Abstract
In vertebrate spindles, most microtubules are formed via branching microtubule nucleation, whereby microtubules nucleate along the side of pre-existing microtubules. Hepatoma up-regulated protein (HURP) is a microtubule-associated protein that has been implicated in spindle assembly, but its mode of action is yet to be defined. In this study, we show that HURP is necessary for RanGTP-induced branching microtubule nucleation in Xenopus egg extract. Specifically, HURP stabilizes the microtubule lattice to promote microtubule formation from γ-TuRC. This function is shifted to promote branching microtubule nucleation through enhanced localization to TPX2 condensates, which form the core of the branch site on microtubules. Lastly, we provide a high-resolution cryo-EM structure of HURP on the microtubule, revealing how HURP binding stabilizes the microtubule lattice. We propose a model in which HURP stabilizes microtubules during their formation, and TPX2 preferentially enriches HURP to microtubules to promote branching microtubule nucleation and thus spindle assembly.
Subject terms: Microtubules, Cryoelectron microscopy, Mitotic spindle
Many proteins work together to promote spindle microtubule formation during cell division. Here, the authors find the protein HURP to be essential for microtubule formation by stabilizing the microtubule lattice and working synergistically with the spindle assembly factor TPX2.
Introduction
Cell division relies on the formation of a bipolar spindle to orient and faithfully segregate chromosomes into two daughter cells. The spindle is made up of many microtubules (MTs), filaments composed of tubulin dimers, that must be nucleated and organized in a regulated manner. Additionally, MT dynamics (e.g., growth rate, catastrophe, and rescue) are regulated by MT-associated proteins (MAPs) to facilitate proper spindle assembly. Dysregulation of MT nucleation, dynamics, and organization can lead to improper chromosome segregation and result in apoptosis or diseases such as cancer1. Therefore, dissecting the complexity of how MTs are formed and regulated is crucial to understanding how cell division is continuously executed in a reliable manner.
In vertebrate spindles, branching MT nucleation forms most spindle MTs2–5. In this process, new MTs nucleate at a shallow angle on the side of pre-existing MTs, resulting in exponential self-amplification6. In vitro, the minimal branching components are tubulin, the gamma-tubulin ring complex (γ-TuRC) which serves as the universal MT nucleation template, and the augmin complex which localizes γ-TuRC to a pre-existing MT7–9. In Xenopus laevis egg extract, the TPX2 protein is required for branching MT nucleation, along with the general nucleation factor XMAP215/chTOG6,10. TPX2 forms a co-condensate with tubulin on a MT and recruits the augmin complex, which in turn recruits γ-TuRC7,11,12. Branching MT nucleation is regulated via the small GTPase Ran. Both the augmin complex and TPX2 are spindle assembly factors (SAF), meaning that they are inhibited by binding of importins and activated by RanGTP-driven importin release13,14. Given that RanGTP is present in a gradient centered around chromatin, branching MT nucleation is spatially regulated to promote MT nucleation near chromosomes5,15,16.
Here, we hypothesized that the hepatoma upregulated protein (HURP) plays a role in branching MT nucleation. HURP is another SAF17,18, first identified in human hepatocellular carcinoma, and has been implicated in a variety of cancers as a putative oncogene19–23. Studies in human cells have demonstrated that HURP is a MT-bundling protein that localizes predominantly to kinetochore fibers (k-fibers) and is important in k-fiber stability and chromosome congression18,24,25. Interestingly, in acentrosomal systems, such as Drosophila oocytes, mouse oocytes, and Xenopus egg extract, loss of HURP leads to spindle assembly defects and a reduction of spindle MT density17,26–28. In Drosophila embryo extract, several augmin subunits were identified as possible interacting partners of HURP29. Additionally, HURP indirectly interacts with TPX2 in Xenopus egg extract and human cells as part of a larger complex28,30. Taken together, this prompted us to investigate HURP’s role in branching MT nucleation.
In this work, we uncover that HURP is integral to branching MT nucleation. By combining live imaging in Xenopus egg extract with in vitro assays and single particle cryo-electron microscopy (cryo-EM), we reveal how HURP promotes MT formation. Altogether, this work uncovers how HURP promotes branching MT nucleation and thereby spindle assembly.
Results
HURP is required for branching MT nucleation in Xenopus egg extract
To investigate whether HURP plays a role in branching MT nucleation, we first developed a strategy to remove HURP from Xenopus egg extract. We generated a HURP-specific antibody and coupled it to beads. The conjugated beads were incubated with egg extract and then removed to immunodeplete endogenous HURP (present at ~320 nM31) (Fig. 1a). When compared to mock-depleted extract, Western blot analysis revealed no co-depletion of other MT nucleation and branching factors such as γ-tubulin, XMAP215, TPX2, and augmin in the HURP-depleted extract. This indicates that the immunodepletion was specific and HURP does not strongly associate with any of these proteins under these conditions (no RanGTP added). This is consistent with previous work suggesting that HURP interacts with TPX2 and XMAP215 in a RanGTP-dependent manner28.
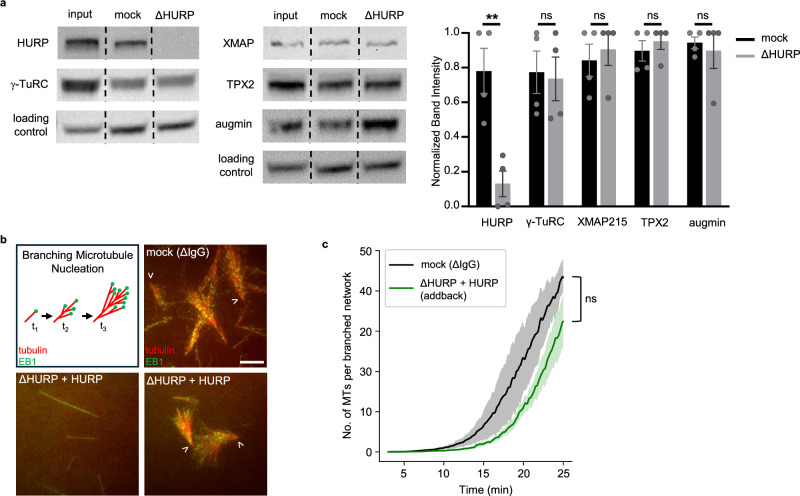
Fig. 1. HURP is necessary for branching microtubule nucleation in Xenopus egg extract.
a Western blot of Xenopus egg extract input, mock-depleted extract, and HURP-depleted extract probed for HURP, γ-TuRC (γ-tubulin), TPX2, and augmin (HAUS1). α-tubulin was probed as a loading control. Rearrangement of nonadjacent lanes (for display purposes) are indicated by dotted lines. Uncropped blots are available in the Source Data file. Band intensities, normalized to the input lane, were averaged across four replicates and plotted in the bar graph (mean ± SEM) for mock-depleted and HURP-depleted extract. HURP quantification p value = 0.005. b RanGTP-induced branching microtubule (MT) nucleation assay in mock-depleted extract, HURP-depleted extract, and HURP-depleted extract plus addback of 250 nM purified HURP construct. MTs (AlexaFluor647-tubulin, pseudo-colored red) and mCherry-EB1 (pseudo-colored green) were imaged for each reaction over 30 min. Representative cropped images (51.54 μm x 51.54 μm) at a timepoint of ~22 min are displayed. Scale bar = 10 µm. c Plot of average MT number (EB1 foci ± SEM) over time (min) in a 110.03 μm x 93.41 μm crop, normalized by the number of branched structures, across replicates in (b) (n = 3). Significance for (c) was calculated by comparing the number of MTs at the final timepoint of the mock and addback conditions. Significance for panels a and c were defined as a p value < 0.5 determined by two-tailed unpaired Student’s t-test. ns = not significant. Double asterisks (**) indicate a p value < 0.01.
Next, we directly observed branching MT nucleation in the presence and absence of HURP in Xenopus egg extract using total internal reflection fluorescence (TIRF) microscopy. In this assay, MTs are visualized via fluorescent tubulin and their growing plus ends via fluorescent end-binding protein 1 (EB1)6. Branching MT nucleation is initiated by the addition of constitutively active RanGTP(Q69L). Addition of the ATPase inhibitor vanadate inhibits motor activity to prevent reorganization of the resulting fan-like branched MT networks6,32, a necessary step to clearly observe and measure MT nucleation over time. As expected, the mock-depleted extract yielded branched, fan-like MT structures in the presence of RanGTP(Q69L). However, no branched MT structures formed in the absence of HURP (Fig. 1b and Supplementary Movie 1). Instead, only single MTs were nucleated and polymerized. This is consistent with the observation that HURP is necessary for the assembly of chromatin-induced MTs17,27.
To verify that the observed phenotype was specific for HURP, we purified full-length HURP (Supplementary Fig. S1) and added it back to HURP-depleted extract, which rescued the formation of branched MT structures and thus indicates that the observed phenotype is a direct consequence of HURP depletion (Fig. 1b and Supplementary Fig. S2). We measured the number of MTs (counted as EB1 foci) over time, normalized to the number of fans present (Supplementary Fig. S3), and found that the rescue assay had similar nucleation kinetics as the mock depletion (Fig. 1c). In conclusion, HURP is necessary for branching MT nucleation in Xenopus egg extract.
HURP promotes MT formation and works together with TPX2
Some MAPs, such as XMAP215 and TPX2, are required for and induce single MT nucleation or branching MT nucleation, respectively, when added in excess to meiotic Xenopus egg extract6,10. To assess the effect of HURP on MT formation, we added excess amounts of purified HURP to Xenopus egg extract and visualized the reaction via TIRF microscopy. Purified HURP was added at final concentrations of 125 nM and 250 nM, resulting in a ~ 1.4-fold and ~1.8-fold increase in total HURP concentration, respectively (Supplementary Fig. S4). We find that the addition of excess HURP promotes the formation of single MTs (Fig. 2a, b, Supplementary Movie 2, and Supplementary Fig. S5). The formation of single MTs is similar to what is observed in the presence of increasing XMAP215 concentration10, but differs from the formation of branched MT networks observed when an excess of TPX2 is added to extract6,33. The different outcomes of excess HURP and excess TPX2 are interesting given that both HURP and TPX2 are SAFs18,29, suggesting that HURP is working in a manner distinct from TPX2 to form MTs.
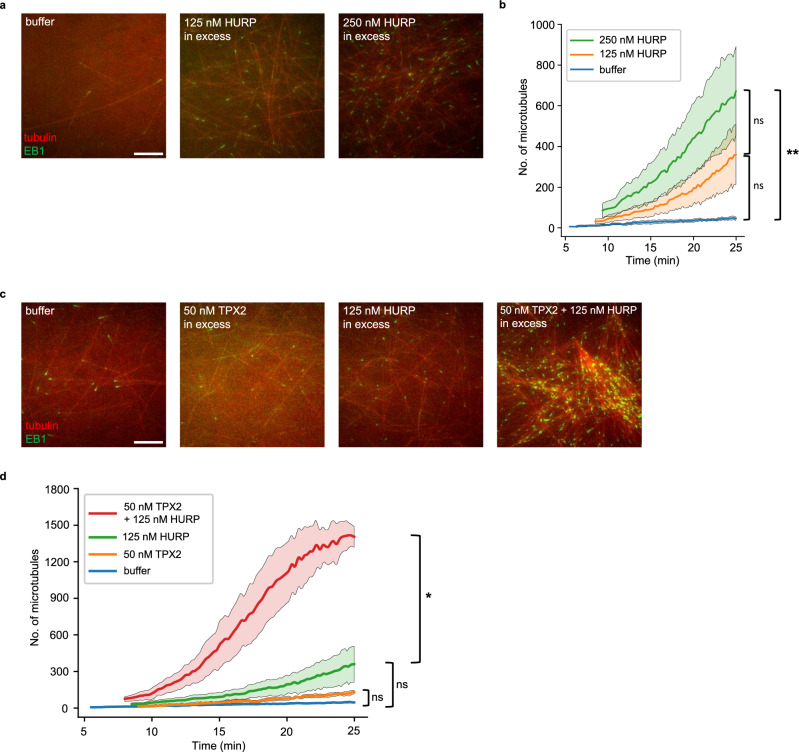
Fig. 2. Excess HURP induces microtubule nucleation in Xenopus egg extract.
a Microtubule (MT) nucleation assay in Xenopus egg extract induced by addition of purified HURP at various concentrations (no added RanGTP). CSF-XB buffer served as the negative control. MTs (AlexaFluor647-tubulin, pseudo-colored red) and mCherry-EB1 (pseudo-colored green) were imaged over 30 minutes. Representative cropped images (51.54 μm x 51.54 μm) at a timepoint of ~15 min are displayed. Scale bar = 10 µm. b Plot of average MT number (EB1 foci ± SEM) over time (min) in a 110.03 μm x 93.41 μm crop across replicates in (a) (n = 3). p value = 0.008 for buffer to 250 nM HURP comparison. c MT nucleation assay in Xenopus egg extract induced by adding purified 50 nM TPX2, 125 nM HURP, or both (no added RanGTP). CSF-XB buffer served as the negative control. MTs and EB1 were imaged over 30 minutes. Representative cropped images (51.54 μm x 51.54 μm) at a timepoint of ~15 min are displayed. The brightness and contrast for the buffer condition was adjusted individually for display purposes. Scale bar = 10 µm. d Plot of average MT number (EB1 foci ± SEM) over time (min) in a 110.03 μm x 93.41 μm crop across replicates of (c) (n = 3). p value = 0.02 for 125 nM HURP comparison to 125 nM HURP + 50 nM TPX2. Significance for panels b and d were calculated by comparing the number of MTs at the final timepoint between conditions. Significance was determined by ordinary one-way ANOVA. ns = not significant. Single asterisk (*) indicates a p value < 0.05. Double asterisks (**) indicate a p value < 0.01.
