Abstract
Objectives
We propose and validate a domain knowledge-driven classification model for diagnosing post-acute sequelae of SARS-CoV-2 infection (PASC), also known as Long COVID, using Electronic Health Records (EHRs) data.
Materials and Methods
We developed a robust model that incorporates features strongly indicative of PASC or associated with the severity of COVID-19 symptoms as identified in our literature review. The XGBoost tree-based architecture was chosen for its ability to handle class-imbalanced data and its potential for high interpretability. Using the training data provided by the Long COVID Computation Challenge (L3C), which was a sample of the National COVID Cohort Collaborative (N3C), our models were fine-tuned and calibrated to optimize Area Under the Receiver Operating characteristic curve (AUROC) and the F1 score, following best practices for the class-imbalanced N3C data.
Results
Our age-stratified classification model demonstrated strong performance with an average 5-fold cross-validated AUROC of 0.844 and F1 score of 0.539 across the young adult, mid-aged, and older-aged populations in the training data. In an independent testing dataset, which was made available after the challenge was over, we achieved an overall AUROC score of 0.814 and F1 score of 0.545.
Discussion
The results demonstrated the utility of knowledge-driven feature engineering in a sparse EHR data and demographic stratification in model development to diagnose a complex and heterogeneously presenting condition like PASC. The model’s architecture, mirroring natural clinician decision-making processes, contributed to its robustness and interpretability, which are crucial for clinical translatability. Further, the model’s generalizability was evaluated over a new cross-sectional data as provided in the later stages of the L3C challenge.
Conclusion
The study proposed and validated the effectiveness of age-stratified, tree-based classification models to diagnose PASC. Our approach highlights the potential of machine learning in addressing the diagnostic challenges posed by the heterogeneity of Long-COVID symptoms.
Keywords: PASC, Long COVID, electronic health records, N3C, clinical decision model
Background and significance
After the acute phase of a COVID-19 infection, many people report new, lasting, and/or worsening symptoms. These symptoms are often unrelated to the symptoms experienced during the acute infection phase and occur even after testing negative for COVID-19. This phenomenon is known as post-acute sequelae of SARS-CoV-2 infection (PASC), otherwise commonly known as Long COVID, and is thought to have affected as many as 15% of adults in the United States.1 Research to understand who will go on to develop PASC, its prognosis, and recommended care paths has been challenging due to the wide range and inconsistency of PASC manifestation across multiple organ systems.
Current studies estimate that the prevalence of PASC diagnosis falls between 10% and 30% of those previously infected with COVID-19, and that the risk of developing PASC ranges anywhere from 10% to 70% based on certain risk factors.2 Although the risk of developing PASC is positively correlated with the severity of the acute COVID-19 infection, 10%-35% of people who had mild responses to the infection also reported developing PASC.3,4
At the time of writing, PASC has been tied to more than 200 symptoms.2 Although the most common symptoms of COVID-19 are respiratory, the symptoms of PASC can be multisystemic, affecting the heart, lungs, immune system, pancreas, gastrointestinal tract, neurological system, blood vessels, reproductive system, etc.2,3 While common symptoms of PASC are being identified,5 the ability to predict the development of PASC prior to its onset remains elusive. In an effort to centralize clinical knowledge about COVID-19 and its complications, the National Institutes of Health (NIH) National Center for Advancing Translational Sciences (NCATS) built the National COVID Cohort Collaborative (N3C) Data Enclave,6 a national multi-institutional database populated with electronic health records (EHR) data from COVID-19 patients that currently contains data from upwards of 60 health organizations in the United States. With this data, the NIH posed the Long COVID Computational Challenge (L3C), with dual goals to develop an algorithm to predict PASC before it occurs and to learn what characteristics predispose individuals to PASC risk. Through our participation in the L3C challenge (September-December 2022), we developed and validated an explainable machine learning (ML) model to predict the likelihood of developing PASC from prior EHR data and to translate that likelihood to a binary label (PASC diagnosis or not) that can be validated against real-world data.
Given the heterogeneity of PASC, successes have been seen in model building on stratified subsets of the N3C cohort. For example, Pfaff et al. and Socia et al. trained separate XGBoost models based on hospitalization due to COVID-19, stratifying those who had and had not been hospitalized with COVID-19.7,8 Both studies reported differences in feature importance and performances among the stratified models. In addition to hospitalization status, a later study by Pfaff et al. also suggested that PASC manifests differently across age groups as “a collection of subphenotypes”.9 Moreover, the value of incorporating vital signs, demographics, conditions, and procedures into models to characterize and/or predict PASC have been uncovered by previous studies.7–10 Based on these works, here we adopted the approach of building separate XGBoost models on a population stratified by age. We hypothesized that stratifying the N3C population by age would also capture the phenotypic differences between the hospitalized patients vs the nonhospitalized patients given that age and hospitalization rate are positively correlated.11 Further, we hypothesized engineering clinically relevant features according to findings from previous literature will enhance the performance and explainability of our model.
Methods
Description of L3C cohort
The dataset provided for the L3C challenge was a subset of the N3C data enclave,6 a collaborative effort to harmonize data regarding COVID-19, following the Observational Medical Outcomes Partnership v 5.3 Common Data Model for data schema and storage. Specifically, the L3C challenge dataset, which was curated from the N3C dataset for the purpose of the challenge, contained both censored (data available only up to 4 weeks after initial diagnosis of COVID-19) and uncensored (all available data without a time cutoff) demographic and clinical information from the EHR for 57 624 patients. All data in the N3C enclave are deidentified.
The final dataset released by the L3C consortium contained all PASC patients (diagnosed with U09.9) from the N3C data enclave, and the non-PASC patient cohort was randomly downsampled to match a 1:4 ratio of PASC to non-PASC patients. The 1:4 case-to-control ratio was established by the L3C challenge organizers as part of the study design. While we adhered to this predetermined ratio, the specific rationale behind this choice was not disclosed to us and falls outside the scope of our paper. In addition, the censored and uncensored datasets with 57 624 patients were the only training dataset available to us for the entire duration of the challenge timeline.
Definition of PASC
The outcome labels provided by the L3C consortium follow a “silver standard” because the negatively labeled outcomes are not rigorously validated as true negatives. Patients who have an International Classification of Diseases (ICD-10) code U09.9 are labeled as true positives for PASC and are otherwise labeled as negative. However, individuals may not seek clinical care despite presenting symptoms of PASC, may not be aware of having PASC, and/or may not have received the U09.9 ICD-10 code during their clinical encounter. For these various reasons, the N3C database’s negatively labeled data most likely contains patients who have actually had PASC (ie, false negatives).
Feature selection and feature engineering
We leveraged domain knowledge gathered from the literature to select relevant features for PASC and reduce mutual information by combining features that we would expect to have a similar role in the context of COVID-19.12 For example, we combined the use of BiPAP (reverse transcription polymerase chain reaction) or CPAP (continuous positive airway pressure) as a binary indicator for noninvasive ventilation and categorized the use of endotracheal intubation and tracheostomy as invasive ventilation. Our final transformed dataset consisted of 184 features (Table S2) that included information about patient demographics, symptoms, clinical conditions, vaccination status, lab testing results, procedures, and administered medical devices such as different types of ventilators. We used a binary coding system to simplify these features: categorical features are coded with one-hot-encoding, and continuous variables and quantitative variables are coded as 0 (corresponding to False) if within the normal range or 1 (corresponding to True) if outside of normal range. Normal ranges for lab testing results and other continuous measurements are found on the Mayo Clinic’s public database.13 We also created a combined binary indicator for race/ethnicity, including an “unknown” category for patients with unknown or unreported race information.
