at two different time points (lanes 4 and 6). Interestingly, only stimulation of cells with db-cAMP for 15 min before TCDD reduced the intensity of the TCDD-dependent signal (lane 5) but not when the cells were stimulated with both substances simultanously (lane 7). These results suggest that AHR and ARNT are not complexed when AHR is driven to the nucleus through intracellular elevation of cAMP, and possibly that cAMP-mediated signaling interferes with TCDD-dependent AHR/ARNT interaction. MM, molecular mass markers.
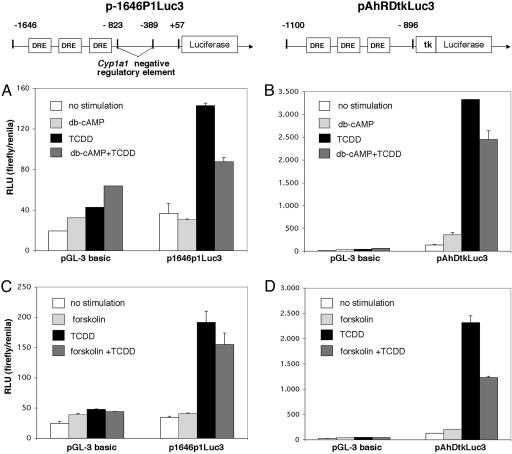
We examined, therefore, in Hepa1 cells the activity of the luciferase gene fused to the Cyp1A1 natural promoter and the endogenous enhancer, containing six DRE copies and also a negative regulatory element (NRE) (28, 29), by using the p1646P1Luc3 reporter plasmid. Db-cAMP or forskolin in contrast to TCDD did not significantly influence luciferase activity (Fig. 4 A and C). However, stimulation with db-cAMP or forskolin for 15 min before TCDD resulted in a moderate but clear and reproducible down-regulation of TCDD-dependent induction of the reporter gene (Fig. 4 A and C). Thus, cAMP or an event downstream of cAMP may, although leading to nuclear translocation of AHR, act as a repressor rather than an activator of AHR-dependent gene expression. To determine whether the NRE of Cyp1A1 is involved in cAMP-dependent reduction of luciferase activity, we used a second reporter construct, the pAhRDtkLuc3, lacking the NRE but containing three copies of DRE. Absence of NRE allowed for a much higher TCDD-dependent induction of the reporter gene that was reduced by db-cAMP or forskolin by ≈30-50% (Fig. 4 B and D). These results suggest that intracellular elevation of cAMP generates a signal that is inhibitory to the response to dioxin, and that for this inhibitory modulation, the NRE is not required. Thus, depending on the type of cellular signaling (cAMP vs. dioxin), the DRE-driven target gene expression may recruit different regulators, thereby mediating different effects on transcription.
Fig. 4.
db-cAMP and forskolin do not activate transcription of Cyp1a1 but interfere with TCDD-dependent Cyp1a1 induction. Reporter constructs for Cyp1a1 were as follows: p1646P1Luc3 (A and C), containing the Cyp1a1 natural promoter with six copies of DRE and the negative regulatory element, and pAhRDtkLuc3 (B and D), lacking the negative regulatory element but containing three copies of DRE. These constructs were transfected into Hepa1 cells. At 24 h posttransfection, cells were stimulated as indicated, and reporter activity was determined. RLU, relative light units; pGL-3 basic, empty vector.
