Abstract
Microtubule-associated proteins (MAPs) play a pivotal role in the assembly and stabilization of microtubules, which are essential for plant cell growth, development, and morphogenesis. A class of plant-specific MAPs, MAP65, plays largely unexplored roles in moso bamboo (Phyllostachys edulis). This study identified 19 PeMAP65 genes in moso bamboo, systematically examining their phylogenetic relationships, conserved motifs, gene structures, collinearity, and cis-acting elements. Analysis of gene expression indicated that PeMAP65s exhibit tissue-specific expression patterns. Functional differentiation was investigated among the members of different PeMAP65 subfamilies according to their expression patterns in different development stages of bamboo shoots. The expression of PeMAP65-18 was positively correlated with the expression of genes involved in secondary cell wall (SCW) biosynthesis. Y1H and Dual-LUC assays demonstrated that the transcription of PeMAP65-18 was upregulated by PeMYB46, a key transcription factor of SCW biosynthesis. The result of subcellular localization showed that PeMAP65-18 was located in cortical microtubules. We speculate that PeMAP65-18 may play a crucial role in the SCW deposition of moso bamboo. This comprehensive analysis of the MAP65 family offers novel insights into the roles of PeMAP65s in moso bamboo, particularly in relation to the formation of SCWs.
Keywords: Phyllostachys edulis, microtubule-associated proteins, secondary cell walls, cellulose, gene family
1. Introduction
Bamboo, a perennial herbaceous plant, is widely distributed across temperate and tropical forests, characterized by lignified secondary cell walls (SCWs) similar to those of woody trees. Moso bamboo (Phyllostachys edulis), in particular, dominates nearly 73% of China’s bamboo forests, covering approximately 6.4 million hectares [1]. As a fast-growing species, moso bamboo reaches maturity within about four months, with its hollow internodal structure and layered cell walls conferring excellent mechanical properties [2]. It is highly valued in industries such as construction, papermaking, and furniture owing to its strong fibers and unique material properties [3]. The cell wall, as the primary load-bearing component of bamboo fiber cells, features a unique structure comprising primary walls and alternating thin–thick multilayered secondary walls. This configuration imparts high rigidity, straight morphology, and stable performance, resulting in exceptional physical and mechanical properties [4]. Given the growing demand for non-timber resources due to wood scarcity and energy depletion, understanding bamboo’s developmental processes, especially the regulation of SCW formation, is essential for enhancing its industrial applications.
The process of cell wall formation involves not only the synthesis of macromolecules but also the transport of their precursors to the deposition site, where they are eventually integrated and assembled [5]. Microtubules (MTs) and microfilaments within the cytoskeleton regulate the spatiotemporal transport and deposition of cell wall materials, thereby promoting overall cell wall formation in plant cells [6,7]. Organized into parallel arrays along the plasma membrane, microtubules control the pattern of cellulose deposition. Numerous MAPs function as linkers between cellulose synthase complexes (CSCs) and microtubules, playing a pivotal role in the early stages of cellulose synthesis [8]. In recent years, with the identification and functional research of cortical microtubule-associated proteins, the mechanism by which cortical microtubules mediate the thickening pattern of SCWs has been partially elucidated [9]. As one of the important components of the cytoskeleton, MTs play an extremely important role in eukaryotic life. In many physiological activities, MTs regulate their own dynamic changes and the array of different cell stages by interacting with MAPs, and they participate in maintaining cell morphology and structure, constructing cell walls and regulating cytoplasmic flow, polar cell growth, cell mitosis, cell differentiation, and intracellular signal transduction [10,11]. Therefore, the regulatory mechanism of MAPs on MTs has been widely investigated in plant biology.
At present, the MAPs discovered in plants have been divided into 12 families, including SPR1, WVD2, and MAP65 [12]. Among them, MAP65 has the most members and is a plant-specific MAP [13]. MAP65 proteins are typically localized to one or more microtubule arrays, where they act as microtubule-binding proteins that cross-link with microtubules and promote the formation of microtubule bundles [14]. MAP65s were first discovered in tobacco (Nicotiana tabacum) [15]. Arabidopsis thaliana contains nine MAP65 family members. [16]. In addition to tobacco and Arabidopsis, nine and eleven members of MAP65s were identified in Populus trichocarpa and Oryza sativa, respectively [17]. Five MAP65s were cloned in Physcomitrella patens [18]. MAP65s were also reported in carrot (Daucus carota) [19], zinnia (Zinnia elegans) [20], cowpea (Vigna sinensis) [21], maize (Zea mays) [22], and grapevine (Vitis vinifera) [23]. Early studies on MAP65s mainly focused on in vitro experiments to analyze how to regulate the polymerization, assembly, alignment, and stability of microtubules [24]. Phenotypic analysis has also been reported, and relationships have been established between microtubules and plant resistance [25], axial root growth, cytokinesis [26], embryo sac development [27], and so on. However, few studies have reported on how microtubule-associated proteins participate in the formation of the SCW.
In this study, MAP65 proteins were identified on a genome-wide scale in moso bamboo. Phylogenetic relationships, conserved motifs, intron-exon distribution, collinearity, and cis-acting elements were investigated. The expression profiles of PeMAP65s across varying tissues and developmental stages were scrutinized. Co-expression and hierarchical gene regulatory networks were constructed. The upstream transcription factors that may regulate PeMAP65s were also assessed, and their regulatory relationship was experimentally verified. Further investigation on the potential role of PeMAP65-18 in microtubule bundling was performed by analyzing its subcellular localization. The results offer an all-encompassing assessment of the MAP65 gene family in moso bamboo, thereby enriching our comprehension of the role of MAP65s in SCW formation.
2. Results
2.1. Identification of the MAP65s in Moso Bamboo
In this study, 19 PeMAP65s were identified and designated PeMAP65-1 through PeMAP65-19 based on their chromosomal positions (Table 1). The molecular weights of PeMAP65 proteins varied, with PeMAP65-13 having the smallest size at 510 amino acids and PeMAP65-10 the largest at 687 amino acids. Additionally, the relative molecular masses varied between 58.70 kDa (PeMAP65-13) and 78.07 kDa (PeMAP65-10). The theoretical isoelectric points (pI) for the PeMAP65 proteins spanned from 5.07 (PeMAP65-6) to 8.2 (PeMAP65-12). All PeMAP65 proteins were unstable proteins (stability coefficient ≥ 40) with stability coefficients ranging from 43.80 (PeMAP65-11) to 63.80 (PeMAP65-13). The subcellular localization prediction indicated that eleven PeMAP65 proteins are located in the cytoplasm, five in the nucleus, two in the chloroplast, and one in the endoplasmic reticulum.
Table 1.
Basic characteristic of PeMAP65s.