Addressing missing values
The EHR is sparse in nature with a large number of missing values, and the proportion of missingness also varies per individual.14 Following our feature engineering method, we replaced all missing values with 0 (corresponding to False). This was done under the assumption that any test, procedure, or device usage was not needed by the patient or was within normal ranges if it was not ordered/prescribed by a clinician (Figure S1). For conditions, it was assumed that the patient did not present with the condition at evaluation if that condition was not recorded in the EHR.
Model development
Given the heterogeneity of PASC, successes have been seen in model building on stratified subsets of the N3C cohort.7,8 As we found differences in PASC prevalence and medical conditions across age groups,15,16 we stratified the cohort into four age groups and trained four independent XGBoost classification models. The age groups were defined as pediatrics <15 years, young adults (15-44), mid adults (45-64), and older adults ≥ 65. For each of our XGBoost classifiers, we implemented a nested cross-validation method to tune hyperparameters and evaluate the performance of our classifiers on each age group. The parameters tuned included the maximum depth of the trees (“max_depth,” ranging from 1 to 9), the minimum loss reduction required for further splitting (“gamma,” ranging from 1 to 9), both L1 and L2 regularization terms (“reg_alpha,” ranging from 40 to 180, and “reg_lambda,” ranging from 0.2 to 1, respectively), the subsample ratio of columns for each tree (“colsample_bytree,” ranging from 0.5 to 1), the minimum sum of instance weight needed in a child node (“min_child_weight,” ranging from 0 to 10), the learning rate (eta, ranging from 0.005 to 0.5), and the number of trees (“n_estimators,” ranging from 50 to 500), and subsample ratio of training instances (“subsample,” ranging from 0.5 to 1). These parameters were selected based on their importance in controlling the complexity and generalization of the model. In each of the 5 outer folds, we tuned the parameters for each of the four XGBoost models through an inner-fold 5-fold cross-validation and also calibrated the classifier using sklearn’s implementation of CalibrateClassifierCV with 3-fold cross-validation.17 To aid the model’s utility and explainability, we also evaluated the relative importance of features using Shapley values.18,19
For training, we used the censored training set provided by the N3C as part of the L3C challenge. The dataset contained 57 624 patients who tested positive for COVID-19 on RT-PCR (reverse transcription polymerase chain reaction)-based lab tests within 7 days of an inpatient or outpatient visit (Table 1).
Table 1.
Demographic distribution of participants included in the study (n = 57 624).
| Age group |
||||
|---|---|---|---|---|
| Characteristics | Pediatrics (0-14) | Young adults (15-44) | Mid adults (45-64) | Older adults (≥65) |
| No. (%) | 4811 (8) | 25 768 (45) | 17 005 (30) | 10 040 (17) |
| Gender, no. (%) | ||||
| Male | 2550 (53) | 9741 (38) | 6808 (40) | 4533 (45) |
| Female | 2261 (47) | 16 027 (62) | 10 197 (60) | 5507 (55) |
| Age (years), mean (SD) | 8.1 (4) | 31.7 (8.7) | 54.6 (5.7) | 73.6 (6.4) |
| Race/Ethnicity, no. (%) | ||||
| American Indian or Alaska Native/non-Hispanic or unknown | <20 | 85 (<1) | 85 (<1) | <50 (<1) |
| Asian/non-Hispanic or unknown | 100 (2) | 574 (2) | 261 (2) | 158 (2) |
| Black or African American/non-Hispanic or unknown | 1184 (25) | 5637 (22) | 3417 (20) | 1466 (15) |
| Native Hawaiian or other Pacific Islander/non-Hispanic or unknown | <20 | 49 (<1) | 30 (<1) | <20 |
| White/non-Hispanic or unknown | 2064 (43) | 12 678 (49) | 9975 (59) | 6882 (69) |
| Other/non-Hispanic or unknown | 64 (1) | 330 (1) | 102 (<1) | 61 (<1) |
| Unknown/non-Hispanic or unknown | 738 (15) | 3552 (14) | 1618 (10) | 861 (9) |
| All race/Hispanic or Latino | 646 (13) | 2863 (11) | 1517 (9) | 556 (6) |
| PASC, no. (%) | ||||
| PASC | 142 (3) | 2742 (11) | 3806 (22) | 2341 (23) |
| No PASC | 4669 (97) | 23 026 (89) | 13 199 (78) | 7699 (77) |
To protect person privacy, we suppress cell sizes less than 20 (unless it is actually 0) according to N3C reporting policy. We further obfuscated the American Indian or Alaska Native, no. for older adults to prevent computation from the marginal totals.
Abbreviation: PASC, post-acute sequelae of SARS-CoV-2 infection.
Adhering to the requirements of the L3C challenge, we define the day of the positive test of the initial acute SARS-CoV-2 infection as the patient’s COVID-19 index date. Using the censored training set, our model was built to predict an individual’s likelihood of developing PASC at ≥4 weeks past their COVID-19 index date using EHR data up to and before that 4-week mark. A PASC diagnosis that occurred sooner than 4 weeks after the COVID-19 index date was not treated as a positive label (relabeled as false for our training) unless a subsequent PASC diagnosis was issued after the 4 weeks past COVID-19 index data. The scope of the L3C challenge strictly required PASC codes past 4 weeks after COVID-19 index, so PASC codes issued prior to 4 weeks past COVID-19 index were irrelevant to our analysis.
Evaluation approach/study design
Our model was developed to predict whether a PASC code appears as soon as 4 weeks after an initial COVID-19 infection (ie, the COVID-19 index date). We evaluated our model based on the AUROC and F1 score using 5-fold cross-validation on the training set.20,21 AUROC was chosen to assess the model’s ability to differentiate PASC vs non-PASC patients. Given the imbalance in the given dataset, the F1 score was considered to be a more relevant metric than accuracy.
Further, we constructed calibration curves (also known as reliability diagrams) to compare the model’s predicted probability of PASC code against the true frequency of the positive label.22 Matching the calibration curve was essential to ensure that the model’s predicted probabilities of PASC correspond to the expected distribution of probabilities based on the real training data.