Taken together, in this study we identified the second messenger cAMP as a mediator of AHR intracellular relocalization, including its nuclear accumulation, but with fundamentally different consequences as opposed to those initiated by the nuclear translocation of TCDD-activated AHR. We propose that the AHR, in response to cAMP, adopts a unique structure and new protein-protein interactions, such that it does not require exogenous ligand to reach the nucleus. Interestingly, the cAMP-dependent form of AHR is not a partner for productive interaction with ARNT. This divergence suggests that cAMP-mediated AHR activation might prevent the formation of the AHR/ARNT complex, which is responsible for the toxic effects of dioxin and other environmental pollutants. Instead of, or in addition to, an interaction with a putative endogenous ligand, the AHR may in response to cAMP adopt novel properties allowing for ligand-independent protein-protein interactions. A similar mechanism has been postulated for the orphan receptors [e.g., the interaction of steroidogenic factor 1 (SF1) with homeodomain protein Ptx1 or Drosophila homolog Fushi-Tarazu factor 1 (FTZ1) with homeodomain protein Ftz (30, 31). The facts identified in this study suggest an interesting possibility that AHR participates in a biochemical signaling cascade that has been conserved and independently coopted by evolution for a role in development and differentiation, and additionally, to balance the induction of carcinogen-activating enzymes such as cytochrome P450s of family 1 (32). Such a signaling in contrast with dioxin-activated AHR may be a measure of the organism's well-being by facilitating the connections between hormones, neurotransmitters, prostaglandins, and their transcriptional targets. It will, therefore, be of utmost importance to now investigate the functional consequences of the cAMP/forskolin-dependent translocation of the AHR to the nucleus, the exact sequence requirement for the binding of the cAMP/forskolin-dependent complex to DNA, and the role of additional factors such as the AHR repressor.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (B.O.-B.).
Author contributions: B.O.-B. designed research; B.O.-B., A.H., O.W., and P.A.-L. performed research; B.O.-B., C.D., M.A., C.W., E.B., and F.O. analyzed data and critically read the manuscript; and B.O.-B. wrote the paper.
Abbreviations: AHR, aryl hydrocarbon receptor; TCDD/dioxin, 2,3,7,8-tetrachlorodibenzo-p-dioxin; db-cAMP, N6-O2′-dibutyryl-cAMP; DRE, dioxin responsive element; mDRE, mutated DRE; PKA, protein kinase A; CYP, cytochrome P450; NRE, negative regulatory element.
References
- 1.Hahn, M. E. (2002) Chem. Biol. Interact. 141, 131-160. [DOI] [PubMed] [Google Scholar]
- 2.Gu, Y. Z., Hogenesch, J. B. & Bradfield, C. A. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 519-561. [DOI] [PubMed] [Google Scholar]
- 3.Rowlands, J. C & Gustafsson, J. A. (1997) Crit. Rev. Toxicol. 27, 109-134. [DOI] [PubMed] [Google Scholar]
- 4.Poellinger, L. (1995) in Inducible Gene Expression, ed. Baeuerle, P. A. (Birkhäuser, Boston), Vol. 1, pp. 177-205. [Google Scholar]
- 5.Emmons, R. B., Duncan, D., Estes, P. A., Kiefel, P., Mosher, J. T., Sonnenfeld, M., Ward, M. P., Dunkan, I. & Crews, S. T. (1999) Development (Cambridge, U.K.) 126, 3937-3945. [DOI] [PubMed] [Google Scholar]
- 6.Poland, A. & Knutson, J. C. (1982) Annu. Rev. Pharmacol. 22, 517-554. [DOI] [PubMed] [Google Scholar]