| Gene Name | Gene ID | ORF (aa) | MW (kDa) | pI | Sub Localization |
|---|---|---|---|---|---|
| PeMAP65-1 | PH02Gene22090.t1 | 595 | 67.26 | 7.56 | nucleus |
| PeMAP65-2 | PH02Gene30151.t3 | 593 | 66.28 | 5.08 | chloroplast |
| PeMAP65-3 | PH02Gene42885.t1 | 583 | 65.59 | 5.34 | cytoplasm |
| PeMAP65-4 | PH02Gene22734.t4 | 676 | 76.84 | 5.74 | cytoplasm |
| PeMAP65-5 | PH02Gene01427.t1 | 617 | 70.27 | 6.41 | cytoplasm |
| PeMAP65-6 | PH02Gene18145.t1 | 582 | 65.06 | 5.07 | cytoplasm |
| PeMAP65-7 | PH02Gene25608.t1 | 577 | 65.03 | 5.52 | cytoplasm |
| PeMAP65-8 | PH02Gene18421.t1 | 561 | 63.74 | 6.26 | cytoplasm |
| PeMAP65-9 | PH02Gene25799.t1 | 590 | 68.31 | 6.97 | cytoplasm |
| PeMAP65-10 | PH02Gene20031.t1 | 687 | 78.07 | 5.83 | cytoplasm |
| PeMAP65-11 | PH02Gene19121.t2 | 610 | 69.57 | 6.27 | nucleus |
| PeMAP65-12 | PH02Gene25122.t1 | 614 | 68.5 | 8.2 | nucleus |
| PeMAP65-13 | PH02Gene27598.t2 | 510 | 58.7 | 5.33 | endoplasmic reticulum |
| PeMAP65-14 | PH02Gene14605.t1 | 636 | 71.55 | 5.35 | nucleus |
| PeMAP65-15 | PH02Gene39808.t1 | 591 | 67 | 6.78 | nucleus |
| PeMAP65-16 | PH02Gene10237.t1 | 584 | 66.63 | 5.15 | cytoplasm |
| PeMAP65-17 | PH02Gene34993.t1 | 641 | 73.39 | 6.41 | chloroplast |
| PeMAP65-18 | PH02Gene23444.t3 | 577 | 65.09 | 7.9 | cytoplasm |
| PeMAP65-19 | PH02Gene45477.t1 | 591 | 68.42 | 6.4 | cytoplasm |
2.2. Phylogenetic Analysis of PeMAP65s
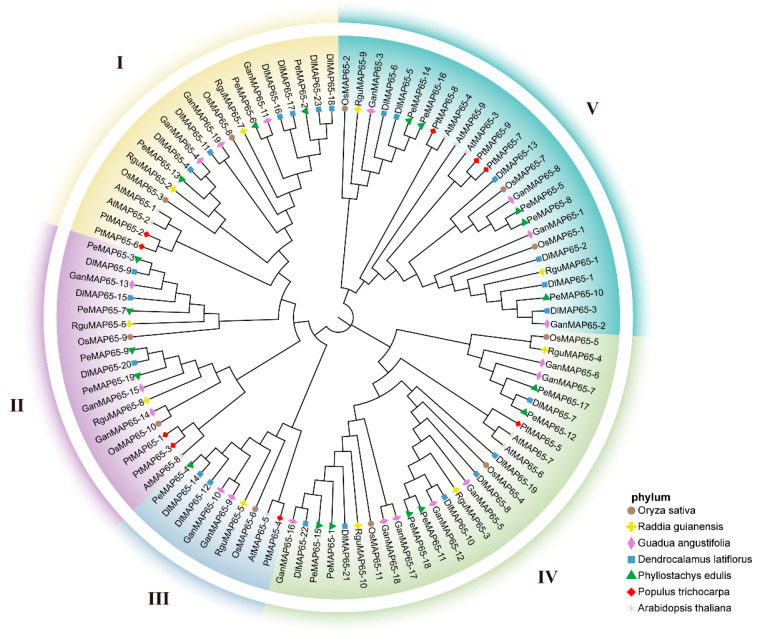
A phylogenetic analysis was performed to investigate the relationship of MAP65s in moso bamboo and other plant species, including three model plants (A. thaliana, O. sativa, and P. trichocarpa) and three other bamboo species (D. latiflorous, G. angustifolia, and R. guianensis) (Figure 1). The phylogenetic tree was divided into five subclades, with MAP65s from these species present in nearly all branches, suggesting the conservation of this gene family across different species throughout evolution. A total of 100 genes were classified into five distinct subfamilies, with the subfamily IV containing the largest number of members, amounting to 28. This subfamily comprised two AtMAP65s, two OsMAP65s, one PtMAP65, six PeMAP65s, three RguMAP65s, seven GanMAP65s, and six DlMAP65s. The subfamily V followed with 26 members, comprising 3 AtMAP65s, 3 OsMAP65s, 3 PtMAP65s, 5 PeMAP65s, 2 RguMAP65s, 4 GanMAP65s, and 6 DlMAP65s. The subfamily III contained the fewest genes, with only nine members. In this subfamily, two tropical woody bamboos, D. latiflorus and G. angustifolia, each had two members, while the other species possessed only a single member. In subfamily I, both PeMAP65s and GanMAP65s had an equal number of members, with three each, while D. latiflorus had six members, and R. guianensis contained only two members.
Figure 1.
Phylogenetic tree of MAP65 gene family in seven species. Phylogenetic analysis of MAP65s across Phyllostachys edulis, Raddia guianensis, Dendrocalamus latiflorus, Guadua angustifolia, Oryza sativa, Populus trichocarpa, and Arabidopsis thaliana. Roman numerals I, II, III, IV, and V denote distinct gene clusters (groups). Variations in symbol shapes and colors represent different species.
2.3. Analysis of Gene Structure and Conserved Motifs
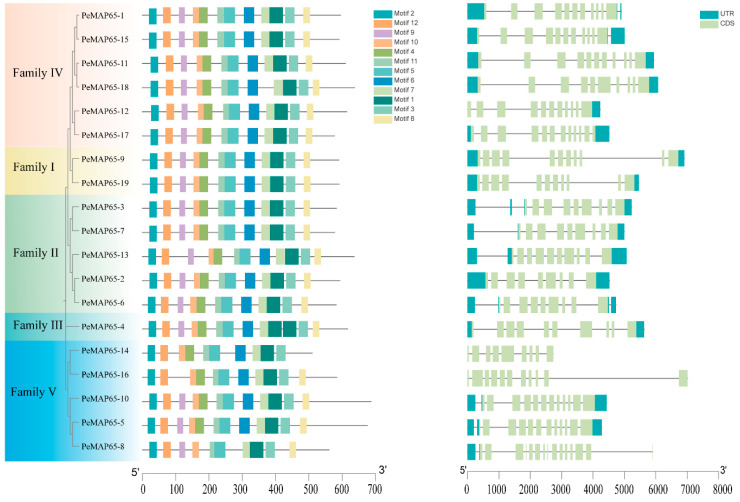
To further clarify the characteristics of the MAP65 family in moso bamboo, 12 conserved motifs were identified across 19 PeMAP65s (Figure 2). In subfamilies I, II, and IV, PeMAP65s contained these 12 motifs. One repetition of motif1 occurred in PeMAP65-4, which belonged to subfamily III. In subfamily V, motif8 and motif9 were absent in PeMAP65-14, motif6 was absent in PeMAP65-8, and motif9 was absent in PeMAP65-16. The genomic DNA sequence lengths of PeMAP65s varied from 2735 bp (PeMAP65-14) to 7016 bp (PeMAP65-16). PeMAP65s contained 9 to 15 exons (Figure 2). PeMAP65s grouped within the same subfamily exhibited similar gene structures; for instance, the two members in subfamily I contained 11 exons, while members in subfamily II typically had 9–10 exons, and the number of exons in subfamily V ranged from 9 to 15.
Figure 2.
Conserved motifs and gene structure of PeMAP65s. The motif distribution of MAP65s (left). A total of 12 motifs are represented, each within differently colored boxes. Gene structures of PeMAP65s (right). The light green box represents the exon, the blue-green box represents the UTR, and the black line indicates the intron.
2.4. Analysis of Cis-Acting Elements in the Promoters of PeMAP65s
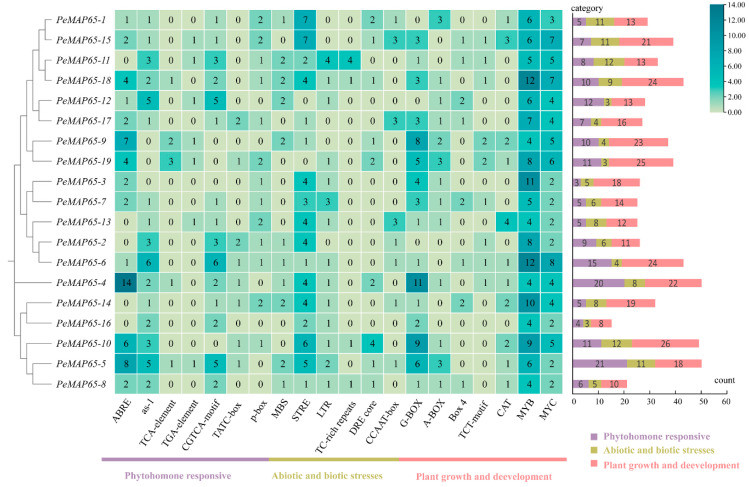
As shown in Figure 3, the promoters of PeMAP65s contain a wide variety of cis-acting elements involved in phytohormone responses, abiotic and biotic stresses, and plant growth and development. Four PeMAP65s contain more than five ABRE elements that respond to abscisic acid (ABA). Stress-responsive elements (STREs) are distributed in the promoter regions of 17 PeMAP65s. MYB and MYC elements related to development and cell cycle regulation were found in all PeMAP65s. Four PeMAP65s, PeMAP65-3, PeMAP65-4, PeMAP65-14, and PeMAP65-18, contain 10 or more MYB elements. These findings suggest that PeMAP65s may exert their biological functions through various signaling pathways.