Results
Five-fold cross-validation on training dataset
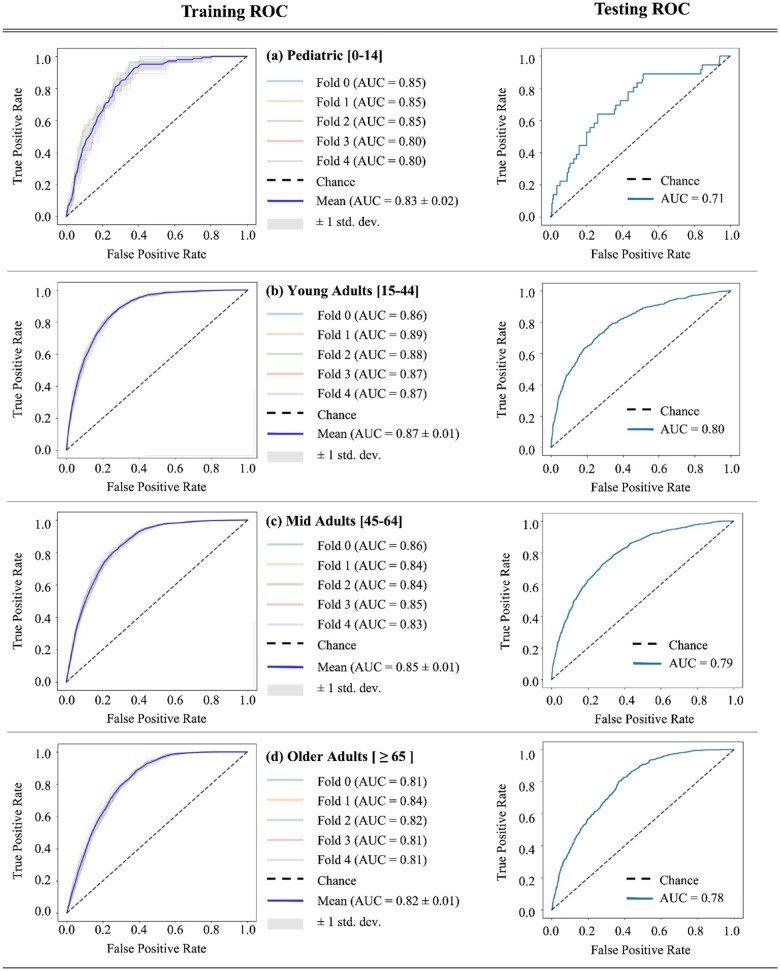
Based on our hypothesis that PASC and health care utilization would manifest differently by age group, we stratified the classification task among pediatric, young, mid, and older age groups. Models for the young, mid, and older age groups had AUROC >0.80 using 5-fold cross-validation (Table 2). From the 5-fold cross-validation on the training dataset, good performances (AUROC ≥0.80) were achieved in the young adults (mean AUROC = 0.872, mean F1 = 0.468), mid adults (mean AUROC = 0.844, mean F1 = 0.585) and the older adults groups (mean AUROC = 0.815, mean F1 = 0.565). The performance, however, was inferior in the pediatric group (mean AUROC = 0.833, mean F1 = 0.024). Particularly, we found a steep decline in the pediatric group’s F1 score compared to that of the other age groups (Figure 1 and Table 2). This drop could be due to the particularly low prevalence of PASC in the pediatric population (Table 1) as indicated by the high “negative to positive support ratio,” which is calculated as the average of the ratio between the number of PASC negative cases and PASC positive cases, to quantify the class imbalance in each age group. In particular, there are 33 negative PASC cases for each positive PASC incidence in the pediatric group from the training dataset.
Table 2.
5-fold XGBoost cross-validation results (training data) and test dataset.
| Evaluation | Age group | Accuracy | F1 | AUROC | Sensitivity | Specificity | Precision | Negative to positive support ratio |
|---|---|---|---|---|---|---|---|---|
| Training (5-fold CV results) | Pediatric (0-14) | 0.969 ±0.001 | 0.024±0.047 | 0.833±0.022 | 0.014±0.029 | 0.998±0.002 | 0.067±0.133 | 33 |
| Young(15-44) | 0.861±0.003 | 0.468±0.017 | 0.872±0.008 | 0.577±0.028 | 0.894±0.003 | 0.394±0.012 | 8.5 | |
| Mid (45-64) | 0.742±0.008 | 0.585±0.009 | 0.844±0.009 | 0.813±0.021 | 0.722±0.014 | 0.457±0.009 | 3.3 | |
| Older ≥ 65 | 0.699±0.012 | 0.565±0.006 | 0.815±0.011 | 0.838±0.023 | 0.657±0.021 | 0.426±0.010 | 3.5 | |
| Test dataset | Pediatric (0-14) | 0.956 | 0.000 | 0.718 | 0.000 | 0.998 | 0.000 | 20 |
| Young (15-44) | 0.791 | 0.482 | 0.796 | 0.609 | 0.825 | 0.399 | 5.3 | |
| Mid (45-64) | 0.671 | 0.564 | 0.798 | 0.822 | 0.619 | 0.429 | 2.9 | |
| Older ≥ 65 | 0.666 | 0.582 | 0.790 | 0.827 | 0.602 | 0.449 | 2.6 | |
| Overall (all groups combined) | 0.736 | 0.545 | 0.814 | 0.752 | 0.731 | 0.427 | 3.8 |
Figure 1.
The resulting receiver operating characteristic (ROC) curves and the respective area under the ROC curve (AUROC) from 5-fold cross-validation training (left column) and testing (right column) are shown for each age stratum.
From all experiments on the training data, our model achieved AUROC scores above 0.80, yet the F1 score was not as high. This indicates that our model’s precision and recall are not balanced, suggesting that the model is either biased toward generating false positives or false negatives. Indeed, our model’s performance has a high number of false positives as indicated by the relatively low precision (Table 2), which indicates that a significant proportion of the model’s identified PASC patients are not actually true PASC patients.
We analyzed the feature importance using Shapley values on the training dataset and found that cough (condition) and respiratory rate (measurement) were important predictive features for all age groups (Table S3 and Figure S3). The SHAP plot provides a visual representation of how each feature contributes to the model’s predictions across different age groups. Specifically, the color gradient in the plot represents feature values (blue for low and red for high), while the SHAP values indicate whether a feature increases (positive x-axis) or decreases the likelihood of a PASC diagnosis (negative x-axis). The presence of these features indicate higher probability of PASC (Figure S3). Dyspnea (condition), ECG (electrocardiogram) (procedure), and number of level 1 visits (procedure) were 3 of the most predictive features in 3 of the 4 age groups. The conditions and measurements identified in Table S3 are all closely related to respiratory symptoms of COVID-19. Our findings agree with prior literature that showed recurrent visits (eg, level 1 and level 2 visits) or high-severity emergency department visits to be associated with PASC.23
Within the demographics category, true age (continuous numerical value) was one of the most important features for the pediatric and the young adult groups. In addition, White/non-Hispanic is another key feature in the model’s PASC prediction. The similarities in the top 10 important features (Table S3) between the features of false positives and true positives were analyzed. The values of the features for the 2 groups were found to be comparable (Figure S4). For example, a high proportion of false positive patients reported COVID-19 related symptoms such as cough, dyspnea, and fatigue. Also, the false positives in the older age group had a high number of severe emergency department visits. Therefore, it can be concluded that the false positive patients may have been in critical condition or in need of chronic clinical management.
