Abstract
Huanglongbing (HLB) is the most serious citrus disease in Vietnam. For the first time, this paper reported root microbial data of HLB-infected Citrus nobilis grown in Dak Lak Province, Vietnam, for further work toward controlling this disease. Roots of HLB-infected C. nobilis were collected, and the genomic DNA was isolated. The Illumina platform was used to sequence the shotgun metagenomic library, and bioinformatic tools were used to analyze sequenced data. We found that 4 kingdoms, 27 phyla, 57 classes, 124 orders, 246 families, 722 genera, and 1758 species of the root microbiome were identified from the sample. Actinomycetota was the predominant phylum (47.37 %), and biosynthesis was the primary function (57.38 %) of the microbiome.
Keywords: Huanglongbing-infected citrus, Root microbiome, Shotgun metagenomics sequencing data, Actinomycetota
Specifications Table
| Subject | Microbiology |
| Specific subject area | The root microbiome of diseased citrus |
| Data format | Raw (fastq.gz files), Filtered, and Analyzed |
| Type of data | Figures and Fastq files |
| Data collection | Root samples of HLB-infected C. nobilis L., which showed Huanglongbing symptoms in leaves, were collected from three positions in Dak Lak. The surface of the roots was sterilized and used to isolate the genomic DNA using DNeasy PowerSoil Pro kit. The shotgun metagenomic library was created using the NEBNext Ultra II DNA Library Prep Kit for Illumina. The DNBSeq-G99 machine was utilized to sequence the shotgun metagenomic library with the Illumina platform. Bcl2fastq 2.20 was used to demultiplex the raw data. Trimmomatic 0.39 and Cutadapt 2.10 were used to filter the sequence data. Kraken2 was used to assess taxonomic profiles, and the MetaCyc database was utilized to analyze functional characteristics. |
| Data source location | • Institution: Institute of Biotechnology and Environment, Tay Nguyen University • District/Province/Country: Buon Don/Dak Lak/ Vietnam • Latitude and longitude coordinates for collected samples: 12°42′59′′N, 107°57′36′′E; 12°43′00′′N, 107°57′32′′E; 12°43′04′′N, 107°57′33′′E |
| Data accessibility | Raw sequences (fastq.gz files) Repository name: Mendeley Data Data identification number: doi: 10.17632/cmvmnnmn4t.1 Direct URL to data: https://data.mendeley.com/datasets/cmvmnnmn4t/1 |
1. Value of the Data
-
•
Data provided valuable taxonomic and functional profiles of the root microbiome of HLB-infected C. nobilis cultivated in Dak Lak, Vietnam.
-
•
Data provided a valuable background of root microbial resources, especially new ones.
-
•
Data could be valuable for further experiments concerning the application of the microbiome for green citrus production.
2. Background
Citrus is one of the main exporters of fruits in Vietnam. Of the citrus cultivars, C. nobilis L. was the main cultivar cultivated in the Dak Lak Province of the country. Currently, citrus plants in this province and Vietnam face the HLB disease caused by unculturable bacteria from the genus Candidatus Liberibacter. HLB is the most serious threat in the citrus industry worldwide [1]. Some ways to control HLB-infected citrus have been used, including using free HLB-infected citrus, cultivation techniques, and controlling citrus psyllid insect vectors [[2], [3], [4]]; however, this disease has not been cured. To our knowledge, data on the composition and functional profiles of the healthy C. nobilis root microbiome have been demonstrated [5]; however, the data on HLB-infected C. nobilis are still unknown. This work aimed to establish data on the root microbiome of HLB-infected C. nobilis using shotgun metagenomics to explore novel secondary metabolites further and apply microorganisms for green citrus production.