Figure 3.
Cis-acting elements predicted in the promoter of PeMAP65s. All cis-acting elements were categorized into three categories. The heatmap demonstrates the number of cis-acting elements with the higher number in blue and the lower number in yellow. On the right, the number in the differently colored boxes represents the count of cis-acting elements in each category.
2.5. Chromosomal Location, Gene Duplication, and Synteny Analysis
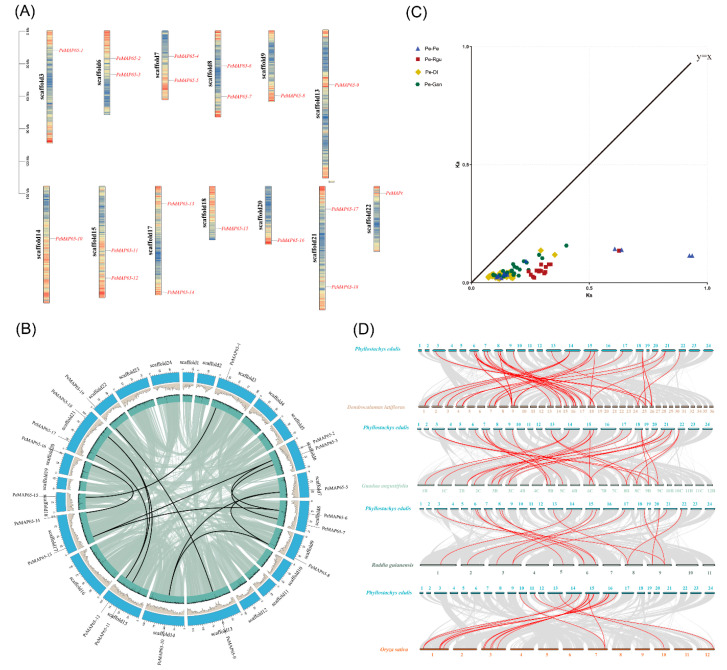
These 19 PeMAP65s were distributed among 13 chromosomes, each containing one or two PeMAP65s, and no tandem repeat events were found (Figure 4A). The evolutionary time of homologous gene pairs was calculated based on Ks values (Table S2). The earliest gene duplication of PeMAP65 occurred around 51.43 million years ago, and the most recent around 5.86 million years ago. The divergence time for MAP65s between P. edulis and R. guianensis was estimated to be between 34.40 million years (PeMAP65-16/RguMAP65-9) and 13.31 million years (PeMAP65-9/RguMAP65-8) ago. Between P. edulis and G. angustifolia, divergence ranges from 22.11 million years (PeMAP65-16/GanMAP65-3) to 5.09 million years (PeMAP65-9/GanMAP65-15). For D. latiflorus, divergence ranges from 19.43 million years (PeMAP65-16/DlMAP65-8) to 3.97 million years (PeMAP65-9/DlMAP65-42). The Ka/Ks ratios between P. edulis and all other bamboo species were less than one, indicating that the MAP65 gene family has undergone purifying selection during its evolution (Figure 4B). Except for PeMAP65-4, a total of 13 putative paralogous gene pairs were identified among 18 PeMAP65s by collinearity analysis in P. edulis (Figure 4B). The analysis of the collinearity relationships among MAP65s in P. edulis, R. guianensis, G. angustifolia, and D. latiflorus revealed that all bamboo species possessed corresponding orthologous genes. The results showed that P. edulis shared 15 homologous gene pairs with R. guianensis, 25 with G. angustifolia, and 53 with D. latiflorus (Figure 4C). The MAP65 gene family was relatively conserved and structurally stable across these bamboo species, suggesting similar functional roles.
Figure 4.
Chromosomal location and synteny analysis of PeMAP65s. (A) The distribution of the PeMAP65s located on the chromosomes in P. edulis. The genetic distance of 13 chromosomes are represented by the scale in megabases (Mb) on the left. Black lines represent the location of the gene on each chromosome. (B) Collinearity analysis of the PeMAP65 gene family. Each PeMAP65 is marked with a short black line on the chromosome, and collinear gene pairs are represented by a black curve. (C) The Ka (non-synonymous substitution) and Ks (synonymous substitution) values of MAP65s were calculated. Pe, P. edulis, Rgu, R. guianensis, Dl, D. latiflorus, Gan, G. angustifolia. (D) Collinearity analysis of MAP65s between P. edulis and three other bamboo species and rice. The gray lines indicate collinear blocks within the P. edulis genome and other plant genomes, and the red curves indicate MAP65s with collinearity.
2.6. Expression Patterns of PeMAP65s
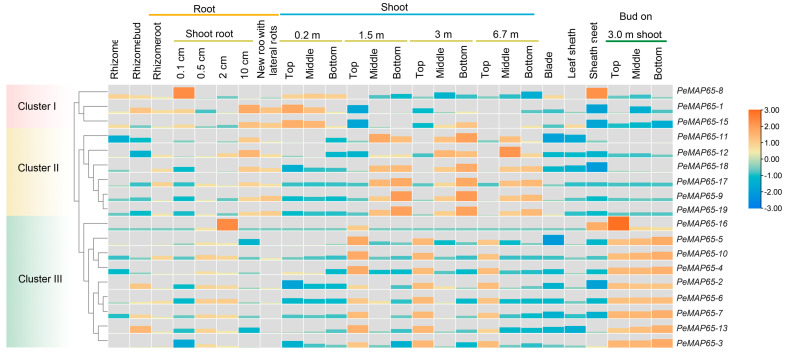
The expression patterns of PeMAP65s in 26 diverse tissues and developmental phases were analyzed according to RNA-seq data from moso bamboo (Figure 5). Except for PeMAP65-14, which was not expressed in any tissue, most PeMAP65s exhibited tissue specificity in their expression and were classified to three clusters. In cluster I, the expression levels of PeMAP65-1, PeMAP65-8, and PeMAP65-15 in the shoots at 0.2 m height were higher than those at greater heights. In cluster II, PeMAP65s were highly expressed in the middle and lower parts of the shoot, while PeMAP65s in cluster III were highly expressed in the upper parts of shoots and shoot buds.
Figure 5.
The expression patterns of PeMAP65s in 26 tissues. The heatmap represents the expression levels of PeMAP65s calculated by Log2FPKM.
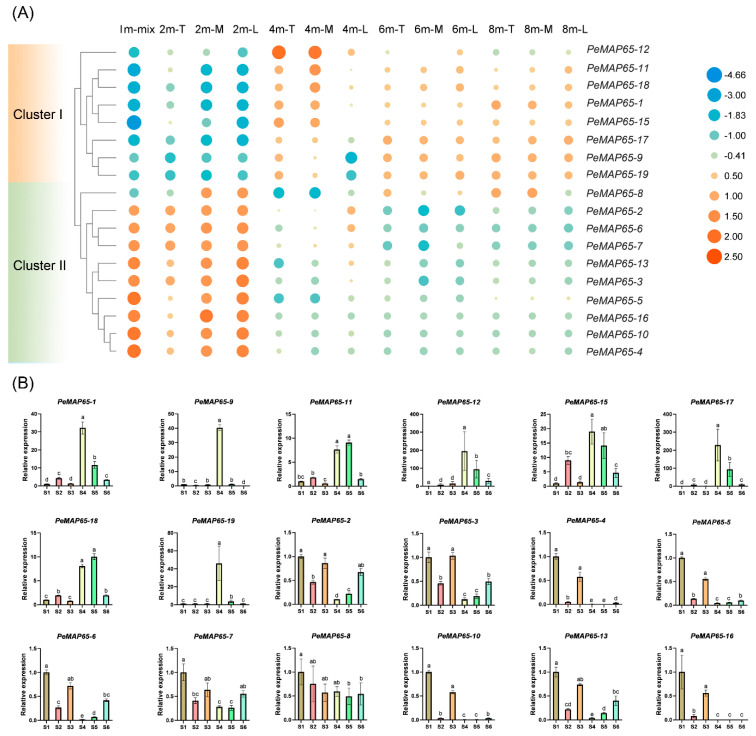
It could be found that PeMAP65s may be related to the growth and lignification process of bamboo shoots. Consequently, the expression of PeMAP65s was further characterized by investigating their expression profiles within the 13th internode of shoots at different heights. It can be clearly observed that the expression of PeMAP65s was mainly divided into two clusters (Figure 6A). Cluster I contained eight PeMAP65s, whose expression was upregulated along with the lignification process of bamboo shoots. However, the 10 PeMAP65s in cluster II showed opposite expression trends, mainly being highly expressed in 1 m and 2 m bamboo shoots. These results indicated that different PeMAP65 members played different roles in different developmental stages of bamboo shoots. The relative expression of PeMAP65s were validated by qRT-PCR (Figure 6B). During rapid bamboo growth, the degree of lignification varies across different internodes within the same bamboo shoot, as well as within different sections of the same internode. The samples used in qRT-PCR were ranked in terms of lignification degree from low to high, denoted as S1, S3, S2, S6, S5, and S4. The results revealed that these PeMAP65s exhibited similar expression patterns consistent with the findings of the transcriptome analysis.