Testing dataset
The unseen testing set (n = 10 580) consisted of a slightly higher proportion of patients in the pediatric (test: 4%, train: 3%), mid adults group (test: 36%, train: 30%), and older adults group (test: 20% train: 17%) (Table S1). Conversely, the proportion of young adults in the test dataset (35.81%) was lower than in the training dataset (45%). Moreover, the overall proportions of most minority racial groups (Asian, American Indian or Alaska Native, Native Hawaiian, and Other) were roughly similar between the two datasets, but there was an increase in the representation of patients identifying as White (overall percentage from train: 55% vs test: 64%) and a decrease in those identifying as Black or African American (train: 20% vs test: 15%) or missing racial information (train: 12% vs 5%). Also, there was an increase in the prevalence of PASC across all age groups (an overall increase of 6%).
In the testing dataset (n = 10 580), best performances were achieved in the young adult population (AUROC = 0.796, F1 = 0.482), mid aged (AUROC = 0.798, F1 = 0.564), and the older aged groups (AUROC = 0.790, F1 = 0.582). The performance was lowest in the pediatric group (AUROC = 0.718, F1 = 0). We also see drops in the specificity and precision with the test set compared to our 5-fold validation results reported in Table 1, suggesting that our model may have overfit to the training dataset. Across all age groups and demographics, our dataset was successful in distinguishing PASC vs non-PASC with AUROC of 0.814 and F1 score of 0.545.
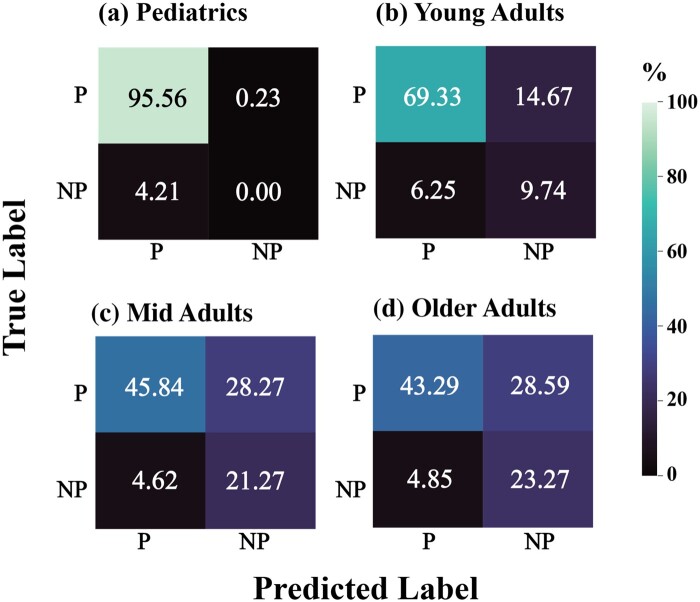
We also investigated the distribution of the true positives, true negatives, false positives, and false negatives of our predicted labels compared to the silver standard. We observe a high false positive rate of our model from the confusion matrices, particularly in the mid adult and older adult groups in the testing dataset (Figure 2).
Figure 2.
Classification performance on the test data where P = PASC and NP = No PASC. While all models perform well on true negatives, false positives are notably high for mid adults and older adults.
Discussion
While data-driven approaches can be powerful for discovering patterns, such methods can generate overly complex solutions that require high computational power and training time. In the context of health care, models that depend on a wide range of variables can also be difficult to explain or interpret in a clinical setting, and it may be impractical to obtain all dependent features for each patient. Hence, our method confined our feature space based on the findings from our literature review, including features that have been shown to be highly predictive of severe instances of COVID-19 infection and PASC. In addition, we stratified our population into subsets of age groups as the phenotypic presentation of PASC and the required clinical testing and interventions vary across ages. Our knowledge-driven feature selection and utilization of an age-stratified classification model allowed us to validate previously identified risk factors while revealing their importance and/or prevalence by age group.
To extract features from the L3C EHR data, we utilized a straightforward and comprehensible method of feature aggregation: collapsing continuous values into categorical values of normal (ie, values within range or values not taken) vs abnormal. The use of these binary indicators allowed us to handle the sparsity of the dataset, imputing the missing values as 0 (corresponding to normal) under the assumption that the feature was not clinically necessary for diagnosis or treatment. By implementing this feature engineering process, we significantly simplified the dataset.
Training and validating models separately for different age groups significantly improved algorithm performance when compared to training on the entire population without age-based stratification. Age-based stratification can yield clinically relevant insights such as important PASC risk factors to look for by age group, enhancing the interpretability of our model and its potential role in clinical decision-making.
While our model has low performance in the pediatric population (<15 years old) in both the training and testing datasets, our model performs well in all other age groups in both the training set under 5-fold cross-validation as well as the hold-out testing dataset. We did observe a decrease in classification performance for all groups with the hold-out test set when compared to the cross-validation results from the training set. Notably, we observed high false positives across all age groups, which may be attributed to potential overfitting during the training phase and/or the difference in the data source in terms of time and location. Also, the higher PASC incidences in the hold-out testing dataset (ie, lower negative to positive support ratio) compared to the training dataset could have led to the lower performance scores in the hold-out testing dataset compared to the mean of our 5-fold cross-validation results. As our model was trained and calibrated on a dataset with a certain class imbalance (ie, higher negative cases), the model simply may not have seen enough examples of the minority class (ie, positive for PASC) to learn from during training.
Overall, our model exhibits high performance in predicting PASC for the young, mid, and older adult age groups. Our model bases its prediction on variables commonly present in the EHR such as a patient’s medical history and list of symptoms commonly associated with COVID-19, and hence can be easily deployed for assessing patients’ risk of PASC after an acute COVID-19 infection. The feature engineering method and modeling techniques we employed allow the model to predict PASC occurrence without requiring numerical lab test results, but rather, can leverage binary features that indicate abnormality. The use of clinically relevant features and transforming continuous variables to binary indicators enhance the model’s interpretability and can, therefore, potentially be deployed as a simple PASC risk prediction tool (eg, a brief self-administered survey for patients).
Limitations
The L3C dataset was curated from multiple hospitals across the United States. The data inevitably contains inherent bias. For example, individuals who cannot easily access health care, whether due to economic constraints or the environment, are underrepresented in the data. This bias may affect the generalizability of these findings to the broader population. Specifically, we found that a patient identifying themselves as “White non-Hispanic” can significantly influence the model’s decision on whether someone develops PASC. Various unobserved confounders could have contributed to this observation: the “White non-Hispanic” population may have higher access to resources and health care, leading to more frequent visits for acute and chronic care management compared to other racial/ethnic groups. Without the consideration of confounding factors and meeting all criteria for causal inference, our current model cannot offer insight as to whether the “White non-Hispanic” population is indeed more likely to develop PASC. However, its feature importance highlights an example of potential bias in the data.
While our defined age ranges for middle-aged adults and older adults agree with clinical age designations,24 we experimentally determined the lower age for the young adult population. This decision was made by observing the distribution of the prevalence of PASC when grouped by age. We observed a drastic decrease in PASC prevalence below the age of 15 (Figure S2). The concept of PASC in the pediatric population may be inherently challenging to define,25 making reliable assessment of long-term outcomes within this demographic difficult. However, within the scope of the L3C challenge, it was important to include populations across the lifespan, and thus pediatric populations were included despite their low representation in the dataset.