3. Data Description
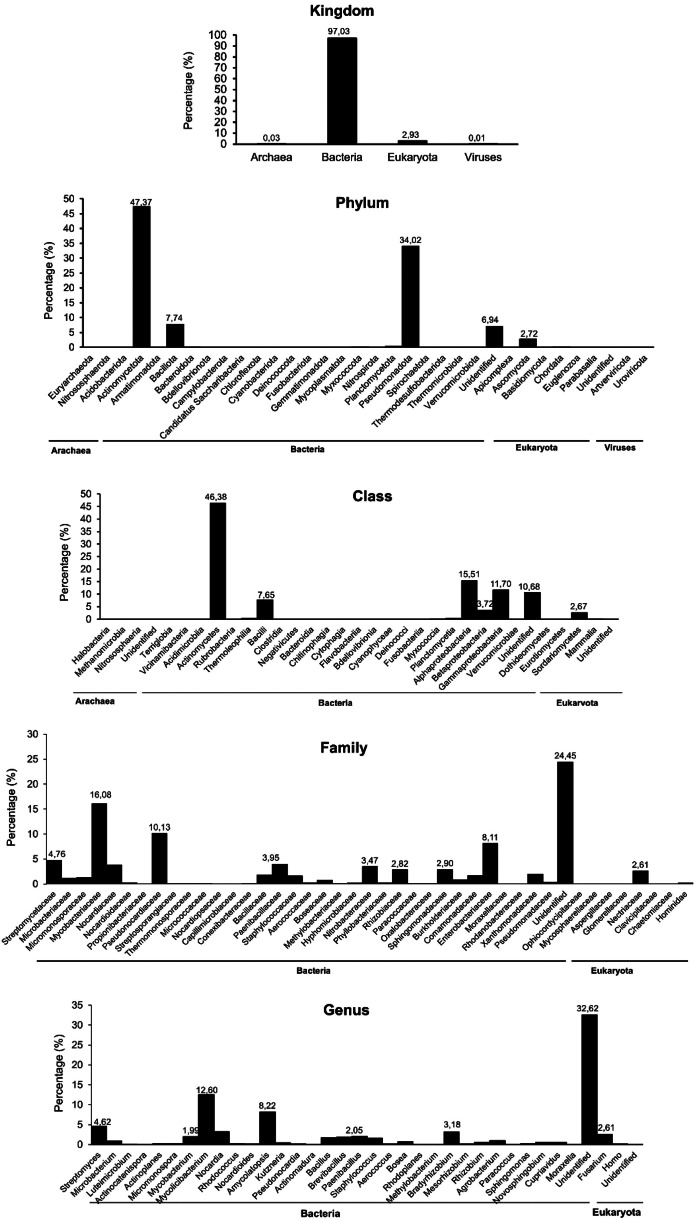
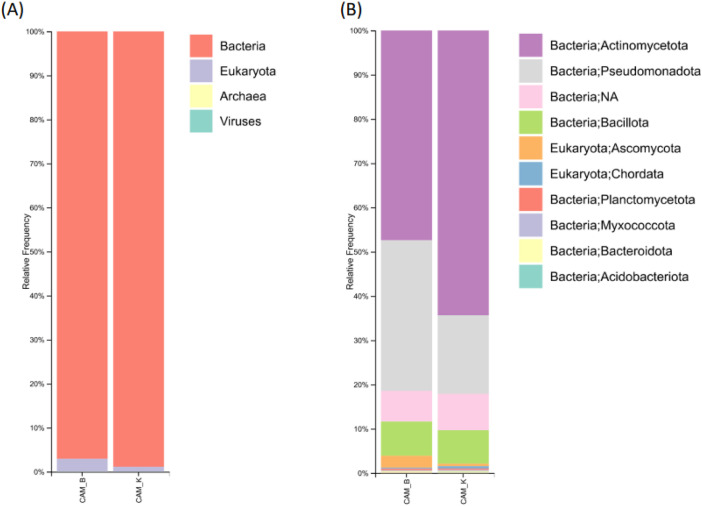
In this experiment, 3190,632 reads were filtered from 32,926,765 raw reads and used to analyze the taxonomic and functional profiles. As shown in Tables S1–S7 and Fig. 1, we identified 4 kingdoms of root microorganisms, among them, bacteria accounted for (97.03 %), followed by eukaryota (2.93 %), archaea (0.03 %), and viruses (0.006 %). Of the 27 identified phyla, Actinomycetota (47.37 %) was the main phylum, followed by Pseudomonadota (34.02 %). Among 57 classes, Actinomycetes (46.38 %), Alphaproteobacteria (15.51 %), Gammaproteobacteria (11.69 %), and Bacilli (7.65 %) were shown to be the predominant classes. Of 124 orders, Mycobacteriales (20.85 %), Pseudonocardiales (10.13 %), Hyphomicrobiales (9.91 %), Enterobacterales (8.60 %), and Bacillales (7.39 %) were the most dominant. Mycobacteriaceae (18.08 %), Pseudonocardiaceae (10.13 %), Enterobacteriaceae (8.11 %), Streptomycetaceae (4.74 %), Paenibacillaceae (3.95 %), and Rhizobiaceae (2.82 %) were the predominant of 246 identified families. Of 722 genera, Mycolicibacterium (12.60 %) was the primary genus, followed by Amycolatopsis (8.22 %), Enterobacter (4.88 %), Streptomyces (4.62 %), and Bradyrhizobium (3.18 %). Finally, 1758 species of the microbiome were identified from the sample; among them, Mycolicibacterium rhodesiae (4.92 %), Mycolicibacterium mucogenicum (2.58 %), and Brevibacillus brevis (1.81 %) were the most abundant. Moreover, Candidatus Liberibacter asiaticus (0.071 %), a causal agent of Huanglongbing [1], was identified from the microbiome. Total unidentified families and genera of the root microbiome were calculated to be 24.72 % and 32.35 %, respectively. Furthermore, we identified numerous bacterial genera and species belonging to Actinomyces, Streptomyces, Bacillus, Brevibacillus, Paenibacillus, and Pseudomonas, which were reported to play an important role in agricultural cultivation [[6], [7], [8], [9], [10], [11], [12]] and suppressing citrus Huanglongbing [[13], [14], [15]], from the microbiome. A comparison (Fig. 2) shows that taxonomic profiles of the root microbiome of healthy and HLB-infected C. nobilis cultivated in Dak Lak differed at kingdom and phylum levels. For example, the kingdom bacteria of healthy C. nobilis was more abundant than that of the HLB-infected C. nobilis. The phylum Actinomycetota of healthy C. nobilis was more predominant than that of the HLB-infected C. nobilis. Raw sequences (fastq.gz files) were deposited in Mendeley Data and can be downloaded at https://data.mendeley.com/datasets/cmvmnnmn4t/1.
Fig. 1.
Taxonomic profiles of the predominant microbiome of HLB-infected Citrus nobilis roots at levels of kingdom, phylum, class, family, and genus.
Fig. 2.
Comparison of taxonomic profiles of the root microbiome of healthy and HLB-infected Citrus nobilis at kingdom (A) and phylum (B) levels. CAM_B, HLB-infected Citrus nobilis (this study); CAM_B, healthy Citrus nobilis (data referred from Tran et al., [5]. NA, unidentified phylum.
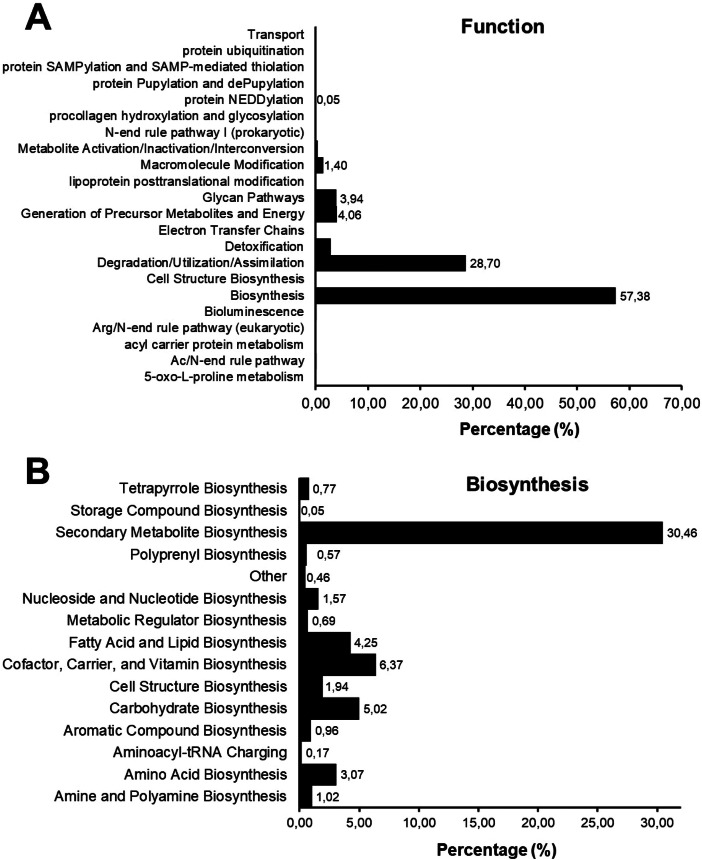
As exhibited in Fig. 3A, biosynthesis (57.38 %) was found to be the primary function of the C. nobilis root microbiome, degradation/utilization/assimilation (28.7 %) was the second most abundant function, followed by generation of precursor metabolites and energy (4.06 %), and glycan pathways (3.94 %). Of the functions concerning biosynthesis, secondary metabolite biosynthesis (30.46 %) was the most predominant, followed by cofactor, prosthetic group, electron carrier, and vitamin biosynthesis (6.37 %); carbohydrate biosynthesis (5.02 %); fatty acid and lipid biosynthesis (4.25 %); amino acid biosynthesis (3.07 %); cell structure biosynthesis (1.94 %); and nucleoside and nucleotide biosynthesis (1.57 %) (Fig. 3B). Raw sequences (fastq.gz files) were deposited in Mendeley Data and can be downloaded at https://data.mendeley.com/datasets/cmvmnnmn4t/1.