Figure 6.
The expression pattern of PeMAP65s in bamboo shoots with different degrees of lignification. (A) The heatmap represents the expression level (Log2FPKM) of PeMAP65s at the 13th internode of bamboo shoots with different heights. T, M, and L represent the top, middle, and lower portions of the 13th internode. (B) The relative expression levels of PeMAP65s in different internodes of moso bamboo shoots with a height of 2.5 m. S1 represents the 22nd internode. S2 and S3 represent the top and lower portions of the 18th internode. S4 and S6 represent the top, middle, and lower portions of the 13th internode. Data represent means (±SD) of three biological replicates. The different letters indicate significant differences, for which the relative expression level between two samples was greater than or equal to 2.
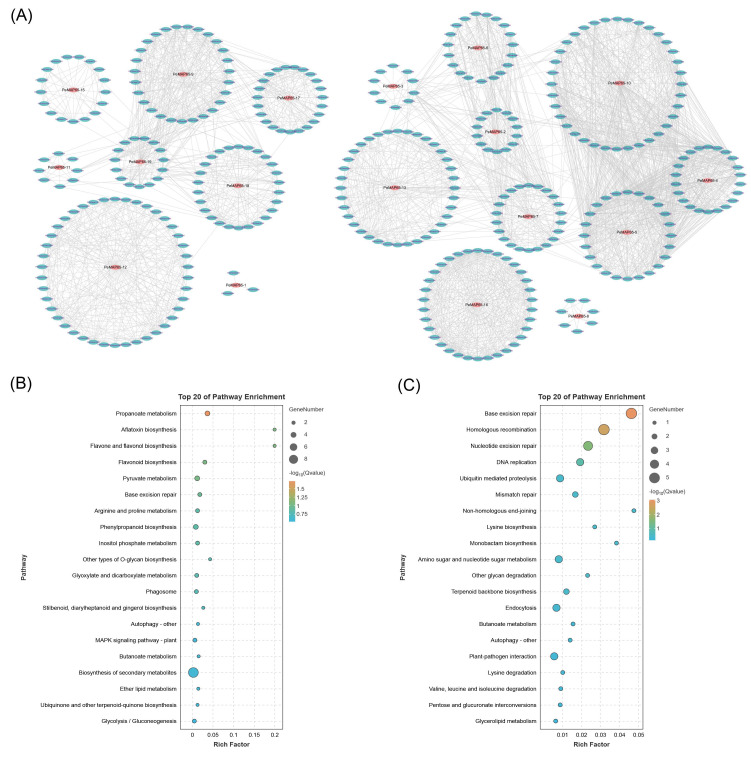
2.7. Co-Expression Network
Using PeMAP65s as the node genes, a co-expression network was constructed based on the database of co-expression networks with functional modules for moso bamboo. A total of 1297 genes exhibited co-expression with 18 PeMAP65s, resulting in the sharing of 2872 edges (Figure 7A). Based on the expression patterns of PeMAP65s in different lignified shoots, we conducted KEGG functional enrichment analysis on the genes co-expressed with PeMAP65s in two clusters. The KEGG functional enrichment results indicate that the genes in cluster I were mainly enriched in biosynthesis of secondary metabolites (Figure 7B), with significant enrichment in phenylpropanoid biosynthesis, while the genes in cluster II were mainly enriched in areas such as base excision repair, homologous recombination, DNA replication, and so on (Figure 7C). These results indicated that PeMAP65s in these two clusters played different functions in bamboo growth and development.
Figure 7.
Co-expression networks of PeMAP65s. (A) Co-expression networks constructed by the database of co-expression networks with functional modules for moso bamboo. Red ellipses represent PeMAP65s, green ellipses represent the co-expressed genes with PeMAP65s, and gray lines represent the edges between these co-expressed genes. (B,C) KEGG functional enrichment on the genes co-expressed with PeMAP65s in two clusters based on the expression patterns of PeMAP65s in different lignified shoots.
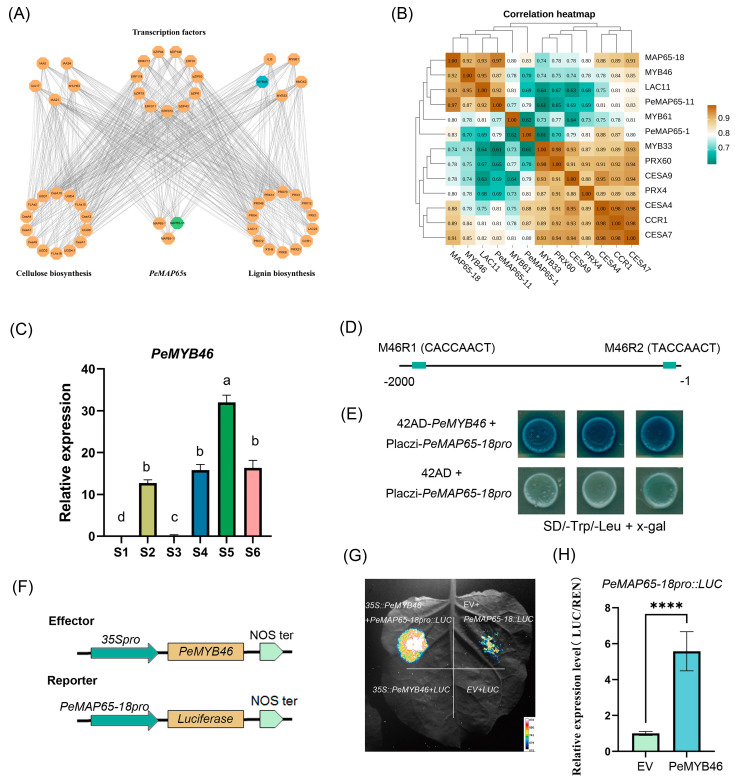
2.8. The Transcriptional Level of MAP65-18 Was Upregulated by PeMYB46
Utilizing a bottom-up Gaussian graphical model (GGM) algorithm, a hierarchical regulatory network was constructed from transcriptomic data (Figure 8A). Based on the hierarchical regulatory network, three PeMAP65s (PeMAP65-1, PeMAP65-11, and PeMAP65-18) were co-expressed with numerous genes involved in lignin and cellulose biosynthesis. Based on the expression levels within the 13th internode of shoots at different heights, ten genes including three transcription factors (MYB33, MYB46, and MYB61), four phenylpropanoid biosynthesis pathway genes (CCR1 and PRX43, PRX60 and LAC11), and three cellulose synthase genes (CESA4, CESA7, and CESA9) were screened and performed to correlation analysis with the three PeMAP65s. The results indicated that the Pearson correlation coefficient between MAP65-18 and SCW biosynthesis genes was 0.74–0.97, which was higher than those of PeMAP65-1 and PeMAP65-11 (Figure 8B). Among them, PeMYB46, as a homolog of the switch transcription factor OsMYB46 contributing to the SCW biosynthesis in rice [28], has a high correlation coefficient of 0.92 with PeMAP65-18, potentially serving as a direct upstream transcription factor for PeMAP65-18. The result of qRT-PCR also confirmed that the expression pattern of PeMYB46 across various internodes of moso bamboo shoots corresponded with the degree of lignification (Figure 8C). Two elements, M46R1 and M46R2, that could be bounded by MYB46 were discovered in the PeMAP65-18 promoter (Figure 8D). The results of Y1H showed that PeMYB46 could bind to the PeMAP65 promoter (Figure 8E). Furthermore, PeMYB46 could enhance the expression of PeMAP65-18, as verified by Dual-LUC assay (Figure 8F–H).