In our feature engineering approach for the L3C EHR data analysis, we transformed continuous variables into binary categories and imputed missing values as normal (ie, within normal range for lab values or patient not presenting condition). While this strategy improved the sparsity of our data, it likely resulted in the loss of granular information, potentially leading to oversimplified analyses that might not adequately capture subtle but critical clinical variations. Similarly, due to the relatively low incidence of missing ethnicity data, we imputed the missing ethnicity information with “non-Hispanic,” reflecting the majority in the dataset. We also aggregated all racial categories for Hispanic or Latino participants given the small sample size. These approaches may have oversimplified the demographic nuances and introduce potential biases in our analysis.
Our primary goal for conducting the SHAP analysis was to provide qualitative insights into feature importance, thus we did not generate CIs for the SHAP values. However, for applications such as health care where decision-making needs to be transparent and reliable, obtaining CIs could be crucial for justifying the model’s predictions and further determining whether observed differences in feature importance between models or features are statistically significant.
In addition, the L3C data was randomly time-shifted by a uniform sampling of 1∼180 days to protect patient privacy. Clinical understanding of both COVID-19 and PASC has been continuously evolving with new studies revealing risk factors for prognosis and updated diagnostic and treatment guidelines for clinicians. Depending on when the patient was seen in the clinic or was hospitalized, the clinical guidelines for diagnosis and management for either acute COVID-19 infection or PASC may have been different. As ML models can be sensitive to such nuances, the lack of consideration for the timing of the data may have impacted the performance of our model. The lack of standardization in the definition of PASC and standards of care has been a challenge across the nation. Given that our understanding of PASC has evolved over time and clinicians in different locations follow different guidelines for diagnosing, testing, and managing PASC, our model may have been able to account for all variations in care.
Lastly, our model was designed to predict PASC diagnosis strictly 4 weeks after the initial acute COVID-19 infection diagnosis according to the solicitation of the L3C competition. From our exploratory data analysis, we found that there was a small percentage (4%) of patients with a PASC diagnosis (U09.9) that occurred before 4 weeks had passed since their initial COVID-19 diagnosis. We regarded these cases as false positives based on current clinical guidelines which define PASC as “patients who, four weeks after the diagnosis of SARS-Cov-2 infection, continue to have signs and symptoms not explainable by other causes.”26 While the percentage of false positive diagnoses from the silver standard labels was insignificant (4%), our decision to relabel these patients as true negative cases could have introduced noise in the data and harmed the precision of our model’s predictions. In addition, using a single COVID index date (ie, initial COVID-19 positive test from the EHR record) per person may affect the precision of our labels, as subsequent infections and their potential impact on PASC development were not accounted for in the provided data.
Conclusion
In this paper, we present an age-stratified XGBoost model for predicting PASC ≥ 4 weeks after an initial COVID-19 infection date. By selecting clinically relevant features per literature review, we reduced the features space and enhanced clinical interpretability of our model. Although limitations exist in our proposed approach, our model successfully demonstrated strong performances for the young adults, mid adults, and older adults age groups. For future studies, we aim to perform a detailed analysis of the false positives and incorporate time-domain in our model building (eg, dynamically taking account of the recurrent encounters) to enhance the precision of our prediction.
Supplementary Material
Acknowledgments
N3C attribution: The analyses described in this publication were conducted with data or tools accessed through the NCATS N3C Data Enclave https://covid.cd2h.org and N3C Attribution & Publication Policy v 1.2-2020-08-25b supported by NCATS Contract No. 75N95023D00001, Axle Informatics Subcontract: NCATS-P00438-B. This research was possible because of the patients whose information is included within the data and the organizations (https://ncats.nih.gov/n3c/resources/data-contribution/data-transfer-agreement-signatories) and scientists who have contributed to the on-going development of this community resource (https://doi.org/10.1093/jamia/ocaa196).
Disclaimer: Authorship was determined using ICMJE recommendations. The N3C Publication committee confirmed that this manuscript msid: 1946.264 is in accordance with N3C data use and attribution policies; however, this content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the N3C program.
Institutional Review Board (IRB): The N3C data transfer to NCATS is performed under a Johns Hopkins University Reliance Protocol # IRB00249128 or individual site agreements with NIH. The N3C Data Enclave is managed under the authority of the NIH; information can be found at https://ncats.nih.gov/n3c/resources.
Individual acknowledgements for core contributors: We gratefully acknowledge the following core contributors to N3C: Adam B. Wilcox, Adam M. Lee, Alexis Graves, Alfred (Jerrod) Anzalone, Amin Manna, Amit Saha, Amy Olex, Andrea Zhou, Andrew E. Williams, Andrew Southerland, Andrew T. Girvin, Anita Walden, Anjali A. Sharathkumar, Benjamin Amor, Benjamin Bates, Brian Hendricks, Brijesh Patel, Caleb Alexander, Carolyn Bramante, Cavin Ward-Caviness, Charisse Madlock-Brown, Christine Suver, Christopher Chute, Christopher Dillon, Chunlei Wu, Clare Schmitt, Cliff Takemoto, Dan Housman, Davera Gabriel, David A. Eichmann, Diego Mazzotti, Don Brown, Eilis Boudreau, Elaine Hill, Elizabeth Zampino, Emily Carlson Marti, Emily R. Pfaff, Evan French, Farrukh M. Koraishy, Federico Mariona, Fred Prior, George Sokos, Greg Martin, Harold Lehmann, Heidi Spratt, Hemalkumar Mehta, Hongfang Liu, Hythem Sidky, J.W. Awori Hayanga, Jami Pincavitch, Jaylyn Clark, Jeremy Richard Harper, Jessica Islam, Jin Ge, Joel Gagnier, Joel H. Saltz, Joel Saltz, Johanna Loomba, John Buse, Jomol Mathew, Joni L. Rutter, Julie A. McMurry, Justin Guinney, Justin Starren, Karen Crowley, Katie Rebecca Bradwell, Kellie M. Walters, Ken Wilkins, Kenneth R. Gersing, Kenrick Dwain Cato, Kimberly Murray, Kristin Kostka, Lavance Northington, Lee Allan Pyles, Leonie Misquitta, Lesley Cottrell, Lili Portilla, Mariam Deacy, Mark M. Bissell, Marshall Clark, Mary Emmett, Mary Morrison Saltz, Matvey B. Palchuk, Melissa A. Haendel, Meredith Adams, Meredith Temple-O’Connor, Michael G. Kurilla, Michele Morris, Nabeel Qureshi, Nasia Safdar, Nicole Garbarini, Noha Sharafeldin, Ofer Sadan, Patricia A. Francis, Penny Wung Burgoon, Peter Robinson, Philip R.O. Payne, Rafael Fuentes, Randeep Jawa, Rebecca Erwin-Cohen, Rena Patel, Richard A. Moffitt, Richard L. Zhu, Rishi Kamaleswaran, Robert Hurley, Robert T. Miller, Saiju Pyarajan, Sam G. Michael, Samuel Bozzette, Sandeep Mallipattu, Satyanarayana Vedula, Scott Chapman, Shawn T. O’Neil, Soko Setoguchi, Stephanie S. Hong, Steve Johnson, Tellen D. Bennett, Tiffany Callahan, Umit Topaloglu, Usman Sheikh, Valery Gordon, Vignesh Subbian, Warren A. Kibbe, Wenndy Hernandez, Will Beasley, Will Cooper, William Hillegass, Xiaohan Tanner Zhang. Details of contributions available at covid.cd2h.org/core-contributors.