To further understand the relationship between HURP and TPX2, we assessed whether HURP and TPX2 work together to promote the formation of MTs. A low level of excess TPX2 alone (50 nM excess) is not sufficient to promote the formation of branched MT networks. However, when we added excess HURP to Xenopus egg extract containing a low level of excess TPX2, we observed a large increase in the number of MTs formed (Figs. 2c, d, Supplementary Movie 3, and Supplementary Fig. S6). In fact, the number of MTs formed over time increased exponentially, a hallmark of branching MT nucleation6, and some branched MT networks could be observed. This suggests that the presence of TPX2 influences the mechanism through which HURP promotes MT formation. In addition, this data suggests that HURP enhances the ability of TPX2 to promote the formation of branched MT networks.
TPX2 recruits additional HURP to MTs in vitro
To investigate how TPX2 and HURP influence each other on the MT, we performed sequential MT binding reactions in vitro using MTs stabilized with the slowly hydrolyzable GTP analog, GMPCPP. We find that when GMPCPP-stabilized MTs are pre-incubated with varying concentrations of TPX2, the subsequent MT binding of a constant concentration of HURP (100 nM) increases until saturation (Fig. 3a, b). However, we found that pre-incubation of MTs with HURP does not affect the subsequent recruitment of TPX2 to the MT (Supplementary Fig. S7). Previous literature has found that although HURP and TPX2 may interact on the MT, they do not directly interact in Xenopus egg extract in the absence of MTs27. We conducted an in vitro pull-down and also found that HURP and TPX2 do not directly interact in solution (Supplementary Fig. S8).
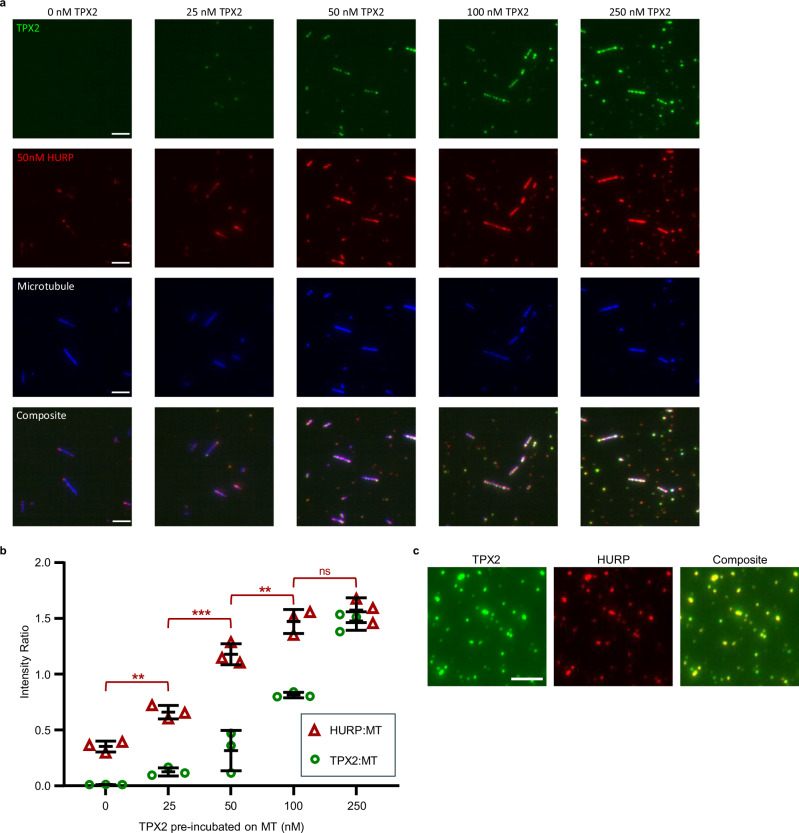
Fig. 3. TPX2 enriches HURP to microtubules in vitro.
a Sequential in vitro microtubule (MT) binding reactions using biotinylated, GMPCPP-stabilized MTs (ATTO647N-tubulin, pseudocolored blue) bound to functionalized coverslips. Each column represents a reaction in which MTs were pre-incubated with the indicated concentration of GFP-TPX2 (or CSF-XB buffer for 0 nM TPX2 condition), followed by 100 nM mCherry-HURP. Representative cropped images (39.77 µm x 39.77 µm) are displayed. Scale bars = 5 µm. b Plot of average HURP:MT and TPX2:MT intensity ratios across replicates (n = 3 per condition) for each sequential MT binding reaction in panel a. Error bars represent standard deviation. Individual points represent the average of each replicate (≥150 MTs per replicate). Significant p values are as follows: 0 nM to 25 nM (p = 0.007), 25 nM to 50 nM (p = 0.0001), and 50 nM to 100 nM (p = 0.009). Significance determined by ordinary one-way ANOVA and defined at a p value < 0.05. ns represents no significance. Double asterisks (**) represent a p value < 0.01. Triple asterisks (***) represents a p value < 0.001. c Bulk co-condensation assay in BRB80 buffer with 1 µM GFP-TPX2 and 50 nM mCherry-HURP, imaged using total internal reflection microscopy (n = 3). Scale bar = 5 µm.
TPX2 enhances the formation of branched MT structures by forming a biomolecular condensate on the MT surface, which recruits soluble tubulin11. To explain how TPX2 can enrich HURP on the MT, we hypothesized that condensed TPX2 can enhance the localization of HURP to the MT. We performed an in vitro bulk phase separation assay, in which TPX2 forms condensates in the absence of MTs, to test whether HURP can enter these condensates. We find that HURP is recruited to TPX2 condensates, showing that HURP is a client of TPX2 condensates (Fig. 3c).
Because this recruitment is dependent on the formation of a condensate, we tested whether HURP can also form condensates in our assay. We find that unlike TPX2, which forms condensates at ~50 nM, HURP at high concentrations of up to 1 µM in low-salt buffer (BRB80) is incapable of forming condensates (Supplementary Fig. S9a, b). Furthermore, in the presence of equimolar tubulin, which lowers the phase boundary of TPX211, HURP remains incapable of forming condensates (Supplementary Fig. S9c). The inability of HURP to form a condensate may explain why TPX2 pre-incubation on the MT enriches HURP, but the reverse is not true. Altogether, these data show that HURP is a client of TPX2 condensates. Because TPX2 condensates preferentially form on MTs, this in turn enhances localization of HURP to the MT.
HURP facilitates MT formation from γ-TuRC in vitro by stabilizing the MT lattice
Having established that HURP is required for Ran-induced branching MT nucleation and that it promotes the formation of single MTs in Xenopus extract, we next addressed how HURP accomplishes these actions. Because nucleation of single MTs requires γ-TuRC as the universal MT template34, we directly investigated how HURP may influence γ-TuRC-mediated MT nucleation. In our minimal in vitro assay, biotinylated γ-TuRC is tethered to the surface of a biotinylated and passivated glass coverslip via neutravidin. Upon introducing fluorescently labeled tubulin, the formation of dynamic MTs from γ-TuRC can be observed in real time using TIRF microscopy35–37. Interestingly, when 50 nM HURP was added, we observed a ~ 3-fold increase in the number of γ-TuRC-mediated MTs nucleated in comparison to the buffer control (Fig. 4a, b, and Supplementary Movie 4). By averaging the number of MTs over time, and fitting to a saturating exponential function, we obtained initial nucleation rates and observed a ~ 2.4-fold increase in the presence of 50 nM HURP (0.47 ± 0.008 MTs/sec) in comparison to the buffer control (0.20 ± 0.005 MTs/sec). Thus, HURP directly increases the number of MTs nucleated from γ-TuRC.
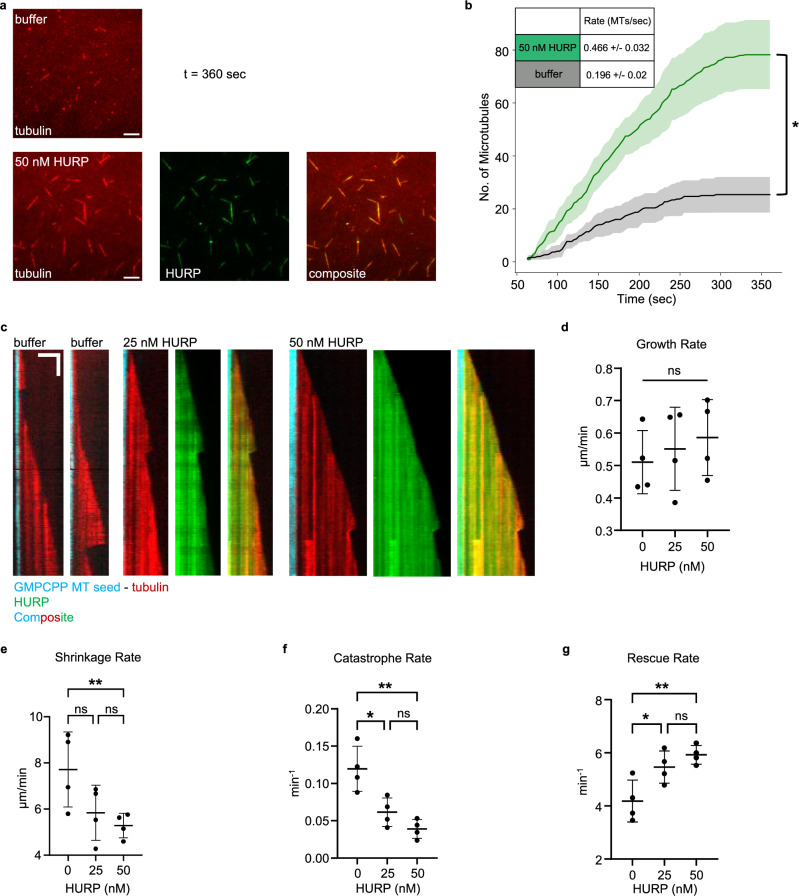
Fig. 4. HURP facilitates nucleation from γ-TuRC in vitro and stabilizes the microtubule lattice.
a In vitro γ-TuRC microtubule (MT) nucleation assay in the presence of 10 μM Alexa Fluor 568-tubulin plus BRB80 buffer (negative control, n = 3) or 50 nM GFP-HURP (n = 4). Representative cropped images (39.77 µm x 39.77 µm) displayed at the final timepoint (360 seconds). Scale bars = 5 µm. b Line graph depicting average MT number over time (sec) ± SEM for each condition in (a). Initial nucleation rates (MTs/sec ± SD, denoted in inset) calculated by fitting data to a saturating exponential function (Eq. (1) in Methods). p value = 0.02. Significance (p value < 0.05) was calculated by comparing the final timepoint of the buffer (25 ± 6.7 MTs; mean ± SEM) and 50 nM HURP (78 ± 13 MTs) conditions via two-tailed Student’s t-test. c Representative kymographs of dynamic MT growth (Alexa568-tubulin, red) from immobilized, GMPCPP-stabilized MT seeds (ATTO647N-tubulin, pseudo-colored cyan) with varying levels of GFP-HURP present (green). Vertical scale bar = 1 minute. Horizontal scale bar = 3 µm. d Scatter plot of average MT growth rate (µm/min ± SD) from four experimental replicates (n = 4) of the dynamic MT growth from seeds assay conditions presented in (c). e Scatter plot of the average MT shrinkage rate (µm/min ± SD) (n = 4). p value = 0.045 for 0 nM to 50 nM comparison. f Scatter plot of the average MT catastrophe rate (events/min ± SD) (n = 4). Significant p values are as follows: 0 nM to 25 nM (p = 0.01), 0 nM to 50 nM (p = 0.001). g Scatter plot of the average MT rescue rate (events/min ± SD) (n = 4). Significant p values are as follows: 0 nM to 25 nM (p = 0.04), 0 nM to 50 nM (p = 0.007). Each individual point in (d–g) represents the average per replicate (≥ 14 MTs measured per replicate). Significance for were calculated by ordinary one-way ANOVA with significance defined as a p value < 0.05. ns = not significant. A single asterisk (*) indicates a p value < 0.05. A double asterisk (**) indicates a p value < 0.01.