Figure 8.
The transcriptional level of PeMAP65-18 was upregulated by PeMYB46. (A) A hierarchical gene regulatory network. At the top level, MYB46 (blue) is shown as a direct target of MAP65-18 (green). (B) The correlation heatmap of PeMAP65s with the genes related to SCW biosynthesis. The data represent the Pearson coefficient. (C) The relative expression level of PeMYB46 in different internodes. The different letters indicated significant differences, which the relative expression level between two samples was greater than or equal to 2. (D) Schematic diagram of MYB binding sites on the PeMAP65-18 promoter. (E) Y1H validation of the interaction between PeMYB46 and PeMAP65-18 promoter. (F) Schematic diagrams of the effector (PeMYB46) and reporter (PeMAP65-18pro) constructs were used in the dual-luciferase reporter assay. (G) An image of luciferase activity. (H) Relative luciferase activity was measured. Error bars represent SD. Statistical analysis was performed using the t test (n = 7, ****, p < 0.0001).
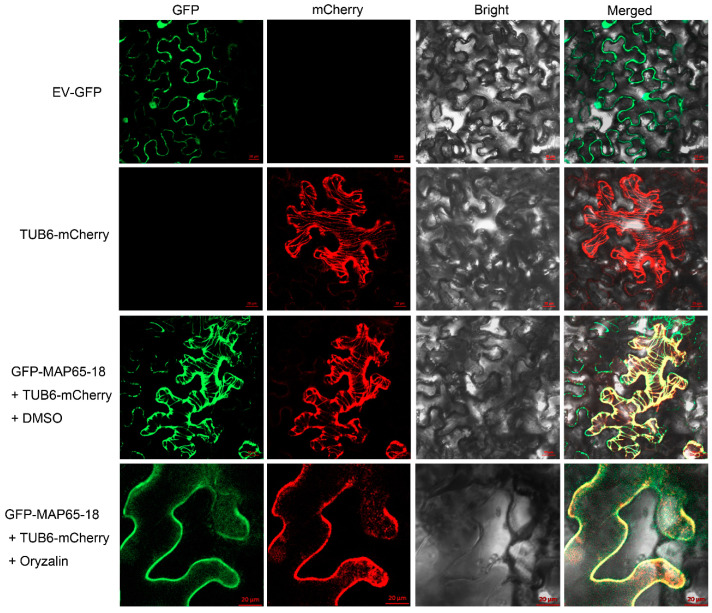
2.9. PeMAP65-18 Was Located in Microtubules
To detect the subcellular localization of PeMAP65-18, the 35S::eGFP-PeMAP65-18 vector was expressed in tobacco leaves. Through laser confocal microscopy observation, the fluorescence signal of eGFP-PeMAP65-18 showed a filamentous distribution, which was consistent with the signal of the microtubule localization marker AtTBU6-mCherry (Figure 9) and conformed to the distribution characteristics of cortical microtubules. Therefore, it was speculated that PeMAP65-18 was located in cortical microtubules. Further supporting this, the filamentous fluorescence signals of both eGFP-PeMAP65-18 and TUB6-mCherry were degraded following treatment with oryzalin, a microtubule depolymerizing agent, indicating that PeMAP65-18 is primarily associated with microtubules.
Figure 9.
Subcellular localization of PeMAP65-18 in tobacco leaves. EV-GFP represents the empty vector containing 35S::GFP. AtTUB6-mCherry used for microtubule localization marker. DMSO or oryzalin (100 μM) treatment for 30 min. Scale bars = 20 µm.
3. Discussion
3.1. Hybridization Events and Chromosome Doubling Drove the Expansion of the PeMAP65 Gene Family
In this study, nineteen MAP65s were identified in moso bamboo. Phylogenetic relationship analysis found that the four bamboo species and rice belonging to the Poaceae family had close genetic relationships. Collinear analysis in the genome of moso bamboo showed that there were 13 pairs of replication genes in the PeMAP65 gene family, and all of them were segmented replications, which indicated that fragment replication was the main driving force for the evolution and expansion of the PeMAP65 gene family. Herbaceous bamboos have the fewest homologous gene pairs with P. edulis due to their independent evolution. The divergence time between P. edulis and R. guianensis for MAP65 genes was estimated to be as early as 34.40 million years ago, which aligns with the divergence of the four ancestral woody bamboo groups (ABCD) [29]. The whole-genome duplication in P. edulis occurred between 7 and 12 million years ago, indicating that the amplification of homologous genes between the genus R. guianensis and P. edulis occurred prior to this event. Around 19.40 million years ago, some tropical woody bamboos underwent two rounds of hybridization, resulting in a hexaploid genome (AABBCC), leading to the highest number of homologous gene pairs between P. edulis and D. latiflorous. The earliest divergence of MAP65 genes between P. edulis and D. latiflorous occurred approximately 19.43 million years ago, which coincides with the formation of neotropical woody bamboo [30]. These results suggest that hybridization events among ancestral woody bamboos and chromosome doubling drove the expansion of the PeMAP65 gene family.
3.2. Functional Differentiation Existed Among the Members of Different MAP65 Subfamilies
Microtubule-associated proteins, specifically MAP65s, play different roles in various biological processes. AtMAP65-1 functions in the early stages of cell division by interacting with microtubules in a sequential stepwise manner to form a fortress-like structure, thereby maintaining microtubule stability [31]. Conversely, AtMAP65-2 stabilizes cytoskeletal organization by promoting MT activity [32]. PeMAP65-13, PeMAP65-6, and PeMAP65-2 cluster with AtMAP65-1 and AtMAP65-2 in subfamily I, suggesting that these PeMAP65s may be broadly involved in mitosis and play critical roles in stabilizing microtubule structures. During mitosis, AtMAP65-4 is localized at the cortical division site and the central region of the spindle, where it regulates the formation of the spindle and kinetochore fibers [33]. The loss of ATMAP65-3 function may result in the failure to initiate cytokinesis [34]. It was speculated that the members of the PeMAP65 subfamily V might play a critical role primarily in the late stages of mitosis, specifically during cytokinesis. A previous study has shown that AtMAP65-6 and AtMAP65-7 can associate with cortical microtubules [35]. PeMAP65-18 was clustered in subfamily VI and located in cortical microtubules, consistent with the localization of AtMAP65-6 and AtMAP65-7, indicating that they may have similar functions.
MAP65 is mainly located on microtubules in the cytoplasm and participates in the dynamic assembly and structural regulation of microtubule skeletons through specific binding to microtubules. In this study, 11 PeMAP65s were predicted to be in the cytoplasm, two in the chloroplast, and one in the endoplasmic reticulum. Although MAP65 is not directly associated with chloroplasts and endoplasmic reticulum, microtubules have been observed to interact with chloroplasts and endoplasmic reticulum in plants [36,37], and MAP65 proteins may be involved in modulating these interactions. However, five PeMAP65s were predicted to be in the nucleus. The results suggested that these PeMAP65s may be related to MT-organizing centers (MTOCs), which are situated on or near the nuclear envelope and which are derived from material related to the nuclear matrix. It may be generalized that some type of nucleus-related MT-organizing structure is basic to all mitotically dividing eukaryotic cells [38]. To fully understand the implications of these predictions, future studies should aim to experimentally validate the localization of these MAP65 proteins and investigate their potential roles in various cellular components.
The expression patterns of PeMAP65s were distinctly classified into two groups in shoots with different development stages. Cluster II primarily included members from subfamilies I and V, which were predominantly expressed in the early stage of shoot development, indicating that these PeMAP65s may be related to cell division in bamboo shoots. In contrast, cluster I contained most members of subfamily IV, which showed high expression in rapidly lignifying tissues. In the hierarchical regulatory network, the three identified PeMAP65s, all belonging to subfamily IV, were co-expressed with genes involved in cellulose and lignin biosynthesis and regulated by multiple transcription factors. These results suggest that PeMAP65s from subfamily IV may be involved in SCW formation by influencing the orientation of cortical MTs, particularly PeMAP65-1, PeMAP65-11, and PeMAP65-18.