Data partners with released data: The following institutions whose data is released or pending: Available: Advocate Health Care Network—UL1TR002389: The Institute for Translational Medicine (ITM) • Aurora Health Care Inc—UL1TR002373: Wisconsin Network For Health Research • Boston University Medical Campus—UL1TR001430: Boston University Clinical and Translational Science Institute • Brown University—U54GM115677: Advance Clinical Translational Research (Advance-CTR) • Carilion Clinic—UL1TR003015: iTHRIV Integrated Translational health Research Institute of Virginia • Case Western Reserve University—UL1TR002548: The Clinical & Translational Science Collaborative of Cleveland (CTSC) • Charleston Area Medical Center—U54GM104942: West Virginia Clinical and Translational Science Institute (WVCTSI) • Children’s Hospital Colorado—UL1TR002535: Colorado Clinical and Translational Sciences Institute • Columbia University Irving Medical Center—UL1TR001873: Irving Institute for Clinical and Translational Research • Dartmouth College—None (Voluntary) Duke University—UL1TR002553: Duke Clinical and Translational Science Institute • George Washington Children’s Research Institute—UL1TR001876: Clinical and Translational Science Institute at Children’s National (CTSA-CN) • George Washington University—UL1TR001876: Clinical and Translational Science Institute at Children’s National (CTSA-CN) • Harvard Medical School—UL1TR002541: Harvard Catalyst • Indiana University School of Medicine—UL1TR002529: Indiana Clinical and Translational Science Institute • Johns Hopkins University—UL1TR003098: Johns Hopkins Institute for Clinical and Translational Research • Louisiana Public Health Institute—None (Voluntary) • Loyola Medicine—Loyola University Medical Center • Loyola University Medical Center—UL1TR002389: The Institute for Translational Medicine (ITM) • Maine Medical Center—U54GM115516: Northern New England Clinical & Translational Research (NNE-CTR) Network • Mary Hitchcock Memorial Hospital & Dartmouth Hitchcock Clinic—None (Voluntary) • Massachusetts General Brigham—UL1TR002541: Harvard Catalyst • Mayo Clinic Rochester—UL1TR002377: Mayo Clinic Center for Clinical and Translational Science (CCaTS) • Medical University of South Carolina—UL1TR001450: South Carolina Clinical & Translational Research Institute (SCTR) • MITRE Corporation—None (Voluntary) • Montefiore Medical Center—UL1TR002556: Institute for Clinical and Translational Research at Einstein and Montefiore • Nemours—U54GM104941: Delaware CTR ACCEL Program • NorthShore University HealthSystem—UL1TR002389: The Institute for Translational Medicine (ITM) • Northwestern University at Chicago—UL1TR001422: Northwestern University Clinical and Translational Science Institute (NUCATS) • OCHIN—INV-018455: Bill and Melinda Gates Foundation grant to Sage Bionetworks • Oregon Health & Science University—UL1TR002369: Oregon Clinical and Translational Research Institute • Penn State Health Milton S. Hershey Medical Center—UL1TR002014: Penn State Clinical and Translational Science Institute • Rush University Medical Center—UL1TR002389: The Institute for Translational Medicine (ITM) • Rutgers, The State University of New Jersey—UL1TR003017: New Jersey Alliance for Clinical and Translational Science • Stony Brook University—U24TR002306 • The Alliance at the University of Puerto Rico, Medical Sciences Campus—U54GM133807: Hispanic Alliance for Clinical and Translational Research (The Alliance) • The Ohio State University—UL1TR002733: Center for Clinical and Translational Science • The State University of New York at Buffalo—UL1TR001412: Clinical and Translational Science Institute • The University of Chicago—UL1TR002389: The Institute for Translational Medicine (ITM) • The University of Iowa—UL1TR002537: Institute for Clinical and Translational Science • The University of Miami Leonard M. Miller School of Medicine—UL1TR002736: University of Miami Clinical and Translational Science Institute • The University of Michigan at Ann Arbor—UL1TR002240: Michigan Institute for Clinical and Health Research • The University of Texas Health Science Center at Houston—UL1TR003167: Center for Clinical and Translational Sciences (CCTS) • The University of Texas Medical Branch at Galveston—UL1TR001439: The Institute for Translational Sciences • The University of Utah—UL1TR002538: Uhealth Center for Clinical and Translational Science • Tufts Medical Center—UL1TR002544: Tufts Clinical and Translational Science Institute • Tulane University—UL1TR003096: Center for Clinical and Translational Science • The Queens Medical Center—None (Voluntary) • University Medical Center New Orleans—U54GM104940: Louisiana Clinical and Translational Science (LA CaTS) Center • University of Alabama at Birmingham—UL1TR003096: Center for Clinical and Translational Science • University of Arkansas for Medical Sciences—UL1TR003107: UAMS Translational Research Institute • University of Cincinnati—UL1TR001425: Center for Clinical and Translational Science and Training • University of Colorado Denver, Anschutz Medical Campus—UL1TR002535: Colorado Clinical and Translational Sciences Institute • University of Illinois at Chicago—UL1TR002003: UIC Center for Clinical and Translational Science • University of Kansas Medical Center—UL1TR002366: Frontiers: University of Kansas Clinical and Translational Science Institute • University of Kentucky—UL1TR001998: UK Center for Clinical and Translational Science • University of Massachusetts Medical School Worcester—UL1TR001453: The UMass Center for Clinical and Translational Science (UMCCTS) • University Medical Center of Southern Nevada—None (voluntary) • University of Minnesota—UL1TR002494: Clinical and Translational Science Institute • University of Mississippi Medical Center—U54GM115428: Mississippi Center for Clinical and Translational Research (CCTR) • University of Nebraska Medical Center—U54GM115458: Great Plains IDeA-Clinical & Translational Research • University of North Carolina at Chapel Hill—UL1TR002489: North Carolina Translational and Clinical Science Institute • University of Oklahoma Health Sciences Center—U54GM104938: Oklahoma Clinical and Translational Science Institute (OCTSI) • University of Pittsburgh—UL1TR001857: The Clinical and Translational Science Institute (CTSI) • University of Pennsylvania—UL1TR001878: Institute for Translational Medicine and Therapeutics • University of Rochester—UL1TR002001: UR Clinical & Translational Science Institute • University of Southern California—UL1TR001855: The Southern California Clinical and Translational Science Institute (SC CTSI) • University of Vermont—U54GM115516: Northern New England Clinical & Translational Research (NNE-CTR) Network • University of Virginia—UL1TR003015: iTHRIV Integrated Translational health Research Institute of Virginia • University of Washington—UL1TR002319: Institute of Translational Health Sciences • University of Wisconsin-Madison—UL1TR002373: UW Institute for Clinical and Translational Research • Vanderbilt University Medical Center—UL1TR002243: Vanderbilt Institute for Clinical and Translational Research • Virginia Commonwealth University—UL1TR002649: C. Kenneth and Dianne Wright Center for Clinical and Translational Research • Wake Forest University Health Sciences—UL1TR001420: Wake Forest Clinical and Translational Science Institute • Washington University in St Louis—UL1TR002345: Institute of Clinical and Translational Sciences • Weill Medical College of Cornell University—UL1TR002384: Weill Cornell Medicine Clinical and Translational Science Center • West Virginia University—U54GM104942: West Virginia Clinical and Translational Science Institute (WVCTSI)