In our in vitro γ-TuRC-mediated MT nucleation assay, only γ-TuRC and soluble tubulin are present at timepoint zero. To test if HURP may act directly on γ-TuRC to promote MT formation, we performed an in vitro pull-down assay and found that HURP does not interact with γ-TuRC (Supplementary Fig. S10a). Additionally, we tested if HURP may localize γ-TuRC to pre-existing GMPCPP-stabilized MTs and found that HURP could not localize γ-TuRC to the MT (Supplementary Fig. S10b and c). These results suggest that HURP does not directly interact with γ-TuRC, either in solution or on the MT, to enhance MT nucleation. We also performed these interaction assays with soluble tubulin and found that although HURP binds weakly to soluble tubulin in the in vitro pull-down assay, HURP does not recruit soluble tubulin to pre-existing MTs (Supplementary Fig. S11). This suggests that HURP may be able to bind soluble tubulin to facilitate MT formation, but that it has a much higher affinity for the MT lattice.
Since HURP has a higher affinity for the MT lattice, we investigated how HURP influences MT dynamics by measuring dynamic growth from immobilized, GMPCPP-stabilized MT seeds in vitro (Fig. 4c and Supplementary Movie 5). We find that, in agreement with a previous finding38, the presence of HURP (25 nM and 50 nM) has no significant effect on MT growth rates (Fig. 4d). However, we do find that shrinkage rates during MT depolymerization are decreased in the presence of 50 nM HURP (5.28 ± 0.53 µm/min) when compared to the buffer control (7.71 ± 1.62 µm/min) (Fig. 4e). This finding suggests that HURP may help to promote the stability of the MT lattice. In accordance with this, we see that the presence of HURP greatly decreases the rate of catastrophe (0.039 ± 0.013 events/min; 50 nM HURP) when compared to the buffer control (0.120 ± 0.015 events/min) (Fig. 4f). We also observe a significant increase in the rate of rescue in the presence of HURP (5.93 ± 0.35 events/min; 50 nM HURP) compared to the buffer control (4.19 ± 0.79 events/min) (Fig. 4g). These MT dynamics data show that HURP stabilizes the MT lattice, specifically by preventing catastrophes and promoting rescue when catastrophes do occur. This stabilizing function is consistent with previous studies characterizing cold-resistance of k-fibers in HURP-depleted cells, in that loss of HURP resulted in less stable (cold-resistant) k-fibers18. Altogether, HURP stabilization of nascent MTs may result in the increase of optically observable MTs over time in our in vitro assay with γ-TuRC nucleated MTs.
A bipartite MT binding mode of HURP
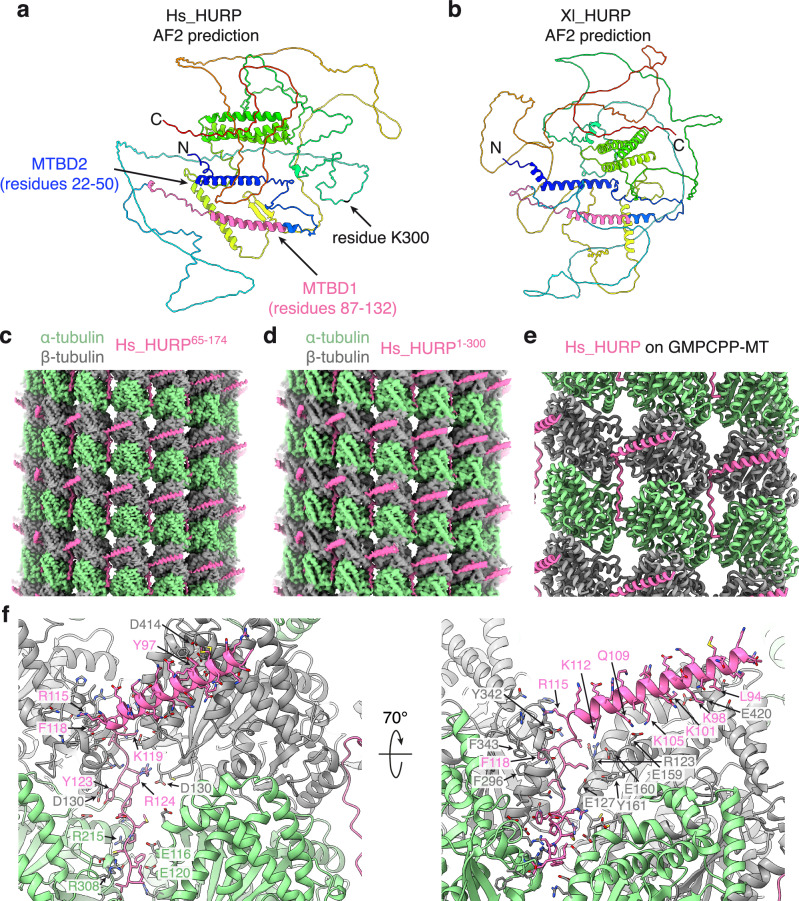
To better understand how HURP binding to the MT lattice influences MT dynamics, we performed single particle cryo-EM of HURP bound to GMPCPP-stabilized MTs. Following a previous study39, we made a human HURP65-174 construct that includes MT-binding domain 1 (MTBD1) and a HURP1-300 construct that includes both MTBD1 and MT-binding domain 2 (MTBD2). These two regions are highly conserved between human and Xenopus laevis (Fig. 5a, b, and Supplementary Fig. S12). We added these two protein constructs, HURP65-174 and HURP1-300, to GMPCPP-stabilized MTs in an over-saturated condition and determined their cryo-EM structures using a newly developed MT data processing pipeline based on CryoSPARC40 (Supplementary Fig. S13) that utilized the results from MT seam search41. Both structures (HURP65-174 and HURP1-300) revealed nearly identical MAP densities (Fig. 5c, d). The structure of the HURP65-174-decorated MT at 2.8 Å local resolution allowed us to perform unbiased and automated protein identification of the non-tubulin densities using the software ModelAngelo42. A search of the ModelAngelo-derived protein sequence against the entire human proteome produced a single hit of HURP protein with high confidence. The hit was further verified by careful visual inspection of the fit between the atomic model (Fig. 5e) and cryo-EM density map.
Fig. 5. Cryo-EM structure of human HURP constructs on GMPCPP-stabilized microtubule lattice.
a Alphafold2 prediction of the human HURP structure, colored in rainbow from C- to N- terminus. The microtubule-binding domain 1 (MTBD1) identified by our cryo-EM structure is colored in pink. The microtubule-binding domain 2 (MTBD2) identified in a previous study39 is colored in blue. b Alphafold2 prediction of the Xenopus laevis HURP structure, colored in rainbow. Its putative MTBD1 based on sequence alignment is colored in pink. c Cryo-EM structure of HURP65-174 bound to the GMPCPP-MT lattice at 2.8 Å local resolution. d Cryo-EM structure of HURP1-300 bound to the GMPCPP-MT at 3.8 Å local resolution. e Atomic model of HURP bound to GMPCPP-MT. f Detailed interactions between human HURP and the MT lattice. All conserved HURP residues engaged in the HURP:MT interaction are labeled in pink. α- and β-tubulin residues within 5 Å distance of HURP were displayed and labeled in green and grey, respectively.
The resolved cryo-EM density of HURP corresponds to residues 87-132 (Fig. 5f), which largely overlaps with the previously identified MTBD139 (Supplementary Fig. S12). The fact that both HURP65-174 and HURP1-300 constructs produce nearly identical MAP densities is consistent with the previous report that MTBD1 is the constitutive MT lattice binding site39. We also purified a HURP1-69 construct that contains only MTBD2. However, this construct showed only weak binding to GMPCPP-MTs in a co-pelleting assay. Therefore, we didn’t proceed with the cryo-EM study.
As revealed by our cryo-EM structure of HURP65-174 bound to a GMPCPP-MT (Fig. 5f), an N-terminal helix of HURP (residues 82-114) binds the external surface of β-tubulin, while a loop following the helix (residues 115-132) binds deeply at the groove between two neighboring protofilaments and simultaneously interacts with four tubulin subunits (Fig. 5e, f). Such interaction with the MT lattice may explain our observations that HURP reduces the frequency of catastrophes and increases the frequency of rescue events (Fig. 4f, g), as HURP binding could strengthen the lateral interaction between protofilaments, including at the MT plus ends. Our atomic model of the HURP-MT complex revealed that their interactions are largely electrostatic, involving conserved basic residues of HURP and acidic residues of tubulin (Fig. 5f and Supplementary Fig. S12). Some conserved aromatic residues of HURP such as F118 insert into a hydrophobic pocket on the tubulin lattice and contribute to the overall HURP-MT binding affinity.
Discussion
In this study, we show that HURP is necessary for RanGTP-induced branching MT nucleation in Xenopus egg extract. We uncovered the mechanism through which HURP facilitates MT generation, namely by bridging lateral tubulin subunits within the MT lattice. There are several other MAPs that bind between neighboring protofilaments and simultaneously interact with four tubulin subunits, including TPX243, EB family proteins44,45, doublecortin (DCX)46, and CAMSAP47 (Supplementary Fig. S14). Those that bind at the inter-dimer interface such as TPX2, EB, and DCX typically have stronger influence on MT growth rate, while they all influence other MT dynamic properties (e.g., shrinkage, catastrophe, and rescue rates) to some extent48–51. Interestingly, so far only HURP and TPX2 have been found to possess a bipartite MT binding mode, in that they both have one element anchored onto the ridge of one protofilament, and another element bound deeply at the groove between protofilaments (Supplementary Fig. S14). These bipartite binding modules with a flexible linker in between provide intrinsic capacity to accommodate different curvatures between neighboring protofilaments once they bind to the MT lattice. It is tempting to speculate that HURP may serve to stabilize lateral tubulin interactions that assemble on γ-TuRC to form the critical nucleus or stabilize the nascent microtubule lattice nucleated by γ-TuRC. Thus, HURP serves as a stapler or glue that keeps protofilaments laterally associated and prevents them from falling apart.
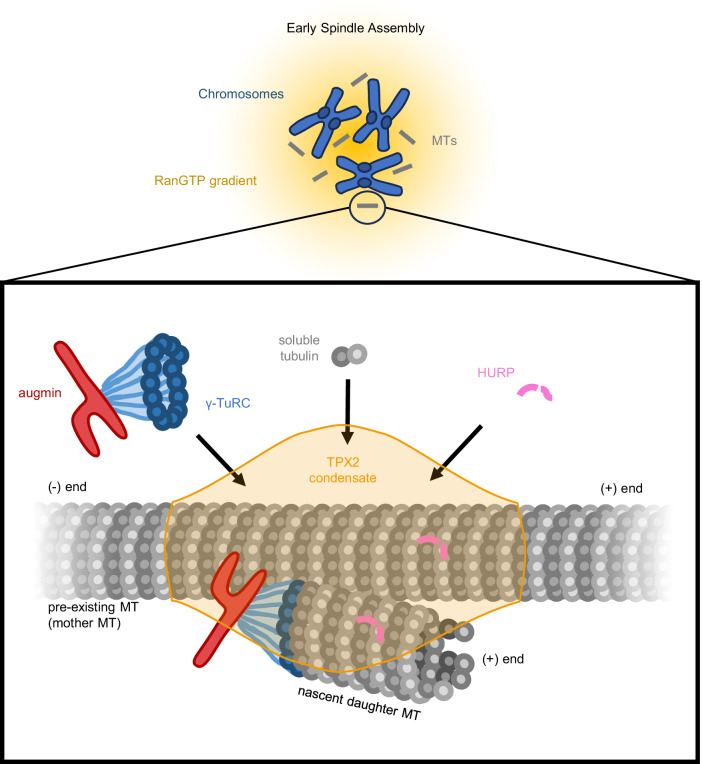
In the presence of TPX2, we found that the MT-stabilizing function of HURP favors branching MT nucleation as opposed to the formation of single MTs. We showed that this occurs by formation of a TPX2 condensate that recruits HURP as a client protein to the MT surface. Therefore, we propose a model in which TPX2, released from inhibitory importins by RanGTP, preferentially enriches HURP to pre-existing MTs through formation of a co-condensate to promote MT formation via branching MT nucleation (Fig. 6). TPX2 also enriches γ-TuRC, augmin, and soluble tubulin to MTs7,11, all necessary for branching MT nucleation. Therefore in this model, TPX2 serves to efficiently localize all components together to facilitate branching MT nucleation during early spindle assembly, with HURP serving as a critical stabilizing protein for daughter MTs. The data presented here utilizes GMPCPP-stabilized MT seeds. Previous work suggests that HURP prefers binding to GDP-MT lattice when compared to GMPCPP-MTs38. This contrasts with TPX2, which binds preferentially to GMPCPP-MTs48. Although we do not observe a lattice preference for HURP in our experiments (Fig. 4c and Movie 5), it is possible that TPX2 may enrich HURP on lower affinity portions of the MT lattice during early spindle assembly, thus amplifying HURP’s MT-stabilizing function.