Fig. 3.
Functional profiles of the root microbiome of HLB-infected Citrus nobilis.
4. Experimental Design, Materials and Methods
Three root samples (about 70 g each) of HLB-infected C. nobilis L., which showed Huanglongbing symptoms in leaves, were collected from three positions (12°42′59′′N, 107°57′36′′E; 12°43′00′′N, 107°57′32′′E; 12°43′04′′N, 107°57′33′′E) in Dak Lak. The leaves exhibited non-symmetrical mottling, thickened, expanded, corky midribs, and indications of zinc insufficiency, which included erect leaves about the shoot [4] (Fig. 4). The samples were combined to generate the representative sample. The root was washed with tap water, then treated with tween 80 for 10 min, and washed again with sterilized water. After that, the surface of the root was sterilized with 2 % sodium hypochlorite solution for 10 min and then 70 % ethanol for 1 min. Finally, the root was washed six times with sterilized distilled water [16]. DNeasy PowerSoil Pro kit (Qiagen, Germany) was used to isolate the genomic DNA from the sample. Using the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, USA), the shotgun metagenomic library was created. The DNBSeq-G99 (MGI) machine was utilized to sequence the shotgun metagenomic library with the Illumina platform (2 × 150 PE). Bcl2fastq 2.20 was used to demultiplex the raw data [17]. Trimmomatic 0.39 [18] and Cutadapt 2.10 [19] were used to filter the sequence data. Kraken2 [20] was used to assess taxonomic profiles, while the MetaCyc database [21] was utilized to analyze functional characteristics.
Fig. 4.
Citrus nobilis exhibiting Huanglongbing symptoms in leaves cultivated in Dak Lak, Vietnam.
Limitations
Not applicable.
Ethics Statement
The current work does not involve human subjects, animal experiments, or any data collected from social media platforms.
CRediT Author Statement
Dinh Minh Tran: Conceptualization, Methodology, Investigation, Formal analysis, Software, Data curation, Validation, Visualization, Writing, Review and Editing. Thi Huyen Nguyen: Investigation, Formal analysis. Anh Dzung Nguyen: Conceptualization, Sampling, Data curation, Validation, Visualization.
Acknowledgments
Tay Nguyen University supported this work under grant number T2024-49CBTĐ.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2024.111061.