Submitted: Icahn School of Medicine at Mount Sinai—UL1TR001433: ConduITS Institute for Translational Sciences • The University of Texas Health Science Center at Tyler—UL1TR003167: Center for Clinical and Translational Sciences (CCTS) • University of California, Davis—UL1TR001860: UCDavis Health Clinical and Translational Science Center • University of California, Irvine—UL1TR001414: The UC Irvine Institute for Clinical and Translational Science (ICTS) • University of California, Los Angeles—UL1TR001881: UCLA Clinical Translational Science Institute • University of California, San Diego—UL1TR001442: Altman Clinical and Translational Research Institute • University of California, San Francisco—UL1TR001872: UCSF Clinical and Translational Science Institute
NYU Langone Health Clinical Science Core, Data Resource Core, and PASC Biorepository Core—OTA-21-015A: Post-Acute Sequelae of SARS-CoV-2 Infection Initiative (RECOVER)
Pending: Arkansas Children’s Hospital—UL1TR003107: UAMS Translational Research Institute • Baylor College of Medicine—None (Voluntary) • Children’s Hospital of Philadelphia—UL1TR001878: Institute for Translational Medicine and Therapeutics • Cincinnati Children’s Hospital Medical Center—UL1TR001425: Center for Clinical and Translational Science and Training • Emory University—UL1TR002378: Georgia Clinical and Translational Science Alliance • HonorHealth—None (Voluntary) • Loyola University Chicago—UL1TR002389: The Institute for Translational Medicine (ITM) • Medical College of Wisconsin—UL1TR001436: Clinical and Translational Science Institute of Southeast Wisconsin • MedStar Health Research Institute—None (Voluntary) • Georgetown University—UL1TR001409: The Georgetown-Howard Universities Center for Clinical and Translational Science (GHUCCTS) • MetroHealth—None (Voluntary) • Montana State University—U54GM115371: American Indian/Alaska Native CTR • NYU Langone Medical Center—UL1TR001445: Langone Health’s Clinical and Translational Science Institute • Ochsner Medical Center—U54GM104940: Louisiana Clinical and Translational Science (LA CaTS) Center • Regenstrief Institute—UL1TR002529: Indiana Clinical and Translational Science Institute • Sanford Research—None (Voluntary) • Stanford University—UL1TR003142: Spectrum: The Stanford Center for Clinical and Translational Research and Education • The Rockefeller University—UL1TR001866: Center for Clinical and Translational Science • The Scripps Research Institute—UL1TR002550: Scripps Research Translational Institute • University of Florida—UL1TR001427: UF Clinical and Translational Science Institute • University of New Mexico Health Sciences Center—UL1TR001449: University of New Mexico Clinical and Translational Science Center • University of Texas Health Science Center at San Antonio—UL1TR002645: Institute for Integration of Medicine and Science • Yale New Haven Hospital—UL1TR001863: Yale Center for Clinical Investigation.
Contributor Information
Will Ke Wang, Department of Biomedical Engineering, Duke University, Durham, NC 27708, United States.
Hayoung Jeong, Department of Biomedical Engineering, Duke University, Durham, NC 27708, United States.
Leeor Hershkovich, Department of Biomedical Engineering, Duke University, Durham, NC 27708, United States.
Peter Cho, Department of Biomedical Engineering, Duke University, Durham, NC 27708, United States.
Karnika Singh, Department of Biomedical Engineering, Duke University, Durham, NC 27708, United States.
Lauren Lederer, Department of Biomedical Engineering, Duke University, Durham, NC 27708, United States.
Ali R Roghanizad, Department of Biomedical Engineering, Duke University, Durham, NC 27708, United States.
Md Mobashir Hasan Shandhi, School of Electrical, Computer, and Energy Engineering, Arizona State University, Tempe, AZ 85281, United States; Biodesign Institute Center for Bioelectronics and Biosensors, Arizona State University, Tempe, AZ 85281, United States.
Warren Kibbe, Department of Biostatistics and Bioinformatics, Duke University School of Medicine, Durham, NC 27708, United States.
Jessilyn Dunn, Department of Biomedical Engineering, Duke University, Durham, NC 27708, United States; Department of Biostatistics and Bioinformatics, Duke University School of Medicine, Durham, NC 27708, United States.
National COVID Cohort Collaborative (N3C) Consortium:
Adam B Wilcox, Adam M Lee, Alexis Graves, Alfred (Jerrod) Anzalone, Amin Manna, Amit Saha, Amy Olex, Andrea Zhou, Andrew E Williams, Andrew Southerland, Andrew T Girvin, Anita Walden, Anjali A Sharathkumar, Benjamin Amor, Benjamin Bates, Brian Hendricks, Brijesh Patel, Caleb Alexander, Carolyn Bramante, Cavin Ward-Caviness, Charisse Madlock-Brown, Christine Suver, Christopher Chute, Christopher Dillon, Chunlei Wu, Clare Schmitt, Cliff Takemoto, Dan Housman, Davera Gabriel, David A Eichmann, Diego Mazzotti, Don Brown, Eilis Boudreau, Elaine Hill, Elizabeth Zampino, Emily Carlson Marti, Emily R Pfaff, Evan French, Farrukh M Koraishy, Federico Mariona, Fred Prior, George Sokos, Greg Martin, Harold Lehmann, Heidi Spratt, Hemalkumar Mehta, Hongfang Liu, Hythem Sidky, J W Awori Hayanga, Jami Pincavitch, Jaylyn Clark, Jeremy Richard Harper, Jessica Islam, Jin Ge, Joel Gagnier, Joel H Saltz, Joel Saltz, Johanna Loomba, John Buse, Jomol Mathew, Joni L Rutter, Julie A McMurry, Justin Guinney, Justin Starren, Karen Crowley, Katie Rebecca Bradwell, Kellie M Walters, Ken Wilkins, Kenneth R Gersing, Kenrick Dwain Cato, Kimberly Murray, Kristin Kostka, Lavance Northington, Lee Allan Pyles, Leonie Misquitta, Lesley Cottrell, Lili Portilla, Mariam Deacy, Mark M Bissell, Marshall Clark, Mary Emmett, Mary Morrison Saltz, Matvey B Palchuk, Melissa A Haendel, Meredith Adams, Meredith Temple-O’Connor, Michael G Kurilla, Michele Morris, Nabeel Qureshi, Nasia Safdar, Nicole Garbarini, Noha Sharafeldin, Ofer Sadan, Patricia A Francis, Penny Wung Burgoon, Peter Robinson, Philip R O Payne, Rafael Fuentes, Randeep Jawa, Rebecca Erwin-Cohen, Rena Patel, Richard A Moffitt, Richard L Zhu, Rishi Kamaleswaran, Robert Hurley, Robert T Miller, Saiju Pyarajan, Sam G Michael, Samuel Bozzette, Sandeep Mallipattu, Satyanarayana Vedula, Scott Chapman, Shawn T O’Neil, Soko Setoguchi, Stephanie S Hong, Steve Johnson, Tellen D Bennett, Tiffany Callahan, Umit Topaloglu, Usman Sheikh, Valery Gordon, Vignesh Subbian, Warren A Kibbe, Wenndy Hernandez, Will Beasley, Will Cooper, William Hillegass, and Xiaohan Tanner Zhang
Supplementary material
Supplementary material is available at JAMIA Open online.