Fig. 6. Model for HURP-TPX2 relationship in branching microtubule nucleation.
During early spindle assembly, RanGTP produced by chromosomes creates a gradient of RanGTP. RanGTP releases spindle assembly factors augmin, TPX2, and HURP from inhibitory importins, allowing them to individually bind to forming microtubules (MTs). Upon binding to a MT, TPX2 forms a condensate and recruits soluble tubulin to the lattice of the pre-existing MT. Additionally, the TPX2 condensate enriches for augmin, γ-TuRC, and HURP, forming a site enriched in components necessary for branching MT nucleation. Being enriched at this site, HURP will bind and stabilize the pre-existing mother MT, as well as nascent daughter MTs that form via branching MT nucleation.
Apart from RanGTP, other studies have determined that the function of human HURP is also positively regulated by Aurora A phosphorylation. Phosphorylation of the C-terminus of human HURP is necessary for localization to spindle MTs30,52. Furthermore, this phosphorylation is hypothesized to contribute to its dynamic localization to the MTs near chromosomes during metaphase30. At metaphase, active Aurora A and TPX2 are bound and localized to spindle poles53. Given our data, we suggest that if TPX2 is sufficiently concentrated at the poles, TPX2 may condense and enrich HURP, resulting in more efficient phosphorylation of HURP by Aurora A and thus maintenance of HURP localization in the metaphase spindle.
Here, we determined that TPX2 and HURP fulfill independent functions, both crucial to branching MT nucleation, and work synergistically to accomplish one output. We show that unlike TPX2, HURP is incapable of forming condensates or localizing tubulin and γ-TuRC to the MT. And unlike HURP, we have previously shown that TPX2 has little effect on the number of MTs formed from γ-TuRC in vitro35. Our results are consistent with previous findings in Xenopus egg extract, which determined that TPX2 and HURP cannot compensate for the loss of one another in chromatin-mediated MT formation27,28.
However, in Drosophila oocytes, TPX2 is dispensable for chromatin-dependent MT generation, while HURP is still required17. This deviation is consistent with differences in the Xenopus and Drosophila proteins. We have previously found that the N-terminal portion of TPX2 drives TPX2 condensation11, yet Drosophila TPX2 only shares homology with the C-terminal domain17,54. Since our model relies on the ability of TPX2 to efficiently form condensates, it is reasonable that the role of TPX2 in Drosophila would not be entirely consistent with Xenopus. In human cells, the role of TPX2 in branching MT nucleation has yet to be fully studied. However, a recent study revealed that HURP interacts with TPX2 in HeLa cells30, suggesting that our model could be conserved in human cells.
Given the localization of HURP to k-fibers18,28, our findings explain how HURP may actively contribute to augmin-mediated k-fiber amplification and maintenance during spindle assembly4. It was previously suggested that HURP confers k-fiber MT stability through its ability to bundle MTs together18,28. In our study, we find that HURP binding to a single, unbundled MT is sufficient to confer stability that results in an increase of MTs being formed from γ-TuRC in vitro. However, MT bundling (e.g., bundling of a nascent daughter MT to the mother MT) could still provide additional stability for MT formation. If true, this would be impactful for k-fiber formation and maintenance, as these MTs are highly bundled.
Just like the Arp2/3 complex, formins, and Spire proteins that were discovered to seed and bring together actin filaments55, their MT counterparts during mitosis seem to be γ-TuRC, XMAP215/chTOG, and HURP. The fact that HURP is locally regulated helps explain how it functions specifically in branching MT nucleation. We believe this concept helps further our understanding of how the cell can form the MT cytoskeleton at the right time and location to enable cell function.
Methods
Ethics
Xenopus laevis husbandry was done in accordance with the NIH Guide for the Care and Use of Laboratory Animals and the approved Institutional Animal Care and Use Committee (IACUC) protocol 1941 of Princeton University.
Cloning
The full-length Xenopus HURP sequence (NCBI Reference Sequence: XP_018087757.1) was synthesized (Genscript) with codon optimization for expression in Escherichia coli in a pUC57 vector. This sequence was inserted into a pST50 vector56 containing N-terminal Strep-6xHis-TEV-GFP-PreScission tags using restriction site cloning using BamHI and HindIII restriction sites and enzymes (NEB Cat #: R3136S and R3104S). The assembled construct was transformed into chemically competent DH5ɑ Escherichia coli cells (NEB Cat #: C2987I), spread on selective Luria broth (LB) agar plates containing 50 μg/mL carbenicillin (GoldBio Cat #: C-103-25), and grown in a 37 °C incubator overnight. Single colonies from the selection plates were grown in selective LB media containing 50 μg/mL ampicillin (Goldbio Cat #: A-301-55) overnight in a 37 °C shaking incubator (180 rpm). Plasmids were purified from overnight cultures using the QIAprep Spin MiniPrep Kit (Qiagen Cat #: 27106). Proper assembly of the construct was screened for by restriction digest of the purified plasmids followed by gel electrophoresis. Final confirmation of the construct was done by Sanger sequencing using T7 and T7-Term sequencing primers (Genewiz). An identical strategy was used to create a full-length HURP construct with N-terminal Strep-6xHis-TEV-mCherry-PreScission tags. To create the Strep-6xHis-TEV tagged construct of the HURP(462-576) fragment used for antibody production, Gibson assembly was used. For Gibson Assembly, the HURP(462-576) fragment was PCR-amplified from the full-length construct, adding overlap sequence to each 5’ end (Forward primer: 5’-ccagggatccGGTGTGCCATATTTCAGAGATATTC-3’, Reverse primer: 5’-agccggatcaTTTGGTTGGGGCTGTTTTG-3’). A pST50 vector56, pre-modified to contain N-terminal Strep-6xHis-TEV tags was PCR-amplified for insertion of the HURP fragment, adding overlap sequence to each 5’ end (Forward primer: 5’-cccaaccaaaTGATCCGGCTGCTAACAAAG-3’, Reverse primer: 5’-atggcacaccGGATCCCTGGAAGTACAG-3’). PCR-amplified fragments were assembled into the final construct using the Gibson Assembly Master Mix (NEB Cat #: E2611L) and verified by Sanger sequencing using the T7 sequencing primer.
Expression and protein purification
Both HURP plasmids were transformed into Rosetta2 (DE3) Escherichia coli cells (NEB: C29871) and grown in 2 L of LB media containing 50 μg/mL ampicillin and 25 μg/mL chloramphenicol at 37 °C in a shaking incubator (200 rpm). After reaching an OD600 of 0.6, protein expression was induced by addition of 0.5 mM isopropyl-β-D-thiogalactoside (IPTG). The full-length HURP construct was expressed for 4 h at room temperature in a shaking incubator (200 rpm). The HURP(462-576) fragment was expressed for 16–18 h at 16 °C in a shaking incubator (200 rpm). After expression, the cultures were pelleted, flash-frozen, and stored at −80 °C.
Purification of the full-length HURP constructs was done by thawing a 2 L cell pellet on ice and resuspended up to a volume of 40 mL in His lysis buffer (50 mM NaH2PO4, pH=8.0, 500 mM NaCl, 20 mM Imidazole, 1 mM MgCl2, 6 mM β-mercaptoethanol (BME), 200 μM phenylmethylsulfonyl fluoride (PMSF), 10 μg/mL DNase I) containing a single dissolved cOmplete EDTA-free Protease Inhibitor Cocktail tablet (Roche: 11873580001). The mixture was first gently homogenized with a Biospec Tissue Tearor (Dremel, Racine, WI). The cell mixture was then lysed by high pressure homogenization in an Emulsiflex C3 (Avestin, Ottawa, Canada) by passing the lysate through the Emulsiflex four times at 10,000–15,000 psi. Cell lysate was spun at 105,000 x g for 30 min at 2 °C in a 45 Ti rotor using a Beckman Optima-XE 100 ultracentrifuge. Supernatant was passed through a 5 mL column volume (CV) of Ni-NTA agarose resin (Qiagen: 30250). The column was then washed with 10 CV of His binding buffer (50 mM NaH2PO4, pH = 8.0, 500 mM NaCl, 20 mM Imidazole, 1 mM MgCl2, 6 mM BME, 200 μM PMSF). Elution of the protein was eluted by flowing 1.5 CV of His elution buffer (50 mM NaH2PO4, pH = 8.0, 500 mM NaCl, 250 mM Imidazole, 1 mM MgCl2, 6 mM BME, 200 μM PMSF) through the resin and collecting the flow-through.
The eluate was further purified using size exclusion chromatography. Eluate was concentrated to a volume of ~750 μL using a 50 kDa molecular weight cut-off (MWCO) spin concentrator (Millipore Sigma: UFC905024). The concentrated eluate was divided into three, 250 μL samples and individually run through a Superdex 200 Increase 10/300 GL column (Cytiva, Marlborough, MA) hooked up to an ӒKTA Pure system. A 500 μL manual injection loop was used for loading and the run was performed in Cytostatic Factor Extract Buffer (CSF-XB) (10 mM HEPES, pH 7.7 using KOH, 100 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2, 5 mM EGTA, 10% v/v sucrose) with a flow rate of 0.5 mL/min. Eluate was collected in 0.5 mL fractions and absorbance at 280 nm was used to track the protein peak. Yield and purity of fractions were assessed by SDS-PAGE gel and Coomassie stain. Fractions with high yield and purity for the full-length HURP construct (MW: 127 kDa) were pooled, spin-concentrated as described above, and snap-frozen in 5 μL aliquots at ~15 μM. Protein concentration was determined via densitometry using BSA standards on an SDS-PAGE gel stained with Sypro Protein Gel Stain (ThermoFisher: S12000).
The HURP(462-576) construct used in generation of the HURP antibody was purified by His and Strep affinity chromatography. The cell lysis and His purification is carried out the same as the full-length HURP construct, except that buffers contained 300 mM NaCl instead of 500 mM. Eluate from the His purification was then diluted ~7-fold with Strep binding buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 1 mM MgCl2, 6 mM BME, 200 μM PMSF) before flowing over 5 mL CV of Strep-Tactin Superflow resin (Neuromics: 2-1206-025). The column was then washed with 10 CV of Strep binding buffer before being eluted with 1.5 CV of elution buffer (Strep binding buffer + 3.3 mM D-desthiobiotin). Yield and purity of the HURP(462-576) construct (MW: 17 kDa) was assessed by SDS-PAGE gel and Coomassie stain. A concentration of ~1 mg/mL was determined by Bradford assay.
Existing constructs of full-length Xenopus laevis TPX2 (Strep-6xHis-GFP-TPX2 and GST-TPX2) were expressed and purified as previously described11. Briefly, expression was induced with 0.75 mM IPTG and expressed for 7 h at 27 °C. The GFP construct was first purified from lysate by His affinity chromatography in a Tris-based buffer, pH 7.75, notably containing 750 nM NaCl. Eluate from the His purification was then further purified by size-exclusion chromatography using a Superdex 200 HiLoad 16/600 column (GE Healthcare: 28-9893-35) in CSF-XB buffer. The GST construct was purified by GST affinity chromatography in a Tris-based buffer, pH 7.75, at 200 mM NaCl, washed with 1 M NaCl prior to eluting, and dialyzed overnight in CSF-XB buffer. Peak fractions were then pooled, concentrated by spin centrifugation (Millipore Sigma: UFC905024), flash frozen, and stored at −80 °C.
Bands of interest (Supplementary Fig. 2) were cut, pre-processed, and sent for LC-MS/MS analysis using a Fusion Lumos mass spectrometer. Results were validated in Scaffold_5.3.357 against a custom database containing the Xenopus laevis and E. coli proteome. Peptide identifications were accepted if established at > 95.0% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if established at > 99.9% probability and contained at least 5 identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm58.
Octameric augmin complex (HAUS subunits 1-8, with the HAUS6 construct lacking its C-terminus and containing N-terminal ZZtag-PreScission tags, HAUS3 and HAUS8 containing N-terminal Strep-GFP tags, and HAUS2 containing C-terminal GFP-6xHis tags) was expressed and purified from Sf9 cells as described previously59. Briefly, the complex was first purified by IgG Sepharose affinity chromatography in a Tris-based buffer (pH 7.7), eluted by PreScission cleavage. Eluate was then spin concentrated and further purified by size-exclusion chromatography using a Superose 6 Increase 10/300 in CSF-XB with 1 mM dithiothreitol (DTT). Purity of peak fractions were analyzed by SDS-PAGE, pooled, flash frozen, and stored in −80 °C.