3.3. PeMAP65-18 Regulated by PeMYB46 Has a Potential Role in SCW Formation
Current research has revealed the presence of a complex hierarchical transcriptional network within plants, mainly consisting of NAC and MYB transcription factors, which regulate the thickening of the SCW [39]. As secondary transcription factors, MYB46 and MYB83 directly activate downstream genes, including MYB20, MYB42, MYB43, and MYB85, as well as structural genes involved in cellulose and lignin biosynthesis [40,41]. Homologous genes of AtMYB46/AtMYB83, PtrMYB3, and PtrMYB20 also participate in the regulation of SCW biosynthesis in poplar [42]. Similarly, overexpression of OsMYB46 or ZmMYB46 in Arabidopsis can directly induce SCW biosynthesis [28]. This multi-tiered, feed-forward transcriptional regulatory structure ensures precise control over SCW biosynthesis, and this regulatory network is highly conserved. In this study, the hierarchical regulatory network indicated that PeMYB46, a crucial transcription factor for SCW biosynthesis, may serve as an upstream transcription factor of PeMAP65-18. Y1H and Dual-LUC assays confirmed that PeMYB46 could bind to the promoter and directly activate the transcription of PeMAP65-18. It is suggested that PeMAP65-18 might play a crucial role in SCW formation of moso bamboo.
Cytoplasmic microtubules can be mainly divided into two subpopulations: cortical microtubules and perinuclear microtubules. Cortical microtubules are located beneath the plasma membrane and arranged parallel along its inner side, forming a network that maintains the cell shape, regulates the direction of cell growth, and plays a critical role in cell wall formation [38]. Perinuclear microtubules, on the other hand, surround the nucleus and other cytoplasmic structures, extending outward from the MTOC in a radial arrangement. These perinuclear microtubules are primarily responsible for intracellular material transport and play a vital role during cell division [38]. Cortical microtubules are essential for guiding the deposition of SCWs, as they regulate the shape, orientation, and pattern of cellulose microfibril synthesis. Early studies showed that cortical microtubules are aligned parallel to cellulose microfibrils, and disrupting these microtubules altered the orientation of the microfibrils, leading to the hypothesis that cortical microtubules mediate CSC movement [43]. Recent findings support this, showing that CSCs move along the trajectory of cortical microtubules [44]. Additionally, proteins like CELLULOSE SYNTHASE INTERACTING1/POM-POM2 (CSI1/POM2) act as bridges between the CSCs and microtubules during cellulose biosynthesis, further highlighting the role of cortical microtubules in SCW thickening [45]. Given the role of cortical microtubules in regulating SCW deposition and based on the distribution characteristics of PeMAP65-18-GFP fluorescence signals, it is speculated that PeMAP65-18 may be involved in bundling cortical microtubules and guiding cellulose microfibril synthesis during SCW formation. Previous studies have demonstrated that transient overexpression of ZeMAP65-1 in Arabidopsis induced the formation of super-bundled microtubules, similar to patterns observed in xylem cells during differentiation [20]. These findings suggest that PeMAP65-18 may similarly play a role in microtubule organization and cellulose synthesis. Future research will investigate the function of PeMAP65s in bamboo through genetic transformation to further elucidate their roles in SCW deposition.
4. Materials and Methods
4.1. Identification of PeMAP65 Members in Moso Bamboo
The genome sequence and annotation of moso bamboo were obtained from BambooGDB (http://bamboo.bamboogdb.org/, accessed on 21 April 2024). The PFAM database (http://pfam.xfam.org/, accessed on 21 April 2024) was utilized to identify proteins containing the MAP65 conservative domain (PF03999) in moso bamboo. The HMMER 3.0 program was then used to search for proteins with the MAP65 domain with a threshold of E < 1 × 10−10. The NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd, accessed on 21 April 2024) was used to analyze the protein domain (E < 0.01). The molecular weight (MW) and isoelectric point (pI) of PeMAP65 proteins were obtained from ExPaSy (https://web.expasy.org/compute_pi/, accessed on 21 April 2024). The subcellular localization was provided by Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi, accessed on 21 April 2024).
4.2. Sequence Alignment and Phylogenetic Tree Construction
MAP65 proteins from six species (Arabidopsis, O. sativa, P. trichocarpa, Dendrocalamus latiflorous, Guadua angustifolia, Raddia guianensis) were aligned using Clustal W. Phylogenetic analysis was then performed using MEGA X (version 10.1.18) with the maximum likelihood method and bootstrap analysis with 1000 replicates. The phylogenetic tree was visually enhanced by Chiplot (https://www.chiplot.online/, accessed on 23 April 2024).
To compare MAP65 proteins in moso bamboo with those in other plant species (Arabidopsis, O. sativa, P. trichocarpa, D. latiflorus, G. angustifolia, guianensis), protein sequences were aligned using Clustal W. Phylogenetic analysis was conducted using MEGA X with the maximum likelihood method and 1000 bootstrap replicates, and the phylogenetic tree was refined using Evolview (https://evolgenius.info//evolview-v2, accessed on 23 April 2024).
4.3. Gene Structure, Chromosomal Location, and Motif Identification
The gene structure and chromosomal location of PeMAP65s were analyzed based on the genome annotation of moso bamboo. The Multiple EM for Motif Elicitation program (MEME) was employed to identify conserved motifs of PeMAP65 protein sequences (http://meme-suite.org/tools/meme, accessed on 25 April 2024). The results were visualized using TBtools-II (v2.119).
4.4. Prediction of Cis-Acting Regulatory Elements in PeMAP65 Promoters
The cis-acting elements of PeMAP65s promoter (2000 bp) were predicted by the PlantCARE online tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 28 April 2024). The heatmap was generated using TBtools-II (v2.119).
4.5. Synteny and Gene Duplication Analysis
For analyzing scaffold distribution and syntenic relationships of PeMAP65s, the wyp1125/MCScanX software package was used. The Ka (non-synonymous substitution) to Ks (synonymous substitution) ratios were calculated. The syntelog analysis of PeMAP65s was performed according to the online database (https://bamboo.genobank.org/, accessed on 28 April 2024). The evolutionary time (T) was calculated using the Ks value with the formula: T = Ks/2λ, λ = 9.1 × 10−9 [46].
4.6. Expression Analysis of MAP65 Genes in Moso Bamboo
For the expression in different tissues and developmental stages, transcriptome data of 26 samples were obtained from the NCBI Sequence Read Archive (SRA) (accession number: SRX2408703-SRX2408728 and SRR13201212-SRR13201244) [47]. To explore the expression patterns in different internodes of bamboo shoots at various heights, transcriptome data were obtained from NCBI SRA with the BioProject IDs: PRJNA673565 and PRJNA682693.
4.7. qRT-PCR for PeMAP65s
Samples for qRT-PCR were collected in mid-April in Hangzhou, Zhejiang Province, China. Bamboo shoots measuring 2.5 m in height were chosen, and specific internodes were selected. The 22nd internode was labeled as S1, while the 18th internode was divided into two parts: upper and lower, labeled as S2 and S3. Similarly, the 13th internode was divided into three parts: upper, middle, and lower, labeled as S4, S5, and S6, respectively.
The total RNA of plant samples was extracted using the RNAprep Pure kit (Tiangen, Beijing, China). The PrimeScript 1st Strand cDNA Synthesis kit (Takara, Dalian, China) was employed to reverse transcribe to cDNA. The primers for quantitative real-time PCR (qRT-PCR) were designed using the Primer5 software (Table S1). The TB Green Premix Ex TaqTM Kit (TaKaRa, Kusatsu, Japan) and QuantStudioTM7 Flex Real-time PCR instrument (Applied Biosystems, Carlsbad, CA, USA) were used. Results were calculated using the 2−ΔΔCt method [48]. The tonoplast intrinsic protein 41 (TIP41) was used as a reference gene [49].
4.8. Construction of Co-Expression and Hierarchical Gene Regulatory Networks
The co-expression relationships between PeMAP65s and their edge genes were obtained through the BambooNET database (http://bioinformatics.cau.edu.cn/bamboo/, accessed on 23 April 2024) and visualized using Cytoscape software v3.10.2. The co-expressed genes were enriched by the GO and KEGG annotation.
From the 2097 genes exhibiting similar or opposing expression trends to PeMAP65s based on the transcriptomic data, 23 genes related to SCW biosynthesis were selected as the lowest tier of the regulatory network. Using the bottom-up GGM algorithm [50], a hierarchical regulatory map was generated encompassing PeMAP65s and its upstream genes using Cytoscape (v3.8.2).