- 7.Ikuta, T., Tachibana, T., Watanabe, J., Yoshida, M., Yoneda, Y. & Kawajiri, K. (2000) J. Biochem. (Tokyo) 127, 503-509. [DOI] [PubMed] [Google Scholar]
- 8.Pollenz, R. S., Sattler, C. A. & Poland, A. (1994) Mol. Pharmacol. 45, 428-438. [PubMed] [Google Scholar]
- 9.Singh, S. S., Hord, N. G. & Perdew, G. H. (1996) Arch. Biochem. Biophys. 329, 47-55. [DOI] [PubMed] [Google Scholar]
- 10.Nebert, D. W. (1994) Biochem. Pharmacol. 47, 25-37. [DOI] [PubMed] [Google Scholar]
- 11.Hankinson, O. (1995) Annu. Rev. Pharmacol. Toxicol. 35, 307-340. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Salguero, P., Pineau, T., Hilbert, D. M., McPhail, T., Lee, S. S., Kimura, S., Nebert, D. W., Rudikoff, S., Ward, J. M. & Gonzalez, F. J. (1995) Science 268, 722-726. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt, J. V., Su, G. H., Reddy, J. K., Simon, M. C. & Bradfield, C. A. (1996) Proc. Natl. Acad. Sci. USA 93, 6731-6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez, F. J. & Fernandez-Salguero, P. (1998) Drug Metab. Dispos. 26, 1194-1198. [PubMed] [Google Scholar]
- 15.Bunger, M. K., Moran, S. M., Glover, E., Thomae, T. L., Lahvis, G. P., Lin, B.C. & Bradfield, C. A. (2003) J. Biol. Chem. 278, 17767-17774. [DOI] [PubMed] [Google Scholar]
- 16.Falahatpisheh, M. H. & Ramos, K. S. (2003) Oncogene 22, 2160-2171. [DOI] [PubMed] [Google Scholar]
- 17.Abbott, B. D., Birnbaum, L. S. & Perdew, G. H. (1995) Dev. Dyn. 204, 133-143. [DOI] [PubMed] [Google Scholar]
- 18.Campbell, S. J., Henderson, C. J., Anthony, D. C., Davidson, D., Clark, A. J. & Wolf, C. R. (2005) J. Biol. Chem. 280, 5828-5835. [DOI] [PubMed] [Google Scholar]
- 19.Chang, C. Y. & Puga, A. (1998) Mol. Cell. Biol. 18, 525-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denison, M. S., Fisher, J. M. & Whitlock, J. P., Jr. (1988) Proc. Natl. Acad. Sci. USA 85, 2528-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen, E. S., Elferink, C. J. & Whitlock, J. P., Jr. (1991) Methods Enzymol. 206, 403-405. [DOI] [PubMed] [Google Scholar]
- 22.Denison, M. S., Fisher, J. M. & Whitlock, J. P., Jr. (1988) J. Biol. Chem. 263, 17221-17224. [PubMed] [Google Scholar]
- 23.Song, Z. & Pollenz, R. S. (2002) Mol. Pharmacol. 62, 806-816. [DOI] [PubMed] [Google Scholar]
- 24.Santiago-Josefat, B., Pozo-Guisado, E., Mulero-Navarro, S. & Fernandez-Salguero, P. M. (2001) Mol. Cell. Biol. 21, 1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollenz, R. S. & Barbour, E. R. (2000) Mol. Cell. Biol. 20, 6095-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter, C. A., Tillitt, D. E. & Hannink, M. (2001) Arch. Biochem. Biophys. 389, 207-217. [DOI] [PubMed] [Google Scholar]
- 27.Pimental, R. A., Liang, B., Yee, G. K., Wilhelmsson, A., Poellinger, L. & Paulson K. E. (1993) Mol. Cell. Biol. 13, 4365-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lusska, A., Shen, E. & Whitlock, J. P., Jr. (1993) J. Biol. Chem. 268, 6575-6580. [PubMed] [Google Scholar]
- 29.Nebert, D. W. & Gonzalez, F. J. (1985) Nucleic Acids Res. 13, 7269-7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tremblay, J. J., Marcil, A., Gauthier, Y. & Drouin, J. (1999) EMBO J. 18, 3431-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu, Y. Li, W., Su, K., Yussa, M., Han, W., Perrimon, N. & Pick., L. (1997) Nature 385, 552-555. [DOI] [PubMed] [Google Scholar]
- 32.Nebert, D. W., Dalton, T. P., Okey, A. B. & Gonzalez, F. J. (2004) J. Biol. Chem. 279, 23847-23850. [DOI] [PubMed] [Google Scholar]