An existing construct of EB1-mCherry (EB1-6xHis-mCherry in a pET21a backbone) was expressed and purified as previously described previously60. Briefly, the protein was purified by His affinity chromatography in sodium phosphate buffers, pH 7.4. Eluate was purified further by size-exclusion chromatography using a Superdex 200 pg 16/600 (GE Healthcare) in CSF-XB. Purity of peak fractions were analyzed by SDS-PAGE, pooled, flash frozen, and stored in −80 °C.
Expression and purification of Ran(Q69L) (Strep-6xHis-TEV-BFP-Ran(Q69L)) was done similarly to a previously published protocol60. Specific to this study, expression of the construct was induced with 0.5 mM IPTG after an OD600 of 0.8 was reached in Terrific Broth (TB) media. Expression was carried out for 18 h at 16 °C. Strep affinity chromatography and subsequent dialysis into CSF-XB was carried out as described previously in ref. 60.
Purification of GST-importin-β was done via GST affinity chromatography and gel filtration, as previously described in ref. 13.
Purification and biotinylation of the native Xenopus laevis γ-TuRC complex was done as described previously36. Briefly, Halo-Magne beads (Promega: G7287) were coated in purified HaloTag-PreScission-humanγTuNA. The beads were then incubated with thawed Xenopus laevis extract to allow for γTuNA to bind to native γ-TuRC. After incubation, beads were washed and incubated with NHS-PEG4-Biotin (ThermoFisher: A39259). After incubation, beads were then resuspended and incubated overnight in a PreScission protease cleavage buffer to allow for cleavage of γTuNA-γTuRC from the HaloTag-bound beads. After cleavage, the supernatant was obtained, concentrated, and spun through a sucrose gradient. Peak fractions from the sucrose gradient were determined by Western blot, then flash frozen and stored at −80 °C. Concentration of γTuRC was assumed to be between 150-200 nM, as previously determined36.
Different human HURP constructs covering MTBD1 (residues 65-174), MTBD2 (residues 1-69) and MTBD1-2 (residues 1-300) were cloned into pMBP vector with an N-terminal MBP tag. The recombinant proteins were overexpressed in E. coli BL21(DE3) at 16 °C overnight after induction by 0.2 mM IPTG at OD600 of 0.8. The proteins were purified by Amylose resin (NEB) and eluted with 10 mM maltose. Then the eluate was further purified by gel filtration (Superdex 200, GE Healthcare) in a buffer containing 25 mM Tris (pH 8.0), 150 mM NaCl, 1 mM DTT. The peak fraction was collected and concentrated to ~5 mg/mL, then stored at −80 °C with aliquots of 20 μL.
Generation of HURP antibody and purification
A total of ~10 mg of purified HURP(462-576) at ~1 mg/mL was sent to the Covance company for inoculation of two rabbits to produce polyclonal antibodies against HURP. Antibody was provided by Covance in the form of antisera. The HURP antibody was then purified by first conjugating 10 mg of HURP(462-576) antigen to 0.5 mL of Affi-gel 15 (Bio-Rad: 1536051) resin (brought up to 10 mL with PBS buffer), rotating overnight at 4 °C. Antigen-coupled beads were then washed sequentially with 0.1 M sodium phosphate buffers (pH 7.2), 0.1 M glycine (pH 2.5), then 0.1 M sodium phosphate buffers of pH 7.2, 11, and 7. After washes, the resin was incubated with 3 mL of antisera, brought up to 10 mL with PBS, rotating overnight at 4 °C. Resin was then washed sequentially with sodium phosphate buffers of pH 7, pH 7 with 0.5 M NaCl, and pH 7 again to remove the salt. Antibody was eluted with 0.1 M glycine, pH 2.5, and quickly neutralized with 1 M HEPES buffer, pH 7.7, at a 1:10 glycine:HEPES ratio. Elution was done in 1 mL fraction volumes (after neutralization with HEPES buffer). Yield was determined by A280 via NanoDrop. Peak fractions were combined, flash frozen in 40 μg aliquots, and stored at −80 °C.
Tubulin labeling and GMPCPP-stabilized microtubules
Unlabeled cycled tubulin purified from bovine brain was purchased from PurSolutions (Cat#: 032005). Bovine brain tubulin was labeled following previously described methods61. Labeling with Cy5-NHS ester (GE Healthcare, PA15101) yielded 54–70% labeling efficiency. Labeling efficiency with Alexa-568 NHS ester (Invitrogen: A20003) was 36–40%. Labeling efficiency with ATTO647N (ATTO-TEC, AD 647N) was 62%. Labeling efficiency with biotin-PEG4-NHS (Thermo Scientific: A39259) was not calculated.
Fluorescent and biotinylated double-cycled GMPCPP-stabilized MTs were made similarly to a previously published protocol62. Tubulin was polymerized (40 μL reaction) at a total tubulin concentration of 20 μM in BRB80 buffer (80 mM K-PIPES at pH 6.8, 1 mM EGTA, 1 mM MgCl2) containing 1 mM GMPCPP. Of the 20 μM total tubulin, 2 μM is composed of ATTO647N-labeled tubulin, 2 μM is composed of biotin-labeled tubulin, and the rest is composed of unlabeled tubulin. The reaction was assembled over ice. Polymerization was performed in 37 °C water bath for 30 min, protecting the reaction from light. MTs were then pelleted by centrifugation in a tabletop centrifuge at 17,000 x g for 15 minutes (room temp). MTs were then depolymerized by resuspending and incubating the pelleted MTs in ice-cold BRB80 buffer for 20 minutes in a volume totaling 80% of the original volume. GMPCPP was then added to 1 mM, and MTs were again polymerized and pelleted as before. This time, MTs were resuspended in warm BRB80 buffer in a volume totaling 80% of the volume prior to pelleting. MTs were flash frozen in 2 μL aliquots and quickly thawed before use. MTs were diluted 1:100 to 1:1000 prior to use in experiments.
Immunodepletion of Xenopus laevis egg extract and western blot analysis
Meiotic Xenopus laevis extract was performed as previously described63 and assayed for quality (described below in the section titled: TIRF imaging of MT formation in Xenopus laevis egg extract). For each extract experiment, three adult Xenopus laevis female frogs (sourced from Nasco/Xenopus1) were induced to ovulate. All steps for the immunodepletion were done on ice and in a 4 °C cold room. For immunodepletions, 300 μL of Dynabeads protein A slurry (Thermofisher: 10001D) was washed thrice 600 μL TBS-T buffer. A magnetic strip was used to separate the beads from the solution for washes. Half of the beads were then resuspended in a HURP antibody solution for HURP depletion. The other half were resuspended in an IgG antibody solution for the mock depletion. Antibody solutions consisted of 40 μg of antibody in a total volume of 180 μL using Tris-buffered saline with Tween-20 (TBS-T) buffer (50 mM Tris, pH 8.0, 138 mM NaCl, 2.7 mM KCl, 0.1% w/v Tween-20). Antibody-bead mixtures were incubated overnight at 4 °C on a rotator. After incubation, beads were washed twice with 600 μL TBS-T and thrice with 600 μL CSF-XB. Both sets of beads were split into three tubes for three rounds of immunodepletion, keeping in CSF-XB. To immunodeplete the extract, consecutive rounds of immunodepletion were performed by first aspirating the CSF-XB from the beads and resuspending gently in 60 μL of prepared extract using a wide bore 100 μL pipette tip. The mixture was incubated on ice for 60 min, resuspending gently every 20 min with a fresh wide bore pipette tip. After 60 min, the process was repeated with the remaining tubes. At the end of the third depletion, extract was collected and placed in a fresh tube to be used for experiments.
For Western blotting, extract samples of the undepleted (input), mock-depleted, and HURP-depleted extract were collected (2 μL of extract in 48 μL 1X SDS Sample Loading Buffer + 50 mM DTT), boiled, and 15 μL of each sample was run on two separate 4–12% Bis-TRIS SDS-PAGE gels (ThermoFisher: NP0321BOX). Proteins were then transferred to a nitrocellulose membrane using the Invitrogen iBlot 2 transfer device (ThermoFisher: IB21001), blocked for 1 hour at room temperature, and cut with a razor blade to allow for probing of multiple factors by Western blot. All probing and blocking of membranes was done in TBS-T + 10% w/v non-fat dairy milk. One membrane was used to probe for HURP (HURP antibody described in this paper, 4 μg/mL) and γ-tubulin (Sigma: T6557, 1:1000). The second membrane was used to probe for XMAP215 (Abcam: ab86073, 1:2000), TPX2 (custom antibody described in Alfaro Aco et al. 2017, 4 μg/mL), HAUS1 (custom antibody described in Song et al. 2018, 3.6 μg/mL), and α-tubulin (Invitrogen: 62204, 1:1000). Membranes were incubated in primary antibody overnight at 4 °C on a shaker, washed thrice with TBS-T on a shaker over the course of 45 min, and then incubated for 1 h at room temperature with either Mouse-IgG, HRP linked secondary antibody (Amersham: NA931-1ML, 1:2000) or Rabbit-IgG, HRP linked secondary antibody (Amersham: NA934-1ML, 1:2000). Membranes were then washed thrice with TBS-T as before, and then incubated with Amersham ECL Western Blotting Detection Reagents (Cytiva: RPN2232) for chemiluminescence detection. Images were taken using the iBright Imaging system, ensuring that no bands were overexposed. The γ-tubulin membrane was stripped with Stripping Buffer (ThermoFisher:46430), reblocked, and probed for the loading control α-tubulin as described above.
To analyze the Western blot for depletion of factors, images were loaded into the Fiji software64 (ImageJ v1.54 f; RRID:SCR_002285) and converted to 8-bit (0-255 intensity range) for ease of processing. The mean intensity of each band was measured by selecting a box that encapsulated each band. Boxes were kept an equal size for each measurement. Additionally, the background mean intensity was also measured for each band by selecting a nearby region of the membrane just above or below the band, again keeping the box size consistent. The mean intensity for each band was subtracted from the background mean intensity and standardized to the respective loading control (α-tubulin band) to account for errors in loading the gel. After standardization, the intensities of the mock- and HURP-depleted bands for each blotted protein of interest were used to calculate percent intensity relative to the input lane. For Fig. 1a, the proportion of intensity relative to the input lane (scaled 0-1) was averaged across the four biological replicates and plotted. Values less than 0 and greater than 1 were converted to 0 and 1, respectively, for plotting and statistical analysis. Statistical significance was determined by unpaired two-tailed t-tests comparing the mock- and HURP-depleted conditions for each protein. Plotting and statistics were performed using GraphPad Prism.
In vitro pull-downs
For each pull-down reaction, 20 µL of GFP-Trap Magnetic Agarose bead slurry (ChromoTek: gtma) were first washed thrice with TBS-T + 5 mg/mL κ-casein and equilibrated into binding buffer (CSF-XB, no sucrose + 0.1% Tween-20 and 5 mg/mL κ-casein). The beads were then incubated with the binding reaction for 2 h at 4 °C on a rotator. Binding reactions (30 µL) contained a final concentration of either 2 µM GFP-HURP or 2 µM His-GFP as the bait and a 2 µM final concentration of the prey protein (soluble tubulin or GST-TPX2) diluted in binding buffer. A final concentration of ~40 nM of γ-TuRC was used. For pull-downs containing tubulin and γ-TuRC, 1 mM GTP was supplemented into the binding reaction to maintain protein stability. Additionally, 100 µM chymostatin was supplemented into the γ-TuRC pull-down assay to inhibit residual PreScission cleavage from its purification. After incubation, the beads were collected using a magnetic strip and washed thrice with binding buffer prior to addition of 4X SDS Sample Loading buffer.
Input (~8%) and bound fractions were run on an SDS-PAGE gel, transferred to a nitrocellulose membrane, and western blotted as described in the above section titled: Immunodepletion of Xenopus laevis egg extract and western blot analysis. In addition to the α-tubulin and γ-tubulin primary antibodies used in the mentioned section, 1:1000 dilution of KPNB1 antibody (ABclonal: A8610), 1:500 dilution of HURP antibody (proteintech: 12038-1-AP), 1:1000 dilution of GFP antibody (Abcam: AB290), 1:1000 dilution of GST antibody (Abcam: ab92), and 1:1000 GCP5 antibody (Santa Cruz Biotechnology: sc-365837) were used to detect GST-importin-β, GFP-HURP, His-GFP, GST-TPX2, and γ-TuRC, respectively. All pull-down reactions were performed three times with similar results.