4.9. Yeast Assays
One-hybrid yeast assays were conducted to determine the interaction between PeMYB46 and the promoter of PeMAP65-18. The recombined plasmid pB42AD-PeMYB46 and pLaczi-PeMAP65-18pro were co-transformed into yeast EYG48 strains. The transformed yeast cells were then grown on selective medium, specifically SD/-Trp-Ura, for three days at 30 °C. The positive colonies were selected and then plated on SD/-Trp/-Ura medium supplemented with X-gal (SD/-Trp/-Ura, 20% galactose, 20% raffinose, 20 mg/mL X-gal).
4.10. Dual-Luciferase Assays
The CDS of PeMYB46 was cloned into the effector vector pGreenII 62-SK, while the promoter (2000 bp) of PeMAP65-18 was cloned into the reporter vector pGreenII 0800-Luc and then transfected into leaves of 4-week-old tobacco. The activities of firefly luciferase (LUC) and renillia luciferase (REN) were measured using the Dual-Luciferase Reporter Assay System (Yeasen, Shanghai, China). Additionally, the fluorescence emitted from luciferase was observed using a low-light cooled CCD imaging apparatus (Lumazone, Singapore) after a Luciferin (100 μM) spray. The images were then processed and analyzed using Image J (Java 1.8.0_172).
4.11. Subcellular Locolization
The full-length coding sequence (CDS) of PeMAP65-18 was cloned and then fused with the enhanced green fluorescent protein (eGFP) under the control of the CaMV35S promoter. The recombinant vectors containing the eGFP-PeMAP65 fusion construct were transformed into Agrobacterium tumefaciens strain GV3101 and infected the tobacco leaves. β-tubulin (TUB6) of Arabidopsis was fused with mCherry as a tubulin marker. After 36 h of infection, tobacco leaves were treated with DMSO or oryzalin for 30 min, respectively. Fluorescence signals were observed on a laser confocal microscope from ZEISS (model LSM 880).
4.12. Data Collection and Statistical Analysis
The figures delineated in this research are conveyed as means ± standard deviation (SD), derived from no less than three independent trials. Statistical significance of the outcomes was ascertained via one-way analysis of variance (ANOVA). GraphPad Prism 12.0 software was utilized for the execution of these statistical examinations.
5. Conclusions
In this study, 19 MAP65s were identified from moso bamboo, and a phylogenetic tree of MAP65s from multiple species was constructed, dividing them into five subgroups. The PeMAP65s within the same subgroup shared similar gene structures and conserved domains. Segmental duplication appeared to be the primary driving force behind the expansion of the MAP65 gene family, which has been subject to purifying selection pressure during evolution. Additionally, chromosome doubling and hybridization events between woody bamboos contributed to the expansion of the PeMAP65s in moso bamboo. The functional differentiation was investigated in the PeMAP65s based on the evolutionary relationship and expression patterns, which could be categorized into two clusters: one related to cell division and the other involved in SCW formation. The expression of PeMAP65-18 was positively correlated with the expression of genes related to SCW biosynthesis and activated by PeMYB46, a pivotal transcription factor involved in SCW formation. Subcellular localization has confirmed that MAP65-18 is located in microtubules, and it was speculated that PeMAP65-18 was responsible for bundling cortical microtubules and guiding cellulose microfibril synthesis during SCW deposition. In summary, the comprehensive analysis of the PeMAP65 family offers novel insights into the roles of MAP65s in moso bamboo, particularly regarding SCW formation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants13213000/s1, Table S1: Primer sequences for qRT-PCR. Table S2: Ks and Ka analysis of duplicated gene pairs.
Author Contributions
Y.J.: Conceptualization, Software, Writing—original draft, Methodology. S.C.: Software. M.L. and L.O. contributed to conducting the study and collecting the data. X.H., J.X. and W.Q.: Formal analysis. Z.L. and R.Z.: Methodology. G.Q.: Conceptualization, Supervision, Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This work was supported by the National Key Research Development Program of China (Grant No. 2021YFD2200504) and the National Non-profit Institute Research Grant of Chinese Academy of Forestry (Grant No. CAFYBB2020ZB004).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Liu L., Chang S.X., Huang C., Yu X., Zhi Y., Jiang P. Controls for phytolith accumulation in Moso bamboo leaves across China. Sci. Total Environ. 2023;893:164886. doi: 10.1016/j.scitotenv.2023.164886. [DOI] [PubMed] [Google Scholar]
- 2.Chen M., Guo L., Ramakrishnan M., Fei Z., Vinod K.K., Ding Y., Jiao C., Gao Z., Zha R., Wang C., et al. Rapid growth of Moso bamboo (Phyllostachys edulis): Cellular roadmaps, transcriptome dynamics, and environmental factors. Plant Cell. 2022;34:3577–3610. doi: 10.1093/plcell/koac193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu D., He S., Leng W., Chen Y., Wu Z. Replacing Plastic with Bamboo: A Review of the Properties and Green Applications of Bamboo-Fiber-Reinforced Polymer Composites. Polymers. 2023;15:4276. doi: 10.3390/polym15214276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiyas N., Sasidharan I., Bindu Kumar K., Gopakumar B., Dan M., Sabulal B. Mechanical superiority of Pseudoxytenanthera bamboo for sustainable engineering solutions. Sci. Rep. 2023;13:18169. doi: 10.1038/s41598-023-45523-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann N., King S., Samuels A.L., McFarlane H.E. Subcellular coordination of plant cell wall synthesis. Dev. Cell. 2021;56:933–948. doi: 10.1016/j.devcel.2021.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z., Persson S., Zhang Y. The connection of cytoskeletal network with plasma membrane and the cell wall. J. Integr. Plant Biol. 2015;57:330–340. doi: 10.1111/jipb.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tobias L.M., Spokevicius A.V., McFarlane H.E., Bossinger G. The cytoskeleton and its role in determining cellulose microfibril angle in secondary cell walls of woody tree species. Plants. 2020;9:90. doi: 10.3390/plants9010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei L., Li S., Gu Y. Cellulose synthase complexes: Composition and regulation. Front. Plant Sci. 2012;3:75. doi: 10.3389/fpls.2012.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krtková J., Benáková M., Schwarzerová K. Multifunctional microtubule-associated proteins in plants. Front. Plant Sci. 2016;7:474. doi: 10.3389/fpls.2016.00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sedbrook J.C., Kaloriti D. Microtubules, MAPs and plant directional cell expansion. Trends Plant Sci. 2008;13:303–310. doi: 10.1016/j.tplants.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Sedbrook J.C. MAPs in plant cells: Delineating microtubule growth dynamics and organization. Curr. Opin. Plant Biol. 2004;7:632–640. doi: 10.1016/j.pbi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 12.Struk S., Dhonukshe P. MAPs: Cellular navigators for microtubule array orientations in Arabidopsis. Plant Cell Rep. 2014;33:1–21. doi: 10.1007/s00299-013-1486-2. [DOI] [PubMed] [Google Scholar]
- 13.Gardiner J. The evolution and diversification of plant microtubule-associated proteins. Plant J. 2013;75:219–229. doi: 10.1111/tpj.12189. [DOI] [PubMed] [Google Scholar]
- 14.Portran D., Zoccoler M., Gaillard J., Stoppin-Mellet V., Neumann E., Arnal I., Martiel J.L., Vantard M. MAP65/Ase1 promote microtubule flexibility. Mol. Biol. Cell. 2013;24:1964–1973. doi: 10.1091/mbc.e13-03-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawano M., Shimmen T., Sonobe S. Possible involvement of 65 kda MAP in elongation growth of azuki bean epicotyls. Plant Cell Physiol. 2000;41:968–976. doi: 10.1093/pcp/pcd022. [DOI] [PubMed] [Google Scholar]