Funding
None declared.
Conflicts of interest
None declared.
Data availability
The N3C Data Enclave (covid.cd2h.org/enclave) is accessible by investigators at institutions that have signed a Data Use Agreement with National Center for Advancing Translational Sciences (NCATS). Researchers seeking access must submit a Data Use Request (DUR) through the N3C Data Enclave. All DURs are subject to review and approval by the N3C Data Access Committee.
References
- 1. Burns A. Long COVID: what do the latest data show? KFF. Accessed April 8, 2023. https://www.kff.org/policy-watch/long-covid-what-do-latest-data-show/
- 2. Davis HE, McCorkell L, Vogel JM, et al. Long COVID: major findings, mechanisms and recommendations. Nat Rev Microbiol. 2023;21:133-146. 10.1038/s41579-022-00846-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lai C-C, Hsu C-K, Yen M-Y, et al. Long COVID: an inevitable sequela of SARS-CoV-2 infection. J Microbiol Immunol Infect. 2023;56:1-9. 10.1016/j.jmii.2022.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Kessel SAM, Olde Hartman TC, Lucassen PLBJ, et al. Post-acute and long-COVID-19 symptoms in patients with mild diseases: a systematic review. Fam Pract. 2021;39:159-167. 10.1093/fampra/cmab076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O’Mahoney LL, Routen A, Gillies C, et al. The prevalence and long-term health effects of long covid among hospitalised and non-hospitalised populations: a systematic review and meta-analysis. eClinicalMedicine. 2022;55:101762. 10.1016/j.eclinm.2022.101762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haendel MA, Chute CG, Bennett TD, et al. ; N3C Consortium. The National COVID Cohort Collaborative (N3C): rationale, design, infrastructure, and deployment. J Am Med Inform Assoc. 2021;28:427-443. 10.1093/jamia/ocaa196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pfaff ER, Girvin AT, Bennett TD, et al. Identifying who has Long COVID in the USA: a machine learning approach using N3C data. Lancet Digit Health. 2022;4:e532-e541. 10.1016/S2589-7500(22)00048-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Socia D, Larie D, Feuerwerker S, et al. Prediction of Long COVID based on severity of initial COVID-19 infection: differences in predictive feature sets between hospitalized versus non-hospitalized index infections. MedRxiv, 10.1101/2023.01.16.23284634, 2023. [DOI]
- 9. Pfaff ER, Madlock-Brown C, Baratta JM, et al. ; RECOVER Consortium. Coding Long COVID: characterizing a new disease through an ICD-10 lens. BMC Med. 2023;21:58. 10.1186/s12916-023-02737-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang H, Zang C, Xu Z, et al. Data-driven identification of post-acute SARS-CoV-2 infection subphenotypes. Nat Med. 2023;29:226-235. 10.1038/s41591-022-02116-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. CDC. Risk for COVID-19 infection, hospitalization, and death by age group. Centers for Disease Control and Prevention. 2020. Accessed March 7, 2023. https://www.cdc.gov/coronavirus/2019-ncov/covid-data/investigations-discovery/hospitalization-death-by-age.html
- 12. Wang L, Foer D, MacPhaul E, et al. PASCLex: a comprehensive post-acute sequelae of COVID-19 (PASC) symptom lexicon derived from electronic health record clinical notes. J Biomed Inform. 2022;125:103951. 10.1016/j.jbi.2021.103951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tests and Procedures. Mayo Clinic. Accessed April 16, 2023. https://www.mayoclinic.org/tests-procedures
- 14. Holmes JH, Beinlich J, Boland MR, et al. Why is the electronic health record so challenging for research and clinical care? Methods Inf Med. 2021;60:32-48. 10.1055/s-0041-1731784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Unim B, Palmieri L, Lo Noce C, et al. Prevalence of COVID-19-related symptoms by age group. Aging Clin Exp Res. 2021;33:1145-1147. 10.1007/s40520-021-01809-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kompaniyets L. Post–COVID-19 symptoms and conditions among children and adolescents–United States, March 1, 2020–January 31, 2022. MMWR Morb Mortal Wkly Rep. 2022;71:996. 10.15585/mmwr.mm7131a3 [DOI] [PMC free article] [PubMed]
- 17. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. https://scikit-learn.org/ [DOI] [PMC free article] [PubMed]
- 18. Lundberg SM, Lee S-I.. A unified approach to interpreting model predictions. In: Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS'17). Curran Associates, Inc.; 2017. 10.48550/arXiv.1705.07874 [DOI] [Google Scholar]
- 19. Lundberg SM, Erion G, Chen H, et al. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell. 2020;2:56-67. 10.1038/s42256-019-0138-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997;30:1145-1159. 10.1016/S0031-3203(96)00142-2 [DOI] [Google Scholar]
- 21. Powers DM. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv preprint arXiv:2010.16061. 2020;41. [Google Scholar]
- 22. Niculescu-Mizil A, Caruana R. Predicting good probabilities with supervised learning. In: Proceedings of the 22nd International Conference on Machine Learning. Association for Computing Machinery; 2005:625-632.
- 23. Jones R, Davis A, Stanley B, et al. Risk predictors and symptom features of Long COVID within a broad primary care patient population including both tested and untested patients. Pragmat Obs Res. 2021;12:93-104. 10.2147/POR.S316186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Estiri H, Strasser ZH, Brat GA, et al. ; The Consortium for Characterization of COVID-19 by EHR (4CE). Evolving phenotypes of non-hospitalized patients that indicate Long COVID. BMC Med. 2021;19:249. 10.1186/s12916-021-02115-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rao S, Gross RS, Mohandas S, et al. Post-acute sequelae of SARS-CoV-2 in children. Pediatrics. 2024;153:e2023062570. 10.1542/peds.2023-062570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sisó-Almirall A, Brito-Zerón P, Conangla Ferrín L, et al. Long covid-19: proposed primary care clinical guidelines for diagnosis and disease management. Int J Environ Res Public Health. 2021;18:4350. 10.3390/ijerph18084350 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The N3C Data Enclave (covid.cd2h.org/enclave) is accessible by investigators at institutions that have signed a Data Use Agreement with National Center for Advancing Translational Sciences (NCATS). Researchers seeking access must submit a Data Use Request (DUR) through the N3C Data Enclave. All DURs are subject to review and approval by the N3C Data Access Committee.