TIRF imaging of microtubule formation in Xenopus laevis egg extract
Meiotic Xenopus laevis egg extracts were prepared as previously described63. For each extract experiment, three adult female Xenopus laevis frogs (sourced from Nasco/Xenopus1) were induced to ovulate. Extracts were either used immediately for experiments or subjected to immunodepletion prior to experiments. Quality of every prepared extract was ensured by performing the branching assay (with Ran(Q69L)), described in detail below, and a negative control of the branching assay (no Ran(Q69L)). A quality extract was defined as forming characteristically dense fan-like structures in the presence of Ran(Q69L) and forming few MTs (<10 MTs in 10 min) when no Ran(Q69L) is added.
For branching MT nucleation assays, 7.5 µL of extract was incubated on ice with 0.5 µL of 10 mM vanadate (0.5 mM final), 0.5 µL of 1 mg/mL end-binding 1 (EB1)-mCherry protein (0.05 mg/mL final), 0.5 µL of 1 mg/mL Cy5-tubulin (0.05 mg/mL final), 0.5 µL of 200 μM Ran(Q69L) (10 μM final), and 0.5 µL of CSF-XB. For addback of purified HURP to HURP-depleted extract, CSF-XB was replaced with 0.5 µL of 5 μM purified HURP (250 nM final). If necessary, purified proteins were previously diluted in CSF-XB to obtain the stock concentrations listed.
For assaying the effect of excess HURP on the number of MTs, 7.5 µL of extract was incubated on ice with 0.5 µL of 10 mM vanadate (0.5 mM final), 0.5 µL 1 mg/mL end-binding 1 (EB1)-mCherry protein (0.05 mg/mL final), 0.5 µL of 1 mg/mL Cy5-tubulin (0.05 mg/mL final), 0.5 µL of CSF-XB, and 0.5 µL of purified HURP. HURP protein was diluted in CSF-XB prior to addition into the reaction mix, such that the final concentrations in the reaction mix were as stated in Fig. 2e, c. A similar approach was used for the addition of 50 nM TPX2 to the extract for Fig. 2c. For the negative control, addition of HURP was replaced with additional CSF-XB. For reactions containing both HURP and TPX2, all CSF-XB was omitted from the reaction mix and was replaced with 0.5 µL of 2.5 μM HURP (125 nM final) and 0.5 µL of 1 μM TPX2 (50 nM final).
All extract reactions (10 µL total) were gently mixed by pipetting twice with a wide-bore 100 µL pipette tip before adding to a flow-channel at 18–20 °C. Flow channels were assembled by evenly spacing thin strips of double sided tape onto a microscope slide and gently pressing a 20 mm × 20 mm coverslip on top of the tape. In this manner, 2-3 flow channels were created to support the number of reactions being imaged. Reactions loaded in multi-flow channel slides (different conditions) were continuously and sequentially imaged for ~30 minutes. Time = 0 is defined as the moment the reaction was loaded into the flow-channel.
Multi-channel fluorescence images for all experiments were acquired with no delay between acquisitions using the NIS-Elements AR program (NIKON, ver. 5.02.01-Build 1270; RRID:SCR_014329). The 647 nm/Cy5 channel (excitation: 678 nm, emission: 694 nm) was used for imaging MTs and the 561 nm channel for EB1 localized to MT plus-tips (ex: 587 nm, em: 610 nm). The images were captured on a Nikon Ti-E inverted system (RRID:SCR_021242), with an Apo TIRF 100 x oil objective (NA = 1.49), and an Andor Neo Zyla (VSC-04209) camera with 2 × 2 binning. Exposure settings for all channels were kept consistent across conditions. Resulting images were 164.92 µm x 139.15 µm. After time-lapse acquisition, a large 3 × 3 multi-channel fluorescence image was acquired to ensure that all time-lapse FOV were representative of the reaction. Stitched 3 × 3 images are provided as Supplementary Figs. for each extract reaction provided. 3 × 3 images were 491.55 µm x 414.75 µm. For all the extract images displayed, the 647 nm channel was pseudo-colored red and the 561 nm channel was pseudo-colored green. Dimensions of representative cropped images are listed in the respective figure legends.
Analysis of microtubule formation over time in Xenopus laevis egg extract
To quantify MT nucleation levels, we extracted the number of MT plus ends (tracked by EB1 spots) for each condition using the 561 nm/EB1-mCherry channel. For early timepoints, the number of MTs were recorded manually in the Fiji software at increments of every 10 frames (~80 s). Manual counting of early timepoints was done due to the inaccuracy of the automated counting system given the inaccurate auto-thresholding when few EB1 foci were present. For later timepoints, we wrote a macro in Fiji to automate counting EB1 spots. Briefly, 110 μm x 93 µm representative windows were cropped from each field of view for the EB1 channel image stack. The image stack was duplicated twice and the Gaussian Blur function was applied to each (one with a sigma(radius) of 1, the other with 5). The 5 radius blur image was subtracted from the 1 radius blur image using the Image Calculator. The subtracted image stack was then autothresholded using the default option, and ‘Analyze Particles’ was used to count the number of thresholded EB1 foci (minimum size = 0.1 μm2). Manual and auto-counts were combined and plotted over time (based on the framerate and time imaging began) using GraphPad Prism version 10.0.3 for Windows (GraphPad Software, www.graphpad.com). Each experiment was performed three times. Statistics were performed by comparing the number of average MTs (± SD) at the final timepoint of each condition. For comparisons between two conditions, two-tailed Student’s t-test was used. For comparisons between more than two conditions, ordinary one-way ANOVA was used.
Preparation of functionalized coverslips and imaging chambers
Functionalized glass coverslips used in all in vitro γ-TuRC nucleation and MT nucleation assays were generated using a previously published method36. Briefly, coverslips were cleaned by sonication in a series of 3 M NaOH followed by Piranha solution (2:3 rate of 30% w/w H2O2 to sulfuric acid), rinsed with milliQ water, dried, then treated with 3-glycidyloxypropyl trimethoxysilane (Sigma: 440167) at 75 °C for 30 min. Following treatment, coverslips were washed with acetone, dried with nitrogen gas, and functionalized with a 9:1 mixture of HO-PEG-NH2 to biotin-CONH-PEG-NH2 by weight (Rapp Polymere: 103000–20 and 133000-25-20) at 75 °C overnight. After incubation, coverslips were washed with MilliQ water, dried, and stored at 4 °C for up to 3 months.
For the γ-TuRC nucleation assays, imaging chambers were made by making a channel on a glass slide with double-sided tape, coating the channel with 2 mg/mL of PLL-g-PEG (in MilliQ water), letting air dry, rinsing with MilliQ water, then drying with nitrogen gas. Functionalized coverslips were then placed on the double-sided tape (functionalized side facing down). Chambers were used within 24 h. Imaging chambers for the MT localization assays were made similarly except that multi-flow channel slides (2-3 flow channels) were made with the double-sided tape, not coated with PLL-g-PEG, and used within one week stored at 4 °C.
In vitro γ-TURC-mediated microtubule nucleation assay
Imaging chambers were blocked with room temperature 5% w/v Pluronic F-127, followed by cold assay buffer (BRB80 with 30 mM KCl, 0.075% w/v methylcellulose 4000 cp, 1% w/v D-glucose, 0.02% w/v Brij-35, and 5 mM BME). If performing the γ-TuRC nucleation assay, the assay mix was also supplemented with 1 mM GTP. After blocking for 5 min, the imaging chambers were washed with cold 0.05 mg/ml κ-casein in assay buffer, then incubated with cold 0.05 mg/mL NeutrAvidin (ThermoFisher: A2666) on a cold block for 1.5 min, then washed with cold BRB80 buffer.
A nucleation mix containing 12.5 μM bovine tubulin (5% AlexaFluor 568-tubulin and the remaining unlabeled) and 1 mg/mL BSA (Sigma: A7906) in cold assay buffer was centrifuged at 280,000 x g in a TLA-100 rotor (Beckman Coulter) for 12 min at 2 °C (20% loss of tubulin assumed, final tubulin concentration = 10 μM). During the spin, an aliquot of biotinylated γ-TuRC was diluted 1/100 in cold BRB80 buffer, flowed into the imaging chamber, and incubated for 7 min at room temperature in a dark humidity chamber. One diluted aliquot was used for all experiments in a given day. After centrifugation of the nucleation mix, glucose oxidase (SERVA GmbH: SE22778, final concentration of 0.68 mg/mL), catalase (Sigma: SRE0041, final concentration of 0.16 mg/mL), and either GFP-HURP (final concentration of 50 nM, pre-diluted in CSF-XB; n = 4) or an equal volume of CSF-XB (n = 3) was added to the nucleation mix. The imaging chamber was then washed with cold BRB80 and the nucleation mix was loaded into the imaging chamber (t = 0) and imaged for 5 min (1 frame per 2-3 s), starting at t = 60 s. We used the same imaging set-up as our previous work35,36, notably a Nikon Ti-E inverted stand (RRID:SCR_021242) with an Apo TIRF 100 x oil objective (NA = 1.49) and an Andor iXon DU-897 EM-CCD camera with EM gain set to 300. The 561 nm channel was used to image Alexa Fluor 568-tubulin and the 488 nm channel was used to visualize GFP-HURP. Exposure settings for all channels were kept consistent across conditions. We used an objective heater collar (Bioptechs: model 150819–13) to maintain 33.5 °C for our experiments. Resulting images were 81.45 μm x 81.45 μm. Dimensions of representative cropped images in figures are listed in the figure legends.
Analysis of in vitro γ-TuRC-mediated microtubule nucleation assay data
For each movie, we started by processing the entire field of view (81.45 μm x 81.45 μm) for the 5 min movies using Fiji. The movies were corrected for thermal drift (translation) by using the StackReg plugin65 (RRID:SCR_003070). To correct for severe drift, we cropped the corrected stack to a 75.09 μm x 75.09 μm square, allowing us to perform the StackReg function again. We wrote a macro in Fiji to semi-automate data analysis. The macro generates kymograph (time-space) plots, using the KymoResliceWide plugin based on manual tracing of individual MTs, and prompts the user to extract relevant parameters from the kymographs.
For each kymograph produced, the user is first prompted to determine if the MT is spontaneously nucleated or γ-TuRC mediated. MT kymographs where the tubulin signal formed a right triangle were considered γ-TuRC-mediated. Random appearances of a long MT or a MT that grew from both ends, indicating that the minus end is not capped by γ-TuRC, were marked spontaneous. The location of each MT was recorded and numbered on a separate Z-projected map to prevent double-counting. If a MT was determined to be γ-TuRC-mediated, we proceeded collecting measurements. We manually recorded the nucleation point (origin) for each MT.
Time of nucleation for each MT was analyzed using a Python script in Jupyter Notebook66 and averaged across all reactions for each condition. The mean and standard error of the mean (SEM) for the number of MTs over time were plotted as shown in Fig. 4b. To determine the rate of γ-TuRC-mediated MTs formed, we fit the MT number curve for each condition to the following condition as done previously36:
| 1 |
In Eq. (1), = the number of MTs nucleated at a given timepoint (t), = maximum number of MTs at the final timepoint (360 s), and = the rate of MT formation. Fitting was done using the scipy.optimize curve_fit function67 to determine the and . The standard deviations of the fit for each condition are provided in Fig. 4b. Additionally, two-tailed Student’s t-test was performed on the average MTs (± SD) at the final timepoint of each condition to determine significance.
Dynamic microtubule growth from seeds
This assay was conducted very similarly to the section titled: in vitro γ-TuRC-mediated microtubule nucleation assay. However, the nucleation mix contained 7.2 μM bovine tubulin (8.7% AlexaFluor 568-tubulin and the remaining unlabeled) and 1 mg/mL BSA (Sigma: A7906) in cold assay buffer. Additionally, instead of incubating the channel with biotinylated γ-TuRC, an aliquot of biotinylated and fluorescent double-cycled GMPCPP-stabilized MTs (ATTO647N-tubulin) were thawed and diluted 1/100 in warm BRB80 buffer, flowed into the imaging chamber, and incubated for 5 minutes at room temperature in a dark humidity chamber. A new aliquot was used for each experiment. All other steps leading up to the imaging were followed as stated in the section titled: in vitro γ-TuRC-mediated microtubule nucleation assay. The reaction was imaged for 10 min (1 frame per 3 seconds). The images were captured on a Nikon Ti-E inverted system (RRID:SCR_021242), with an Apo TIRF 100 x oil objective (NA = 1.49), and an Andor Neo Zyla (VSC-04209) camera with no binning. The 561 nm channel was used to image Alexa Fluor 568-tubulin, the 647 nm channel was used to image ATTO647N-tubulin seeds, and the 488 nm channel was used to visualize GFP-HURP. Exposure settings for all channels were kept consistent across conditions. We used an objective heater collar (Bioptechs: model 150819–13) to maintain 33.5 °C for our experiments. Resulting images were 164.92 µm x 139.15 µm. Dimensions of representative cropped movies are listed in the figure legends.