- 16.Smertenko A.P., Chang H.Y., Wagner V., Kaloriti D., Fenyk S., Sonobe S., Lloyd C., Hauser M.T., Hussey P.J. The Arabidopsis microtubule-associated protein AtMAP65-1: Molecular analysis of its microtubule bundling activity. Plant Cell. 2004;16:2035–2047. doi: 10.1105/tpc.104.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo L., Ho C.M., Kong Z., Lee Y.R., Qian Q., Liu B. Evaluating the microtubule cytoskeleton and its interacting proteins in monocots by mining the rice genome. Ann. Bot. 2009;103:387–402. doi: 10.1093/aob/mcn248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosetsu K., de Keijzer J., Janson M.E., Goshima G. MICROTUBULE-ASSOCIATED PROTEIN65 is essential for maintenance of phragmoplast bipolarity and formation of the cell plate in Physcomitrella patens. Plant Cell. 2013;25:4479–4492. doi: 10.1105/tpc.113.117432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan J., Mao G., Smertenko A., Hussey P.J., Naldrett M., Bottrill A., Lloyd C.W. Identification of a MAP65 isoform involved in directional expansion of plant cells. FEBS Lett. 2003;534:161–163. doi: 10.1016/S0014-5793(02)03848-6. [DOI] [PubMed] [Google Scholar]
- 20.Mao G., Buschmann H., Doonan J.H., Lloyd C.W. The role of MAP65-1 in microtubule bundling during Zinnia tracheary element formation. J. Cell Sci. 2006;119:753–758. doi: 10.1242/jcs.02813. [DOI] [PubMed] [Google Scholar]
- 21.Panteris E., Komis G., Adamakis I.D., Samaj J., Bosabalidis A.M. MAP65 in tubulin/colchicine paracrystals of Vigna sinensis root cells: Possible role in the assembly and stabilization of atypical tubulin polymers. Cytoskeleton. 2010;67:152–160. doi: 10.1002/cm.20432. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y., Zuo M., Liang Y., Jiang M., Zhang J., Scheller H.V., Tan M., Zhang A. MAP65-1a positively regulates H2O2 amplification and enhances brassinosteroid-induced antioxidant defence in maize. J. Exp. Bot. 2013;64:3787–3802. doi: 10.1093/jxb/ert215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parrotta L., Faleri C., Cresti M., Cai G. Proteins immunologically related to MAP65-1 accumulate and localize differentially during bud development in Vitis vinifera L. Protoplasma. 2017;254:1591–1605. doi: 10.1007/s00709-016-1055-y. [DOI] [PubMed] [Google Scholar]
- 24.Fache V., Gaillard J., Van Damme D., Geelen D., Neumann E., Stoppin-Mellet V., Vantard M. Arabidopsis kinetochore fiber-associated MAP65-4 cross-links microtubules and promotes microtubule bundle elongation. Plant Cell. 2010;22:3804–3815. doi: 10.1105/tpc.110.080606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardham A.R. Microtubules and biotic interactions. Plant J. 2013;75:278–289. doi: 10.1111/tpj.12171. [DOI] [PubMed] [Google Scholar]
- 26.Lucas J.R. Appearance of microtubules at the cytokinesis to interphase transition in Arabidopsis thaliana. Cytoskeleton. 2021;78:361–371. doi: 10.1002/cm.21689. [DOI] [PubMed] [Google Scholar]
- 27.Chettoor A.M., Yang B. Evans MMS: Control of cellularization, nuclear localization, and antipodal cell cluster development in maize embryo sacs. Genetics. 2023;225:iyad101. doi: 10.1093/genetics/iyad101. [DOI] [PubMed] [Google Scholar]
- 28.Zhong R., Lee C., McCarthy R.L., Reeves C.K., Jones E.G., Ye Z.H. Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 2011;52:1856–1871. doi: 10.1093/pcp/pcr123. [DOI] [PubMed] [Google Scholar]
- 29.Guo Z.H., Ma P.F., Yang G.Q., Hu J.Y., Liu Y.L., Xia E.H., Zhong M.C., Zhao L., Sun G.L., Xu Y.X., et al. Genome Sequences Provide Insights into the Reticulate Origin and Unique Traits of Woody Bamboos. Mol. Plant. 2019;12:1353–1365. doi: 10.1016/j.molp.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Ma P.F., Liu Y.L., Guo C., Jin G., Guo Z.H., Mao L., Yang Y.Z., Niu L.Z., Wang Y.J., Clark L.G., et al. Genome assemblies of 11 bamboo species highlight diversification induced by dynamic subgenome dominance. Nat. Genet. 2024;56:710–720. doi: 10.1038/s41588-024-01683-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smertenko A.P., Chang H.Y., Sonobe S., Fenyk S.I., Weingartner M., Bögre L., Hussey P.J. Control of the AtMAP65-1 interaction with microtubules through the cell cycle. J. Cell Sci. 2006;119:3227–3237. doi: 10.1242/jcs.03051. [DOI] [PubMed] [Google Scholar]
- 32.Li H., Zeng X., Liu Z.Q., Meng Q.T., Yuan M., Mao T.L. Arabidopsis microtubule-associated protein AtMAP65-2 acts as a microtubule stabilizer. Plant Mol. Biol. 2009;69:313–324. doi: 10.1007/s11103-008-9426-1. [DOI] [PubMed] [Google Scholar]
- 33.Van Damme D., Van Poucke K., Boutant E., Ritzenthaler C., Inzé D., Geelen D. In vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiol. 2004;136:3956–3967. doi: 10.1104/pp.104.051623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quentin M., Baurès I., Hoefle C., Caillaud M.C., Allasia V., Panabières F., Abad P., Hückelhoven R., Keller H., Favery B. The Arabidopsis microtubule-associated protein MAP65-3 supports infection by filamentous biotrophic pathogens by down-regulating salicylic acid-dependent defenses. J. Exp. Bot. 2016;67:1731–1743. doi: 10.1093/jxb/erv564. [DOI] [PubMed] [Google Scholar]
- 35.Mao T., Jin L., Li H., Liu B., Yuan M. Two microtubule-associated proteins of the Arabidopsis MAP65 family function differently on microtubules. Plant Physiol. 2005;138:654–662. doi: 10.1104/pp.104.052456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zang J., Klemm S., Pain C., Duckney P., Wang P. A novel plant actin-microtubule bridging complex regulates cytoskeletal and er structure at er-pm contact sites. Curr. Biol. 2021;31:1251–1260.e4. doi: 10.1016/j.cub.2020.12.009. [DOI] [PubMed] [Google Scholar]
- 37.Meier N.D., Seward K., Caplan J.L., Dinesh-Kumar S.P. Calponin homology domain containing kinesin, KIS1, regulates chloroplast stromule formation and immunity. Sci. Adv. 2023;9:eadi7407. doi: 10.1126/sciadv.adi7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baluka F., Barlow P.W., Lichtscheidl I.K., Volkmann D. The plant cell body: A cytoskeletal tool for cellular development and morphogenesis. Protoplasma. 1998;202:1–10. doi: 10.1007/BF01280869. [DOI] [Google Scholar]
- 39.Paredez A.R., Persson S., Ehrhardt D.W., Somerville C.R. Genetic evidence that cellulose synthase activity influences microtubule cortical array organization. Plant Physiol. 2008;147:1723–1734. doi: 10.1104/pp.108.120196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong R., Ye Z.H. MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 2012;53:368–380. doi: 10.1093/pcp/pcr185. [DOI] [PubMed] [Google Scholar]
- 41.Geng P., Zhang S., Liu J., Zhao C., Wu J., Cao Y., Fu C., Han X., He H., Zhao Q. MYB20, MYB42, MYB43, and MYB85 Regulate Phenylalanine and Lignin Biosynthesis during Secondary Cell Wall Formation. Plant Physiol. 2020;182:1272–1283. doi: 10.1104/pp.19.01070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye Z.H. The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 2010;51:1084–1090. doi: 10.1093/pcp/pcq064. [DOI] [PubMed] [Google Scholar]
- 43.Baskin T.I. On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma. 2001;215:150–171. doi: 10.1007/BF01280311. [DOI] [PubMed] [Google Scholar]
- 44.Paredez A.R., Somerville C.R., Ehrhardt D.W. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 45.Bringmann M., Li E., Sampathkumar A., Kocabek T., Hauser M.T., Persson S. POM-POM2/cellulose synthase interacting1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell. 2012;24:163–177. doi: 10.1105/tpc.111.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lynch M., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 47.Zhao H., Gao Z., Wang L., Wang J., Wang S., Fei B., Chen C., Shi C., Liu X., Zhang H., et al. Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis) Gigascience. 2018;7:giy115. doi: 10.1093/gigascience/giy115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 49.Fan C., Ma J., Guo Q., Li X., Wang H., Lu M. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis) PLoS ONE. 2013;8:e56573. doi: 10.1371/journal.pone.0056573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu S., Li Q., Wei H., Chang M.J., Tunlaya-Anukit S., Kim H., Liu J., Song J., Sun Y.H., Yuan L., et al. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. USA. 2013;110:10848–10853. doi: 10.1073/pnas.1308936110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article and Supplementary Materials.