Appendix. Supplementary materials
Data Availability
References
- 1.da Graça J.V., Douhan G.W., Halbert S.E., Keremane M.L., Lee R.F., Vidalakis G., Zhao H. Huanglongbing: an overview of a complex pathosystem ravaging the world's citrus. J. Integr. Plant Biol. 2016;58(4):373–387. doi: 10.1111/jipb.12437. [DOI] [PubMed] [Google Scholar]
- 2.C. Yang, V. Ancona, An overview of the mechanisms against ``Candidatus Liberibacter asiaticus'': virulence targets, citrus defenses, and microbiome. Front. Microbiol. 13 (2022) 850588. doi: 10.3389/fmicb.2022.850588. [DOI] [PMC free article] [PubMed]
- 3.Li J., Li L., Pang Z., Kolbasov V.G., Ehsani R., Carter E.W., Wang N. Developing citrus huanglongbing (HLB) management strategies based on the severity of symptoms in HLB-endemic citrus-producing regions. Phytopathology. 2019;109(4):582–592. doi: 10.1094/PHYTO-08-18-0287-R. [DOI] [PubMed] [Google Scholar]
- 4.Li X., Ruan H., Zhou C., Meng X., Chen W. Controlling Citrus Huanglongbing: green sustainable development route is the future. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.760481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tran D.M., Nguyen D.S., Nguyen T.H., Tran T.P.H., Nguyen A.D. Shotgun metagenomic dataset of root endophytic microbiome of citrus (Citrus Nobilis L.) Data Br. 2024;56:110777. doi: 10.1016/j.dib.2024.110777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gousia J., Ishfaq S., Uqab B., Mudasir S. In: Microbiomes for the Management of Agricultural Sustainability. Dar G.H., Bhat R.A., Mehmood M.A., editors. Springer; Cham: 2023. Actinomycetes as biofertilisers for sustainable agriculture; pp. 183–192. [Google Scholar]
- 7.Olanrewaju O.S., Babalola O.O. Streptomyces: implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 2019;103:1179–1188. doi: 10.1007/s00253-018-09577-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ejaz U., Sohail M., Ghanemi A. Cellulases: from bioactivity to a variety of industrial applications. Biomimetics (Basel) 2021;6:44. doi: 10.3390/biomimetics6030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saxena A.K., Kumar M., Chakdar H., Anuroopa N., Bagyaraj D.J. Bacillus species in soil as a natural resource for plant health and nutrition. J. Appl. Microbiol. 2020;128:1583–1594. doi: 10.1111/jam.14506. [DOI] [PubMed] [Google Scholar]
- 10.Panda A.K., Bisht S.S., DeMondal S., Senthil Kumar N., Gurusubramanian G., Panigrahi A.K. Brevibacillus as a biological tool: a short review. Antonie Van Leeuwenhoek. 2014;105:623–639. doi: 10.1007/s10482-013-0099-7. [DOI] [PubMed] [Google Scholar]
- 11.Lal S., Chiarini L., Tabacchioni S. In: Bacilli and Agrobiotechnology. Islam M., Rahman M., Pandey P., Jha C., Aeron A., editors. Springer; Cham: 2016. New insights in plant-associated Paenibacillus species: biocontrol and plant growth-promoting activity; pp. 237–279. [Google Scholar]
- 12.Bhat B.A., Tariq L., Nissar S., Islam S.T., Islam S.U., Mangral Z., Ilyas N., Sayyed R.Z., Muthusamy G., Kim W., Dar T.U.H. The role of plant-associated rhizobacteria in plant growth, biocontrol and abiotic stress management. J. Appl. Microbiol. 2022;133:2717–2741. doi: 10.1111/jam.15796. [DOI] [PubMed] [Google Scholar]
- 13.Dominguez J., Jayachandran K., Stover E., Krystel J., Shetty K.G. Endophytes and plant extracts as potential antimicrobial agents against Candidatus Liberibacter asiaticus, causal agent of Huanglongbing. Microorganisms. 2023;11(6):1529. doi: 10.3390/microorganisms11061529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., Chen Y., Ma J., Zhao D., Wang Y., Yan L., Wu L., He L. Controlling citrus Huanglongbing based on soil remediation and biocontrol. Eur. J. Plant Pathol. 2024;169:379–393. doi: 10.1007/s10658-024-02835-y. [DOI] [Google Scholar]
- 15.Poveda J., Roeschlin R.A., Marano M.R., Favaro M.A. Microorganisms as biocontrol agents against bacterial citrus diseases. Biol. Control. 2021;158 doi: 10.1016/j.biocontrol.2021.104602. [DOI] [Google Scholar]
- 16.Nguyen T.H., Tran D.M. Root endophytic microbiome dataset of sugarcane (Saccharum officinarum L.) cultivated in the Central Highlands, Vietnam, established by the 16S rRNA metagenomics. Data Br. 2023;48 doi: 10.1016/j.dib.2023.109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Illumina, bcl2fastq2 Conversion Software v2.20. (2017). https://support.illumina.com/downloads/bcl2fastq-conversion-software-v2-20.html.
- 18.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 20.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caspi R., Billington R., Ferrer L., Foerster H., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Mueller L.A., Ong Q., Paley S., Subhraveti P., Weaver D.S., Karp P.D. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44:D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.