Analysis of dynamic growth from microtubule seeds
Movie pre-processing and generation of kymographs was done as stated in the section titled: Analysis of in vitro γ-TuRC-mediated microtubule nucleation assay data. We then wrote an ImageJ macro to manually record the line coordinates manually traced on the kymographs to measure growth speeds and depolymerization speeds. Additionally, the user was prompted to input the number of catastrophes and rescues in each kymograph. A catastrophe was recorded if the microtubule depolymerized by three or more pixels (~0.5 µm) and a rescue recorded if depolymerization was stalled for three or more frames (~ 9 s). Growth speeds of each line were calculated, weighted based on total polymerization time of all measured MTs in the experiment, and averaged to obtain the average MT growth rate. Depolymerization speeds were calculated similarly. Catastrophe and rescue rates were calculated by measuring the total number of events per total polymerization (or depolymerization) time measured. About 20 or more MTs were analyzed per experiment. Each condition was repeated four times. Measurements from the four replicates of each condition were averaged and statistically analyzed by ordinary one-way ANOVA.
Microtubule localization assays
Imaging chambers were prepped the same as in the first paragraph of the section titled: in vitro γ-TuRC-mediated microtubule nucleation assay. After the NeutrAvidin incubation and cold BRB80 wash, the localization assays were performed as follows:
For the MT localization assay, fluorescent and biotinylated GMPCPP-stabilized MTs were quickly thawed and diluted 1:100 to 1:1000 in warm BRB80, flowed into the imaging chamber(s), and incubated for 7 min at room temperature in a dark humidity chamber. A fresh dilution of MTs was used for each slide. ATTO647N-labeled MTs were used for all MT recruitment assays, except for the γ-TuRC recruitment assay which utilized Alexa Fluor 568-labeled MTs. After incubation/binding of MTs in the imaging chamber, all following incubations were performed at room temperature in a dark humidity chamber. MT localization assays were performed as follows:
For recruitment of γ-TuRC to the MT, an estimated ~ 70 nM of purified γ-TuRC (assuming 175 nM concentration for undiluted γ-TuRC) was pre-incubated on ice with either 150 nM GFP-augmin, 100 nM GFP-HURP, or an equal volume of CSF-XB for 5 minutes. After incubation of imaging chambers with MTs as stated above, the imaging chambers were washed with 0.05 mg/ml κ-casein in cold assay buffer, then incubated with the pre-incubated protein mix for 5 min. After incubation, the chambers were washed with BRB80 buffer and then incubated with 2 μg/mL Alexa Fluor 647-conjugated γ-tubulin (XenC) antibody (described previously60) for 10 min. After incubation, unbound protein was washed away with cold BRB80 buffer supplemented with 0.68 mg/ml glucose oxidase and 0.16 mg/ml glucose oxidase. Reactions were imaged immediately.
For recruitment of soluble tubulin to the MT, 100 nM of Alexa Fluor 568-tubulin was pre-incubated on ice with either 100 nM GFP-TPX2 or 100 nM GFP-HURP for 5 min. After incubation of imaging chambers with MTs as stated above, the imaging chambers were washed with 0.05 mg/mL κ-casein in cold assay buffer, incubated with the pre-incubated protein mix for 5 min. After incubation, unbound protein was washed away with cold BRB80 buffer supplemented with 0.68 mg/mL glucose oxidase and 0.16 mg/mL glucose oxidase. Reactions were imaged immediately.
For sequential binding reactions, MT-containing imaging chambers were washed with 0.05 mg/ml κ-casein in cold assay buffer, then the first protein (or CSF-XB for the negative control) was flowed in (diluted in CSF-XB to the concentrations indicated in each figure) and incubated for 5 min. Unbound proteins were washed with BRB80 buffer then the second protein was flowed in and incubated for 5 additional minutes. After incubation, unbound protein was washed away with cold BRB80 buffer supplemented with 0.68 mg/mL glucose oxidase and 0.16 mg/mL glucose oxidase. Reactions were imaged immediately.
Multi-channel fluorescence images for all experiments were acquired using the NIS-Elements AR program (NIKON, ver. 5.02.01-Build 1270; RRID:SCR_014329). For each assay, a 3 × 3 field of view image was acquired to obtain more MTs for analysis. The 647 nm channel was used for imaging the Alexa647-XenC antibody or ATTO647N-labeled MTs. The 561 nm channel was used to visualize mCherry-HURP, Alexa Fluor 568-tubulin, or Alexa Fluor 568-labeled MTs. The 488 nm channel was used to visualize GFP-HURP, GFP-TPX2, or GFP-augmin. Within each experiment, laser power and exposure settings were kept consistent across conditions, except in Supplementary Fig. S5 where exposure differences are specified in the figure legend. The images were captured on a Nikon Ti-E inverted system (RRID:SCR_021242), with an Apo TIRF 100 x oil objective (NA = 1.49), and an Andor Neo Zyla (VSC-04209) camera with 2 × 2 binning. Resulting 3 × 3 stitched images were 491.55 µm x 414.75 µm.
Analysis of microtubule binding/recruitment assays
3 × 3 images were analyzed in Fiji by first isolating the MT channel, auto-thresholding the image using the default setting, and then performing the ‘Open’ and ‘Dilate’ functions sequentially to remove background. ‘Analyze Particles’ of particles greater than 100 pixel units (1.66 μm2) was applied and particles added to the ROI manager. The ROIs were then used to measure the intensity of each individual ROI for the MT and protein of interest. Any ROIs associated with an aggregate were excluded. The inverse ROI was used to measure the average background signal. For the inverse ROI, any protein aggregates or isolated phase condensates were excluded from the measurement. Intensity measurements were imported into a spreadsheet, corrected by subtracting the background intensity, and the protein:MT intensity ratios calculated. The average protein:MT intensity for each replicate was calculated and averaged within each condition. Averages for each condition in a given experiment were plotted and statistically analyzed by either Welch’s two-tailed t-test or ordinary one-way ANOVA using GraphPad Prism to determine if conditions were statistically different. All MT binding assays for any given experiment were either done on the same day or on two separate days. All assays were repeated three times. Reaction mixes for replicates performed on the same day were prepared independently. Purified proteins used for all replicates in any given experiment were derived from a single protein prep.
Bulk phase and co-condensate assay
Purified GFP-TPX2 and ATTO647N-labeled tubulin stocks were precleared of aggregates by centrifugation at 280,000 x g for 10 min at 4 °C in a TLA-100 rotor. The supernatants were collected and kept on ice. A flow chamber was then washed with 40 μL of cold assay buffer (BRB80 + 50 μg/mL κ-casein). Purified GFP-TPX2, ATTO647N-labeled tubulin, and mCherry-HURP were diluted to their target concentrations in assay buffer, mixed thoroughly, and pipetted into the flow chamber. The flow chamber was sealed with nail polish and incubated coverslip side down for 10 min to allow condensates to settle. Condensates were imaged in bulk and on the coverslip surface using epifluorescence on a Nikon Ti-E microscope with a 100x objective and 1.49 numerical aperture. Exposure times and LED power were consistent across all tested concentrations for each protein. An ORCA-Fusion BT digital CMOS camera was used for acquisition. Condensation was determined visually by assessing whether each condition promoted the formation of mesoscale droplets. The TPX2 phase boundary was assessed by measuring the average % Area Fraction of thresholded images in ImageJ for each TPX2 concentration (n ≥ 5 for each concentration), using the average auto-threshold value of the most concentrated condition, determined by the Triangle method in FIJI software. These points were plotted (TPX2 concentration vs Area Fraction) and fit to a line using the scipy.stats.linregress() function, with the phase boundary being determined as the x-intercept of the fitted line.
Cryo-EM sample preparation
GMPCPP-MTs were prepared using an established protocol44. Polymerized GMPCPP-MTs were diluted to 0.1 mg/mL concentration and applied to a glow-discharged C-flat 1.2/1.3-4 C holey carbon EM grid (Protochips) and incubated inside a Vitrobot (Thermo Fisher Scientific) set at 25 °C and 95% humidity for 1 min. Then the grid was washed twice with 3 μL of room temperature HURP proteins (at 1 mg/mL concentration) to allow maximum decoration, before blotting and vitrification in liquid ethane.
Cryo-EM data collection
Cryo-EM data were collected using a 300 keV Titan Krios microscope equipped with a Cs-corrector (Thermo Fisher Scientific) and a Bioquantum Energy Filter (Gatan) at the Center for Cellular Imaging (WUCCI) in Washington University in St. Louis. In total around 2000 movies were collected using a K2 Summit direct electron detector (Gatan) in counting mode and an exposure rate of 8.5 electrons/pixel/s on the detector camera. The images were recorded at a nominal magnification of 81,000X, corresponding to a calibrated pixel size of 1.39 Å, with a defocus range from −1.0 to −2.5 μm. A total exposure time of 9 s, corresponding to a total dose of 39.6 electrons/Å2 on the specimen, was fractionated into 30 movie frames. The data were collected automatically using EPU software (Thermo Fisher Scientific). Data collection statistics are reported in Supplementary Table 1.
Cryo-EM data processing
MTs were automatically picked from 1728 motion-corrected micrographs using template based ‘Filament Tracer’ in CryoSPARC v.4.640. The templates were a few 2D class averages from a small subset of MTs that were picked manually. The step size between adjacent MT particle boxes was set to be 83 Å, which is the length of an α,β-tubulin heterodimer. Two-dimensional classification was used to remove junk particles and off-centered MT particles (Supplementary Fig. S8). In the next step, we performed supervised 3D classification of the MT particles using two reference models (13-PF and 14-PF MTs), and around 80% of the MT particles are 14-PF MTs, which is typical for GMPCPP-MTs. We used only the 14-PF MTs for subsequence data processing in 5 major steps. In step 1, we used ‘Helical Refinement’ function in CryoSPARC to obtain a 3D reconstruction by specifying the 14-start pseudo-helical symmetry (rise 83.0 Å, twist 0°), and then fed the outputs to ‘Local Refinement’ function to further refine the structure with a cylindrical mask covering most of the MT density. In step 2, we used pyem (10.5281/zenodo.3576630) to convert the alignment parameters from CryoSPARC to Relion v4.0 format, and then to FREALIGN v9.11 format using customized python scripts. In step 3, we used a previously established protocol to determine the seam location for each MT particle41, and imported the alignment parameters with the correct seam location back to CryoSPARC. In step 4, we performed local refinement (using a cylindrical mask covering most of the MT density), local CTF refinement and another round of local refinement, resulting in a MT C1 reconstruction at 3.3 Å resolution. In step 5, to fully utilize the pseudo-helical symmetry of MT, we did symmetry expansion using the 3-start symmetry (rise 8.9 Å, twist −25.8°) and helical order of 11, resulting in 11-fold increase of the total number of particles (in theory one can set the helical order to be 14), and performed local refinement of all the symmetry expanded MT particles using a cylindrical mask covering most of the MT density. In step 5, we continued the local refinement using a shorter cylindrical mask (200 Å in length) that covers only 2 protofilaments, resulting in the best local resolution of 2.8 Å.
Protein identification, model building, and refinement
We used the cryo-EM density map of HURP65-174 decorated MT at 2.8 Å local resolution as the input for a deep-learning based software ModelAngelo v1.042, which automatically determines the protein sequence for a density and builds tentative atomic models. A search of ModelAngelo derived protein sequence against the entire human proteome produced a single hit of HURP protein with high confidence. The hit was further verified by careful visual inspection of the fit between the atomic model and cryo-EM density map using Coot v0.9.868. The atomic models of α/β-tubulin were adapted from PDB 6DPU69, and adjusted in Coot based on the current density map.
Atomic model for a patch of 3 × 3 tubulin dimers were built and refined into the sharpened cryo-EM density map using Phenix.real_space_refine v1.20.1-4487. Secondary structures, Ramachandran, and rotamer restraints were applied during refinement. The quality of the refined model was assessed by MolProbity70, with statistics reported in Supplementary Table 1.
Statistics & reproducibility
For all results, statistical significance is defined as a p value ≤ 0.05 and is denoted by a single asterisk (*). p values ≤ 0.01 were denoted with a double asterisk (**). p values ≤ 0.001 were denoted with a triple asterisk (***). Ns denotes results that were not significant (p value > 0.05). Statistical methods for determining significance were reported in each individual figure legend and in the methods for each assay. No statistical method was used to predetermine sample size. No data was excluded from the analyses and fluorescence image data analysis was done blind.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Information
Source data
Acknowledgements
We especially thank Jodi Kraus, Collin McManus, and Sophie Travis of the Petry Lab for providing reagents for this work and feedback on the manuscript. We also thank Matthew Black for help in data pre-processing. We thank all former and current members of the Petry and Zhang Lab for any training, discussion, and advice pertaining to this work. V.A.V. was supported by NIH Training Grant (T32GM007388) and the Princeton University Charlotte Elizabeth Procter Fellowship. B.G. was supported by the NSF GRFP DGE-2039656. R.Z. is supported by NIGMS grant 1R01GM138854. S.P. is supported by NIGMS grant 1R01GM141100-01A1.
Author contributions
V.A.V. conceptualized and designed the study with supervision from S.P. V.A.V. performed and analyzed all MT localization assays, nucleation assays, and all experiments in Xenopus egg extract. V.A.V. performed and analyzed all revision experiments. M.M. purified the human HURP constructs and prepared the cryo-EM samples. M.M. and R.Z. collected and processed the cryo-EM data. R.Z. developed the new CryoSPARC-based pipeline for MT data processing. B.G. designed and performed phase assays in the initial submission V.A.V. and R.Z. wrote the first draft of the manuscript. All authors contributed to manuscript editing and data interpretation. S.P. and R.Z. supervised the project and acquired funding.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The single-particle cryoEM structure of HURP65-174-decorated GMPCCP-MT has been deposited in the Electron Microscopy Data Bank (EMDB) with accession code EMD-47173. The refined atomic model for a patch (3 × 3) of tubulin dimers with HURP decoration has been deposited in the Protein Data Bank (PDB) with ID code 9DUQ. Source data are provided with this paper.
Code availability
Python scripts needed for the new CryoSPARC- and FREALIGN-based MT data processing pipeline is available on Github [https://github.com/rui--zhang/Microtubule].
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rui Zhang, Email: zhangrui@wustl.edu.
Sabine Petry, Email: spetry@princeton.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53630-6.
References
- 1.Compton, D. A. Spindle assembly in animal cells. Annu Rev. Biochem69, 95–114 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Lawo, S. et al. HAUS, the 8-Subunit Human Augmin Complex, Regulates Centrosome and Spindle Integrity. Current Biol.19, 816–826 (2009). [DOI] [PubMed]
- 3.Uehara, R. et al. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. USA106, 6998–7003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David, A. F. et al. Augmin accumulation on long-lived microtubules drives amplification and kinetochore-directed growth. J. Cell Biol.218, 2150–2168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gouveia, B. et al. Acentrosomal spindles assemble from branching microtubule nucleation near chromosomes in Xenopus laevis egg extract. Nat. Commun.14, 1–15 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petry, S., Groen, A. C., Ishihara, K., Mitchison, T. J. & Vale, R. D. Branching Microtubule Nucleation in Xenopus Egg Extracts Mediated by augmin and TPX2. Cell152, (2013). [DOI] [PMC free article] [PubMed]
- 7.Alfaro-Aco, R., Thawani, A. & Petry, S. Biochemical reconstitution of branching microtubule nucleation. Elife9, e49797 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tariq, A., Green, L., Jeynes, J. C. G., Soeller, C. & Wakefield, J. G. In vitro reconstitution of branching microtubule nucleation. Elife9, e49769 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang, Y., Hong, X., Hua, S. & Jiang, K. Reconstitution and mechanistic dissection of the human microtubule branching machinery. J.Cell Biol.221, e202109053 (2022). [DOI] [PMC free article] [PubMed]
- 10.Thawani, A., Kadzik, R. S. & Petry, S. XMAP215 is a microtubule nucleation factor that functions synergistically with the γ-tubulin ring complex. Nat. Cell Biol.20, 575–585 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King, M. R. & Petry, S. Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat. Commun.11, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song, J. G. et al. Mechanism of how augmin directly targets the γ-tubulin ring complex to microtubules. J. Cell Biol.217, 2417–2428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraus, J., Travis, S. M., King, M. R. & Petry, S. Augmin is a Ran-regulated spindle assembly factor. J. Biol. Chem.299, 104736 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ustinova, K., Ruhnow, F., Gili, M. & Surrey, T. Microtubule binding of the human augmin complex is directly controlled by importins and Ran-GTP. J Cell Sci.136, (2023). [DOI] [PMC free article] [PubMed]
- 15.Kalab, P., Weis, K. & Heald, R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science (1979)295, 2452–2456 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Kaláb, P., Pralle, A., Isacoff, E. Y., Heald, R. & Weis, K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature440, 697–701 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Hayward, D., Metz, J., Pellacani, C. & Wakefield, J. G. Synergy between Multiple Microtubule-Generating Pathways Confers Robustness to Centrosome-Driven Mitotic Spindle Formation. Dev. Cell28, 81–93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silljé, H. H. W., Nagel, S., Körner, R. & Nigg, E. A. HURP Is a Ran-Importin β-Regulated Protein that Stabilizes Kinetochore Microtubules in the Vicinity of Chromosomes. Curr. Biol.16, 731–742 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Tsou, A. P. et al. Identification of a novel cell cycle regulated gene, HURP, overexpressed in human hepatocellular carcinoma. Oncogene22, 298–307 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Kuo, T. C., Chang, P. Y., Huang, S. F., Chou, C. K. & Chao, C. C. K. Knockdown of HURP inhibits the proliferation of hepacellular carcinoma cells via downregulation of gankyrin and accumulation of p53. Biochem Pharm.83, 758–768 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Chen, J. et al. Hepatoma upregulated protein expression is involved in the pathogenesis of human breast carcinogenesis. Oncol. Lett.8, 2543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eissa, S. et al. Evaluation of urinary HURP mRNA as a marker for detection of bladder cancer: relation to bilharziasis. Med. Oncol.31, 804 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Hassan, M. et al. Elevated Expression of Hepatoma Up-Regulated Protein Inhibits γ-Irradiation-Induced Apoptosis of Prostate Cancer Cells. J. Cell Biochem117, 1308–1318 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Wong, J. & Fang, G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol.173, 879–891 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuchiya, K. et al. Ran-GTP Is Non-essential to Activate NuMA for Mitotic Spindle-Pole Focusing but Dynamically Polarizes HURP Near Chromosomes. Curr. Biol.31, 115–127.e3 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Breuer, M. et al. HURP permits MTOC sorting for robust meiotic spindle bipolarity, similar to extra centrosome clustering in cancer cells. J. Cell Biol.191, 1251–1260 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casanova, C. M. et al. Hepatoma up-regulated protein is required for chromatin-induced microtubule assembly independently of TPX2. Mol. Biol. Cell19, 4900–4908 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koffa, M. D. et al. HURP Is Part of a Ran-Dependent Complex Involved in Spindle Formation. Curr. Biol.16, 743–754 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Hayward, D. & Wakefield, J. G. Chromatin-mediated microtubule nucleation in Drosophila syncytial embryos. Commun Integr. Biol.7, e28512 (2014). [DOI] [PMC free article] [PubMed]
- 30.Didaskalou, S. et al. HURP localization in metaphase is the result of a multi-step process requiring its phosphorylation at Ser627 residue. Front Cell Dev. Biol.11, 981425 (2023). [DOI] [PMC free article] [PubMed]
- 31.Wühr, M. et al. Deep Proteomics of the Xenopus laevis Egg using an mRNA-derived Reference Database. Curr. Biol.24, 1467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scrofani, J. et al. Branched microtubule nucleation and dynein transport organize RanGTP asters in Xenopus laevis egg extract. Mol. Biol. Cell35, 12–13 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alfaro-Aco, R., Thawani, A. & Petry, S. Structural analysis of the role of TPX2 in branching microtubule nucleation. J. Cell Biol.216, 983–997 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiese, C. & Zheng, Y. Microtubule nucleation: γ-tubulin and beyond. J. Cell Sci.119, 4143–4153 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Thawani, A. et al. The transition state and regulation of γ-TuRC-mediated microtubule nucleation revealed by single molecule microscopy. Elife9, e54253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rale, M. J., Romer, B., Mahon, B. P., Travis, S. M. & Petry, S. The conserved centrosomin motif, γTuNA, forms a dimer that directly activates microtubule nucleation by the γ-tubulin ring complex (γTuRC). Elife11, e80053 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Consolati, T. et al. Microtubule Nucleation Properties of Single Human γTuRCs Explained by Their Cryo-EM Structure. Dev. Cell53, 603–617.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castrogiovanni, C. et al. Evidence for a HURP/EB free mixed-nucleotide zone in kinetochore-microtubules. Nat. Commun. 202213, 1–16 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song, L., Craney, A. & Rape, M. Microtubule-Dependent Regulation of Mitotic Protein Degradation. Mol. Cell53, 179–192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Zhang, R. & Nogales, E. A new protocol to accurately determine microtubule lattice seam location. J. Struct. Biol.192, 245–254 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jamali, K. et al. Automated model building and protein identification in cryo-EM maps. Preprint at bioRxiv10.1101/2023.05.16.541002 (2023). [DOI] [PMC free article] [PubMed]
- 43.Zhang, R., Roostalu, J., Surrey, T. & Nogales, E. Structural insight into TPX2-stimulated microtubule assembly. Elife6, (2017). [DOI] [PMC free article] [PubMed]
- 44.Zhang, R., Alushin, G. M., Brown, A. & Nogales, E. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell162, 849–859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maurer, S. P., Fourniol, F. J., Bohner, G., Moores, C. A. & Surrey, T. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell149, 371–382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moores, C. A. et al. Mechanism of Microtubule Stabilization by Doublecortin. Mol. Cell14, 833–839 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Atherton, J. et al. A structural model for microtubule minus-end recognition and protection by CAMSAP proteins. Nat. Struct. Mol. Biol.24, 931–943 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roostalu, J., Cade, N. I. & Surrey, T. Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat. Cell Biol.17, 1422–1434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maurer, S. P. et al. EB1 Accelerates Two Conformational Transitions Important for Microtubule Maturation and Dynamics. Curr. Biol.24, 372–384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bechstedt, S., Lu, K. & Brouhard, G. J. Doublecortin Recognizes the Longitudinal Curvature of the Microtubule End and Lattice. Curr. Biol.24, 2366–2375 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Jiang, K. et al. Microtubule Minus-End Stabilization by Polymerization-Driven CAMSAP Deposition. Dev. Cell28, 295–309 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Wong, J., Lerrigo, R., Jang, C. Y. & Fang, G. Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Mol. Biol. Cell19, 2083–2091 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bayliss, R., Sardon, T., Vernos, I. & Conti, E. Structural Basis of Aurora-A Activation by TPX2 at the Mitotic Spindle. Mol. Cell12, 851–862 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Goshima, G. Identification of a TPX2-Like Microtubule-Associated Protein in Drosophila. PLoS One6, e28120 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goode, B. L. & Eck, M. J. Mechanism and Function of Formins in the Control of Actin Assembly. Annu Rev. Biochem76, 593–627 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Tan, S., Kern, R. C. & Selleck, W. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif.40, 385–395 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Searle, B. C. Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics10, 1265–1269 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Nesvizhskii, A. I., Keller, A., Kolker, E. & Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem.75, 4646–4658 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Travis, S. M. et al. Integrated model of the vertebrate augmin complex. Nat. Commun.14, 1–10 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thawani, A., Stone, H. A., Shaevitz, J. W. & Petry, S. Spatiotemporal organization of branched microtubule networks. Elife8, e43890 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hyman, A. et al. [39] Preparation of modified tubulins. Methods Enzymol.196, 478–485 (1991). [DOI] [PubMed] [Google Scholar]
- 62.Setru, S. U. et al. A hydrodynamic instability drives protein droplet formation on microtubules to nucleate branches. Nat. Phys.17, 493–498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Good, M. C. & Heald, R. Preparation of Cellular Extracts from Xenopus Eggs and Embryos. Cold Spring Harb Protoc2018, pdb.prot097055 (2018). [DOI] [PMC free article] [PubMed]
- 64.Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thévenaz, P., Ruttimann, U. E. & Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process.7, 27–41 (1998). [DOI] [PubMed] [Google Scholar]
- 66.Kluyver, T. et al. Jupyter Notebooks – a publishing format for reproducible computational workflows. Positioning and Power in Academic Publishing: Players, Agents and Agendas - Proceedings of the 20th International Conference on Electronic Publishing, ELPUB 2016 87–90 (2016) 10.3233/978-1-61499-649-1-87.
- 67.Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. Biol. Crystallogr60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Zhang, R., LaFrance, B. & Nogales, E. Separating the effects of nucleotide and EB binding on microtubule structure. Proc. Natl. Acad. Sci. USA115, E6191–E6200 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen, V. B. et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D. Biol. Crystallogr66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Information
Data Availability Statement
The single-particle cryoEM structure of HURP65-174-decorated GMPCCP-MT has been deposited in the Electron Microscopy Data Bank (EMDB) with accession code EMD-47173. The refined atomic model for a patch (3 × 3) of tubulin dimers with HURP decoration has been deposited in the Protein Data Bank (PDB) with ID code 9DUQ. Source data are provided with this paper.
Python scripts needed for the new CryoSPARC- and FREALIGN-based MT data processing pipeline is available on Github [https://github.com/rui--zhang/Microtubule].